Abstract
Interphasic chromatin condenses into the chromosomes in order to facilitate the correct segregation of genetic information. It has been previously reported that the phosphorylation and methylation of the N-terminal tail of histone H3 are responsible for chromosome condensation. In this study, we demonstrate that the deacetylation and methylation of histone H3 lysine 9 (H3K9) are required for proper chromosome condensation. We confirmed that H3K9ac levels were reduced, whereas H3K9me3 levels were increased in mitotic cells, via immunofluorescence and Western blot analysis. Nocodazole treatment induced G2/M arrest but co-treatment with TSA, an HDAC inhibitor, delayed cell cycle progression. However, the HMTase inhibitor, AdoX, had no effect on nocodazole-induced G2/M arrest, thereby indicating that sequential modifications of H3K9 are required for proper chromosome condensation. The expression of SUV39H1 and SETDB1, H3K9me3-responsible HMTases, are specifically increased along with H3K9me3 in nocodazole-arrested buoyant cells, which suggests that the increased expression of those proteins is an important step in chromosome condensa-tion. H3K9me3 was highly concentrated in the vertical chromosomal axis during prophase and prometaphase. Collectively, the results of this study indicate that sequential modifications at H3K9 are associated with correct chromosome condensation, and that H3K9me3 may be relevant to the condensation of chromosome length.
Keywords: chromosome condensation, deacetylation, H3K9, methylation, SETDB1, SUV39H1
INTRODUCTION
High ordered chromatin is important for the storage of the genetic material in the interphase nucleus (Campos and Reinberg, 2009). During cell cycle progression, chromosome condensation is an indispensable process that facilitates the resolution of sister chromatids in mitosis and the maintenance of genomic stability. The results of some recent studies have shown that histone modifications within the nucleosome induce important structural changes for chromosome condensation (Duan et al., 2008; Trojer and Reinberg, 2007).
The phosphorylation of H3S10 (p-H3S10) by Aurora B kinase regulates the dynamic condensation/relaxation of chromosomes (Hirota et al., 2005; Prigent and Dimitrov, 2003). Although p-H3S10 is detected only at minimal levels in interphase, p-H3S10 is initiated at pericentromeric regions in late G2 and increased along the chromosomal arms during mitosis (Nowak and Corces, 2004; Van Hooser et al., 1998). As it persists during prophase and metaphase, this modification has been regarded as a marker for mitosis. The tri-methylation of H3K9 (H3K9me3) was initially studied as a marker of epigenetic silencing and constitutive heterochromatin which is found close to the centromeres (Schotta et al., 2004; Stewart et al., 2005). The results of functional analyses revealed that H3K9-me3 is tightly regulated in a cell cycle-dependent manner (Heit et al., 2009; Melcher et al., 2000). As compared to the modification of p-H3S10, H3K9me3 was increased in G2, reached a maximum at metaphase, and declined rapidly during entry into the next interphase (Duan et al., 2008; McManus et al., 2006). The inhibition of H3K9me3 or p-H3S10 perturbed chromosome condensation during mitosis (Kondo et al., 2008; Lv et al., 2010; Monier et al., 2007). Moreover, p-H3S10 could influence H3K9 methylation in a selective manner, suggesting the complexity of histone modifications in mitosis (Duan et al., 2008).
Since H3K9 is a residue that can be acetylated or me-thylated, many possibilities have been explored in terms of the progression of modificational changes at H3K9, and what proteins coordinate modifications at H3K9 in various model systems such as embryonic development or gene expression models (Heit et al., 2009; Wang et al., 2007). SUV39H1 performs a crucial role in the initial steps of heterochromatin formation in mammals by selective H3K9me3 (Rea et al., 2000; Rice et al., 2003). SUV39H1-deficient mice were subjected to chromosomal instability (Peters et al., 2001). Additionally, SETDB1 methylates H3K9me3 for stable heterochromatin formation during cell cycle progression (Ryu et al., 2006; Sarraf and Stancheva, 2004). Interestingly, the complete loss of H3K9me3 might have no effect on mitosis, if a specific depletion of histone deacetylase HDAC was induced by the drugs (Warrener et al., 2010), thereby suggesting the possible existence of sequential steps necessary for precise cell cycle progression. Therefore, many H3K9me3-responsible HMTases may play an important role in mitotic progression and chromosome segregation, and there is an increased possibility that cross-talk of histone modifications is necessary for correct chromosome condensation.
Chromosome condensation is a complex mechanism that ensures cell population integrity. Since it can be acetylated and methylated at H3K9, histone deacetylation is required prior to H3K9 methylation (Stewart et al., 2005). In this study, we have determined that sequential modifications at H3K9 are required for proper chromosome condensation. Additionally, we have demonstrated that H3K9me3 was stained at the vertical axis during chromosome condensation.
MATERIALS AND METHODS
Cell culture, antibodies, and reagents
A549 lung carcinoma cells and HeLa cervical carcinoma cells were obtained from the ATCC (USA). Cells were cultured in Dulbecco’s modified Eagle’s medium plus 10% fetal bovine serumand penicillin-streptomycin (50 U/ml) in an incubator at 37℃ with 5% CO2. SUV39H1, SETDB1, G9a, and acetylation- or methylation-specific antibodies were purchased from Abcam (USA). Antibodies against cyclin B1 and actin were obtained from Santa Cruz (USA). Rhodamine-tagged secondary antibody was purchased from Chemicon (USA). Trichostatin A (TSA) was obtained from Sigma (USA), and adenosine dialdehyde (AdoX) was a gift from Dr. Huang (The Burnham institute, USA).
Histone extraction
Histones were extracted from cells via standard acid extraction procedures, as described previously (Park and Kim, 2008). In brief, after the removal of the cytoplasmic fraction, the nuclear pellets were extracted with 0.2 M H2SO4 solution. The next day, 100% TCA solution was added to the nuclear pellet, followed by acetone/HCl washing. The final white pellet was dried and dissolved in distilled water. Histone concentrations were determined via a Bio-Rad DC protein assay (USA). The histones were separated via 15% SDS-PAGE.
Immunoblot
Immunoblot analysis was conducted as described previously (Kim et al., 2006). The harvested cells were lyzed in nuclear isolation buffer (250 mM sucrose, 200 mM NaCl, 10 mM Tris-HCl pH 8.0, 2 mM MgCl2, 1 mM CaCl2, 1% Triton X-100and 1 mM phenylmethanesulfonyl fluoride) and separated via SDS-PAGE. The gels were transferred to PVDF membranes, followed by blocking with 5% non-fat dry milk in TBS-T (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 0.1% Tween20). The membranes were incubated with primary antibodies, followed by treatment with HRP-conjugated secondary antibodies. The immunoblots were visualized via ECL.
FACS analysis
The 5 × 105 cells were cultured in a 60 mm culture dish. After drug treatment, the cells were harvested, washed twice in PBS, and fixed for 24 h in 75% ethanol. The cells were stained for 30 min with propidium iodide (50 mg/ml). Cell cycle distribution was analyzed via flow cytometric analysis according to Becton Dickinson’s recommended protocols (USA).
Metaphase chromosome preparation
To obtain metaphase chromosomes, A549 cells were treated for 2 h with 0.1% colcemid. The chromosomeswere prepared with hypotonic buffer (50 mM KCl, 10 mM MgSO4, and 5 mM HEPES pH 8.0) and fixed in methanol. The lysates were dropped gently onto the slides. The slides were subsequently processed for immunostaining and DAPI staining as described below.
Immunostaining
Cells at 50-80% confluence were plated onto sterilized glass coverslips. The slides were washed in phosphate-buffered saline (PBS) and fixed for 2 min in 4% paraformaldehyde on ice, followed by 0.1% Triton X-100 treatment. The slides were then incubated with antibodies for 1 h in PBS plus 4% milk, followed by incubation with rhodamine-conjugated anti-rabbit IgG or fluorescein isothiocyanate (FITC)-conjugated anti-mouse IgG. The slides were then stained briefly with DAPI (0.5 mg/ml), washed with PBS,mounted, and analyzed via confocal microscopy.
RESULTS
Reduced acetylation and increased trimethylation of H3K9 during cell cycle progression
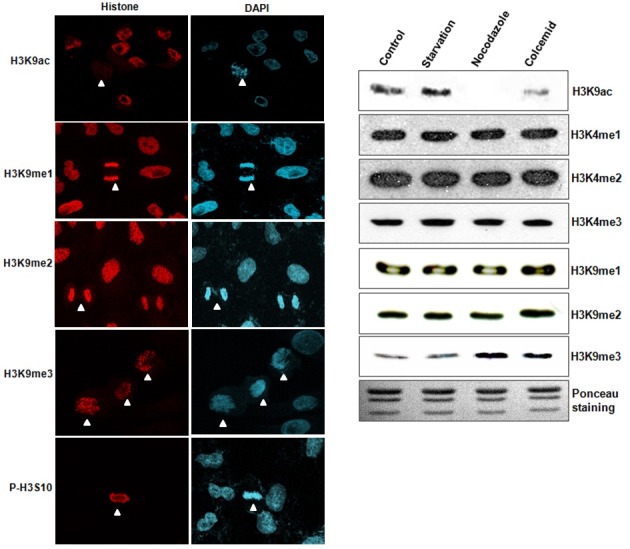
Although various histone modifications are important for correct chromosome condensation, we focused herein on the manner in which modifications at H3K9 might be altered during cell cycle progression. We conducted immunostaining in asynchronous A549 cells using specific antibodies for H3K9 modification. Cell cycle stages were assumed from the DAPI staining. The phosphorylation of H3 serine 10 (p-H3S10) was employed as a positive marker, as compared with other types of modifications. Acetylation of H3K9 (H3K9ac) was stained in most attached interphase cells, but disappeared in the mitotic cells (Fig. 1A, the first row), thereby indicating that deacetylation of H3K9 plays a role in chromosomal condensation. H3K9me3, like p-H3S10, was increased significantly in the mitotic cells (Fig. 1A, the fourth row). However, H3K9me1 and H3K9me2 evidenced no changes in staining during cell cycle progression (Fig. 1A, the second and third row). Other types of histone modifications including H3K4me1, H3K4me2, and H3K4me3 were not altered (data not shown). In order to confirm this phenomenon using different cell systems, HeLa cells were arrested in specific cell stages via serum starvation, nocodazole, or colcemid treatments. Consistent with our other findings, we determined that H3K9ac was reduced but H3K9me3 was increased in G2/M-arrested cells using Western blot analysis, whereas other types of histone modifications were unchanged (Fig. 1B). These data demonstrate that the deacetylation and methylation of H3K9 are involved in chromosome condensation during cell cycle progression in a variety of cell types.
Fig. 1. Changes of histone modifications during cell cycle progression. (A) Histone modifications (red) were determined by immunostaining in culturing A549 cells. DNA was DAPI-stained for assumptions of mitotic stage. Acetylation of H3K9 (H3K9ac) was reduced in the mitotic cells, but trimethyation of H3K9 (H3K9me3) was increased in the mitotic cells. However, monomethylation of H3K9 (H3K9me1) and dimethylation of H3K9 (H3K9me2) were not changed under identical conditions. P-H3S10 is shown as a mitotic control. White arrow head; mitotic cells. (B) Western blot analyses were conducted using synchronized HeLa cells. H3K9me3 and H3K9ac expressions were altered in the nocodazole and colcemid treatment groups, but not in the serum star-vation group.
Sequential histone modifications of H3K9 for chromosome condensation
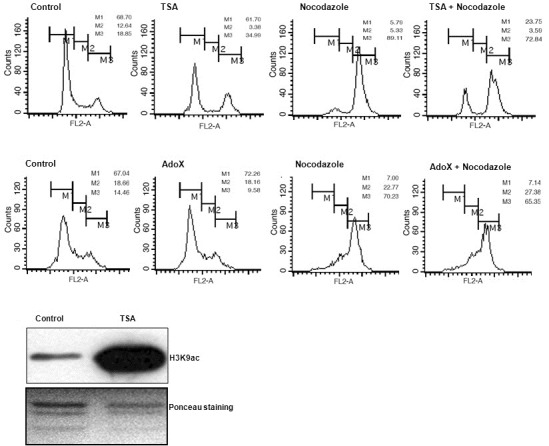
Because acetylation and methylation at H3K9 could not be simultaneously modified, we hypothesized that deacetylation of H3K9 proceeds prior to H3K9me3 in the process of chromosome condensation. To confirm this, we introduced a combination treatment of nocodazole and TSA HDAC inhibitor in A549 cells, and the cell cycle populations were evaluated via FACS analysis. Nocodazole treatment induced G2/M phase arrest in approximately 70-89% cells among the entirety of the cell population. TSA treatment induced a reduction in the G1 cell population and an increase in the G2/M phase cell population (Fig. 2A). However, combination treatment with TSA and nocodazole yielded no synergistic increase in G2/M phase, but significantly delayed the entry of the G2/M cell population. AdoX, an inhibitor of HMTases, modulates histone methylation including H3K9 (Chen et al., 2004; Park and Kim, 2008). We applied AdoX treatment to inhibit HMTase activity in a similar fashion. However, no specific changes in cell cycle distribution were observed, thereby implying that the inhibition of HMTase activity exerts no specific effect on cell cycle progression (Fig. 2B). However, although TSA treatment appears to affect chromosome condensation, it does not guarantee the deacetylation of H3K9. Therefore, we attempted to determine whether TSA re-gulates H3K9ac. As shown in Fig. 2C, TSA treatment dramatically induced H3K9ac, thus implying that H3K9 is a control point for TSA inhibition. Therefore, the data collected in this study sug-gest that the deacetylation of H3K9 is a prerequisite of, and H3K9me3 is also required for chromosome condensation.
Fig. 2. Delay of cell cycle progression in deacetylation inhibition, not in methylation inhibition. (A) Nocodazole treatment induced G2/M phase arrest in A549 cells. TSA treatment increased G2/M phase. However, combination treatment of TSA and nocodazole evidenced a delayed G2/M phase. Each cell population was measured and expressed as a number. (B) AdoX was treated for HMTase inhibition. AdoX had no effect on nocodazole-induced G2/M arrest. (C) Western blot analyses were conducted after TSA treatment in HeLa cells. H3K9ac levels were increased in the TSA treatment group.
Increased expression of H3K9me3-responsible SUV39H1 and SETDB1 HMTases
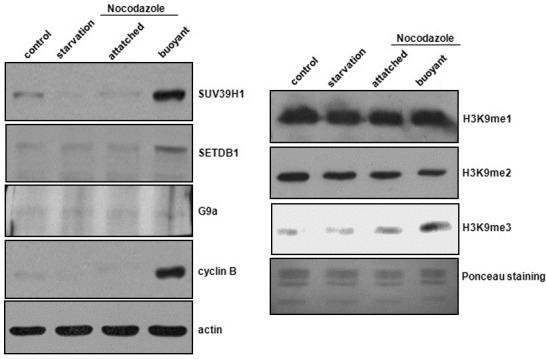
There is a possibility that H3K9me3-responsible proteins may be regulated for an increased level of H3K9me3 during cell cycle progression. We evaluated the expression levels of SUV39H1 and SETDB1, which have been recognized as H3K9me3-responsible HMTases, after nocodazole treatment in the HeLa cells. We conducted Western blot analysis after the separation of the attached and buoyant HeLa cells for the quantitative analysis of G2/M cell population. Over93% of the buoyant cells werein G2/M phase, whereas no significant G2/M arrest was noted in the attached cells (data not shown). Cyclin B expression was examined as a G2/M-specific marker. Expression levels of SUV39H1 and SETDB1 were increased only in the buoyant cells, and not in the attached cells (Fig. 3A). Consistent with this finding, H3K9me3 levels were also increased in the buoyant cells (Fig. 3B). We also conducted Western blot analysis on G9a, which is an HMTase responsible for H3K9me2, but no changes in its expression were detected. H3K9me1 and H3K9me2 were also unchanged. Collectively, our findings suggest that the increased expression of H3K9-me3-responsible HMTases might constitute an important step for chromosome condensation.
Fig. 3. Increased expression of H3K9me3-responsible HMT-ases. Nocodazole was applied for 12 h, and the at-tached cells and buoyant cells, respectively, were harvested. (A) Western blotting was conducted with various antibodies including SUV39H1, SETDB1, and G9a. Cyclin B was employed as a marker for G2/M phase. (B) Western blot analyses were conducted in order to detect histone modifications. H3K9me3 was increased in the buoyant cells, whereas nei-ther H3K9me1 or H3K9me2 were altered.
Cooperative modificational changes of methylation at H3K9 during mitotic progression
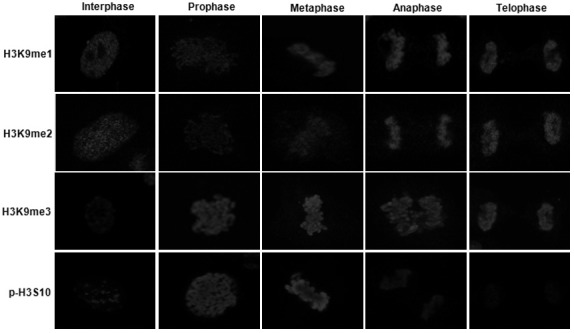
We attempted to analyze the mechanisms underlying the sequential modifications of H3K9 occurring during mitotic progression. We carried out immunostaining in asynchronous A549 cells, and captured cells in different stages of mitosis by postulating their chromosomal shapes. Since acetylation of H3K9 was observed only in interphase, we were unable to detect it in late G2 (data not shown). H3K9me3 was observed only at minimal levels in interphase, increased in late G2, and sustained to telophase in the mitotic cells (Fig. 4). Interestingly, modifications of H3K9me1 and H3K9me2 were noted in interphase, temporalily reduced in prophase and metaphase, and then resumed in anaphase, thus suggesting that H3K9me1 and H3K9me2 might be involved in different ways with H3K9me3 during chromosomal condensation. Unlike H3K9me1 and H3K9me2, p-H3S10 was observed at minimal levels in inter-phase, increased dramatically in prophase and metaphase, and disappeared in anaphase. These results indicate that mono-, di-, and tri-methylation at H3K9 might be involved in chromosomal condensation during mitosis.
Fig. 4. Histone modification profiles during mitotic stages. Histone modifications were determined by immunostaining in cultured A549 cells. The histone modifications evidenced distinct patterns relative to interphase cells. H3K9me1 and H3K9me2 were temporarily decreased in mitotic entry, but restored at anaphase. H3K9me3 levels were increased during mitotic stages. As a control, p-H3S10 was also increased at prophase and metaphase, and reduced after anaphase.
Distinct pattern of H3K9me3 at the vertical axis in prophase and prometaphase chromosomes
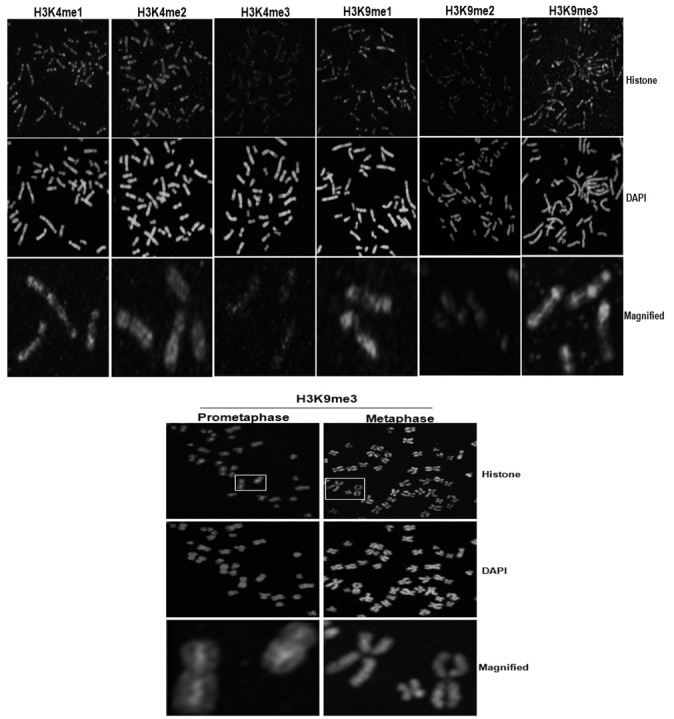
To analyze modifications of H3K9 occurring at the chromosome level, we prepared mitotic chromosomes via colcemid treatment in the A549 cells. As shown in Fig. 5, H3K4me1, H3K4me2, H3K4me3, H3K9me1, and H3K9me2 evidenced similar staining patterns in chromosomes in prophase. Those modifications were shown to be dispersed in the chromosome arms but were not observed in centromeric regions, thus suggesting that these are important modifications for the structural condensation of chromosomes. However, H3K9me3 was shown to be strong at the vertical axis in prophase chromosomes. This staining pattern was also observed at pro-metaphase, thereby indicating that H3K9me3 may be involved in the condensation of chromosome length at the beginning of mitosis. The H3K9me3 of the metaphase chromosomes stained evenly. This result may then reflect an association between H3K9me3 and chromosome condensation length.
Fig. 5. Analyses of histone modifications at the chromosome levels. To extract mitotic chromosomes, A549 cells were arrested by colcemid and spread onto the slides with hypotonic solution, followed by immunostaining with the indicated histone antibodies. Histone modifications were detected in the chromosome arms, where-as H3K9me3 was detected at the vertical axis of the chromosome during prophase and prometaphase.
DISCUSSION
In this study, we emphasized that sequential modifications of H3K9 (deacetylation and methylation) are associated with chromosome condensation during cell cycle progression. We determined that H3K9ac was reduced dramatically during chromosome condensation, whereas H3K9me3 was concomitantly increased under identical conditions. Increased expression of H3K9me3-responsible HMTases is a requirement for increased H3K9me3 during mitotic stages. Moreover, we also determined that modifications of H3K9me1 and H3K9me2 were temporarily reduced at prophase and reappeared at metaphase. However, histone modifications at H3K4 were not altered in our analyses. These results reveal that progressive methylations after deacetylation at H3K9 could be indispensable histone modifications for proper chromosome condensation.
TSA, a well-known HDAC inhibitor, can inhibit cell growth via G2/M arrest and can also induce terminal differentiation (Blagosklonny et al., 2002; Yao et al., 2006). However, entry into G2/M phase was delayed by a combination treatment of TSA and nocodazole, even though TSA itself appears to influence G2/M arrest. Class I HDACs, including HDAC 1, 2, and 8, are detected primarily in the nucleus and their activities are inhibited by TSA, whereas NAD+-dependent HDAC family proteins are unaffected by TSA (de Ruijter et al., 2003). In this regard, class I HDACs might be proteins responsible for chro-mosome condensation. In fact, the Class I HDACs are involved in the development of the transcriptionally repressive state to produce high-order chromosomal structures (Ma and Schultz, 2008). At the protein levels, the expression or activity of HDACs could be increased in chromosome condensation during cell cycle progression (Li et al., 2006; Magnaghi-Jaulin and Jaulin, 2006; Parker et al., 2007; Shin et al., 2003). Moreover, as TSA treatment obviously induced H3K9 acetylation in our Western blot analysis, we assumed that the delay of G2/M phase due to co-treatment with TSA and nocodazole might have resulted in a deficiency of deacetylation of H3K9, as at least one control point. However, it still remains possible that the acetylation/ deacetylation of other histone residues are involved in chromosome condensation. In contrast with this phenomenon, AdoX, an inhibitor of histone methylation, has no effect on nocodazole-induced G2/M arrest. This may be no obstacle to prerequisites such as the deacetylation step for chromosome conden-sation, strongly suggesting that deacetylation of H3K9 might function as a cue for chromosome condensation (Warrener et al., 2010). Some research groups have also suggested that AdoX induced apoptosis via G2/M arrest in leukemia cells (Dasgupta et al., 2008; Schwerk and Schulze-Osthoff, 2005). The effects of AdoX might vary in different cell contexts, or may be concentration-dependent. Although the inhibitory effect of AdoX may vary, it was interesting to note that the inhibition of histone methyla-tion by AdoX had no effect on G2/M entry.
It is conceivable that mono, di, tri-methylation of H3K9 is known to localize at repressive heterochromatin (Fodor et al., 2006). Our Western blot analysis showed that SUV39H1 and SETDB1 were dramatically increased in the G2/M cells. Because SUV39H1 and SETDB1 is H3K9me3-responsible HMT-ases, its expression could be correlated with increased enzyme activity. Indeed, we have seen many evidences for increased H3K9me3 in our various analyses. Therefore, we strongly suggest that expression of H3K9me3-responsible HMTases might be collectively involved in chromosome condensation. However, H3K9me2-responsible G9a was not changed in same experiment. Interestingly, our mitotic profiles on histone modifications have shown that H3K9me1 and H3K9me2 were disappeared in prophase, the beginning point of the mitosis, and reappeared from metaphase. In this meaning, although G9a expression has no change in G2/M cells, it remains possible that different types of regulatory mechanisms on H3K9me1 or H3K9me2 might exist (Fig. 4). It was also interesting to note that modifications between p-H3S10 and H3K9me1 or H3K9me were opposite in mitosis entry. A kind of stereotypic hindrance may plausibly exist between p-H3S10 and H3K9me1or H3K9me2 (Duan et al., 2008). However, the increased levels of H3K9me3 observed herein were maintained from prophase to telophase, thereby indicating that H3K9me3 might constitute a key modification event occurring after deacetylation of H3K9 during chromosomal condensation. Our analysis demonstrates that only H3K9-me3 was strong at the vertical axis in prophase and pro-metaphase chromosomes. Chromatin condenses into discrete chromosomes at prophase (Kim et al., 2009; Neitzel et al., 2002). H3K9me3 may be involved in the condensation of chromosome length at the beginning of mitosis. In other words, H3K9me3 is an indispensable modification for longitudinal chromosomal condensation. After determining that H3K9me3 might be associated with condensation for chromosome length and sister chromatid pairing, some proteins could be evaluated for possible functional interactions during cell cycle progression.
Chromosome condensation is a critically important step in cell division in mammals. It is imperative that various histone modifications are required for this process. Although we demonstrate sequential modifications at H3K9 in this paper, future studies will be necessary to elucidate the epigenetic relationships among other types of histone modifications in the context of chromosomal condensation.
Acknowledgments
The authors appreciate the Korea Basic Science Institute (KBSI) for technical assistance regarding microscopic image analysis. This work was supported by grants from the Korean Research Foundation (KRF C00141), and the Ministry of Education, Science and Technology (the Regional Research Universities Program/Medical and Bio-Materials Research Center), S. Korea. This work was also supported by the visiting scholar program of Kangwon National University.
References
- 1.Blagosklonny M.V., Robey R., Sackett D.L., Du L., Traganos F., Darzynkiewicz Z., Fojo T., Bates S.E. Histone deacetylase inhibitors all induce p21 but differentially cause tubulin acetylation, mitotic arrest, and cytotoxicity. Mol. Cancer Ther. (2002);1:937–941. [PubMed] [Google Scholar]
- 2.Campos E.I., Reinberg D. Histones: annotating chromatin. Annu. Rev. Genet. (2009);43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- 3.Chen D.H., Wu K.T., Hung C.J., Hsieh M., Li C. Effects of adenosine dialdehyde treatment on in vitro and in vivo stable protein methylation in HeLa cells. J. Biochem. (2004);136:371–376. doi: 10.1093/jb/mvh131. [DOI] [PubMed] [Google Scholar]
- 4.Dasgupta A., Jung K.J., Jeong S.J., Brady J.N. Inhi-bition of methyltransferases results in induction of g2/m checkpoint and programmed cell death in human T-lymphotropic virus type 1-transformed cells. J. Virol. (2008);82:49–59. doi: 10.1128/JVI.01497-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Ruijter A.J., van Gennip A.H., Caron H.N., Kemp S., van Kuilenburg A.B. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem. J. (2003);370:737–749. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duan Q., Chen H., Costa M., Dai W. Phosphorylation of H3S10 blocks the access of H3K9 by specific antibodies and histone methyltransferase. Implication in regulating chromatin dynamics and epigenetic inheritance during mitosis. J. Biol. Chem. (2008);283:33585–33590. doi: 10.1074/jbc.M803312200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fodor B.D., Kubicek S., Yonezawa M., O’Sullivan R.J., Sengupta R., Perez-Burgos L., Opravil S., Mechtler K., Schotta G., Jenuwein T. Jmjd2b antagonizes H3K9 trimethylation at pericentric heterochromatin in mammalian cells. Genes Dev. (2006);20:1557–1562. doi: 10.1101/gad.388206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heit R., Rattner J.B., Chan G.K., Hendzel M.J. G2 histone methylation is required for the proper segregation of chromosomes. J. Cell Sci. (2009);122:2957–2968. doi: 10.1242/jcs.045351. [DOI] [PubMed] [Google Scholar]
- 9.Hirota T., Lipp J.J., Toh B.H., Peters J.M. Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature. (2005);438:1176–1180. doi: 10.1038/nature04254. [DOI] [PubMed] [Google Scholar]
- 10.Kim K.C., Geng L., Huang S. Inactivation of a histone methyltransferase by mutations in human cancers. Cancer Res. (2003);63:7619–7623. [PubMed] [Google Scholar]
- 11.Kim K.C., Jung C.S., Choi K.H. Overexpression of p73 enhances cisplatin-induced apoptosis in HeLa cells. Arch. Pharm. Res. (2006);29:152–158. doi: 10.1007/BF02974277. [DOI] [PubMed] [Google Scholar]
- 12.Kim H., Lee S., Park B., Koh W., Lee D.J., Lim D.S., Lee S. Cancer-upregulated gene 2 (CUG2), a new component of centromere complex, is required for kinetochore function. Mol. Cells. (2009);27:697–701. doi: 10.1007/s10059-009-0083-2. [DOI] [PubMed] [Google Scholar]
- 13.Kondo Y., Shen L., Ahmed S., Boumber Y., Sekido Y., Haddad B.R., Issa J.P. Downregulation of histone H3 lysine 9 methyltransferase G9a induces centrosome disruption and chromosome instability in cancer cells. PLoS One. (2008);3:e2037. doi: 10.1371/journal.pone.0002037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y., Kao G.D., Garcia B.A., Shabanowitz J., Hunt D.F., Qin J., Phelan C., Lazar M.A. A novel histone deacetylase pathway regulates mitosis by modulating Aurora B kinase activity. Genes Dev. (2006);20:2566–2579. doi: 10.1101/gad.1455006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lv S., Bu W., Jiao H., Liu B., Zhu L., Zhao H., Liao J., Li J., Xu X. LSD1 is required for chromosome segregation during mitosis. Eur. J. Cell Biol. (2010);89:557–563. doi: 10.1016/j.ejcb.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 16.Ma P., Schultz R.M. Histone deacetylase 1 (HDAC1) regulates histone acetylation, development, and gene expression in preimplantation mouse embryos. Dev. Biol. (2008);319:110–120. doi: 10.1016/j.ydbio.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Magnaghi-Jaulin L., Jaulin C. Histone deacetylase activity is necessary for chromosome condensation during meiotic maturation in Xenopus laevis. Chromosome Res. (2006);14:319–332. doi: 10.1007/s10577-006-1049-2. [DOI] [PubMed] [Google Scholar]
- 18.McManus K.J., Biron V.L., Heit R., Underhill D.A., Hendzel M.J. Dynamic changes in histone H3 lysine 9 methylations: identification of a mitosis-specific function for dynamic methylation in chromosome congression and segregation. J. Biol. Chem. (2006);281:8888–8897. doi: 10.1074/jbc.M505323200. [DOI] [PubMed] [Google Scholar]
- 19.Melcher M., Schmid M., Aagaard L., Selenko P., Laible G., Jenuwein T. Structure-function analysis of SUV39H1 reveals a dominant role in heterochromatin organization, chromosome segregation, and mitotic progression. Mol. Cell. Biol. (2000);20:3728–3741. doi: 10.1128/mcb.20.10.3728-3741.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Monier K., Mouradian S., Sullivan K.F. DNA methylation promotes Aurora-B-driven phosphorylation of histone H3 in chromosomal subdomains. J. Cell Sci. (2007);120:101–114. doi: 10.1242/jcs.03326. [DOI] [PubMed] [Google Scholar]
- 21.Neitzel H., Neumann L.M., Schindler D., Wirges A., Tonnies H., Trimborn M., Krebsova A., Richter R., Sperling K. Premature chromosome condensation in humans associated with microcephaly and mental retardation: a novel autosomal recessive condition. Am. J. Hum. Genet. (2002);70:1015–1022. doi: 10.1086/339518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nowak S.J., Corces V.G. Phosphorylation of histone H3: a balancing act between chromosome condensation and transcriptional activation. Trends Genet. (2004);20:214–220. doi: 10.1016/j.tig.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 23.Park J.A., Kim K.C. A close link between meta-bolic pathway and methylation of histone H3 lysine 9 (H3K9). Korean J. Genet. (2008);30:11–16. [Google Scholar]
- 24.Parker K., Maxson J., Mooney A., Wiley E.A. Class I histone deacetylase Thd1p promotes global chromatin conden-sation in Tetrahymena thermophila. Eukaryot. Cell. (2007);6:1913–1924. doi: 10.1128/EC.00217-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peters A.H., O’Carroll D., Scherthan H., Mechtler K., Sauer S., Schofer C., Weipoltshammer K., Pagani M., Lachner M., Kohlmaier A., et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. (2001);107:323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- 26.Prigent C., Dimitrov S. Phosphorylation of serine 10 in histone H3, what for? J. Cell Sci. (2003);116:3677–3685. doi: 10.1242/jcs.00735. [DOI] [PubMed] [Google Scholar]
- 27.Rea S., Eisenhaber F., O’Carroll D., Strahl B.D., Sun Z.W., Schmid M., Opravil S., Mechtler K., Ponting C.P., Allis C.D., et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. (2000);406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 28.Rice J.C., Briggs S.D., Ueberheide B., Barber C.M., Sha-banowitz J., Hunt D.F., Shinkai Y., Allis C.D. Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol. Cell. (2003);12:1591–1598. doi: 10.1016/s1097-2765(03)00479-9. [DOI] [PubMed] [Google Scholar]
- 29.Ryu H., Lee J., Hagerty S.W., Soh B.Y., McAlpin S.E., Cormier K.A., Smith K.M., Ferrante R.J. ESET/SETDB1 gene expression and histone H3 (K9) trimethylation in Huntington’s disease. Proc. Natl. Acad. Sci. USA. (2006);103:19176–19181. doi: 10.1073/pnas.0606373103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sarraf S.A., Stancheva I. Methyl-CpG binding protein MBD1 couples histone H3 methylation at lysine 9 by SETDB1 to DNA replication and chromatin assem-bly. Mol. Cell. (2004);15:595–605. doi: 10.1016/j.molcel.2004.06.043. [DOI] [PubMed] [Google Scholar]
- 31.Schotta G., Lachner M., Sarma K., Ebert A., Sengupta R., Reuter G., Reinberg D., Jenuwein T. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. (2004);18:1251–1262. doi: 10.1101/gad.300704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwerk C., Schulze-Osthoff K. Methyltransferase inhibition induces p53-dependent apoptosis and a novel form of cell death. Oncogene. (2005);24:7002–7011. doi: 10.1038/sj.onc.1208855. [DOI] [PubMed] [Google Scholar]
- 33.Shin H.J., Baek K.H., Jeon A.H., Kim S.J., Jang K.L., Sung Y.C., Kim C.M., Lee C.W. Inhibition of histone deacetylase activity increases chromosomal instability by the aberrant regulation of mitotic checkpoint activation. Oncogene. (2003);22:3853–3858. doi: 10.1038/sj.onc.1206502. [DOI] [PubMed] [Google Scholar]
- 34.Stewart M.D., Li J., Wong J. Relationship between histone H3 lysine 9 methylation, transcription repression, and heterochromatin protein 1 recruitment. Mol. Cell. Biol. (2005);25:2525–2538. doi: 10.1128/MCB.25.7.2525-2538.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trojer P., Reinberg D. Facultative heterochromatin: is there a distinctive molecular signature? Mol. Cell. (2007);28:1–13. doi: 10.1016/j.molcel.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 36.Van Hooser A., Goodrich D.W., Allis C.D., Brinkley B.R., Mancini M.A. Histone H3 phosphorylation is required for the initiation, but not maintenance, of mammalian chromosome condensation. J. Cell Sci. (1998);111:3497–3506. doi: 10.1242/jcs.111.23.3497. [DOI] [PubMed] [Google Scholar]
- 37.Wang F., Kou Z., Zhang Y., Gao S. Dynamic reprogramming of histone acetylation and methylation in the first cell cycle of cloned mouse embryos. Biol. Re-prod. (2007);77:1007–1016. doi: 10.1095/biolreprod.107.063149. [DOI] [PubMed] [Google Scholar]
- 38.Warrener R., Chia K., Warren W.D., Brooks K., Gabrielli B. Inhibition of histone deacetylase 3 produces mitotic defects independent of alterations in histone H3 lysine 9 acetylation and methylation. Mol. Pharmacol. (2010);78:384–393. doi: 10.1124/mol.109.062976. [DOI] [PubMed] [Google Scholar]
- 39.Yao J., Duan L., Fan M., Wu X. NF-kappaB sig-naling pathway is involved in growth inhibition, G2/M arrest and apoptosis induced by Trichostatin A in human tongue carcinoma cells. Pharmacol. Res. (2006);54:406–413. doi: 10.1016/j.phrs.2006.08.003. [DOI] [PubMed] [Google Scholar]


