Abstract
Degradation of maternally stored mRNAs after fertilization is an essential process for mammalian embryogenesis. Maternal mRNA degradation depending on deadenylases in mammalian early embryos has been mostly speculated, rather than directly demonstrated. Previously, we found that gene expression of nocturnin, which functions as a circadian clock-controlled deadenylase in mammalian cells, was clearly changed during the maternal-to-zygotic transition (MZT). Here, we investigated the possible role of nocturnin during mouse MZT. First, we examined the expression profile and localization of nocturnin in mouse oocytes and early embryos. The abundance of Nocturnin mRNA level was significantly decreased from the MII to 4-cell stages and slightly increased from the 8-cell to blastocyst stages, whereas the Nocturnin protein level was almost stable in all examined cells including GV and MII oocytes and early embryos. Nocturnin was localized in both the cytoplasm and the nucleus of all examined cells. We then examined the effect of loss or gain of Nocturnin function on early embryonic development. Knockdown of Nocturnin by injection of Nocturnin antisense expression vector into 1-cell embryos resulted in the delay of early embryonic development to the early blastocyst stage. Moreover, Nocturnin-overexpressed embryos by injection of Nocturnin expression vector impaired their development from the 1-cell to 2-cell or 4-cell stages. These results suggest that precise expression of nocturnin is critical to proper development of early mouse embryos. Functional analysis of nocturnin may contribute to the understanding of the possible role of the deadenylase at mouse MZT.
Keywords: Deadenylase, Early embryonic development, Mouse, Nocturnin
Maternal-to-zygotic transition (MZT) is an essential process for erasure of the oogenic program by establishing the embryonic program in totipotent zygotes [1, 2]. MZT is regulated by maternally inherited transcripts and proteins that have been accumulated in the oocyte and activation of the embryonic genome [3, 4]. In mice, maternal mRNA and proteins stored in ovulated oocytes with a transcriptionally quiescent state are immediately degraded after fertilization, and a large number of them are degraded by the 2-cell stage, which is when major zygotic gene activation (ZGA) occurs.
The ubiquitin-proteasome system (UPS), which is involved in oocyte meiosis resumption, spindle assembly and polar body emission [5,6,7], has a key role in the degradation of stored maternal proteins after fertilization [8, 9]. Moreover, a zygote-specific UPS pathway mediated by ZPAC protein, which we have recently identified as a zygote-specific proteasome chaperone, is important for removal of maternal proteins in mouse MZT [10]. Meanwhile, the molecular mechanisms underlying the degradation of stored maternal mRNAs in zygotes are not well understood. In fact, microRNAs, which are necessary for destabilization of mRNAs, are not essential during preimplantation embryonic development [11].
In mammalian cells, changes in mRNA poly(A) tail length are linked to the regulation of mRNA stabilization, translation and localization [12, 13]. In particular, the first step of mRNA degradation is the poly(A) tail-shortening process, referred to as deadenylation. Deadenylation-dependent mRNA degradation is the major pathway in mammalian cells. One or a combination of many deadenylases including POP2 (also known as CAF1), CAF1Z, poly(A)-specific ribonuclease (PARN), PAN2, CCR4, Nocturnin (also known as Ccrn4l), ANGEL and 2'PDE families facilitates this process in mammalian cells [12]. Although the occurrence of mRNA deadenylation during maternal mRNA degradation has been directly demonstrated in Xenopus [14] and Drosophila embryos [15, 16], the dependence of maternal mRNA degradation on deadenylases in mammalian oocytes has been mostly inferred, rather than explicitly assessed.
We have previously performed mRNA differential display analysis to compare embryos at the late 1-cell stage with oocytes at the MII stage in order to understand the mechanisms governing the transition of oocytes into totipotent zygotes during the mouse MZT [10, 17, 18]. In line with this, we found that Nocturnin gene expression changed during the mouse MZT. Interestingly, nocturnin in mammalian cells has been shown to be a clock-controlled deadenylase [19, 20]. Thus, we focused our attention on the role of nocturnin in mouse oocytes and early embryos. Herein, we show that that the Nocturnin mRNA level in oocytes drastically decreased after fertilization until the late 2-cell stage, although the Nocturnin protein level remained mostly stable during preimplantation development. Furthermore, Nocturnin knockdown seemed to be the cause of delayed early embryonic development to the blastocyst stage, whereas Nocturnin-overexpressed embryos were severely impaired in their development after the 2-cell stage. These findings support the possible role of nocturnin during the mouse MZT.
Materials and Methods
Animals
All mice (ICR) were purchased from Kiwa Experimental Animals (Wakayama, Japan) and maintained in light-controlled, air-conditioned rooms. This study was carried out in strict accordance with the recommendations in the Guidelines of Kinki University for the Care and Use of Laboratory Animals. The protocol was approved by the Committee on the Ethics of Animal Experiments of Kinki University. All mice were sacrificed by cervical dislocation, and all efforts were made in order to minimize suffering and to reduce the number of animals used in the present study.
Collection of oocytes, in vitro fertilization (IVF) and embryo culture
Collection of oocytes, IVF and embryo culture were performed according to previously described procedures [18]. In brief, spermatozoa were collected from the cauda epididymis of a male mouse. The sperm suspension was incubated for 1.5 h to allow for capacitation at 37 C under 5% CO2 in air. Oocytes were collected from the excised oviducts of female mice (2–3 months old) that had been superovulated with pregnant mare serum gonadotropin (PMSG; PEAMEX, ZENOAQ, Fukushima, Japan) followed 48 h later by treatment with human chorionic gonadotropin (hCG: Gonatropin, ASKA Pharmaceutical, Tokyo, Japan). Cumulus-oocyte complexes were recovered in a pre-equilibrated HTF medium. The sperm suspension was added to the oocyte-containing medium. Morphologically normal fertilized oocytes with a second polar body were collected from the sperm suspension 1 h after insemination. The eggs were cultured in a K-SOM medium at 37 C under 5% CO2 in air [21]. In addition, we only used fertilized oocytes with male and female pronuclei at 6 hours post insemination (hpi) for further experiments.
Quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) analysis
Quantitative RT-PCR analyses were performed as previously described [22]. In brief, total RNA was isolated from 20 pooled oocytes or embryos by using RNAqueous-Micro Kits (Ambion, Austin, TX, USA). cDNA was synthesized from each total RNA by using SuperScript III Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA), respectively. Prepared cDNA samples were amplified using the specific primers (Table 1) and analyzed by quantitative RT-PCR. Real-Time quantitative PCR was performed using an ABI 7300 instrument and TaqMan Gene Expression Assays (Applied Biosystems, Foster City, CA, USA). To express the data concisely, the duplicated data were averaged. The averaged data of the transcripts of each gene from each of three independent samples were analyzed.
Table 1. PCR primers.
| Primer name | Sequences (5'–3') | Applications |
| Noc_F | ACCTGAGGACAGATTGCTCCA | Quantitative RT-PCR for Noc |
| Noc_R | AGGATCAGGCACTTCCTCTCTTC | |
| Actin_F | ACCACAGTCCATGCCATCAC | Quantitative RT-PCR for Actin |
| Actin_R | TCCACCACCCTGTTGCTGTA |
Generation of anti-Nocturnin antiserum
The design of peptide synthesis of Nocturnin (QSEPPYTTWKIR, residues 354-365 a.a.) was based on the relative hydrophilicity and flexibility of regions analyzed by a computer program (GENETYX-Mac Ver. 12.0.3, GENETYX, Tokyo, Japan). The region used as the synthetic peptide has high hydrophilicity and no putative site of modification (data not shown). Anti-Nocturnin antiserum was obtained by injection of the peptide–KLH (keyhole limpet hemocyanin) complex followed by booster injections at one-week intervals, six times in total, into New Zealand white rabbits (Kitayama LABES, Nagano, Japan). ELISA was used to compare the serum titer from the rabbits before and after immunization with the peptides. Finally, anti-Nocturnin antibody was fractionated with 40% ammonium sulphate and used throughout this study.
Western blot analysis
The procedures for immunoblot analysis were essentially those reported previously [10, 17, 18]. In brief, in Western blot analysis, protein was isolated from 30 embryos in each experimental group. We used the following antibodies: anti-Nocturnin (1:300) or anti-Actin (1:7,500; Santa Cruz Biotechnology, Santa Cruz, CA, USA; SC-1616) as a loading control. We used a goat anti-rabbit IgG HRP conjugate (1:100,000; Millipore, Billerica, MA, USA; AP180P) and/or donkey anti-mouse IgG HRP conjugate (1:100,000; Santa Cruz Biotechnology; A-0545) as a second antibody. As the positive control of anti-Nocturnin antibody, the N-terminal 6xHis-tagged recombinant Nocturnin was used. At least three independent experiments were performed for each group.
Immunocytochemical analysis
Immunocytochemical analysis was performed as described previously [23]. In brief, oocytes and embryos were fixed in 4% PFA (Nacalai Tesque, Kyoto, Japan) in phosphate-buffered saline (PBS) for 30 min at room temperature and then incubated in PBS containing 0.1–0.2% Triton X-100 (Nacalai Tesque) overnight at 4 C. The samples were treated with 1% acid tyrode in PBS for 3 min at room temperature to remove the zona pellucida and then washed thoroughly in PBS including 0.01% polyvinyl alcohol (PVA; Sigma-Aldrich, St. Louis, Mo, USA; P8136). Then, the samples were incubated with anti-Nocturnin antibody (1:3,000) in PBS containing 30 mg/ml bovine serum albumin overnight at 4 C. After incubation, the samples were reacted with Alexa Flour 594-labeled donkey anti-rabbit IgG (Invitrogen, 1:1,000) for 1 h at room temperature. Specimens were mounted on glass slides in a Vectashield mounting medium (Vector Laboratories, Burlingame, CA, USA) containing 2–5 μg/ml DAPI (Invitrogen). The fluorescence images of oocytes and embryos were obtained using a fluorescence microscope (BIOREVO BZ-9000; KEYENCE, Tokyo, Japan). Three independent experiments were performed.
Microinjection of Nocturnin antisense or sense expression vectors
The procedure for microinjection of expression vectors was essentially as described previously [10]. We subcloned full-length Noc cDNA from an ovary cDNA library using pGEM T-easy vector (Epicentre Technologies, Madison, WI, USA). To investigate the effects of knockdown or overexpression of Nocturnin on the development of early embryos, Nocturnin antisense expression vector (pβ–actin promoter/antisense Nocturnin/IRES/luc+/SV40) with bicistronic expression of both Nocturnin antisense RNA and humanized firefly codon-optimized luciferase (luc+) gene or Nocturnin expression vector (pβ–actin promoter/sense Nocturnin/IRES/luc+/SV40) with bicistronic expression of both Nocturnin cDNA and humanized firefly codon-optimized luciferase (luc+) gene was injected into the pronucleus of zygotes at 6 to 8 hpi. The injected zygotes showing luciferase activity at 16 h after microinjection were selected and then cultured to examine the effect of antisense Nocturnin expression or the effect of Nocturnin expression on subsequent embryonic development to blastocyst stage. In these experiments, pβ–actin promoter/IRES/luc+/SV40 was used as a control expression vector.
Statistical analysis
For statistical analysis, we used StatView version 5.0 (SAS Institute, Cary, NC, USA) and Microsoft Excel, and performed analysis of variance (ANOVA) with an α level of 0.05 to determine possible statistically significant differences between group means.
Results
Expression of nocturnin during MZT
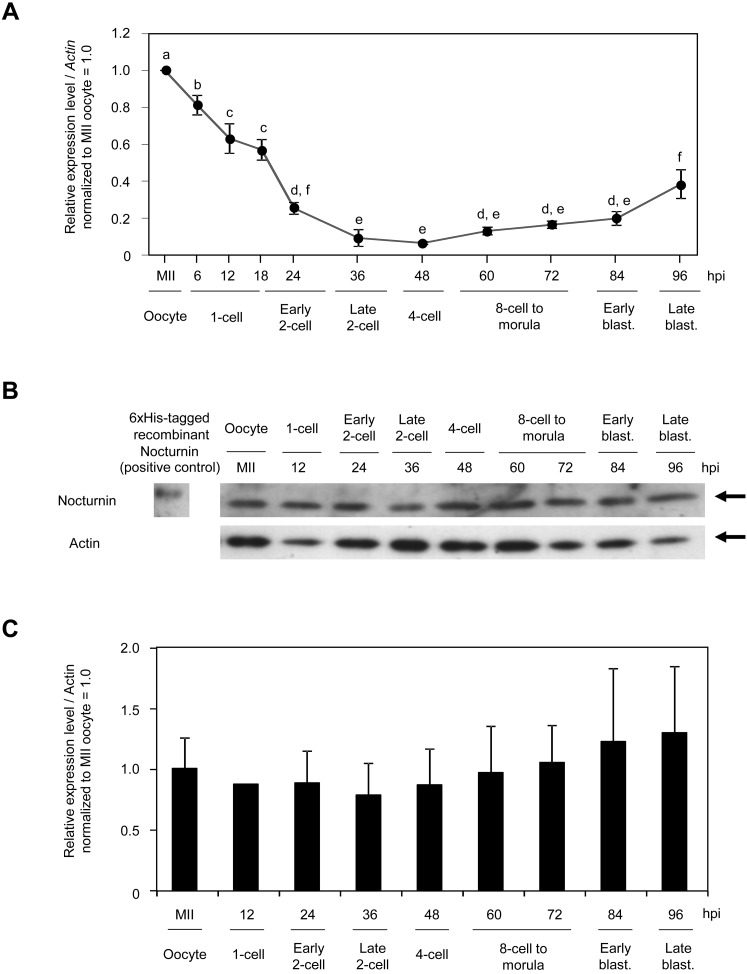
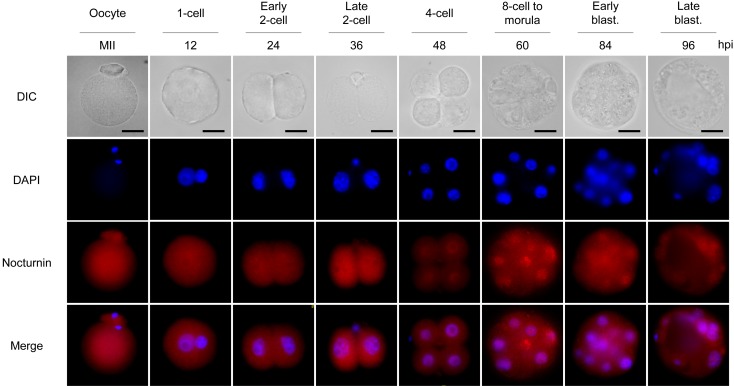
Since the Nocturnin gene was predicted to be maternally expressed in early mouse embryos according to its EST profile described in the UniGene database, we examined gene expression profiles and subcellular localization of nocturnin during early mouse embryogenesis. In early mouse embryos, the Nocturnin gene showed a unique maternal gene expression profile. The amount of Nocturnin mRNA, which was highest in oocytes, began to decrease after fertilization by the 4-cell stage and then slightly increased up to the blastocyst stage (Fig. 1A). However, the abundance of Nocturnin protein was maintained during preimplantation development, even after its mRNA level was markedly decreased (Fig. 1B, C), suggesting posttranscriptional regulation of Nocturnin gene expression. Consistent with the immunoblot analysis, intense signals of Nocturnin protein were diffusely detected in the cytoplasm and nucleus at all embryonic developmental stages (Fig. 2). As development proceeded, the Nocturnin signals, which decreased at the 4-cell stage, were clearly detectable at the 8-cell stage. Taken together, these results raise the possibility that nocturnin is functioning in the cytoplasm and nucleus during the MZT of early mouse embryos.
Fig. 1.
Expression profiles of nocturnin in mouse early embryos. A: Quantitative RT-PCR for the expression profile of Nocturnin mRNA in mouse oocytes and early mouse embryos. Different lowercase letters indicate statistical significance (P <0.05). Actin was used as a control. B, C: Immunoblot analysis for the expression profile of Nocturnin protein in mouse oocytes and early embryos. Different lowercase letters indicate statistical significance (P <0.05). Actin was used as a loading control. Moreover, 6xHis-tagged recombinant Nocturnin was used as positive control for Nocturnin antibody. The experiments were performed three times. Different superscripts indicate statistical differences (P <0.05).
Fig. 2.
Subcellular localization of Nocturnin in mouse oocytes and early mouse embryos. Subcellular localization of Nocturnin was observed during mouse preimplantation embryo development. Scale bars represent 20 μm.
Effect of Nocturnin loss of function on early mouse embryonic development
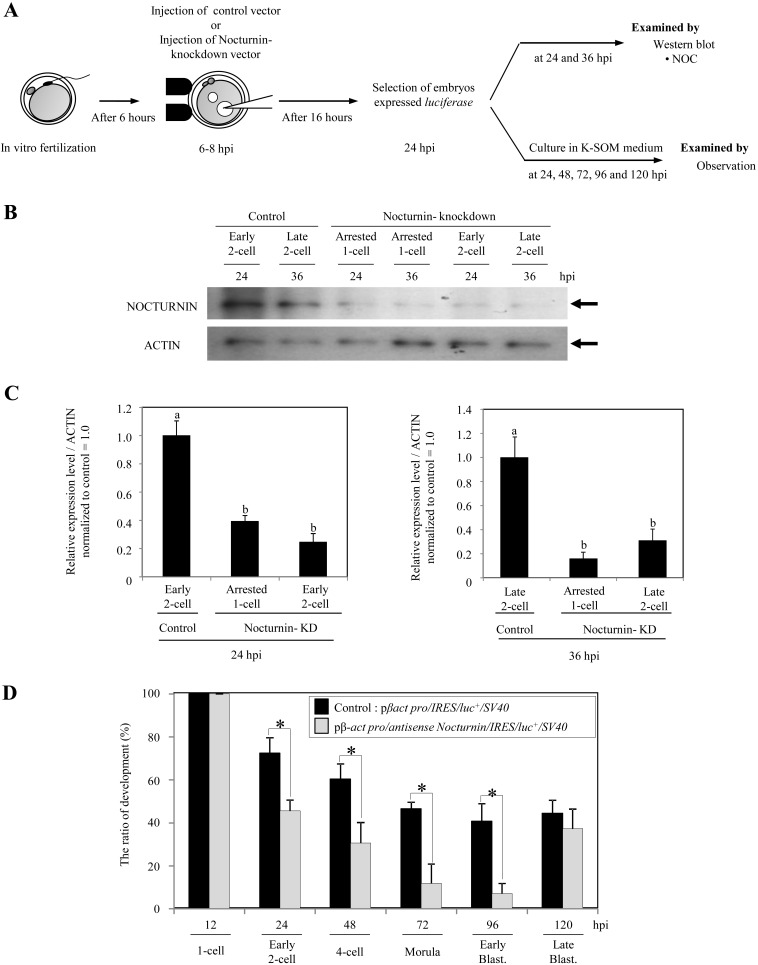
In order to clarify the role of nocturnin in the development of early mouse embryos, Nocturnin knockdown embryos were generated by pronuclei injection of Nocturnin antisense DNA with internal ribosomal entry sites (IRES)-luciferase as a marker for successful expression (Fig. 3A). At 24 hpi, immunoblot analysis revealed more than 61% loss of Nocturnin proteins in the embryos showing luciferase expression that were injected with Nocturnin antisense DNA (arrested 1-cell embryos vs. control, 0.39 ± 0.04 vs. 1.00 ± 0.1, P <0.05; early 2-cell embryos vs. control, 0.25 ± 0.06 vs. 1.00 ± 0.1, P <0.05; ave. ± S.E.) (Fig. 3B and 3C, left). Also, at 36 hpi more than 70% reduction of Nocturnin proteins was found in the Nocturnin antisense DNA injected embryos showing the luciferase activity (arrested 1-cell embryos vs. control, 0.16 ± 0.06 vs. 1.00 ± 0.17, P <0.05; early 2-cell embryos vs. control, 0.31 ± 0.1 vs. 1.00 ± 0.17, P <0.05; ave. ± S.E.) (Fig. 3B and 3C, right). As shown in Fig. 3D, in vitro development of luciferase-positive embryos injected with Nocturnin antisense expression vector to the early blastocyst stage was significantly delayed compared with that of control embryos injected with the control expression vector (Nocturnin knockdown embryos vs. control embryos: the early 2-cell stage at 24 hpi, 46 vs. 72%, P <0.05; the 4-cell stage at 48 hpi, 30 vs. 60%, P <0.05; the morula stage at 72 hpi, 12 vs. 47%, P <0.05; the early blastocyst stage at 96 hpi, 7 vs. 41%, P <0.05). Interestingly, it is reported that Nocturnin-knockout mice exhibit no obvious abnormalities in development or reproduction [24, 25]. In accordance with this, the luciferase-positive Nocturnin knockdown embryos developed to each stage (2-cell, 4-cell, 6–8-cell, morula and blastocyst) as well as the control embryos (Table 2).
Fig. 3.
Nocturnin knockdown embryos showed delayed early embryo development. A: Schematic diagram of experiments using fertilized oocytes injected with control expression vector (pβ–actin promoter/IRES/luc+/SV40) or Nocturnin antisense expression vector (pβ–actin promoter/antisense Nocturnin/IRES/luc+/SV40) from 6–8 hpi. B, C: Nocturnin-knockdown by a Nocturnin antisense expression vector was confirmed by immunoblot analysis. Actin was used as a control. The experiments were performed three times. D: Effect of Nocturnin knockdown on the development of early mouse embryos. An asterisk indicates that the value is statistically different (P <0.05).
Table 2. Effect of Nocturnin knockdown on the development of early mouse embryos.
| Injected vectors | No. (%) of embryos | No. (%) of embryos developed to | ||||||||
| Injected | Survived | Expressed | 1-cell | 2-cell | 4-cell | 6 to 8-cell | Morulae | Blastocysts | Degenerated | |
| Control | 341 | 252 (74) | 204 (81) | 204 (100) | 156 (76) | 130 (64) | 114 (56) | 99 (49) | 83 (41) | 77 (38) |
| Antisense Nocturnin | 545 | 324 (59) | 88 (27) | 88 (100) | 61 (69) | 52 (59) | 43 (49) | 33 (38) | 28 (32) | 34 (39) |
Effect of Nocturnin gain of function on early mouse embryonic development
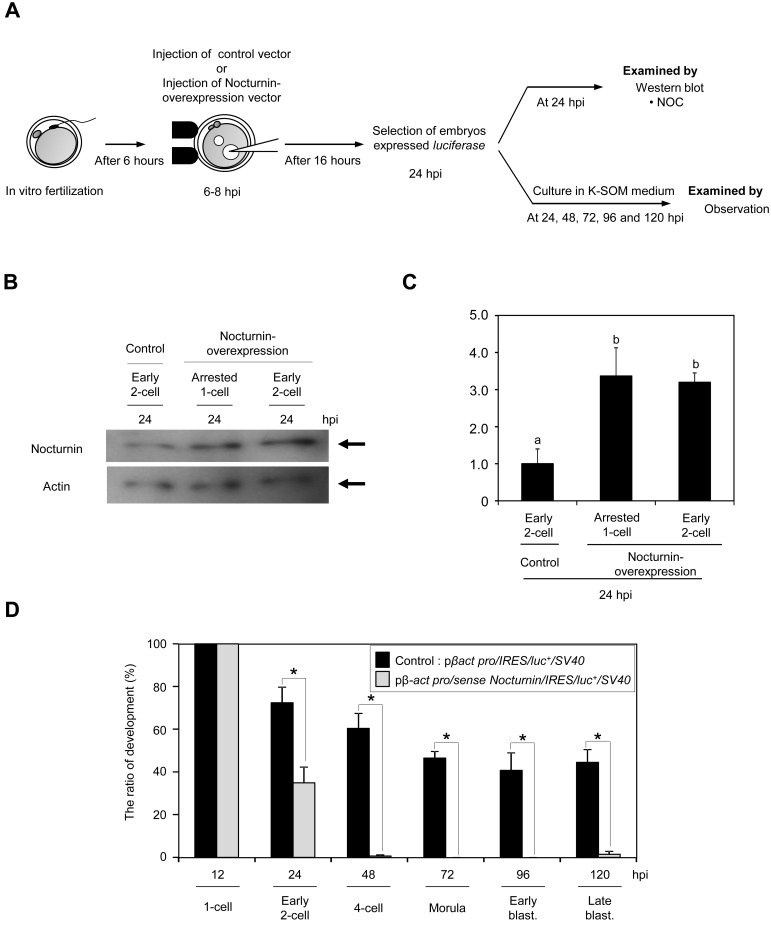
Next, we examined the effect of Nocturnin overexpression on the development of early mouse embryo. Embryos overexpressing Nocturnin were generated by pronuclei injection of Nocturnin cDNA with internal ribosomal entry sites (IRES)-luciferase as a marker for successful expression (Fig. 4A). At 24 hpi, the amount of Nocturnin proteins was significantly increased by approximately 336% or 320% in arrested 1-cell or early 2-cell embryos compared with that of the control embryos, respectively (Fig. 4B, C). The majority of the luciferase-positive embryos injected with Nocturnin expression vector were arrested at the 1-cell to 2-cell stages, and only 4% of them developed to the 4-cell stage, whereas 61% of control embryos developed normally to the 4-cell stage (Table 3). Moreover, the luciferase-positive Nocturnin-overexpressed embryos failed to develop beyond the 1-cell or 2-cell stages; only 35% and 0.6% of the overexpressed embryos developed to the 2-cell and 4-cell stages, respectively (Fig. 4D).
Fig. 4.
Embryos with overexpression of Nocturnin were arrested at the 1-cell to 2-cell stages. A: Schematic diagram of experiments using fertilized oocytes injected with control expression vector (pβ–actin promoter/IRES/luc+/SV40) or Nocturnin expression vector (pβ–actin promoter/sense Nocturnin/IRES/luc+/SV40) from 6–8 hpi. B,C: Nocturnin-overexpression by a Nocturnin expression vector was confirmed by immunoblot analysis. Actin was used as a control. The experiments were performed three times. D: Effect of Nocturnin overexpression on the development of early mouse embryos. An asterisk indicates that the value is statistically different (P <0.05).
Table 3. Effect of Nocturnin-overexpression on the development of early mouse embryos.
| Injected vectors | No. (%) of embryos | No. (%) of embryos developed to | |||||||||
| Injected | Survived | Expressed | 1-cell | 2-cell | 4-cell | 6 to 8-cell | Morulae | Blastocysts | Degenerated | ||
| Control | 341 | 252 (74) | 204 (81) | 204 (100) | 156 (76) | 130 (64) | 114 (56) | 99 (49) | 83 (41) | 77 (38) | |
| Sense Nocturnin | 193 | 151 (78) | 113 (75) | 113 (100) | 47 *(42) | 5 *(4) | 1 *(1) | 1* (1) | 1 *(1) | 108 *(96) | |
* Significant difference from control (P<0.05).
Discussion
In this study, we showed that the circadian deadenylase nocturnin is expressed in mouse MII oocytes and early embryos and that precise expression of nocturnin is essential for normal preimplantation development of embryos.
In early mammalian embryogenesis, degradation of stored maternal mRNAs and proteins is one of the critical events that govern the MZT for embryonic development [26]. Also, correct regulation of the MZT is an important process for remodeling of an oocyte into a totipotent zygote. Recently, we reported that proteasomal degradation of maternal proteins is implicated in establishment of the embryonic program during the MZT [10, 27]. Our data may provide evidence for the possible involvement of a circadian deadenylase, nocturnin, in maternal mRNA degradation during the mouse MZT.
Circadian genes expressed in most organs, tissues and cells encode transcription factors that regulate the circadian clock in mammals [28]. Nocturnin also has rhythmic expression in multiple mouse tissues under the control of the circadian clock [29]. However, we have previously demonstrated that circadian genes in mammalian oocytes and early embryos are not involved in the ticking of the circadian clock [22, 28]. Indeed, we observed no rhythmic expression of the circadian deadenylase nocturnin in preimplantation embryos. Thus, nocturnin likely plays a role as a deadenylase in early mouse embryos beyond circadian control, although nocturnin expressed in early mouse embryos was not directly proven to possess deadenylase activity in this study.
In the course of functional analysis by loss- and gain-of-function approaches, we noticed that precise expression of nocturnin in early embryos is critical for normal preimplantation development. Since it is known that there is functional redundancy among deadenylases including nocturnin in eukaryotic cells [12], moderate loss of nocturnin seems to result in delayed embryonic development. Meanwhile, the high amount of nocturnin expression was severely harmful to early embryonic development. In mammalian cells, nocturnin overexpression significantly increases the expression of fatty acid-binding protein 4 (ap2) and peroxisome proliferator-activated receptor-γ2 (PPAR-γ2) [30], whether bona fide target mRNAs directly depend on Nocturnin for deadenylation remains unknown in early mouse embryos. Therefore, these results suggest that exogenously overexpressed nocturnin may perturb regulated posttranscriptional expression of some target maternal mRNAs in early mouse embryos.
Understanding the function of deadenylases including nocturnin in degradation of stored maternal mRNAs during the mouse MZT helps to elucidate molecular mechanisms governing the remodeling of the oocyte into a totipotent zygotes.
Acknowledgment
We thank N Backes-Kamimura and J Walhelm-Kimura for manuscript support and editing. This study was supported in part by a Grant-in-Aid for the 21st Century COE Program of the Ministry of Education, Culture, Sports, Science and Technology of Japan (to SN), Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science 23658292 and 23380164 (to KM) and a grant from the INAMORI Foundation (to KM).
References
- 1.Bettegowda A, Smith GW. Mechanisms of maternal mRNA regulation: implications for mammalian early embryonic development. Front Biosci 2007; 12: 3713–3726 [DOI] [PubMed] [Google Scholar]
- 2.Stitzel ML, Seydoux G. Regulation of the oocyte-to-zygote transition. Science 2007; 316: 407–408 [DOI] [PubMed] [Google Scholar]
- 3.Schultz RM. The molecular foundations of the maternal to zygotic transition in the preimplantation embryo. Hum Reprod Update 2002; 8: 323–331 [DOI] [PubMed] [Google Scholar]
- 4.Minami N, Suzuki T, Tsukamoto S. Zygotic gene activation and maternal factors in mammals. J Reprod Dev 2007; 53: 707–715 [DOI] [PubMed] [Google Scholar]
- 5.Huo LJ, Fan HY, Zhong ZS, Chen DY, Schatten H, Sun QY. Ubiquitin-proteasome pathway modulates mouse oocyte meiotic maturation and fertilization via regulation of MAPK cascade and cyclin B1 degradation. Mech Dev 2004; 121: 1275–1287 [DOI] [PubMed] [Google Scholar]
- 6.Huo LJ, Yu LZ, Liang CG, Fan HY, Chen DY, Sun QY. Cell-cycle-dependent subcellular localization of cyclin B1, phosphorylated cyclin B1 and p34cdc2 during oocyte meiotic maturation and fertilization in mouse. Zygote 2005; 13: 45–53 [DOI] [PubMed] [Google Scholar]
- 7.Huo LJ, Zhong ZS, Liang CG, Wang Q, Yin S, Ai JS, Yu LZ, Chen DY, Schatten H, Sun QY. Degradation of securin in mouse and pig oocytes is dependent on ubiquitin-proteasome pathway and is required for proteolysis of the cohesion subunit, Rec8, at the metaphase-to-anaphase transition. Front Biosci 2006; 11: 2193–2202 [DOI] [PubMed] [Google Scholar]
- 8.Evsikov AV, de Vries WN, Peaston AE, Radford EE, Fancher KS, Chen FH, Blake JA, Bult CJ, Latham KE, Solter D, Knowles BB. Systems biology of the 2-cell mouse embryo. Cytogenet Genome Res 2004; 105: 240–250 [DOI] [PubMed] [Google Scholar]
- 9.Solter D, Hiiragi T, Evsikov AV, Moyer J, De Vries WN, Peaston AE, Knowles BB. Epigenetic mechanisms in early mammalian development. Cold Spring Harb Symp Quant Biol 2004; 69: 11–17 [DOI] [PubMed] [Google Scholar]
- 10.Shin SW, Shimizu N, Tokoro M, Nishikawa S, Hatanaka Y, Anzai M, Hamazaki J, Kishigami S, Saeki K, Hosoi Y, Iritani A, Murata S, Matsumoto K. Mouse zygote-specific proteasome assembly chaperone important for maternal-to-zygotic transition. Biol Open 2013; 2: 170–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suh N, Blelloch R. Small RNAs in early mammalian development: from gametes to gastrulation. Development 2011; 138: 1653–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldstrohm AC, Wickens M. Multifunctional deadenylase complexes diversify mRNA control. Nat Rev Mol Cell Biol 2008; 9: 337–344 [DOI] [PubMed] [Google Scholar]
- 13.Wu X, Brewer G. The regulation of mRNA stability in mammalian cells: 2.0. Gene 2012; 500: 10–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Radford HE, Meijer HA, de Moor CH. Translational control by cytoplasmic polyadenylation in Xenopus oocytes. Biochim Biophys Acta2008; 1779: 217–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Semotok JL, Cooperstock RL, Pinder BD, Vari HK, Lipshitz HD, Smibert CA. Smaug recruits the CCR4/POP2/NOT deadenylase complex to trigger maternal transcript localization in the early drosophila embryo. Curr Biol 2005; 15: 284–294 [DOI] [PubMed] [Google Scholar]
- 16.Rouget C, Papin C, Boureux A, Meunier AC, Franco B, Robine N, Lai EC, Pelisson A, Simonelig M. Maternal mRNA deadenylation and decay by the piRNA pathway in the early Drosophila embryo. Nature 2010; 467: 1128–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mizuno S, Sono Y, Matsuoka T, Matsumoto K, Saeki K, Hosoi Y, Fukuda A, Morimoto Y, Iritani A. Expression and subcellular localization of GSE protein in germ cells and preimplantation embryos. J Reprod Dev 2006; 52: 429–438 [DOI] [PubMed] [Google Scholar]
- 18.Matsuoka T, Sato M, Tokoro M, Shin SW, Uenoyama A, Ito K, Hitomi S, Amano T, Anzai M, Kato H, Mitani T, Saeki K, Hosoi Y, Iritani A, Matsumoto K. Identification of ZAG1, a novel protein expressed in mouse preimplantation, and its putative roles in zygotic genome activation. J Reprod Dev 2008; 54: 192–197 [DOI] [PubMed] [Google Scholar]
- 19.Baggs JE, Green CB. Nocturnin, a deadenylase in Xenopus laevis retina: a mechanism for posttranscriptional control of circadian-related mRNA. Curr Biol 2003; 13: 189–198 [DOI] [PubMed] [Google Scholar]
- 20.Garbarino-Pico E, Niu S, Rollag MD, Strayer CA, Besharse JC, Green CB. Immediate early response of the circadian polyA ribonuclease nocturnin to two extracellular stimuli. RNA 2007; 13: 745–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lawitts JA, Biggers JD. Culture of preimplantation embryos. Guide to techniques in mouse development1993; 225: 153–164. [DOI] [PubMed] [Google Scholar]
- 22.Amano T, Matsushita A, Hatanaka Y, Watanabe T, Oishi K, Ishida N, Anzai M, Mitani T, Kato H, Kishigami S, Saeki K, Hosoi Y, Iritani A, Matsumoto K. Expression and functional analyses of circadian genes in mouse oocytes and preimplantation embryos: Cry1 is involved in the meiotic process independently of circadian clock regulation. Biol Reprod 2009; 80: 473–483 [DOI] [PubMed] [Google Scholar]
- 23.Tokoro M, Shin SW, Nishikawa S, Lee HH, Hatanaka Y, Amano T, Mitani T, Kato H, Anzai M, Kishigami S, Saeki K, Hosoi Y, Iritani A, Matsumoto K. Deposition of acetylated histones by RNAP II promoter clearance may occur at onset of zygotic gene activation in preimplantation mouse embryos. J Reprod Dev 2010; 56: 607–615 [DOI] [PubMed] [Google Scholar]
- 24.Stubblefield JJ, Terrien J, Green CB. Nocturnin: at the crossroads of clocks and metabolism. Trends Endocrinol Metab 2012; 23: 326–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Green CB, Douris N, Kojima S, Strayer CA, Fogerty J, Lourim D, Keller SR, Besharse JC. Loss of Nocturnin, a circadian deadenylase, confers resistance to hepatic steatosis and diet-induced obesity. Proc Natl Acad Sci USA 2007; 104: 9888–9893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li L, Zheng P, Dean J. Maternal control of early mouse development. Development 2010; 137: 859–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shin SW, Tokoro M, Nishikawa S, Lee HH, Hatanaka Y, Nishihara T, Amano T, Anzai M, Kato H, Mitani T, Kishigami S, Saeki K, Hosoi Y, Iritani A, Matsumoto K. Inhibition of the ubiquitin-proteasome system leads to delay of the onset of ZGA gene expression. J Reprod Dev 2010; 56: 655–663 [DOI] [PubMed] [Google Scholar]
- 28.Amano T, Tokunaga K, Kakegawa R, Yanagisawa A, Takemoto A, Tatemizo A, Watanabe T, Hatanaka Y, Matsushita A, Kishi M, Anzai M, Kato H, Mitani T, Kishigami S, Saeki K, Hosoi Y, Iritani A, Matsumoto K. Expression analysis of circadian genes in oocytes and preimplantation embryos of cattle and rabbits. Anim Reprod Sci. 2010; 121: 225–235 [DOI] [PubMed] [Google Scholar]
- 29.Wang Y, Osterbur DL, Megaw PL, Tosini G, Fukuhara C, Green CB, Besharse JC. Rhythmic expression of Nocturnin mRNA in multiple tissues of the mouse. BMC Dev Biol 2001; 1: 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kawai M, Green CB, Lecka-Czernik B, Douris N, Gilbert MR, Kojima S, Ackert-Bicknell C, Garg N, Horowitz MC, Adamo ML, Clemmons DR, Rosen CJ. A circadian-regulated gene, Nocturnin, promotes adipogenesis by stimulating PPAR-gamma nuclear translocation. Proc Natl Acad Sci USA 2010; 107: 10508–10513 [DOI] [PMC free article] [PubMed] [Google Scholar]