Abstract
Chemotaxis is the common way of flagellated bacteria to direct their locomotion to sites of most favourable living conditions, that are sites with the highest concentrations of energy sources and the lowest amounts of bacteriotoxic substances. The general prerequisites for chemotaxis are chemoreceptors, a chemosensory signal-transduction system and the flagellar apparatus.
Epsilonproteobacteria like Campylobacter sp. show specific variations of the common chemotaxis components. CheV, a CheW-like linking-protein with an additional response regulator (RR) domain, was identified as commonly used coupling scaffold protein of Campylobacter jejuni. It attaches the histidine autokinase (CheAY), which also has an additional RR-domain, to the chemoreceptors signalling domains. These additional RR-domains seem to play an important role in the regulation of the CheAY-phosphorylation state and thereby in sensory adaptation.
The Campylobacter-chemoreceptors are arranged into the three groups A, B, and C. Group A contains membrane-anchored receptors sensing periplasmic signals, group B consists only of one receptor with two cytoplasmic ligand-proteins representing a bipartite energy taxis system that senses pyruvate and fumarate, and group C receptors are cytoplasmic signalling domains with mostly unknown cytoplasmic ligand-binding proteins as sensory constituents. Recent findings demonstrating different alleles of the TLP7 chemoreceptor, specific for formic acid, led to an amendment of this grouping.
Keywords: Campylobacter jejuni, chemotaxis, Epsilonproteobacteria, receptors, review, signal transduction, Tlp groups
Introduction
The Epsilonproteobacterium Campylobacter jejuni is, due to recent epidemiological data, the most common germ causing bacterial foodborne enteritis worldwide [1–3]. Campylobacter enteritis can lead to various post-infectious complications like the Guillain–Barré syndrome (GBS), reactive arthritis, reactive myositis and idiopathic peripheral neuropathy [4–6].
The bacterium’s flagellar motility is an important factor for the intestinal colonization of its avian and mammalian hosts but also for the invasion of intestinal epithelial cells and for overcoming the mucosal barrier [7–12].
Chemotaxis, presumably the most ancient form of signal processing, is the way of archaea and bacteria to direct their flagellar locomotion according to the presence of specific chemoeffectors. This phenomenon was already observed by Engelmann and Pfeffer at the end of the nine-teenth century [13–15].
Chemoeffectors attracting bacterial cells to a point of high effector-concentration are called attractors and chemotaxis in the direction of increasing attractor concentrations is defined as “positive”. In contrast, chemorepellent-gradients cause bacteria to swim to points of lower chemoeffector concentrations and this is considered as “negative” chemotaxis. Typically, energy sources act as chemoattractors while toxic substances function as chemorepellents. In this way, bacteria are trying to fathom the most favorable niches with relatively high chemoattractor and low chemorepellent concentrations. Temporal sensing is the fundamental principle that underlies this process of decision-making. The overall bacterial motion is principally a series of alternating phases of straight swimming and tumbling. If a bacterium is moving along a chemical gradient of a chemoattractor, the straight swimming phases last longer, but if the bacterium gets “abroad” and swims accidentally in regions of lower attractor-concentration, it starts sooner to tumble and tries to reorientate depending on the chemotaxins concentration [16]. The structural prerequisites for chemotaxis are simply at least one flagellum, a set of different chemoreceptors and a chemosensory signal-transduction system connecting both.
Experimental approaches
There are principally two major techniques to study chemotaxis and flagellar motility: microscopy and different variations of chemotaxis assays.
While conventional light microscopy is not sufficient to document the fast movement of C. jejuni or even to visualize the extremely thin flagellar filaments, dark field microscopy [17] and video-enhanced differential interference-contrast microscopy [18] coupled with computer ized image analysis [19, 20] are widely used in the field of bacterial taxis. Fluorescence microscopy is another useful tool especially to study protein-protein-interactions within the chemoreceptor-signalling cascade [21, 22]. A special fluorescence microscopy technique called stimulated emission depletion (STED) microscopy is able to achieve an effective resolution of 15 nm in biological samples. Recently, it has been increasingly used to visualize subcellular structures and advanced into an important tool in this field [23, 24].
Different kinds of chemotaxis assays were established in order to screen for substances with chemoeffector activity with reference to a specific bacterial species [25]. For Campylobacter a semiquantitative version, the so-called chemical-in-plug assay, based on opalescence changes of a (semi-)solid agar due to the concentration of bacterial cells is commonly used [26, 27]. Recently, serious concerns for this method were considered as chemotaxis-independent responses were observed using non-chemotactic and non-motile mutants in this assay [28].
Chemoreceptor signal transduction in C. jejuni
C. jejuni detects the variations of chemotaxin-concentrations in its environment by a set of different chemoreceptors, that share a common two-component system architecture for signal transduction (see Fig. 1) [25].
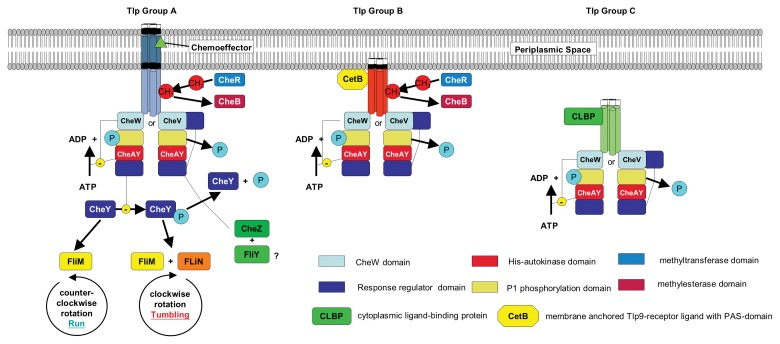
Fig. 1.
Chemotaxis signal transduction cascades for the three Tlp-chemoreceptor groups: The signalling domains forming trimers of dimers are depicted in steel blue (group A), Carnelian red (group B) and light green (group C); CheW and the CheW-homologue domain of CheV are depicted in light blue; CheV owns an additional response regulator (RR) domain, which is also part of CheAY and CheY. In contrast to Helicobacter pylori, CheA is composed of not only a His-autokinase domain but also a P1 phosphorylation domain. It has an additional RR-domain and that is why it is named CheAY in C. jejuni. For further details, see text.
These two-component systems consist commonly of a membrane-associated histidine autokinase (CheA) and a cytoplasmic RR (CheY) [29].
These chemoreceptors, belonging to the group of methyl-accepting chemotaxis proteins (MCPs), exist as either integral membrane or free drifting cytoplasmic proteins [30]. The first sense the environmental signals via their N-terminal periplasmic sensory domain to their C-terminal cytoplasmic signalling domain. The second interact with cytoplasmic proteins, which detect chemoeffector signals and sense this intracellular signal likewise to their C-terminal cytoplasmic signalling domain. The MCP-monomers have an average molecular mass of 60 kDa. They arrange to very stable homodimers, which usually form groups of three [29]. A coupling scaffold protein called CheW interacts with the MCPs cytoplasmic signalling domains. When CheW attaches the histidine kinase, CheA the final ternary signalling complex is compiled [25, 31–34]. Thus CheA autophosphorylation, which primarily occurs at the P1 phosphorylation domain, is inhibited. Hence, the phosphorylation of the CheY RR-domain is reduced.
Most members of the Epsilonproteobacteria like Campylobacter sp. show specific modifications of this general scheme [35]. The preferentially and parallel to CheW involved coupling scaffold protein of C. jejuni is CheV. CheV is a CheW-like linking-protein with an additional RR-domain that may play a role in mediating adaptation to attractants [36, 37]. A second modification is an additional RR-domain (homologue to CheY) at the C-terminus of the CheA-protein, which is termed CheAY [30, 38]. The characteristic module of these RR-domains is the CheY-like phosphoacceptor domain commonly designated as receiver (REC) domain [39]. Typically, CheA proteins lacking a (secondary) P2 phosphorylation domain (besides the P1 phosphorylation domain) are CheAY proteins [40]. It is not finally clear, which role this additional RR-domain plays in the function of CheAY. CheAY is able to phosphorylate its RR-domain but CheY seems to be the preferred substrate [41]. Otherwise, CheY can phos phorylate CheAY at its RR-domain [41]. Thus, it was assumed that CheAY might remove phosphoryl groups from CheY and functions in that way as a so-called phosphate sink [35].
However, increasing ligand occupancy results in higher amounts of non-phosphorylated CheY. Non-phosphorylated CheY interacts weakly with the FliM switch proteins of the flagellar motor, which are the final effectors of the sensory transduction chain [42–45]. Because of the only weak interaction between unphosphorylated CheY and FliM, CheY is not able to break up the FliN– FliN interaction within the FliN-dimer via the N-terminal hydrophobic patch [46]. Thus, a CheY–FliN interaction does not take place and a counterclockwise rotation of the flagella is induced leading to a fast straightforward movement of the bacterial cell, a so-called “run”. While moving along a gradient of a chemoattractant, the intracellular concentration of phosphorylated CheY inside the bacterial cytoplasm decreases. Therefore, the rate of site directed “runs” along the gradient increases [47].
In the case of decreasing ligand occupancy of the TLP-receptors CheAY autophosphorylation is less inhibited, resulting in a higher amount of CheAY-P. CheAY-P inhibits the dephosphorylation of CheY, which results in higher quantities of phosphorylated CheY and CheB. CheB is a receptor-demethylating enzyme, which is also activated by phosphorylation (see below). CheY-P interacts as well with the N-terminal parts of the FliM proteins of the motor switch complex but in a stronger way than non-phosphorylated CheY [43]. This stronger binding tethers CheY-P to FliM so that it is able to break up FliN-dimers to bind to FliN [46]. Thus, it triggers a clockwise flagellar rotation resulting in bacterial “tumbling” [44, 46].
The cytoplasmic phosphatase CheZ dephosphorylates phosphate-activated CheY and quenches thereby the signal for bacterial “tumbling” [48–50]. Additionally, it is assumed that the flagellar switch protein FliY, which has proven phosphatase activity in Bacillus subtilis also dephosphorylates CheY-P. In C. jejuni FliY the CheY binding domain is missing, but the active site consensus sequence of the CheC phosphatase family is present in FliY [35, 51]. Until the bacterium reaches the place of a balance between the varying chemoeffectors, this chemosensing will result in a stereoscopic “zigzag” path through the three-dimensional spatial gradients of different chemoattractors and chemorepellents [25, 52].
Different mechanisms of sensory adaptation
Sensory adaption in general is defined as the restoration of the prestimulus state in the continual presence of the chemoeffector. Epsilonproteobacteria like Campylobacter sp. seem to have two different mechanisms to achieve this. The first mechanism bases on the methylation of certain sites of the MCP receptor signalling domains (see Fig. 1). The second is based on the modification of the CheAY coupling by CheV.
Adaptation due to methylation/demethylation of methyl-accepting domains of the chemoreceptors signalling domains was recently shown to work at least for Tlp1 and Tlp4/DocC (but not Tlp9/CetA) [53]. The crucial methylation adaptation system proteins are the methyltransferase CheR and the methylesterase CheB. Further, central parts are the cytoplasmic domains of the MCP-receptors that have adjacent to the CheA(Y)- and CheV/CheW-binding sites, methyl-accepting domains with methylable glutamyl-side chains [54].
The methylation of the specific glutamyl-residues on the chemoreceptor signalling domains is a S-adenosylmethionine-consuming process catalysed by CheR [47]. Methylation is triggered by attracting stimuli. It promotes the autophosphorylation of CheA(Y) favouring clockwise flagellar rotation (tumbling) [55, 56]. It was demonstrated that the signalling domains possess a specific site, which can be methylated by CheR, and a further distinct CheR-binding site that is formed by a pentapeptide, which is specific for high-abundance receptors. CheR is able to methylate the designated sites of the low-abundance receptors while binding to these specific binding sites onto the high-abundance receptors [57, 58].
The methylesterase CheB is CheR’s functional antagonist. Firstly, it demethylates the Tlp-signalling domains during adaptation to repelling stimuli. CheB proteins usually consist of a methylesterase domain and a RR-domain that controls, depending on its phosphorylation state, the demethylation activity. In C. jejuni, however, this CheB RR-domain is absent [30, 53]. Secondly, CheB possesses an amidase activity. Thus, CheB can catalyse the conversion of glutamine residues into glutamate on the Tlp-receptor signalling domains. The liberation of glutamate residues inhibits the autophosphorylation of CheA(Y), which favours a counterclockwise flagellar rotation [59]. Thirdly, CheB is regulated by CheA(Y)-mediated phosphorylation. Phospho-CheB has a significantly increased methylesterase activity, while the unphosphorylated enzyme has less methylesterase activity [60, 61]. Fourthly, the CheA(Y) binding sites for CheY and CheB are identical. Thus, CheB and CheY compete with each other in this regard [62].
Finally, a high methylation rate of the signalling domains decreases the Tlp-receptors affinity to chemoactive ligands [63, 64].
The second sensory adaptation mechanism involves the CheW-like coupling protein CheV. CheV and CheW are expressed in parallel in C. jejuni and other Epsilonproteobacteria. The noticeable differences between both proteins are the higher affinity to the Tlp-receptors signalling domains (at least for these of Tlp1) [65] and the additional RR-domain of CheV [30].
The varying binding strength of CheV and CheW to different chemoreceptor signalling domains suggests that both proteins may out-compete each other depending on the specific receptor and the protein concentration [33, 66]. Thus, CheW preferentially binds Tlp4, whereas CheV demonstrates a high affinity to Tlp1, Tlp4, Tlp6 as well as Tlp8 [65, 66]. It is possible that this competition is part of a kind of sensory adaptation. It could also be that alternative pathways are controlled by either protein [33].
For B. subtilis, an adaptation mechanism was demonstrated requiring CheV phosphorylation [37]. It is assumable that a similar mechanism could also exist in C. jejuni, but the phosphorylation status of Campylobacter-CheV is not finally clear [37]. CheV catalyses the dephosphorylation of CheA(Y), but the halftime of phosphorylated CheV in Epsilonproteobacteria seems to be remarkably short [41, 67].
It was also suggested that a delayed activation of CheZ that results in an enhanced CheY dephosphorylation, plays a role in sensory adaptation to chemoeffector stimulation [68].
All these parallel-acting adaptational mechanisms determine the extent of reaction to a fireworks of various attracting and repelling chemoeffectors and help to maintain the bacterial cells orientation in a steady-state of conflicting and cumulative signals [69].
Specific Campylobacter chemoreceptors
The genome of C. jejuni encodes for altogether ten chemoreceptors, designated as “Tlps” for transducer-like proteins. These chemoreceptors can be categorized into three different groups (A–C) with respect to structural homologies. With regard to their individual molecular composition, they are either integral membrane proteins or proteins that reside in the cytoplasm as shown in Fig. 2 [30]. Thereby, group A receptors possess the similar composition as the MCPs of Escherichia coli and as the family A transducers of Halobacterium salinarium [70]. In general, two amino-terminal located transmembrane domains and a periplasmic ligand-binding domain is followed by a highly conserved carboxy-terminal signalling domain [71]. In addition, a structurally conserved HAMP-domain (a linker domain present in histidine-kinases, adenyl-cyclases, methyl-accepting-proteins and phosphatases) connects the transmembrane helices with the signalling domain. Thereby, the HAMP-domain, which is conserved in histidine kinases, adenylyl cyclases, MCPs and phosphatases, converts ligand-induced conformational changes into kinase-controlling signals [72–75]. Finally, within the signalling domain reside glutamyl-residues for the reversible methylation or demethylation of the chemoreceptor by CheR and CheB, respectively (see above) [76].
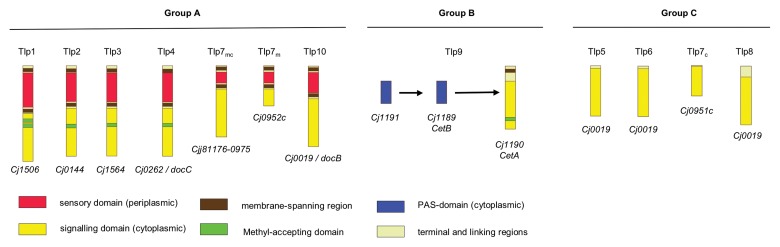
Fig. 2.
Domain organization of the three different C. jejuni Tlp-chemoreceptor groups: group A receptors are anchored by membrane-spanning regions in the inner and obviously also in the outer membrane, have a periplasmic sensory and a cytoplasmic signalling domain; the only group B receptor Tlp9 (CetA), anchored in the inner membrane, interacts with CetB triggering pyruvate and fumarate signals [84]; group C chemoreceptors consist of a single cytoplasmic signalling domain; the cytoplasmic Tlp7c (encoded by cj0951c) receptor is assigned to group C, while it was shown that there is no read-through mechanism [77], whereas the membrane associated forms Tlp7m (encoded by cj0952c) and Tlp7mc (encoded by cjj81176-0975 according to the nomenclature of the 81-176 sequence) are assigned to group A (depiction modified after Marchant et al. [30])
The group A receptors of C. jejuni are Tlp1, Tlp2, Tlp3, Tlp4 and Tlp10, whereby the signalling domains of the Tlps 2, 3 and 4 are identical [38]. The situation for chemoreceptor Tlp7 is inconsistent. Tlp7 belongs to the group A receptors if encoded by one single gene (cjj81176-0975) as in the reference strains 81-176 and 81116. But in the C. jejuni strains NCTC 11168 and B2, the sections for periplasmic binding and transmembrane localization are encoded on gene cj0952c, whereas the signalling domain is encoded by the adjacent gene cj0951c [30, 77]. Thus, the membrane-associated partial receptor encoded by cj0952 (termed Tlp7m) belongs like the cjj81176-0975-encoded receptor, consisting of the membrane-associated and the cytoplasmatic domains (termed Tlp7mc), to group A, whereas the cytoplasmic part, encoded by cj0951c (Tlp7c), should be considered as group C chemoreceptor. However, up to now it was not shown that Tlp7m and Tlp7c sense together. It is assumable that Tlp7c could also interact and sense with other ligands than Tlp7m. Additionally, the two-gene receptor variant could be significantly more often detected in C. jejuni isolates of bovine origin as well as of the multilocus sequence types ST-21 and ST-61 [78]. The most extensive studies about the function of these chemoreceptors and how signals are transduced have been carried out in E. coli [79, 80]. The chemoreceptors are as mentioned above composed as transmembrane homodimers that form trimers of dimers and extended clusters. The development of nanodiscs, small particles of lipid bilayers containing a defined number of chemoreceptors, allowed to characterize the interaction of chemoreceptors with regard to ligand binding, transmembrane signalling and activation of the chemotaxis histidine kinase CheA(Y). Thereby, it has been demonstrated that dimers of receptors are capable of binding ligands and to perform transmembrane signalling. Maximal activation of the histidine kinase was in turn preferentially observed in nanodiscs that contained trimers of dimers [81]. Transmembrane chemoreceptors together with CheW/CheV and CheA(Y) aggregate in patches predominantly at the cell poles, whereby clustering depends not solely on the chemoreceptor, but also on the presence of the associated CheW/CheV and CheA(Y) proteins [82]. Moreover, it can be speculated that these patches are assembled by different trimers of different homodimers in a way that allows the regulation of CheA(Y) proteins via interactions with the chemoreceptors [83].
In addition to the MCPs of chemoreceptor group A, the genome of C. jejuni encodes for four chemoreceptors Tlp5, Tlp6, Tlp7c and Tlp8, which represent the receptor group C [30]. In contrast to group A receptors, these molecules contain neither a periplasmic binding domain nor transmembrane regions and, thus, are believed to reside in the cytosol. However, despite their homology to signalling domains of other chemoreceptors, nothing is known about their biological function, but the estimated cytosolic localisation may be a hint that they sense intracellular signals in C. jejuni.
Chemoreceptor group B is comprised only by Tlp9 (CetA), which together with CetB, possesses structural similarities to the energy taxis receptor Aer of E. coli. CetB has a PAS (Per, ARNT and Sim) domain while CetA includes two transmembrane helices, a HAMP-domain and a highly conserved domain (HCD). Both proteins together represent a bipartite energy taxis system; whereas dimers of CetB are only peripherally associated with the membrane, CetA is membrane-anchored by its transmembrane domains. As in the case of E. coli energy taxis receptor Aer, experimental data suggest that CetB is a signal sensing protein that conveys the signals to CetA. CetA in turn transmits the energy taxis signals to core signal transduction proteins of the chemotactic system CheW/ CheV, CheA(Y) and, thus, to CheY [84, 85]. Based on genome sequence analysis data, there is a potential second cytoplasmic ligand-binding protein for Tlp9 encoded by cj1191 [30, 38].
The chemotactical behaviour of C. jejuni was initially reported by Hugdahl and co-workers [26]. They described the chemotactical response of the bacterium with respect to different amino acids, carbohydrates, organic acids and components of mucin and bile. In more recent investigations the panel of substances, which affect the chemotactic competence could be enlarged and it was shown that chemoattractants most notably serve as respiratory electron donors and electron acceptors contributing to the maintenance of energy metabolism [27]. Our current knowledge about the respective receptors of C. jejuni that interact with particular chemoattractants or chemorepellents is still poor. However, it could be shown that Tlp9 (CetA, CetB) triggers the response of C. jejuni towards pyruvate and fumarate, whereby the former serves as an electron donor and the latter is known to be a respiratory electron acceptor [84]. In addition, Tlp7 (cj0952c/cj0951c, cjj81176-0975) could be linked to the sensing of electron donor formate, and Tlp1 was identified to represent the chemoreceptor for aspartate [65, 77]. Despite the progress that has been achieved to match the chemoreceptors of C. jejuni to its particular ligand, the overall picture of receptors and their respective chemoeffectors is still incomplete and awaits further examinations.
The role of chemotaxis in host colonization and the pathogenesis of campylobacteriosis
The direct association of the chemotaxis system with the flagellar apparatus affects apparently the bacterial motility of C. jejuni. Motility in turn is an essential factor for the pathogenicity of campylobacteriosis, as non-motile strains possess a decreased capacity to infect host cells [7–9]. Consequently, the loss of chemoreceptor functions can be associated with reduced motility and, thus, reduced infectivity as it has been demonstrated by transposon insertion for the chemoreceptors Tlp3 (cj1564) as well as CetB (cj1189) and CetA (cj1190) representing the bipartite receptor Tlp9 [84, 86]. Vegge and co-workers could add Tlp4 (cj0262c) to the pool of chemoreceptors contributing to a complete C. jejuni-motility [27]. Recently, it was also shown that a transposon insertion in Tlp 7 (cj0951c/cj0952c or cjj81176-0975) is accompanied by reduced motility of C. jejuni [77]. Finally, since all signals, which are sensed by the chemoreceptors, are transmitted to flagellum via the chemotaxis core proteins it is unsurprising that the loss of proteins such as CheA and CheY also results in a motility-deficient phenotype [86].
In addition to the motility affecting phenotype mentioned above, some chemoreceptors are also involved in the commensal colonization of the avian intestine by C. jejuni. Tlp1 (cj1506c), Tlp4 (cj0262c) and Tlp10 (cj0019c) could be demonstrated, in contrast to Tlp7 (cj0951c/cj0952c or cjj81176-0975) and Tlp9 (cj1189), to be relevant for chicken intestinal colonization since a mutation within the corresponding genes significantly decreased the number of bacteria in the faeces. Interestingly, a transposon insertion in gene Tlp10 (cj0019) did not affect the in vitro motility of the bacterium indicating that the lack of a chemoreceptor is not mandatorily linked to the motility of C. jejuni [65, 87]. The in vivo examination of the intestinal pathology induced by C. jejuni was not feasible for decades since an appropriate vertebrate model was not available. Recently, Bereswill and co-workers established a gnotobiotic mouse model with a complete human intestinal microflora [88]. These intestinal “humanized” mice could be stably colonized by the pathogen, which was accompanied by a pro-inflammatory immune response and, therefore, are suitable as an infection and inflammation model. Using this model it could be shown that a Tlp7-deficient C. jejuni-mutant strain colonizes the “humanized” mice with similar bacterial loads compared to the wild type strain but does not induce immunopathology regarding the number of apoptotic cells and the number of T-lymphocytes within the colon mucosa.
Acknowledgements
The authors’ work was supported by the Deutsche Forschungsgemeinschaft (GR 906/13-1) and the Forschungsförderungsprogramm of the Universitätsmedizin Göttingen, Germany.
Contributor Information
A. E. Zautner, Universitätsmedizin Göttingen, Abteilung für Medizinische Mikrobiologie, Göttingen, Germany.
A. Malik Tareen, Universitätsmedizin Göttingen, Abteilung für Medizinische Mikrobiologie, Göttingen, Germany.
U. Groß, Universitätsmedizin Göttingen, Abteilung für Medizinische Mikrobiologie, Göttingen, Germany
R. Lugert, Universitätsmedizin Göttingen, Abteilung für Medizinische Mikrobiologie, Göttingen, Germany
References
- 1.Butzler JP. Campylobacter, from obscurity to celebrity. Clin Microbiol Infect. 2004 Oct;10(10):868–876. doi: 10.1111/j.1469-0691.2004.00983.x. [DOI] [PubMed] [Google Scholar]
- 2.European Food Safety Authority – EFSA. Campylobacter and Salmonella prevalence estimates. EFSA J. 2010;8:1503. [Google Scholar]
- 3.Robert Koch Institut – RKI. Aktuelle Daten und Informationen zu Infektionskrankheiten und Public Health. Epidemiologisches Bulletin. 2007;36:331–334. [Google Scholar]
- 4.Allos BM. Campylobacter jejuni Infections: update on emerging issues and trends. Clin Infect Dis. 2001 Apr 15;32(8):1201–1206. doi: 10.1086/319760. [DOI] [PubMed] [Google Scholar]
- 5.Schmidt-Ott R, Schmidt H, Feldmann S, Brass F, Krone B, Gross U. Improved serological diagnosis stresses the major role of Campylobacter jejuni in triggering Guillain-Barré syndrome. Clin Vaccine Immunol. 2006 Jul;13(7):779–783. doi: 10.1128/CVI.00065-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zautner AE, et al. Campylobacter jejuni – The Search for virulence-associated factors. Arch für Lebensmittelhyg. 2010;61:91–101. [Google Scholar]
- 7.Morooka T, Umeda A, Amako K. Motility as an intestinal colonization factor for Campylobacter jejuni. J Gen Microbiol. 1985 Aug;131(8):1973–1980. doi: 10.1099/00221287-131-8-1973. [DOI] [PubMed] [Google Scholar]
- 8.Nachamkin I, Yang XH, Stern NJ. Role of Campylobacter jejuni flagella as colonization factors for three-day-old chicks: analysis with flagellar mutants. Appl Environ Microbiol. 1993 May;59(5):1269–1273. doi: 10.1128/aem.59.5.1269-1273.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pavlovskis OR, Rollins DM, Haberberger RL, Jr., Green AE, Habash L, Strocko S, Walker RI. Significance of flagella in colonization resistance of rabbits immunized with Campylobacter spp. Infect Immun. 1991 Jul;59(7):2259–2264. doi: 10.1128/iai.59.7.2259-2264.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Szymanski CM, King M, Haardt M, Armstrong GD. Campylobacter jejuni motility and invasion of Caco-2 cells. Infect Immun. 1995 Nov;63(11):4295–4300. doi: 10.1128/iai.63.11.4295-4300.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wassenaar TM, Bleumink-Pluym NM, van der Zeijst BA. Inactivation of Campylobacter jejuni flagellin genes by homologous recombination demonstrates that flaA but not flaB is required for invasion. EMBO J. 1991 Aug;10(8):2055–2061. doi: 10.1002/j.1460-2075.1991.tb07736.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yao R, Burr DH, Doig P, Trust TJ, Niu H, Guerry P. Isolation of motile and non-motile insertional mutants of Campylobacter jejuni: the role of motility in adherence and invasion of eukaryotic cells. Mol Microbiol. 1994 Dec;14(5):883–893. doi: 10.1111/j.1365-2958.1994.tb01324.x. [DOI] [PubMed] [Google Scholar]
- 13.Engelmann TW. Neue Methode zur Untersuchung der Sauerstoffausscheidung pflanzlicher und thierischer Organismen. Pflügers Arch. 1881;25:285–292. [Google Scholar]
- 14.Pfeffer W. Locomotorische Richtungsbewegungen durch chemische Reize. Unt Bot Inst Tübingen. 1884;1(III):363–482. [Google Scholar]
- 15.Pfeffer W. Über chemotaktische Bewegungen von Bakterien, Flagellaten und Volvocineen. Unt Bot Inst Tübingen. 1888;2(III):582–661. [Google Scholar]
- 16.Adler J. Chemotaxis in bacteria. Annu Rev Biochem. 1975;44:341–356. doi: 10.1146/annurev.bi.44.070175.002013. [DOI] [PubMed] [Google Scholar]
- 17.Macnab RM. Examination of bacterial flagellation by dark-field microscopy. J Clin Microbiol. 1976 Sep;4(3):258–265. doi: 10.1128/jcm.4.3.258-265.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Block SM, Fahrner KA, Berg HC. Visualization of bacterial flagella by video-enhanced light microscopy. J Bacteriol. 1991 Jan;173(2):933–936. doi: 10.1128/jb.173.2.933-936.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Staropoli JF, Alon U. Computerized analysis of chemotaxis at different stages of bacterial growth. Biophys J. 2000 Jan;78(1):513–519. doi: 10.1016/S0006-3495(00)76613-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lauga E, DiLuzio WR, Whitesides GM, Stone HA. Swimming in circles: motion of bacteria near solid boundaries. Biophys J. 2006 Jan 15;90(2):400–412. doi: 10.1529/biophysj.105.069401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pierce DW, Vale RD. Single-molecule fluorescence detection of green fluorescence protein and application to single-protein dynamics. Methods Cell Biol. 1999 Xxx;58(000):49–73. doi: 10.1016/s0091-679x(08)61948-2. [DOI] [PubMed] [Google Scholar]
- 22.Khan S, Pierce D, Vale RD. Interactions of the chemotaxis signal protein CheY with bacterial flagellar motors visualized by evanescent wave microscopy. Curr Biol. 2000 Jul-Aug;10(15):927–930. doi: 10.1016/s0960-9822(00)00629-1. [DOI] [PubMed] [Google Scholar]
- 23.Hell SW, Wichmann J. Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Opt Lett. 1994 Jun 1;19(11):780–782. doi: 10.1364/ol.19.000780. [DOI] [PubMed] [Google Scholar]
- 24.Donnert G, Keller J, Medda R, Andrei MA, Rizzoli SO, Lührmann R, Jahn R, Eggeling C, Hell SW. Macromolecular-scale resolution in biological fluorescence microscopy. Proc Natl Acad Sci U S A. 2006 Aug 1;103(31):11440–11445. doi: 10.1073/pnas.0604965103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller LD, Russell MH, Alexandre G. Diversity in bacterial chemotactic responses and niche adaptation. Adv Appl Microbiol. 2009;66:53–75. doi: 10.1016/S0065-2164(08)00803-4. [DOI] [PubMed] [Google Scholar]
- 26.Hugdahl MB, Beery JT, Doyle MP. Chemotactic behavior of Campylobacter jejuni. Infect Immun. 1988 Jun;56(6):1560–1566. doi: 10.1128/iai.56.6.1560-1566.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vegge CS, Brøndsted L, Li YP, Bang DD, Ingmer H. Energy taxis drives Campylobacter jejuni toward the most favorable conditions for growth. Appl Environ Microbiol. 2009 Aug;75(16):5308–5314. doi: 10.1128/AEM.00287-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanungpean D, Kakuda T, Takai S. False positive responses of Campylobacter jejuni when using the chemical-in-plug chemotaxis assay. J Vet Med Sci. 2011 Mar;73(3):389–391. doi: 10.1292/jvms.10-0396. [DOI] [PubMed] [Google Scholar]
- 29.Lux R, Shi W. Chemotaxis-guided movements in bacteria. Crit Rev Oral Biol Med. 2004 Jul 1;15(4):207–220. doi: 10.1177/154411130401500404. [DOI] [PubMed] [Google Scholar]
- 30.Marchant J, Wren B, Ketley J. Exploiting genome sequence: predictions for mechanisms of Campylobacter chemotaxis. Trends Microbiol. 2002 Apr;10(4):155–159. doi: 10.1016/s0966-842x(02)02323-5. [DOI] [PubMed] [Google Scholar]
- 31.Gegner JA, Graham DR, Roth AF, Dahlquist FW. Assembly of an MCP receptor, CheW, and kinase CheA complex in the bacterial chemotaxis signal transduction pathway. Cell. 1992 Sep 18;70(6):975–982. doi: 10.1016/0092-8674(92)90247-a. [DOI] [PubMed] [Google Scholar]
- 32.Boukhvalova MS, Dahlquist FW, Stewart RC. CheW binding interactions with CheA and Tar. Importance for chemotaxis signaling in Escherichia coli. J Biol Chem. 2002 Jun 21;277(25):22251–2259. doi: 10.1074/jbc.M110908200. [DOI] [PubMed] [Google Scholar]
- 33.Korolik V. Aspartate chemosensory receptor signalling in Campylobacter jejuni. Virulence. 2010 Sep-Oct;1(5):414–417. doi: 10.4161/viru.1.5.12735. [DOI] [PubMed] [Google Scholar]
- 34.Wuichet K, Zhulin IB. Origins and diversification of a complex signal transduction system in prokaryotes. Sci Signal. 2010 Jun 29;3(128):ra50. doi: 10.1126/scisignal.2000724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lertsethtakarn P, Ottemann KM, Hendrixson DR. Motility and chemotaxis in Campylobacter and Helicobacter . Annu Rev Microbiol. 2011;65:389–410. doi: 10.1146/annurev-micro-090110-102908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pittman MS, Goodwin M, Kelly DJ. Chemotaxis in the human gastric pathogen Helicobacter pylori: different roles for CheW and the three CheV paralogues, and evidence for CheV2 phosphorylation. Microbiology. 2001 Sep;147(Pt 9):2493–2504. doi: 10.1099/00221287-147-9-2493. [DOI] [PubMed] [Google Scholar]
- 37.Alexander RP, Lowenthal AC, Harshey RM, Ottemann KM. CheV: CheW-like coupling proteins at the core of the chemotaxis signaling network. Trends Microbiol. 2010 Nov;18(11):494–503. doi: 10.1016/j.tim.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parkhill J, Wren BW, Mungall K, Ketley JM, Churcher C, Basham D, Chillingworth T, Davies RM, Feltwell T, Holroyd S, Jagels K, Karlyshev AV, Moule S, Pallen MJ, Penn CW, Quail MA, Rajandream MA, Rutherford KM, van Vliet AH, Whitehead S, Barrell BG. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature. 2000 Feb 10;403(6770):665–668. doi: 10.1038/35001088. [DOI] [PubMed] [Google Scholar]
- 39.Galperin MY. Structural classification of bacterial response regulators: diversity of output domains and domain combinations. J Bacteriol. 2006 Jun;188(12):4169–4182. doi: 10.1128/JB.01887-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wuichet K, Alexander RP, Zhulin IB. Comparative genomic and protein sequence analyses of a complex system controlling bacterial chemotaxis. Methods Enzymol. 2007;422:1–431. doi: 10.1016/S0076-6879(06)22001-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiménez-Pearson MA, Delany I, Scarlato V, Beier D. Phosphate flow in the chemotactic response system of Helicobacter pylori. Microbiology. 2005 Oct;151(Pt 10):3299–3311. doi: 10.1099/mic.0.28217-0. [DOI] [PubMed] [Google Scholar]
- 42.Sockett H, Yamaguchi S, Kihara M, Irikura VM, Macnab RM. Molecular analysis of the flagellar switch protein FliM of Salmonella typhimurium. J Bacteriol. 1992 Feb;174(3):793–806. doi: 10.1128/jb.174.3.793-806.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Welch M, Oosawa K, Aizawa S, Eisenbach M. Phosphorylation-dependent binding of a signal molecule to the flagellar switch of bacteria. Proc Natl Acad Sci U S A. 1993 Oct 1;90(19):8787–8791. doi: 10.1073/pnas.90.19.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bren A, Eisenbach M. The N terminus of the flagellar switch protein, FliM, is the binding domain for the chemotactic response regulator, CheY. J Mol Biol. 1998 May 8;278(3):507–514. doi: 10.1006/jmbi.1998.1730. [DOI] [PubMed] [Google Scholar]
- 45.Szurmant H, Ordal GW. Diversity in chemotaxis mechanisms among the bacteria and archaea. Microbiol Mol Biol Rev. 2004 Jun;68(2):301–319. doi: 10.1128/MMBR.68.2.301-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sarkar MK, Paul K, Blair D. Chemotaxis signaling protein CheY binds to the rotor protein FliN to control the direction of flagellar rotation in Escherichia coli. Proc Natl Acad Sci U S A. 2010 May 18;107(20):9370–9375. doi: 10.1073/pnas.1000935107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bren A, Eisenbach M. How signals are heard during bacterial chemotaxis: protein-protein interactions in sensory signal propagation. J Bacteriol. 2000 Dec;182(24):6865–6873. doi: 10.1128/jb.182.24.6865-6873.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bourret RB, Stock AM. Molecular information processing: lessons from bacterial chemotaxis. J Biol Chem. 2002 Mar 22;277(12):9625–9628. doi: 10.1074/jbc.R100066200. [DOI] [PubMed] [Google Scholar]
- 49.Zhao R, Collins EJ, Bourret RB, Silversmith RE. Structure and catalytic mechanism of the E. coli chemotaxis phosphatase CheZ. Nat Struct Biol. 2002 Aug;9(8):570–575. doi: 10.1038/nsb816. [DOI] [PubMed] [Google Scholar]
- 50.Terry K, Go AC, Ottemann KM. Proteomic mapping of a suppressor of non-chemotactic cheW mutants reveals that Helicobacter pylori contains a new chemotaxis protein. Mol Microbiol. 2006 Aug;61(4):871–882. doi: 10.1111/j.1365-2958.2006.05283.x. [DOI] [PubMed] [Google Scholar]
- 51.Lowenthal AC, Hill M, Sycuro LK, Mehmood K, Salama NR, Ottemann KM. Functional analysis of the Helicobacter pylori flagellar switch proteins. J Bacteriol. 2009 Dec;191(23):7147–7156. doi: 10.1128/JB.00749-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Berg HC, Brown DA. Chemotaxis in Escherichia coli analysed by three-dimensional tracking. Nature. 1972 Oct 27;239(5374):500–504. doi: 10.1038/239500a0. [DOI] [PubMed] [Google Scholar]
- 53.Kanungpean D, Kakuda T, Takai S. Participation of CheR and CheB in the chemosensory response of Campylobacter jejuni. Microbiology. 2011 May;157(Pt 5):1279–1289. doi: 10.1099/mic.0.047399-0. [DOI] [PubMed] [Google Scholar]
- 54.Macnab RM. How bacteria assemble flagella. Annu Rev Microbiol. 2003;57:77–100. doi: 10.1146/annurev.micro.57.030502.090832. [DOI] [PubMed] [Google Scholar]
- 55.Borkovich KA, Alex LA, Simon MI. Attenuation of sensory receptor signaling by covalent modification. Proc Natl Acad Sci U S A. 1992 Aug 1;89(15):6756–6760. doi: 10.1073/pnas.89.15.6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ninfa EG, Stock A, Mowbray S, Stock J. Reconstitution of the bacterial chemotaxis signal transduction system from purified components. J Biol Chem. 1991 May 25;266(15):9764–9770. [PubMed] [Google Scholar]
- 57.Le Moual H, Quang T, Koshland DE., Jr. Methylation of the Escherichia coli chemotaxis receptors: intra- and interdimer mechanisms. Biochemistry. 1997 Oct 28;36(43):13441–13448. doi: 10.1021/bi9713207. [DOI] [PubMed] [Google Scholar]
- 58.Li J, Li G, Weis RM. The serine chemoreceptor from Escherichia coli is methylated through an inter-dimer process. Biochemistry. 1997 Sep 30;36(39):11851–11857. doi: 10.1021/bi971510h. [DOI] [PubMed] [Google Scholar]
- 59.Djordjevic S, Stock AM. Structural analysis of bacterial chemotaxis proteins: components of a dynamic signaling system. J Struct Biol. 1998 Dec 15;124(2-3):189–200. doi: 10.1006/jsbi.1998.4034. [DOI] [PubMed] [Google Scholar]
- 60.Hess JF, Oosawa K, Kaplan N, Simon MI. Phosphorylation of three proteins in the signaling pathway of bacterial chemotaxis. Cell. 1988 Apr 8;53(1):79–87. doi: 10.1016/0092-8674(88)90489-8. [DOI] [PubMed] [Google Scholar]
- 61.Lupas A, Stock J. Phosphorylation of an N-terminal regulatory domain activates the CheB methylesterase in bacterial chemotaxis. J Biol Chem. 1989 Oct 15;264(29):17337–17342. [PubMed] [Google Scholar]
- 62.Li J, Swanson RV, Simon MI, Weis RM. The response regulators CheB and CheY exhibit competitive binding to the kinase CheA. Biochemistry. 1995 Nov 14;34(45):14626–14636. doi: 10.1021/bi00045a003. [DOI] [PubMed] [Google Scholar]
- 63.Bornhorst JA, Falke JJ. Attractant regulation of the aspartate receptor-kinase complex: limited cooperative interactions between receptors and effects of the receptor modification state. Biochemistry. 2000 Aug 8;39(31):9486–9493. doi: 10.1021/bi0002737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li G, Weis RM. Covalent modification regulates ligand binding to receptor complexes in the chemosensory system of Escherichia coli. Cell. 2000 Feb 4;100(3):357–365. doi: 10.1016/s0092-8674(00)80671-6. [DOI] [PubMed] [Google Scholar]
- 65.Hartley-Tassell LE, Shewell LK, Day CJ, Wilson JC, Sandhu R, Ketley JM, Korolik V. Identification and characterization of the aspartate chemosensory receptor of Campylobacter jejuni. Mol Microbiol. 2010 Feb;75(3):710–730. doi: 10.1111/j.1365-2958.2009.07010.x. [DOI] [PubMed] [Google Scholar]
- 66.Parrish JR, Yu J, Liu G, Hines JA, Chan JE, Mangiola BA, Zhang H, Pacifico S, Fotouhi F, DiRita VJ, Ideker T, Andrews P, Finley RL., Jr. A proteome-wide protein interaction map for Campylobacter jejuni. Genome Biol. 2007;8(7):R130. doi: 10.1186/gb-2007-8-7-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lertsethtakarn P, Ottemann KM. A remote CheZ orthologue retains phosphatase function. Mol Microbiol. 2010 Jul 1;77(1):225–235. doi: 10.1111/j.1365-2958.2010.07200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Blat Y, Gillespie B, Bren A, Dahlquist FW, Eisenbach M. Regulation of phosphatase activity in bacterial chemotaxis. J Mol Biol. 1998 Dec 11;284(4):1191–1199. doi: 10.1006/jmbi.1998.2224. [DOI] [PubMed] [Google Scholar]
- 69.Alon U, Surette MG, Barkai N, Leibler S. Robustness in bacterial chemotaxis. Nature. 1999 Jan 14;397(6715):168–171. doi: 10.1038/16483. [DOI] [PubMed] [Google Scholar]
- 70.Zhang W, Brooun A, McCandless J, Banda P, Alam M. Signal transduction in the archaeon Halobacterium salinarium is processed through three subfamilies of 13 soluble and membrane-bound transducer proteins. Proc Natl Acad Sci U S A. 1996 May 14;93(10):4649–4654. doi: 10.1073/pnas.93.10.4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Morgan DG, Baumgartner JW, Hazelbauer GL. Proteins antigenically related to methyl-accepting chemotaxis proteins of Escherichia coli detected in a wide range of bacterial species. J Bacteriol. 1993 Jan;175(1):133–140. doi: 10.1128/jb.175.1.133-140.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Le Moual H, Koshland DE., Jr. Molecular evolution of the C-terminal cytoplasmic domain of a superfamily of bacterial receptors involved in taxis. J Mol Biol. 1996 Aug 30;261(4):568–585. doi: 10.1006/jmbi.1996.0483. [DOI] [PubMed] [Google Scholar]
- 73.Butler SL, Falke JJ. Cysteine and disulfide scanning reveals two amphiphilic helices in the linker region of the aspartate chemoreceptor. Biochemistry. 1998 Jul 28;37(30):10746–10756. doi: 10.1021/bi980607g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Aravind L, Ponting CP. The cytoplasmic helical linker domain of receptor histidine kinase and methyl-accepting proteins is common to many prokaryotic signalling proteins. FEMS Microbiol Lett. 1999 Jul 1;176(1):111–116. doi: 10.1111/j.1574-6968.1999.tb13650.x. [DOI] [PubMed] [Google Scholar]
- 75.Williams SB, Stewart V. Functional similarities among two-component sensors and methyl-accepting chemotaxis proteins suggest a role for linker region amphipathic helices in transmembrane signal transduction. Mol Microbiol. 1999 Sep;33(6):1093–1102. doi: 10.1046/j.1365-2958.1999.01562.x. [DOI] [PubMed] [Google Scholar]
- 76.Weis RM, Koshland DE., Jr. Reversible receptor methylation is essential for normal chemotaxis of Escherichia coli in gradients of aspartic acid. Proc Natl Acad Sci U S A. 1988 Jan;85(1):83–87. doi: 10.1073/pnas.85.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tareen AM, Dasti JI, Zautner AE, Gross U, Lugert R. Campylobacter jejuni proteins Cj0952c and Cj0951c affect chemotactic behaviour towards formic acid and are important for invasion of host cells. Microbiology. 2010 Oct;156(Pt 10):3123–3135. doi: 10.1099/mic.0.039438-0. [DOI] [PubMed] [Google Scholar]
- 78.Zautner AE, Herrmann S, Corso J, Tareen AM, Alter T, Gross U. Epidemiological association of different Campylobacter jejuni groups with metabolism-associated genetic markers. Appl Environ Microbiol. 2011 Apr;77(7):2359–2365. doi: 10.1128/AEM.02403-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sourjik V. Receptor clustering and signal processing in E. coli chemotaxis. Trends Microbiol. 2004 Dec;12(12):569–576. doi: 10.1016/j.tim.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 80.Parkinson JS, Ames P, Studdert CA. Collaborative signaling by bacterial chemoreceptors. Curr Opin Microbiol. 2005 Apr;8(2):116–121. doi: 10.1016/j.mib.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 81.Boldog T, Grimme S, Li M, Sligar SG, Hazelbauer GL. Nanodiscs separate chemoreceptor oligomeric states and reveal their signaling properties. Proc Natl Acad Sci U S A. 2006 Aug 1;103(31):11509–11514. doi: 10.1073/pnas.0604988103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Maddock JR, Shapiro L. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science. 1993 Mar 19;259(5102):1717–1723. doi: 10.1126/science.8456299. [DOI] [PubMed] [Google Scholar]
- 83.Wadhams GH, Armitage JP. Making sense of it all: bacterial chemotaxis. Nat Rev Mol Cell Biol. 2004 Dec;5(12):1024–1037. doi: 10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- 84.Hendrixson DR, Akerley BJ, DiRita VJ. Transposon mutagenesis of Campylobacter jejuni identifies a bipartite energy taxis system required for motility. Mol Microbiol. 2001 Apr;40(1):214–224. doi: 10.1046/j.1365-2958.2001.02376.x. [DOI] [PubMed] [Google Scholar]
- 85.Elliott KT, Dirita VJ. Characterization of CetA and CetB, a bipartite energy taxis system in Campylobacter jejuni. Mol Microbiol. 2008 Sep;69(5):1091–1103. doi: 10.1111/j.1365-2958.2008.06357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Golden NJ, Acheson DW. Identification of motility and autoagglutination Campylobacter jejuni mutants by random transposon mutagenesis. Infect Immun. 2002 Apr;70(4):1761–1771. doi: 10.1128/IAI.70.4.1761-1771.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hendrixson DR, DiRita VJ. Identification of Campylobacter jejuni genes involved in commensal colonization of the chick gastrointestinal tract. Mol Microbiol. 2004 Apr;52(2):471–484. doi: 10.1111/j.1365-2958.2004.03988.x. [DOI] [PubMed] [Google Scholar]
- 88.Bereswill S, Fischer A, Plickert R, Haag LM, Otto B, Kühl AA, Dasti JI, Zautner AE, Muñoz M, Loddenkemper C, Gross U, Göbel UB, Heimesaat MM. Novel murine infection models provide deep insights into the "ménage à trois" of Campylobacter jejuni, microbiota and host innate immunity. PLoS One. 2011;6(6):e20953. doi: 10.1371/journal.pone.0020953. [DOI] [PMC free article] [PubMed] [Google Scholar]