Abstract
The neural crest (NC) is a multipotent population of migratory cells unique to the vertebrate embryo, contributing to the development of multiple organ systems. Transcription factors pax3 and zic1 are among the earliest genes activated in NC progenitors, and they are both necessary and sufficient to promote NC fate. In order to further characterize the function of these transcription factors during NC development we have used hormone inducible fusion proteins in a Xenopus animal cap assay, and DNA microarray to identify downstream targets of Pax3 and Zic1. Here we present the results of this screen and the initial validation of these targets using quantitative RT-PCR, in situ hybridization and morpholinos-mediated knockdown. Among the targets identified we found several well-characterized NC-specific genes, including snail2, foxd3, gbx2, twist, sox8 and sox9, which validate our approach. We also obtained several factors with no known function in Xenopus NC, which represent novel regulators of NC fate. The comprehensive characterization of Pax3 and Zic1 targets function in the NC gene regulatory network, are essential to understanding the mechanisms regulating the emergence of this important cell population.
Keywords: Xenopus, Neural crest, Pax3, Zic1, Microarray, Gene regulatory network
Introduction
The neural crest (NC) is a multipotent population of migratory cells unique to the vertebrate embryo. The NC arises from the lateral edge of the neural plate, in a region known as the neural plate border (NPB). By the end of neurulation NC progenitors reside in the most dorsal aspect of the neural tube. They eventually delaminate from the neuroepthelium and migrate to different locations in the embryo where they differentiate into a large variety of cell types including peripheral neurons and glia, craniofacial cartilage and bone, pigment cells and portions of the cardiovascular system (reviewed in LeDouarin et al., 2004). Because of its contribution to multiple lineages, abnormal development of the NC often results in a broad range of clinical manifestations, affecting multiple organ systems (reviewed in Bolande 1997).
In Xenopus the NPB is a heterogeneous tissue that can produce three distinct cell types: the NC, the pre-placodal ectoderm (PE) and the hatching gland (HG). Experimental evidence from several laboratories indicate that the segregation of these cell populations depends largely on the early activation of two transcription factors: Pax3 and Zic1 (Monsoro-Burq et al., 2005; Sato et al., 2005, Hong and Saint-Jeannet, 2007; Garnett et al., 2012). pax3 and zic1 are initially broadly expressed at the NPB and become progressively restricted to different regions of the ectoderm. pax3 is expressed in the presumptive HG cells, and zic1 marks the prospective PE, while both factors are co-expressed in the NC forming region. (Hong and Saint-Jeannet, 2007). Using gain of function and knockdown approaches in whole embryos we and others have shown that Pax3 and Zic1 are necessary and sufficient to promote HG and PE fates, respectively, while their combined activity is essential to specify the NC (Monsoro-Burq et al., 2005; Sato et al., 2005, Hong and Saint-Jeannet, 2007). Moreover, by manipulating the expression levels of Pax3 and Zic1 in naïve ectoderm it is possible to generate a fairly pure population of NC progenitors, in absence of other NPB cell types (Hong and Saint-Jeannet, 2007), further demonstrating the importance of the cooperation between Pax3 and Zic1 in promoting NC fate.
To fully understand the role of Pax3 and Zic1 during NC formation, it is critical to characterize their downstream targets. To this end we have used hormone-inducible fusion proteins to simultaneously express Pax3 and Zic1 in Xenopus animal caps to convert this tissue to NC, and used this sample to screen a DNA microarray. From the large data set generated, selected candidate genes were validated by quantitative RT-PCR (qPCR), in situ hybridization and morpholino-mediated knockdown. The genes recovered in the screen include several known NC-specific transcription factors, which validate our approach, but also a number of genes representing novel players in the gene regulatory network underlying NC development.
Materials and Methods
Embryo injections, explants culture and dexamethasone treatment
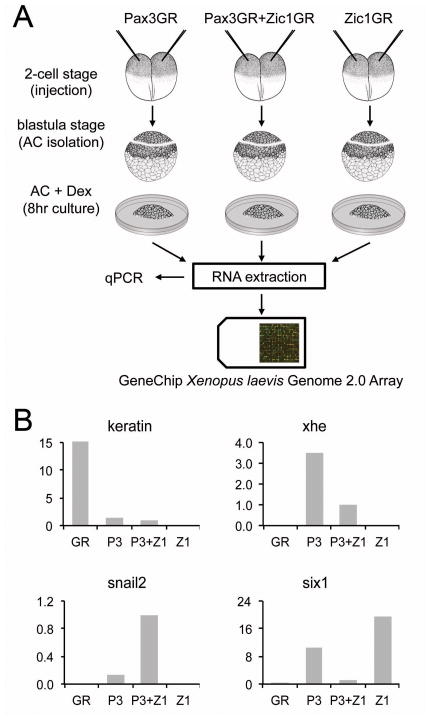
Xenopus laevis embryos were staged as previously described (Nieuwkoop, 1967). Pax3GR and Zic1GR constructs were generated as previously described (Kolm and Sive, 1995; Hong and Saint-Jeannet, 2007). RNAs encoding Pax3GR and Zic1GR were synthesized in vitro with the Message Machine kit (Ambion, Austin, TX). Both blastomeres of 2-cell stage embryos were injected in the animal pole region, with 250 pg of Pax3GR RNA or 250 pg of Zic1GR RNA, or both. At late blastula stage (stage 9), animal cap (AC) were dissected and cultured for 8 hour (equivalent stage 13.5) in 0.5X NAM containing 10 μM Dexamethasone (Dex; Sigma-Aldrich, St. Louis, MO) to activate Pax3GR or/and Zic1GR. The animal explants were subsequently analyzed by qPCR. Pax3 (Pax3MO; TCTCAGTTCCCTTGC CAAGTATTAA; Monsoro-Burq et al., 2005), Zic1 (Zic1MO; AAGTCTTCCAACAATGG GCAGCGAA; Sato et al., 2005) and Wnt8 (Wnt8MO; AAAGTGGTGTTTTGCAT GATGAAGG; Park and Saint-Jeannet, 2008) morpholino antisense oligonucleotides were purchased from GeneTools (Philomath, OR). In whole embryos antisense oligonucleotides were injected into the animal pole region of one blastomere at the 2-cell stage and embryos analyzed by in situ hybridization at stage 15. To identify the injected side, 500 pg of β-galactosidase mRNA was coinjected as a lineage tracer. For animal cap experiments, both blastomeres at the 2-cell stage were injected in the animal pole region, with Fgf8a mRNA (5pg; Christen and Slack, 1997) and Chordin DNA (10 pg; Sasai et al., 1994) alone or in combination with Wnt8MO, explants were then dissected at the late blastula stage and immediately cultured in vitro for several hours in 0.5X NAM (Hong et al., 2008). Animal explants were subsequently analyzed by qPCR as described (Hong and Saint-Jeannet, 2007).
RNA preparation and microarray analysis
Individual animal cap explants were homogenized and total RNA extracted using RNeasy Micro Kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. During the extraction procedure, the samples were treated with DNase I to eliminate possible genomic DNA contamination. The amount of isolated RNA was quantified using a spectrophotometer. Extracted RNAs (five replicate for each injection set) were used for the microarray experiment. All subsequent steps including probe labeling, hybridization, and initial data analysis was performed at the University of Pennsylvania Microarray Facility. Briefly, around 0.1–0.2 μg total RNA was converted into first-strand cDNA using Superscript II reverse transcriptase primed by a poly(T) oligomer that incorporates the T7 promoter. Second-strand cDNA synthesis is followed by in vitro transcription for linear amplification of each transcript and incorporation of biotinylated nucleotides. The cRNA products were fragmented to 200 nucleotides or less, heated at 99°C for 5 min and hybridized for 16 hr at 45°C to a GeneChip Xenopus laevis Genome 2.0 Array (Affymetrix, Cleveland, OH). Hybridized arrays were processed by the GeneChip Fluidics system, and scanned in the GeneChip Scanner. Probes intensities were exported in cel files using GCOS software (Affymetrix, Clevland, OH). Subsequent quantification of expression levels and statistical identification of differentially regulated genes was performed using Partek Genomics Suite (Partek, Inc., St Louis, MO). Normalized, log2-transformed probeset intensities were calculated using GC-RMA. ANOVA with multiple testing corrections was used to determine p-values for likelihood of differential expression, based on reproducibility in five replicates to identify cDNAs activated in response to Pax3GR+Zic1GR. Pairwise comparisons yielding fold changes were performed between all conditions.
Quantitative RT-PCR
cDNA synthesis was performed from 1 μg total RNA using Superscript VILO cDNA Synthesis Kit (Invitrogen, Valencia, CA) according to the manufacturer’s instructions. Quantitative PCR was performed using the primers shown in Table 4 and the Power SYBR Green PCR Master Mix (Invtrogen, Valencia, CA) on an Eco Real-Time PCR System (Illumina, San Diego, CA). The reaction mixture consisted of 1X Power SYBR Green PCR Master Mix, 200 nM primers, and 1 μl of cDNA in a total volume of 10 μl. The cycling conditions used were as follow: incubation at 50°C for 2 min; activation at 95°C for 10 min; 40 cycles at 95°C for 10 s and 60°C for 30 s; melt curve at 95°C for 15 s, 55°C for 15 s, and at 95°C for 15 s. The ΔΔCT method was used to quantify the results. Each reaction included a control without template and a standard curve of serial dilution points in 10 fold increments. In each case a reference gene was used for normalization (elongation factor 1 alpha, ef1α or ornithine decarboxylase, odc).
Table 4.
Primers for quantitative RT-PCR
| Unigene | Gene | Forward primer | Reverse primer |
|---|---|---|---|
| Xl.53491 | hoxd1 | 5′-CGTCCAGAGGAAAAGACCAG-3′ | 5′-CATCCCCGCAAGAAGTGTAT-3′ |
| Xl.6159 | mfap2 | 5′-TAGAGGCCCACCAGCTCTTA-3′ | 5′-GCCTGGTAGAAAGTGGAGGA-3′ |
| Xl.20029 | pdgfra | 5′-CAAAGGCAGCACTTTCCTTC-3′ | 5′-ATCATGCCAGGGTATGGTGT-3′ |
| Xl.46706 | MGC82246 | 5′-CGTCTAACCGCAAGATGGAT-3′ | 5′-GTTTGTCATCTTCCCGCTGT-3′ |
| Xl.802 | LOC397753 | 5′-AACAGGAGTAGGCAGCCTTTAGCA-3′ | 5′-AATGTGCCTAGCCAGATACCAGCA-3′ |
| Xl.76016 | fhl3 | 5′-TATGCCAAGAAATGTGCAGGGTGC-3′ | 5′-ACGGGAACAATTGAAGCAGCTGTG-3′ |
| Xl.47341 | ggh | 5′-GCTTTGCTGCTTCTTCTGCT-3′ | 5′-TTGCTCCAGCAGATTCAATG-3′ |
| Xl.49589 | abtb2 | 5′-GGCAACTTAAGCCAGAGTGC-3′ | 5′-TGAGATCTGTCCTGCCACAG-3′ |
| Xl.14729 | pmp22 | 5′-GCCCTGAAGCAGTTGAGAAC-3′ | 5′-GCAGATGGGACACATGACAC-3′ |
| Xl.14741 | LOC100137640 | 5′-TGAGCTGCTGATCGTGTGGAAGAT-3′ | 5′-AGATTTCTGAAGGCCGAGGCAGAT-3′ |
| Xl.76806 | MGC53056 | 5′-ACTACTTTGCTGAGCTGGCTGACT-3′ | 5′-CGTGTGCAGGGAATTGTGAGGTTT-3′ |
| Xl.78260 | slit2 | 5′-GGGCATAATGTTGCTGAGGT-3′ | 5′-TTATTGTCTCCGGGAGGTTG-3′ |
| Xl.368 | cer1 | 5′-GAATGGAGCCCCACAGAATA-3′ | 5′-TCGTCGATCTTGCTGATTTG-3′ |
| Xl.50785 | tfap2e | 5′-ATCACTGGTGGAAGGTGAGG-3′ | 5′-TCTGTGGGATCAGAGTGCTG-3′ |
| Xl.1269 | p2ry4 | 5′-GTGCAGCACAGGAGAATGAA-3′ | 5′-ACATTCTTGGCCAACCTGTC-3′ |
| Xl.17302 | ppm1k | 5′-AAGGAATGGACGGTGTCAAG-3′ | 5′-ACTGCCATCCAGGTCAAAAC-3′ |
| Xl.47031 | LOC431899 | 5′-AGAGAGCAGACAGCATTTGTCCGT-3′ | 5′-TTGCTGAACCCGCTTAAGTTCTGC-3′ |
| Xl.689 | nprc | 5′-GTGAAAAACTGGGGCACTGT-3′ | 5′-TGTTGTTTCGTTTCCCATGA-3′ |
| Xl.697 | nr1i2 | 5′-GGTGTCTGCTGGTTGGTTTT-3′ | 5′-AGTTGTGGGGCTTGATTTTG-3′ |
| Xl.1405 | LOC100036942 | 5′-TGTGTTGTGGGAGAGATGCTGGAT-3′ | 5′-AGGTTTGGAAGGGAGACTGGCTTT-3′ |
| Xl.25175 | tspan18 | 5′-TGCAAGGGAGGTGAAGAGGCATTA-3′ | 5′-CCATTTACACCGCAGCATCCGAAA-3′ |
| Xl.47735 | hoxb2 | 5′-TCCAGAACCGAAGGATGAAGCACA-3′ | 5′-TCATTGCTGTCCAATCCCTGGTGA-3′ |
| Xl.12464 | lmx1b.1 | 5′-TCCTTATGGCAATGACACCA-3′ | 5′-ACAGTCGGTCAATGGGATTC-3′ |
| Xl.30849 | LOC495203 | 5′-AGCGCCTCTGGAGAAAGAACAAGA-3′ | 5′-ATCGTTTCAGCCTGTCGAACTGGA-3′ |
| Xl.67432 | trhd-b | 5′-GGGGTGACAACCATCATTTC-3′ | 5′-TTTGTGCTGACTGGCTCATC-3′ |
| Xl.17336 | rassf10 | 5′-TCCAAGAAACCTACTTGGTGGGCA-3′ | 5′-TCGTCGCACTTCTGTGCATAGTCA-3′ |
| Xl.46804 | emp3 | 5′-AATGGCAGCAATGGATTTTC-3′ | 5′-TCCTGTTGTGGGTGCTGATA-3′ |
| Xl.34931 | LOC398346 | 5′-AGGGCAAACAAGAGGTCAGTGAGT-3′ | 5′-AGCGATTAGCCAATGAAACGGCAC-3′ |
| Xl.15219 | b4galt4 | 5′-TAGCTGAGACCAAGCAGCAA-3′ | 5′-AAGGGCTGTTTGAACCTGTG-3′ |
| Xl.23480 | pnhd | 5′-TAGGGCTCTGGCACAAATG-3′ | 5′-GCTCACAATGTCACAAGGAATG-3′ |
| Xl. 6133 | keratin | 5′-CACCAGAACACAGAGTAC-3′ | 5′-CAACCTTCCCATCAACCA-3′ |
| Xl.120 | xhe | 5′-CATGTCTAATGGCGGTTGTG-3′ | 5′-TGCTGGATGATCCCCATATT-3′ |
| Xl.3818 | snail2 | 5′-CATGGGAATAAGTGCAACCA-3′ | 5′-AGGCACGTGAAGGGTAGAGA-3′ |
| Xl.683 | six1 | 5′-CTGGAGAGCCACCAGTTCTC-3′ | 5′-AGTGGTCTCCCCCTCAGTTT-3′ |
| Xl.17432 | ef1α | 5′-ACCCTCCTCTTGGTCGTTTT-3′ | 5′-TTTGGTTTTCGCTGCTTTCT-3′ |
| Xl.4223 | odc | 5′-ACATGGCATTCTCCCTGAAG-3′ | 5′-TGGTCCCAAGGCTAAAGTTG-3′ |
Whole-mount in situ hybridization
The full coding sequence of each candidate gene was amplified by PCR, ligated into pGEM-T Easy Vector (Invitrogen, Valencia, CA), and sequenced. Sense control and antisense digoxigenin-labeled probes were synthesized for each gene using a DIG RNA Labeling Kit (Roche Diagnostics, Indianapolis, IN). Embryos at the neurula and tailbud stages were fixed in MEMFA and processed for whole-mount in situ hybridization (ISH) as described (Harland, 1991). For injected embryos β-galactosidase activity was detected using Red-Gal (Research Organics, Cleveland, OH) prior to the ISH procedure.
Results
DNA microarray screen
At the end of gastrulation pax3 and zic1 are co-expressed in the NC forming region. In this tissue Pax3 and Zic1 synergize to specify the NC (Monsoro-Burq et al., 2005; Sato et al., 2005; Hong and Saint-Jeannet, 2007; Garnett et al., 2012). In isolated animal cap explants the co-expression of Pax3 and Zic1 is sufficient to convert this tissue into NC, in the absence of other NPB cell types (Hong and Saint-Jeannet, 2007). Taking advantage of this unique explant preparation we performed a microarray analysis to identify genes synergistically activated by Pax3 and Zic1.
Xenopus embryos were injected at the 2-cell stage with mRNA encoding Pax3GR (250 pg) and Zic1GR (250 pg), the hormone-inducible version of Pax3 and Zic1, alone or in combination as previously described (Hong and Saint-Jeannet, 2007). At the blastula stage (stage 9), animal pole regions were dissected and cultured for 8 hours in the presence of dexamethasone (Fig 1A). A glucocorticoid receptor (GR) mRNA was also injected as negative control. Animal explants for each treatment group (GR, Pax3GR, Zic1GR and Pax3GR+Zic1GR) were collected for RNA extraction, and an aliquot of each sample was assayed by quantitative RT-PCR (qPCR) for the expression of epidermis (keratin), NC (snail2), PE (six1) and HG (xhe) specific genes. Each treatment is expected to lead to a specific pattern of gene expression as summarized in Table 1 (Hong and Saint-Jeannet, 2007). After confirmation by qPCR that the expected pattern of gene expression was observed in each injection group (Fig 1B), the samples were used to screen the Affymetrix GeneChip Xenopus laevis Genome 2.0 Array. For each treatment group five independent samples were analyzed. All subsequent steps including probe labeling, hybridization and initial data analysis were performed at the University of Pennsylvania Microarray Facility (see Experimental Procedures).
Figure 1. Strategy to isolate Pax3-Zic1 targets.
(A) Procedures used to identify Pax3-Zic1 targets in the developing NC. Xenopus embryos were injected at the 2-cell stage with mRNA encoding GR (not shown), Pax3GR and Zic1GR (250 pg each), alone or in combination. At the blastula stage (stage 9), animal cap explants (AC) were dissected and cultured for 8 hours in the presence of dexamethasone. RNA were extracted from each sample, analyzed by qPCR and subsequently used to screen a GeneChip Xenopus laevis Genome 2.0 Array (Affymetrix). (B) RNA extracted from GR, Pax3GR, Zic1GR, or Pax3GR+Zic1GR injected animal cap explants, were analyzed by qPCR for the expression of various marker genes, keratin (epidermis), xhe (HE), snail2 (NC), and six1 (PE) to confirm that the expected pattern of gene expression was observed in each injection group. The relative expression levels have been normalized to ef1α. P3, Pax3GR; Z1, Zic1GR.
Table 1.
Expected pattern of gene expression in GR, Pax3GR and Zic1GR injected animal caps.
| keratin (epidermis) | snail2 (neural crest) | six1 (placode) | xhe (hatching gland) | |
|---|---|---|---|---|
| GR | +++ | + | + | + |
| Pax3GR | + | + | + | +++ |
| Zic1GR | + | + | +++ | + |
| Pax3GR+Zic1GR | + | +++ | + | + |
(+), low expression levels; (+++), high expression levels
Selection of Pax3-Zic1 targets
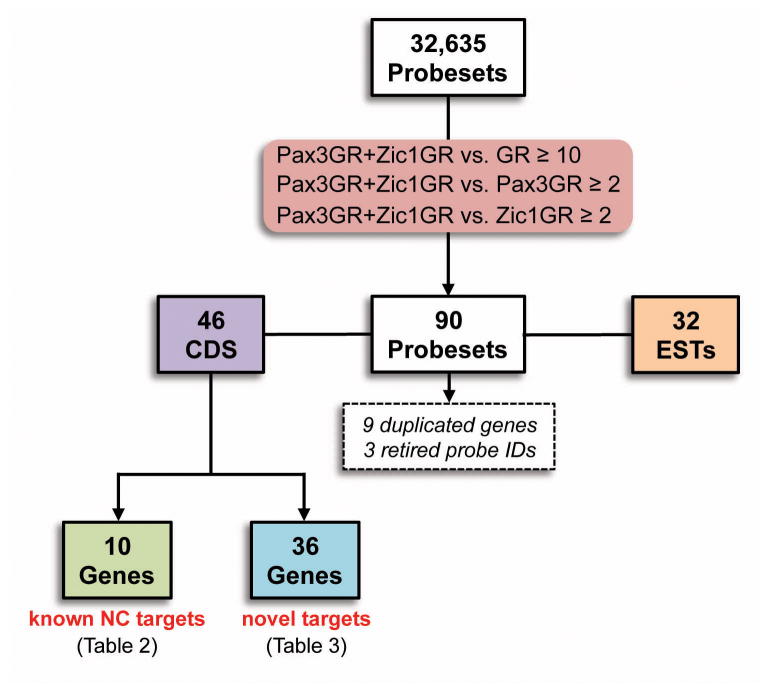
Among the 32,635 probesets included on the GeneChip Xenopus laevis Genome 2.0 Array we selected genes specifically activated by Pax3 and Zic1 based on the following criteria: probesets in the Pax3GR+Zic1GR sample that have a signal intensity at least (i) ten fold above the level seen in the GR sample, and (ii) two fold above the level seen in the Pax3GR and Zic1GR groups. This initial selection resulted in 90 probe IDs (Fig 2), 12 of which corresponded to duplicated genes or retired probe IDs. Among the remaining probe IDs 46 were fully annotated genes with complete coding sequence (CDS) available in Genebank. This group included 10 well-characterized NC-specific genes: ednra, foxd3, gbx2, olig4, snail2, sox8, sox9, tcf7, twist and zic5 (Table 2). The recovery of these genes was an important validation of our experimental approach. The other positives corresponded to 36 genes with no reported function in Xenopus NC (Table 3). For the 32 remaining probe IDs (Fig 2), only partial sequence information was available for these genes (EST), most of them with no apparent functional domain.
Figure 2.
Flow chart and criteria for the selection of Pax3-Zic1 targets.
Table 2.
Known NC-specific genes targets of Pax3-Zic1 in the DNA microarray
| Unigene | Gene
|
Relative expression in microarray
|
References | |||
|---|---|---|---|---|---|---|
| Symbol | Full name | P | P+Z | Z | ||
| Xl.56668 | ednra | Endothelin receptor type A | 0.1 | 1.0 | 0.5 | (Bonano et al., 2008) |
| Xl.973 | gbx2 | Gastrulation brain homeobox 2 | 0.4 | 1.0 | 0.0 | (Li et al., 2009) |
| Xl.525 | foxd3 | Forkhead box D3 | 0.3 | 1.0 | 0.0 | (Pohl and Knochel, 2001) |
| Xl.72230 | olig4 | Oligodendrocyte transcription factor 4 | 0.4 | 1.0 | 0.1 | (Bronchain et al., 2007) |
| Xl.3818 | snail2 | Snail homolog 2 | 0.2 | 1.0 | 0.2 | (Mancilla and Mayor, 1996) |
| Xl.29789 | sox8 | SRY (sex determining region Y)-box 8 | 0.1 | 1.0 | 0.3 | (O’Donnell et al., 2006) |
| Xl.1690 | sox9 | SRY (sex determining region Y)-box 9 | 0.2 | 1.0 | 0.1 | (Spokony et al., 2002) |
| Xl.3962 | tcf7 | Transcription factor 7 (T-cell specific) | 0.4 | 1.0 | 0.1 | (Klingel et al., 2012) |
| Xl.879 | twist | Twist homolog 1 | 0.2 | 1.0 | 0.1 | (Linker et al., 2000) |
| Xl.1043 | zic5 | Zic family member 5 | 0.3 | 1.0 | 0.4 | (Nakata et al., 2000) |
P, Pax3-GR; Z, Zic1-GR
Table 3.
Pax3-Zic1 targets and their validation by qPCR
| Unigene | Gene
|
Relative expression
|
||||||
|---|---|---|---|---|---|---|---|---|
| Microarray
|
qPCR
|
|||||||
| Symbol | Full name | P | P+Z | Z | P | P+Z | Z | |
| Xl.53491 | hoxd1 | Homeobox D1 | 0.3 | 1.0 | 0.0 | 0.6 | 1.0 | 0.0 |
| Xl.6159 | mfap2 | Microfibrillar-associated protein 2 | 0.4 | 1.0 | 0.0 | 0.9 | 1.0 | 0.1 |
| Xl.20029 | pdgfra | Platelet-derived growth factor receptor alpha | 0.1 | 1.0 | 0.3 | 0.2 | 1.0 | 0.1 |
| Xl.46706 | ak3 | Adenylate kinase 3 | 0.3 | 1.0 | 0.2 | 0.4 | 1.0 | 0.3 |
| Xl.802 | LOC397753 | Hypothetical protein LOC397753 | 0.5 | 1.0 | 0.0 | 1.0 | 1.0 | 0.0 |
| Xl.76016 | fhl3 | Four and a half LIM domain 3 | 0.1 | 1.0 | 0.2 | 1.3 | 1.0 | 0.6 |
| Xl.47341 | ggh | Gamma-glutamyl hydrolase | 0.1 | 1.0 | 0.3 | 0.6 | 1.0 | 0.4 |
| Xl.49589 | abtb2 | Ankyrin repeat and BTB domain containing 2 | 0.1 | 1.0 | 0.4 | 0.4 | 1.0 | 0.8 |
| Xl.14729 | pmp22 | Peripheral myelin protein 22 | 0.1 | 1.0 | 0.3 | 0.2 | 1.0 | 0.5 |
| Xl.14741 | LOC100137640 | Hypothetical protein LOC100137640 | 0.3 | 1.0 | 0.1 | 0.7 | 1.0 | 0.6 |
| Xl.53219 | MGC116527 | Hypothetical protein MGC116527 | 0.3 | 1.0 | 0.3 | n/a | n/a | n/a |
| Xl.76806 | MGC53056 | Similar to selenoprotein T | 0.4 | 1.0 | 0.4 | 1.9 | 1.0 | 1.2 |
| Xl.78260 | slit2 | Slit homolog 2 | 0.3 | 1.0 | 0.4 | 1.1 | 1.0 | 0.4 |
| Xl.40286 | foxa2 | Forkhead box A2 | 0.0 | 1.0 | 0.0 | n/a | n/a | n/a |
| Xl.368 | cer1 | Cerberus 1 | 0.1 | 1.0 | 0.0 | 0.1 | 1.0 | 0.0 |
| Xl.50785 | tfap2e | Transcription factor AP-2 epsilon | 0.4 | 1.0 | 0.1 | n/a | n/a | n/a |
| Xl.1269 | p2ry4 | Pyrimidinergic receptor P2Y, G-protein coupled, 4 | 0.4 | 1.0 | 0.4 | 0.6 | 1.0 | 0.5 |
| Xl.17302 | ppm1k | Protein phosphatase, Mg2+/Mn2+ dependent, 1K | 0.4 | 1.0 | 0.1 | 0.4 | 1.0 | 0.2 |
| Xl.47031 | LOC431899 | Hypothetical protein LOC431899 | 0.4 | 1.0 | 0.1 | 0.8 | 1.0 | 0.1 |
| Xl.689 | nprc | Natriuretic peptide receptor C | 0.2 | 1.0 | 0.1 | 0.3 | 1.0 | 0.2 |
| Xl.697 | nr1i2 | Nuclear receptor subfamily 1, group I, member 2 | 0.2 | 1.0 | 0.0 | 1.2 | 1.0 | 0.0 |
| Xl.1405 | LOC100036942 | Hypothetical protein LOC100036942 | 0.3 | 1.0 | 0.0 | 0.1 | 1.0 | 0.0 |
| Xl.25175 | tspan18 | Tetraspanin 18 | 0.0 | 1.0 | 0.2 | 0.1 | 1.0 | 0.6 |
| Xl.54392 | hoxb4 | Homeobox B4 | 0.2 | 1.0 | 0.0 | n/a | n/a | n/a |
| Xl.47735 | hoxb2 | Homeobox B2 | 0.3 | 1.0 | 0.0 | 1.1 | 1.0 | 0.0 |
| Xl.12464 | lmx1b.1 | LIM homeobox transcription factor 1, beta, gene 1 | 0.4 | 1.0 | 0.2 | 0.8 | 1.0 | 0.3 |
| Xl.30849 | LOC495203 | Hypothetical LOC495203 | 0.1 | 1.0 | 0.2 | 0.2 | 1.0 | 0.8 |
| Xl.67432 | trhd | Tumorhead | 0.2 | 1.0 | 0.2 | 2.6 | 1.0 | 2.0 |
| Xl.17336 | rassf10 | Ras association domain-contain protein 10 | 0.1 | 1.0 | 0.3 | 0.3 | 1.0 | 0.3 |
| Xl.46804 | emp3 | Epithelial membrane protein 3 | 0.2 | 1.0 | 0.1 | 0.2 | 1.0 | 0.2 |
| Xl.34931 | itga5 | Integrin alpha 5 subunit | 0.2 | 1.0 | 0.2 | 0.7 | 1.0 | 0.7 |
| Xl.15219 | b4galt4 | Beta 1,4-galactosyltransferase 4 | 0.3 | 1.0 | 0.2 | 0.7 | 1.0 | 0.2 |
| Xl.23480 | pnhd | Pinhead | 0.3 | 1.0 | 0.4 | n/a | n/a | n/a |
| Xl.47587 | c10orf54 | Chromosome 10 open reading frame 54 | 0.2 | 1.0 | 0.1 | n/a | n/a | n/a |
| Xl.52914 | MGC115333 | Hypothetical protein MGC115333 | 0.3 | 1.0 | 0.4 | n/a | n/a | n/a |
| Xl.50785 | MGC131123 | Hypothetical protein MGC131123 | 0.4 | 1.0 | 0.1 | n/a | n/a | n/a |
P, Pax3-GR; Z, Zic1-GR; n/a, not analyzed. For the qPCR the expression level was normalized to the expression of ef1α.
Validation of Pax3-Zic1 targets
The candidate genes with complete CDS were first validated by qPCR on Pax3GR and Zic1GR injected animal caps. Among them we found 7 genes (LOC397753, fhl3, MGC53056, slit2, nrli2, hoxb2, and trhd) showing higher expression levels in the Pax3GR group as compared to Pax3GR+Zic1GR group (Table 3), and therefore were excluded from further study.
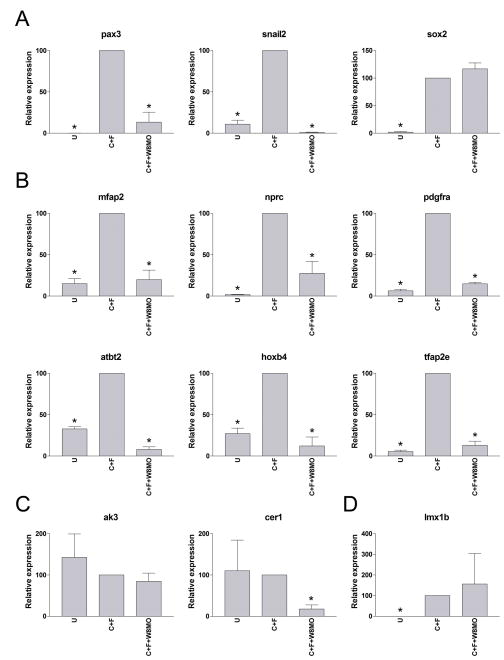
Pax3 and Zic1 are activated at the NPB in response to a specific set of inductive signals: first a Bmp signal, which must be partially attenuated by Bmp antagonists, and then a separate signal mediated by either a canonical Wnt or Fgf (reviewed in Stuhlmiller and Garcia-Castro, 2012). While Fgf8 and Wnt8 are both required to induce the NC, the NC-inducing activity of Fgf8 in Xenopus appears to be largely indirect through the activation of Wnt8 (Hong et al., 2008). In animal cap explants, expression of Chordin (to attenuate Bmp signaling) and Fgf8a induce strong expression of pax3 and snail2 (Fig 3A). However this activity requires active canonical Wnt signaling since co-injection of Wnt8 morpholino antisense oligonucleotide blocks the induction of these genes, without affecting the expression of sox2, a neural plate marker (Fig 3A; Hong et al., 2008). Using this NC-inducing assay we analyzed the expression of a subset of Pax3-Zic1 targets for further validation. We predict that genes activated by Pax3-Zic1 should respond to these inducing signals in a similar manner as snail2. Indeed, we found that a vast majority of these Pax3-Zic1 targets were following the same pattern of gene induction (Fig 3B) suggesting that these factors may have an important role in NC formation downstream of Pax3 and Zic1. However, we also found a few outliers that did not follow this pattern of induction (Fig 3C–D). These targets may represent genes that require additional signals for their maintenance, signals that may not be present in this explant system. Alternatively, these targets may represent genes regulated by Pax3 and Zic1 in other tissues besides the NC - for example, Pax3 and Zic1 have been shown to directly regulate myogenesis (Bajard et al., 2006; Pan et al., 2011)
Figure 3. Regulation of Pax3-Zic1 targets expression by NC-inducing signals.
(A) In animal caps, induction of snail2 and pax3 by co-expression of Chordin (10 pg) and Fgf8a (5 pg) (C+F) is dramatically reduced in the context of embryos injected with Wnt8MO (W8MO; 50 ng). Interference with Wnt signaling pathway did not affect sox2 expression. (B) A majority of Pax3-Zic1 targets followed a pattern of activation in response to NC-inducing signals similar to that of snail2 and pax3, while a limited number of genes did not (C–D). The data are an average of at least three independent experiments. The values were normalized to odc and presented as mean ± s.e.m.; (*), p<0.05 versus C+F.
Developmental expression pattern of selected Pax3-Zic1 targets
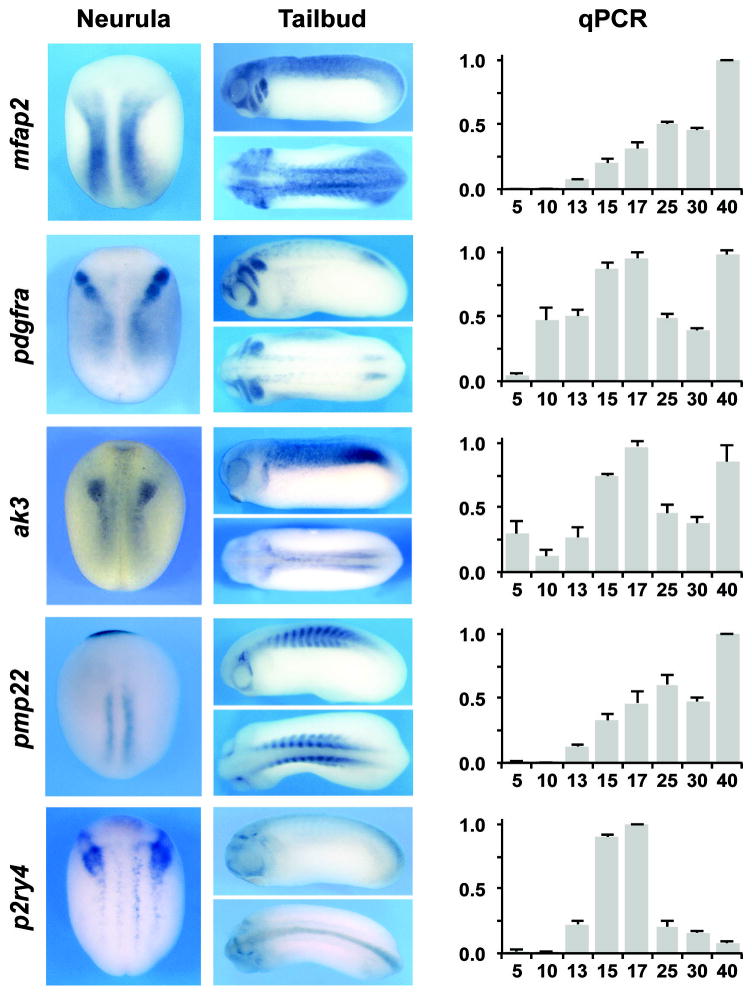
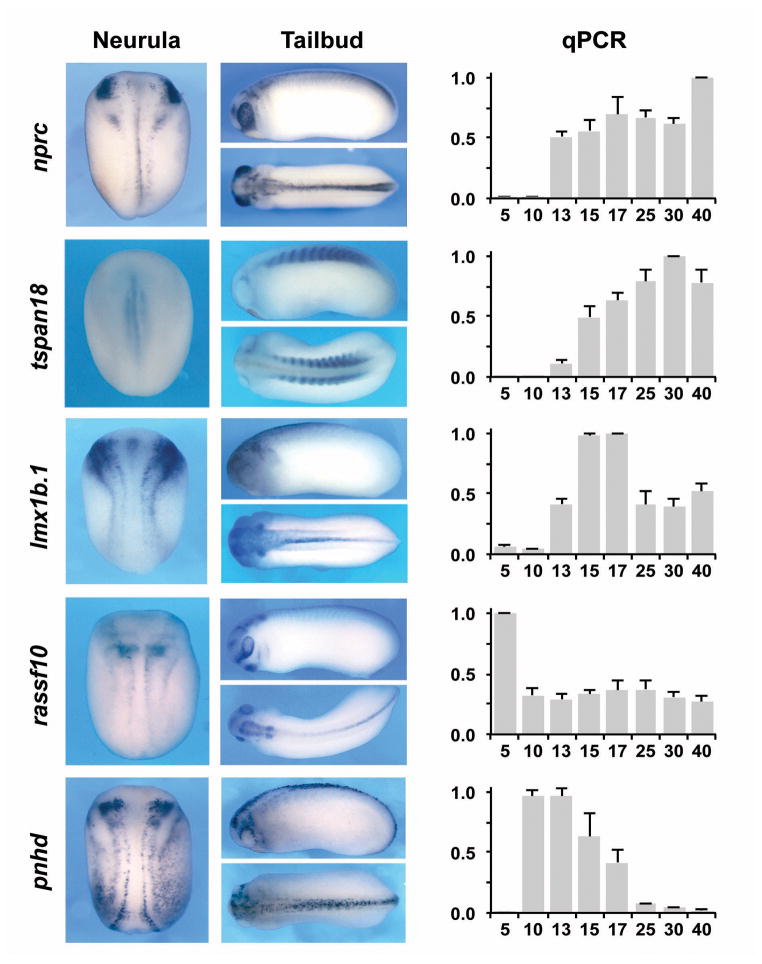
As a proof of principle we started to analyze the developmental expression of ten of these targets at various stages by qPCR using specific primer sets (Table 4). In addition we performed whole mount in situ hybridization (ISH) at neurula (stage 15) and early tailbud (stage 25) stages to define the spatial expression of these genes at stages relevant to NC development. The results are described below and presented in Fig 4.
Figure 4. Developmental expression pattern of selected Pax3-Zic1 targets.
The Spatiotemporal expression pattern of 10 Pax3-Zic1 targets was analyzed by qPCR and whole-mount ISH. Expression levels of the target genes were analyzed by qPCR at the indicated stage (right panels). The values were normalized to odc and presented as mean ± s.d. The data are an average of at least three sets of embryos. For ISH embryos were analyzed at the neurula (left panels; dorsal view, anterior to top) and tailbud (middle panels; lateral and dorsal views, anterior to the left) stages.
mfap2, encodes microfibrillar-associated protein 2, a major antigen of elastin-associated microfibrils and a candidate gene for the etiology of inherited connective tissue diseases. It is a regulator of bone remodeling and its absence has been associated with osteopenia (Weinbaum et al., 2008; Craft et al., 2012). The function of Mfap2 during embryonic development has not been studied. In Xenopus embryos, mfap2 was activated around stage 13 and increased progressively up to stage 25. Its expression was maintained at relatively high level up to stage 40. By ISH, mfap2 was highly expressed in a region that corresponds to the prospective trunk NC at neurula stage, and was later confined to the branchial arches, the central nervous system (CNS) and the somites.
pdgfr, encodes the platelet-derived growth factor receptor alpha, a tyrosine-protein kinase that acts as a receptor for Pgdfa, Pgdfb and Pgdfc ligands. In mouse conditional inactivation of Pgdfra in the NC resulted in neonatal lethality due to aortic arch defects and cleft palate (Tallquist and Soriano, 2003), indicative of an important function of this signaling pathway in the cardiac and cranial NC. In Xenopus pdgfra was activated at stage 10 and was maintained at relatively constant level throughout development. By ISH, pdgfra was highly expressed in cranial NC progenitors at neurula stage and in the branchial arches and the presomitic mesoderm at the tailbud stage, as previously reported (Ho et al., 1994). pdfgra expression has been reported in Xenopus mesoderm and its disruption resulted in randomized migration of mesodermal cells, suggesting a role for Pdgfra in the guided migration of these cells (Nagel et al., 2004). The specific function of pdgfra in the NC has not been studied in frogs.
ak3, encodes adenylate kinase 3, a mitochondrial GTP:AMP phosphotransferase, specific for the phosphorylation of AMP using either GTP or ITP as a substrate (Noma et al., 2001). In rat, Ak3 is expressed throughout embryogenesis (Tanabe et al., 1993) however its role during development has not been studied. In Xenopus ak3 appeared to be maternally expressed, with a peak of expression at stage 17, and was maintained up to stage 40. By ISH, ak3 was detected in the most posterior domain of the cranial NC, as well as in the trunk region. This expression in the NC region was confirmed by histology (not shown). At tailbud stage, ak3 expression was more diffuse and restricted dorsally.
pmp22, encodes peripheral myelin protein 22, a component of myelin in the adult peripheral nervous system (PNS). In zebrafish, pmp22 is expressed in the NC and the sclerotome and has been implicated in early PNS development (Wulf et al., 1999), although its precise function remains unknown. In human, micro-mutations of PMP22 have been associated with Charcot-Marie-Tooth disease, a condition affecting peripheral nerves (Koutsis et al., 2012). In Xenopus, pmp22 is activated at stage 13 and increases throughout development. Spatially, at stage 15, pmp22 was detected in the mid trunk region in the somitic mesoderm, as well as in the prospective cement gland. At tailbud stage, this gene was strongly expressed in the somite and trigeminal ganglion.
p2ry4, encodes the pyrimidinergic receptor P2Y, G-protein coupled 4 receptor, a class of receptors responsive to uridine nucleotides (UDP, UTP and UMP) and partially responsive to ATP. In the adult, p2ry4 mRNA have been detected in human placenta (Communi et al., 1995) and in murine stomach, intestine, and liver (Suarez-Huerta et al., 2001). P2ry4 function in development has not been studied. In Xenopus p2ry4 was strongly activated at stage 13 to reach a maximum expression at stage 17. In subsequent stages its expression rapidly declined over time. By ISH, p2ry4 was specifically expressed in the NC progenitors at the NPB at the early neurula stage. Later its expression was more diffuse and restricted to discrete domains of the CNS as well as in the first branchial arch.
nprc, encodes the natriuretic peptide receptor C. Natriuretic peptides are highly conserved hormones regulating body fluid balance and blood pressure (Lelievre et al., 2006; Potter et al., 2006). Nprc has been proposed to act mainly as a clearance receptor, however there is also evidence that Nprc may activate intracellular signaling pathways. Osteocrin (also known as musclin) produced by bone forming cells and skeletal muscle has been recently identified as a specific ligand for Nprc (Moffatt and Thomas, 2009). Chemically induced mutant mice and targeted mutant mice of the nprc gene displayed skeletal overgrowth (Jaubert et al., 1999; Matsukawa et al., 1999). In Xenopus nprc is activated at the neurula stage (stage 13) and was maintained at relatively high levels up to stage 40. Spatially nprc is restricted to the most anterior domain of the cranial NC, the mandibular NC, and was more weakly expressed in the posterior region of the NPB. At the tailbud stage, its expression domain included the developing eyes, the first branchial arch and the spinal cord.
tspan18, encodes tetraspanin 18, a member of a large family of transmembrane proteins that have been implicated in fundamental biological processes, including cell adhesion, motility and proliferation (Charrin et al., 2009). The specific function of Tspan18 in development has not been studied. One member of this family, Tspan1, has been shown to regulate gastrulation movement and primary neurogenesis in Xenopus (Yamamoto et al., 2007). Another member, tspan24, expressed in the trigeminal placode was required to maintain trigeminal placode progenitors in chick embryos (McCabe and Bronner, 2011). Human TSPAN18 was identified as a susceptibility locus for schizophrenia in Han Chinese by genome wide association (Yue et al., 2011). In Xenopus tspan18 was strongly activated at stage 13 and its expression was maintained at a relatively high level up to stage 40. By ISH, at the neurula stage, tspan18 was detected in the mid-trunk region in the presomitic mesoderm, and at the tailbud stage was confined to the developing somites.
lmx1b.1, encodes a LIM homeodomain transcription factor. Lmx1b is essential for dorso-ventral limb patterning and the specification of serotonergic and dopaminergic progenitors in the brain (Li et al., 2010; Demarque and Spitzer, 2010; Yan el al., 2011). Human mutations in the LMX1B gene have been associated with Nail-patella syndrome, a genetic disorder that results in poorly developed nails and kneecaps, but can also affect many other areas of the body (McIntosh et al., 1998). In Xenopus, lmx1b.1 was activated at stage 13, and this expression was substantially increased by stage 15. By ISH, lmx1b.1 was broadly expressed at the NPB including the NC forming region at the neurula stage. At the tailbud stage the expression was more diffuse, with strong expression in the otic vesicle. A previous study also reported lmx1b.1 expression in the cranial NC, neural plate, placodal region and the pronephros (Haldin et al., 2003). The function of lmx1b.1 in NC progenitors has been not analyzed.
rassf10, encodes the Ras association domain protein 10. The Ras association domain family is a group of tumor suppressors that are frequently epigenetically inactivated in various tumors. They are linked to apoptosis, cell cycle control and microtubule stability (Damman et al., 2000). In Xenopus, rassf10 is expressed maternally, and was maintained at all stages examined. By ISH, rassf10 was spatially restricted to the NPB and the neural plate at neurula stage. At stage 25, rassf10 was detected in the brain, posterior spinal cord, cement gland, branchial arches and the eye, consistent with a recent report (Hill et al., 2011). Nothing is known about the biological properties of this protein during embryogenesis.
pnhd, encodes pinhead, a protein with no obvious orthologs out-side frogs, fish, and the invertebrate Ciona intestinalis (Kenwrick et al., 2004). It was first discovered in Xenopus tropicalis in a functional screen for genes involved in neural development. It is essential for head development as Pnhd knockdown induced severe microcephaly, whereas pnhd overexpression resulted in enlarged heads (Kenwrick et al., 2004). pnhd expression was activated at stage 10 and then decreased gradually, to be essentially undetectable at stage 40. By ISH, at the neurula stage pnhd is expressed in the anterior neural plate into 2 discrete domains medial to the NC, as well as in the epidermis outlining the NC forming region (Kenwrick et al., 2004). At the tailbud stage pnhd was detected in the trigeminal ganglion and in the dorsal spinal cord.
Among these ten targets, eight had specific expression in NC progenitors suggesting an important function in the NC lineage, while two of them, pmp22 and tspan18, were confined to the mesoderm and developing somites. The recovery of genes expressed in the mesoderm is not completely unexpected since a synergistic interaction between Pax3 and Zic1 has been recently described in the control of myogenesis (Himeda et al., 2013).
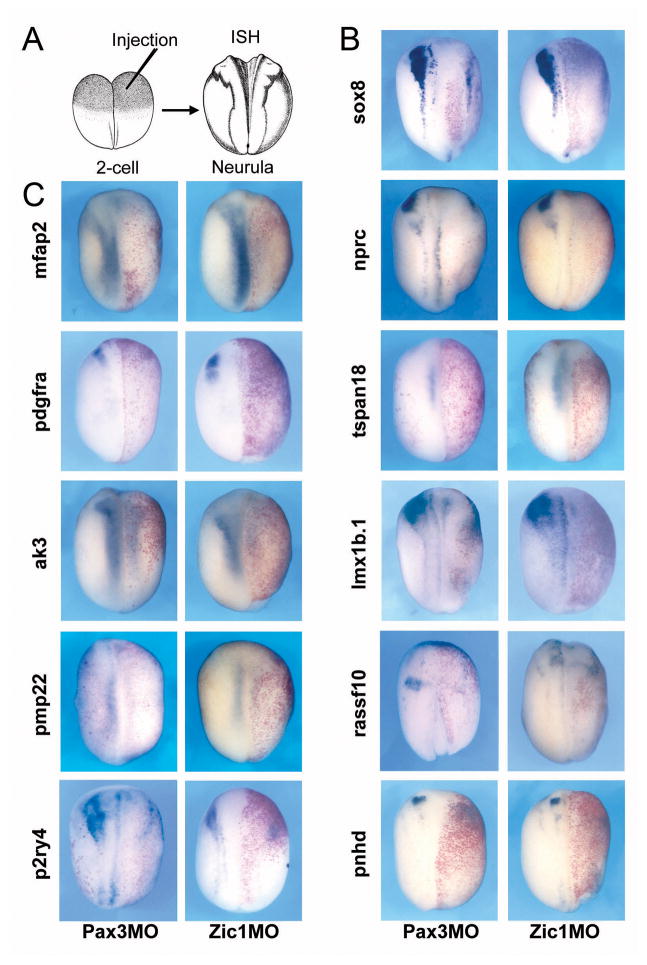
The expression of Pax3-Zic1 targets require Pax3 and Zic1 function
The expression of bona fide downstream targets of Pax3 and Zic1 is expected to depend on Pax3 and Zic1 function. To test this we blocked Pax3 or Zic1 function by injection at the 2-cell stage of morpholino antisense oligonucleotides (Fig 5A–B; Monsoro-Burq et al., 2005; Sato et al., 2005; Hong and Saint-Jeannet, 2007). For all ten Pax3-Zic1 target genes analyzed in this study we observed a loss of expression (Fig. 5C). This result further establishes the position of these targets downstream of Pax3 and Zic1 in the gene regulatory cascade leading to NC formation.
Figure 5. Pax3 or Zic1 morpholino mediated knockdown blocked Pax3-Zic1 targets expression at the neurula stage.
(A) Pax3MO (40 ng) or Zic1MO (40 ng) were injected in one blastomere at the 2-cell stage embryos. Embryos were collected at the neurula stage (stage 15–18) and analyzed by ISH (upper left). (B) ISH with a sox8 probe was used as a positive control for the activity of each morpholino. (C) The 10 Pax3-Zic1 targets analyzed showed reduced expression upon Pax3 or Zic1 knockdown. All panels are showing dorsal views, anterior to top. The injected side is on the right as indicted by the presence of the lineage tracer (red staining).
Discussion
The characterization of the regulatory inputs controlling NC formation is critical to understand how these processes may be altered in pathological situations. Because Pax3 and Zic1 have been defined as important upstream regulators of NC fate (Monsoro-Burq et al., 2005; Sato et al., 2005, Hong and Saint-Jeannet, 2007; Garnett et al., 2012), we predicted that analysis of Pax3-Zic1-induced genes will lead to the identification of novel factors critically required for NC formation, that will expand our understanding of the gene regulatory network underlying NC formation. Therefore the fundamental knowledge resulting from these studies is expected to provide important insights into NC pathologies arising from the defective regulation of various components of this regulatory cascade.
We have used hormone-inducible fusion proteins to simultaneously express Pax3 and Zic1 in Xenopus animal caps to convert this tissue to NC (Hong and Saint-Jeannet, 2007), and used this sample to screen a DNA microarray. Among the genes isolated we found several well-characterized NC-specific genes including snail2, foxd3, gbx2, sox9, sox8, twist, zic5 and ednra, which is an important validation of our approach (Table 2). We have also identified several genes that have not been previously implicated in NC formation. They are potential regulators of gene transcription, signal transduction and cell signaling, as well as genes with yet uncharacterized function. The preliminary validation of a subset of these targets, confirmed their position in the NC gene regulatory network downstream of Pax3 and Zic1. We propose that these factors are integral parts of the NC molecular signature, and represent novel regulators of NC fate.
An important task ahead is to evaluate the functional significance of these targets and to define their position in the NC gene regulatory network. This will be addressed using gain-of-function and loss-of-function approaches in the context of the whole embryo and in animal cap explants. Furthermore, the epistatic relationship between these targets and known NC-specific genes can be easily tested in rescue assays. These perturbation studies will be the primary basis to establish functional linkages between these regulatory genes. Further validation of these interactions will be needed to separate direct from indirect targets through the identification and characterization of cis-regulatory sequences using Chromatin Immunoprecipitation (ChIP) and ChIP-Sequence.
Our screen was designed to select for genes that are synergistically upregulated by Pax3 and Zic1 as compared to each factor individually, therefore we expect to recover target genes that are co-regulated by Pax3 and Zic1. It is still unclear whether the cooperative ability of Pax3 and Zic1 to activate these target genes is mediated by binding directly and independently to the regulatory regions of these putative target genes, or whether Zic1 and Pax3 interact with one another to regulate gene expression. It has been shown that Sox10, in synergy with Pax3, strongly activates Mitf expression by direct binding to a proximal region of the Mitf promoter that contains binding sites for both factors (Bondurand et al., 2000). In the context of another known target of Pax3, the cRet gene, Pax3 and Sox10 have been shown to physically interact and this interaction is believed to contribute to Pax3 and Sox10 synergistic activation, as Sox10 mutants that cannot bind to DNA retain the ability to activate cRet enhancer in the presence of Pax3 (Lang and Epstein, 2003). Therefore, multiple scenarios can be expected with regard to the molecular mechanisms by which Pax3 and Zic1 may regulate these target genes. Identification and characterization of the regulatory sequence of the various targets should provide important information on the type of interaction Pax3 and Zic1 establish with these targets, and the mechanism by which they regulate target gene expression.
In conclusion, we used a microarray-based approach to identify Pax3 and Zic1 targets during NC development. We found that Pax3 and Zic1 activate the expression of genes with a broad range of functions, from transcription to cell signaling. The preliminary characterization of a number of these targets indicates that we have identified several novel genes expressed in NC progenitors that are potentially important to regulate NC formation. This study provides a valuable resource to further build and investigate the NC gene regulatory network.
Highlights.
Pax3 and Zic1 are necessary and sufficient to promote neural crest (NC) fate
Using DNA microarray we identify downstream targets of Pax3 and Zic1
We obtained several novel factors expressed in NC progenitors
These targets were validated by qPCR and morpholino-mediated knockdown
These factors represent novel players in the NC gene regulatory network
Acknowledgments
We thank Dr. Jane McCutcheon for comments on the manuscript. This work was supported by a grant from the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology to C-S H (2010-0025108), and a grant from the National Institutes of Health to J-P S-J (RO1-DE014212).
Grant Sponsor: National Institutes of Heath; R01DE014212
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bajard L, Relaix F, Lagha M, Rocancourt D, Daubas P, Buckingham ME. A novel genetic hierarchy functions during hypaxial myogenesis: Pax3 directly activates Myf5 in muscle progenitor cells in the limb. Genes Dev. 2006;20:2450–2464. doi: 10.1101/gad.382806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolande RP. Neurocristopathy: its growth and development in 20 years. Pediatr Pathol Lab Med. 1997;17(1):1–25. [PubMed] [Google Scholar]
- Bonano M, Tríbulo C, De Calisto J, Marchant L, Sánchez SS, Mayor R, Aybar MJ. A new role for the Endothelin-1/Endothelin-A receptor signaling during early neural crest specification. Dev Biol. 2008;323(1):114–29. doi: 10.1016/j.ydbio.2008.08.007. [DOI] [PubMed] [Google Scholar]
- Bondurand N, Pingault V, Goerich DE, Lemort N, Sock E, Le Caignec C, Wegner M, Goossens M. Interaction among SOX10, PAX3 and MITF, three genes altered in Waardenburg syndrome. Hum Mol Genet. 2000;9(13):1907–17. doi: 10.1093/hmg/9.13.1907. [DOI] [PubMed] [Google Scholar]
- Bronchain OJ, Pollet N, Ymlahi-Ouazzani Q, Dhorne-Pollet S, Helbling JC, Lecarpentier JE, Percheron K, Wegnez M. The olig family: phylogenetic analysis and early gene expression in Xenopus tropicalis. Dev Genes Evol. 2007;217(7):485–97. doi: 10.1007/s00427-007-0158-z. [DOI] [PubMed] [Google Scholar]
- Charrin S, le Naour F, Silvie O, Milhiet PE, Boucheix C, Rubinstein E. Lateral organization of membrane proteins: tetraspanins spin their web. Biochem J. 2009;420(2):133–54. doi: 10.1042/BJ20082422. [DOI] [PubMed] [Google Scholar]
- Christen B, Slack JMW. FGF-8 is associated with anteroposterior patterning and limb regeneration in Xenopus. Dev Biol. 1997;192:455–66. doi: 10.1006/dbio.1997.8732. [DOI] [PubMed] [Google Scholar]
- Craft CS, Broekelmann TJ, Zou W, Chappel JC, Teitelbaum SL, Mecham RP. Oophorectomy-induced bone loss is attenuated in MAGP1-deficient mice. J Cell Biochem. 2012;113(1):93–9. doi: 10.1002/jcb.23331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Communi D, Pirotton S, Parmentier M, Boeynaems JM. Cloning and functional expression of a human uridine nucleotide receptor. J Biol Chem. 1995;270(52):30849–52. doi: 10.1074/jbc.270.52.30849. [DOI] [PubMed] [Google Scholar]
- Dammann R, Li C, Yoon JH, Chin PL, Bates S, Pfeifer GP. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat Genet. 2000;25(3):315–9. doi: 10.1038/77083. [DOI] [PubMed] [Google Scholar]
- Demarque M, Spitzer NC. Activity-dependent expression of Lmx1b regulates specification of serotonergic neurons modulating swimming behavior. Neuron. 2010;67(2):321–34. doi: 10.1016/j.neuron.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnett AT, Square TA, Medeiros DM. BMP, Wnt and FGF signals are integrated through evolutionarily conserved enhancers to achieve robust expression of Pax3 and Zic genes at the zebrafish neural plate border. Development. 2012 doi: 10.1242/dev.081497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldin CE, Nijjar S, Masse K, Barnett MW, Jones EA. Isolation and growth factor inducibility of the Xenopus laevis Lmx1b gene. Int J Dev Biol. 2003;47(4):253–62. [PubMed] [Google Scholar]
- Harland RM. In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods Cell Biol. 1991;36:685–95. doi: 10.1016/s0091-679x(08)60307-6. [DOI] [PubMed] [Google Scholar]
- Hill VK, Underhill-Day N, Krex D, Robel K, Sangan CB, Summersgill HR, Morris M, Gentle D, Chalmers AD, Maher ER, et al. Epigenetic inactivation of the RASSF10 candidate tumor suppressor gene is a frequent and an early event in gliomagenesis. Oncogene. 2011;30(8):978–89. doi: 10.1038/onc.2010.471. [DOI] [PubMed] [Google Scholar]
- Himeda CL, Barro MV, Emerson CP., Jr Pax3 synergizes with Gli2 and Zic1 in transactivating the Myf5 epaxial somite enhancer’. Dev Biol. 2013 doi: 10.1016/j.ydbio.2013.09.006. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Symes K, Yordan C, Gudas LJ, Mercola M. Localization of PDGF A and PDGFR alpha mRNA in Xenopus embryos suggests signalling from neural ectoderm and pharyngeal endoderm to neural crest cells. Mech Dev. 1994;48(3):165–74. doi: 10.1016/0925-4773(94)90057-4. [DOI] [PubMed] [Google Scholar]
- Hong CS, Park BY, Saint-Jeannet JP. Fgf8a induces neural crest indirectly through the activation of Wnt8 in the paraxial mesoderm. Development. 2008;135(23):3903–10. doi: 10.1242/dev.026229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong CS, Saint-Jeannet JP. The activity of Pax3 and Zic1 regulates three distinct cell fates at the neural plate border. Mol Biol Cell. 2007;18(6):2192–202. doi: 10.1091/mbc.E06-11-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaubert J, Jaubert F, Martin N, Washburn LL, Lee BK, Eicher EM, Guenet JL. Three new allelic mouse mutations that cause skeletal overgrowth involve the natriuretic peptide receptor C gene (Npr3) Proc Natl Acad Sci U S A. 1999;96(18):10278–83. doi: 10.1073/pnas.96.18.10278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenwrick S, Amaya E, Papalopulu N. Pilot morpholino screen in Xenopus tropicalis identifies a novel gene involved in head development. Dev Dyn. 2004;229(2):289–99. doi: 10.1002/dvdy.10440. [DOI] [PubMed] [Google Scholar]
- Klingel S, Morath I, Strietz J, Menzel K, Holstein TW, Gradl D. Subfunctionalization and neofunctionalization of vertebrate Lef/Tcf transcription factors. Dev Biol. 2012;368(1):44–53. doi: 10.1016/j.ydbio.2012.05.012. [DOI] [PubMed] [Google Scholar]
- Kolm PJ, Sive HL. Efficient hormone-inducible protein function in Xenopus laevis. Dev Biol. 1995;171(1):267–72. doi: 10.1006/dbio.1995.1279. [DOI] [PubMed] [Google Scholar]
- Koutsis G, Pandraud A, Polke JM, Wood NW, Panas M, Karadima G, Houlden H. Novel peripheral myelin protein 22 (PMP22) micromutations associated with variable phenotypes in Greek patients with Charcot-Marie-Tooth disease. Brain. 2012;135(Pt 8):e217, 1–6. doi: 10.1093/brain/aws034. author reply e218, 1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang D, Epstein JA. Sox10 and Pax3 physically interact to mediate activation of a conserved c-RET enhancer. Hum Mol Genet. 2003;12(8):937–45. doi: 10.1093/hmg/ddg107. [DOI] [PubMed] [Google Scholar]
- Le Douarin NM, Creuzet S, Couly G, Dupin E. Neural crest cell plasticity and its limits. Development. 2004;131(19):4637–50. doi: 10.1242/dev.01350. [DOI] [PubMed] [Google Scholar]
- Lelievre V, Hu Z, Ioffe Y, Byun JY, Flores A, Seksenyan A, Waschek JA. Paradoxical antagonism of PACAP receptor signaling by VIP in Xenopus oocytes via the type-C natriuretic peptide receptor. Cell Signal. 2006;18(11):2013–21. doi: 10.1016/j.cellsig.2006.03.013. [DOI] [PubMed] [Google Scholar]
- Li B, Kuriyama S, Moreno M, Mayor R. The posteriorizing gene Gbx2 is a direct target of Wnt signaling and the eraliest factor in neural crest induction. Development. 2009;136:3267–78. doi: 10.1242/dev.036954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Qiu Q, Watson SS, Schweitzer R, Johnson RL. Uncoupling skeletal and connective tissue patterning: conditional deletion in cartilage progenitors reveals cell-autonomous requirements for Lmx1b in dorsal-ventral limb patterning. Development. 2010;137(7):1181–8. doi: 10.1242/dev.045237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linker C, Bronner-Fraser M, Mayor R. Relationship between gene expression domains of Xsnail, Xslug, and Xtwist and cell movement in the prospective neural crest of Xenopus. Dev Biol. 2000;224(2):215–25. doi: 10.1006/dbio.2000.9723. [DOI] [PubMed] [Google Scholar]
- Mancilla A, Mayor R. Neural crest formation in Xenopus laevis: mechanisms of Xslug induction. Dev Biol. 1996;177(2):580–9. doi: 10.1006/dbio.1996.0187. [DOI] [PubMed] [Google Scholar]
- Matsukawa N, Grzesik WJ, Takahashi N, Pandey KN, Pang S, Yamauchi M, Smithies O. The natriuretic peptide clearance receptor locally modulates the physiological effects of the natriuretic peptide system. Proc Natl Acad Sci U S A. 1999;96(13):7403–8. doi: 10.1073/pnas.96.13.7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe KL, Bronner M. Tetraspanin, CD151, is required for maintenance of trigeminal placode identity. J Neurochem. 2011;117(2):221–30. doi: 10.1111/j.1471-4159.2011.07190.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh I, Dreyer SD, Clough MV, Dunston JA, Eyaid W, Roig CM, Montgomery T, Ala-Mello S, Kaitila I, Winterpacht A, et al. Mutation analysis of LMX1B gene in nail-patella syndrome patients. Am J Hum Genet. 1998;63(6):1651–8. doi: 10.1086/302165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelsen JW, Schmeichel KL, Beckerle MC, Winge DR. The LIM motif defines a specific zinc-binding protein domain. Proc Natl Acad Sci U S A. 1993;90(10):4404–8. doi: 10.1073/pnas.90.10.4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt P, Thomas GP. Osteocrin--beyond just another bone protein? Cell Mol Life Sci. 2009;66(7):1135–9. doi: 10.1007/s00018-009-8716-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsoro-Burq AH, Wang E, Harland R. Msx1 and Pax3 cooperate to mediate FGF8 and WNT signals during Xenopus neural crest induction. Dev Cell. 2005;8(2):167–78. doi: 10.1016/j.devcel.2004.12.017. [DOI] [PubMed] [Google Scholar]
- Nagel M, Tahinci E, Symes K, Winklbauer R. Guidance of mesoderm cell migration in the Xenopus gastrula requires PDGF signaling. Development. 2004;131(11):2727–36. doi: 10.1242/dev.01141. [DOI] [PubMed] [Google Scholar]
- Nakata K, Koyabu Y, Aruga J, Mikoshiba K. A novel member of the Xenopus Zic family, Zic5, mediates neural crest development. Mech Dev. 2000;99(1–2):83–91. doi: 10.1016/s0925-4773(00)00480-9. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. Normal table of Xenopus laevis (Daudin) North Holland Publishing Company; 1967. [Google Scholar]
- Noma T, Fujisawa K, Yamashiro Y, Shinohara M, Nakazawa A, Gondo T, Ishihara T, Yoshinobu K. Structure and expression of human mitochondrial adenylate kinase targeted to the mitochondrial matrix. Biochem J. 2001;358(Pt 1):225–32. doi: 10.1042/0264-6021:3580225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell M, Hong CS, Huang X, Delnicki RJ, Saint-Jeannet JP. Functional analysis of Sox8 during neural crest development in Xenopus. Development. 2006;133(19):3817–26. doi: 10.1242/dev.02558. [DOI] [PubMed] [Google Scholar]
- Pan H, Gustafsson MK, Aruga J, Tiedken JJ, Chen JC, Emerson CP., Jr A role for Zic1 and Zic2 in Myf5 regulation and somite myogenesis. Dev Biol. 2011;351:120–127. doi: 10.1016/j.ydbio.2010.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl BS, Knöchel W. Overexpression of the transcriptional repressor FoxD3 prevents neural crest formation in Xenopus embryos. Mech Dev. 2001;103(1–2):93–106. doi: 10.1016/s0925-4773(01)00334-3. [DOI] [PubMed] [Google Scholar]
- Potter LR, Abbey-Hosch S, Dickey DM. Natriuretic peptides, their receptors, and cyclic guanosine monophosphate-dependent signaling functions. Endocr Rev. 2006;27(1):47–72. doi: 10.1210/er.2005-0014. [DOI] [PubMed] [Google Scholar]
- Sato T, Sasai N, Sasai Y. Neural crest determination by co-activation of Pax3 and Zic1 genes in Xenopus ectoderm. Development. 2005;132(10):2355–63. doi: 10.1242/dev.01823. [DOI] [PubMed] [Google Scholar]
- Spokony RF, Aoki Y, Saint-Germain N, Magner-Fink E, Saint-Jeannet JP. The transcription factor Sox9 is required for cranial neural crest development in Xenopus. Development. 2002;129(2):421–32. doi: 10.1242/dev.129.2.421. [DOI] [PubMed] [Google Scholar]
- Stuhlmiller TJ, Garcia-Castro MI. Current perspectives of the signaling pathways directing neural crest induction. Cell Mol Life Sci. 2012;69(22):3715–37. doi: 10.1007/s00018-012-0991-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez-Huerta N, Pouillon V, Boeynaems J, Robaye B. Molecular cloning and characterization of the mouse P2Y4 nucleotide receptor. Eur J Pharmacol. 2001;416(3):197–202. doi: 10.1016/s0014-2999(01)00875-5. [DOI] [PubMed] [Google Scholar]
- Tallquist MD, Soriano P. Cell autonomous requirement for PDGFRalpha in populations of cranial and cardiac neural crest cells. Development. 2003;130(3):507–18. doi: 10.1242/dev.00241. [DOI] [PubMed] [Google Scholar]
- Tanabe T, Yamada M, Noma T, Kajii T, Nakazawa A. Tissue-specific and developmentally regulated expression of the genes encoding adenylate kinase isozymes. J Biochem. 1993;113(2):200–7. doi: 10.1093/oxfordjournals.jbchem.a124026. [DOI] [PubMed] [Google Scholar]
- Weinbaum JS, Broekelmann TJ, Pierce RA, Werneck CC, Segade F, Craft CS, Knutsen RH, Mecham RP. Deficiency in microfibril-associated glycoprotein-1 leads to complex phenotypes in multiple organ systems. J Biol Chem. 2008;283(37):25533–43. doi: 10.1074/jbc.M709962200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulf P, Bernhardt RR, Suter U. Characterization of peripheral myelin protein 22 in zebrafish (zPMP22) suggests an early role in the development of the peripheral nervous system. J Neurosci Res. 1999;57(4):467–78. [PubMed] [Google Scholar]
- Yamamoto Y, Grubisic K, Oelgeschlager M. Xenopus Tetraspanin-1 regulates gastrulation movements and neural differentiation in the early Xenopus embryo. Differentiation. 2007;75(3):235–45. doi: 10.1111/j.1432-0436.2006.00134.x. [DOI] [PubMed] [Google Scholar]
- Yan CH, Levesque M, Claxton S, Johnson RL, Ang SL. Lmx1a and lmx1b function cooperatively to regulate proliferation, specification, and differentiation of midbrain dopaminergic progenitors. Neurosci. 2011;31(35):12413–25. doi: 10.1523/JNEUROSCI.1077-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue WH, Wang HF, Sun LD, Tang FL, Liu ZH, Zhang HX, Li WQ, Zhang YL, Zhang Y, Ma CC, et al. Genome-wide association study identifies a susceptibility locus for schizophrenia in Han Chinese at 11p11.2. Nat Genet. 2011;43(12):1228–31. doi: 10.1038/ng.979. [DOI] [PubMed] [Google Scholar]