Abstract
We demonstrate the application of super-localization microscopy to identify sequence-specific portions of single-stranded DNA (ssDNA) with sequence resolution of 50 nucleotides, corresponding to a spatial resolution of 30 nm. Super-resolution imaging was achieved using a variation of a single-molecule localization method, termed as ‘motion blur’ point accumulation for imaging in nanoscale topography (mbPAINT). The target ssDNA molecules were immobilized on the substrate. Short, dye-labeled, and complementary ssDNA molecules stochastically bound to the target ssDNA, with repeated binding events allowing super-resolution. Sequence specificity was demonstrated via the use of a control, non-complementary probe. The results support the possibility of employing relatively inexpensive short ssDNAs to identify gene sequence specificity with improved resolution in comparison to the existing methods.
Keywords: super-resolution microscopy, single-stranded DNA, optical mapping, mbPAINT, biosensing, materials and biointerfaces
INTRODUCTION
Genome optical mapping has been a great tool for whole genome sequencing,1–5 but can be even more powerful if two aspects are improved: (1) increasing the mapping resolution to a scale below the diffraction limit of light, which currently limits the maximum mapping resolution to ~0.8 kilobase pair for stretched double-stranded DNA (dsDNA); (2) improving the sequence specificity beyond the current limitation of the available sequence specific enzymes for dsDNA molecules, which are often the target molecules in optical mapping. Utilizing a super-resolution imaging method is a key for increasing the resolution, and optical mapping of single-stranded DNA (ssDNA) instead of dsDNA is a possible solution to improve the sequence specificity because ssDNAs can be used as labeling probes with no sequence limit.
Several super-resolution imaging technologies have been established to study important biological and physical problems,6,7 such as living cell imaging,8,9 kinesin walking,10 synaptic vesicle diffusion in neuron cells,11 virus-cell interaction,12 and surface reaction site mapping of nano-materials.13,14 There are hardware, software, and chemically-assisted routes to achieve super-localization/resolution conditions.11,13–30 These diverse super-localization/resolution methods share the common principle that only one molecule is detected at a time within the distance of the diffraction limit (~250 nm for green light),31 often with the help of statistical methods during data analysis.32,33 Traditionally, dsDNA and DNA super-structures have been used as molecular rulers to demonstrate the resolution of super-resolution imaging technologies.3,24,34,35 Very recently, super-resolution mapping of dsDNA3,36,37 and localization of ssDNA38 were reported. These works open new directions for the genome optical mapping.
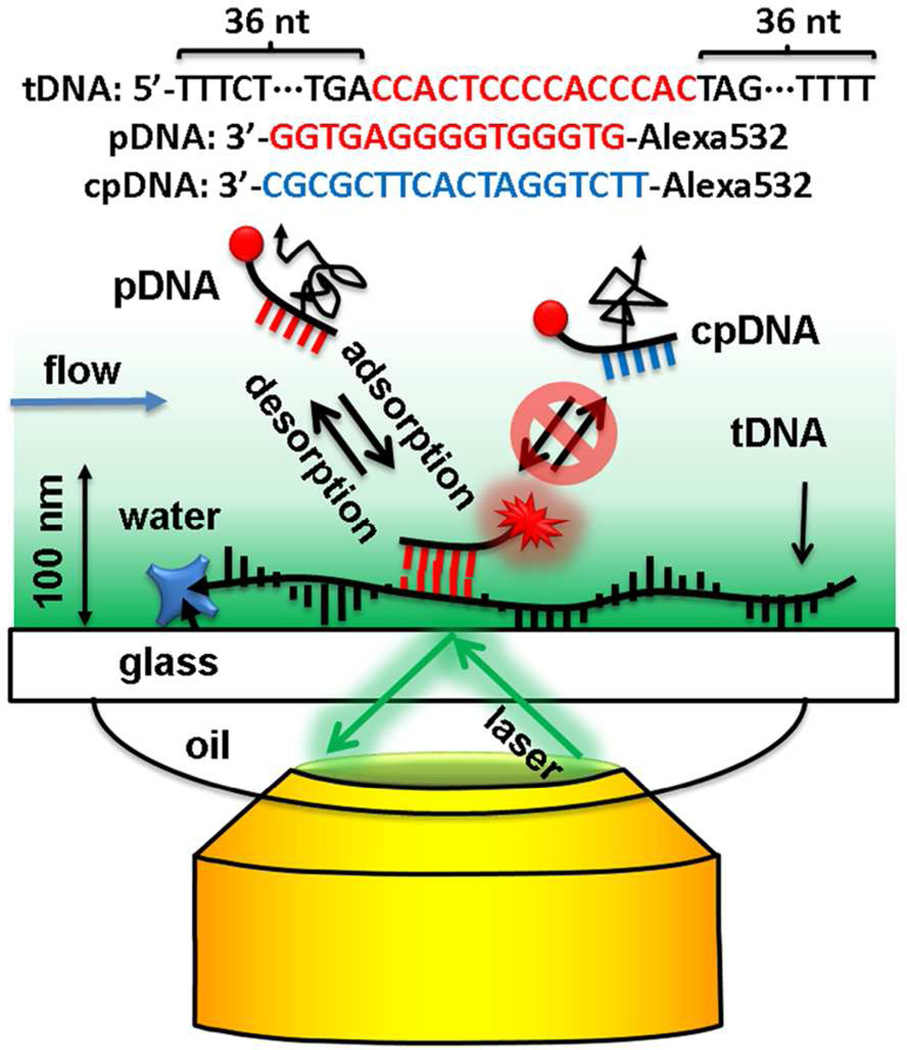
In this work, we focus on the super-resolution imaging of ssDNA with stochastic binding of complementary ssDNA,38 a technique we term motion blur point accumulation for imaging in nanoscale topography (mbPAINT). This is achieved with similar strategy as previously reported38,39 and the influence of experimental conditions such as incident angles and salt concentrations are studied. We immobilized the target ssDNA (tDNA) on a substrate (Fig. 1). The substrate was passivated by polyethylene glycol (PEG) molecules and was confirmed to have no affinity to the probe ssDNA (pDNA). Then we monitored the dynamic adsorption of the pDNA to the tDNA. Each binding molecule induced a diffraction-limited fluorescence image, which was fitted with a 2D Gaussian function for its center location and a super-resolution image was generated with the molecule locations. The diffraction limit of light was thus broken by the single-molecule localization method mbPAINT.
Figure 1.
Scheme of super-resolution imaging of immobilized target tDNA, with a fluorescently labeled probe pDNA, using mbPAINT and a TIRF microscope. Only the molecules within the evanescent field of the excitation light were excited and only those localized were observed. The negative control probe cpDNA, does not bind to the tDNA, so it diffuses freely in the solution and cannot be observed. Drawing not to scale.
RESULTS AND DISCUSSION
mbPAINT, depicted in Figure 1, is an extension of PAINT, which traditionally uses the unique properties of fluorescent dyes that only emit fluorescence upon binding to the target, due to the changes in their chemical structures or environments.13,27 In our case, fluorescently tagged pDNA molecules were detectable under total internal reflection fluorescence (TIRF) microscopy when they were reversibly adsorbed onto the immobilized tDNA at the interface, but were unobservable if they were freely diffusing near the interface (Fig. 1, 2, and 3). The sequence of tDNA was 5’-CCACTCCCCACCCAC, which was inserted into a random sequence with total length of 87 nucleotides (nt) to mimic the environment of a real target sequence. The tDNA was functionalized with biotin at the 5’ end and was attached to the surface via biotin-streptavidin interaction.40–42 The pDNA molecules, Alexa532-5’-GTGGGTGGGGAGTGG were labeled with Alexa Fluor 532 fluorescent dye at the 5’ end. A non-complementary negative control probe DNA (cpDNA) with sequence Alexa532-5’-TTCTGGATCACTTCGCGC oligomer was used to test for non-specific interactions with the target. Buffer flow was introduced to stretch the ssDNA on the substrate.43
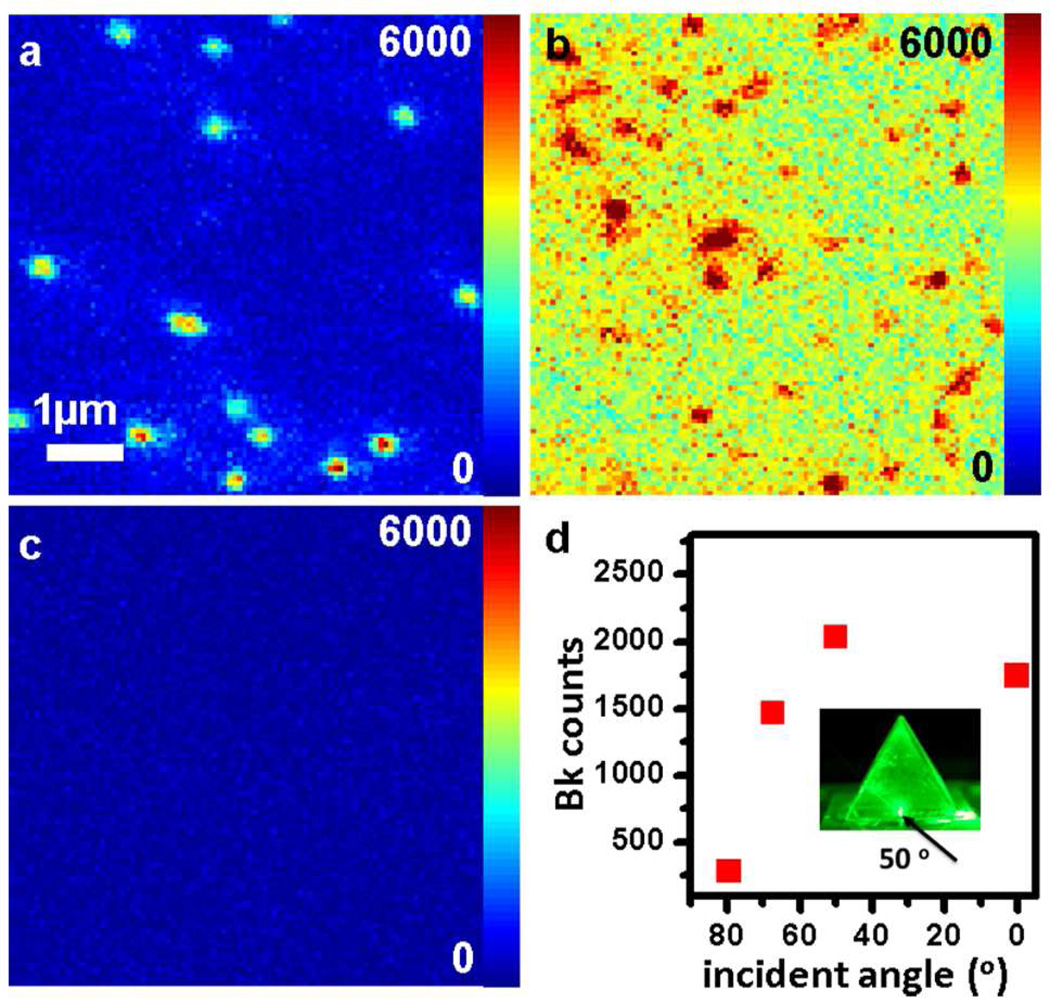
Figure 2.
Typical fluorescence images of (a) dye-pDNA (1 nM) binding to the glass surface at TIRF mode, (b) at epifluorescence mode, and (c) water at epifluorescence mode that confirms the background and noise are dominated by the dye molecules. (d) Average background (Bk) counts at different incident angles with 1 nM dye-cpDNA in the solution (inset shows measuring incident angle with a glass prism).
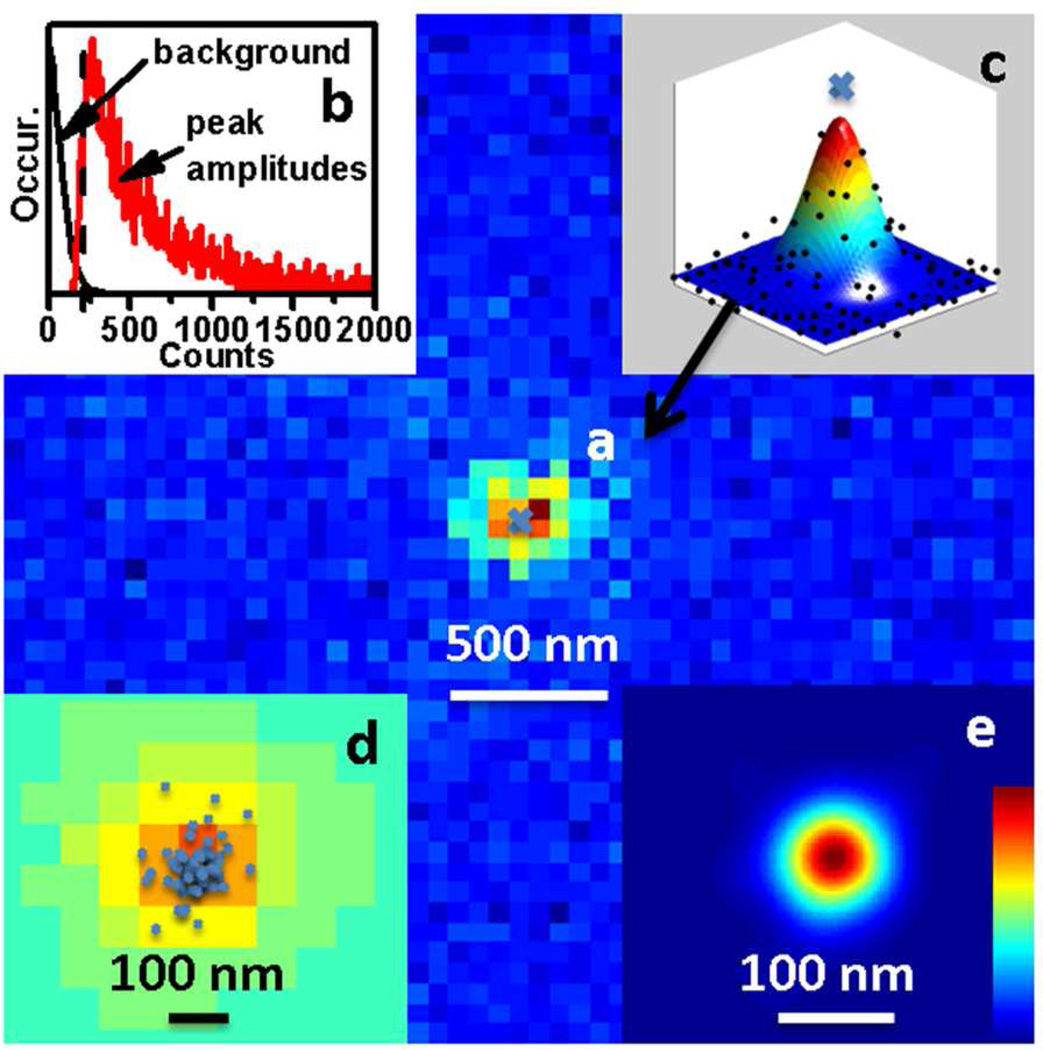
Figure 3.
Data analysis and resolution determination. (a) A fluorescence event is defined on this frame with peak amplitude at least 3 times the background noise. (b) Histograms of background and the amplitudes of the peaks. The dashed line indicates the threshold chosen to distinguish between the background and an adsorption event. (c) The peak is fitted with a 2D Gaussian function for its center position and peak amplitude. (d) The peak centers on all the frames that have events are summarized for this area. (e) Each center is defined as an event and is converted to a virtual Gaussian peak with amplitude = 1 and standard deviation = 30 nm and is converted to a super-resolution image.
The dwell times of freely diffusing molecules were much faster than our observation time per frame (32 ms), rendering them undetectable in TIRF mode until they stochastically bound to the target, tDNA (Fig. 2a). Although these bulk molecules contribute to the background counts, in TIRF mode the contribution of background fluorescence to the image is negligible at the low concentrations of the dye-labeled probe DNA molecules required for single-molecule analysis (~0.6 molecule/µm3, or 1 nM). This is because under our TIRF conditions, in which the penetration depth of the excitation light is ~60 nm from the interface at incident angle of 79°,44,45 the diffusing pDNA and cpDNA have a diffusion coefficient estimated by the Stokes-Einstein relation on the order of 107 nm2/s for small bio-molecules (~10 kd) under normal conditions in water.46 Thus, the average dwell time of diffusing pDNA and cpDNA in-and-out of the excitation volume is on the order of 10−4 s with a diffusion length according to Fick’s law,47 where D is the diffusion coefficient and t is the time. In contrast, outside the TIRF conditions, when the excitation optics are in epifluorescence mode, diffusing molecules are excited and observed in the background of the diffraction-limited area of a fixed molecule, which increases the background level and brings extra noise when diffusing fluorescent probes are present (Fig. 2b) in comparison to pure solvent under the same epifluorescence excitation conditions (Fig. 2c). Fig. 2d quantifies the background counts as a function of incident angle to demonstrate the value of performing the DNA localization measurements under TIRF conditions.
The dynamic adsorption of dye-labeled pDNA molecules on an immobilized tDNA was imaged and the binding events were used to generate super-resolution images (Fig. 3). The dynamic binding events of pDNA to the tDNA were observed as a movie with a frame rate of 16 frames/s. The movie was analyzed with a home-made MATLAB program. The binding events were defined as the fluorescent spots in each frame (Fig. 3a), observed as increased counts in the form of a point spread function in the frames of the movie. A fluorescence event was defined if the maximum pixel intensity of a point spread function was larger than 3 times the background noise on a frame (Fig. 3b) to satisfy >99% confidence for event identification.48 The binding events were then isolated from each frame and fitted with 2D Gaussian functions for their centers (Fig. 3c). The typical size of the point spread function had a standard deviation of ~120 nm, which was defined by the diffraction limit of light and the optics. The centers from all frames were recorded and plotted in one combined image (Fig. 3d). A super-resolution image was generated with the sum of all the centers that were converted to virtual Gaussian peaks with amplitude 1 binding event and standard deviation 30 nm, ~50 nt assuming 0.6 nm/nt49 (Fig. 3e). The actual localization precision could be better than 30 nm because the length of the PEG and DNA linker was over 60 nm, which was dangling on the substrate surface and contributed to the 30 nm uncertainty. This super-resolution image was then analyzed to find the locations of tDNA on the surface. Confidence on the location precision was established when there were multiple events on the same location in multiple frames. These events were either due to the blinking of the bound molecule or desorption and adsorption of molecules on the same location. As such the super-localization of tDNA molecules on the substrate was achieved by fitting the centers of the events.
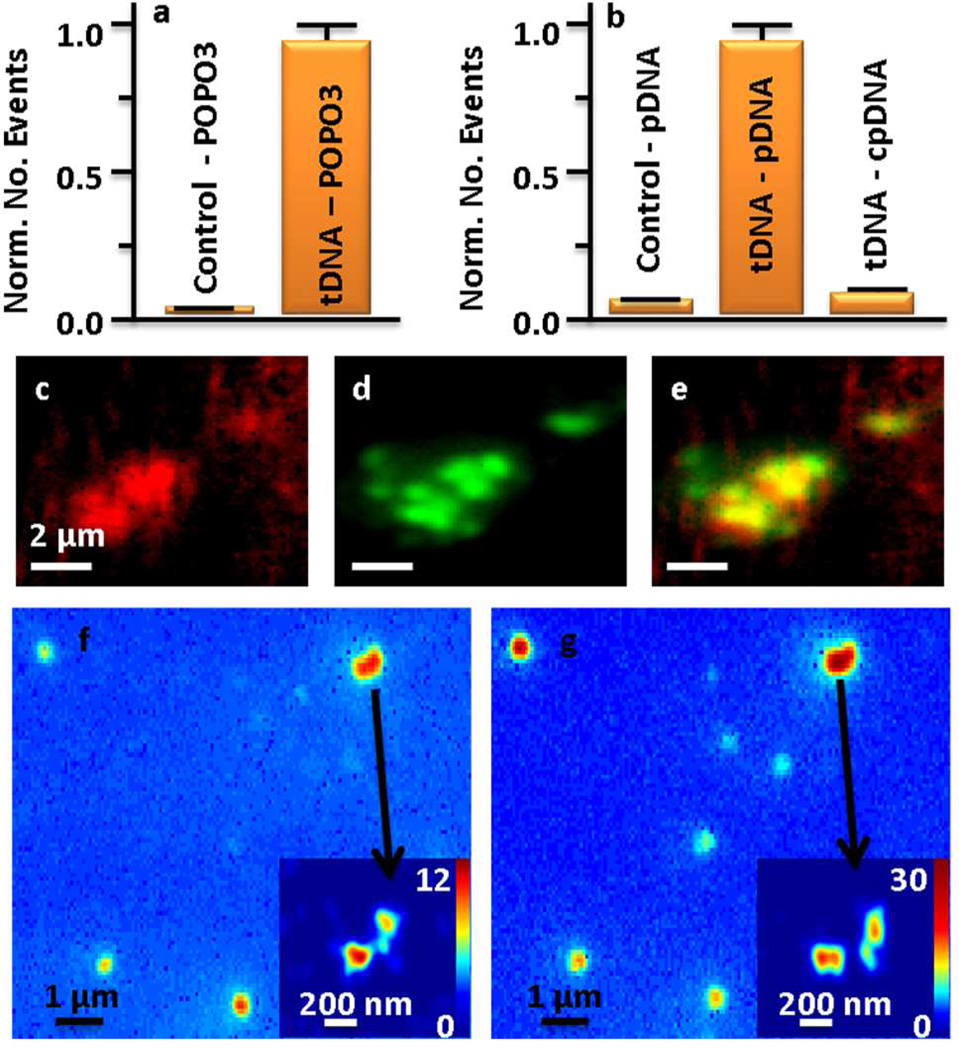
To confirm that tDNA molecules have been successfully grafted onto the substrate and the rest of the substrate surface has been appropriately passivated, three control experiments were carried out. The main concerns are that the pDNA could bind to streptavidin or the defects of the passivation, where positive charges may occur. (1) The successful immobilization of tDNA on the substrate was demonstrated in Fig. 4 by the specific nucleic acid stain dye POPO-3 (Invitrogen) and the pDNA. The POPO-3 dye did not stain the streptavidin modified PEG-biotin control glass substrate (Fig. 4a). The same sample was then exposed to tDNA-biotin for 10 min, washed, and then stained by POPO-3. This confirmed the immobilization of tDNA on the substrate. (2) Because POPO-3 and pDNA could bind to defects on the substrate, the specific binding of the pDNA to the tDNA was also confirmed (Fig. 4b). The PEG substrates to immobilize tDNA molecules were made from grafting of PEG molecules to the aminosilane modified glass slides. The small adsorption number of the pDNA to the PEG substrates confirmed that the amine groups had been well passivated by the PEG molecules. Amine groups, if not passivated, adsorb DNA molecules due to electrostatic interactions. The pDNA that was complementary to the tDNA on the surface, bound well to the tDNA immobilized substrate. The cpDNA that was not complementary to the tDNA, bound an order of magnitude less to the substrate under the same conditions. This value was similar to the amount of events found on an unfunctionalized substrate. Thus, the pDNA was indeed bound to the tDNA through specific complimentary base sequence interactions. (3) The binding sites observed with pDNA were further confirmed by the co-localization of pDNA and TOTO-3 (Fig. 4c–e). TOTO-3 is a nucleic acid stain dye that emits in the red. The adsorption behaviors of the two emitting molecules were monitored in the green and red color channels at the same time. The green Alexa-532 labeled pDNA (1 nM) and the red DNA stain dye TOTO-3 (10 nM) were mixed together in the solution and introduced onto the tDNA modified substrate. The two dyes were excited with different lasers and imaged at two different channels. High levels of co-localization of the two channels (Fig. 4e) were observed, which provides evidence for the specific binding of both molecules to the tDNA.
Figure 4.
Demonstration of the specific adsorption of pDNA to tDNA and super-localization of sequence specific tDNA. (a) Histograms of number of events observed for the adsorption of DNA stain dye POPO-3 to substrates before (control) and after immobilization of tDNA. (b) Histograms of number of events observed for the adsorption of pDNA to substrates before (control) and after immobilization of tDNA, and the minimal adsorption of cpDNA to the tDNA substrate. (c) Sum fluorescence image of the adsorption of TOTO-3 and (d) pDNA to tDNA substrate and (e) overlap of the two images. (f) Sum fluorescence image and (inset) super-resolution image of POPO-3 to tDNA substrate. (g) Sum fluorescence image and (inset) super-resolution image of pDNA to tDNA substrate. The numbers indicated in the super-resolution image color scale bar, quantify the number of adsorption events at each location.
Super-resolution imaging of a specific ssDNA sequence was also demonstrated in areas that had more than one tDNA molecule within the diffraction limit of light. Diffraction-limited images were acquired of both pDNA (Fig. 4f) and POPO-3 (Fig. 4g) bound stochastically to the tDNA, and super-resolution images of both processes (insets) were achieved. The regular diffraction-limited fluorescence images do not distinguish multiple tDNA molecules that were in the area of the diffraction limit of light. However, the super-resolution images (insets) clearly reveal that these sites have multiple tDNA molecules. It is worth noting that pDNA is sequence specific and POPO-3 is not. As such, for a very long tDNA, combinations of POPO-3 and other nucleic acid stains can be used to image the whole tDNA, with pDNA used for optical mapping.
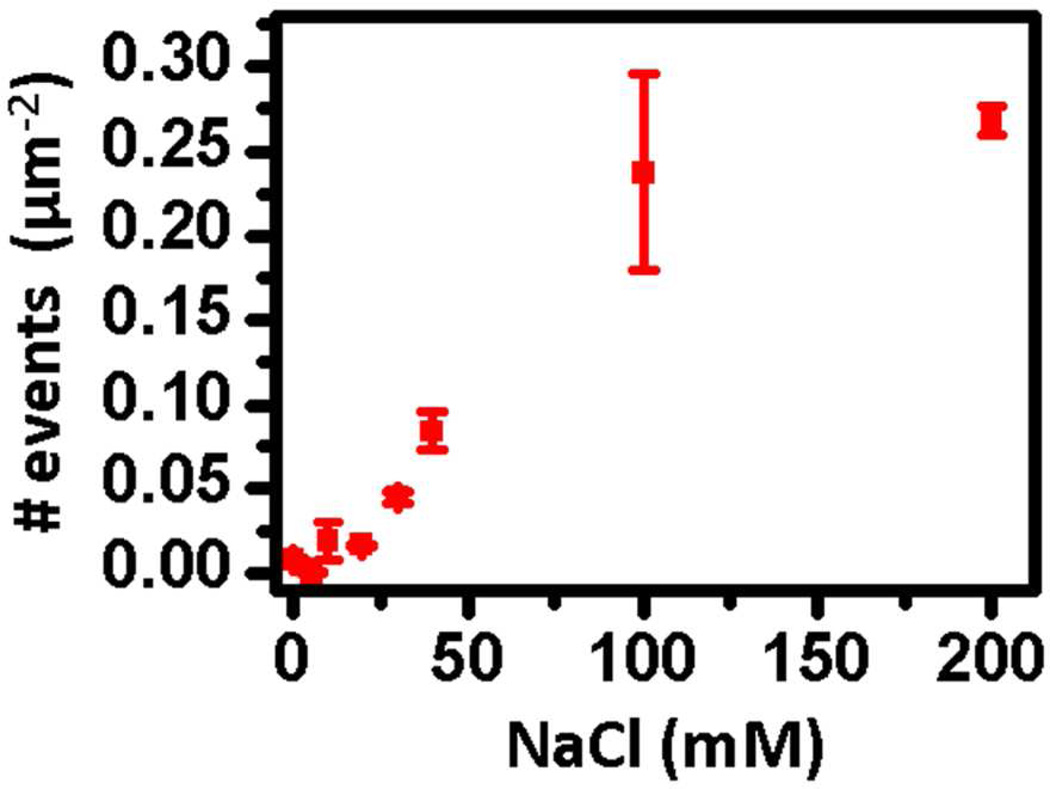
Establishing the minimum DNA sequence length that can be resolved by mbPAINT analysis is an exciting question that is larger than the scope of the current proof-of-concept demonstration, but is only limited by the ability to detect enough photons from a stochastic reversible hybridization event such that it is possible to statistically distinguish the binding from the background. Although a systematic benchmarking characterization will be presented elsewhere, here we demonstrate the possibility of using the solution ionic conditions to tune the hybridization event dynamics for our pDNA over a broad range. Because the hybridization probability of pDNA to tDNA is screened by the electrostatic repulsion between them, the hybridization dynamics can be tuned by controlling the solution ionic strength, and will be required as the probe sequence is systematically reduced. As shown, the number of detected events on each frame increases within 0–100 mM NaCl and approaching saturation after 100 mM NaCl (Fig. 5), but there is a broad tunable dynamic range accessible via this simple method.
Figure 5.
Number of new events per frame as a function of salt concentration in the presence of 25 mM HEPES buffer.
In summary, we have successfully demonstrated that the super-resolution method mbPAINT can be used to identify ssDNA sequences as short as 50 nt. These proof-of-concept experiments could lead to further development of super-resolution mapping of long ssDNA, where further optimization of experimental conditions might be required. In addition to establishing the lower bound of sequence resolution as discussed above, future experiments will include expanding to multi-color parallel sequence determination, quantitative investigation of the binding kinetics, and immobilizing and stretching a longer bacteriophage lambda ssDNA on the substrate for optical mapping.
EXPERIMENTAL
Sample preparation
DNA samples were obtained from Eurofins MWG Operon. The target sequence (tDNA) is: Amino-C6-5’-TTTCTTATGACAGAATGATTCAACTAACATACTTGA-CCACTCCCCACCCAC-AGCTTATGACAGAATGAT TCAACTAACATACTTTT-C7-Amino. The probe sequence (pDNA) is: Alexa532-5’-GTGGGTGGGGAGTGG. The control probe sequence (cpDNApDNA) is: Alexa532-5’-TTCTGGATCACTTCGCGC.
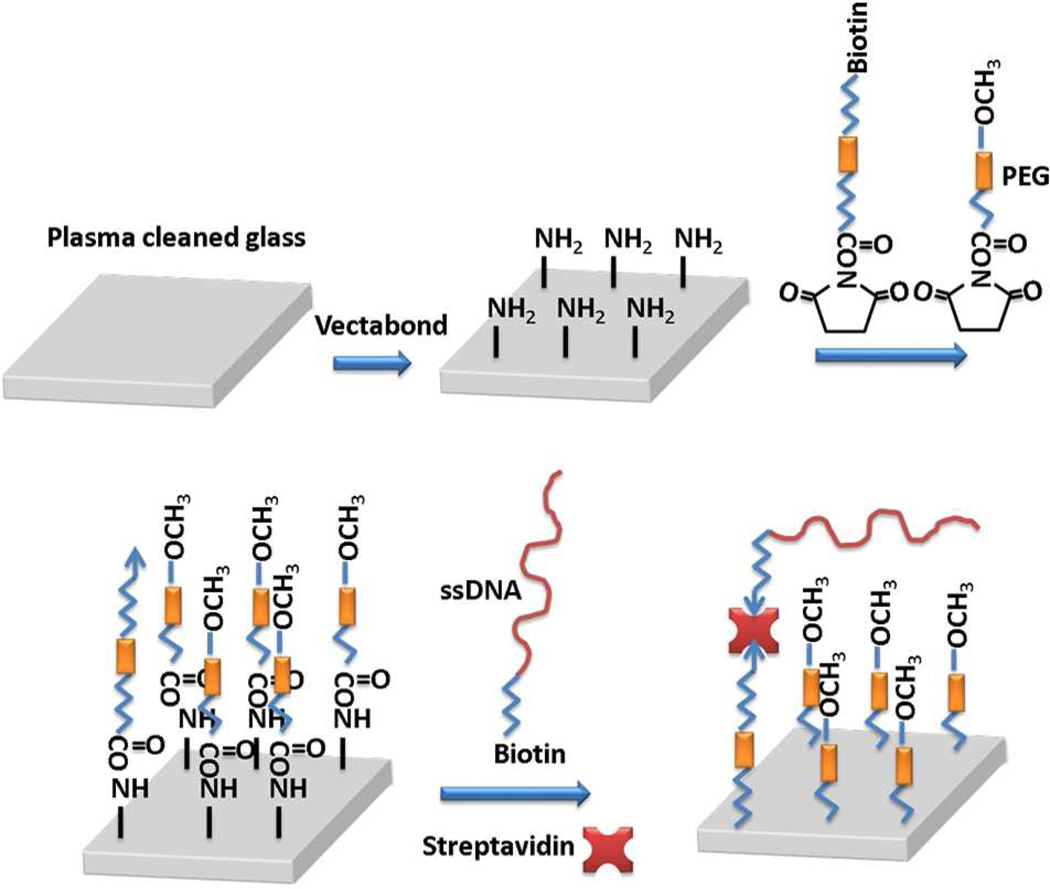
Glass cover slides were cleaned and modified with amine group (Fig. 6). First, glass slides (VWR) were sonicated in 1% detergent water (Liquinox) for 20 min followed by copious rinsing of Millipore DI water. Then the glass slides were soaked in 80 °C NH4OH:H2O2:H2O (1:1:5) solution for 5 min and rinsed with water and blown dry with nitrogen. The slides were oxygen plasma activated for 2 min and grafted in 1% (v/v) aminosilane reagent (Vectabond, Vectorlabs) acetone solution for 5 min followed by soaking in water for 1 min. The glass slides were then rinsed with water and dried by nitrogen for further grafting.
Figure 6.
Substrate preparation and grafting of tDNA.
Biotinylated glass slides were grafted starting from aminosilane functionalized glass slides. Grafting solution was an aqueous mixture of 25% (w/w, mass fraction) methoxypolyethylene glycol 5,000 propionic acid N-succinimidyl ester (>80%, Sigma-Aldrich), 0.25 % (w/w) SUNBRIGHT BI-050TS (Biotin-PEG-COO-MAL, Mw 5000, NOF America Corporation), and 0.8 % (w/w) NaHCO3 (Sigma-Aldrich). Silicone molds (Grace Bio-Labs) were used to shape the target area and the solution was allowed to dry overnight. The excess solution was rinsed off with water, and then the slides were dried with nitrogen. The biotinylated glass slides were sealed with custom hybriwell chambers (Grace Bio-Labs) and dual silicon press-fit tubing connectors (Grace Bio-Labs). Teflon tubing (Tub FEP Red 1685, Western Analytical Products), super flangeless ferrule (P-248X, Western Analytical Products), super flangeless gray delrin (P-252X, Western Analytical Products), female luer (P-628, West Analytical Producets), 1 cc luer slip syringe (MVI), and Genie Plus syringe pump (Kent Scientific) were used to construct a flow system that was attached to each biotin-PEGylated slide.
The tDNA molecules were immobilized onto the biotinylated glasses via biotin-streptavidin interaction. The tDNA was functionalized with biotin by EDC-NHS chemistry.50 Mixture (1 mL) of 1 µM biotin (Sigma-Aldrich), 2 mM 1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC, Sigma-Aldrich), 5 mM N-hydroxysulfosuccinimide (sulfo-NHS, Sigma-Aldrich), 100 mM NaCl (Sigma-Aldrich) and 100 mM HEPES buffer (pH~6, Sigma-Aldrich) were incubated at room temperature for 15 min. Then 100 µL of 10 µM tDNA was added to the solution and the pH value was adjusted to ~8 by adding 10 µL 7.5% NaHCO3 solution. The final solution was reacted at room temperature for ~3 hours. The biotinylated glass with flow cell was incubated with mixture of 0.25 mg/mL streptavidin (Invitrogen), 25 mM HEPES buffer (pH ~7), and 40 mM NaCl for 2 min, and flushed with buffer solution (25 mM HEPES, 40 mM NaCl) three times. The as-synthesized biotin-tDNA (biotin concentration 0.1 nM) in buffer solution was injected into the flow cell, incubated for 1 min, and was flushed with buffer solution three times again.
Fluorescence Imaging
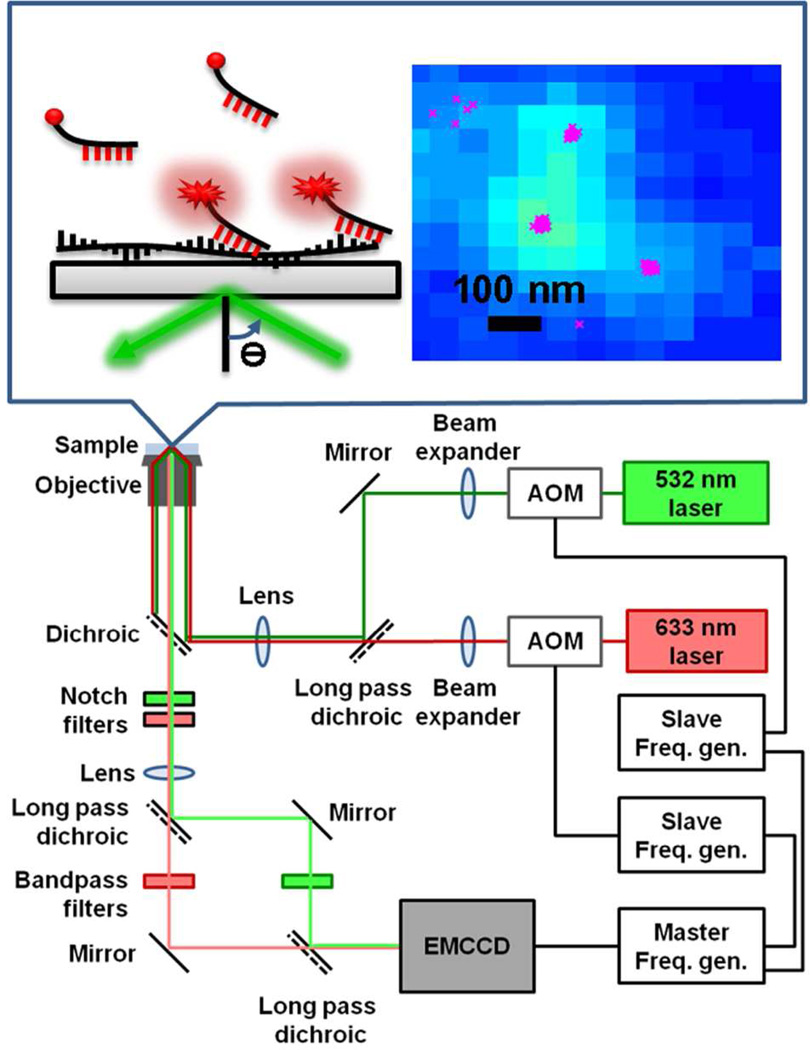
The wide field fluorescence images were obtained with a home-build TIRF microscope (Fig. 7). A 532 nm solid state laser (Coherent, Compass 315M-100SL) was used to excite the Alexa Fluor 532 dye and POPO-3 dye. The objective used was 1.45 NA, 100×, oil-immersion objective (Carl-Zeiss, alpha Plan-Fluar). The TIRF excitation penetrated a 1/e depth of ~85 nm at an intensity of ~5 or 10 W/cm2. A 633 nm HeNe laser (CVI Melles Griot, 25-LHP-925-249) with intensity ~5 W/cm2 was used to excite the TOTO-3 dye. Emission was collected in an epi-fluorescence setup and was detected on an electron-multiplied charge coupled device (Andor, iXon 897) at an integration time of 30 ms, frame rate of 16.1 Hz, and electron multiplying gain of 300.
Figure 7.
Scheme of the TIRF microscope, including acousto-optic modulator (AOM) controlled by frequency generators (Freq. gen.) and electron-multiplying charge coupled device (EMCCD) detector. The top scheme shows the experimental geometry and a cumulative fluorescence image of 1000 frames with the x-dots representing the fitted centers of individual fluorescence binding events on separated frames.
Before imaging, the detection solutions were flowed into the sample chamber at 15 µL/min for ~5 min using Genie plus flow system. The Alexa532-pDNA solution is a mixture of 1 nM pDNA, 25 mM HEPES buffer, 40 mM NaCl, 1% (v/v, volume fraction) 2-mercaptoethanol and ~70% (v/v) pre-saturated 6-hydroxy-2,5,7,8-tetramethylchroman-2-carboxylic acid (Trolox, Sigma-Aldrich) solution. The DNA stain dye solution is 10 nM POPO-3 or TOTO-3 (Invitrogen) in 25 mM of HEPES buffer, 40 mM NaCl, 1% (v/v) mercaptoethanol and ~70% (v/v) pre-saturated Trolox solution. The solution flow rate was 15 µL/min during imaging. The incident angle of the laser was measured with a glass prism instead of the sample such that the laser transmitted out of the prism.
ACKNOWLEDGMENT
C.F.L. thanks National Science Foundation (NSF) grants CBET-1134417, and CHE-1151647; the Welch Foundation grant C-1787; and the National Institutes of Health (NIH) grant GM94246-01A1. L.K. thanks the NSF Graduate Research Fellowship Grant 0940902. We also thank David Cooper, Joey Tauzin, Chad Byers, Stephan Link and his group for helpful discussions.
Footnotes
The authors declare no competing financial interests.
REFERENCES
- 1.Lin J, Qi R, Aston C, Jing J, Anantharaman TS, Mishra B, White O, Daly MJ, Minton KW, Venter JC, Schwartz DC. Science. 1999;285:1558–1562. doi: 10.1126/science.285.5433.1558. [DOI] [PubMed] [Google Scholar]
- 2.Jo K, Dhingra DM, Odijk T, de Pablo JJ, Graham MD, Runnheim R, Forrest D, Schwartz DC. Proc. Natl. Acad. Sci. USA. 2007;104:2673–2678. doi: 10.1073/pnas.0611151104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neely RK, Dedecker P, Hotta JI, Urbanaviciute G, Klimasauskas S, Hofkens J. Chem. Sci. 2010;1:453–460. [Google Scholar]
- 4.Dong Y, Xie M, Jiang Y, Xiao N, Du X, Zhang W, Tosser-Klopp G, Wang J, Yang S, Liang J, Chen W, Chen J, Zeng P, Hou Y, Bian C, Pan S, Li Y, Liu X, Wang W, Servin B, Sayre B, Zhu B, Sweeney D, Moore R, Nie W, Shen Y, Zhao R, Zhang G, Li J, Faraut T, Womack J, Zhang Y, Kijas J, Cockett N, Xu X, Zhao S, Wang J, Wang W. Nat. Biotechnol. 2013;31:135–141. doi: 10.1038/nbt.2478. [DOI] [PubMed] [Google Scholar]
- 5.Lam ET, Hastie A, Lin C, Ehrlich D, Das SK, Austin MD, Deshpande P, Cao H, Nagarajan N, Xiao M, Kwok PY. Nat. Biotechnol. 2012;30:771–776. doi: 10.1038/nbt.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fernández-Suárez M, Ting AY. Nat. Rev. Mol. Cell Biol. 2008;9:929–943. doi: 10.1038/nrm2531. [DOI] [PubMed] [Google Scholar]
- 7.van de Linde S, Heilemann M, Sauer M. Annu. Rev. Phys. Chem. 2012;63:519–540. doi: 10.1146/annurev-physchem-032811-112012. [DOI] [PubMed] [Google Scholar]
- 8.Huang B, Babcock H, Zhuang X. Cell. 2010;143:1047–1058. doi: 10.1016/j.cell.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biteen JS, Thompson MA, Tselentis NK, Bowman GR, Shapiro L, Moerner WE. Nat. Methods. 2008;5:947–949. doi: 10.1038/NMETH.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kural C, Kim H, Syed S, Goshima G, Gelfand VI, Selvin PR. Science. 2005;308:1469–1472. doi: 10.1126/science.1108408. [DOI] [PubMed] [Google Scholar]
- 11.Willig KI, Rizzoli SO, Westphal V, Jahn R, Hell SW. Nature. 2006;440:935–939. doi: 10.1038/nature04592. [DOI] [PubMed] [Google Scholar]
- 12.Baumgartel V, Muller B, Lamb DC. Viruses. 2012;4:777–799. doi: 10.3390/v4050777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou X, Andoy NM, Liu G, Choudhary E, Han KS, Shen H, Chen P. Nat. Nanotechnol. 2012;7:237–241. doi: 10.1038/nnano.2012.18. [DOI] [PubMed] [Google Scholar]
- 14.Roeffaers MB, De Cremer G, Libeert J, Ameloot R, Dedecker P, Bons AJ, Buckins M, Martens JA, Sels BF, De Vos DE, Hofkens J. Angew. Chem., Int. Ed. 2009;48:9285–9289. doi: 10.1002/anie.200904944. [DOI] [PubMed] [Google Scholar]
- 15.Hell SW, Wichmann J. Opt. Lett. 1994;19:780–782. doi: 10.1364/ol.19.000780. [DOI] [PubMed] [Google Scholar]
- 16.Hell SW, Kroug M. Appl. Phys. B. 1995;60:495–497. [Google Scholar]
- 17.Heintzmann R, Jovin TM, Cremer C. J. Opt. Soc. Am. A. 2002;19:1599–1609. doi: 10.1364/josaa.19.001599. [DOI] [PubMed] [Google Scholar]
- 18.Gustafsson MG. Proc. Natl. Acad. Sci. USA. 2005;102:13081–13086. doi: 10.1073/pnas.0406877102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dertinger T, Colyer R, Iyer G, Weiss S, Enderlein J. Proc. Natl. Acad. Sci. USA. 2009;106:22287–22292. doi: 10.1073/pnas.0907866106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bornfleth H, Satzler K, Eils R, Cremer C. J. Microsc. 1998;189:118–136. doi: 10.1006/excr.1998.3934. [DOI] [PubMed] [Google Scholar]
- 21.Lemmer P, Gunkel M, Weiland Y, Muller P, Baddeley D, Kaufmann R, Urich A, Eipel H, Amberger R, Hausmann M, Cremer C. J. Microsc. 2009;235:163–171. doi: 10.1111/j.1365-2818.2009.03196.x. [DOI] [PubMed] [Google Scholar]
- 22.Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 23.Hess ST, Girirajan TP, Mason MD. Biophys. J. 2006;91:4258–4272. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rust MJ, Bates M, Zhuang X. Nat. Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heilemann M, van de Linde S, Schuttpelz M, Kasper R, Seefeldt B, Mukherjee A, Tinnefeld P, Sauer M. Angew. Chem., Int. Ed. 2008;47:6172–6176. doi: 10.1002/anie.200802376. [DOI] [PubMed] [Google Scholar]
- 26.Wombacher R, Heidbreder M, van de Linde S, Sheetz MP, Heilemann M, Cornish VW, Sauer M. Nat. Methods. 2010;7:717–719. doi: 10.1038/nmeth.1489. [DOI] [PubMed] [Google Scholar]
- 27.Sharonov A, Hochstrasser RM. Proc. Natl. Acad. Sci. USA. 2006;103:18911–18916. doi: 10.1073/pnas.0609643104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hofmann M, Eggeling C, Jakobs S, Hell SW. Proc. Natl. Acad. Sci. USA. 2005;102:17565–17569. doi: 10.1073/pnas.0506010102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walder R, Nelson N, Schwartz DK. Nat. Commun. 2011;2:515. doi: 10.1038/ncomms1530. [DOI] [PubMed] [Google Scholar]
- 30.Cheng M-C, Leske AT, Matsuoka T, Kim BC, Lee J, Burns MA, Takayama S, Biteen JS. J. Phys. Chem. B. 2012;117:4406–4411. doi: 10.1021/jp307635v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson RE, Larson DR, Webb WW. Biophys. J. 2002;82:2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simonson PD, Rothenberg E, Selvin PR. Nano Lett. 2011;11:5090–5096. doi: 10.1021/nl203560r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cox S, Rosten E, Monypenny J, Jovanovic-Talisman T, Burnette DT, Lippincott-Schwartz J, Jones GE, Heintzmann R. Nat. Methods. 2012;9:195–200. doi: 10.1038/nmeth.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baday M, Cravens A, Hastie A, Kim H, Kudeki DE, Kwok PY, Xiao M, Selvin PR. Nano Lett. 2012;12:3861–3866. doi: 10.1021/nl302069q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Flors C, Ravarani CN, Dryden DT. ChemPhysChem. 2009;10:2201–2204. doi: 10.1002/cphc.200900384. [DOI] [PubMed] [Google Scholar]
- 36.Schoen I, Ries J, Klotzsch E, Ewers H, Vogel V. Nano Lett. 2011;11:4008–4011. doi: 10.1021/nl2025954. [DOI] [PubMed] [Google Scholar]
- 37.Baday M, Cravens A, Hastie A, Kim H, Kudeki DE, Kwok P-Y, Xiao M, Selvin PR. Nano Lett. 2012;12:3861–3866. doi: 10.1021/nl302069q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jungmann R, Steinhauer C, Scheible M, Kuzyk A, Tinnefeld P, Simmel FC. Nano Lett. 2010;10:4756–4761. doi: 10.1021/nl103427w. [DOI] [PubMed] [Google Scholar]
- 39.Kastantin M, Walder R, Schwartz DK. Langmuir. 2012;28:12443–12456. doi: 10.1021/la3017134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Darugar Q, Kim H, Gorelick RJ, Landes CF. J. Virol. 2008;82:12164–12171. doi: 10.1128/JVI.01158-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taylor NJ, Darugar Q, Kourentzi K, Willson RC, Landes CF. Biochem. Biophys. Res. Commun. 2008;373:213–218. doi: 10.1016/j.bbrc.2008.05.191. [DOI] [PubMed] [Google Scholar]
- 42.Cooper D, Uhm H, Tauzin LJ, Poddar N, Landes CF. ChemBioChem. 2013;14:1075–1080. doi: 10.1002/cbic.201300030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bell JC, Plank JL, Dombrowski CC, Kowalczykowski SC. Nature. 2012;491:274–278. doi: 10.1038/nature11598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Axelrod D, Burghardt TP, Thompson NL. Annu. Rev. Biophys. Bioeng. 1984;13:247–268. doi: 10.1146/annurev.bb.13.060184.001335. [DOI] [PubMed] [Google Scholar]
- 45.Gell C, Berndt M, Enderlein J, Diez S. J. Microsc. 2009;234:38–46. doi: 10.1111/j.1365-2818.2009.03147.x. [DOI] [PubMed] [Google Scholar]
- 46.Young ME, Carroad PA, Bell RL. Biotechnol. Bioeng. 1980;22:947–955. [Google Scholar]
- 47.Fick A. Philos. Mag. 1855;10:30–39. [Google Scholar]
- 48.Peterson EM, Harris JM. Anal. Chem. 2010;82:189–196. doi: 10.1021/ac901710t. [DOI] [PubMed] [Google Scholar]
- 49.Zhang Y, Zhou H, Ou-Yang ZC. Biophys. J. 2001;81:1133–1143. doi: 10.1016/S0006-3495(01)75770-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pieper JS, Hafmans T, Veerkamp JH, van Kuppevelt TH. Biomaterials. 2000;21:581–593. doi: 10.1016/s0142-9612(99)00222-7. [DOI] [PubMed] [Google Scholar]