Abstract
In mice, one of the major epigenetic errors associated with somatic cell nuclear transfer (SCNT) is ectopic expression of Xist during the preimplantation period in both sexes. We found that this aberrant Xist expression could be impeded by deletion of Xist from the putative active X chromosome in donor cells. In male clones, it was also found that prior injection of Xist-specific siRNA could significantly improve the postimplantation development of cloned embryos as a result of a significant repression of Xist at the morula stage. In this study, we examined whether the same knockdown strategy could work as well in female SCNT-derived embryos. Embryos were reconstructed with cumulus cell nuclei and injected with Xist-specific siRNA at 6–7 h after oocyte activation. RNA FISH analysis revealed that siRNA treatment successfully repressed Xist RNA at the morula stage, as shown by the significant decrease in the number of cloud-type Xist signals in the blastomere nuclei. However, blastomeres with different sizes (from “pinpoint” to “cloud”) and numbers of Xist RNA signals remained within single embryos. After implantation, the dysregulated Xist expression was normalized autonomously, as in male clones, to a state of monoallelic expression in both embryonic and extraembryonic tissues. However, at term there was no significant improvement in the survival of the siRNA-injected cloned embryos. Thus, siRNA injection was largely effective in repressing the Xist overexpression in female cloned embryos but failed to rescue them, probably because of an inability to mimic consistent monoallelic Xist expression in these embryos. This could only be achieved in female embryos by applying a gene knockout strategy rather than an siRNA approach.
Keywords: Knockdown, Mouse, Nuclear transfer, RNA FISH, Xist
Since the first birth of mammals from somatically cloned embryos in the 1990s, cloning mammals by somatic cell nuclear transfer (SCNT) has been very inefficient mostly because of the loss of embryos transferred during pregnancy or from abnormal delivery at term [1,2,3]. Although the abnormal development of SCNT-derived embryos might be attributable to incomplete or erroneous reprogramming of the donor genome, very little information is available for understanding the molecular mechanisms of such reprogramming, making technical improvement of SCNT very difficult. One of the successful technical breakthroughs for SCNT is the treatment of reconstructed embryos with histone deacetylase inhibitors (HDACis), which are expected to enhance the accessibility of the putative ooplasmic reprogramming factors by loosening the chromatin structure [4,5,6]. Their effects are most profound in mouse SCNT experiments. For example, the birthrates of cloned mice increased twofold to fivefold, depending on the donor mouse strain and the type of HDACi used [7]. Currently, many laboratories routinely use trichostatin A, a potent HDACi, to obtain consistent results from mouse cloning [7].
Another promising approach for improvements in SCNT is targeting specific genes that may be the keys for normal development of cloned embryos. Recently, we performed genome-wide gene expression analysis using SCNT-generated blastocysts and found that X-linked genes were specifically downregulated in these cloned embryos [8]. At least two causes were responsible for this X-linked gene downregulation. One was the chromosome-wide downregulation by ectopic expression of Xist, a noncoding RNA gene responsible for the onset of X chromosome inactivation (XCI). Another was the repression of two discrete gene families, Magea and Xlr, which are enriched with a somatic cell type repressive histone modification, the dimethylation of histone H3 at lysine 9 [8, 9]. Of these, the ectopic expression of Xist, at least, was proven to be one of the major causes of inefficient SCNT in mice, because deletion of Xist from the active X chromosome in the donor genome resulted in about a 10-fold increase in the birthrates of cloned offspring in both sexes [8]. To gain better insight into the mechanisms underlying the developmental arrest of SCNT-generated embryos caused by Xist upregulation, Xist was repressed temporally in SCNT-generated embryos by the injection of specific short interfering (si) RNA. Prior siRNA injection efficiently repressed the ectopic Xist expression in male SCNT-generated embryos at least up to the morula stage [10]. After transfer of these embryos to recipient foster-mothers, they showed 10 times higher survival than controls. Interestingly, RNA fluorescent in situ hybridization (FISH) analysis revealed that aberrant Xist expression was autonomously corrected in both embryonic and extraembryonic tissues irrespective of the siRNA treatment [10]. Consequently, the expression pattern of Xist in male cloned embryos after implantation was indistinguishable from that of normally fertilized embryos. These results indicated that the fate of SCNT-generated embryos is determined during the very short period before implantation.
In the above-mentioned siRNA study, only male SCNT-generated embryos were used because it was anticipated that exact quantitative adjustment of the Xist expression level for female embryos by simple siRNA injection would be difficult. However, we injected Xist-specific siRNA into fertilized embryos and found that the sex ratio of the resultant offspring was 1:1 [10]. This result indicated that temporally limited downregulation of Xist in female preimplantation embryos would not be harmful for their postimplantation development to term. If this is true, we may expect that downregulation of Xist by siRNA injection might be also beneficial for development of female SCNT-generated embryos. In the male Xist-knockdown study, the keys for the success were the downregulation of Xist at the morula stage and the autonomous correction of Xist regulation after implantation [10]. These are also important issues for improving our understanding of XCI regulation during the pre- and peri-implantation periods in mouse embryos.
In the present study, we investigated the developmental potential of cumulus-derived female SCNT-generated embryos following injection with Xist-specific siRNA at the pronuclear stage. In some experiments, we investigated Xist expression during the pre- and postimplantation stages by RNA FISH to see whether there is any sex-specific Xist regulation in cloned embryos.
Materials and Methods
Animals
B6D2F1 strain female mice (Japan SLC, Shizuoka, Japan) were used to collect oocytes and donor cumulus cells. For embryo transfer experiments, ICR strain female mice (CLEA Japan, Tokyo, Japan) were used as pseudopregnant recipients after being mated with vasectomized ICR strain male mice. All mice were maintained under specific-pathogen-free conditions, provided with water and commercial laboratory mouse chow ad libitum and housed under controlled lighting conditions (light: 0700–2100 h). All animal experiments described here were approved by the Animal Experimentation Committee at the RIKEN Tsukuba Institute and were performed in accordance with the committee's guiding principles.
Preparation of embryos
Nuclear transfer was carried out as described previously [11,12]. For oocyte collection, B6D2F1 females were induced to superovulate by an injection of 7.5 IU equine chorionic gonadotropin (eCG; Peamex, Sankyo, Tokyo, Japan), followed 48–50 h later by 7.5 IU human chorionic gonadotropin (hCG; Puberogen, Sankyo). Cumulus-oocyte complexes were placed in KSOM medium [13] containing 0.1% hyaluronidase (Sigma-Aldrich, St. Louis, MO, USA) to remove the cumulus cells and matrix. After washing with fresh KSOM, oocytes were moved to Hepes-buffered KSOM with 7.5 μg/ml cytochalasin B (Calbiochem, San Diego, CA, USA) on the stage of an inverted microscope, and nuclei were removed with a small amount of cytoplasm using a piezo-driven micromanipulator (Prime Tech, Ibaraki, Japan). Enucleated oocytes were cultured in KSOM in an incubator for 30–60 min to allow the oolemma to recover. The nuclei of cumulus cells obtained from superovulated B6D2F1 females were injected into the enucleated oocytes using a piezo-driven micromanipulator. After nuclear transfer, reconstructed oocytes were cultured with KSOM and transferred into Ca2+-free KSOM including 2.5 mM SrCl2, 5 μg/ml cytochalasin B and 0, 5 or 50 nM trichostatin A (Sigma-Aldrich). One hour later, activated oocytes were transferred into KSOM containing 5 μg/ml cytochalasin B and 50 nM trichostatin A and were cultured further for 5 h. After washing, the oocytes were cultured in KSOM containing 50 nM trichostatin A for 2 h and then cultured in fresh KSOM for 24 h. For postimplantation embryo collection experiments, we used 5 μM latrunculin A instead of cytochalasin B, as described previously [14]. Embryo culture was performed at 37.5 C under 5% CO2 in air throughout all experiments.
In some experiments, female embryonic stem (ES) cells were used as nuclear donors, and experiments were performed as described above. The donor ES cell line used was BRC6, which was derived from blastocysts of the C57BL/6NCrSlc strain and maintained at the RIKEN BioResource Center (catalogue number: AES0010). ES cells after thawing were cultured for 4 days according to Ying [15] but without any passages to achieve the confluent state before nuclear transfer.
Normally fertilized embryos were generated by conventional in vitro fertilization (IVF), as described previously [8]. Oocytes from C57BL/6NCrSlc females (Japan SLC) and epididymal spermatozoa from DBA/2CrSlc males (Japan SLC) were used to obtain IVF embryos with the same B6D2F1 genetic background as cloned embryos. Only female embryos were used for analysis after sexing embryos by genomic polymerase chain reaction (PCR) amplification for the Eif2s3y gene. The primer sequences used were 5′–AGCCGACGAGTTGAGAAACAC–3′ and 5′–TCACGCCTCTTCTTATCTGGC–3′.
siRNA injection
When reconstructed oocytes formed distinct pseudopronuclei (at 6–7 h after activation), siRNA (5 μM final concentration) against Xist was injected using a piezo-driven micropipette, as described previously [10]. Some reconstructed oocytes were injected with nonspecific siRNA and used as controls.
Embryo transfer
Reconstructed embryos that reached the 2-cell stage after 24 h of culture in KSOM were transferred into the oviducts of pseudopregnant ICR strain female mice mated with vasectomized males the day before. On day 20, the recipients were examined for the presence of fetuses, and live pups were nursed by lactating ICR foster-mothers. In some experiments, postimplantation embryos were recovered from the uteri at day 5.5 under a stereomicroscope.
Quantitative RT–PCR (QPCR)
cDNAs of single embryos were synthesized with Cells-to-cDNA II Kits (Ambion). QPCR was performed using QuantiTect SYBR Green PCR kits (Qiagen, Tokyo, Japan) and an ABI Prism 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA, USA). All PCR runs were performed at an annealing temperature of 60 C for 50 cycles. The primer sequences were as follows: 5′–GTCAGCAAGAGCCTTGAATTG–3′ and 5′–TTTGCTGAGTCTTGAGGAGAATC–3′ for Xist and 5′–CAACAGCAACTCCCACTCTTC–3′ and 5′–CCTGTTGCTGTAGCCGTATTC–3′ for Gapdh.
RNA FISH
A probe to detect Xist RNA was prepared by nick translation with Cy3-dCTP (GE Healthcare, Buckinghamshire, UK) from a Xist genomic clone. Embryos were incubated in 0.1% Triton X-100 in phosphate buffered saline for 10 sec on ice and fixed with 4% paraformaldehyde for 10 min at room temperature. Hybridization was carried out at 37 C overnight. The nuclei of embryos were stained with DAPI. Xist signals were observed using a confocal scanning laser microscope (Digital Eclipse C1; Nikon Instruments, Tokyo, Japan).
Statistical analysis
For statistical analysis of the RNA FISH data, the percentages of blastomeres with no cloud-type Xist signals (2 pinpoint + 1 pinpoint + no signal) in individual cloned embryos were transformed using arcsine transformation and then analyzed by one-way analysis of variance (ANOVA). For quantitative RT–PCR, the values were analyzed by one-way ANOVA. The developmental efficiencies of embryos were analyzed by Chi-squared test with Yates' correction. A probability of P<0.05 was considered statistically significant.
Results
Development of cumulus cell-derived cloned embryos injected with Xist-specific siRNA
About 90% of cumulus cell-derived cloned embryos had developed to the 2-cell stage by 24 h, irrespective of the day of experiment or treatment with siRNA injection. When embryos were treated with 0 or 5 nM TSA, the rates of implantation and full-term development were slightly higher in the siRNA-injected group than in the control group, but they were not significantly different (P>0.05; Table 1). As expected, the control cloned embryos showed improved development with 50 nM trichostatin A treatment, but the siRNA-injected cloned embryos did not (Table 1). These results indicate that the prior Xist-siRNA injection into cloned embryos reconstructed using cumulus cell nuclei had no beneficial effect on the in vivo development of cloned embryos.
Table 1. In vivo development of cumulus cell-derived cloned embryos with or without Xist-siRNA injection.
| Trichostatin A (nM) | Treatment | No. embryos transferred | No. (%) implanted | No. (%) born | No. (%) placenta only |
| 0 | Xist siRNA | 91 | 42 (46) | 4 (4) | 0 (0) |
| Control siRNA | 48 | 17 (35) | 1 (2) | 1 (2) | |
| 5 | Xist siRNA | 67 | 32 (48) | 1 (2) | 1 (2) |
| Control siRNA | 46 | 16 (35) | 0 (0) | 1 (2) | |
| 50 | Xist siRNA | 150 | 39 (26) | 4 (3) | 5 (3) |
| Control siRNA | 169 | 66 (39) | 13 (8) | 4 (2) |
There was no significant difference in the results between the Xist-specific siRNA-injected and control groups with the same trichostatin A treatment (Chi-squared test with Yates' correction).
Effect of siRNA treatment on Xist expression pattern in female cloned embryos at the morula and blastocyst stages
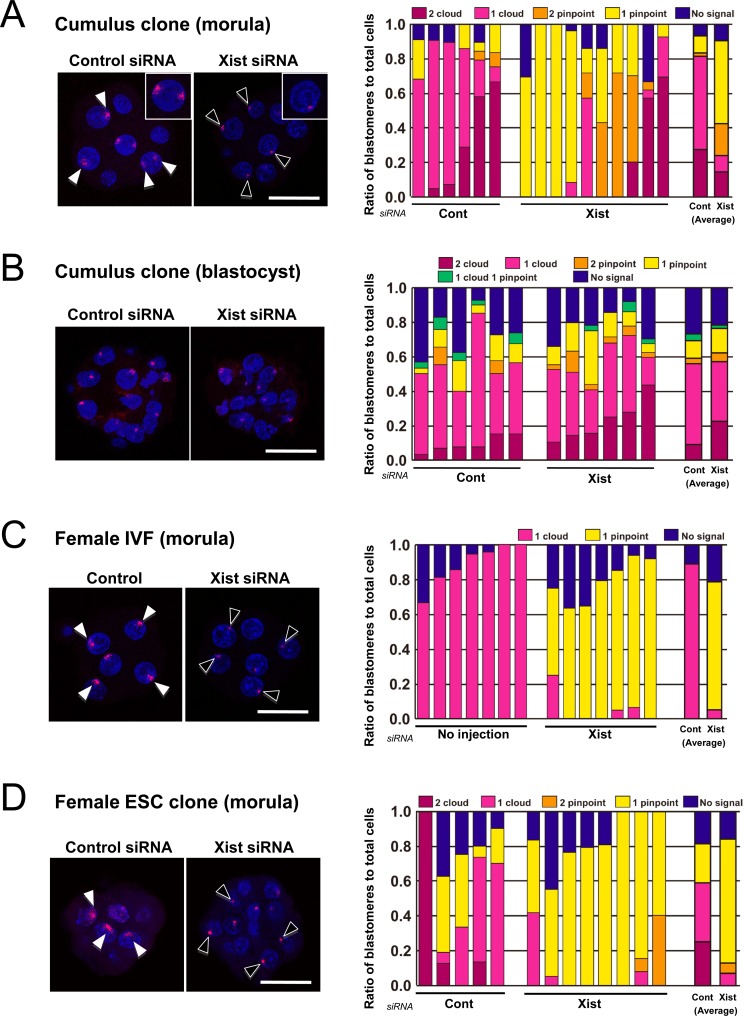
To evaluate the effect of Xist-specific siRNA on the expression of Xist in female cloned embryos, we observed Xist RNA deposition in cumulus cell-derived cloned embryos with or without Xist-siRNA treatment using RNA FISH. At the morula stage (72 h after oocyte activation), two “cloud” Xist RNA signals, a hallmark of extensive Xist RNA coating [16], were found in more than half (5/6) of the control cloned embryos, indicating that Xist had been expressed ectopically from the putative active X chromosome in these blastomeres (Fig. 1A). Following Xist-specific siRNA injection, the proportion of embryos having ectopic Xist expression was reduced to 3/10. Furthermore, Xist-specific siRNA treatment increased the percentage of blastomeres with no cloud signal from 19 to 76% (P<0.005; Fig. 1A). At the blastocyst stage (96 h after activation), all the cloned embryos showed blastomeres with ectopic Xist expression, irrespective of the siRNA treatment (Fig. 1B). In the Xist-specific siRNA group, the average percentage of blastomeres without a cloud signal was 41%, which was not significantly different from that in the control group (40%; P>0.05; Fig. 1B). These findings collectively indicated that in cumulus cell-derived cloned embryos, the Xist expression was effectively repressed by Xist-specific siRNA at the morula stage, but its efficacy diminished thereafter. This stage-dependent effect of siRNA on Xist expression was very similar to that found in male cloned embryos [10]. However, the patterns of Xist signals were very complicated in female cloned embryos because of the presence of two Xist alleles, each of which can show either no, pinpoint or cloud signals even after Xist-specific siRNA treatment (Fig. 1A and B). By contrast, in normally fertilized female embryos, the Xist signal pattern was simple, usually with one pinpoint and one cloud signal in each embryo treated with Xist-specific siRNA and control siRNA, respectively (Fig. 1C).
Fig. 1.
RNA-FISH analyses of Xist in cumulus cell-derived cloned embryos (A and B), female IVF embryos (C), and female ES cell-derived embryos (D). The right panels show the ratios of blastomeres classified according to the number of cloud or pinpoint signals of Xist RNA. A: At the morula stage (72 h after activation), the Xist siRNA treatment significantly increased the proportion of blastomeres without cloud signals (counting those with two pinpoints + one pinpoint + no signal) (19 vs. 76% on average; P<0.005). B: At the blastocyst stage (96 h after activation), there was no significant difference in the proportion of blastomeres without cloud signals between the control and Xist-specific siRNA groups (40 vs. 41%; P>0.05). C: In normally fertilized female embryos, the majority of blastomeres showed expression of one cloud signal in controls and one pinpoint signal in Xist-specific siRNA-injected embryos. D: In embryos cloned from female ES cells, the expression patterns or Xist with or without Xist-specific siRNA treatment were similar to those found in cumulus cell-derived cloned embryos (see A). (Scale bar, 50 μm).
To exclude the possibility that the results observed above were specific to embryos cloned from cumulus cells, we also reconstructed cloned embryos using female ES cells and examined them for the Xist expression pattern by RNA FISH at the morula stage. The percentage of blastomeres lacking a cloud signal was increased from 41 to 93% by siRNA treatment (P<0.005; Fig. 1D). This clearly indicated that siRNA was also effective in repressing the Xist RNA in ES-derived cloned embryos.
The effects of Xist-siRNA on the Xist expression levels in cumulus cell-derived cloned embryos
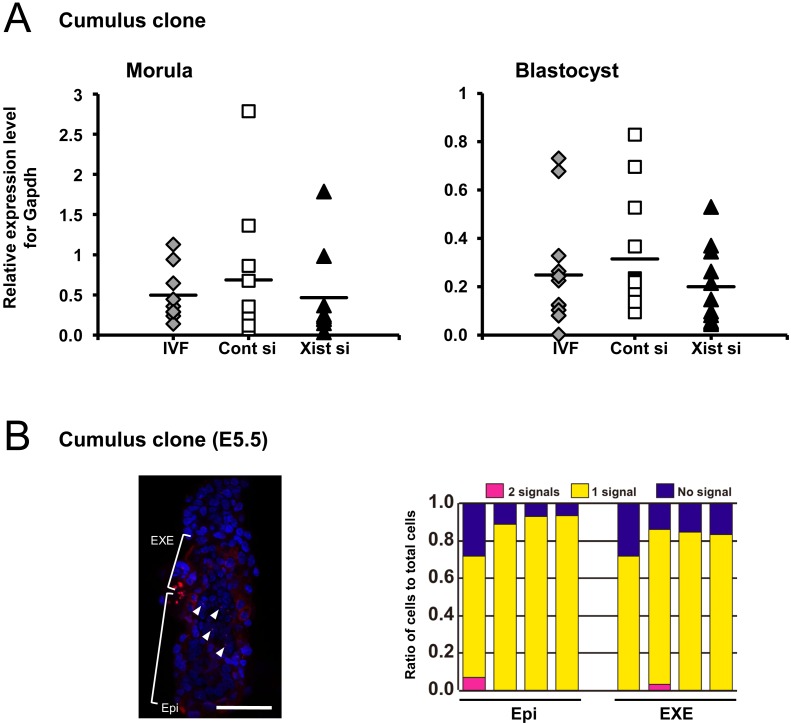
We then examined the effect of Xist-siRNA injection on the Xist gene expression levels by QPCR using single embryos. At the morula stage, the control cloned embryos unexpectedly showed Xist expression at the same level as fertilized embryos (P>0.05). Xist-specific siRNA injection decreased the Xist expression level of cloned embryos, but the effect was not statistically significant (P>0.05; Fig. 2A). At the blastocyst stage, upregulation of Xist in control cloned embryos was observed, but here again, there were no significant differences among the three experimental groups (P>0.05; Fig. 2A).
Fig. 2.
A: QPCR for Xist in cumulus cell-derived cloned embryos injected with Xist-specific siRNA and cultured for 72 h (morula) and 96 h (blastocyst). There were no differences among the three experimental groups, including IVF-derived embryos, at either the morula or blastocyst stages (P>0.05). B: The XCI status of cloned embryos at embryo day 5.5. A representative image of RNA FISH for Xist (red) with nuclear staining (blue) in cumulus cell-derived cloned embryos (left), and the ratio of cells classified according to the number of Xist RNA signals (right). Most cells in the epiblast (Epi) and extraembryonic ectoderm (EXE) showed a single Xist signal in the nucleus. (Scale bar, 50 μm).
Xist expression in cumulus cell-derived cloned embryos at day 5.5
We examined the Xist expression pattern in cumulus cell-derived cloned embryos at day 5.5 by RNA FISH to determine whether ectopic Xist expression seen during the preimplantation stage would be corrected autonomously after implantation, as in male cloned embryos without siRNA treatment [10]. In all the embryos we examined (n = 4), the majority of cells in both the embryonic and extraembryonic tissues had one Xist signal (Fig. 2B), indicating that Xist expression was normalized, as in male cloned embryos reported previously [10].
Discussion
The primary purpose of this study was to determine whether downregulation of Xist in female cloned embryos by specific siRNA treatment could improve their postimplantation development to term. Although Xist-specific siRNA significantly repressed the Xist RNA coating on the X chromosome at the morula stage, it had no or very little beneficial effect on the embryos' subsequent development. This result contrasts sharply with that of our previous study using male cloned embryos [10]. The survival rates of the transferred embryos that had been injected with Xist-specific siRNA were consistently greater than 10%, reaching about 20% in combination with trichostatin A treatment [10]. It is known that female embryos that carry a paternal mutant Xist die during the early postimplantation stage because of failure of XCI in the extraembryonic ectoderm [17]. Therefore, it was possible that Xist knockdown could also affect development of the extraembryonic tissues, resulting in the low birthrate of female clones. According to our observations, however, this scenario was found to be unlikely because the Xist expression was normalized by the postimplantation stage, as shown by RNA FISH for Xist at day 5.5.
Another possible cause of this failure by Xist-specific siRNA to rescue female cloned embryos might have been the great variability in Xist expression patterns between blastomeres. As far as we observed by RNA FISH, the repressing effect of Xist siRNA in female cloned embryos was very similar to that in male cloned embryos, because the number of blastomeres without cloud signals increased at the morula stage, but not at the blastocyst stage. However, female cloned embryos were inherently prone to double patterns of Xist expression, and the extent of this can vary between embryos and even between blastomeres in the same embryos [8]. As shown by RNA FISH analysis in the present study, this variability could not be rescued by Xist-specific siRNA injection. Because Xist exerts its effect in cis, the abnormal Xist RNA levels in each allele might lead to catastrophic X-linked gene dysregulation. The coexistence of blastomeres with such different gene expression patterns might have reduced the developmental potential of the embryos. Our quantitative Xist expression analysis failed to show any significant downregulation of the Xist expression level in individual embryos as a whole. This might also reflect the variation in Xist expression pattern between embryos and between blastomeres within embryos. In theory, it is technically difficult with the conventional siRNA strategy to induce normal monoallelic expression. By contrast, complete deletion of one of the Xist alleles from the donor genome allowed the cloned embryos to mimic the normal pattern, resulting in remarkable improvements in the birthrates of cloned offspring [8].
We clearly demonstrated that the aberrant Xist expression pattern in cumulus cell-derived cloned embryos was corrected at implantation in both embryonic and extraembryonic tissues, as in Sertoli cell-derived cloned embryos [10]. It is well known that in female embryos, both X chromosomes become active in the inner cell mass of the late blastocyst stage, followed by new random XCI initiating in the epiblast [18]. Thus, the imprint mark of imprinted XCI during preimplantation periods is thought to be erased on the embryonic side. This was confirmed experimentally by the random XCI pattern in female placental cells derived from ES cells [19]. It has been reported that paternal Xist deletion resulted in reversal of XCI with maternal X inactivation in the extraembryonic ectoderm, indicating that the initial imprint that establishes imprinted XCI is also erased in trophectoderm cells [20]. Our present study and these previous findings collectively indicate that the XCI regulation machinery was altered before implantation in both embryonic and extraembryonic cell lineages.
The status of XCI can change dramatically during mammalian development through the complex interactions of many different molecules in a stage-specific manner [21]. Whereas high-resolution epigenetic analyses using female ES cells have revealed the molecular mechanisms of XCI in depth [22], nuclear transfer studies have provided invaluable information on the dynamics of XCI throughout the life cycle [8, 10, 23,24,25]. One of the advantages of such techniques is the easy generation of a variety of embryos with donor-specific epigenetic features. We suspect that epigenetic dynamics, including XCI, genomic imprinting and other long-lasting epigenetic memories, have evolved specifically for regulating development in mammals and that these can be unraveled by nuclear transfer techniques.
Acknowledgment
This research was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology (Japan) (SM, KI and AO) and the NOVARTIS Foundation (Japan) for the Promotion of Science (KI).
References
- 1.Ogura A, Inoue K, Ogonuki N, Lee J, Kohda T, Ishino F. Phenotypic effects of somatic cell cloning in the mouse. Cloning Stem Cells 2002; 4: 397–405 [DOI] [PubMed] [Google Scholar]
- 2.Meissner A, Jaenisch R. Mammalian nuclear transfer. Dev Dyn 2006; 235: 2460–2469 [DOI] [PubMed] [Google Scholar]
- 3.Thuan NV, Kishigami S, Wakayama T. How to improve the success rate of mouse cloning technology. J Reprod Dev 2010; 56: 20–30 [DOI] [PubMed] [Google Scholar]
- 4.Kishigami S, Mizutani E, Ohta H, Hikichi T, Thuan NV, Wakayama S, Bui HT, Wakayama T. Significant improvement of mouse cloning technique by treatment with trichostatin A after somatic nuclear transfer. Biochem Biophys Res Commun 2006; 340: 183–189 [DOI] [PubMed] [Google Scholar]
- 5.Rybouchkin A, Kato Y, Tsunoda Y. Role of histone acetylation in reprogramming of somatic nuclei following nuclear transfer. Biol Reprod 2006; 74: 1083–1089 [DOI] [PubMed] [Google Scholar]
- 6.Van Thuan N, Bui HT, Kim JH, Hikichi T, Wakayama S, Kishigami S, Mizutani E, Wakayama T. The histone deacetylase inhibitor scriptaid enhances nascent mRNA production and rescues full-term development in cloned inbred mice. Reproduction 2009; 138: 309–317 [DOI] [PubMed] [Google Scholar]
- 7.Ogura A, Inoue K, Wakayama T. Recent advancements in cloning by somatic cell nuclear transfer. Philos Trans R Soc Lond B Biol Sci 2013; 368: 20110329; DOI . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Inoue K, Kohda T, Sugimoto M, Sado T, Ogonuki N, Matoba S, Shiura H, Ikeda R, Mochida K, Fujii T, Sawai K, Otte AP, Tian XC, Yang X, Ishino F, Abe K, Ogura A. Impeding Xist expression from the active X chromosome improves mouse somatic cell nuclear transfer. Science 2010; 330: 496–499 [DOI] [PubMed] [Google Scholar]
- 9.Wen B, Wu H, Shinkai Y, Irizarry RA, Feinberg AP. Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat Genet 2009; 41: 246–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matoba S, Inoue K, Kohda T, Sugimoto M, Mizutani E, Ogonuki N, Nakamura T, Abe K, Nakano T, Ishino F, Ogura A. RNAi-mediated knockdown of Xist can rescue the impaired postimplantation development of cloned mouse embryos. Proc Natl Acad Sci USA 2011; 108: 20621–20626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wakayama T, Perry AC, Zuccotti M, Johnson KR, Yanagimachi R. Full-term development of mice from enucleated oocytes injected with cumulus cell nuclei. Nature 1998; 394: 369–374 [DOI] [PubMed] [Google Scholar]
- 12.Ogura A, Inoue K, Ogonuki N, Noguchi A, Takano K, Nagano R, Suzuki O, Lee J, Ishino F, Matsuda J. Production of male cloned mice from fresh, cultured, and cryopreserved immature Sertoli cells. Biol Reprod 2000; 62: 1579–1584 [DOI] [PubMed] [Google Scholar]
- 13.Lawitts JA, Biggers JD. Culture of preimplantation embryos. Methods Enzymol 1993; 225: 153–164 [DOI] [PubMed] [Google Scholar]
- 14.Terashita Y, Wakayama S, Yamagata K, Li C, Sato E, Wakayama T. Latrunculin A can improve the birth rate of cloned mice and simplify the nuclear transfer protocol by gently inhibiting actin polymerization. Biol Reprod 2012; 86: 180 [DOI] [PubMed] [Google Scholar]
- 15.Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. The ground state of embryonic stem cell self-renewal. Nature 2008; 453: 519–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams LH, Kalantry S, Starmer J, Magnuson T. Transcription precedes loss of Xist coating and depletion of H3K27me3 during X-chromosome reprogramming in the mouse inner cell mass. Development 2011; 138: 2049–2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marahrens Y, Panning B, Dausman J, Strauss W, Jaenisch R. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev 1997; 11: 156–166 [DOI] [PubMed] [Google Scholar]
- 18.Okamoto I, Otte AP, Allis CD, Reinberg D, Heard E. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science 2004; 303: 644–649 [DOI] [PubMed] [Google Scholar]
- 19.Murakami K, Araki K, Ohtsuka S, Wakayama T, Niwa H. Choice of random rather than imprinted X inactivation in female embryonic stem cell-derived extra-embryonic cells. Development 2011; 138: 197–202 [DOI] [PubMed] [Google Scholar]
- 20.Mugford JW, Yee D, Magnuson T. Failure of extra-embryonic progenitor maintenance in the absence of dosage compensation. Development 2012; 139: 2130–2138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barakat TS, Gribnau J. X chromosome inactivation in the cycle of life. Development 2012; 139: 2085–2089 [DOI] [PubMed] [Google Scholar]
- 22.Pollex T, Heard E.Recent advances in X-chromosome inactivation research. Curr Opin Cell Biol 2012; pii S0955-0674(12)00170-6. [DOI] [PubMed] [Google Scholar]
- 23.Bao S, Miyoshi N, Okamoto I, Jenuwein T, Heard E, Azim Surani M. Initiation of epigenetic reprogramming of the X chromosome in somatic nuclei transplanted to a mouse oocyte. EMBO Rep 2005; 6: 748–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Senda S, Wakayama T, Yamazaki Y, Ohgane J, Hattori N, Tanaka S, Yanagimachi R, Shiota K. Skewed X-inactivation in cloned mice. Biochem Biophys Res Commun 2004; 321: 38–44 [DOI] [PubMed] [Google Scholar]
- 25.Nolen LD, Gao S, Han Z, Mann MR, Gie Chung Y, Otte AP, Bartolomei MS, Latham KE. X chromosome reactivation and regulation in cloned embryos. Dev Biol 2005; 279: 525–540 [DOI] [PubMed] [Google Scholar]