Abstract
Conserved human cytochrome b5 (b5) residues D58 and D65 are critical for interactions with CYP2E1 and CYP2C19, whereas E48 and E49 are essential for stimulating the 17,20-lyase activity of CYP17A1. Here, we show that b5 mutations E48G, E49G, D58G, and D65G have reduced capacity to stimulate CYP3A4-catalyzed progesterone and testosterone 6β-hydroxylation or nifedipine oxidation. The b5 double mutation D58G/D65G fails to stimulate these reactions, similar to CYP2E1 and CYP2C19, whereas mutation E48G/E49G retains 23–42% of wild-type stimulation. Neither mutation impairs the activity stimulation of wild-type b5, nor does mutation D58G/D65G impair the partial stimulation of mutations E48G or E48G/E49G. For assays reconstituted with a single phospholipid, phosphatidyl serine afforded the highest testosterone 6β-hydroxylase activity with wild-type b5 but the poorest activity with b5 mutation E48G/E49G, and the activity stimulation of mutation E48G/E49G was lost at [NaCl] > 50 mM. Cross-linking of CYP3A4 and b5 decreased in the order wild-type > E48G/E49G > D58G/D65G and varied with phospholipid. We conclude that two b5 acidic surfaces, primarily the domain including residues D58-D65, participate in the stimulation of CYP3A4 activities. Our data suggest that a minor population of CYP3A4 molecules remains sensitive to b5 mutation E48G/E49G, consistent with phospholipid-dependent conformational heterogeneity of CYP3A4.
Keywords: Cytochrome b5, testosterone, CYP3A4, allostery, cytochrome P450, drug oxidation
Introduction
Cytochrome P450 3A4 (CYP3A4) constitutes 30 and 70% of the total CYPs in human liver and intestine, respectively, and metabolizes nearly 60% of the drugs currently in use (1). In addition to its important function in xenobiotic metabolism, CYP3A4 also metabolizes endogenous steroids, including cortisol, testosterone, 17β-estradiol, progesterone, and androstenedione (2–6). The ability of CYP3A4 to interact simultaneously with multiple substrate molecules and its conformational heterogeneity leads to non-Michaelis-Menten kinetics (6,7). Crystallographic studies of human CYP3A4 in complex with ketoconazole and erythromycin (8) have shown that the protein undergoes dramatic conformational changes upon ligand binding and have revealed the remarkable flexibility of CYP3A4 active site, which provides a structural basis for its substrate promiscuity. Furthermore, significant progress has been made recently (9–11) to confirm a pivotal role of CYP3A4 oligomerization in its functional heterogeneity.
One particular feature rather unique to CYP3A4 is the “Phe-cluster” of seven phenylalanine residues, which form a hydrophobic core pointing towards the active site (12). Interactions with redox partners (12,13) might trigger conformational movements in the Phe-cluster, including substrate recognition sequence 1 (SRS-1) residue Phe-108 and SRS-4 residue Phe-304. The challenge in understanding the catalytic mechanisms of such a promiscuous enzyme as CYP3A4 requires a description of how these structural elements mediate the interactions with redox partners and substrates.
Cytochrome b5 (b5) is an important component in most CYP3A4-catalyzed reactions, including testosterone 6β-hydroxylation and nifedipine oxidation. In reconstituted assays with purified proteins, b5 stimulates these activities 4–5 fold (14,15). Perret et al. (16) demonstrated electron shuttles between CYP3A4 and b5 and reported the critical roles of the structure and dynamics of the CYP3A4•POR•b5 complexes on catalysis. Furthermore, studies of 7-benzyloxyquinoline debenzylation in the presence of α-naphthoflavone have identified 2 pools of CYP3A4 conformers with different susceptibility to H2O2-dependent heme loss in the presence of b5 (17). Comparable results using holo-b5 and redox-inactive apo-b5 in several studies suggest that an allosteric effect on the CYP3A4•POR complex dominates the stimulatory mechanism over influence on electron transfer steps (5,18).
Despite great interest and considerable progress in the allosteric modulation of CYP3A4, its specific interaction sites of b5 are yet to be determined. Cross-linking studies have identified residues on CYP3A4 and b5, which come into close proximity in vitro (19); however, the contributions of these interactions to substrate oxidation are not known. Our previous study of b5 mutations with CYP17A1, CYP2E1, and CYP2C19 has generated a model in which b5 interacts with P450 or P450•POR complexes via at least two critical surface domains, which incorporate the acidic residues E48-E49 for CYP17A1 or D58-D65 for CYP2E1 and CYP2C19 (20,21). Because CYP3A4 exhibits a broad substrate profile, high conformational flexibility, cooperative kinetics, and prominent sensitivity to phospholipid and buffer conditions (22), its interactions with b5 might be more complex than these other P450s. Consequently, we assessed b5 stimulation of CYP3A4 catalysis in reconstituted assays with POR and various phospholipids.
Material and methods
Materials
PMSF, 5-aminolevulinic acid, Nonidet P-40, DTT, ampicillin, isopropyl-β-D-thiogalactopyranoside (IPTG), 1,2-didodecanoyl-sn-glycero-3-phosphocholine (DLPC), 1,2-diacyl-sn-glycero-3-phosphoethanolamine (PE), 1,2-diacyl-sn-glycero-3-phospho-L-serine (PS), were obtained from Sigma (St. Louis, MO). 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) was obtained from Cayman (Ann Arbor, MI). Brain PC L-α-phosphatidylcholine (PC(mix)) was from Avanti Polar Lipids (Alabaster, AL). Ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) was obtained from Pierce (Rockford, IL), PVDF membrane was obtained from Millipore (Billerica, MA), Clarity Western ECL reagent was obtained from Bio-Rad (Richmond, CA), and Blue Devil autoradiography film was acquired from Genesee Scientific (San Diego, CA). The antibodies rabbit anti-human CYP3A4 was obtained from Enzo Life Sciences (Farmingdale, NY) and used at 1:1500 dilution; rabbit anti-cytochrome b5 was used at 1:9000 dilution; and secondary antibody goat anti-rabbit IgG-HRP conjugate was used at 1:9000 dilution. Complete mini-protease inhibitor was obtained from Roche Diagnostics (Indianapolis, IN). Bactotryptone and yeast extract were purchased from Difco (Detroit, MI). Molecular biology reagents, including restriction enzymes, ligases, and Phusion polymerase, were obtained from New England BioLabs (Beverly, MA). Progesterone was obtained from Calbiochem (San Diego, CA). Control yeast microsomal lipid (CYMS) was prepared as described (23).
Plasmids
The expression plasmids were generous gifts obtained from the following investigators: human CYP3A4 in pKK3A4HIS from Professor James R. Halpert (University of California, San Diego, CA); human POR in pET22 from Professor Walter L. Miller (University of California, San Francisco, CA); human b5 in pLW01-b5H4 from Professor Lucy Waskell (University of Michigan). The b5 mutation plasmids were prepared as described previously (20).
Expression and purification of recombinant proteins in E. coli
The plasmids encoding modified CYP3A4 (24), POR (25), or b5 protein were individually transformed in E. coli strain C41(DE3) cells (OverExpress, Lucigen, Middleton, WI). For all cases, 1 liter of Terrific Broth (supplemented with 0.5 mM 5-aminolevulinic acid for CYP3A4) and ampicillin was inoculated with 20 mL of an overnight pre-culture. The cells were grown at 37 °C with shaking at 250 rpm until the A600 reached 0.6–1.2 AU, at which time the culture was induced with IPTG (0.4 mM for CYP3A4 and POR, 10 μM for b5) and grown for 48 h at 25–27°C. After cell lysis with a French Press, the recombinant proteins were solubilized using mild non-denaturing detergents: 0.5% Nonidet P-40 for CYP3A4 and 0.2% cholate/0.2% Triton X-100 for POR (25). After centrifugation at 100,000 × g for 45 min, the supernatant was mixed with Ni-NTA affinity resin and purified to homogeneity in a single step upon elution with 300 mM imidazole, followed by buffer exchange using PD-10 columns. Yields averaged approximately 100 nmol pure protein per liter of culture. The protocol for expression and reconstitution of recombinant human b5 was based on the procedure of Mulrooney and Waskell (26). Control microsomes without human P450 enzymes were prepared from strain YiV(B) transformed with native V60 plasmids as described (23).
Reconstituted enzyme assays
In a 2 mL polypropylene tube, purified human CYP3A4 (30 pmol) was mixed with a 2- or 3-fold molar excess of POR, various amounts of b5, and CYMS (20 μg protein) or other specified phospholipid (10 μg, prepared using a probe Sonifier, Branson Ultrasonics Corporation, Danbury, CT) in less than 10 μL volume and incubated for 5 min. The reaction mixture was then diluted to 0.2 mL with 40 mM HEPES buffer (pH = 7.4), 30 mM MgCl2, 2.4 mM glutathione, and substrates progesterone (50 μM), testosterone (50 μM), or nifedipine (200 μM) added from methanol stock solutions diluted to 0.5–2%. Amber vials were used for nifedipine oxidation assay as described (27) because nifedipine is light sensitive. The resulting mixture was pre-incubated at 37 °C for 3 min before adding NADPH (1 mM) and incubating at 37 °C for another 20 min. The reaction mixture was extracted with 1 mL methylene chloride (with 5% 1M Na2CO3 buffer containing 2M NaCl for nifedipine), and the organic phase was dried under nitrogen flow.
Chromatography, data acquisition, and determination of kinetic constants
Reaction products were analyzed using the Agilent 1260 Infinity HPLC system with UV detector. Incubation extracts were dissolved in 20 μL of methanol, and 2–5 μL injections were resolved with a 50 × 2.1 mm, 2.6 μm, C8 Kinetex column (Phenomenex, Torrance, CA) equipped with a guard column at a flow rate of 0.4 mL/min. Aqueous methanol linear gradients were employed as follows: for progesterone and testosterone 6β-hydroxylation assay, 27% methanol from 0 to 0.5 min, jump to 39% methanol, and gradient from 39% to 75% methanol over 30 min; for nifedipine oxidation assay, 50% methanol for 4 min, gradient to 100% methanol over 1 min. Products were identified by retention times of external standards chromatographed at the beginnings and ends of the experiments using absorbance at 254 nm (A254) for progesterone and testosterone and 230 nm (A230) for nifedipine. The data were processed with Laura4 software (LabLogic) and graphed with GraphPad Prism 6 (GraphPad Software, San Diego, CA).
Each experiment was performed at least three times, and all data are presented as mean ± standard deviation. Kinetic analysis of CYP3A4 oxidations was done using a allosteric sigmoidal model (Graphpad Prism 6 software). Values of v at various substrate concentrations were fitted by nonlinear regression to the equation:
where h is the Hill coefficient, Vmax is the maximal velocity, and K0.5 is the substrate concentration that gives the half-maximum velocity.
Spectroscopic methods
UV/Vis spectra were recorded on a Shimadzu UV-Vis spectrophotometer (UV-2600) with UV-Probe software using a quartz cuvette with a path length of 10 mm. Circular dichroism (CD) spectra were recorded on an Aviv Model 202 spectrometer. Cell path length was 10 mm for measurements above 240 nm and 0.5 mm for measurements in the 190–240 nm regions.
Cross-linking and immunoblot experiments
Purified human CYP3A4 (40 pmol), b5 (WT or variants) and phospholipids were reconstituted at a molar ratio of 1:10:100. After incubation for 30 min at 25°C, freshly prepared EDC was added to a final concentration of 2 mM in a total volume of 30 μl containing 50 mM potassium phosphate (pH 7.0). The mixture was shaken gently at room temperature for 2 h, followed by adding an equal volume of Laemmli sample buffer and boiling for 5 min. Control incubations without EDC were conducted in parallel. The cross-linked proteins were resolved by electrophoresis with 7.5% SDS-polyacrylamide gels and electroblotted onto PVDF membrane. The membranes were blocked for 1 h at room temperature in PBS containing 5% skim milk plus 0.1% Tween-20 and was incubated overnight at 4°C with rabbit anti-human CYP3A4 antibody or rabbit anti-human b5, followed by incubation with secondary antibody for 1 h at room temperature. The blots were incubated for 1 min with ECL reagent and exposed to photographic film. Immunoblot species were quantified by measuring the scanned band intensities using ImageJ software (http://rsb.info.nih.gov/ij/index.html).
Results
Progesterone 6β-hydroxylation
To probe the interaction of CYP3A4 with b5, a series of b5 mutations was generated, expressed, and purified as previously (20,21). All mutations displayed absorption spectra comparable to that of the wild-type b5 in the reduced and oxidized state (not shown), indicating structural and electronic integrity.
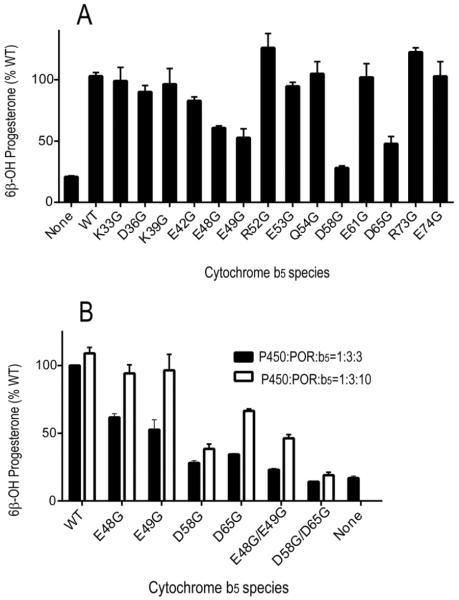
Each b5 mutation was tested for its capacity to increase the rate of progesterone 6β-hydroxylation (Figure 1A). In a reconstituted system containing purified CYP3A4, POR and CYMS as lipid source, the presence of wild-type b5 increased the rate of progesterone metabolism 5-fold, with P450:POR:b5 molar ratio of 1:3:3. The activity in the presence of b5 mutations E48G, E49G, D58G, and D65G was reduced to less than 60% that observed with wild-type b5, suggesting that acidic residues at these positions might be important for the association of b5 with the CYP3A4•POR complex. In contrast, all other mutations stimulated progesterone 6β-hydroxylation similar to wild-type b5 (Figure 1A). To further assess the interaction between adjacent residues on the surface of b5, we tested the capacity of double mutations E48G/E49G and D58G/D65G to stimulate CYP3A4-catalyzed progesterone 6β-hydroxylation. The double mutation D58G/D65G did not enhance CYP3A4-catalyzed progesterone 6β-hydroxylation with P450:POR:b5 molar ratio of 1:3:3 or 1:3:10 (Figure 1B). The b5 double mutation E48G/E49G minimally stimulated activity with a P450:POR:b5 molar ratio of 1:3:3, and a 2-fold stimulation was observed when the P450:b5 molar ratio was increased to 1:10. This shift in the dose-response relationship and reduction in maximal stimulation suggests that either the affinity of the E48G/E49G mutation for the CYP3A4•POR complex is lower than for wild-type b5 and/or that this mutation acts on only a subpopulation of CYP3A4 molecules.
FIGURE 1.
Stimulation of CYP3A4-catalyzed progesterone 6β-hydroxylation by b5 and b5 mutations. (A) Effect of b5 and b5 mutations on catalytic activities of CYP3A4 in the presence of CYMS as lipid for reconstitution. Incubations contained 50 μM progesterone and a molar ratio of P450:POR:b5 at 1:3:3. Results are shown as the percentage activity compared to wild-type b5 values (=100%) from triplicate determinations, means ± standard deviations. (B) Comparison of wild-type b5 and selected b5 mutations on CYP3A4 catalysis with molar ratio of P450:POR:b5 at 1:3:3 (black bars) or 1:3:10 (white bars). Results are shown as the percentage activity compared to wild-type b5 values with 1:3:3 ratio of P450:POR:b5 (=100%) from triplicate determinations, means ± standard deviations.
Testosterone 6β-hydroxylation and nifedipine oxidation
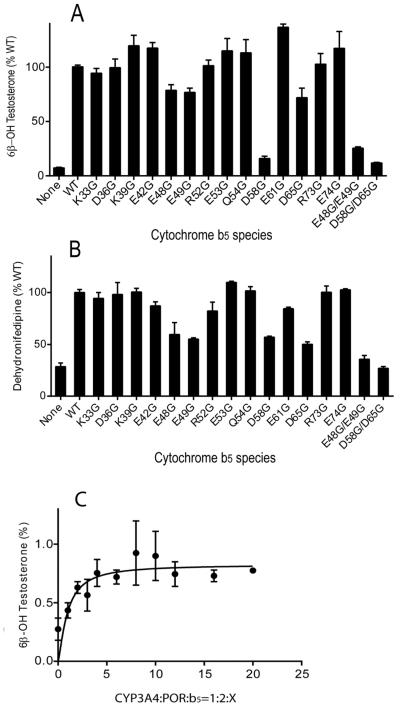
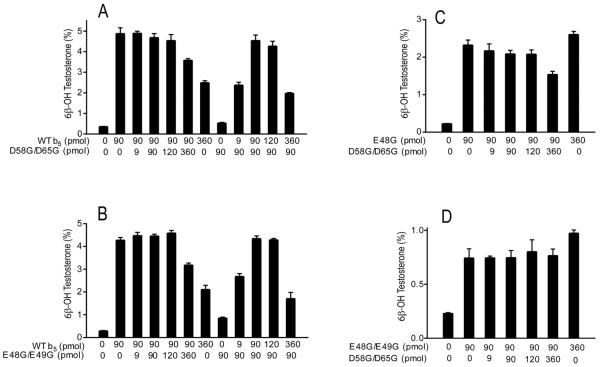
In the reconstituted system, wild-type b5 increased the rate of testosterone metabolism up to 12-fold with a P450:POR:b5 molar ratio of 1:2:3. Similar to results with progesterone, the same four mutations E48G, E49G, D58G, and D65G afforded lower activity compared to wild-type b5 (Figure 2A). The double mutation D58G/D65G completely abolished the ability of b5 to enhance CYP3A4-catalyzed testosterone 6β-hydroxylation, whereas E48G/E49G retained only 25% of wild-type activity (Figure 2A). The 6β-testosterone hydroxylation in the presence of mutation E48G/E49G was not increased at higher P450:b5 molar ratios (Figure 2C). An equivalent decrease in function of double mutations D58G/D65G and E48G/E49G was also observed for nifedipine oxidation with a P450:POR:b5 molar ratio of 1:2:4 (Figure 2B). These data show that b5 residues D58 and D65 are critical for stimulating these CYP3A4-catalyzed oxygenations, while residues E48 and E49 also contribute significantly.
FIGURE 2.
Stimulation of CYP3A4-catalyzed testosterone 6β-hydroxylation and nifedipine oxidation by b5 and b5 mutations. Purified CYP3A4 (30 pmol) was reconstituted with POR, various b5 mutations, and CYMS as lipid for reconstitution. Catalytic activities toward (A) testosterone at 50 μM substrate concentration at a P450:POR:b5 molar ratio of 1:2:3 and (B) nifedipine at 200 μM substrate concentration at a P450:POR:b5 molar ratio of 1:2:4 were analyzed by HPLC. Results are shown as the percentage activity compared to wild-type b5 values (=100%) from duplicate determinations, means ± standard deviations. (C) Titration of CYP3A4-catalyzed testosterone 6β-hydroxylation with human cytochrome b5 mutation E48G/E49G. The stimulation of activity with P450:POR molar ratio of 1:2 reaches a maximum at a 3–5 molar excess of the b5 mutation and does not increase further.
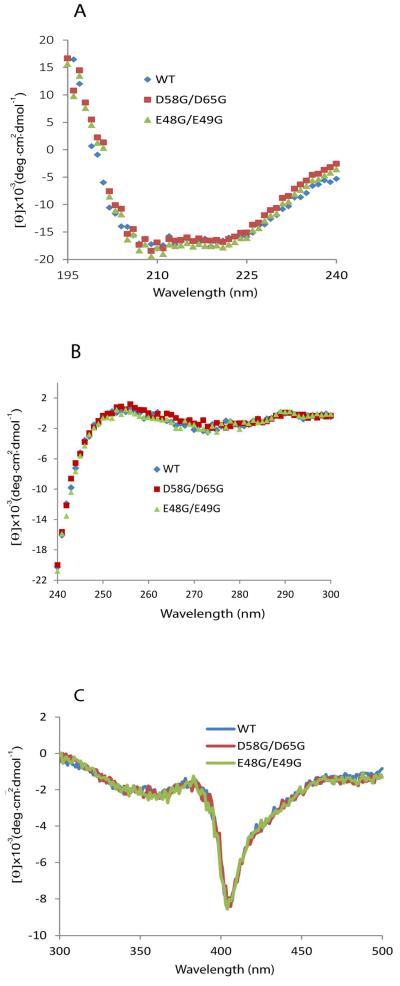
The b5 double mutations E48G/E49G and D58/D65 both exhibit visible absorption and circular dichroism (CD) spectra in the far UV and near UV range indistinguishable from wild type b5, which indicates that these mutations do not introduce gross changes in polypeptide backbone or tertiary structure (Figure 3A, 3B). CD spectra in the visible range are also equivalent, which indicates that these mutations do not significantly alter the environment around the heme center (Figure 3C). These CD spectra, however, do not exclude subtle changes in unstructured regions or altered conformational dynamics in these b5 mutations.
FIGURE 3.
Circular dichroism (CD) spectra of cytochrome b5 and mutations. CD spectra in (A) the far-UV, (B) the near UV, and (C) visible range for wild-type, D58G/D65G, and E48G/E49G double mutations are shown. Samples contained 10 μM b5 in 100 mM potassium phosphate, pH 7.4.
Kinetics and influence of ionic strength
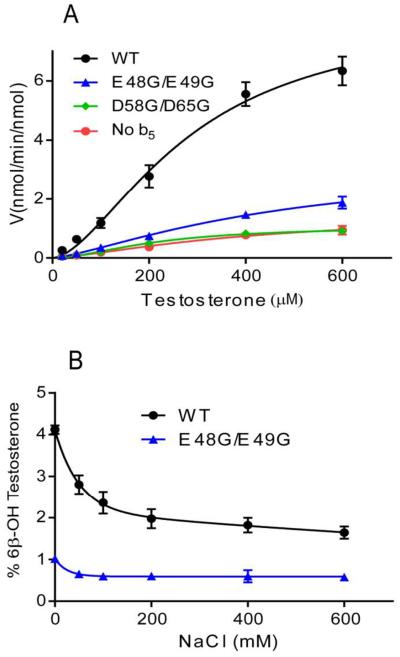
For most activities, the presence of b5 increases Vmax of CYP3A4, but b5 can also change Km, as observed for some CYP2E1-catalyzed reactions (28,29). Because CYP3A4 activity obeys sigmoidal rather than Michaelis-Menten kinetics, v vs. [S] data were fit to an allosteric sigmoidal model, to determine changes in Vmax and K0.5. Vmax values in the presence of the wild-type b5 and E48G/E49G were approximately 5.5-fold and 2.1-fold greater than assays conducted in the absence of b5. In contrast, the K0.5 value decreased slightly for wild-type b5 and did not change for mutation E48G/E49G (Figure 4A). The CYP3A4 kinetic constants with b5 mutation D58G/D65G were indistinguishable from control assays without b5. Thus, we found that wild-type b5 and mutation E48G/E49G increased Vmax but did not change K0.5 for testosterone 6β-hydroxylation.
FIGURE 4.
(A) Steady-state kinetics of testosterone 6β-hydroxylation by CYP3A4 in the absence or presence of wild type b5 or mutations. Incubations contained 20 to 600 μM testosterone and a molar ratio of P450:POR:b5 at 1:2:3. The allosteric sigmoidal model was fit to the data points as described under Material and Methods. Kinetic parameters for CYP3A4 with wild-type b5: the Hill coefficient h = 1.7 ± 0.2, Vmax = 8.3 ± 1.0 nmol/min/nmol, and K0.5 = 284 ± 52 μM; with b5 mutation E48G/E49G, h = 1.4 ± 0.2, Vmax = 3.2 ± 0.8 nmol/min/nmol, and K0.5 = 467 ± 174 μM; with b5 mutation D58G/D65G, h = 1.8 ± 0.2, Vmax = 1.1 ± 0.1 nmol/min/nmol, and K0.5 = 215 ± 26 μM; without b5, h is 1.4 ± 0.3, Vmax = 1.5 ± 0.3 nmol/min/nmol, and K0.5 = 409 ± 170 μM. (B) Effect of [NaCl] on CYP3A4 testosterone 6β-hydroxylation in the presence of wild-type b5 or b5 mutation E48G/E49G. Reaction conditions were equivalent to those described in (A) with 200 μM testosterone.
The effects of ionic strength on the CYP3A4 activity in the presence of wild-type b5 or mutation E48G/E49G are shown in Figure 4B. The loss of activity observed with both b5 species at high NaCl concentrations suggests that electrostatic forces contribute to these interactions; however, the activity stimulation with b5 mutation E48G/E49G is much more sensitive to salt disruption than with wild-type b5. With b5 mutation E48G/E49G, CYP3A4-catalyzed testosterone 6β-hydroxylation is reduced to basal values at NaCl concentrations above 50 mM, whereas 200 mM NaCl reduces wild-type b5 stimulation only ~50%, with slight further loss at higher NaCl concentrations. These results suggest that wild-type b5 interactions with the CYP3A4•POR complex are more stable and therefore more catalytically productive than for b5 mutation E48G/E49G, possibly due to greater hydrophobic interactions, which high ionic strength conditions do not disrupt.
Competition assays using wild-type b5 and mutations in CYP3A4 testosterone 6β-hydroxylation
To determine whether b5 mutation D58G/D65G binds poorly to the CYP3A4•POR catalytic complex or binds similarly to wild-type b5 but fails to elicit the conformational changes necessary to form a fully functional catalytic complex, we performed competition assays as described previously (20). In the first experiment, the concentration of wild-type b5 was held constant at the optimum 3:1 b5:CYP3A4 molar ratio, and increasing amounts of mutation D58G/D65G were added. As shown in Figure 5A, b5 mutation D58G/D65G did not decrease testosterone 6β-hydroxylation activity until a 4-fold molar excess was added and the total b5:CYP3A4 molar ratio reached 15:1. Turnover also declined when the molar ratio of wild-type b5 to CYP3A4 reached 12:1, consistent with competition between b5 and the P450 for limiting POR, as we have suggested previously (20,21). In the reverse scenario, the presence of mutation D58G/D65G did not impair the capacity of wild-type b5 to stimulate CYP3A4-catalyzed testosterone 6β-hydroxylation at wild-type b5/CYP3A4 molar ratios as low as 0.3:1 (Figure 5A).
FIGURE 5.
Competition assays. CYP3A4-catalyzed formation of 6β-hydroxytestosterone in the presence of (A) wild-type b5 and D58G/D65G double mutation or (B) wild-type b5 and E48G/E49G double mutation. Reactions were conducted with a constant P450:POR molar ratio of 1:2 in the presence of constant wild-type b5 and varying amounts of double mutation, or in the presence of constant double mutation and varying amounts of wild-type b5. Competition experiments using b5 mutations E48G (C) and E48G/E49G (D) show no inhibition by double mutation D58G/D65G. Error bars indicate means ± standard deviations of triplicate assays. Due to low conversion, experiments for panels C and D used tracer [3H]-testosterone as in ref. 21.
Competition experiments between wild-type b5 and mutation E48G/E49G yielded qualitatively similar results, except that mutation E48G/E49G stimulated testosterone 6β-hydroxylation activity 2-fold over baseline (Figure 5B). The b5 mutation D58G/D65G did not compromise the residual stimulation of testosterone 6β-hydroxylation activity using b5 mutations E48G and E48G/E49G, similar to results with wild-type b5 (Figure 5C, 5D). These data suggest that b5 interacts with the CYP3A4•POR complex using binding sites that include residues D58 and D65 and that the majority of these complexes also require E48 and E49 for productive interactions.
Effect of phospholipids on b5 stimulation of CYP3A4 activities
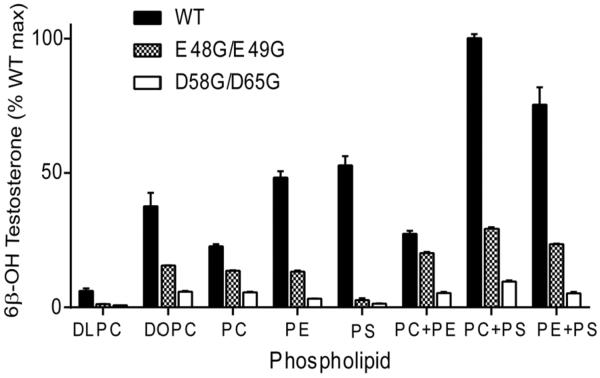
Prior reports found that maximal catalytic activity of purified CYP3A4 was obtained when the P450 is reconstituted with redox partners in the presence of specific anionic phospholipids (20–22) and ionic detergents (22). To explore the contributions of individual phospholipids to b5 action on the CYP3A4•POR complex, experiments were carried out with the purified proteins reconstituted with various phospholipids. Wild-type b5 stimulated CYP3A4-catalyzed testosterone 6β-hydroxylation when reconstituted with all phospholipids in the order PS > PE > PC(mix), and poor activity was seen only with DLPC (Figure 6). Combinations of two phospholipids gave similar or higher activity than either phospholipid alone. Consistent with previous reports, reconstitution with PC+PS gave the highest testosterone 6β-hydroxylation activity with wild-type b5, about 4-fold higher than assays using PC alone (Figure 6). Neither CHAPS nor sodium cholate at 0.1 μg/μl influenced CYP3A4 catalysis (data not shown) in our experimental conditions.
FIGURE 6.
Effect of phospholipids on b5-stimulated catalysis of CYP3A4. Purified CYP3A4 (30 pmol) and POR were reconstituted with wild-type b5 or mutations D58G/D65G and E48G/E49G in the presence of various phospholipids at a P450:POR:b5 molar ratio of 1:2:3. Activities with reconstituted assays using two types of purified phosphatidyl choline (DLPC and DOPC) and three types of natural phospholipids (PC from porcine brain, containing fatty acyl groups 18:1 (33%), 16:0 (31%) and 18:0 (17%); PE from bovine brain, containing fatty acyl groups 18:1 (24%), 20:4 (19%) and 18:0 (16%); and PS from bovine brain, containing fatty acyl groups 18:0 (40%) and 18:1 (29%)) are shown. Incubations were carried out with 50 μM testosterone, and the formation of 6β-hydroxytestosterone was determined by HPLC analysis. Results are shown as the percentage activity compared to wild-type b5 values in PC+PS (=100%), means ± standard deviations from triplicate determinations. In the absence of b5, conversion to 6β-hydroxytestosterone is 0.4±0.1% under these conditions.
In marked contrast to results with wild-type b5, the b5 mutation E48G/E49G did not stimulate CYP3A4-catalyzed testosterone 6β-hydroxylation in assays reconstituted with PS but gave similar activity with all other lipids and combinations (within a factor of 2), except for being inactive with DLPC (Figure 6). Finally, b5 mutation D58G/D65G failed to stimulate testosterone 6β-hydroxylation activity in assays reconstituted with any phospholipids alone or in combination, as observed with yeast microsomal lipids. These data support the model that b5 residues D58 and D65 are essential for stimulating CYP3A4-catalyzed testosterone 6β-hydroxylation, whereas residues E48 and E49 are dispensable under some conditions or for a subpopulation of CYP3A4 molecules, which might reflect the conformational flexibility and heterogeneity of CYP3A4 and/or b5.
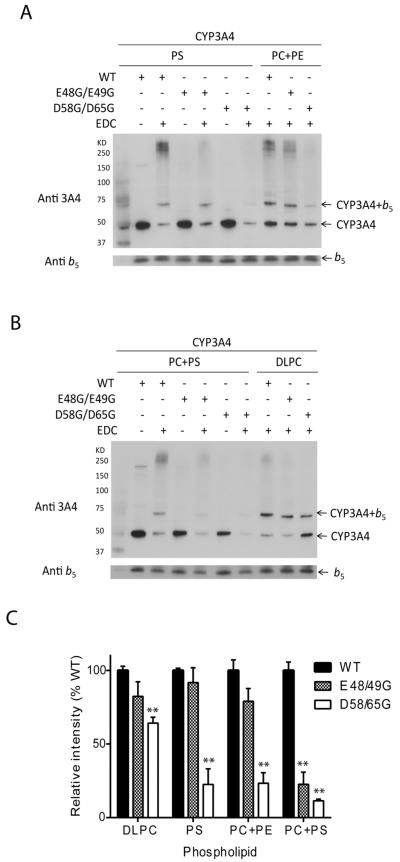
Cross-linking CYP3A4 with wild-type b5 or b5 mutations
Previous studies using cross-linking, mass spectrometry, and computer modeling have suggested that the α2–loop–α3 and α4–loop–α5 segments of b5 including residues E61 and E42 or E48 (human b5 numbering) interact with CYP3A4 (19). The contributions of b5 residues D58 and D65, however, were not explored, and these interactions were not investigated in different lipid environments. EDC, a zero-length cross-linker, covalently links the amino groups of lysines to the carboxyl groups of aspartic or glutamic acids. The amount of cross-linked complex between b5 and CYP3A4 correlates with the capacity of each b5 species to stimulate CYP3A4 activity: wild-type ≥ E48G/E49G > D58G/D65G in the presence of all phospholipids studied (Figure 7). Immunoblot analyses demonstrate that the formation of CYP3A4-b5 cross-links is phospholipid-dependent. CYP3A4 forms cross-links with wild-type b5 and b5 mutation E48G/E49G in all phospholipid conditions studied, but the degree of complex formation varies in the order DLPC > PC+PE > PS > PC+PS (Figure 7A, B), which does not match the pattern of activity stimulation. The inactive b5 mutation D58G/D65G only forms cross-links with CYP3A4 during incubations with DLPC and to a lesser extent in PC+PE, despite the fact that this mutation fails to augment CYP3A4 activity under all conditions. These results indicate that not all interactions between b5 and CYP3A4 lead to activity stimulation, particularly those in the presence of DLPC. Nevertheless, reduced complex formation appears to be the primary reason why b5 mutation D58G/D65G fails to stimulate CYP3A4 activities. In contrast, b5 mutation E48G/E49G interacts productively with at least a subset of CYP3A4•POR complexes.
FIGURE 7.
Immunoblot analysis of CYP3A4 and b5 cross-linked with EDC, and the effect of phospholipid on the complex formation. Reactions were performed in 50 μmM potassium phosphate buffer pH 7.0, containing 2 μM CYP3A4, 20 μM b5, 200 μM phospholipid, and 2 mM EDC. (A) CYP3A4 (50 kDa) and wild-type b5 or mutations (16 kDa) incubated with PS or PC+PE (1:1) in the absence of presence of EDC (B) CYP3A4 and b5 incubated with PC+PS (1:1) or DLPC in the absence of presence of EDC. (C) The immunoreactive bands corresponding to CYP3A4-b5 (1:1) complex were quantified by densitometric scanning. The bar graphs show the relative densities of bands for two experiments expressed as a percentage of the condition with wild-type b5. Mean ± standard error, **, p < 0.01.
Discussion
Our previous mutagenesis studies have identified two anionic surfaces of b5, which are required for stimulation of specific cytochrome P450 activities. The b5 residues E48 and E49 are essential for the b5 stimulation of CYP17A1-catalyzed steroid 17,20-lyase activity but not for CYP2E1-catalyzed chlorzoxazone 6-hydroxylation or for CYP2C19-catalyzed (S)-mephenytoin 4-hydroxylation and progesterone 21-hydroxylation. Conversely, the b5 residues D58 and D65 are required for the b5 stimulation of these CYP2E1 and CYP2C19 activities but not for CYP17A1-catalyzed steroid 17,20-lyase activity. The major finding of this study is that the promiscuous enzyme CYP3A4, which catalyzes many b5-stimulated reactions, also requires the b5 residues D58 and D65, similar to CYP2E1 and CYP2C19 (Figures 1 and 2). In contrast, the b5 double mutation E48G/E49G stimulates progesterone or testosterone 6β-hydroxylation and nifedipine oxidation with 23–42% the effect of wild-type b5. The inactive or less active b5 mutations do not impair stimulation of wild-type b5 or active b5 mutations (Figure 5), consistent with poor competition for binding to the CYP3A4•POR complex. Several interpretations of these data are plausible, including a model in which b5 residues D58 and D65 participate directly in a critical and catalytically relevant interaction with CYP3A4 and that b5 residues E48 and E49 are required for productive interactions with most but not all CYP3A4 molecules. Our data do not exclude additional mechanisms or alternative explanations, such as a weaker effect of b5 mutation E48G/E49G on the rate-limiting step (kcat) than for wild-type b5.
Further support for this model derives from the known conformational flexibility of the CYP3A4 protein and from the influence of phospholipid on catalysis. Whereas the b5 double mutation D58G/D65G fails to stimulate any of the assayed CYP3A4 activities under all reconstitution conditions tested, the action of the b5 double mutation E48G/E49G is highly dependent on phospholipid composition. When assays are reconstituted with PC+PE, the b5 double mutation E48G/E49G supports CYP3A4-catalyzed testosterone 6β-hydroxylation at a rate 74% that of the reaction with wild-type b5. In marked contrast, for assays reconstituted with PS alone, CYP3A4-catalyzed testosterone 6β-hydroxylation activity with wild-type b5 is even higher than in PC+PE, but the b5 double mutation E48G/E49G affords negligible stimulation with PS alone (Figure 6).
The phospholipid composition markedly influences P450 turnover and b5 stimulation (30,31), and CYP3A family activities are highest when reconstituted with acidic phospholipids such as PS (22,32). PS with unsaturated fatty acids improves interactions between the P450 and POR (32–34) and binds to CYP3Al, which alters its conformation (35). In our assay system, CYP3A4-catalyzed testosterone 6β-hydroxylation decreases with phospholipids in the order PS > PE > PC. This pattern of activity is similar to the phospholipid effect we observed for CYP17A1, CYP2E1, and CYP2C19 stimulation with wild-type b5 (21). In contrast, the influence of different phospholipids on CYP3A4 activities in the presence of partially active b5 mutation E48G/E49G is variable and unpredictable. Phospholipid membranes are known to influence the tertiary structure of the heme-binding domain of soluble b5 (36), and the membrane-spanning C-terminal helix of b5—which also interacts with phospholipids—is required to influence P450 activities (37). Consequently, the influence of these phospholipid interactions with not only the P450 and POR but with b5 as well is undoubtedly critical for regulation of the activities of these complex catalytic systems.
Our data suggest that CYP3A4 molecules are conformationally heterogeneous and differentially responsive to b5 mutations, as was suggested from studies of α-naphthoflavone influence on CYP3A4 chemistry (17). The major CYP3A4 species under most conditions require b5 residues E48 and E49 for activity stimulation, but the minor species are insensitive to glycine substitutions at these positions. The composition of phospholipid, in addition to regulating global activity, might also shift the proportion of these two conformational variants and thus the fractional stimulation afforded by b5 mutation E48G/E49G (Figure 6). Our results cannot distinguish whether the influence of substitutions at b5 residues E48 and E49 derive from changes in direct interactions with CYP3A4 and/or POR or from transmission of conformational changes to other parts of the b5 molecule.
Our data are consistent with the studies of Zhao et al (19), in which acidic residues within α-helix 4 are critical for binding to CYP3A4, as we found for CYP2E1 and CYP2C19. Using EDC and mass spectrometry, lysine residues 96, 127 and 421 on the proximal surface of CYP3A4 were cross-linked primarily with b5 residues E56 and E43 (19), which correspond to E61 on α-helix 4 and E48 in our numbering. We interpret our cross-linking data cautiously, because a chemical cross-link implies only a close proximity between 2 side-chains but not necessarily a catalytically important interaction. In our hands, cross-linking was most efficient in assays reconstituted with DLPC, in which CYP3A4 activities were lowest. Furthermore, the b5 mutation E61G stimulates CYP3A4-catalyzed reactions equivalent to wild-type b5, even though E61 engages in cross-linking chemistry with CYP3A4 (19).
The lack of competition with wild-type b5 stimulation by mutations D58G/D65G and E48G/E49G and the loss of E48G/E49G stimulation at [NaCl] > 50 mM are consistent with loss of electrostatic forces generated by these surface regions, which are involved in stabilizing the binding of b5 to CYP3A4•POR complexes. The contribution of residues E48 and E49 on the surface of the adjacent α-helix 3 appears to be minor, unlike the case for CYP17A1 (20). The involvement of two acidic surfaces of b5 in CYP3A4, whose relative importance varies with phospholipid, appears to reflect the large and plastic active site of CYP3A4 and the existence of functionally significant conformational variants (12). Among all the human P450 enzymes, CYP3A4 features the most diverse and complex interactions among ligands, substrates, and effectors, which all regulate CYP3A4 activity (6,11,38,39).
Several lines of evidence support a variety of mechanisms with which b5 might enhance P450-catalyzed reactions, including: (i) direct transfer of a rate-limiting electron, (ii) complexing with P450 to facilitate a two-electron transfer during a single interaction with POR, (iii) decreasing the extent of uncoupling or (iv) direct effector actions, enhancing product formation without redox changes of b5 (40). Since these proposed actions are not mutually exclusive, it is likely that several mechanisms contribute, depending on the specific substrates, enzymes, and assay conditions. In this study, we found that two surface domains, which incorporate the critical residues D58-D65 and E48-E49, contribute to b5 stimulation of CYP3A4 catalysis. Our findings are consistent with two populations of CYP3A4 molecules with distinct conformations and physical properties, but we cannot exclude other mechanisms, including differential participation of these b5 surfaces in the diverse actions of b5 on CYP3A4. Given the prominence of CYP3A4 in the metabolism of xenobiotics and steroids, further biochemical and biophysical studies to define these protein-protein interactions in relevant phospholipid environments might reveal additional mechanisms regulating the metabolism of drugs and endogenous lipids.
Conclusions
Cytochrome b5 residues D58-D65 are critical for stimulating CYP3A4 chemistry; while the contribution of resides E48-E49 varies with phospholipid. These findings suggest the existence of two subpopulations of CYP3A4 molecules which differentially responsive to b5 mutations and phospholipids.
-
-
Cytochrome b5 residues D58 and D65 are required to stimulate CYP3A4 activities
-
-
Cytochrome b5 double-mutation E48G/E49G retains partial activity with CYP3A4
-
-
The magnitude of stimulation by b5 mutation E48G/E49G varies with phospholipid
-
-
Our data suggest two phospholipid-dependent populations of CYP3A4
Acknowledgments
We thank Dr. Jacqueline Naffin-Olivos for constructing the expression plasmids for many of the b5 mutations and Dr. Paul J. Lennon for assistance with CD experiments. We also thank Dr. Paul Hollenberg for critical reading of the manuscript.
Abbreviations
- b5
cytochrome b5
- CYMS
control yeast microsomes
- CYP3A4
cytochrome P450 3A4
- PS
1,2-diacyl-sn-glycero-3-phospho-L-serine
- DLPC
1,2-didodecanoyl-sn-glycero-3-phosphocholine
- DOPC
1,2-dioleoyl-sn-glycero-3-phosphocholine
- PC
L-α-phosphatidylcholine
- PE
1,2-diacyl-sn-glycero-3-phosphoethanolamine
- IPTG
isopropyl-β-D-thiogalactopyranoside
- POR
cytochrome P450 oxidoreductase
- EDC
ethyl-3-(3-dimethylaminopropyl) carbodiimide
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Shimada T, Yamazaki H, Mimura M, Inui Y, Guengerich FP. J Pharmacol Exp Ther. 1994;270:414–423. [PubMed] [Google Scholar]
- [2].Abel SM, Back DJ. J Steroid Biochem Mol Biol. 1993;46:827–832. doi: 10.1016/0960-0760(93)90325-q. [DOI] [PubMed] [Google Scholar]
- [3].Waxman DJ, Attisano C, Guengerich FP, Lapenson DP. Arch Biochem Biophys. 1988;263:424–436. doi: 10.1016/0003-9861(88)90655-8. [DOI] [PubMed] [Google Scholar]
- [4].Kerlan V, Dreano Y, Bercovici JP, Beaune PH, Floch HH, Berthou F. Biochem Pharmacol. 1992;44:1745–1756. doi: 10.1016/0006-2952(92)90068-t. [DOI] [PubMed] [Google Scholar]
- [5].Yamazaki H, Gillam EM, Dong MS, Johnson WW, Guengerich FP, Shimada T. Arch Biochem Biophys. 1997;342:329–337. doi: 10.1006/abbi.1997.0125. [DOI] [PubMed] [Google Scholar]
- [6].Guengerich FP. Annu Rev Pharmacol Toxicol. 1999;39:1–17. doi: 10.1146/annurev.pharmtox.39.1.1. [DOI] [PubMed] [Google Scholar]
- [7].Hutzler JM, Tracy TS. Drug Metab Dispos. 2002;30:355–362. doi: 10.1124/dmd.30.4.355. [DOI] [PubMed] [Google Scholar]
- [8].Ekroos M, Sjogren T. Proc Natl Acad Sci U S A. 2006;103:13682–13687. doi: 10.1073/pnas.0603236103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Davydov DR, Fernando H, Baas BJ, Sligar SG, Halpert JR. Biochemistry. 2005;44:13902–13913. doi: 10.1021/bi0509346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Davydov DR, Sineva EV, Sistla S, Davydova NY, Frank DJ, Sligar SG, Halpert JR. Biochim Biophys Acta. 2010;1797:378–390. doi: 10.1016/j.bbabio.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Davydov DR, Davydova NY, Sineva EV, Kufareva I, Halpert JR. Biochem J. 2013;453:219–230. doi: 10.1042/BJ20130398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Williams PA, Cosme J, Vinkovic DM, Ward A, Angove HC, Day PJ, Vonrhein C, Tickle IJ, Jhoti H. Science. 2004;305:683–686. doi: 10.1126/science.1099736. [DOI] [PubMed] [Google Scholar]
- [13].Yano JK, Wester MR, Schoch GA, Griffin KJ, Stout CD, Johnson EF. J Biol Chem. 2004;279:38091–38094. doi: 10.1074/jbc.C400293200. [DOI] [PubMed] [Google Scholar]
- [14].Yamazaki H, Nakano M, Imai Y, Ueng YF, Guengerich FP, Shimada T. Arch Biochem Biophys. 1996;325:174–182. doi: 10.1006/abbi.1996.0022. [DOI] [PubMed] [Google Scholar]
- [15].Yamazaki H, Johnson WW, Ueng YF, Shimada T, Guengerich FP. J Biol Chem. 1996;271:27438–27444. doi: 10.1074/jbc.271.44.27438. [DOI] [PubMed] [Google Scholar]
- [16].Perret A, Pompon D. Biochemistry. 1998;37:11412–11424. doi: 10.1021/bi980908q. [DOI] [PubMed] [Google Scholar]
- [17].Kumar S, Davydov DR, Halpert JR. Drug Metab Dispos. 2005;33:1131–1136. doi: 10.1124/dmd.105.004606. [DOI] [PubMed] [Google Scholar]
- [18].Yamazaki H, Nakamura M, Komatsu T, Ohyama K, Hatanaka N, Asahi S, Shimada N, Guengerich FP, Shimada T, Nakajima M, Yokoi T. Protein Expr Purif. 2002;24:329–337. doi: 10.1006/prep.2001.1578. [DOI] [PubMed] [Google Scholar]
- [19].Zhao C, Gao Q, Roberts AG, Shaffer SA, Doneanu CE, Xue S, Goodlett DR, Nelson SD, Atkins WM. Biochemistry. 2012;51:9488–9500. doi: 10.1021/bi301069r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Naffin-Olivos JL, Auchus RJ. Biochemistry. 2006;45:755–762. doi: 10.1021/bi051623y. [DOI] [PubMed] [Google Scholar]
- [21].Peng HM, Auchus RJ. Biochemistry. 2013;52:210–220. doi: 10.1021/bi301384n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Imaoka S, Imai Y, Shimada T, Funae Y. Biochemistry. 1992;31:6063–6069. doi: 10.1021/bi00141a015. [DOI] [PubMed] [Google Scholar]
- [23].Sherbet DP, Tiosano D, Kwist KM, Hochberg Z, Auchus RJ. J Biol Chem. 2003;278:48563–48569. doi: 10.1074/jbc.M307586200. [DOI] [PubMed] [Google Scholar]
- [24].Domanski TL, He YA, Khan KK, Roussel F, Wang Q, Halpert JR. Biochemistry. 2001;40:10150–10160. doi: 10.1021/bi010758a. [DOI] [PubMed] [Google Scholar]
- [25].Sandee D, Miller WL. Endocrinology. 2011;152:2904–2908. doi: 10.1210/en.2011-0230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Mulrooney SB, Waskell L. Protein Expr Purif. 2000;19:173–178. doi: 10.1006/prep.2000.1228. [DOI] [PubMed] [Google Scholar]
- [27].Sohl CD, Cheng Q, Guengerich FP. Nat Protoc. 2009;4:1252–1257. doi: 10.1038/nprot.2009.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Patten CJ, Ishizaki H, Aoyama T, Lee M, Ning SM, Huang W, Gonzalez FJ, Yang CS. Arch Biochem Biophys. 1992;299:163–171. doi: 10.1016/0003-9861(92)90258-x. [DOI] [PubMed] [Google Scholar]
- [29].Gillam EM, Guo Z, Guengerich FP. Arch Biochem Biophys. 1994;312:59–66. doi: 10.1006/abbi.1994.1280. [DOI] [PubMed] [Google Scholar]
- [30].Yun CH, Ahn T, Guengerich FP. Arch Biochem Biophys. 1998;356:229–238. doi: 10.1006/abbi.1998.0759. [DOI] [PubMed] [Google Scholar]
- [31].Yun CH, Song M, Kim H. J Biol Chem. 1997;272:19725–19730. doi: 10.1074/jbc.272.32.19725. [DOI] [PubMed] [Google Scholar]
- [32].Kim KH, Ahn T, Yun CH. Biochemistry. 2003;42:15377–15387. doi: 10.1021/bi035280k. [DOI] [PubMed] [Google Scholar]
- [33].Miwa GT, Lu AY. Arch Biochem Biophys. 1981;211:454–458. doi: 10.1016/0003-9861(81)90477-x. [DOI] [PubMed] [Google Scholar]
- [34].French JS, Guengerich FP, Coon MJ. J Biol Chem. 1980;255:4112–4119. [PubMed] [Google Scholar]
- [35].Eberhart DC, Parkinson A. Arch Biochem Biophys. 1991;291:231–240. doi: 10.1016/0003-9861(91)90128-6. [DOI] [PubMed] [Google Scholar]
- [36].Basova LV, Tiktopulo EI, Kutyshenko VP, Mauk AG, Bychkova VE. Biochim Biophys Acta. 2008;1778:1015–1026. doi: 10.1016/j.bbamem.2007.12.028. [DOI] [PubMed] [Google Scholar]
- [37].Lee-Robichaud P, Kaderbhai MA, Kaderbhai N, Wright JN, Akhtar M. Biochem J. 1997;321(Pt 3):857–863. doi: 10.1042/bj3210857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Hosea NA, Miller GP, Guengerich FP. Biochemistry. 2000;39:5929–5939. doi: 10.1021/bi992765t. [DOI] [PubMed] [Google Scholar]
- [39].Khan KK, He YQ, Domanski TL, Halpert JR. Mol Pharmacol. 2002;61:495–506. doi: 10.1124/mol.61.3.495. [DOI] [PubMed] [Google Scholar]
- [40].Schenkman JB, Jansson I. Pharmacol Ther. 2003;97:139–152. doi: 10.1016/s0163-7258(02)00327-3. [DOI] [PubMed] [Google Scholar]