Abstract
Genetic studies have identified dozens of autism spectrum disorder (ASD) susceptibility genes, raising two critical questions: 1) do these genetic loci converge on specific biological processes, and 2) where does the phenotypic specificity of ASD arise, given its genetic overlap with intellectual disability (ID)? To address this, we mapped ASD and ID risk genes onto co-expression networks representing developmental trajectories and transcriptional profiles representing fetal and adult cortical laminae. ASD genes tightly coalesce in modules that implicate distinct biological functions during human cortical development, including early transcriptional regulation and synaptic development. Bioinformatic analyses suggest translational regulation by FMRP and transcriptional co-regulation by common transcription factors connect these processes. At a circuit level, ASD genes are enriched in superficial cortical layers and glutamatergic projection neurons. Furthermore, we show that the patterns of ASD and ID risk genes are distinct, providing a novel biological framework for investigating the pathophysiology of ASD.
Keywords: gene networks, systems biology, exome, rare variants, Intellectual disability, human cortical development, gene expression, FMRP, Satb1, MEF2, RNA-seq
Introduction
Autism Spectrum Disorder (ASD) is a heterogeneous neurodevelopmental disorder, in which hundreds of genes have been implicated (Berg and Geschwind, 2012; Geschwind and Levitt, 2007). Analysis of copy number variation (CNV) and exomesequencing (Iossifov et al., 2012; Neale et al., 2012; O'Roak et al., 2012b; Sanders et al., 2012)have identified rare de novo variants (RDNVs) that alter dozens of protein coding genes in ASD, none of which account for more than 1% of ASD cases(Devlin and Scherer, 2012). This, and the fact that a significant fraction (40-60%) of ASD is explained by common variation(Klei et al., 2012), points to a heterogeneous genetic architecture.
These findings raise several issues. Based on the background human mutation rate (MacArthur et al., 2012), most genes affected by only one observed RDNV to date are likely false positives that do not increase risk for ASD(Gratten et al., 2013). It is therefore essential to develop approaches that prioritize singleton variants, especially missense mutations. Furthermore, given the heterogeneity of ASD, it would valuable to identify common pathways, cell-types, or circuits disrupted within ASD itself. Recent studies combining gene expression, protein-protein interactions (PPIs), and other systematic gene annotation resources suggest some molecular convergence in subsets of ASD risk genes(Ben-David and Shifman, 2012; Gilman et al., 2011; Sakai et al., 2011; Voineagu et al., 2011). Yet, it remains unclear how the large number of genes implicated through different methods may converge to affect human brain development, which is critical to a mechanistic understanding of ASD (Berg and Geschwind, 2012). Additionally, ASD has considerable overlap with ID at the genetic level, so identifying molecular pathways and circuits that confer the phenotypic specificity of ASD would be of considerable utility (Geschwind, 2011; Matson and Shoemaker, 2009).
Here, we took a stepwise approach to determine if genes implicated in ASD affect convergent pathways during in vivo human neural development, and whether they are enriched in specific cells or circuits (Figure 1A). First, we constructed transcriptional networks representing genome-wide functional relationships during fetal and early postnatal brain development and mapped genes from multiple ASD and ID resources to these networks. We then assessed shared neurobiological function among these genes, including co-regulatory relationships and enrichment in layer-specific patterns from micro-dissected human fetal and adult primate cortical laminae. We used validation in independent in vivo and in vitro expression data and additional functional evidence (shared annotated pathways and PPIs) to confirm shared function among genes, and we replicated the enrichment analyses in independent data to ensure robustness. Our integration of an unsupervised network analysis with large gene sets from multiple resources permits rigorous interrogation of biological convergence and specificity in ASD that takes its heterogeneity into consideration and enables comparison of ASD with ID.
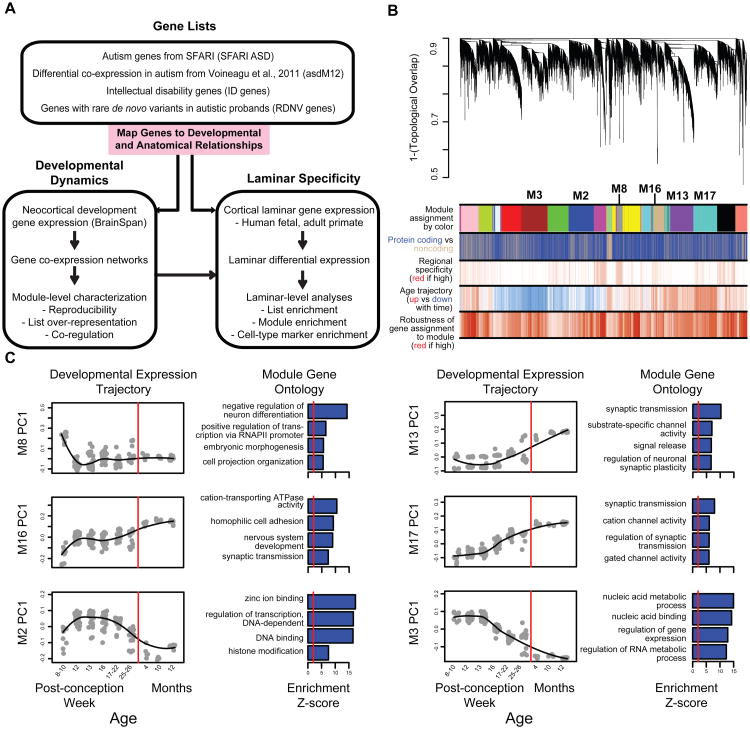
Figure 1. Methodological overview and co-expression network analysis.
(A) Flowchart of the overall approach.
(B) Network analysis dendrogram showing modules based on the co-expression topological overlap of genes throughout development. Color bars below give information on module membership, gene biotype, cortical region specificity, age trajectory, and robustness of module assignment.
(C) Module characterization including GO enrichment and trajectory throughout development. The fit line represents locally weighted scatterplot smoothing (Extended Experimental Methods). GO enrichments are adjusted for multiple comparisons (FDR < 0.01), and reported Z-scores represent relative enrichment, with the red line at Z = 2. See also Table S1 and Figure S1.
Results
Genome-wide co-expression networks reflect biological processes essential to human neocortical development
We reasoned that transcriptomic data from human neocortex would inform our understanding of ASD pathophysiology, as the cerebral cortex has been consistently implicated in ASD pathophysiology by multiple modalities (Amaral et al., 2008; Ecker et al., 2012; Geschwind, 2011; Rubenstein, 2010; Voineagu et al., 2011). We focused on gene expression from cortical development spanning post-conception week (PCW) 8 to 12 months after birth, as this time period reflects many critical molecular processes that orchestrate brain circuit formation that could be disrupted by genetic hits in ASD(Andersen, 2003; Courchesne et al., 2011).
We constructed networks of gene relationships agnostic to ASD candidate genes based on BrainSpan whole-genome transcriptomic data collected by RNA-seq (www.brainspan.org). We applied signed, weighted gene co-expression network analysis (WGCNA, Experimental Procedures; Zhang and Horvath, 2005) and identified17 co-expression modules (labeled numerically, e.g. M8, and by color, e.g. magenta, see Table S1B for module details). These modules represent genes that share highly similar expression patterns during cortical development(Figure 1B), and additional analyses show that these modules identify highly significant shared expression patterns that are replicated in independent data from both in vivo and in vitro human neural development (Figures S1A-C, Extended Experimental Procedures).
First, we investigated each module's developmental trajectory by calculating the module eigengene (ME, the first principal component of the module) and assessed shared function among genes within the module by enrichment for Gene Ontology (GO) annotation terms. Representative examples for up and down-regulated modules are shown in Figure 1C. Module eigengenes for M13, M16, and M17 increase during early cortical development and are each enriched for the GO term synaptic transmission (Figure 1C). M16 is upregulated the earliest, starting at PCW10 and its hubs (most inter-connected genes based on correlation to the ME, kME) include genes coding for the structural synaptic proteins SV2A and NRXN1. M16 GO terms include cation transporter activity, homophillic cell adhesion, and nervous system development, consistent with early development of synaptic ultrastructure. M17 represents a later phase of synaptic maturation, as it is upregulated after PCW 13 and its hubs include CAMK2B and CACNA1C, which are important for calcium-dependent regulation of synaptic activity. M13 increases last, after PCW16, and its hubs include the NMDA and GABA receptor subunits GRIN2A and GABRA1, while GO terms include substrate-specific channel activity and regulation of neuronal synaptic plasticity. These three modules have closely aligned, yet distinct developmental trajectories that likely reflect sequential phases of synaptic development, maturation, and function, all of which are essential to the development of the cerebral cortex.
In contrast, M2 and M3 have anti-correlated trajectories to M13, M16, and M17 (r = -0.46 to -0.96, Table S1B), and are enriched in GO terms associated with DNA binding and transcriptional regulation (Figure 1C). Expression inM3 is initially upregulated and then decreases after PCW 12, suggesting its functions may be most important prior to M2, which is upregulated after PCW10 and peaks later (PCW 12 to PCW 22). Given the GO enrichment and anti-correlation to the synaptic module MEs, genes in these modules may be critical to orchestrating processes such as progenitor proliferation and cell fate specification via initial repression followed by de-repression of neuronal genes (Srinivasan et al., 2012). Furthermore, many of the genes found in M2 and M3 are part of well-studied chromatin remodeling complexes, most notably the BAF complex (ARID1A and SMARCA4 in M2; ARID1B, SMARCB1, SMARCC1, SMARCC2, SMARCD1, ARID2, DPF2, BCL11A, BCL11B, and ACTL6A in M3), that have recently been linked to neural differentiation and neurodevelopmental disorders (Ronan et al., 2013; Yoo et al., 2009).
Since, positive correlations among genes also reflect pair-wise interactions between proteins (Ramani et al., 2008),enrichment for protein-protein interactions within modules provides an independent line of validation for shared function in these modules at the protein level. We combined all known PPIs from In Web (Rossin et al., 2011)and BioGRID (Stark, 2006) into one network,comprising251,881 interactions between 18,384 proteins, and observed that12/17 of all co-expression modules, including all the modules in Figure 1C are enriched for PPI after stringent multiple testing correction (p <0.003, Table S1B). Overall, 10/17 co-expression modules are preserved in independent gene expression data sets, enriched for GO terms, and enriched for PPI, while 2/17 are enriched for two of these three criteria. These results demonstrate the utility of a systems biology approach: instead of analyzing lists of thousands of genes regulated during development, we focused on this set of 12 reproducible and biologically meaningful modules sharing distinct expression patterns and biological functions. An interactive network is available at our website for graphical exploration of individual genes and their relationships.
Genes implicated in ASD are highly co-expressed during human cortical development
We next asked whether genes associated with risk for ASD converge on common biological processes. We compiled a set of 155 ASD genetic risk candidates from the Simons Foundation Autism Research Initiative (SFARI) AutDB database (Basu et al., 2009), which we refer to as SFARI ASD. The SFARI ASD list is a manually curated set of candidate genes implicated by common variant association, candidate gene studies, genes within ASD-associated CNV, and, to a lesser extent, syndromic forms of ASD (Experimental Procedures). We mapped this gene set to the protein coding genes in the developmental co-expression network and observed that SFARI ASD genes are most over-represented in M16 (p = 0.0024, odds-ratio (OR) = 2.9, 95% confidence interval = [1.4-5.5], false discovery rate (FDR) < 0.05), and less so in M13 and M17 (Figure 2A),
Figure 2. Enrichment of SFARI ASD, asdM12 and ID genes in developmental networks.
(A) Module-level enrichment for gene sets from a curated set of ASD risk genes (SFARI ASD), a curated set of ID genes (“ID all”), and an unbiased set of ASD risk genes (asdM12). Overlapping (ASD/ID overlap) and non-overlapping sets (“ASD only”, “ID only”) are also shown. All enrichment values for over-represented lists with p < 0.05, OR > 1 are shown to demonstrate enrichment trends (*p < 0.05, **FDR < 0.05).
(B), (C) and (D) show network plots for M13, M16, and M17 respectively. Most hub genes overlapping with SFARI ASD and asdM12 enrichment are not the same, showing that enrichment of these two sets is not driven by a narrow shared subset of genes.
Network plots comprise the top 200 connected genes (based on kME, a measure of intramodular connectivity) and their top 1000 connections in the sub-network. Genes with membership in SFARI ASD, asdM12, or the “ID all” list are labeled and plotted according to multidimensional scaling (MDS) of gene expression correlations, which graphs genes with similar expression patterns closer to each other. See also Table S2.
We also examined a set of ASD genes previously shown to be dysregulated in postmortem ASD temporal and frontal cortex (asdM12; Voineagu et al., 2011), which represents a shared molecular pathology in ASD brain identified in an unbiased, genome-wide manner. TheasdM12gene set was strongly enriched in the same three modules as SFARI ASD genes, M13, M16 and M17 (asdM12-M13, p = 3.0×10-15, OR 3.6 [2.7-4.8]; asdM12-M16, p = 3.5×10-15, OR 3.9 [2.8-5.3]; asdM12-M17, p = 1.0×10-7, OR 2.5 [1.8-3.4]; each at FDR< 0.05). A remarkable 42% of asdM12 and 25% of the SFARI ASD sets are found in one of these three modules. Our analysis, which uses gene sets identified based on different methods (only 15 genes overlap between SFARI ASD and asdM12), converges onto three modules involved in prenatal and early postnatal synaptic development.
We next hypothesized that mapping ID genes to this network would enable us to assess whether ASD susceptibility genes show any specificity in their developmental expression patterns. We compiled an extensive set of high confidence genes implicated in monogenic forms of ID from multiple publications (In low and Restifo, 2004; Lubs et al., 2012; Ropers, 2008; van Bokhoven, 2011), referred to as “ID all” (see Experimental Procedures). Remarkably, this set of364 genes expressed in human neocortexis not enriched in any of the 12co-expression modules. Importantly, this lack of enrichment is at a relaxed threshold that reduces the risk of false negatives (uncorrected p > 0.05). Removing the small set of 37 genes (<10%) that overlap between ASD and ID to establish exclusive sets (“ASD only”, “ID only”) further confirms that ASD genes exhibit enrichment, while ID genes do not (Figure 2A, Table S2B). Thus, it is genes connected with the ASD phenotype that are enriched in 3 specific transcriptional modules related to synaptic function during development, but not those that have been related solely to ID.
RDNVs are highly enriched in two co-expression modules in early fetal development
Additional evidence implicating specific genes in ASD comes from whole-exome sequencing in families (Iossifov et al., 2012; Neale et al., 2012; O'Roak et al., 2012b; Sanders et al., 2012), which has identified many rare protein disrupting variants (nonsense, splice-site, frameshift) over-represented in individuals with ASD compared to their unaffected siblings (OR >2). This evidence is largely distinct from the evidence implicating genes in SFARI ASD and asdM12, as it is from purely non-inherited, rare variation discovered in an unbiased, genome-wide manner. We therefore asked whether RDNV-affected genes found in ASD probands shared biological function. We also tested silent RDNVs since they should not exhibit a similar pattern of functional enrichment, providing a key control for gene size, GC content, and other features affecting mutability (Michaelson et al., 2012).
We first tested for enrichment using RDNVs from three studies sharing similar coverage criteria and variant calling methodology (Neale et al., 2012; O'Roak et al., 2012b; Sanders et al., 2012), representing 622 ASD probands and 222 unaffected siblings. Strikingly, genes expressed during development and affected by protein disrupting RDNVs in probands (60 genes, Table S2A, Discovery Set) are significantly enriched in two modules, M2 and M3, which exhibithighly similar developmental trajectories and functional enrichment, indicative of remarkable biological specificity. Eight genes harboring protein disrupting RDNVs are enriched in M2 (p = 0.006, OR = 3.2 [1.3-6.8]; FDR < 0.05) and 10 are enriched in M3 (p = 0.0011, OR = 3.6 [1.6-7.2]; FDR < 0.05). A trend for enrichment is observed for M16 as well, but this does not pass the FDR threshold. For comparison, genes affected by RDNVs in unaffected siblings or affected by silent mutations are not enriched in any modules (Table S2B, p > 0.05). Since missense RDNVs are only weakly over-represented in ASD (Sanders et al., 2012),we reasoned that overlap with network modules might prioritize specific subsets of this RDNV class. We find that a subset of missense RDNV affected genes is over-represented in the same pathways as the more deleterious protein disrupting RDNVs (M2 and M3, Table S2B). Taken together, out of 385protein disrupting or missense RDNV affected genes expressed in brain, 34 are found in M2 (p = 2.9 × 10-4, OR = 2.1 [1.4-3.0], FDR < 0.05) and 41 in M3 (p = 2.3 × 10-5, OR = 2.2 [1.5-3.1], FDR < 0.05). There is no enrichment for this combined set in any other modules. Furthermore, the combined set of protein disrupting and missense RDNVs from unaffected siblings was not found enriched any modules (p > 0.05).
We further validated the observed RDNV enrichment pattern in M2 and M3 in an independent set of patients from a study with more stringent RDNV calling criteria (Iossifov et al., 2012). In this additional set of 343 ASD probands and unaffected siblings, we found that the patterns of RDNV enrichment replicated, with the set of protein disrupting and missense RDNVs from ASD probands enriched specifically in M2 and M3 (p < 0.05), and RDNVs from siblings and silent RDNVS not enriched in any set (Table S2B, Replication Set). Combining across all studies, we find that out of 598protein disrupting or missense RDNV affected genes expressed in brain, 52 are in M2 (p = 9.6 × 10-6, OR = 2.0 [1.5-2.8]) and 61 are in M3 (p = 8.5 × 10-7, OR = 2.1 [1.6-2.8]). Importantly, the enrichment pattern across modules is not only replicated in the independent set, but is stronger in the combined set, is robust to perturbations in module composition (Figure S3A), and is not driven by variants from any one study (Table S2C-D). We show the enrichment pattern of this combined set across 965 ASD probands and 565 unaffected siblings in Figure 3A and use this combined set for the remainder of our analyses.
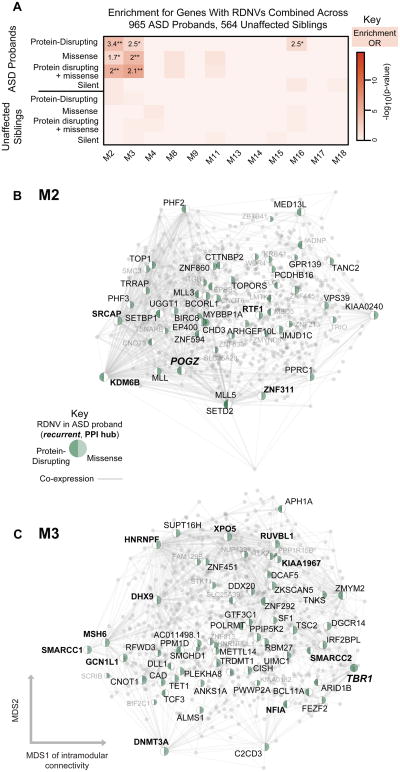
Figure 3. Enrichment of genes affected by RDNVs in developmental networks.
(A) Module-level enrichment for multiple categories of RDNV in ASD affected probands and unaffected siblings combined across four studies. All enrichment values for over represented lists with p < 0.05, OR > 1 are shown to demonstrate enrichment trends (*p < 0.05, **validated in replication set) marked. M2 and M3 are strongly enriched for protein disrupting and missense RDNV affected genes in probands. Enrichment for genes affected by silent RDNVs in probands and RDNV gene sets affected in siblings represent control gene sets and do not show enrichment.
(B) and C) show network plots for M2 and M3, with all genes plotted and all genes carrying RDNVs displayed.
Network plots show all genes in the module with protein disrupting or missense RDNV-affected genes highlighted. Those with high intramodular connectivity (kME > 0.75) are labeled in black, with the rest in grey. The top 1000 connections are shown, and genes are plotted according to the multidimensional scaling (MDS) of co-expression as in Figure 2. See also Figure S2-3 and Table S2.
We next asked whether M2 and M3 prioritized functional subsets of genes with RDNVs. We confirmed that RDNV-affected genes in M2 and M3 are significantly enriched for interactions at a protein level (Figure S2A-D), and highlight genes that are both PPI hubs and co-expression hubs in Figure 3B-C. Furthermore, M2 and M3 genes harboring RDNVs are also more dosage sensitive, as evidenced by the significant increase in the probability of haploinsufficiency (P(HI), Extended Experimental Methods) among genes affected by these mutation classes(Huang et al., 2010; Luo et al., 2012). This is consistent with the heterozygous state of variants observed in ASD probands. Overall, a remarkable proportion, 113/598 (19%) of genes affected by known RDNVs are co-expressed in two modules reflecting similar temporal trends of highexpression in cortex during the neurodevelopmental period of early neuronal fate determination, migration, and cortical lamination. Of note, as with M13, M16, and M17, which were enriched for asdM12 and SFARI ASD, ID genes showed no enrichment in M2 or M3 (p > 0.05).
We also observed that the SFARI ASD genes and asdM12 genes, which are enriched for inherited common variants in ASD (small average effect size), affect the synaptic modules, M13,M16, and M17. In contrast, the non-inherited (larger average effect size) RDNVspreferentially affect the early transcriptional regulation modules (Extended Experimental Methods). We emphasize that this is not absolute, as M16 includes some genes harboring RDNVs (e.g. in SCN2A, SHANK2, NRXN1; Figure 2A). To formally assess common variant enrichment using independent data, we compared ASD GWA signals across these modules (Extended Experimental Methods). Genes inM13 and M16 were more strongly affected by common variation in at least one of two ASD GWA studies(Anney et al., 2012; Wang et al., 2009) than M2 or M3 (Figure S3E). This is consistent with susceptibility of distinct biological processes for different mutational classes, and that in general more severe biological consequences would result from early transcriptional dysregulation during neuronal proliferation and differentiation, compared with later disruption of synaptic development and neuronal function.
ASD gene enriched modules are linked by translational and transcriptional regulation
Upregulated and downregulated modules are highly anti-correlated throughout development, so we hypothesized that common molecular regulatory relationships could potentially link genes within these modules. We first used a set of FMRP-RNA interactors from a cross-linking and immunoprecipitation (CLIP) experiment (Darnell et al., 2011), since Iossifov et al. (2012) had previously shown that RDNVs identified in their exome sequencing study were enriched in this class of genes. Remarkably FMRP targets are specifically enriched in modules that also contain ASD-related genes M2, M16, and M17 (FMRP-M2 p = 1.6×10-13, OR = 3.0 [2.3-3.9]; FMRP-M16 p = 2.4×10-29, OR =5.7 [4.3-7.6]; FMRP-M17 p = 9.3×10-10, OR =2.4 [1.8-3.1]; all at FDR < 0.05; Figure 4A). This provides a strong, independent line of evidence that translational regulation by FMRP not only affects genes harboring RDNVs, but links different molecular pathways that are co-expressed during early fetal cortical development and susceptible to diverse classes of ASD genetic mutation.
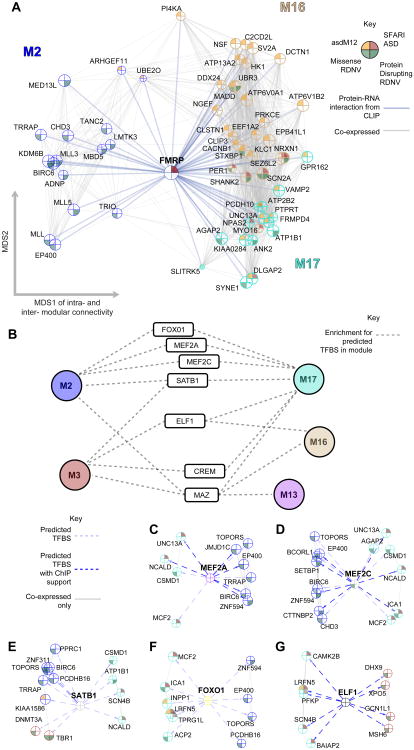
Figure 4. Translational and transcriptional co-regulation connects developmentally distinct ASD-affected modules.
(A) Co-expression based network plot of FMRP interactions with genes in M2, M16, and M17 that are either affected by RDNVs or in an ASD candidate list. Genes are plotted as in Figure 2 and 3, but now across modules, with FMRP placed at the center.
(B) Summary of TF binding site (TFBS) enrichment in modules for TFs that have evidence for function in a neurodevelopmental context. Dashed lines indicate enrichment in the module for predicted binding sites.
(C-G) MEF2A, MEF2C, SATB1, FOXO1, and ELF1 are all enriched for their binding motifs in the upstream regions of ASD gene-enriched modules following anti-correlated developmental patterns. Network plots highlight genes with a predicted binding site (light dashed arrow) contributing to this enrichment that are also affected by RDNVs or in an ASD candidate list. Arrows representing a TFBS found in a ChIP experiment are marked in dark blue.
For network plots, the top 1000 positive connections between genes are plotted and node size is proportional to connectivity within the genes' assigned module, therefore larger nodes are more central hubs. See also Table S3.
We next tested whether ASD associated modules are also linked at the transcriptional level (Experimental Procedures). We found17 TFs that are predicted to link at least one upregulated and one downregulated module based on binding site enrichment (Figure 4B, Table S3A-B). Many of these TFs are expressed during fetal development (Table S1A), have been previously implicated in relevant neuronal functions, and have DNA binding targets have been experimentally characterized (Table S3B). For example, MEF2A and MEF2C, both members of a TF family regulating synaptic plasticity and glutamatergic synapse number (Ebert and Greenberg, 2013), are enriched for binding targets in M2 and M17, which are anti-correlated across development (Figure 4C-D). SATB1, which is required for the development of cortical interneurons(Close et al., 2012), ELF1, which is involved in axonal guidance, and FOXO1 which regulates neuronal polarity(de la Torre-Ubieta and Bonni, 2011) also link these two modules (Figure 4E-F). To provide further evidence that these are experimentally plausible binding sites, we overlaid these bioinformatic predictions with chromatin immunoprecipitation (ChIP) data where available, supporting many of these predicted interactions, including 39% of MEF2A, 23% of MEF2C and 87% of ELF1binding sites (Figure 4C, 4D, 4G; Extended Experimental Procedures). These results implicate existing and novel TFs as putative co-regulators of ASD-associated gene networks during neocortical development.
ASD-associated genes exhibitlaminar and cellular enrichment
Deficits in cortical patterning and layering have been observed in ASD(Voineagu et al., 2011), we therefore tested whether ASD-affected genes are enriched in the developing laminae of fetal cortex and the terminally differentiated laminae of adult cortex (Experimental Procedures). We compared multiple ASD gene lists with the ID gene sets for enrichment in laminae of the developing and adult cortex, and found a sharp contrast in laminar enrichment between ASD and ID genes(Figure 5A-B). Additionally, in adult, asdM12 exhibits strongly significant enrichment in L3 (Z >2.7, FDR < 0.01), while other ASD lists follow a similar trend of superficial layer enrichment (Z>2, p <0.05). In contrast, the “ID all” and “ID only” gene sets follow a trend of lower layer enrichment (Figure 5B), an across-layer pattern that is significantly different from all of the ASD lists (Figure 5C-D, Extended Experimental Procedures).
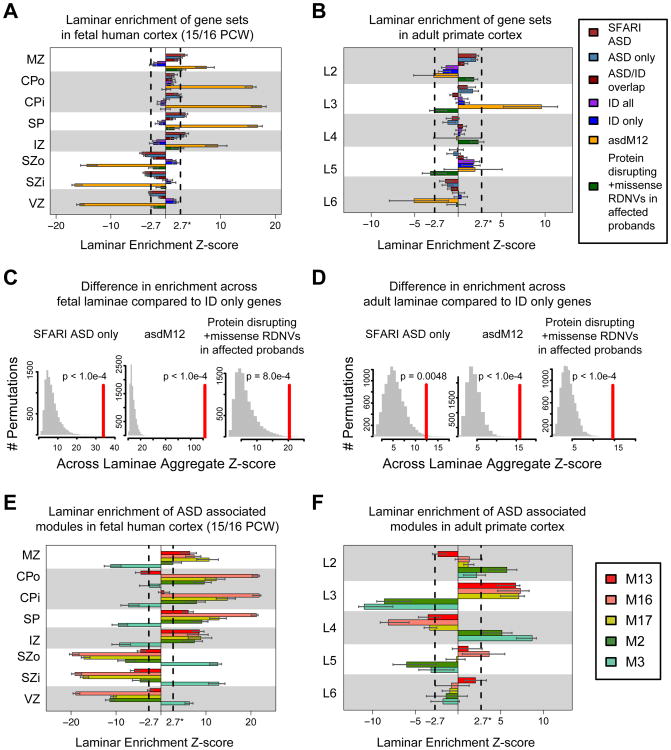
Figure 5. Enrichment for laminar differential expression of gene sets and associated developmental co-expression modules in fetal human and adult primate cortex.
(A) In fetal cortex, ASD sets (SFARI, asdM12, and RDNV-affected) are enriched for differential expression in laminae containing post mitotic neurons, whereas genes implicated in ID are weakly enriched in germinal layers. A high Z-score for a gene set in a layer corresponds to differential expression across the gene set in that layer.
(B) In adult cortex, asdM12sets show strong enrichment in layer 3, where as ID genes are weakly enriched in layer 5.
(C) and (D) Summing the Z-score across layers in A) and B) and comparing to randomly permuted sets of genes of similar size demonstrates that, in both fetal and adult cortex, the laminar distribution of multiple ASD implicated gene sets is significantly distinct from that of genes implicated only in ID.
(E) SFARI/asdM12 associated developmental co-expression modules M13, M16, and M17 follow enrichment trends similar to the SFARI/asdM12 gene set in fetal brain. However, the modules strongly associated with the RDNV affected genes, M2 and M3, show distinct enrichment patterns.
(F) ASD-associated modules are predominantly enriched in superficial layers 2-4 of adult cortex. Additionally, M16 shows weak enrichment in L5. In contrast to fetal cortex, M2 and M3 are in enriched in the same laminae in adult suggesting they serve distinct functions during cortical development that contribute to superficial cortical layers 2-4.
Dashed lines in bar plots indicate Z = 2.7 (equivalent to FDR = 0.01), error bars indicate 95% bootstrapped CIs. Laminae: Marginal Zone (MZ), Outer/Inner Cortical Plate (CPo/CPi), Subplate (SP), Intermediate Zone (IZ), Outer/Inner Subventricular Zone (SZo/SZi), Ventricular Zone (VZ), and adult cortical layers 2-6 (L2-6). See also Figure S4.
We also observed a similar trend in superficial layer enrichment for the modules that are enriched in asdM12 genes (M13, M16, and M17; Figure 5F). M13and M16also exhibit weaker enrichment in L5 and L6. Module-level analysis in fetal brain also highlighted a difference between the RDNV enriched modules, M2 and M3. Although both M2 and M3 are most highly expressed in early human fetal development (prior to PCW 17), M2 reaches its peak later and is enriched in the cortical plate (CPi/CPo), whereas M3peaks earlier, consistent with its enrichment in the germinal zone (VZ, SZi, SZo; Figure 5E). In adult, this distinction is no longer present (Figure 5F), with both M2 and M3 showing enrichment in superficial layers (L2, L4). We also asked whether any of these gene sets or modules were enriched for cell-type specific markers paralleling the observed laminar enrichment. We observed enrichment in this set of well-curated upper layer glutamatergic neuron markers among asdM12, M2, and M3 genes (Extended Experimental Procedures, Figure S4C-D), which agrees with the L2-4enrichment of asdM12 and ASD risk gene modules.
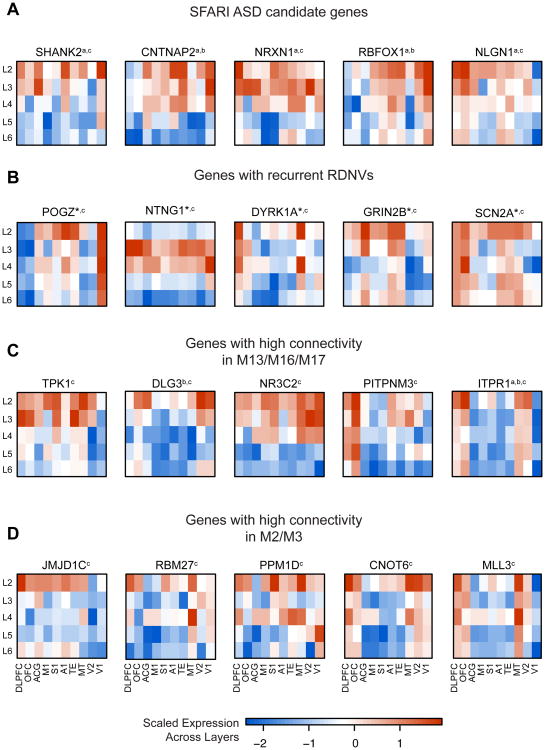
Figure 6A highlights adult layer-level expression patterns of several strong ASD candidate genes with enriched expression in superficial layers (e.g. SHANK2, CNTNAP2) and shows that many genes recurrently affected by protein disrupting RDNVs in the 965 ASD probands and an additional set of patients assessed by targeted sequencing (O'Roak et al., 2012a) also show superficial layer enrichment (e.g. SCN2A, POGZ, Figure 6B). We use these mature laminae for cell-marker enrichment analyses because laminar expression patterns are more clearly delineated relative to PCW 15-21 (Figure 5A and 5E, Figures S4A-B). Furthermore, neuronal migration in humans persists into the third trimester, and upper layer neuronal identity is not finalized until after PCW 28 (Bystron et al., 2008). Out of the 6 genes with recurrent RDNVs in probands in which we can detect layer preference, 5 are predominantly expressed in superficial layers in adult. Some of the genes in Figure 6 also show expression in a lower layer (NLGN1, SCN2A, ITPR1, MLL3), though superficial layer enrichment is stronger (larger differential expression t-value in Table S1A).
Figure 6. Laminar patterns for genes implicated in ASD.
(A) SFARI candidate genes for ASD.
(C) Genes with strong recurrent RDNV evidence across studies. Genes not displayed include TBR1 (lower layer enriched), CHD8 (no layer enrichment detected), CUL3 (no layer enrichment detected), and KATNAL2 (not detected in these data).
(B) Genes with high connectivity in M13, M16, and M17.
(C) RDNV genes with high connectivity in M2 and M3.
a indicates membership in SFARI ASD, b indicates membership in asdM12, c indicates the gene is affected by a RDNV, *indicates recurrent RDNVs
Color bar values represent scaled expression (standard deviation of the mean-centered expression value across layers). All genes shown have t > 2 for enrichment in an upper layer (L2 or L3) over background, and t < 2 for lower layers (L5 or L6). Regions: dorsolateral prefrontal (DLPFC), orbitofrontal (OFC), anterior central gyrus (ACG), primary motor (M1), primary somatosensory (S1), primary auditory (A1), higher-order visual area TE (TE), higher-order visual area MT/5 (MT), secondary visual cortex (V2), primary visual cortex (V1).
Discussion
Our analyses offer a genome-wide neurobiological context to begin to unify the genetics of ASD, providing robust evidence of both molecular pathway and circuit-level convergence (Figure 7A-B). Integration of ASD genes with developmental co-expression networks and laminar expression data connects multiple ASD risk enriched modules to glutamatergic neurons in upper cortical layers (L2-L4), tying ASD risk genes to specific brain circuitry (Figure 7C). The observation of convergent biology in ASD stands in striking contrast with ID, which does not show the same level of developmental or anatomical specificity. Laminar enrichment in the “ASD/ID overlap” genes show a similar pattern as the “ASD only” genes (in L2, Figure 5B). Therefore disruption in ID genes that also cause ASD likely affects superficial layers compared to disruption in genes causing ID only; our analyses lead to the prediction that specific disruption of cortical-cortical connectivity, for example by targeting upper layer glutamatergic neurons which predominantly comprise inter- and intra-hemispheric projections, is more likely to affect core ASD phenotypes such as social behavior, rather than general intellectual ability alone.
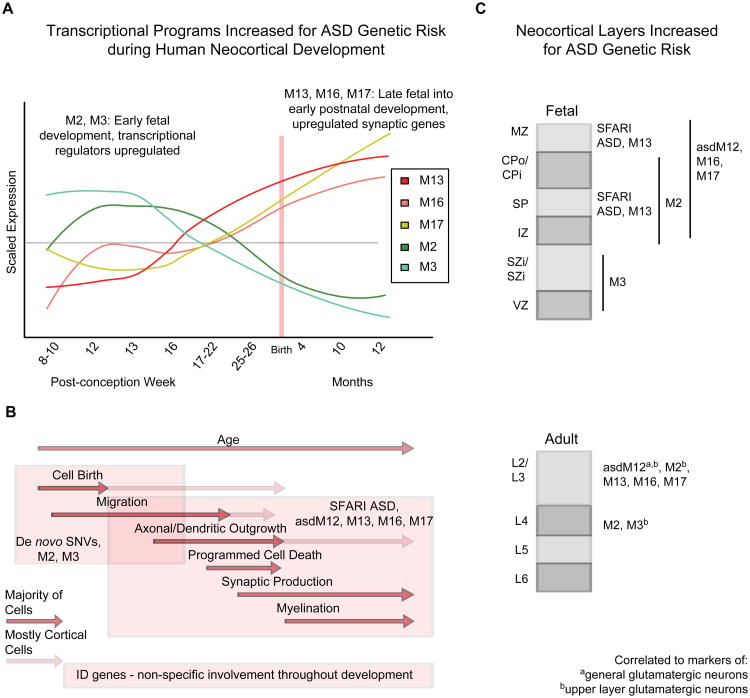
Figure 7. Summary of findings and model for effects of ASD implicated gene sets.
(A) ASD risk genes from multiple sources were enriched in five co-expression modules throughout development, M2, M3, M13, M16, and M17.
(B) Early transcriptional regulators in M2/M3 are enriched for RDNVs, while the later expressed synaptic genes are associated with previously studied ASD genes (Biological processes time periods adopted from Andersen, 2003).
(C) ASD genes are most consistently associated with laminae containing post-mitotic neurons during early fetal development (broadly in IZ, SP, CPo/CPi, and MZ) and superficial layers in adult (L2-4). Multiple modules are also strongly associated with markers of upper layer glutamatergic neurons in adult cortex, suggesting many ASD genes preferentially affect these cell types.
B) and C) also summarize that ID genes are largely distinct from ASD genes in both developmental trajectory and neocortical layer enrichment.
Both figures A and B correspond to the same time scale as marked by the axis on the plot in (A). We summarize the strongly enriched findings, but note that weaker enrichment for other patterns exists that may be important for subsets of ASD. Individual genes can be prioritized for biological validation using a combination of network position, bioinformatic scores, and the biological context highlighted here, as discussed in the text and shown in Table S4.
Our analysis further links specific molecules and pathways to the cortical-cortical intra- and inter-hemispheric disconnection that has been hypothesized as a shared circuit-level deficit unifying diverse ASD etiologies (Belmonte et al., 2004; Geschwind and Levitt, 2007). An illustrative example is the disruption of ARID1B, a BAF complex member that harbors a RDNV and is a hub of M3. Severe mutations in ARID1B cause corpus callosum abnormalities, ID, and ASD (Halgren et al., 2011; Santen et al., 2012). Another BAF complex member, SMARCC2, implicated by RDNVs in probands, controls cortical thickness by repressing the pool of intermediate progenitors, which preferentially contribute to forming cortical layers 2-4 (Tuoc et al., 2013), providing another molecular link to inter- and intra-hemispheric connectivity. These analyses make the first systematic connection between genes disrupted in ASD and this circuit-level disruption. As additional genes in the early fetal co-expression modules are found to harbor recurrent RDNVs, cortical-cortical connectivity will be a valuable phenotype to assess in both animal models and human patients.
Translational regulation by FMRP during fetal cortical development and transcriptional co-regulation of ASD candidate genes provides another level of convergent biology in ASD, and a rich starting point for further experimental investigation. Notable also are TFs that are predicted to drive the transcriptional co-regulation of molecular and circuit-level processes, including MEF2A, MEF2C, and SATB1, which have binding site enrichment in M2 and M17. This is intriguing in light of decreased PVALB expression in ASD brain (Voineagu et al., 2011), the hypothesized convergent mechanism of a shift in the excitation-inhibition balance in ASD (Rubenstein and Merzenich, 2003), and the observation that SATB1 plays a key role in regulating cortical PVALB+ and SST+ interneuron development (Close et al., 2012; Denaxa et al., 2012). We speculate that M2 and M17 reflect processes involved in the migration and differentiation of inhibitory and excitatory cell populations whose balanced co-regulation may be essential to proper cortical development. These analyses underscore the notion that understanding the structure of the transcriptional and chromatin regulatory networks underlying cortical development and their relationship to translational control will better inform the genetic risk architecture of ASD.
In addition to demonstrating biological convergence, network analysis further allowed us to stratify the full set of 684 RDNV-affected genes to a narrower list of 113 genes (Table S1A) that we hypothesize are more likely to confer increased ASD risk based on their enrichment in M2 and M3, and an elevated probability of conferring a phenotype when haploinsufficient. Furthermore, we demonstrate that the observed enrichment is specific by comparison to silent RDNVs and unaffected siblings' RDNVs. As an example of how to prioritize these candidates further based on the functional relationships summarized in Figure 7, we constructed a list of candidates using Table S1A, filtering by expression during development, membership in M2 or M3, high predicted haploinsufficiency (P(HI) > 0.5), protein disrupting or missense mutation in probands, and either a layer preference (t > 2 for a particular layer) or a cell-type preference (r > 0.2 for a cell-type) in Table S4. This yields a set of 24 candidates with a hypothesized layer- or cell-type phenotype for investigation. Among these, TBR1 is known to harbor recurrent mutations, while CHD3 is a member of the same gene family as CHD8, a gene with the strong recurrent de novo mutation evidence (O'Roak et al., 2012a). Additionally, SMARCC1 and SMARCC2 are members of the BAF complex, which is of particular interest since it is statistically associated with ASD: 6/28 BAF complex genes are affected by RDNVs (p = 1.5×10-3, OR = 5.7 [1.9-14.5]).Other RDNV-affected molecular families, including the CCR4-NOT complex members (CNOT family) and chromodomain helicase DNA binding proteins (CHD), are also seen in M2 and M3 and have been linked to the regulation of neuronal proliferation and differentiation(Feng et al., 2013; Potts et al., 2011; Ronan et al., 2013; Zheng et al., 2012).
In parallel work, Willsey et al. 2013, find convergence on fetal cortical developmental networks in frontal lobe by seeding with a subset of high confidence ASD genes identified by exome sequencing. Despite the different analytical approaches, there is remarkable overlap between the developmental processes implicated by the gene networks identified in our studies. Although we see the strongest cell-type and layer enrichment in adult L2-4, we also see a signal in CPi during fetal development and a weaker signal in L5-6 of adult, consistent with a subset of genes affecting lower-layer glutamatergic neurons. Together, our studies highlight the importance of understanding the spatial and temporal context of specific genes for future mechanistic investigation.
We also acknowledge several issues that challenged our approach. Many of the genes we identified as putatively involved in ASD do not have complete PPI data, P(HI) scores, TF binding site information, or are not well studied in brain. This is one reason why we rely most heavily on RNA-seq based transcriptome data, as it comprehensively represents relationships present in the developing human brain in an unbiased manner. We did not assess enrichment of genetic hits in other brain regions across development, as sample size and cell-type heterogeneity make it difficult to interpret co-expression across cytoarchitecturally diverse brain regions such as cerebellum and amygdala, which may also be involved in ASD (Amaral et al., 2008). We also focused on single gene disruption in ASD and did not include CNVs affecting multiple genes to improve signal to noise. Additionally, current genetic approaches favor de novo mutation detection; as different classes of mutations (e.g. inherited rare coding or non-coding regulatory variants) are identified, we speculate that heritable variants will affect genes in the modules related to synaptic development and function, rather than earlier transcriptional regulation. Likewise, It will also be useful to investigate rare, inherited recessive ASD risk variants (Lim et al., 2013; Yu et al., 2013) when sufficient data are available, so as to compare with other forms of genetic variation.
The conclusions summarized in Figure 7 pass a stringent multiple comparisons cut-off; weaker enrichment patterns may become more salient with higher resolution tiling of gene expression during development and increased sample sizes in sequencing studies. To facilitate future studies we have shared the code used in this analysis (Extended Experimental Procedures) and provided a graphical interface for exploring specific genes within the network context. We have shown how an integrative approach, which is not driven by any small set of samples, candidate genes, or candidate hypotheses, can place heterogeneous genetic etiologies into a unifying structure. These analyses provide a working frame work for mechanistic investigation and hypothesis testing, which points to interactions between genes in specific cell types and circuits, as well as the general biological processes in which these genes are implicated.
Experimental Procedures
Developmental expression data
BrainSpan developmental RNA-seqdata (obtained from www.brainspan.org) summarized to GENCODE10 (Harrow et al., 2006) gene-level reads per kilobase million mapped reads (RPKM) values were used (Extended Experimental Procedures for data preprocessing, see Table S1D for sample details). Only neocortical regions were used in our analysis and only genes with a normalized RPKM value of 1 in at least one region at one time point for 80% of the available samples were considered expressed.
Weighted Gene Co-expression Network Analysis
We used the R package WGCNA (Langfelder et al., 2008) to construct co-expression networks, as previously done (Voineagu et al., 2011) and described in detail in Extended Experimental Methods. The modules were characterized using GO Elite to control the network-wide false discovery rate, with all enriched pathways comprising at least 10 genes at Z > 2 and FDR < 0.01 (Zambon et al., 2012). All network plots were constructed using the igraph package in R (Csárdi and Nepusz, 2006).
Protein-protein interaction enrichment
When assessing PPI enrichment in modules, a degree-matched permutation analysis was applied in order to control for biological and methodological biases in PPI data (see Extended Experimental Procedures for details).
Gene sets
The SFARI ASD set was compiled using the online SFARI gene database, AutDB. We used the “Gene Score,” which classifies evidence levels, to restrict our set to those categorized as S (Syndromic) and evidence levels 1-4 (high confidence - minimal evidence). We obtained asdM12 and adsM16 from a prior, independent gene expression study that profiled expression changes in ASD cortex and applied WGCNA to identify modules of dysregulated genes ASD (Voineagu et al., 2011). We curated ID genes from four reviews cataloging genes causing “ID all” ( In low and Restifo, 2004; Lubs et al., 2012; Ropers, 2008; van Bokhoven, 2011) resulting in 401 genes. For candidate lists, we used the HUGO gene nomenclature to find updated gene symbols. We obtained RDNVs from four publications (Iossifov et al., 2012; Neale et al., 2012; O'Roak et al., 2012b; Sanders et al., 2012), and split them into discovery and validation sets as discussed in the results (see Extended Experimental Procedures for further details about gene sets).
Gene set over-representation
All enrichments of gene sets were performed using a two-sided Fisher exact test with 95% confidence calculated according to the R function fisher.test. The false discovery rate was controlled across candidate ASD gene set enrichments, the discovery RDNV set enrichment, and FMRP target enrichment (Table S2B). For RDNV enrichment, we required an OR > 1 and FDR < 0.05 for enrichment in the discovery set, and OR > 1 with p < 0.05 for validation in the replication set. When showing a lack of enrichment, we require p > 0.05 to reduce false negatives, as future studies that add expression time points for networks and genes for enrichment may find enrichment in pathways not enriched here.
Transcription Factor Binding Site Enrichment
The top 200 genes in each module (ranked by kME) were used for TF motif enrichment analysis. Enrichment for each TF motif in TRANSFAC (Matys et al., 2003) was compared to 3 background datasets to ensure robustness: 1000 bp sequences upstream of all human genes, human CpG islands, and the sequence of human chromosome 20 (Extended Experimental Procedures). Only TFs with p < 0.05 across all backgrounds are considered enriched. ChIP data was obtained from ENCODE (Consortium, 2011) and the ChIP enrichment analysis (Lachmann et al., 2010) resource.
Layer-specific and Cell-type Marker Enrichment
We utilized human fetal neocortical laminar gene expression datasets from BrainSpan, at PCW 15/16 and PCW 21 and primate neocortical laminar gene expression data from a published study (Bernard et al., 2012). For laminar specificity, differential expression of each gene in each layer was calculated against background, resulting in t-values for each gene in each layer (Table S1A). We quantified the skew of differential expression t-values of each gene set in each layer, applied aFDR cut-off across all enrichments in all layers (Z = 2.7, FDR = 0.01), and computed bootstrapped confidence intervals to assess enrichment of gene sets in layers. To quantify cell-marker relationships, we used an analogous method, replacing the t-value by the correlation of each gene to a set of known cell marker genes in the adult layer data (Table S1A). Statistical comparison of enrichment trends across layers between ASD and IDgene sets set was performed by comparing the distribution of scores across layers using a permutation analysis (Extended Experimental Methods).
Supplementary Material
Highlights.
ASD genes from multiple sources converge on prenatal neurodevelopmental processes
Transcriptional and translational co-regulation link ASD genes at multiple levels
Multiple ASD risk gene modules are enriched in superficial cortical layers
These patterns highlight features that distinguish ASD from ID
Acknowledgments
We thank the Simons Foundation Autism Research Initiative (www.sfari.org) and the Autism Genetic Resource Exchange (www.agre.org) and all families involved for making this work possible. We gratefully acknowledge data resources from the BrainSpan consortium (NIMH grant 5RC2 MH089921) and the Allen Brain Institute (www.brainspan.org, www.brain-map.org). This work supported by a NIMH Training Grant and NRSA Fellowship (T32MH073526 and F30MH099886, NNP), an R01 (5R01MH060233:05, DHG), an Autism Center for Excellence network grant (5R01MH100027), and the Medical Scientist Training Program at UCLA. We also acknowledge the support of the NINDS Informatics Center for Neurogenetics and Neurogenomics (P30NS062691) at UCLA. We thank Jason Chen, Michael Gandal, and Jason Stein for critically reading the manuscript, as well as Willsey et al. for sharing their results prior to publication.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amaral DG, Schumann CM, Nordahl CW. Neuroanatomy of autism. Trends Neurosci. 2008;31:137–145. doi: 10.1016/j.tins.2007.12.005. [DOI] [PubMed] [Google Scholar]
- Andersen SL. Trajectories of brain development: point of vulnerability or window of opportunity? Neuroscience & Biobehavioral Reviews. 2003;27:3–18. doi: 10.1016/s0149-7634(03)00005-8. [DOI] [PubMed] [Google Scholar]
- Anney R, Klei L, Pinto D, Almeida J, Bacchelli E, Baird G, Bolshakova N, Bölte S, Bolton PF, Bourgeron T, et al. Individual common variants exert weak effects on the risk for autism spectrum disorderspi. Human Molecular Genetics. 2012;21:4781–4792. doi: 10.1093/hmg/dds301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu SN, Kollu R, Banerjee-Basu S. AutDB: a gene reference resource for autism research. Nucleic Acids Res. 2009;37:D832–D836. doi: 10.1093/nar/gkn835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmonte MK, Allen G, Beckel-Mitchener A, Boulanger LM, Carper RA, Webb SJ. Autism and abnormal development of brain connectivity. Journal of Neuroscience. 2004;24:9228–9231. doi: 10.1523/JNEUROSCI.3340-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-David E, Shifman S. Combined analysis of exome sequencing points toward a major role for transcription regulation during brain development in autism. Mol Psychiatry. 2012 doi: 10.1038/mp.2012.148. [DOI] [PubMed] [Google Scholar]
- Berg JM, Geschwind DH. Autism genetics: searching for specificity and convergence. Genome Biol. 2012;13:247. doi: 10.1186/gb-2012-13-7-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard A, Lubbers LS, Tanis KQ, Luo R, Podtelezhnikov AA, Finney EM, McWhorter MME, Serikawa K, Lemon TA, Morgan RJ, et al. Transcriptional Architecture of the Primate Neocortex. Neuron. 2012;73:1083–1099. doi: 10.1016/j.neuron.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bystron I, Blakemore C, Rakic P. Development of the human cerebral cortex: Boulder Committee revisited. Nat Rev Neurosci. 2008;9:110–122. doi: 10.1038/nrn2252. [DOI] [PubMed] [Google Scholar]
- Close J, Xu H, De Marco García N, Batista-Brito R, Rossignol E, Rudy B, Fishell G. Satb1 is an activity-modulated transcription factor required for the terminal differentiation and connectivity of medial ganglionic eminence-derived cortical interneurons. Journal of Neuroscience. 2012;32:17690–17705. doi: 10.1523/JNEUROSCI.3583-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium, E.P. A User's Guide to the Encyclopedia of DNA Elements (ENCODE) PLoS Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courchesne E, Campbell K, Solso S. Brain growth across the life span in autism: Age-specific changes in anatomical pathology. Brain Research. 2011;1380:138–145. doi: 10.1016/j.brainres.2010.09.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csárdi G, Nepusz T. The igraph software package for complex network research. InterJournal, Complex Systems. 2006;1695 [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KYS, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, et al. FMRP Stalls Ribosomal Translocation on mRNAs Linked to Synaptic Function and Autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Torre-Ubieta L, Bonni A. Transcriptional regulation of neuronal polarity and morphogenesis in the mammalian brain. Neuron. 2011;72:22–40. doi: 10.1016/j.neuron.2011.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denaxa M, Kalaitzidou M, Garefalaki A, Achimastou A, Lasrado R, Maes T, Pachnis V. Maturation-Promoting Activity of SATB1 in MGE-Derived Cortical Interneurons. Cell Reports. 2012;2:1351–1362. doi: 10.1016/j.celrep.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin B, Scherer SW. Genetic architecture in autism spectrum disorder. Current Opinion in Genetics & Development. 2012;22:229–237. doi: 10.1016/j.gde.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Ebert DH, Greenberg ME. Activity-dependent neuronal signalling and autism spectrum disorder. Nature. 2013;493:327–337. doi: 10.1038/nature11860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker CC, Suckling JJ, Deoni SCS, Lombardo MVM, Bullmore ETE, Baron-Cohen SS, Catani MM, Jezzard PP, Barnes AA, Bailey AJA, et al. Brain anatomy and its relationship to behavior in adults with autism spectrum disorder: a multicenter magnetic resonance imaging study. Archives of General Psychiatry. 2012;69:195–209. doi: 10.1001/archgenpsychiatry.2011.1251. [DOI] [PubMed] [Google Scholar]
- Feng W, Khan MA, Bellvis P, Zhu Z, Bernhardt O, Herold-Mende C, Liu HK. The Chromatin Remodeler CHD7 Regulates Adult Neurogenesis via Activation of SoxC Transcription Factors. Cell Stem Cell. 2013;13:62–72. doi: 10.1016/j.stem.2013.05.002. [DOI] [PubMed] [Google Scholar]
- Geschwind DH. Genetics of autism spectrum disorders. Trends Cogn Sci (Regul Ed) 2011;15:409–416. doi: 10.1016/j.tics.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geschwind DH, Levitt P. Autism spectrum disorders: developmental disconnection syndromes. Current Opinion in Neurobiology. 2007;17:103–111. doi: 10.1016/j.conb.2007.01.009. [DOI] [PubMed] [Google Scholar]
- Gilman SR, Iossifov I, Levy D, Ronemus M, Wigler M, Vitkup D. Rare de novo variants associated with autism implicate a large functional network of genes involved in formation and function of synapses. Neuron. 2011;70:898–907. doi: 10.1016/j.neuron.2011.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratten J, Visscher PM, Mowry BJ, Wray NR. Interpreting the role of de novo protein-coding mutations in neuropsychiatric disease. Nat Genet. 2013;45:234–238. doi: 10.1038/ng.2555. [DOI] [PubMed] [Google Scholar]
- Halgren C, Kjaergaard S, Bak M, Hansen C, El-Schich Z, Anderson CM, Henriksen KF, Hjalgrim H, Kirchhoff M, Bijlsma EK, et al. Corpus callosum abnormalities, intellectual disability, speech impairment, and autism in patients with haploinsufficiency of ARID1B. Clinical Genetics. 2011;82:248–255. doi: 10.1111/j.1399-0004.2011.01755.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J, Denoeud F, Frankish A, Reymond A, Chen CK, Chrast J, Lagarde J, Gilbert JG, Storey R, Swarbreck D, et al. GENCODE: producing a reference annotation for ENCODE. -Abstract - UK PubMed Central. Genome Biol. 2006;7:S4. doi: 10.1186/gb-2006-7-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang N, Lee I, Marcotte EM, Hurles ME. Characterising and predicting haploinsufficiency in the human genome. PLoS Genet. 2010;6:e1001154. doi: 10.1371/journal.pgen.1001154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inlow JK, Restifo LL. Molecular and comparative genetics of mental retardation. Genetics. 2004;166:835–881. doi: 10.1534/genetics.166.2.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iossifov I, Ronemus M, Levy D, Wang Z, Hakker I, Rosenbaum J, Yamrom B, Lee YH, Narzisi G, Leotta A, et al. De Novo Gene Disruptions in Children on the Autistic Spectrum. Neuron. 2012;74:285–299. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klei L, Sanders SJ, Murtha MT, Hus V, Lowe JK, Willsey AJ, Moreno-De-Luca D, Yu TW, Fombonne E, Geschwind D, et al. Common genetic variants, acting additively, are a major source of risk for autism. Mol Autism. 2012;3:9. doi: 10.1186/2040-2392-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachmann A, Xu H, Krishnan J, Berger SI, Mazloom AR, Ma'ayan A. ChEA: transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics. 2010;26:2438–2444. doi: 10.1093/bioinformatics/btq466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Zhang B, Horvath S. Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut package for R. Bioinformatics. 2008;24:719–720. doi: 10.1093/bioinformatics/btm563. [DOI] [PubMed] [Google Scholar]
- Lim ET, Raychaudhuri S, Sanders SJ, Stevens C, Sabo A, MacArthur DG, Neale BM, Kirby A, Ruderfer DM, Fromer M, et al. Rare Complete Knockouts in Humans: Population Distribution and Significant Role in Autism Spectrum Disorders. Neuron. 2013;77:235–242. doi: 10.1016/j.neuron.2012.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubs HA, Stevenson RE, Schwartz CE. Fragile X and X-Linked Intellectual Disability: Four Decades of Discovery. The American Journal of Human Genetics. 2012;90:579–590. doi: 10.1016/j.ajhg.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R, Sanders SJ, Tian Y, Voineagu I, Huang N, Chu SH, Klei L, Cai C, Ou J, Lowe JK, et al. Genome-wide Transcriptome Profiling Reveals the Functional Impact of Rare De Novo and Recurrent CNVs in Autism Spectrum Disorders. Am J Hum Genet. 2012;91:38–55. doi: 10.1016/j.ajhg.2012.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur DG, Balasubramanian S, Frankish A, Huang N, Morris J, Walter K, Jostins L, Habegger L, Pickrell JK, Montgomery SB, et al. A systematic survey of loss-of-function variants in human protein-coding genes. Science. 2012;335:823–828. doi: 10.1126/science.1215040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson JL, Shoemaker M. Intellectual disability and its relationship to autism spectrum disorders. Research in Developmental Disabilities. 2009;30:1107–1114. doi: 10.1016/j.ridd.2009.06.003. [DOI] [PubMed] [Google Scholar]
- Matys V, Fricke E, Geffers R, Gössling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelson JJ, Shi Y, Gujral M, Zheng H, Malhotra D, Jin X, Jian M, Liu G, Greer D, Bhandari A, et al. Whole-Genome Sequencing in Autism Identifies Hot Spots for De Novo Germline Mutation. Cell. 2012;151:1431–1442. doi: 10.1016/j.cell.2012.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale BM, Kou Y, Liu L, Ma'ayan A, Samocha KE, Sabo A, Lin CF, Stevens C, Wang LS, Makarov V, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012:1–5. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Roak BJ, Vives LL, Fu WW, Egertson JD, Stanaway IB, Phelps IG, Carvill GG, Kumar AA, Lee CC, Ankenman KK, et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012a;338:1619–1622. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, Levy R, Ko A, Lee C, Smith JD, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012b:1–7. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts RC, Zhang P, Wurster AL, Precht P, Mughal MR, Wood WH, III, Zhang Y, Becker KG, Mattson MP, Pazin MJ. CHD5, a Brain-Specific Paralog of Mi2 Chromatin Remodeling Enzymes, Regulates Expression of Neuronal Genes. PLoS ONE. 2011;6:e24515. doi: 10.1371/journal.pone.0024515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramani AK, Li Z, Hart GT, Carlson MW, Boutz DR, Marcotte EM. A map of human protein interactions derived from co-expression of human mRNAs and their orthologs. Mol Syst Biol. 2008;4 doi: 10.1038/msb.2008.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronan JL, Wu W, Crabtree GR. From neural development to cognition: unexpected roles for chromatin. Nat Rev Genet. 2013;14:347–359. doi: 10.1038/nrg3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ropers HH. Genetics of intellectual disability. Current Opinion in Genetics & Development. 2008;18:241–250. doi: 10.1016/j.gde.2008.07.008. [DOI] [PubMed] [Google Scholar]
- Rossin EJ, Lage K, Raychaudhuri S, Xavier RJ, Tatar D, Benita Y, Constortium, I.I.B.D.G. Cotsapas C, Daly MJ. Proteins Encoded in Genomic Regions Associated with Immune-Mediated Disease Physically Interact and Suggest Underlying Biology. PLoS Genet. 2011;7:e1001273. doi: 10.1371/journal.pgen.1001273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubenstein JLR, Merzenich MM. Model of autism: increased ratio of excitation/inhibition in key neural systems. Genes, Brain and Behavior. 2003;2:255–267. doi: 10.1034/j.1601-183x.2003.00037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubenstein JLR. Annual Research Review: Development of the cerebral cortex: implications for neurodevelopmental disorders. Journal of Child Psychology and Psychiatry. 2010;52:339–355. doi: 10.1111/j.1469-7610.2010.02307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai Y, Shaw CA, Dawson BC, Dugas DV, Al-Mohtaseb Z, Hill DE, Zoghbi HY. Protein interactome reveals converging molecular pathways among autism disorders. Science Translational Medicine. 2011;3:86ra49. doi: 10.1126/scitranslmed.3002166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Willsey AJ, Ercan-Sencicek AG, Dilullo NM, Parikshak NN, Stein JL, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santen GWE, Aten E, Sun Y, Almomani R, Gilissen C, Nielsen M, Kant SG, Snoeck IN, Peeters EAJ, Hilhorst-Hofstee Y, et al. Mutations in SWI/SNF chromatin remodeling complex gene ARID1B cause Coffin-Siris syndrome. Nat Genet. 2012;44:379–380. doi: 10.1038/ng.2217. [DOI] [PubMed] [Google Scholar]
- Srinivasan K, Leone DP, Bateson RK, Dobreva G, Kohwi Y, Kohwi-Shigematsu T, Grosschedl R, McConnell SK. A network of genetic repression and derepression specifies projection fates in the developing neocortex. Proceedings of the National Academy of Sciences. 2012;109:19071–19078. doi: 10.1073/pnas.1216793109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark C. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuoc TC, Boretius S, Sansom SN, Pitulescu ME, Frahm J, Livesey FJ, Stoykova A. Chromatin regulation by BAF170 controls cerebral cortical size and thickness. Dev Cell. 2013;25:256–269. doi: 10.1016/j.devcel.2013.04.005. [DOI] [PubMed] [Google Scholar]
- van Bokhoven H. Genetic and epigenetic networks in intellectual disabilities. Annu Rev Genet. 2011;45:81–104. doi: 10.1146/annurev-genet-110410-132512. [DOI] [PubMed] [Google Scholar]
- Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S, Mill J, Cantor RM, Blencowe BJ, Geschwind DH. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Zhang H, Ma D, Bućan M, Glessner JT, Abrahams BS, Salyakina D, Imielinski M, Bradfield JP, Sleiman PMA, et al. Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature. 2009;459:528–533. doi: 10.1038/nature07999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo AS, Staahl BT, Chen L, Crabtree GR. MicroRNA-mediated switching of chromatinremodelling complexes in neural development. Nature. 2009 doi: 10.1038/nature08139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu TW, Chahrour MH, Coulter ME, Jiralerspong S, Okamura-Ikeda K, Ataman B, Schmitz-Abe K, Harmin DA, Adli M, Malik AN, et al. Using Whole-Exome Sequencing to Identify Inherited Causes of Autism. Cell. 2013;77:259–273. doi: 10.1016/j.neuron.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambon AC, Gaj S, Ho I, Hanspers K, Vranizan K, Evelo CT, Conklin BR, Pico AR, Salomonis N. GO-Elite: A Flexible Solution for Pathway and Ontology Over-Representation. Bioinformatics. 2012 doi: 10.1093/bioinformatics/bts366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Horvath S. A General Framework for Weighted Gene Co-Expression Network Analysis. Statistical Applications in Genetics and Molecular Biology. 2005;4 doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- Zheng X, Dumitru R, Lackford BL, Freudenberg JM, Singh AP, Archer TK, Jothi R, Hu G. Cnot1, Cnot2, and Cnot3 Maintain Mouse and Human ESC Identity and Inhibit Extraembryonic Differentiation. Stem Cells. 2012;30:910–922. doi: 10.1002/stem.1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.