Abstract
Previous investigations of the core gene regulatory circuitry that controls embryonic stem cell (ESC) pluripotency have largely focused on the roles of transcription, chromatin and non-coding RNA regulators1–3. Alternative splicing (AS) represents a widely acting mode of gene regulation4–8, yet its role in regulating ESC pluripotency and differentiation is poorly understood. Here, we identify the Muscleblind-like RNA binding proteins, MBNL1 and MBNL2, as conserved and direct negative regulators of a large program of cassette exon AS events that are differentially regulated between ESCs and other cell types. Knockdown of MBNL proteins in differentiated cells causes switching to an ESC-like AS pattern for approximately half of these events, whereas over-expression of MBNL proteins in ESCs promotes differentiated cell-like AS patterns. Among the MBNL-regulated events is an ESC-specific AS switch in the forkhead family transcription factor FOXP1 that controls pluripotency9. Consistent with a central and negative regulatory role for MBNL proteins in pluripotency, their knockdown significantly enhances the expression of key pluripotency genes and the formation of induced pluripotent stem cells (iPSCs) during somatic cell reprogramming.
A core set of transcription factors that includes OCT4, NANOG, and SOX2, together with specific microRNAs and long non-coding RNAs, controls the expression of genes required for the establishment and maintenance of ESC pluripotency1–3,10–12. Alternative splicing (AS), the process by which splice sites in primary transcripts are differentially selected to produce structurally and functionally distinct mRNA and protein isoforms, provides a powerful additional mechanism with which to control cell fate7,8,13, yet its role in the regulation of pluripotency has only recently begun to emerge. In particular, the inclusion of a highly conserved ESC-specific “switch” exon in the FOXP1 transcription factor changes its DNA binding specificity such that it stimulates the expression of pluripotency transcription factors, including OCT4 and NANOG, while repressing genes required for differentiation9. However, the trans-acting regulators of this and other AS events14–16 implicated in ESC biology are not known. These factors are important to identify, as they may control regulatory cascades that direct cell fate, and likewise they may also control the efficiency and kinetics of somatic cell reprogramming.
To identify such factors, we used high-throughput RNA sequencing (RNA-Seq) data to define human and mouse cassette alternative exons that are differentially spliced between ESCs/iPSCs and diverse differentiated cells and tissues, referred to below as “ESC-differential AS”. A splicing code analysis17 was then performed to identify cis-elements that may promote or repress these exons. The RNA-Seq data used to profile AS were also used to detect human and mouse splicing factor genes that are differentially expressed between ESCs/iPSCs and non-ESCs/tissues. By integrating these data sources, we sought to identify differentially expressed splicing regulators with defined binding sites that match cis-elements predicted by the code analysis to function in ESC-differential AS.
We identified 181 human and 103 mouse ESC-differential AS events, with comparable proportions of exons that are ≥25% more included or more skipped in ESCs versus the other profiled cells and tissues (Fig. 1a, Supplementary Figs. 1a, 2 and Supplementary Tables 1, 2). When comparing orthologous exons in both species, 25 of the human and mouse ESC-differential AS events overlapped (p<2.2e−16; hypergeometric test). The human and mouse ESC-differential AS events are significantly enriched in genes associated with the cytoskeleton (e.g. DST, ADD3), plasma membrane (e.g. DNM2, ITGA6) and kinase activity (e.g. CASK, MARK2 and MAP2K7) (Supplementary Table 3). They also include the aforementioned FOXP1 ESC-switch AS event, and previously unknown AS events in other transcription or chromatin regulatory factors (e.g. TEAD1 and MTA1) that have been implicated in controlling pluripotency18,19. These results suggest a considerably more extensive role for regulated AS in ESC biology than previously appreciated.
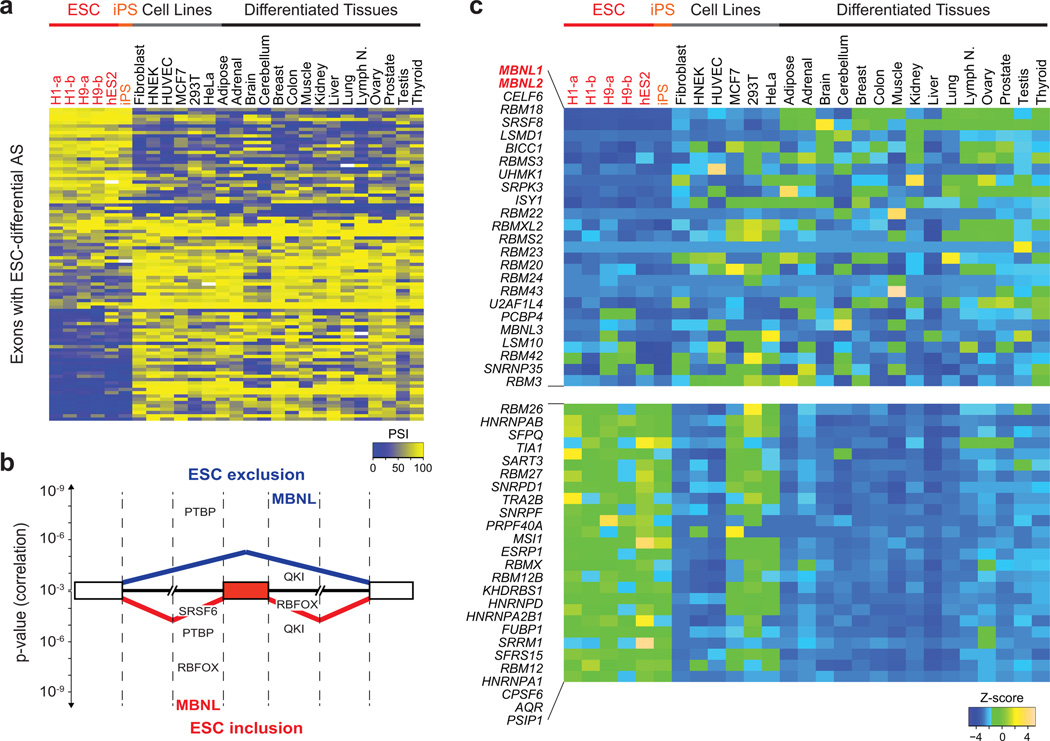
Figure 1. Identification of regulators of ESC-differential alternative splicing.
a) Heatmap of percent spliced in (PSI) values for 95 representative ESC-differential AS events in transcripts that are widely expressed across human ESCs/iPSCs, non-ESC lines and differentiated tissues. b) Splicing code features that are significantly associated with ESC-differential AS. Features are ranked according to Pearson correlation p-values (y-axis) for alternative exons with either lower (top) or higher (bottom) inclusion in ESCs/iPSCs. Dashed lines indicate 300 nucleotide intervals from splice sites. c) Heatmap of Z-scores of mRNA expression (cRPKM) levels for splicing factors. Twenty-five splicing factors with the lowest or highest relative mRNA expression levels in ESCs/iPSCs compared to non-ESCs/tissues are shown. cRPKM, corrected reads per kilobase exon model per million reads30.
The splicing code analysis revealed that motifs corresponding to consensus binding sites of the conserved Muscleblind-like (MBNL) proteins are the most strongly associated with ESC-differential AS in human and mouse. The presence of MBNL motifs in downstream flanking intronic sequences is associated with exon skipping in ESCs, whereas their presence in upstream flanking intronic sequences is associated with exon inclusion in ESCs (Fig 1b, human code; Supplementary Fig. 1b, mouse code). To a lesser extent, features resembling binding sites for other splicing regulators, including Polypyrimidine tract binding protein (PTBP) and RNA-binding fox (RBFOX) proteins, may also be associated with ESC-differential AS.
From RNA-Seq expression profiling 221 known or putative splicing factors, eleven genes showed significant differential expression between human ESCs/iPSCs and other cells and tissues (Bonferroni-corrected p < 0.05, Wilcoxon rank-sum test) (Fig. 1c and Supplementary Table 4). Remarkably, MBNL1 and MBNL2 had the lowest relative mRNA levels in ESCs/iPSCs compared to other cells and tissues (Fig. 1c, Supplementary Fig. 3a and Methods). Quantitative RT-PCR assays confirmed this observation (Supplementary Fig. 3b). Similar results were obtained when analyzing mouse expression data (Supplementary Fig. 3c–e and Supplementary Table 4). PTBP, RBFOX and other splicing factors potentially associated with ESC-differential AS by the splicing code analysis did not exhibit significant differences in mRNA levels between ESCs/iPSCs and other cells or tissues. Collectively, these results suggest a conserved and prominent role for MBNL1 and MBNL2 in ESC-differential AS.
Because MBNL proteins are expressed at minimal levels in ESCs compared to other cell types, we hypothesized that they may repress ESC-differential exons in non-ESCs, and/or activate the inclusion of exons in non-ESCs that are skipped in ESCs. Indeed, previous studies have shown that in differentiated cells, MBNL proteins suppress exon inclusion when they bind upstream flanking intronic sequences, and they promote inclusion when binding to downstream flanking intronic sequences20,21. The results of the splicing code analysis are consistent with this mode of regulation, when taking into account that MBNL proteins are depleted in ESCs relative to differentiated cells and tissues (Fig. 1b and Supplementary Fig. 1b).
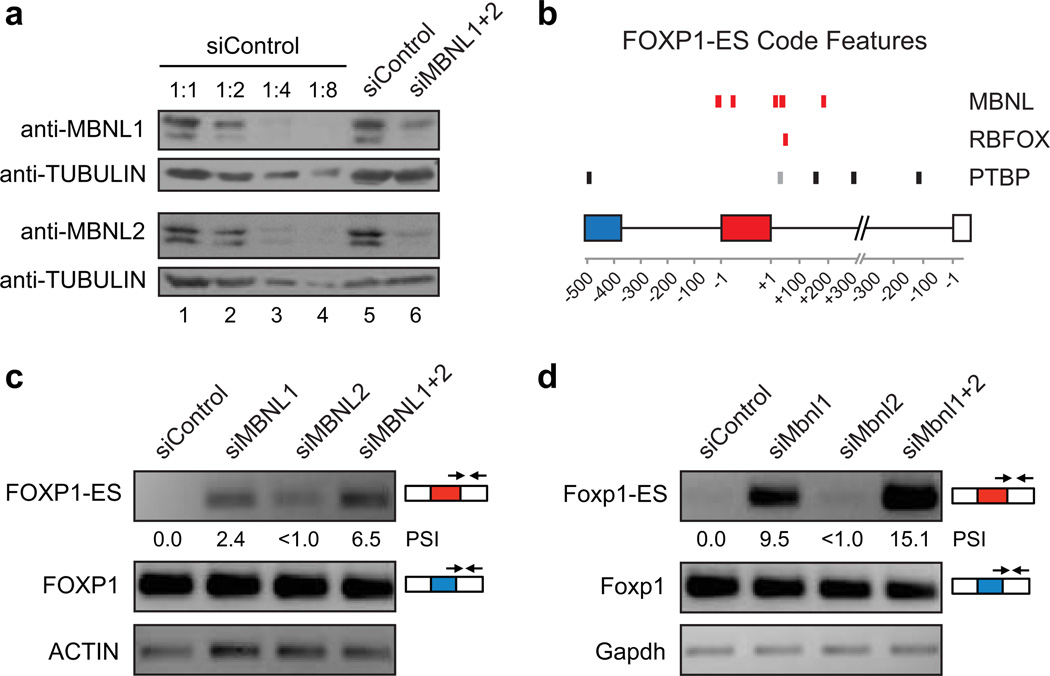
To test the above hypothesis, we used siRNAs to knockdown MBNL1 and MBNL2 (to ~10% of their endogenous levels), individually or together, in human (293T and HeLa) and mouse (neuro2A [N2A]) cells (Fig. 2a and Supplementary Fig. 4a; see below). For comparison, knockdowns were performed in human (H9) and mouse (CGR8) ESCs. RT-PCR assays were used to monitor the ESCswitch exon of FOXP1/Foxp1 (human exon 18b/mouse exon 16b), which is partially included in ESCs and fully skipped in differentiated cell types9. The splicing code analysis suggested that this exon is associated with conserved regulation by MBNL proteins, through possible direct disruption of splice site recognition (Fig. 2b; see legend and below). Knockdown of MBNL2 in 293T or HeLa cells resulted in a <1% increase in FOXP1 exon 18b inclusion, whereas knockdown of MBNL1 alone, or together with MBNL2, resulted in increases in PSI, from zero to 2–2.4% and 6.5–7.1%, respectively (Figs. 2c, 3c and Supplementary Figs. 4b, 5). More pronounced effects were observed for Foxp1 exon 16b in N2A cells (PSI shift from 0 to 15.1 for the double knockdown; Fig. 2d and Supplementary Fig. 4c). Knockdowns in ESCs had modest effects on exon 18b/16b splicing, consistent with the low levels of MBNL/Mbnl expression in these cells (Supplementary Fig. 4d, e). Knockdown of a third MBNL family member, MBNL3, which has a more restricted cell-type distribution compared to MBNL1 and MBNL222, had no detectable effect on exon 18b splicing (Supplementary Fig. 5). These results suggest that MBNL1 and MBNL2 proteins have conserved and partially redundant roles in the negative regulation of FOXP1/Foxp1 exon 18b/16b inclusion.
Figure 2. MBNL proteins regulate ESC-specific alternative splicing.
a) Western blots confirming efficient knockdown of MBNL1 and MBNL2 proteins in human 293T cells transfected with siRNA pools targeting these factors (siMBNL1+2, lane 6). Lane 5, lysate from cells transfected with a non-targeting siRNA pool (siControl). Lanes 1–4, serial dilutions (1:1, 1:2, 1:4 and 1:8) of lysate from cells transfected with siControl. b) Splicing code map highlighting genomic locations of MBNL, RBFOX and PTBP motifs associated with ESC-specific AS of FOXP1/Foxp1 exon 18b/16b, the inclusion of which forms the FOXP1-ES/Foxp1-ES isoform. Human (black), mouse (grey) or conserved features (red) are indicated. Note that conserved MBNL motifs are associated with possible direct interference of exon 18b/16b splice site regulation. c) RT-PCR assays monitoring mRNA levels of FOXP1 canonical (blue exon) and FOXP1-ES (red exon) isoforms in 293T cells transfected with siControl, siMBNL1, siMBNL2 or siMBNL1+2. RT-PCR employed splice junction-specific primers, as indicated. Expression levels of ACTIN are shown as loading controls. d) mRNA levels of murine Foxp1-canonical and Foxp1-ES isoforms were assayed as in (c) in N2A cells. Expression levels of Gapdh are shown as loading controls.
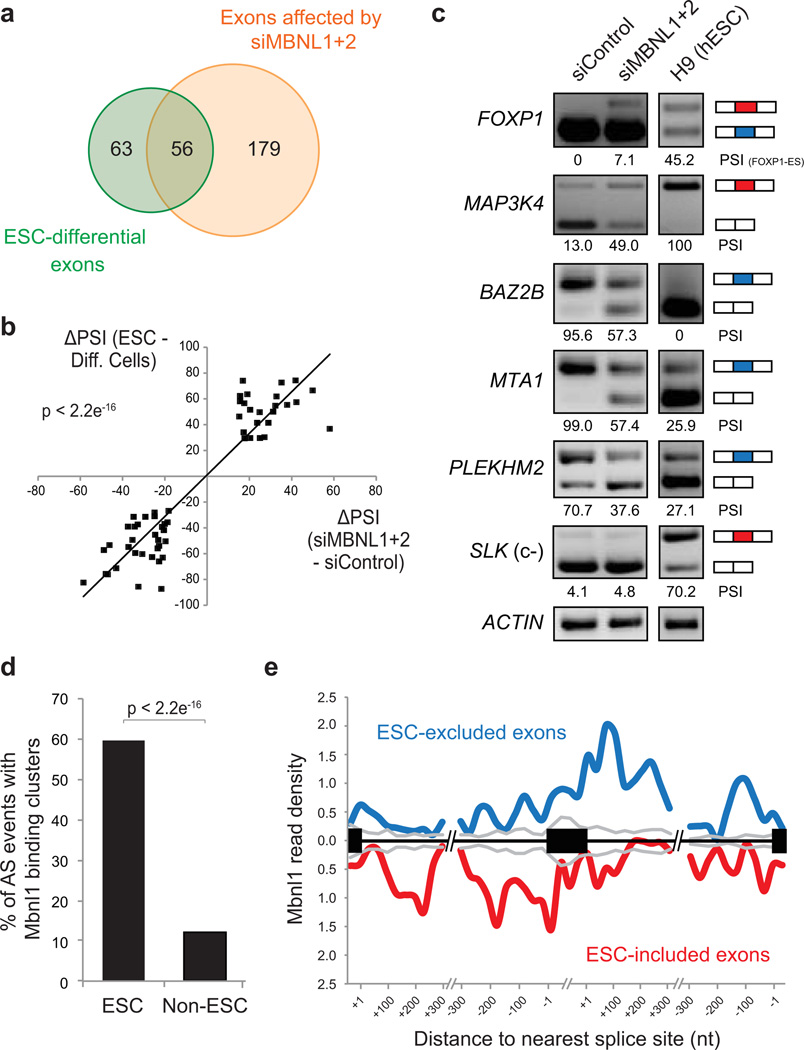
Figure 3. MBNL proteins regulate approximately half of ESC-differential alternative splicing events.
a) Venn diagram showing the proportion of ESC-differential AS events (green) that display ≥15 PSI change between HeLa cells transfected with siRNA pools targeting MBNL1+2 (siMBNL1+2) versus siControl pool (orange). b) High association (p<2.2e−16, one-sided binomial test between quadrants) between differences in PSIs of ESCs versus differentiated cells/tissues, and differences in PSIs of siMBNL1+2 knockdown versus siControl treatments. c) Representative RT-PCR validations for ESC-differential AS events that have PSI changes in HeLa cells following siMBNL1+2 transfection and for ESC-differential AS events that do not change upon siMBNL1+2 knockdown (c-); splicing patterns in human H9 ESCs are shown for comparison. d) Percentage of AS events with overlapping Mbnl1 CLIP-Seq binding clusters20 in C2C12 cells for ESC-differential or non-ESC-regulated alternative exons. e) Merged map of Mbnl binding clusters in transcripts with ESC-differential AS events. Maps of Mbnl1 binding sites with respect to exons that have higher or lower inclusion in ESCs/iPSCs, relative to non-ESCs/tissues.
To assess the extent to which ESC-differential AS events are controlled by MBNL proteins, MBNL1+2 were knocked down in HeLa cells, and RNA-Seq profiling was used to detect AS changes (Fig. 3). Of 119 profiled ESC-differentially spliced exons, nearly half are affected by knockdown of MBNL proteins, with a ≥15 PSI change towards an ESC-like AS pattern (Fig. 3a). A strong overall association (p<2.2e−16, one-sided binomial test) was observed when comparing PSI changes for exons differentially spliced between ESCs and non-ESCs/tissues, and PSI changes for the same exons following knockdown of MBNL proteins (Fig. 3b). RT-PCR experiments confirmed all analyzed MBNL knockdown-dependent and -independent PSI changes (Fig. 3c and Supplementary Fig. 6a). The specificity of the knockdown experiments was further demonstrated by comparing individual siRNAs that target different sequences within MBNL1 transcripts (Supplementary Fig. 6b). Comparable results were observed when MBNL1+2 proteins were simultaneously knocked down in 293T cells, and in undifferentiated C2C12 mouse myoblast cells (Supplementary Fig. 7). Conversely, over-expression of Mbnl1+2 proteins in mouse ESCs promoted differentiated cell-like patterns for all analyzed ESC-differential AS events (Supplementary Fig. 8), including a switch to the exclusive use of the canonical (i.e. non-ESC) exon 16 in Foxp1 transcripts. Consistent with this observation, over-expression of Mbnl proteins in ESCs also led to increased kinetics of silencing of core pluripotency factors upon differentiation, and further promoted the expression of specific lineage markers representative of all three germ layers (Supplementary Fig. 9).
Mapping of Mbnl protein binding to endogenous transcripts using UV-crosslinking coupled to immunoprecipitation and sequencing (CLIP-Seq or HITS-CLIP23) in undifferentiated C2C12 myoblast cells20 confirmed that these proteins directly target ESC-differential AS events, including Foxp1 exon 16b (Fig. 3d and Supplementary Fig. 10a). Of 57 mouse ESC-differential exons expressed in C2C12 cells, ~34 (60%) are associated with overlapping or proximal clusters of Mbnl CLIP-Seq tags (“binding clusters”), whereas binding clusters are associated with 72/601 (12%) of exons that are not differentially regulated in ESCs (p < 2.2e−16, proportion test; Fig. 3d). The binding clusters associated with ESC-differential AS are significantly enriched in consensus binding sites for MBNL proteins (Supplementary Fig. 10b)20,21,24. Moreover, consistent with the splicing code analysis (Fig. 1b and Supplementary Fig. 1b) and previous results20,21, the locations of Mbnl binding clusters correlate with whether the target exons are more or less included in ESCs compared to other cells and tissues (Fig. 3e). Collectively, the results so far demonstrate that MBNL proteins act widely and directly to regulate ESC-differential AS, and consequently pluripotency factor expression.
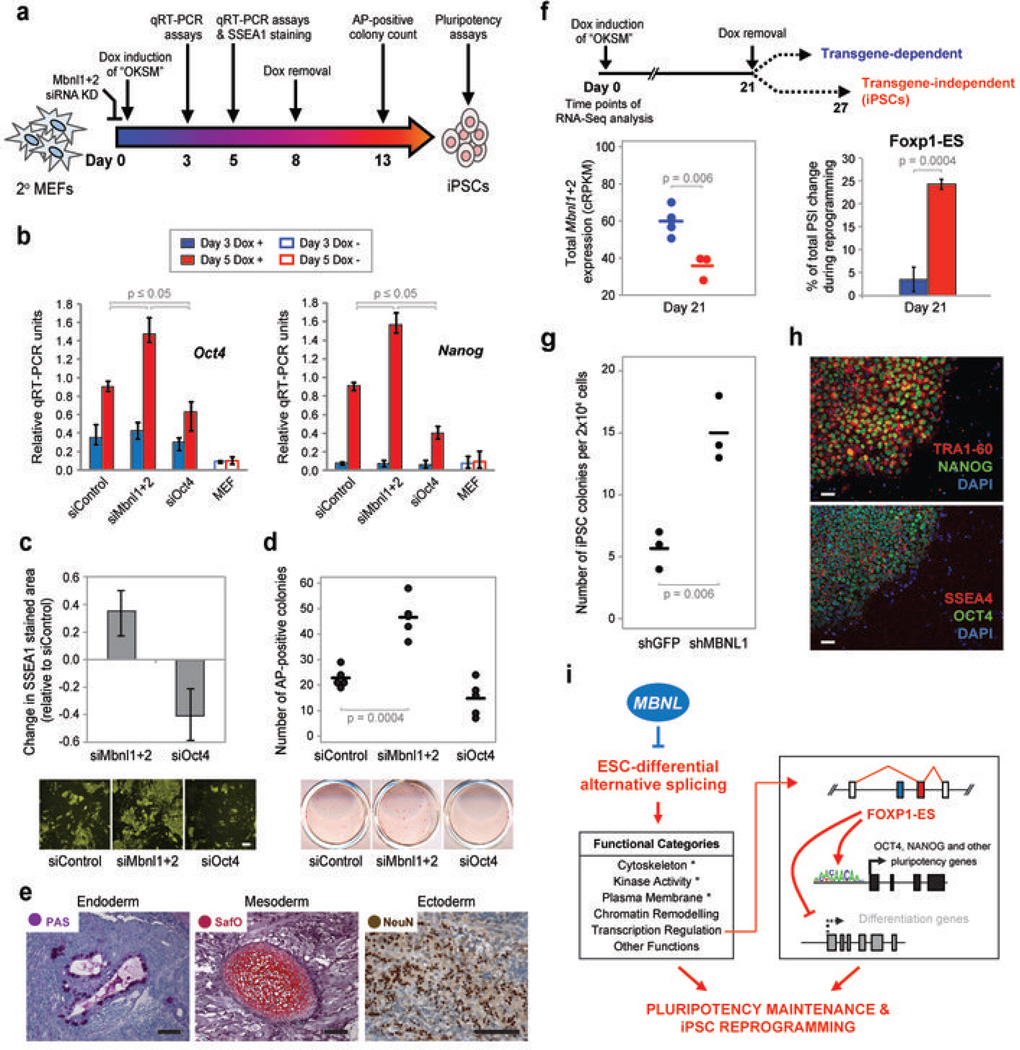
We next asked whether MBNL proteins impact somatic cell reprogramming (Fig 4a). Secondary mouse embryonic fibroblasts (MEFs)25 expressing the “OKSM” factors (Oct4, Klf4, Sox2, c-Myc)26 from transgenes under doxycycline (Dox)-inducible control were transfected with siRNA pools to knockdown Mbnl1 and Mbnl2 (siMbnl1+2), or with a control, non-targeting siRNA pool (siControl). At days 3 and 5 post Dox-induction, mRNA expression of endogenous pluripotency genes, including Oct4, Nanog, Sall4 and Alpl, were assayed by qRT-PCR (Fig. 4b and Supplementary Fig. 11a). None of these genes displayed significant changes in expression at day 3 (Fig. 4a and b); however, at day 5, Mbnl knockdown stimulated their expression by approximately 2-fold over the siControl treatment (Fig. 4b and Supplementary Fig. 11a). Mbnl knockdown also resulted in a ~30% increase in the colony area immunostained for SSEA1, a pluripotency-associated marker expressed early during reprogramming (Fig. 4c). In contrast, knockdown of Oct4 (siOct4) resulted in significant reductions in endogenous pluripotency gene expression and in SSEA1-positive colonies (Fig. 4b,c).
Figure 4. Knockdown of Mbnl proteins enhances reprogramming efficiency and kinetics.
a) Experimental scheme. b) qRT-PCR quantification of mRNA expression levels of endogenous Oct4 and Nanog (data for additional genes in Supplementary Fig. 11a). Secondary MEFs were transfected with control siRNAs (siControl), siRNAs targeting Mbnl1 and Mbnl2 (siMbnl1+2) or Oct4 (siOct4) and treated with doxcycline (Dox) for 3 days (blue bars) or 5 days (red bars) before analysis. Empty bars, secondary MEFs without Dox induction. Values represent Means ± Range (n=3). c) Top, quantification of SSEA1-stained area change relative to siControl at day 5 post Dox-induction; values represent Means ± Range (n=3); bottom, representative images of SSEA1 staining. Scale bar = 100 µm. d) Top, quantification of Dox-independent iPSC colony formation. Secondary MEFs were treated with Dox for 8 days followed by 5 days of Dox withdrawal and counting of alkaline phosphatase (AP)-positive colonies; bottom, representative images of AP staining. e) Teratoma assay assessing the pluripotency potential of iPSCs derived from secondary MEFs following knockdown of Mbnl proteins. Hematoxylin and eosin staining, with additional staining/immunolabeling using periodic acid-Schiff (PAS; for detection of glycogen or glycoprotein producing cells), Safranin O (SafO; for detection of cartilage), or antibody to neuronal nuclear antigen (NeuN); additional teratoma analysis and chimera testing of the pluripotency potential of siMbnl iPSCs in Supplementary Figs. 12 and 13. Scale bar = 100 µm. f) Top, experimental scheme for clonal analysis. Upon Dox removal at day 21, clones derived from single cells either survive and form iPSCs (“transgene-independent”) or do not survive (“transgene-dependent”). Bottom, analysis of total Mbnl1/Mbnl2 mRNA expression (left) and percentage of total PSI change (right) for Foxp1 exon 16b in transgene-independent (red) and transgene-dependent (blue) clones, at day 21, where total PSI change is the PSI difference between MEFs and iPSCs during reprogramming. g) Quantification (by morphological examination) of human iPSC colonies formed by reprogramming BJ fibroblasts expressing shRNA targeting GFP (shGFP) or MBNL1 (shMBNL1). h) Immunostaining of human iPSCs derived from shMBNL-expressing BJ fibroblasts for TRA1-60, NANOG, SSEA4 and OCT4 pluripotency markers. Scale bar = 50 µm. Additional characterization of human iPSCs in Supplementary Fig. 15. p-values of one-sided t-tests shown for all comparisons in Fig 4. i) Model for the role of MBNL proteins in the regulation of ESC-differential AS, pluripotency and iPSC reprogramming.
Successful reprogramming requires that cells transition to an OKSM transgene-independent state27. We therefore asked whether suppression of Mbnl proteins promotes transgene-independence. OKSM transgenes were induced for eight days, then cells were cultured for five days without Dox (Fig. 4a). Whereas knockdown of Oct4 reduced colony formation, knockdown of Mbnl proteins resulted in an approximate 2-fold increase in transgene-independent colonies, as detected by alkaline phosphatase (AP) staining (p = 0.0004; one-sided t-test) (Fig. 4d and Supplementary Fig. 11b). iPSC lines derived from transgene-independent colonies following Mbnl knockdown were pluripotent and contributed to all three germ layers in both teratoma and chimera assays (Fig. 4e and Supplementary Figs. 11–13). Consistent with these results, Mbnl expression is significantly reduced in secondary MEF clones27 cultured in the presence of Dox that are competent to achieve transgene-independence (when Dox is removed) versus those that are not (p = 0.006; one-sided t-test) (Fig. 4f, left). Moreover, the PSI levels of ESC-differential AS events, including Foxp1 exon 16b, significantly correlate with ESC/iPSC AS patterns only in those clones that transition to transgene-independence (Fig. 4f, right; Supplementary Fig. 14 and Supplementary Table 5; r= 0.80, p= 3.2e−12). Strikingly, knockdown of MBNL1 in human fibroblasts expressing OKSM also resulted in an approximate 2-fold increase in the appearance of iPSC colonies (Fig. 4g, h and Supplementary Fig. 15). MBNL proteins thus have a conserved, negative regulatory role in somatic cell reprogramming.
The results of this study reveal that MBNL proteins negatively regulate an ESC-differential AS network that controls pluripotency and reprogramming (Fig. 4i). These proteins likely act in part by directly repressing the ESC-specific splicing switch in FOXP1, which promotes the expression of core pluripotency genes. However, additional genes with MBNL-regulated AS events have been linked to the control of pluripotency, suggesting a more extensive role for the AS network in ESC biology (Fig. 4i). These observations represent the first evidence that trans-acting splicing regulators play a central role in the core circuitry required for ESC pluripotency and reprogramming. Our results further offer a potential new approach for enhancing the production of iPSCs for research and therapeutic applications.
FULL METHODS ONLINE
Cell lines and cell culture
HeLa, 293T and C2C12 cell lines were maintained in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% Fetal Bovine Serum (FBS) and antibiotics (penicillin/streptomycin). Neuro2A (N2A) cells were grown in DMEM supplemented with 10% FBS, sodium pyruvate, MEM non-essential amino acids, and penicillin/streptomycin. H9 human ESCs, CGR8 and R1 mouse ESCs were cultured as described previously31. Secondary mouse embryonic fibroblasts (MEFs) were maintained in DMEM supplemented with 10% FBS, L-glutamine and penicillin/streptomycin on 0.1% gelatin-coated plates. During reprogramming, secondary MEFs were grown in mouse ES media and induced to express OKMS factors using 1.5 µg/mL of doxycycline (Dox) as described previously32.
siRNA transfection
Cells were transfected with SMART-pool siRNAs (Dharmacon) using DharmaFECT1 reagent (Dharmacon), as recommended by the manufacturer. A non-targeting siRNA pool was used as a control. Cells were harvested 48 or 72 hours post transfection. In reprogramming experiments, secondary MEFs were transfected with siRNA pools using Lipofectamine™ RNAiMAX (Invitrogen), as described previously33, and the OKMS transgenes were induced the day after with Dox. Sequences of siRNAs are provided in Supplementary Information.
Protein extraction and western blotting
Cell pellets were lysed in radioimmunoprecipitation assay (RIPA) buffer by brief sonication. 30–150 ug of protein lysate was separated on a 10% SDS-polyacrylamide gel and transferred to a PVDF membrane. The membranes were blotted with the following antibodies: anti-Flag M2 (1:1500, Sigma), anti-MBNL1 (1:500, Abcam), anti-MBNL2 (1:200, Santa Cruz Biotechnology) and anti-β-tubulin (1:5000, Sigma). Secondary antibodies (GE Healthcare) and chemiluminescence reagents (Perkin Elmer) were used as per the manufacturer’s instructions.
RNA extraction and (q)RT-PCR assays
Total RNA was extracted using Tri Reagent (Sigma) or RNeasy columns (Qiagen). RT-PCR assays were performed using the OneStep RT-PCR kit (Qiagen), as per the manufacturer’s instructions. 20 ng total RNA or 1 ng of polyA+ RNA was used per 10 µL reaction. Radiolabeled reactions contained 0.3 µCi of α-32P-dCTP per 10 µL reaction. The number of amplification cycles was 22 for ACTIN and Gapdh, and 27–32 for all other transcripts analyzed. Reaction products were separated on 1–3% agarose gels. Quantification of isoform abundance was performed using either ImageQuant (GE Healthcare) or ImageJ software. To selectively amplify the FOXP1/Foxp1 isoforms (Fig. 2C and 2D), primers specific for splice junctions were used. All primer sequences are available upon request.
For quantitative (q)RT-PCR, first-strand cDNAs were generated from 1–3 µg of total RNA or 100 ng of polyA+ RNA using SuperScript III Reverse Transcriptase (Invitrogen), as per the manufacturer’s recommendations, and diluted to 20 ng/µL and 1ng/µL, respectively. qPCR reactions were performed in a 384 well format using 1 µL of each diluted cDNA and FastStart Universal SYBR Green Master (Roche Applied Science). Primers used for qRT-PCR reactions are available upon request.
Immunofluorescence
For immunofluorescence experiments, cells were fixed in 4% PFA for 10 min at room temperature, washed with PBS, and permeabilized for 10 min at 4°C with 0.1% Triton X100. After one hour of blocking, cells were incubated with primary antibodies overnight at 4°C, and then with secondary antibodies for 1 hour at room temperature. Nuclei were stained with Hoechst 33258 (1:5000, Sigma-Aldrich). Primary antibodies used in this study are: mouse IgM anti-SSEA1 (1:500, BD Biosciences), mouse anti-Oct4 (1:200, BD Biosciences), rabbit anti-Nanog (1:200, Cosmo Bio), goat anti-Dppa4 (1:250, R&D), mouse IgM anti-TRA1-60 (1:100, Invitrogen), rabbit anti-NANOG (1:400, Cell Signalling), mouse anti-SSEA4 (1:100, Invitrogen), rabbit anti-OCT4 (1:200, Abcam), mouse anti-alpha fetoprotein (1:200, R&D), mouse anti-smooth muscle actin (1:200, Invitrogen), and mouse anti-beta-III-tubulin (1:200, Millipore). Secondary antiodies used in this study are: anti-mouse IgM Alexa555 (1:1000, Molecular Probes), anti-mouse IgG Alexa555 (1:1000, Molecular Probes), anti-rabbit IgG Alexa594 (1:1000, Molecular Probes), anti-rabbit IgG Alexa488 (1:500, Molecular Probes), and anti-goat IgG Alexa546 (1:1000, Molecular Probes).
iPSC colony formation assays and imaging from secondary MEF reprogramming
Secondary MEFs were seeded in 12-well plates, transfected with siRNA pools, and treated with Dox for 5 days before fixing and staining. The plates were imaged (for both SSEA1-immunostained and DAPI channels) using an IN Cell Analyzer 2000 (GE Healthcare) with a 4× objective. For each well, 20 non-overlapping fields were captured and images were analyzed using the Columbus System (PerkinElmer). A custom script was generated to identify SSEA1-positive and DAPI-positive colonies. The overall signal in each well was determined using the sum of the overlap area for the 20 fields captured.
To assay the formation of Dox-independent colonies, secondary MEFs transfected with siRNA pools were treated with Dox for 8 days. Cell counting was performed before and at each passage after siRNA transfection and doubling rates were determined not to significantly change (data not shown). At day 8, the same number of cells were passaged into Dox-free mESC media on 12-well plates and cultured until day 13, when they were fixed and stained with alkaline phosphatase for colony counting.
Teratoma Analysis
Cells were suspended in PBS and Matrigel (BD Bioscience) mixed solution, and 1 × 106 cells in 100 µl were injected subcutaneously into both dorsal flanks of nude mice (CByJ.Cg-Foxn1nu/J) anaesthetized with isoflurane. Four to 5 weeks after injection, mice were sacrificed and teratomas were dissected, fixed overnight in 10% buffered formalin phosphate, and embedded in paraffin. Three to 4 µm thick sections were deparaffinized and hydrated in distilled water. Sections were stained either with haematoxylin and eosin for regular histological examination, or with the following dyes; 0.1 % Safranin O solution (cartilage, mesoderm-derived tissue) or 0.5 % Periodic Acid Schiff (PAS) solution (glycoprotein-producing intestinal cell, endoderm-derived tissue). For immunohistochemistry, sections were deparaffinized and hydrated, and antigen retrieval process was performed. After blocking, sections were incubated overnight at 4°C with primary monoclonal antibody (1:100, Millipore MAB377, clone A60) specific for neuronal nuclear antigen (NeuN, ectoderm-derived tissue), followed by washing in PBS. After 1 hour of incubation with secondary anti-mouse-HRP conjugated antibody (1:500, Jackson ImmunoResearch, 115-035-003), signal was visualized by DAB (3,3’-diaminobenzidine; Vector Laboratories, SK-4100) substrate for 5–20 minutes. Sections were counter-stained with haematoxylin.
Chimerism analysis
Chimera aggregation and whole mount staining were performed as described previously34. Chimeras were obtained through aggregation of siMbnl iPS cell clumps with diploid Hsd:ICR(CD-1) embryos. E10.5 embryos were dissected after Dox treatment in utero via ingestion 24 hours prior to dissection. After dissection, embryos were fixed with 0.25% glutaraldehyde, rinsed in wash buffer [2mM MgCl2, 0.01% sodium deoxycholate, and 0.02% Nonidet-P40 in PBS] and then stained overnight in LacZ staining solution [20mM MgCl2, 5mM K3Fe(CN)6, 5mM K4Fe(CN)6 and 1mg/ml X-gal in PBS]. Embryos were embedded in paraffin, sectioned and counterstained with nuclear fast red.
Generation and characterization of human iPS cells
Human BJ foreskin fibroblasts (Stemgent) were reprogrammed using published protocols35,36, with the following modifications. BJ fibroblasts were first infected with lentivirus vectors encoding both a puromycin resistance gene and doxycycline (dox)-inducible shRNA targeting either GFP (negative control, target sequence: 5’ GCAAGCTGACCCTGAAGTTCAT 3’) or MBNL1 shRNA (target sequence: 5’ GCCTGCTTTGATTCATTGAAA 3’). Lentiviral vector preparations and infections were performed as described35. After selection with 1 µg/ml puromycin, shRNA-encoding BJ fibroblasts were infected with a second lentivirus vector (obtained from Addgene) co-expressing both the mouse retrovirus receptor mSlc7a1 and the blasticidin resistance gene37. During transient selection with 5 µg/ml blasticidin, puromycin was reduced to 0.5 µg/ml and maintained at this concentration for 6 days after infection with retroviral reprogramming vectors.
pMXs-based retrovirus vectors encoding the four reprogramming factors hOCT4, hSOX2, hKLF4, hCMYC (OSKM)37, were obtained from Adgene and packaged exactly as described35. Puromycin/blasticidin-resistant BJ fibroblasts were infected in triplicate, using three separate preparations of retrovirus vectors. shRNA expression was induced by treatment with 2 µg/ml Dox, which was initiated contemporaneously with retrovirus vector infection; control cells were treated with vehicle only.
Six days after retrovirus infection, BJ fibroblasts were seeded on a monolayer of feeder cells. Embryonic day 12.5 fibroblasts from Tg(DR4)1Jae/J mice (Jackson Laboratory) were seeded on collagen-coated 6 well plates at a density of 3×105 cells per well as described35; retrovirus-infected BJ fibroblasts were seeded at a density of 2×104 cells per well. At day 28 of reprogramming, quantification (by whole-well morphological examination and by TRA1-60 immunostaining) of human iPSC colonies was performed by investigators who were blinded as to the experimental conditions. To count TRA1-60 positive colonies, the plates were imaged (for both TRA1-60-immunostained and DAPI channels) using an IN Cell Analyzer 2000 (GE Healthcare) with a 4× objective. For each well, 64 non-overlapping fields were captured and images were analyzed using the Columbus System (PerkinElmer). Knockdown of MBNL1 resulted in an approximate two-fold increase in TRA1-60 immunostaining colonies over the control knockdown with GFP-targeting shRNA (Fig. S13 and data not shown). Additional OSKM retrovirus-infected BJ fibroblasts were seeded in parallel; individual colonies from dox-treated plates were manually isolated four weeks post-infection, and seeded on feeders in collagen-coated 24 well plates35. Cells from these colonies were expanded, and subsequently characterized as described38.
Clonal analysis by RNA-Seq during reprogramming
In a single-cell assay, secondary MEFs were plated in individual wells of a 96-well plate, OKSM factors were induced by Dox treatment and the clonal derivatives were cultured for 21 days. Removal of Dox at day 21 revealed that approximately 50% of clones produced abundant AP-positive colonies (transgene-independent clones), while the rest yielded little or no colonies (transgene-dependent clones). RNA-Seq analysis was performed for 3 transgene-independent and 5 transgene-dependent clones at day 21 after Dox induction (Supplementary Table 1).
Using RNA-Seq derived PSI values (see Supplementary Methods Online), the inclusion levels of mouse ESC-differential cassette alternative exons were quantified for each of these clones. 51 ESC-differential AS events with sufficient read coverage in all samples and with a ≥ 25 PSI difference between iPSCs and MEFs were compared between the two types of clones (Supplementary Fig. 14, Supplementary Table 5).
RNA-Seq data and analysis
We used RNA-Seq data from 36 and 32 different human and mouse samples, respectively. Details and sample sources are provided in Supplementary Table 1. The samples comprise, for human: 5 ESC lines (3 different cell lines and 2 replicates), 2 iPSC lines, 7 non-ESC lines, and 22 adult tissues (18 different tissues and 4 replicates); for mouse: 6 ESCs, 2 iPSC colonies, 8 non-ESC lines (5 different cell lines and 3 replicates) and 16 adult tissues (10 different types and 6 replicates). Details of RNA-Seq analysis in Supplementary information.
To identify alternative exons differentially regulated in ESCs, we first calculated a single averaged PSI value for tissues of similar origin (see Supplementary Table 1). Only events with enough coverage in at least two ESC samples and three distinct tissue types were considered. ‘ESC-differential AS events’ were defined as those with a mean PSI difference of ≥25 between ESCs and differentiated tissues. To account for AS events potentially related to cell proliferation, we also required a mean PSI difference of ≥25 between ESC lines and non-ESC lines, when the event had sufficient coverage in at least one cell line sample. The set of background AS events used throughout the study are alternatively spliced exons (defined here as exons with PSI values of <95% and >5% in at least one sample) that meet the same expression requirement (i.e. in ≥ 2 ESCs and ≥ 3 differentiated tissue types) and that show an average difference in PSI level of < 5% between ESCs and differentiated tissue samples, and between the ESC and non-ES cell lines.
Analyses of splicing factor expression
A total of 221 human and 214 mouse genes were selected for analysis based on literature mining for previously described splicing functions, “splicing”- and/or “spliceosome”-associated Gene Ontology (GO) terms, and/or the presence of a PFAM-annotated RNA-binding domain (Supplementary Table 4). To calculate the mRNA expression values for each sample, we used corrected (for mappability) Reads Per Kilobase pair and Million mapped reads values (cRPKMs) of the “stable” (as defined by BioMart) Ensembl transcript for each gene, as previously described39.
Splicing factor genes were ranked according to the relative extent of their differential expression (as determined by cRPKM values) in ESCs and iPSCs versus non-ESC lines and tissues by comparing summed ranks of each gene in all ESC/iPSCs across the full range of samples. Based on this approach, human MBNL1 and MBNL2 showed the 1st and 2nd lowest overall rank in ESCs/iPSCs, respectively, and mouse Mbnl1 and Mbnl2 showed the 1st and 3rd lowest overall rank in ESCs/iPSCs, respectively.
To assess the statistical significance of the differential expression of individual splicing factor genes, we compared their cRPKM values in ESCs/iPSCs to the cRPKMs in all other cell lines and differentiated tissues using a Wilcoxon rank-sum test after quantile normalization. Splicing factors with Bonferroni-corrected p-values < 0.05 were considered significantly differentially expressed (Supplementary Table 4).
Splicing code analyses
The feature vectors for each species were produced by extracting sequence-based features from alternatively spliced exons, their adjacent constitutive exons, and 300 nucleotides of flanking intronic sequence. The features used were a subset of those defined in40, with the following differences: (a) all sequence length features are now in the log domain; (b) due to a lack of comprehensive transcript libraries and the corresponding uncertainty about downstream consequences of frame shifts, premature termination codon (PTC) features were excluded; and (c) conservation scores and conservation-weighted motifs were excluded from the feature set. In addition, related features (i.e. consensus recognition sequences for a given splicing factor inferred by different methods) were combined and included as independent features.
To identify features strongly associated with ESC-differential exon inclusion or exclusion, we compared 172 ESC-differential exons and 908 background (BG) exons for human, along with 102 ESC-differential exons and 811 BG exons for mouse. Associations between features and ESC-differential splicing were detected using Pearson correlation. For each feature, we computed the correlation between its value and the difference in average PSI values in ESCs and non-ESCs, across exons. To obtain more accurate correlation values, we considered two scenarios: (a) a positive scenario in which the differences in average PSI values in ESCs and non-ESCs are larger than 25%,and (b) negative scenario in which the differences in average PSI values in ESCs and non-ESCs are smaller than −25%.
CLIP-Seq analysis
We used recently described Mbnl1 CLIP-Seq data from C2C12 cells41. In order to estimate the fractions of ESC-differential and background AS events that are associated with Mbnl1 binding, we asked whether CLIP binding clusters overlap the alternative exon and/or flanking intron sequences of each event. CLIP binding clusters were defined as previously described41. In short, CLIP-Seq tags were trimmed of adapters and then collapsed to remove redundant sequences. These tags were mapped to genome and a database of splice junctions using Bowtie. To identify CLIP clusters lying within genic regions, gene boundaries were first defined using RefSeq, Ensembl, and UCSC tables. For each window of 30 nucleotides covered by at least one CLIP-Seq tag, a test was performed to assess whether the tag density in the window exceeded that which is predicted by a simple Poisson model which accounts for gene expression and pre-mRNA length. An AS event was considered to have an overlapping Mbnl1 binding cluster if the mid-point of the cluster is located within the alternative exon, within 300 nt of the 5´ or 3´ ends of upstream or downstream flanking introns, and/or within 30 nt within the 3´ end of C1 exon or the 5´ end of C2 exon. Only AS events that had significant read coverage (see above) in at least one of two C2C12 samples used were analyzed. In total, 57 ESC-differential AS events and 601 background AS events were compared.
To generate an RNA regulatory map42 highlighting Mbnl1 binding sites in relation to ESC-differential AS events with either higher (ESC-included) or lower (ESC-excluded) exon inclusion levels in ESCs versus other cell lines and tissues, we applied the following procedure: for each nucleotide position from the regions described above and from three sets of AS events (ESC-included, ESC-excluded and background), we counted the average number of Mbnl1 CLIP-Seq tags. To minimize the impact of outliers with extreme read density, we limit the maximum count per event to an average of 10 reads per position within each region. In order to normalize the length of the AS exon, we divided each exon into 100 bins and uniquely assigned each position to the integer of 100*position/exon_length, with a relative weight inversely related to the length of the AS exon. To draw the map, we used sliding windows of 30nt for the intronic regions and 25nt for the length-corrected exons (total of four windows shown).
Evolutionary conservation of ESC-differential events
We analyzed three different aspects of conservation of the human and mouse ESC-differential alternative exons43. To determine whether the alternative exon is conserved at the genomic level, we performed a lift-over of the exon coordinates using Galaxy (https://main.g2.bx.psu.edu/). Exons with a unique lift-over hit in the other species, and with AG (splicing acceptor) and/or GT (splicing donor) sites were considered to be Genome-conserved in the other species. In addition, if the orthologous exon has a PSI of <95% and >5% in at least one sample from each species, AS of the exon was defined as conserved. Finally, to assess whether ESC-differential regulation is conserved, we applied two criteria: (i) the exons are independently detected as ESC-differential in human and mouse using the criteria as described above (total = 25 AS events), and (ii) the orthologous exons must meet minimal read coverage requirements (also as described above) to afford direct comparison.
Analyses of function and protein domain enrichment
To investigate whether ESC-differential events are significantly enriched in genes with specific functional associations and/or protein domains, we used the online tool DAVID (http://david.abcc.ncifcrf.gov/)44,45 (with annotations from levels 3, 4 and 5 in the GO hierarchy), KEGG pathways and InterPro domains. As background, we used the genes with at least one AS event that met the minimal expression criteria described above (i.e. detection in ≥ 2 ESCs and ≥ 3 differentiated cell/tissue types). The main clusters of functionally related genes enriched in both human and mouse (as well as among the conserved) ESC-differential events (Supplementary Table S3) are associated with: actin cytoskeleton, plasma membrane (including cell junctions), and protein kinase-associated terms.
Supplementary Material
Acknowledgements
The authors thank Ulrich Braunschweig, Jonathan Ellis, Serge Gueroussov and Bushra Raj for helpful comments on the manuscript. We gratefully acknowledge Dax Torti in the Donnelly Sequencing Centre for sequencing samples, Leo Lee for assisting with the splicing code analysis, Jodi Garner (Hospital for Sick Children Embryonic Stem Cell Facility) for preparing feeder cells, Alina Piekna for morphological examination of human iPSC colonies, Masahiro Narimatsu for assisting with chimerism analysis, and Patricia Mero for assisting with cell imaging. This work was supported by grants from the Canadian Institutes of Health Research (CIHR) (to B.J.B., J.L.W., A.N., J.E. and B.J.F.), the Ontario Research Fund (to J.L.W., B.J.B., A.N. and others), the Canadian Stem Cell Network (to A.N. and B.J.B.), and by a grant from the National Institutes of Health (R33MH087908) to J.E. H.H. was supported by a University of Toronto Open Fellowship. P.J.R., M.I. and N.L.B.-M. were supported by postdoctoral fellowships from the Ontario Stem Cell Initiative, Human Frontiers Science Program Organization, and the Marie Curie Actions, respectively.
Footnotes
Full Methods and any associated references are available in the online version of the paper.
Author Contributions
H.H. performed experiments in Figs. 1–4, S2–9 and S11–13. M.I. performed bioinformatic analyses in Figs. 1–4, S1, S3, S7, S10 and S14, with input from N.L.B.-M. L.D., A.G. assisted with secondary MEF reprogramming experiments and clone characterization, and D.T. generated secondary MEF lines and performed chimerism testing. P.J.R., T.T. and M.G. performed human reprogramming experiments and iPSC characterization. H-K. S. performed teratoma assays. B.A. and B.J.F. generated splicing code data. I.P.M., H-K. S. and D.O. assisted with ESC overexpression and differentiation experiments. E.W. and C.B.B. generated and analyzed CLIP-Seq data. E.N. and V.S. performed RT-PCR validation experiments. B.J.B., H.H., and M.I. designed the study, with input from J.L.W., J.E., A.N. and J.M. B.J.B. H.H., and M.I. wrote the manuscript, with input from the other authors.
References
- 1.Young RA. Control of the embryonic stem cell state. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annual review of biochemistry. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bao X, et al. MicroRNAs in somatic cell reprogramming. Current opinion in cell biology. 2013 doi: 10.1016/j.ceb.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 4.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nature Genetics. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 5.Wang ET, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braunschweig U, Gueroussov S, Plocik AM, Graveley BR, Blencowe BJ. Dynamic Integration of Splicing within Gene Regulatory Pathways. Cell. 2013;152:1252–1269. doi: 10.1016/j.cell.2013.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kalsotra A, Cooper TA. Functional consequences of developmentally regulated alternative splicing. Nature reviews. Genetics. 2011;12:715–729. doi: 10.1038/nrg3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gabut M, et al. An alternative splicing switch regulates embryonic stem cell pluripotency and reprogramming. Cell. 2011;147:132–146. doi: 10.1016/j.cell.2011.08.023. [DOI] [PubMed] [Google Scholar]
- 10.Chen X, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 11.Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Silva J, et al. Nanog is the gateway to the pluripotent ground state. Cell. 2009;138:722–737. doi: 10.1016/j.cell.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Irimia M, Blencowe BJ. Alternative splicing: decoding an expansive regulatory layer. Current opinion in cell biology. 2012 doi: 10.1016/j.ceb.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 14.Rao S, et al. Differential roles of Sall4 isoforms in embryonic stem cell pluripotency. Mol Cell Biol. 2010;30:5364–5380. doi: 10.1128/MCB.00419-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salomonis N, et al. Alternative splicing regulates mouse embryonic stem cell pluripotency and differentiation. Proc Natl Acad Sci USA. 2010;107:10514–10519. doi: 10.1073/pnas.0912260107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mayshar Y, et al. Fibroblast growth factor 4 and its novel splice isoform have opposing effects on the maintenance of human embryonic stem cell self-renewal. Stem cells. 2008;26:767–774. doi: 10.1634/stemcells.2007-1037. [DOI] [PubMed] [Google Scholar]
- 17.Barash Y, et al. Deciphering the splicing code. Nature. 2010;465:53–59. doi: 10.1038/nature09000. [DOI] [PubMed] [Google Scholar]
- 18.Liang J, et al. Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nature cell biology. 2008;10:731–739. doi: 10.1038/ncb1736. [DOI] [PubMed] [Google Scholar]
- 19.Lian I, et al. The role of YAP transcription coactivator in regulating stem cell self-renewal and differentiation. Genes & development. 2010;24:1106–1118. doi: 10.1101/gad.1903310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang ET, et al. Transcriptome-wide regulation of pre-mRNA splicing and mRNA localization by muscleblind proteins. Cell. 2012;150:710–724. doi: 10.1016/j.cell.2012.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Charizanis K, et al. Muscleblind-like 2-Mediated Alternative Splicing in the Developing Brain and Dysregulation in Myotonic Dystrophy. Neuron. 2012;75:437–450. doi: 10.1016/j.neuron.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pascual M, Vicente M, Monferrer L, Artero R. The Muscleblind family of proteins: an emerging class of regulators of developmentally programmed alternative splicing. Differentiation. 2006;74:65–80. doi: 10.1111/j.1432-0436.2006.00060.x. [DOI] [PubMed] [Google Scholar]
- 23.Licatalosi DD, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fernandez-Costa JM, Llamusi MB, Garcia-Lopez A, Artero R. Alternative splicing regulation by Muscleblind proteins: from development to disease. Biological reviews of the Cambridge Philosophical Society. 2011;86:947–958. doi: 10.1111/j.1469-185X.2011.00180.x. [DOI] [PubMed] [Google Scholar]
- 25.Woltjen K, et al. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009;458:766–770. doi: 10.1038/nature07863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 27.Golipour A, et al. A late transition in somatic cell reprogramming requires regulators distinct from the pluripotency network. Cell stem cell. 2012;11:769–782. doi: 10.1016/j.stem.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 28.Samavarchi-Tehrani P, et al. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010;7:64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 29.Calarco JA, et al. Global analysis of alternative splicing differences between humans and chimpanzees. Genes Dev. 2007;21:2963–2975. doi: 10.1101/gad.1606907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Labbe RM, et al. A comparative transcriptomic analysis reveals conserved features of stem cell pluripotency in planarians and mammals. Stem cells. 2012;30:1734–1745. doi: 10.1002/stem.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
REFERENCES
- 31.Gabut M, et al. An alternative splicing switch regulates embryonic stem cell pluripotency and reprogramming. Cell. 2011;147:132–146. doi: 10.1016/j.cell.2011.08.023. [DOI] [PubMed] [Google Scholar]
- 32.Polo JM, Hochedlinger K. When fibroblasts MET iPSCs. Cell Stem Cell. 2010;7:5–6. doi: 10.1016/j.stem.2010.05.018. [DOI] [PubMed] [Google Scholar]
- 33.Samavarchi-Tehrani P, et al. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010;7:64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 34.Golipour A, et al. A late transition in somatic cell reprogramming requires regulators distinct from the pluripotency network. Cell stem cell. 2012;11:769–782. doi: 10.1016/j.stem.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 35.Hotta A, et al. EOS lentiviral vector selection system for human induced pluripotent stem cells. Nat Protoc. 2009;4:1828–1844. doi: 10.1038/nprot.2009.201. [DOI] [PubMed] [Google Scholar]
- 36.Hotta A, et al. Isolation of human iPS cells using EOS lentiviral vectors to select for pluripotency. Nat Methods. 2009;6:370–376. doi: 10.1038/nmeth.1325. [DOI] [PubMed] [Google Scholar]
- 37.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 38.Cheung AY, et al. Isolation of MECP2-null Rett Syndrome patient hiPS cells and isogenic controls through X-chromosome inactivation. Hum Mol Genet. 2011;20:2103–2115. doi: 10.1093/hmg/ddr093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Labbe RM, et al. A comparative transcriptomic analysis reveals conserved features of stem cell pluripotency in planarians and mammals. Stem cells. 2012;30:1734–1745. doi: 10.1002/stem.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiong HY, Barash Y, Frey BJ. Bayesian prediction of tissue-regulated splicing using RNA sequence and cellular context. Bioinformatics. 2011;27:2554–2562. doi: 10.1093/bioinformatics/btr444. [DOI] [PubMed] [Google Scholar]
- 41.Wang ET, et al. Transcriptome-wide regulation of pre-mRNA splicing and mRNA localization by muscleblind proteins. Cell. 2012;150:710–724. doi: 10.1016/j.cell.2012.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Licatalosi DD, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Irimia M, Rukov JL, Roy SW, Vinther J, Garcia-Fernandez J. Quantitative regulation of alternative splicing in evolution and development. BioEssays. 2009;31:40–50. doi: 10.1002/bies.080092. [DOI] [PubMed] [Google Scholar]
- 44.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 45.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic acids research. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.