Abstract
Arabidopsis WRKY proteins are plant-specific transcrip-tion factors, encoded by a large gene family, which contain the highly conserved amino acid sequence WRKYGQK and the zinc-finger-like motifs, Cys2His2 or Cys2HisCys. They can recognize and bind the TTGAC(C/T) W-box cis-elements found in the promoters of target genes, and are involved in the regulation of gene expression during pathogen defense, wounding, trichome development, and senescence. Here we investigated the physiological function of the Arabidopsis WRKY22 transcription factor during dark-induced senescence. WRKY22 transcription was suppressed by light and promoted by darkness. In addi-tion, AtWRKY22 expression was markedly induced by H2O2. These results indicated that AtWRKY22 was involved in signal pathways in response to abiotic stress. Dark-treated AtWRKY22 over-expression and knockout lines showed accelerated and delayed senescence phenotypes, respectively, and senescence-associated genes exhibited increased and decreased expression levels. Mutual regulation existed between AtWRKY22 and AtWRKY6, AtWR-KY53, and AtWRKY70, respectively. Moreover, AtWRKY22 could influence their relative expression levels by feedback regulation or by other, as yet unknown mechanisms in response to dark. These results prove that AtWRKY22 participates in the dark-induced senescence signal transduction pathway.
Keywords: abiotic stress, AtWRKY22, dark, senescence
INTRODUCTION
Leaf senescence is a developmentally programmed degeneration process that constitutes the final step of leaf development. Senescence is controlled by multiple developmental and environmental signals (Lim et al., 2003). It often occurs in an age-dependent manner and is affected by complex interaction between developmental age and factors such as shading, extreme temperature, drought, and exposure to nutrient deficiency (Woo et al., 2004). Although leaf senescence is a succession of physiological and molecular events, the process can be split into initiation, degeneration, and terminal phases according to the three-stage theory (Yoshida, 2003). In Arabidopsis the initiation of natural senescence is associated with the developmental aging process, and senescence occurs just after a particular developmental time point (Buchanan-Wollas-ton, 1997). At the time of initiation of leaf senescence, photosynthesis activity begins to decrease and the function of leaves transforms from sink to source (Yoshida, 2003). The degeneration processes are characterized by the degradation of macromolecules and the disassembly of cellular components, which are accompanied by the mobilization and translocation of valuable resources from the leaves to growing organs or reserve organs (Buchanan-Wollaston, 1997). The terminal phase results in the death or abscission of the whole leaf and the symptoms of leaf senescence become visible completely.
To understand the molecular mechanisms of foliar senescence, many genes expressed during senescence have been identified from a variety of plant species, including Arabidopsis, tomato, maize, and rice (Buchanan-Wollaston, 1997; He and Gan, 2002; Hinderhofer and Zentgraf, 2001; Oh et al., 1996; Park et al., 1998; Quirino et al., 2000). For example, specific sets of genes, designated senescence-associated genes (SAGs), are up-regulated during foliar senescence, including proteases, nucleases, lipid-, carbohydrate- and nitrogen-metabolizing enzymes, stress-responsive proteins, and transcriptional regulators, which are involved in the breakdown of cellular compo-nents (Buchanan-Wollaston et al., 2003). Different sets of genes, designated senescence-down-regulated genes, are down-regulated during foliar senescence, including the genes related to photosynthesis (Gan et al., 1997). Other gene products are associated with mobilization of nutrients and minerals from senescing tissue to developing parts of plants (Buchanan-Wollaston, 1997). Current molecular studies on leaf senescence are mainly focused on genes up-regulated during senescence. On the basis of the functions of their protein products, genes related to senescence have been classified into six conceptual categories (Lim et al., 2003). Class I genes control the developmental aging process by affecting metabolic rate of cell. For example, mutation of ORE4 leads to retardation of age-dependent senescence, but dark- and phytohormone-induced leaf senescence remains largely unaffected (Woo et al., 2002). Class II consists of genes related to synthesis and response of phytohormones (Grbic et al., 1995; Lanahan et al., 1994; Oh et al., 1997). These genes can regulate other endogenous biological processes in addition to leaf senescence. Class III genes can alter senescence in response to environmental factors. Class V genes are involved in the degradation process of senescence regulatory factors. For example, ORE9 encodes an F-box protein that is a component of the ubiquitin E3 ligase complex (Woo et al., 2001), and has a role in the degradation of proteins that negatively regulate leaf senescence. Class VI genes operate downstream of the senescence signal transduc-tion pathway that is involved in executing the senescence process. The genes in class I-V are primarily involved in the initiation and/or progression of senescence, and the genes in class VI are largely involved in the progression of senescence (Lim et al., 2003).
Regulatory genes that are classified into class IV have an important role in recognizing and transducing the age or stress information into senescence-related physiology or the regulation of senescence-associated genes (SAGs). Limited numbers of regulating factors involved in leaf senescence have been identified up to now. It is necessary to identify possible candidates for regulatory genes to delineate the molecular mechanism underlying leaf senescence. WRKY proteins are a superfamily of transcription factors with potential regulatory roles related to various biotic and abiotic stress responses. Arabidopsis WRKY proteins participate in regulation of plant devel-opment (Johnson et al., 2002), material metabolism (Devaiah et al., 2007), abiotic stress (Li et al., 2009; 2010; Qiu et al., 2009), seed dormancy and germination (Jiang et al., 2009). They also regulate plant responses to disease resistance and establish the corresponding pathway of signal transduction (Chen et al., 2002; Journot-Catalino et al., 2006; Yu et al., 2001). Expression profiling in Arabidopsis reveals that WRKY transcription factors are the second largest family of transcription factors in the se-nescence transcriptome (Guo et al., 2004). In addition, 36% of Arabidopsis WRKY genes show at least twofold changes in transcription levels after 1- or 5-day dark treatment (Lin, 2004). These results indicate that some WRKY proteins play an important role in the process of dark-induced leaf senescence. WRKY6 and WRKY53 in Arabidopsis appear to be involved in the regulation of senescence. AtWRKY6 controls plant senescence and pathogen defense by specifically binding to the W-box in the promoter of the senescence-induced receptor-like kinase (SIRK) gene in Arabidopsis (Robatzek and Somssich, 2002). The expression of AtWRKY53 increases at the early stages but decreases at the later stages of leaf senescence, suggesting that AtWRKY53 has a regulatory role in the early stages of leaf senescence (Hinderhofer et al., 2001). Although the functions of some Arabidopsis WRKY genes in senescence have come to light, the functions of other leaf senescence-associated Arabidopsis WRKY transcription factors remain to be elucidated in detail.
AtWRKY22 is one target gene of AtWRKY53 (Miao et al., 2004), at the same time we have found that the expression of AtWRKY22 is induced by darkness, indicating that AtWRKY22 has an important role during dark-induced senescence. Thus, it is necessary to elucidate the function of AtWRKY22 during dark-induced senescence to understand the molecular mechanism of senescence. In this study, we use AtWRKY22 over-expression lines and T-DNA insertion mutants to investigate the role of AtWRKY22 during dark-induced senescence. Our results show that in the AtWRKY22 T-DNA insertion mutants, senescence is delayed, whereas in the AtWRKY22 over-expression lines senescence is accelerated, compared with control lines.
MATERIALS AND METHODS
Plant materials and growth conditions
Arabidopsis thaliana ecotype Columbia (Col-0) was used as the wild-type strain. The WRKY22 mutant lines (W22m-1, salk_ 094892; W22m-2, salk_056943) and 35S:W22 plants were obtained using the background of A. thaliana ecotype Columbia (Col-0). Seeds were surface sterilized, sown on Murashige and Skoog medium containing 1% sucrose and cold-treated for three days at 4℃. The plates were illuminated with white fluorescent light in a growth chamber at 22℃ under long-day (16 h light/8 h dark) conditions. After seven days seedlings were trans- ferred to soil, and were grown in an environmentally controlled growth room at 22℃ with a 16 h light/8 h dark photoperiod with moderate light intensity (150 μM m-2 s-1).
Induction treatment
For H2O2 treatment, 3%H2O2 (gram/100 ml) was sprayed onto the rosette leaves of four-week-old wild-type plants and leaf samples were harvested at one hour interval for RNA extraction. For dark treatment, fully developed leaves detached from four-week-old plants were placed on 9-mm-diameter Petri dishes with double-layer Whatman filter papers in the base and containing 15 ml distilled water. The Petri dishes were placed in the dark room at 22℃ (Guo and Crawford, 2005). Detached leaves were harvested every one and half days or three days for RNA extraction. The leaves were sampled after four-day dark treatment for determination of chlorophyll content, staining of dead cells with Evans Blue, and determination of cell death rate.
Over-expression lines and identification of Arabidopsis WRKY22 insertion mutants
To generate the 35S:W22 construct, the AtWRKY22 cDNA was cloned into the transformation vector pOCA30, which contains the modified cauliflower mosaic virus (CaMV) 35S promoter (35S:W22). The resulting plasmid was transformed into Agrobacterium tumefaciens strain LBA4404 and introduced into plants by the floral-dip method (Clough and Bent, 1998). Trans-genic seedlings were selected on kanamycin medium (50 μg.ml-1) to identify T1 transgenic plants. The two WRKY22 T-DNA insertion lines were obtained from the Arabidopsis Biological Resource Center (USA). The two WRKY22 mutants were tracked by genomic polymerase chain reaction (PCR) using the gene-specific primers wrky22m-A (5’-CGAGTTAAA-CAAGATGTATCGAGCT3’) and wrky22m-B (5’-AACCCATC-AAAGGTTCACCATA-3’) and the T-DNA-specific right-border primer KO-1 (5’-AAACGTCCGCAATGTGTTAT-3’).
RNA blotting analysis
Total RNA was isolated from detached leaves, as described by Lagrimini et al. (1987). Twenty micrograms of cellular RNA was size-fractionated by electrophoresis through a 1.2% formaldehyde-agarose gel and then capillary-blotted onto Hybond-N+ nylon membranes. Subsequently, the membranes were hybridized to 32P-dATP-labeled probes at 68℃ according to standard procedures (Sambrook et al., 1989). After hybridization, the membranes were washed with 2× SSC and 0.5% SDS at 68℃ for 10 min, and twice with 0.5× SSC and 0.1% SDS at 68℃ for 20 min every time. Finally, the membranes were washed with 0.1× SSC and 0.1% SDS at 68℃ for 20 min and then exposed to x-ray film.
Quantitative real-time PCR analysis
After the leaves were dark-treated with dark for 36 h or 72 h, samples were frozen immediately in liquid nitrogen, and stored at -80℃. RNA was extracted from these samples using an RNeasy Plant Mini kit (Qiagen, USA). One microgram of DNase- treated total RNA was used as a template for first-strand cDNA synthesis. Reverse transcription was performed with M-MLV Reverse Transcriptase (Sigma-Aldrich) and oligo (dT) primers in a 20 μl reaction volume. One microliter of reverse transcription product was used as template for qPCR. Reactions (20 μl each) were performed using the Lightcycler FastStart DNA Master SYBR Green I kit (Roche, Germany) on a Roche Light-Cycler 480 real-time PCR machine, according to the manufacturer’s instructions. ACT2 (AT3G18780) was used as a control in qPCR. Gene-specific primers for detecting transcripts of ACT2, WRKY22, WRKY6, WRKY53, WRKY70, SAG12, SAG18, SAG20 and SIRK are listed in Table 1. The qPCR reactions (20 μl each) for these genes contained the following: 10 μl 2× TransStart Green SuperMix, 0.5 μM forward and reverse primers, and 1 μl cDNA. The annealing temperature was 60℃ in all cases. A no-template control was routinely included to confirm the absence of DNA or RNA contamination. The mean value of four replicates was normalized using the ACT2 gene as the control. Standard curves were generated using linearized plasmid DNA for each gene of interest. A second set of experiments was conducted on an independent set of tissue as a control.
Table 1.
List of quantitative RT-PCR primer sequences
| Quantitative RT-PCR primers | ||
|---|---|---|
| Gene | Primer sequences (5’ → 3’) | |
| Primer forward | Primer reverse | |
| AtWRKY22 (AT4G01250) | ACT2 AT3G18780 | CGTTTCTGGTTCTGTGGCTTT |
| AtWRKY6 (AT1G62300) | AGGAAGAACAAGATGATCGAACGGACG | TCACCAACTCATTTTTCGCACGCT |
| AtWRKY53 (AT4G23810) | GACGGGGATGCTACGGTTT | TTTTGGGTAATGGCTGGTTTG |
| AtWRKY70 (AT3G56400) | ACTTGAGGACGCATTTTCTTGGAGG | TGCTTTGTTGCCTTGCACCCTT |
| SAG12 (AT5G45890) | TCCAATTCTATTCGTCTGGTGTGT | CCACTTTCTCCCCATTTTGTTC |
| SAG18 (AT1G71190) | GTTTGCGAGGTGAGAAAATAGGA | AGAGTAGCATCGTTTGGGTGAAG |
| SAG20 (AT3G10985) | TCGGTAACGTTGTTGCTGGA | ACCAAACTCTTTCAAATCGCCA |
| SIRK (AF486619) | AGCAGCTCAATTAAGTAAATGGCG | CCCGCAATCTATACTTATGAAACCA |
| ACT2 AT3G18780 | TGTGCCAATCTACGAGGGTTT | TTTCCCGCTCTGCTGTTGT |
Quantitative RT-PCR primer sequences of AtWRKY22, AtWRKY6, AtWRKY53, AtWRKY70, SAG12, SAG18, SAG20, ACT2 and SIRK.
Measurement of chlorophyll content
Chlorophyll was extracted from the fourth fully-grown leaf with 80% (v/v) acetone. Total chlorophyll content was determined spectrophotometrically at 652 nm following the method of Lichtenthaler (1987).
Measurement of cell death
The fourth fully-grown leaves, which were dark-treated for four days, were completely submerged in 0.1% (grams per one hundred milliliters) aqueous Evans Blue dye (Sigma-Aldrich) and subjected to two five-min periods of vacuum followed by 30 min under vacuum. The leaves were then washed three times with distilled water (15 min each). Dye bound to dead cells was solubilized in 50% (v/v) methanol and 1% (w/v) SDS at 60℃ for 30 min and then quantified by absorbance at 600 nm. For 100% cell death, the detached leaves were heated at 100℃ for 5 min before staining. Two-to-three leaves were pooled for each sample (Koch and Slusarenko, 1990). Six samples were analyzed for each data point. This experiment was repeated six times with equivalent results.
RESULTS
cDNA cloning, sequence analysis, and constitutive expression of AtWRKY22 in different Arabidopsis tissues with different developmental ages
The expression of Arabidopsis WRKY22 was induced by pathogen attack (Dong et al., 2003). Therefore, we inoculated four-week-old Arabidopsis rosette leaves with a DC3000 avirulent strain of P. syringae pv. Tomato, and harvested the inoculated-leaves after four hours. Total RNA was extracted in preparation for reverse transcription. A full length AtWRKY22 cDNA sequence was obtained by RT-PCR and comprised 897 bp encoding a predicted 298 amino acid protein. Sequence analysis at the amino acid level showed that the full-length AtWRKY22 protein contained a single WRKY domain (WRKY-GQK) and a characteristic Cys2His2 zinc-finger-like at its C-terminus. WRKY proteins are classified into three groups according to their structures (Eulgem et al., 2000), and AtWR-KY22 belongs to group II.
High sequence similarity and similar expression profiles might indicate similar function; therefore, we aligned the amino acid sequence of four proteins: AtWRKY6, AtWRKY22, OsWRKY23, and AtWRKY53. The first three belong to subgroup II and contain a typical WRKY domain and the zinc-finger-like motif Cys2-His2, while AtWRKY53 belongs to subgroup III and contains a zinc-finger-like motif Cys2-His/Cys in addition to the WRKY domain. BLAST searching with the sequence of AtWRKY22 (At4g01250) as the query found 36%, 55%, and 43% identity at the amino acid level with AtWRKY53 (At4g23810), AtWRKY6 (At1g62300), and OsWRKY23 (Os01g53260), respectively (data not shown).
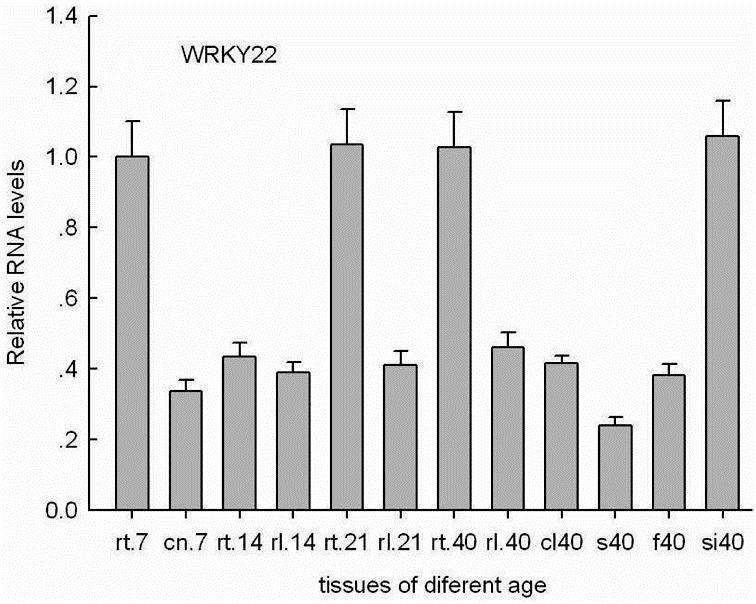
Although the similarity between AtWRKY22 protein and AtWRKY53, AtWRKY6, OsWRKY23 proteins is not high, the relative high expression level of AtWRKY22 was found to be the same as AtWRKY53 and OsWRKY23 under the condition of dark-induced senescence (Jing et al., 2009; Miao et al., 2004) (Figs. 2D and 7D). On the other hand, there was a basal expression of AtWRKY22 in all organs examined under normal conditions, including roots, cotyledons, rosette leaves, cauline leaves, flowers, inflorescence stalks, and siliques. The roots and siliques exhibited higher AtWRKY22 expression than the other organs. Though as the age of the rosette leaves increased, the expression of AtWRKY22 was very slightly enhanced (Fig. 1), statistical test showed that there was no any difference among rosette leaves and roots with different developmental age.
Fig. 2. RNA blot analysis of the WRKY22 expression profiles in Arabidopsis under alternate light-dark treatment, H2O2 application, and continuous dark conditions. (A) The expression profile of Arabidopsis WRKY22 over the two-day experimental period under long-day (16 h light/8 h dark) conditions. Illumination was from 8:00 to 22:00. Sampling under dark conditions was undertaken at 24:00, 3:00, and 6:00. Sampling under light conditions was undertaken at 9:00, 12:00, 15:00, 18:00 and 21:00. (B) The expression profile of Arabidopsis WRKY22 over the two-day experimental period under short-day (8 h light/16 h dark) conditions. Illumination was from 10:00 to 18:00. Sampling under dark conditions was undertaken at 21:00, 24:00, 3:00, 6:00, and 9:00. Sampling under light conditions was under-taken at 12:00, 15:00, and 18:00. (C) Arabidopsis WRKY22 expression profile after treatment with 3% (gram/100 ml) H2O2 for 1-6 h. (D) Arabidopsis WRKY22 expression profile under continuous dark conditions for up to eight days. Ck in Figs. (C and D) represent AtWRKY22 expression levels under normal conditions. As a control for equal loading, ethidium bromide staining of RNA is shown at the bottom; each lane was loaded with 20 μg total RNA.
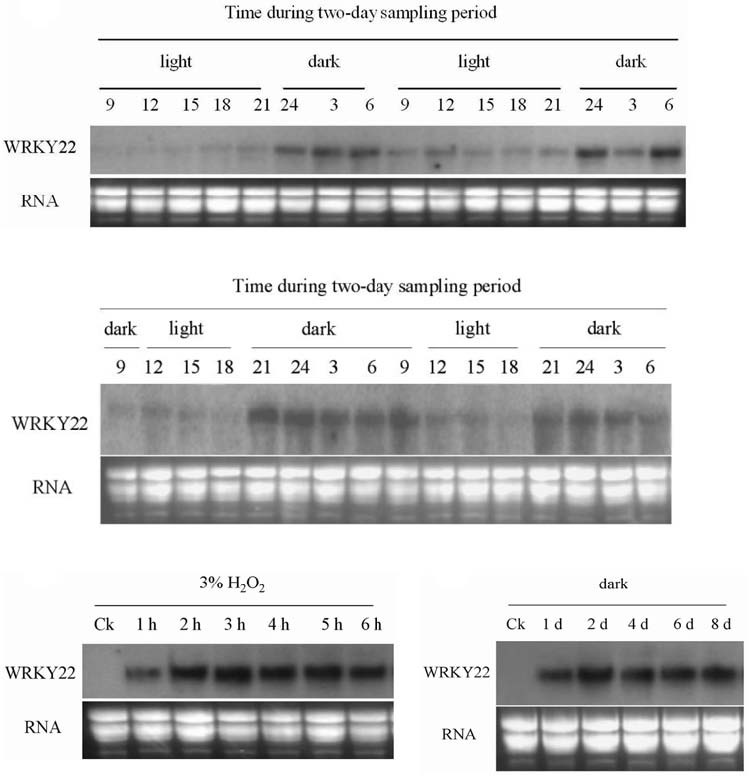
Fig. 7. Real-time PCR analysis using gene-specific primers (Table 1) of four Arabidopsis WRKY genes (AtWRKY22, AtWRKY6, AtWRKY53, and AtWR-KY70) expression under the normal and dark conditions in the different WRKY mutants and AtWRKY22 over-expression plants (35S:W22, L1 and L5). (A) The effect of At-WRKY22 mutation (W22m-1, salk_ 094892; W22m-2, salk_056943) and over-expression (35S:W22, L1 and L5) on the expression of AtWRKY6 under one and half days dark treat-ment and normal conditions. (B and C) The effect of AtWRKY-22 mutation and its over-expression on the expression of AtWRKY53 and AtWRKY70 under three-day dark treatment and normal conditions. (D) The relative expression analysis of AtWRKY22 in the control plant (wt), AtWRKY6 mutant (wrky6m, Salk_012997), and AtWRKY70 mutant (wrky70m, Salk_ 025198) in response to three-day dark treatment and normal conditions. Histograms are the average of triplicate assays and error bars indicate SD (n = 3). Differences between control plants (wt or vector) and mu-tant lines or transgenic lines were tested with one-way ANOVA followed by LSD post hoc test. *Dif-ferences for the expressional levels of WRKY genes in different WRKY mutants or in 35S:W22 plants compared with that in control plants (Wt, Vector) are significant (P < 0.05, n = 3). **Differences for the expressional levels of WRKY genes in different WRKY mutants or in 35S:W22 plants compared with that in control plants (Wt, Vector) are highly significant(P < 0.01,n = 3).
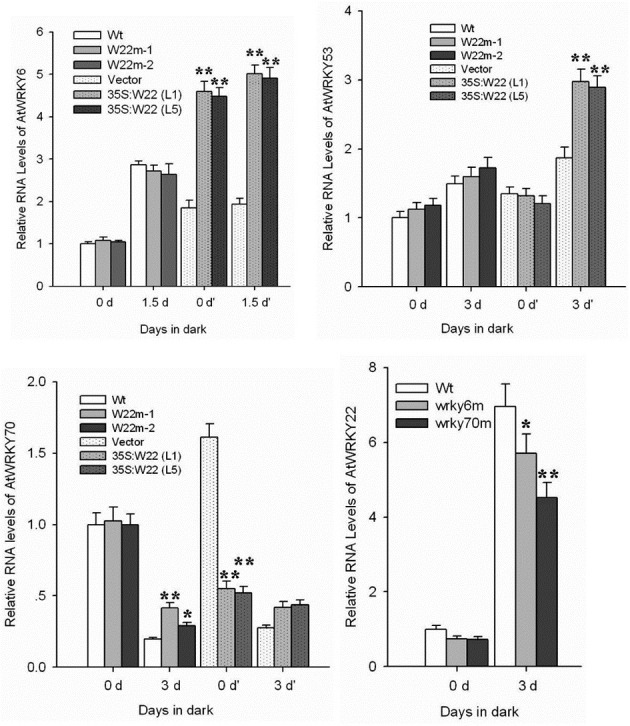
Fig. 1. Relative RNA levels of Arabidopsis WRKY22 were analyzed by real-time PCR using gene-specific primers (Table 1) in different organs (rt, root; cn, cotyledon; rl, rosette leaf; s, inflorescence stalks; cl, cauline leaf; f, flower; si, silique) with different ages (the number after the letters represent development age, for example rt7 denotes roots of seven days developmental age). Histograms are the average of triplicate assays and the bars indicate SD.
Expression profiles of AtWRKY22 in response to light, dark, and H2O2
To confirm whether AtWRKY22 transcription is induced by light and dark signals, we performed Northern blot hybridization using the full-length WRKY22 cDNA as a probe. Wild-type plants were grown in a chamber under long-day (16 h light/8 h dark) or short-day (8 h light/16 h dark) conditions and harvested at three-hour intervals over two days. WRKY22 transcription was suppressed by light but induced by dark treatment (Figs. 2A and 2B). The level of AtWRKY22 transcripts increased gradually when plants were transferred from light to dark conditions, reaching a peak when under continuous dark conditions for two days. Thereafter, a high relative mRNA level of At-WRKY22 was maintained (Fig. 2D). These results indicated that AtWRY22 transcription is regulated by light and dark signals. In addition, AtWRKY22 transcription was maintained at a high level after 3% H2O2 treatment for six hours (Fig. 2C). Both of dark (Keench et al., 2007) and H2O2 (David, 1995) treatment can induce plant senescence, thus AtWRKY22 might participate in senescence-induced signal transduction pathways.
Arabidopsis WRKY22 mutants result in a delayed- senescence phenotype of detached leaves
To better understand the biological function of the AtWRKY22 gene in cell senescence, we first characterized WRKY22 T- DNA insertion mutants (W22m-1, salk_094892; W22m-2, salk_ 056943). The structure of the AtWRKY22 gene and position of the T-DNA insertion in the AtWRKY22 mutants is illustrated in Fig. 3A. The AtWRKY22 gene is located on the Arabidopsis fourth chromosome and contains two introns and three extrons (Fig. 3A). The expression of WRKY22 was clearly suppressed in the W22m-1 mutant plants and decreased to a great extent in the W22m-2 mutant plants (Fig. 3B). Although minor differ-ences in physiological phenotypes existed between the two AtWRKYY22 mutants under dark conditions, the physiological change was roughly similar, indicating that the function of the AtWRKY22 gene is lost completely in W22m-1 and almost completely in W22m-2. The T-DNA sequence was inserted in the 3’ terminal region of AtWRKY22 in the W22m-2 mutant, leading to decreased AtWRKY22 transcription and reduced translational efficiency. Besides the change of AtWRKY22 expression and translation, no other obvious differences in morphology or growth were observed between the wild-type and two mutant plants under normal growth conditions (data not shown).
Fig. 3. Characteristic and delayed-sene-scence phenotypes of the WRKY22 T-DNA insertion mutant plants (W22m-1, salk_094892; W22m-2, salk_056943). (A) Characterization of the Arabidopsis WRKY22 gene and position of the T-DNA insertion sites. (B) PCR screening and examining by hybridization of the Arabidopsis WR-KY22 mutant lines. (C) Senescence symptoms of detached leaves from the WR-KY22 mutant and control plants of different ages after dark treatment for four days. (D) Staining of dead cells with Evans Blue in detached full-grown fourth rosette leaves of the control and WRKY22 mutants. (E) Chlorophyll content in detached full-grown fourth rosette leaves of control and WRK-Y22 mutant plants measured spectropho-tometrically. FW, fresh weight. (F) Cell death measured spectrophotometrically using Evans Blue staining of detached full-grown fourth rosette leaves of control and WRKY22 mutant plants after dark treatment for four days. (E) and (F), Error bars indicate SD (n = 6). Differences between wild type (Wt) plants and mutant lines were tested with one-way ANOVA followed by LSD post hoc test. *Differences for AtWR-KY22 mutants compared with wild-type plants are significant (P < 0.05, n = 6).
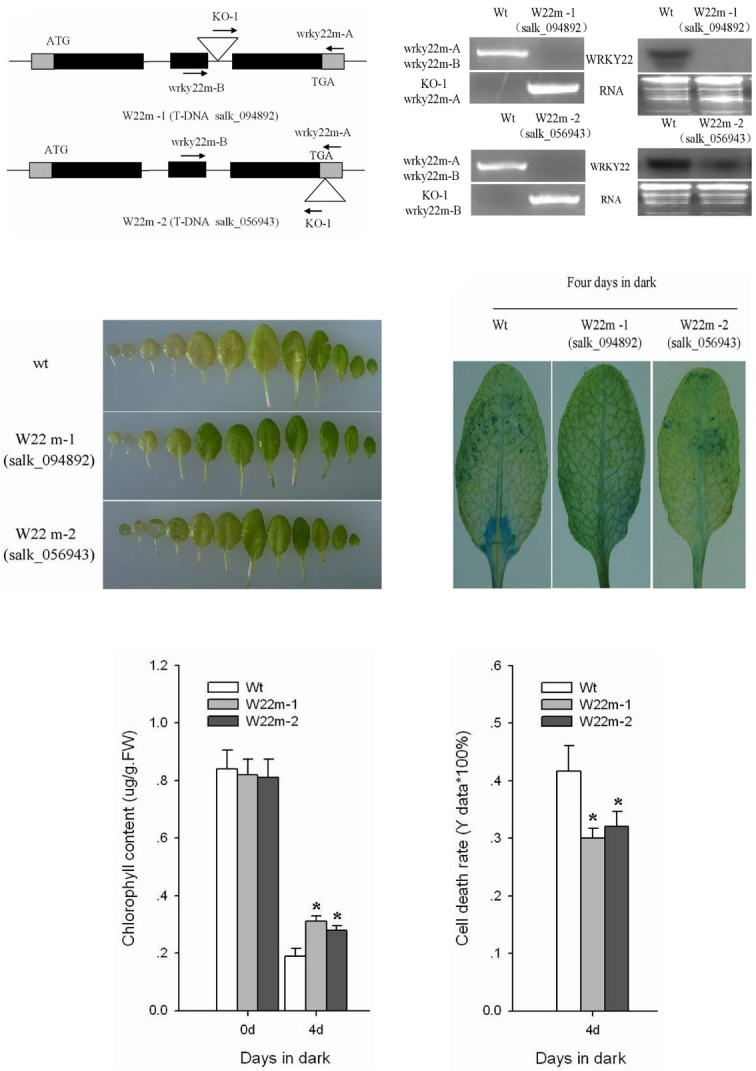
Earlier work demonstrated that both H2O2 (David, 1995) and dark (Keench et al., 2007) could induce plant senescence. AtWRKY22 transcription increased continuously with 3% H2O2 treatment (Fig. 2C) and with continuous dark treatment (Fig. 2D), indicating that AtWRKY22 might have a physiological role in the senescence-related signal transduction pathways. There- fore, we assessed the function of AtWRKY22 in vivo in dark-induced leaf senescence. We first examined the responses to dark of detached leaves of different ages from WRKY22-mutants. The detached leaves from AtWRKY22-mutants showed a moderate delayed-senescence phenotype compared with wild-type plants (Fig. 3C). To minimize any developmental effects, full-grown fourth euphylla (referring to the sixth leaf of each plant from left to right in Fig. 3C) were used in the following experiments. The fourth euphylla of WRKY22 mutants and wild-type plants were detached and treated with continuous dark for four days. The response of detached leaves to dark treatment was monitored by measuring the chlorophyll content and the cell death rate. Staining showed that a small quantity of dead cells occurred at the edge or middle of the leaves from the two mutants, while many dead cells were found in those of wild-type plants (Fig. 3D). Significantly lower levels of cell death were observed in the mutant leaves compared to the wild-type when cell death was measured spectrophotometrically (Fig. 3F). The decrease in leaf chlorophyll content in the wild-type plants (from 0.84 μg.g-1.FW-1 to 0.19 μg.g-1.FW-1) was larger than in the two AtWRKY22 mutants (from 0.82 μg.g-1.FW-1 to 0.31 μg. g-1.FW-1 for W22m-1; from 0.81 μg.g-1.FW-1 to 0.28 μg.g-1.FW-1 for W22m-2) (Fig. 3E). These results showed that mutation of AtWRKY22 resulted in an altered response to dark-induced leaf senescence.
Over-expression of Arabidopsis WRKY22 influences plant morphogenesis
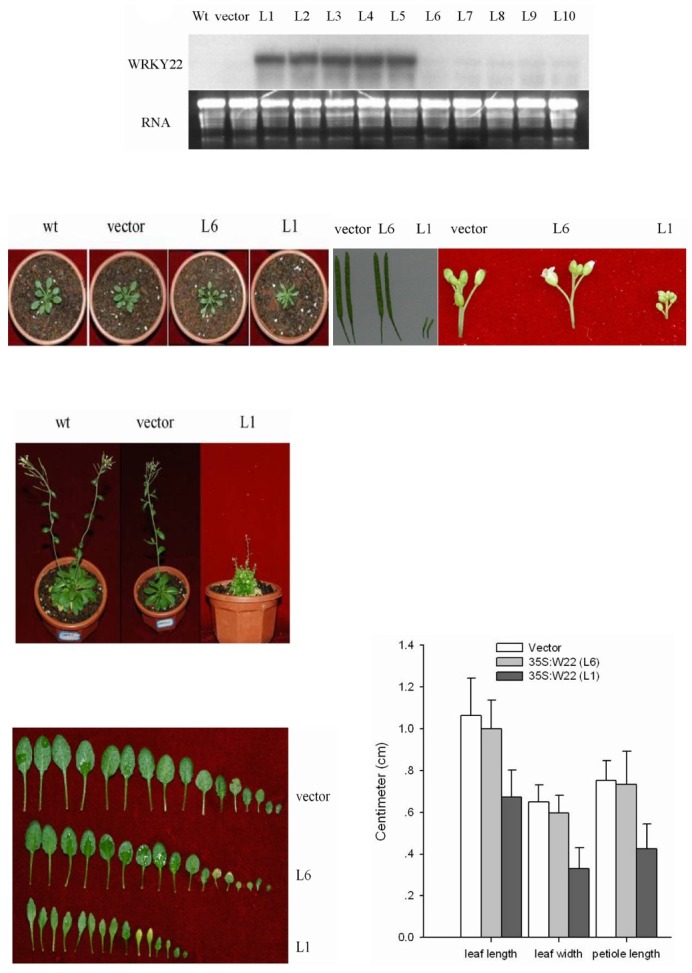
To characterize the physiological function of the AtWRKY22 gene in dark-induced senescence, we generated more than 25 independent transgenic Arabidopsis lines, each harboring the AtWRKY22 cDNA under the control of the CaMV 35S promoter (35S:W22). RNA blotting analysis showed that five transgenic plants contained a high level of the AtWRKY22 transcript under normal conditions (Fig. 4A). These transgenic 35S:W22 plants exhibited serious morphological and developmental defects. During the entire vegetative growth phases, 35S:W22 plants showed stunted growth (Fig. 4C), a more compact growth form (Fig. 4B), and narrower leaves (Figs. 4E and 4F). The most striking phenotype was partial sterility. The young siliques of WRKY22 over-expression transgenic plants were undeveloped and empty (Fig. 4D). Furthermore, the seed content was clearly reduced compared with those of control plants. These results indicated that over-expression of AtWRKY22 is deleterious for normal plant development
Fig. 4. Characteristics of the WRKY22 over-expression plants. (A) RNA blot of 35S: W22 transgenic plants to identify lines with constitutively increased expression of WRKY22. The numbers L1 to L10 above the gel denote independent transgenic plants. (B) WRKY22 over-expres-sion plants (L1 and L6) and wild-type plants (wt) after 15 days. (C) WRKY22 over-expression plants (L1) and control plants (wt and vector) after 45 days. (D) The silique (left) and inflorescence (right) phenotypes of WRKY22 over-expression plants (L1 and L6) and control plants (vector). (E) The rosette leaves of WRKY22 over-expression plants (L1 and L6) and control plants (vector). (F) The full-grown fourth rosette leaves of WRKY22 over-expression plants (L6 and L1) and control plants (vector) with the same developmental age were selected to determine leaf length, leaf width, and petiole length. Error bars indicate SD (n = 30). Vectors represent transgenic seedlings that were obtained by Agrobacterium tumefaciens strain LBA4404-mediated transformation of wild-type plants with plasmid pOCA30.
Over-expression of Arabidopsis WRKY22 accelerates senescence of detached leaves
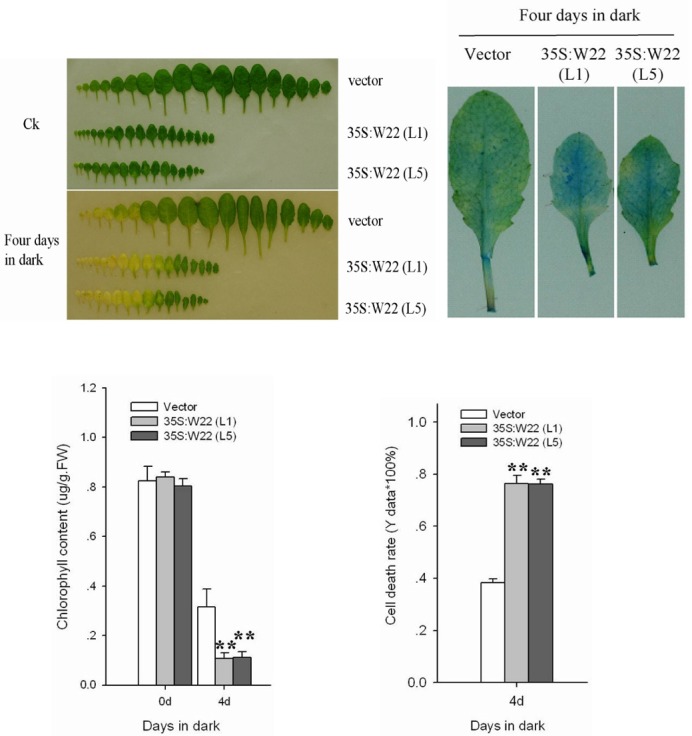
To further analyze the biological functions of the AtWRKY22 gene in leaf senescence, two transgenic lines of 35S:W22 (L1) and 35S:W22 (L5), which exhibited the greatest ectopic expression level of the AtWRKY22 gene, were selected for further analysis of leaf senescence. Detached leaves of different ages from four-week-old plants were dark-treated for four days at 22℃. As shown in Fig. 5A, the two transgenic over-expression AtWRKY22 plants, 35S:W22 (L1) and 35S:W22 (L5), displayed accelerated senescence compared with the control vector transgenic plants. The full-grown fourth rosette leaves of the control vector plant and the two transgenic over-expression AtWRKY22 lines were treated with dark for four days, after which dead cells in the leaves were stained with Evans Blue, total chlorophyll content was determined spectrophotometrically at 652 nm, and the cell death rate was measured. The staining of dead cells and cell death rate in the two transgenic over-expression WRKY22 lines was higher than those in the control vector transgenic lines (Figs. 5B and 5D). The reduction in chlorophyll content of leaves from the over-expression WRKY22 lines was greater (from 0.80-0.84 μg.g-1.FW-1 to 0.107-0.11 μg.g-1. FW-1) than in the control vector transgenic plant (0.82 μg.g-1. FW-1 to 0.315 μg.g-1.FW-1) (Fig. 5C). These results indicated that over-expression of WRKY22 contributed to acceleration of leaf senescence under dark conditions.
Fig. 5. Accelerated-senescence phenotype of 35S:W22 plants. Two 35S:W22 lines (L1 and L5) exhibiting a high level of WRKY22 transcription were selected for analysis of the response to dark treatment. (A) Senescence symptoms in detached leaves of different ages of 35S:W22 plants and the control plant (vector) after four-day dark treatment. (B) Staining of dead cells with Evans Blue in de-tached full-grown fourth rosette leaves of 35S:W22 lines and the control vector transgenic plant (vector). (C) Chlorophyll content in detached fourth rosette leaves of 35S:W22 lines and the control plant (vector). Chlorophyll was extracted and measured spectro-photo-metrically. FW, fresh weight. (D) Cell death in detached full-grown fourth rosette leaves of 35S:W22 and the control plant (vector) after dark treatment for four days. Cell death was measured spectrophotometri-cally after staining with Evans Blue. (C) and (D), Error bars indicate SD (n = 6). Differences between control plants (vector) and transgenic lines were tested with one-way ANOVA followed by LSD post hoc test. **Differences for 35S:W22 plants compared with control vector transgenic plants are highly signifi-cant (P < 0.01, n = 6).
Effects of dark-induced leaf senescence on expression of senescence-associated genes in Arabidopsis WRKY22 mutants and WRKY22 over-expression plants
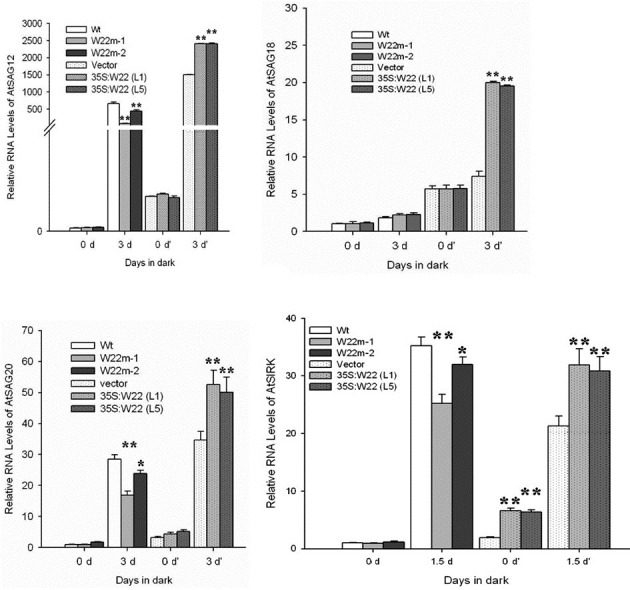
Increasing evidences suggest that many senescence-asso-ciated genes (SAGs) are induced in response to dark (Azumi and Watanabe, 1991). To investigate the molecular changes associated with dark-induced senescence, we evaluated the expression of four genes (SAG12, SAG18, SAG20, SIRK) in control plants (wt, vector), the WRKY22 mutants (W22m-1, salk_094892; W22m-2, salk_056943), and the 35S:W22 transgenic lines (L1 and L5). The increase of relative RNA levels of three senescence-associated genes (SAG12, SAG20, SIRK) in wild-type plants was higher than in WRKY22 mutants (Figs. 6A, 6C, and 6D), while there was no difference of SAG18 relative expression level between the WRKY22 mutants and control plants (Fig. 6B) under dark conditions. Over-expression of AtWRKY22 had no effect on the relative expression levels of three senescence-associated genes (SAG12, SAG18, SAG20) (Figs. 6A-6C), but the relative expression level of SIRK increased when AtWRKY22 was over-expressed under normal conditions. The expression levels of four senescence-associa-ted genes (SAG12, SAG18, SAG20, SIRK) were increased to a greater extent in WRKY22 over-expression lines than in control plants under dark conditions (Figs. 6A-6D). Thus, the molecular data were consistent with the physiological results, indicating that Arabidopsis WRKY22 participated in the dark-induced senescence signal transduction pathway.
Fig. 6. Effect of dark treatment and normal conditions on expression of senescence-associated genes in the WRKY22 mutants (W22m-1, salk_094892; W22m-2, salk_056943) and WRKY22 over-expression plants (35S:W22, L1 and L5) compared with the control plants (Wt, Vector). Relative RNA levels of four se-nescence-associated genes were analyzed by real-time PCR using gene-specific primers (Table 1) after samples were dark-treated at 22℃ for three (SAG12, SAG18, SAG20) or one and half (SIRK) days. Histograms are the average of triplicate assays and error bars indicate SD (n = 3). Differences between control plants (wt or vector) and mutant lines or transgenic lines were tested with one-way ANOVA followed by LSD post hoc test. *Differences for the expressional levels of senescence-associated genes in WRKY22 mutants or in 35S:W22 plants compared with that in control plants (Wt, Vector) are significant (P < 0.05, n = 3), **Differences for the expressional levels of senescence-associated genes in WRKY22 mutants or in 35S:W22 plants compared with that in control plants (Wt, Vector) are highly significant (P < 0.01, n = 3).
Effects of dark treatment on expression of AtWRKY6, AtWRKY22, AtWRKY53, and AtWRKY70 in different Arabidopsis WRKY mutants and WRKY22 over-expression plants
WRKY6 regulates plant senescence by binding to the W-box in the promoter of the senescence-induced receptor-like kinase (SIRK) gene in Arabidopsis (Robatzek and Somssich, 2002). AtWRKY53 protein plays a regulatory role in the early events of leaf senescence (Hinderhofer and Zentgraf, 2001). Arabidopsis WRKY70 is located in the node of convergence for jasmonate-mediated and salicylate-mediated signals (Li et al., 2004) and acts as a negative regulator of senescence (Ulker et al., 2007). These three Arabidopsis WRKY genes are important senescence-associated regulatory genes. We investigated the rela-tionship between WRKY22 and WRKY6, WRKY53, WRKY70 to characterize the molecular mechanism of AtWRKY22’s participation in leaf senescence. Figures 7A and 7B showed that mutation of AtWRKY22 had no effect on the accumulation of AtWRKY6 and AtWRKY53 transcripts compared with control plants under dark conditions. Furthermore, mutation of AtWRKY6 resulted in decreased expression levels of AtWRKYY22 compared with control plant in response to dark treatment (Fig. 7D). However, AtWRKY22 could influence the relative expression level of AtWRKY6 and AtWRKY53 by feedback regulation under certain conditions. For example, over-expression of At-WRKY22 increased the relative expression level of AtWRKY6 (Fig. 7A), but had no effect on the relative expression level of AtWRKY53 (Fig. 7B) compared with vector-transgenic control plants under normal conditions. Under dark conditions, in addition to increasing AtWRKY6 expression, the relative expression of AtWRKY53 in the AtWRKY22 over-expression plant (L5) was larger than the vector-transgenic control plants (Figs. 7A and 7B). These results indicated that AtWRKY22 could regulate the expression of AtWRKY6 and AtWRKY53 through different methods of feedback regulation. Figure 7C shows that dark treatment reduced the expression of AtWRKY70. The decrease in relative expression of AtWRKY70 in AtWRKY22 mutants (W22m-1, salk_094892; W22m-2, salk_056943) was some what lower than in wild-type plant in response to dark (Fig. 7C). On the other hand, over-expression of AtWRKY22 significantly decreased the relative expression of At-WRKY70 under normal conditions (Fig. 7C). Mutation of AtWRKY70 significantly decreased the relative expression level of WR-KY22 compared with wild-type plants in response to dark treatment (Fig. 7D).
DISCUSSION
Leaf senescence is a process of programmed cell death that occurs in an age-dependent manner and is induced by various environmental cues (Yoshida, 2003). Elucidating the molecular mechanism of leaf senescence will not only contribute to our knowledge of this fundamental developmental process, but could also lead to ways of manipulating senescence for agricultural applications (Gan et al., 1997). In recent years, several families of sequence-specific DNA binding transcription factors have been identified, including the two largest groups of senescence-associated transcription factors, the NAC and WRKY superfamilies, which have been implicated in the regulation of leaf senescence (Buchanan-Wollaston et al., 2005; Olsen et al., 2005). Our data on AtWRKY22 offer evidence that this transcription factor participates in regulation of dark-induced senescence in detached leaves of Arabidopsis.
The literature suggests that AtWRKY22 and AtWRKY29 are important downstream components of the mitogen-activated protein kinases (MAPK)-mediated signal transduction pathway, which confers resistance to both bacterial and fungal pathogens and can be up-regulated by a pathogen associated molecular patterns -induced MAPK cascade (Asai et al., 2002). The expression of AtWRKY22 is induced by salicylic acid (SA), pathogen attack (Dong et al., 2003), chitin (Wan et al., 2004), and flagellin (Asai et al., 2002). These results suggest that AtWRKY22 has an important role in the pathogen resistance response. Both pathogen infection and treatment with SA can induce production of reactive oxygen species accompanied by senescence (Lamb and Dixon, 1997). SA is a key mediator of plant stress responses, including disease and systemic acquired resistance, as well as acting as a specific regulator in developmental senescence (Graaff et al., 2006). These data indicate that the senescence-signaling pathway is often linked to pathogen defense (Quirino et al., 1999). According to genevestigator (https://www.genevestigator.com), the expression of WRKY22 is induced by dark and its relative high ex-pressional level is found in senescent leaves. We investigated the constitutive expression level of the Arabidopsis WRKY22 in different organs at different developmental stages. Relatively high expression levels of WRKY22 were found in roots and siliques, and developmental age increased, the relative expression level in adult rosette leaves showed a negligible change (Fig. 1). Furthermore, we found that AtWRKY22 was significantly induced under dark conditions in detached leaves of Arabidopsis (Figs. 2A, 2B, 2D, and 7D). Thus, we deduced that AtWRKY22 was likely to participate in the process of dark-induced leaf senescence.
The results from dark-treated detached leaves of WRKY22 over-expression lines showed that senescence could be accelerated in the Arabidopsis (Fig. 5A), including accelerated loss of chlorophyll (Fig. 5C) and a high cell death rate (Fig. 5D). By contrast senescence was delayed to a certain extent in the Arabidopsis WRKY22 T-DNA insertion mutants (Fig. 3C). Further analysis indicated that the expression levels of four senescence-associated genes (SAG12, SAG18, SAG20, SIRK) in WRKY22 over-expression lines was higher than that in control lines, and the expressions of three senescence-associated genes (SAG12, SAG20, SIRK) in T-DNA insertion mutant lines was lower than that in control lines, in response to dark treat-ment (Figs. 6A-6D). These results support the hypothesis that Arabidopsis WRKY22 participates in the process of leaf senescence.
Functional redundancy within a multigene family often complicates genetic attempts to define the role of individual members (Bouche and Bouchez, 2001). The plant WRKY transcription factor gene family is a superfamily containing numerous members. Phylogenetic analysis indicates that some WRKY gene members are located on the same evolutionary branch (Dong et al., 2003), suggesting that these WRKY genes have a similar physiological functions. Arabidopsis WRKY22 T-DNA insertion mutants only exhibited a moderate delay of leaf senescence, whereas the over-expression lines showed obviously accelerated senescence (Figs. 3C and 5A). The above results imply the existence of other WRKY proteins with similar biological functions that participate in the dark-induced senescence signal transduction pathway together with WRKY22. Arabidopsis WRKY22 and WRKY29 belong to the same phylogenetic lineage (Dong et al., 2003), and exhibit similar expression profiles under various abiotic stresses, such as darkness, wounding, low temperature, and H2O2 (data not shown). Perhaps AtWRKY22 and AtWRKY29 in the MAPK-mediated signal cascade and senescence-associated signal transduction pathway provide redundant functions? No difference in growth phenotype was observed between AtWRKY22 mutant plants and wild-type plants under normal growth conditions (data not shown), indicating the possible presence of redundant genes having similar functions to AtWRKY22 or AtWRKY29, which did not participate in the process of natural senescence.
Our previous work showed that the expression of Oryza sativa WRKY23 was induced by the dark and that heterologous expression of OsWRKY23 enhanced dark-induced leaf senescence in Arabidopsis (Jing et al., 2009). In addition, certain Arabidopsis WRKY genes have important roles in dark-induced senescence signal transduction. For example, the function of senescence-induced receptor-like kinase (SIRK) gene depends on AtWRKY6 protein (Robatzek and Somssich, 2002). At-WRKY6 controls plant senescence and pathogen defense by specifically binding to the W-box in the promoter of the senescence-induced receptor-like kinase (SIRK) gene in Arabidopsis (Robatzek et al., 2002). The Arabidopsis WRKY53 protein plays a regulatory role in the early events of leaf senescence (Miao et al., 2004). AtWRKY53 is a positive regulatory factor and regulates the expression of target genes directly, but an epithiospecifying senescence regulator (ESR) protein that is induced by jasmonic acid (JA) and inhibited by SA can block the process (Miao et al., 2007). AtWRKY70 participates in the regulation of leaf senescence as a negative regulatory factor, and delays the course of leaf senescence by restraining the expression of defense target genes regulated by SA, JA or ethylene (ET) (Ulker et al., 2007). Leaf senescence was delayed in AtWRKY22 T-DNA insertion mutants and accelerated in AtWRKY22 over-expression lines in response to dark treatment. This result suggests that the transcription factor AtWRKY22 functions in the process of leaf senescence as a positive regulator. Over-expression lines of WRKY22 under normal conditions exhibited no changes in the expressions of senescence-associated genes (SAG12, SAG18, SAG20) compared with control plants (Figs. 6A-6C). However, lower relative expression levels of SAG12, SAG20 and SIRK were found in the AtWRKY22 mutants compared with wild-type plants in response to dark treatment (Figs. 6A, 6C and 6D). Thus, there is a possibility that Arabidopsis WRKY22 does not regulate the expression of senescence-associated genes directly. The DNA binding activities of Arabidopsis WRKY18, WRKY40 and WRKY60 proteins depend on their formed homocomplexes and heterocomplexes in response to infection by the hemibiotrophic bacterial pathogen Pseudomonas syringae (Xu et al., 2006). Perhaps the regulation of the expression of WRKY22 target genes also depends on its forming homologous or heterogen-ous complexes with other WRKY transcription factors or other unknown dark-induced factors. Further research is required to identify which factors cooperate with WRKY22 to participate in the senescence signal transduction pathway.
WRKY genes are functionally connected, forming a transcriptional network composed of positive and negative feedback loops and feed-forward modules (Eulgem et al., 2007). Analysis of the expression profiles of AtWRKY6, AtWRKY22, AtWR-KY53, and AtWRKY70 in different Arabidopsis WRKY mutants and AtWRKY22 over-expression lines in response to dark treatments, indicated that there was mutual regulation between AtWRKY22 and AtWRKY6, AtWRKY53, and AtWRKY70. Based on the analysis of the expression profiles of senescence- associated WRKY genes and information related to AtWRKY22 in the literature, we hypothesized that At-WRKY22 participates in regulation of leaf senescence through various signal pathways. At least seven WRKY gene family members, including WRKY22 and WRKY29, are identified as target genes of the WRKY53 protein (Miao et al., 2004). Expression of WRKY22 and WRKY29 are up-regulated in WRKY53 over-expression lines, but are down-regulated in WRKY53 RNAi and knockout lines (Miao et al., 2004). Moreover, mutation of AtWRKY22 had no effect on the relative expression level of AtWRKY53 under normal or dark conditions (Fig. 7B). Therefore, AtWRKY22 might function in leaf senescence as a downstream member of the WRKY53-mediated signal transduction pathway. AtWR-KY22, together with an unknown dark-induced factor, pro-moted expression of AtWRKY53 by positive feedback under dark conditions (Fig. 7B). AtWRKY22 over-expression lines had higher relative expression levels of AtWRKY6 than control plants under dark and normal conditions (Fig. 7A). Therefore, the second possible mechanism by which AtWRKY22 functions in leaf senescence is by activation of AtWRKY6 directly or indi-rectly accompanied by promoting expression of SIRK. However, mutation of AtWRKY22 did not affect the expression level of AtWRKY6 compared with control plants in response to dark and normal conditions (Fig. 7A), indicting the possible presence of a functionally redundant unknown factor. AtWRKY70 is a WRKY transcription factor that functions as a negative regulator of developmental senescence, loss of AtWRKY70 function promotes both developmental and dark-induced leaf senescence (Ulker et al., 2007). Under normal conditions, AtWRKY70 represses both SA and JA/ET mediated defense marker genes expression; leaf senescence initiates when the repression is removed. Over-expression of AtWRKY22 resulted in a significant decrease in the transcription level of AtWRKY70 under normal conditions (Fig. 7C), indicating that plants over-expres-sion AtWRKY22 showed the phenotype of accelerated senescence under normal conditions, although there was no additional experimental data. Figure 7C showed that dark treatment could decrease the expression of AtWRKY70, and this decrease in AtWRKY70 expression level was lower in AtWRKY22 mutant lines than in control plants. Therefore, the third mechanism by which AtWRKY22 functions in leaf senescence could be by suppressing the activity of AtWRKY70. Further experiments are necessary to investigate and compare the expression of genes regulated by AtWRKY70 in AtWRKY22 insertion mutants, AtWRKY22 over-expression lines, and control plants, under dark and normal conditions to confirm whether At-WRKY22 regulates leaf senescence through an AtWRKY70-mediated signal transduction pathway. Thus there are three possible mechanisms of AtWRKY22 involvement in leaf senescence, and it is possible that AtWRKY22 participates in leaf senescence via all three mechanisms. Further investigation is required to identify the main mechanisms or other possible mechanisms of AtWRK-Y22 activity.
In summary, we have shown that a novel transcription factor, AtWRKY22, is involved in regulating senescence of detached leaves in Arabidopsis. The data indicate that AtWRKY22 can regulate the expression of senescence-associated genes indirectly or through other WRKY-mediated senescence signal pathways. There is still much work to do as so to uncover the detailed molecular mechanism of its role in regulating leaf senescence.
Acknowledgments
This study was supported by the Natural Science Foundation of Yunnan Province (grant no. O9SJ041B01), and the Science Foundation of Ministry of Agriculture of the People’s Republic ofChina (2009ZX08009-066B).
References
- 1.Asai T., Tena G., Plotnikova J., Willmann M.R., Chiu W.L., Gomez-Gomez L., Boller T., Ausubel F.M., Sheen J. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature. (2002);415:977–983. doi: 10.1038/415977a. [DOI] [PubMed] [Google Scholar]
- 2.Azumi Y., Watanabe A. Evidence for a senes-cence-associated gene induced by darkness. Plant Physiol. (1991);95:577–583. doi: 10.1104/pp.95.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bouche N., Bouchez D. Arabidopsis gene knockout: phenotypes wanted. Curr. Opin. Plant Biol. (2001);4:111–117. doi: 10.1016/s1369-5266(00)00145-x. [DOI] [PubMed] [Google Scholar]
- 4.Buchanan-Wollaston V. The molecular biology of leaf senescence. J. Exp. Bot. (1997);48:181–199. [Google Scholar]
- 5.Buchanan-Wollaston V., Earl S., Harrison E., Mathas E., Navabpour S., Page T., Pink D. The molecular analysis of leaf senescence-a genomics approach. Plant Biotechnol. J. (2003);1:3–22. doi: 10.1046/j.1467-7652.2003.00004.x. [DOI] [PubMed] [Google Scholar]
- 6.Buchanan-Wollaston V., Page T., Harrison E., Breeze E., Lim P.O., Nam H.G., Lin J.F., Wu S.H., Swidzinski J., Ishizaki K., et al. Comparative transcriptome analysis reveals significant differences in gene expression and signaling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J. (2005);42:567–585. doi: 10.1111/j.1365-313X.2005.02399.x. [DOI] [PubMed] [Google Scholar]
- 7.Chen C.H., Chen Z.X. Potentiation of developmentally regulated plant defence response by AtWRKY18, a pathogen-induced Arabidopsis transcription factor. Plant Physiol. (2002);129:706–716. doi: 10.1104/pp.001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clough S.J., Bent A.F. Floral dip: a simplified method for agrobacterium-mediated transformation of Arabidopsis thali-ana. Plant J. (1998);16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 9.David A.D., Ahmad S. Antioxidant defenses of plants and fungi in oxidative stress and antioxidant defenses in biology. Chapman & Hall; New York: (1995). pp. 298–355. [Google Scholar]
- 10.Devaiah B.N., Karthikeyan A.S., Raghothama K.G. WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol. (2007);143:1789–1801. doi: 10.1104/pp.106.093971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong J., Chen C., Chen Z. Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol. Biol. (2003);51:21–37. doi: 10.1023/a:1020780022549. [DOI] [PubMed] [Google Scholar]
- 12.Eulgem T., Somssich I.E. Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. (2007);10:366–371. doi: 10.1016/j.pbi.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 13.Eulgem T., Rushton P.J., Robatzek S., Somssich I.E. The WRKY superfamily of plant transcriptional factors. Trends Plant Sci. (2000);5:199–206. doi: 10.1016/s1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- 14.Gan S., Amasino R.M. Making sense of senescence. Plant Physiol. (1997);113:313–319. doi: 10.1104/pp.113.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graaff E.V.D., Schwacke R., Schneider A., Desimone M., Flügge U.I., Kunze R. Transcription analysis of Arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiol. (2006);141:776–792. doi: 10.1104/pp.106.079293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grbic V., Bleecker A.B. Ethylene regulates the timing of leaf senescence in Arabidopsis. Plant J. (1995);8:595–602. [Google Scholar]
- 17.Guo F.Q., Crawford N.M. Arabidopsis nitric oxide synthase1 is targeted to mitochondria and protects against oxi-dative damage and dark-induced senescence. Plant Cell. (2005);17:3436–3450. doi: 10.1105/tpc.105.037770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo Y., Cai Z., Gan S. Transcriptome of Arabidopsis leaf senescence. Plant Cell Environ. (2004);27:521–549. [Google Scholar]
- 19.He Y., Gan S. A gene encoding an acyl hydrolase is involved in leaf senescence in Arabidopsis. Plant Cell. (2002);14:805–815. doi: 10.1105/tpc.010422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hinderhofer K., Zentgraf U. Identification of a tran-scription factor specifically expressed at the onset of leaf senescence. Planta. (2001);213:469–473. doi: 10.1007/s004250000512. [DOI] [PubMed] [Google Scholar]
- 21.Jiang W.B., Yu D.Q. Arabidopsis WRKY2 transcription factor mediates seed germination and postgermination arrest of development by abscisic acid. BMC Plant Biol. (2009);9:1–14. doi: 10.1186/1471-2229-9-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jing S., Zhou X., Song Y., Yu D. Heterologous expression of OsWRKY23 gene enhances pathogen defense and dark-induced leaf senescence in Arabidopsis. Plant Growth Regul. (2009);58:181–190. [Google Scholar]
- 23.Johnson C.S., Kolevski B., Smyth D.R. TRANS-PARENT TESTA GLABRA 2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell. (2002);14:1359–1375. doi: 10.1105/tpc.001404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Journot-Catalino N., Somssich I.E., Roby D., Kroj T. The transcription factors WRKY11 and WRKY17 act as negative regulators of basal resistance in Arabidopsis thaliana. Plant Cell. (2006);18:3289–3302. doi: 10.1105/tpc.106.044149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keench O., Pesquet E., Ahad A., Askne A., Nordvall D., Vodnala S.M., Tuominen H., Hurry V., Dizengremel P., Gardestrom P. The different fates of mitochondria and chloroplasts during dark-induced senescence in Arabidopsis leaves. Plant Cell Environ. (2007);30:1523–1534. doi: 10.1111/j.1365-3040.2007.01724.x. [DOI] [PubMed] [Google Scholar]
- 26.Koch E., Slusarenko A. Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell. (1990);2:437–445. doi: 10.1105/tpc.2.5.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lagrimini L.M., Burkhart W., Moyer M., Rothstein S. Molecular cloning of complementary DNA encoding the lignin-forming peroxidase from tobacco: molecular analysis and tissue-specific expression. Proc. Natl Acad. Sci. USA. (1987);84:7542–7546. doi: 10.1073/pnas.84.21.7542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lamb C., Dixon R.A. The oxidative burst in plant disease resistance, Annu. Rev. Plant Physiol. Plant Mol. Biol. (1997);48:251–275. doi: 10.1146/annurev.arplant.48.1.251. [DOI] [PubMed] [Google Scholar]
- 29.Lanahan M.B., Yen H.C., Giovannoni J.J. The never ripe mutation blocks ethylene perception in tomato. Plant Cell. (1994);6:521–530. doi: 10.1105/tpc.6.4.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li J., Brader G., Palva E.T. The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defence. Plant Cell. (2004);16:319–331. doi: 10.1105/tpc.016980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li S., Fu Q., Huang W., Yu D. Functional analysis of an Arabidopsis transcription factor WRKY25 in heat stress. Plant Cell Rep. (2009);28:683–693. doi: 10.1007/s00299-008-0666-y. [DOI] [PubMed] [Google Scholar]
- 32.Li S., Zhou X, Chen L, Huang W., Yu D. Functional characterization of Arabidopsis thaliana WRKY39 in heat stress. Mol. Cells. (2010);29:475–483. doi: 10.1007/s10059-010-0059-2. [DOI] [PubMed] [Google Scholar]
- 33.Lichtenthaler H.K. Chlorophylls and carotenoids-pigments of photosynthetic biomembranes. Methods Enzymol. (1987);148:350–382. [Google Scholar]
- 34.Lim P.O., Woo H.R., Nam H.G. Molecular genetics of leaf senescence in Arabidopsis. Trends Plant Sci. (2003);8:272–278. doi: 10.1016/S1360-1385(03)00103-1. [DOI] [PubMed] [Google Scholar]
- 35.Lin J.F., Wu S.H. Molecular events in senescing Arabidopsis leaves. Plant J. (2004);39:612–628. doi: 10.1111/j.1365-313X.2004.02160.x. [DOI] [PubMed] [Google Scholar]
- 36.Miao Y., Zentgraf U. The antagonist function of Arabidopsis WRKY53 and ESR/ESP in leaf senescence is modulated by the jasmonic and salicylic acid equilibrium. Plant Cell. (2007);19:819–830. doi: 10.1105/tpc.106.042705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miao Y., Laun T., Zimmermamn P., Zentgraf U. Tar- gets of the WRKY53 transcription factor and its role dur-ing leaf senescence in Arabidopsis. Plant Mol. Biol. (2004);55:853–867. doi: 10.1007/s11103-004-2142-6. [DOI] [PubMed] [Google Scholar]
- 38.Oh S.A., Lee S.Y., Chung K.K., Lee C.H., Nam H.G. A senescence-associated gene of Arabidopsis thaliana is distinctively regulated during natural and artificially induced leaf senescence. Plant Mol. Biol. (1996);30:739–754. doi: 10.1007/BF00019008. [DOI] [PubMed] [Google Scholar]
- 39.Oh S.A., Park J.H., Lee G.I., Paek K.H., Park S.K., Nam H.G. Identification of three genetic loci controlling leaf senescence in Arabidopsis thaliana. Plant J. (1997);12:527–535. doi: 10.1046/j.1365-313x.1997.00527.x. [DOI] [PubMed] [Google Scholar]
- 40.Olsen A.N., Ernst H.A., Leggio L.L., Skriver K. NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci. (2005);10:79–87. doi: 10.1016/j.tplants.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 41.Park J.H., Oh S.A., Kim Y.H., Woo H.R., Nam H.G. Differential expression of senescence-associated mRNAs during leaf senescence induced by different senescence-inducing factors in Arabidopsis. Plant Mol. Biol. (1998);37:445–454. doi: 10.1023/a:1005958300951. [DOI] [PubMed] [Google Scholar]
- 42.Qiu Y.P., Yu D.Q. Over-expression of the stress-induced OsWRKY45 enhances disease resistance and drought tolerance in Arabidopsis. Environ. Exp. Bot. (2009);65:35–47. [Google Scholar]
- 43.Quirino B.F., Normanly J., Amasino R.A. Diverse range of gene activity during Arabidopsis thaliana leaf senescence includes pathogen-independent induction of defense-related genes. Plant Mol. Biol. (1999);40:267–278. doi: 10.1023/a:1006199932265. [DOI] [PubMed] [Google Scholar]
- 44.Quirino B.F., Noh Y., Himelblau E., Amasino R.M. Molecular aspects of leaf senescence. Trends Plant Sci. (2000);5:278–282. doi: 10.1016/s1360-1385(00)01655-1. [DOI] [PubMed] [Google Scholar]
- 45.Robatzek S., Somssich I.E. Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Gene Dev. (2002);16:1139–1149. doi: 10.1101/gad.222702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sambrook J., Fritsch E.F., Maniatis T. Molecular Clo-ning: A Laboratory Manual. 2nd ed Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: (1989). [Google Scholar]
- 47.Ulker B., Shahid Mukhtar M., Somssich I.E. The WRKY70 transcription factor of Arabidopsis influences both the plant senescence and defense signaling pathways. Planta. (2007);226:125–137. doi: 10.1007/s00425-006-0474-y. [DOI] [PubMed] [Google Scholar]
- 48.Wan J., Zhang S., Stacey G. Activation of a mitogen- activated protein kinase pathway in Arabidopsis by chitin. Mol. Plant Pathol. (2004);5:125–135. doi: 10.1111/j.1364-3703.2004.00215.x. [DOI] [PubMed] [Google Scholar]
- 49.Woo H.R., Chung K.M., Park J.H., Oh S.A., Ahn T., Hong S.H., Jang S.K., Nam H.G. ORE9, an F-box protein that regulates leaf senescence in Arabidopsis. Plant Cell. (2001);13:1779–1790. doi: 10.1105/TPC.010061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Woo H.R., Goh C.H., Park J.H., de la Serve B.T., Kim J.H., Park Y.I., Nam H.G. Extended leaf longevity in the ore4-1 mutant of Arabidopsis with a reduced expression of a plastid ribosomal protein gene. Plant J. (2002);31:331–340. doi: 10.1046/j.1365-313x.2002.01355.x. [DOI] [PubMed] [Google Scholar]
- 51.Woo H.R., Kim J.H., Nam H.G., Lim P.O. The delayed leaf senescence mutants of Arabidopsis, ore1, ore3, and ore9 are tolerant to oxidative stress. Plant Cell Physiol. (2004);45:923–932. doi: 10.1093/pcp/pch110. [DOI] [PubMed] [Google Scholar]
- 52.Xu X., Chen C., Fan B., Chen Z. Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell. (2006);18:1310–1326. doi: 10.1105/tpc.105.037523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yoshida S. Molecular regulation of leaf senescence. Curr. Opin. Plant Biol. (2003);6:79–84. doi: 10.1016/s1369526602000092. [DOI] [PubMed] [Google Scholar]
- 54.Yu D., Chen C., Chen Z. Evidence for an important role of WRKY DNA binding protein in the regulation of NPR1 gene expression. Plant Cell. (2001);13:1527–1539. doi: 10.1105/TPC.010115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zipfel C., Robatzek S., Navarro L., Oakeley E.J., Jones J.D.G., Felix G., Boller T. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature. (2004);428:764–767. doi: 10.1038/nature02485. [DOI] [PubMed] [Google Scholar]


