Abstract
Successful host defense against numerous pulmonary infections depends on bacterial clearance by polymorphonuclear leukocytes (PMNs); however, excessive PMN accumulation can result in life-threatening lung injury. Local expression of CXC chemokines is critical for PMN recruitment. The impact of chemokine-dependent PMN recruitment during pulmonary Mycobacterium tuberculosis infection is not fully understood. Here, we analyzed expression of genes encoding CXC chemokines in M. tuberculosis–infected murine lung tissue and found that M. tuberculosis infection promotes upregulation of Cxcr2 and its ligand Cxcl5. To determine the contribution of CXCL5 in pulmonary PMN recruitment, we generated Cxcl5–/– mice and analyzed their immune response against M. tuberculosis. Both Cxcr2–/– mice and Cxcl5–/– mice, which are deficient for only one of numerous CXCR2 ligands, exhibited enhanced survival compared with that of WT mice following high-dose M. tuberculosis infection. The resistance of Cxcl5–/– mice to M. tuberculosis infection was not due to heightened M. tuberculosis clearance but was the result of impaired PMN recruitment, which reduced pulmonary inflammation. Lung epithelial cells were the main source of CXCL5 upon M. tuberculosis infection, and secretion of CXCL5 was reduced by blocking TLR2 signaling. Together, our data indicate that TLR2-induced epithelial-derived CXCL5 is critical for PMN-driven destructive inflammation in pulmonary tuberculosis.
Introduction
Tuberculosis (TB) is an aerosol-transmitted chronic infectious disease, which causes close to 1.5 million deaths annually; close to 9 million new cases of TB develop each year (1). The causative agent, Mycobacterium tuberculosis, is capable of persisting in mononuclear phagocytes over prolonged periods of time and is rarely eradicated. Rather, it is contained within macrophages inside granulomatous lesions (2). Persistence is controlled by T lymphocytes, most notably, by CD4 T helper 1 cells (3).
Soon after inhalation, M. tuberculosis bacilli reach the lower respiratory tract where they are sensed by pattern recognition receptors (PRRs) expressed by alveolar macrophages, DCs, and epithelial cells (4). First, PRR sensing results in prompt mobilization of effector functions that limit early invasion by M. tuberculosis (5). Second, it induces proinflammatory cytokines and chemokines, which coordinate recruitment and activation of additional innate immune cells, including polymorphonuclear leukocytes (PMNs) and inflammatory monocytes. Third, PRR sensing sets into motion signals for instruction of the acquired immune response (6, 7).
Pathology in TB is characterized by destructive processes induced and mediated by the immune system (8). Several lines of evidence from experimental M. tuberculosis infection in mice support a correlation between increased PMN influx and pathology in TB (9). During active TB, PMNs are not only the most abundant cell type in sputum and bronchoalveolar lavage (BAL) fluid of patients, but they also carry the highest M. tuberculosis load (10). Investigations on propensities of PMNs to eradicate M. tuberculosis generated conflicting results (11). Some reported mycobactericidal activities of human PMNs (12–15), while others demonstrated that human PMNs phagocytose M. tuberculosis but fail to kill the bacilli (16–20). In the zebrafish model of mycobacterial infection, PMNs kill Mycobacterium marinum through phagocytosis of infected macrophages and thereby contribute to host defense (21). In mice, protective effects of PMNs have been observed only after infection via routes other than aerosol (22, 23). More recently, we and others described an association between TB susceptibility and increased PMN responses in mice (24–27). Susceptible mouse strains, such as DBA/2 and CBA/J, are characterized by increased PMN influx into pulmonary lesions (9). Similarly, susceptible gene deletion mutant strains suffer from increased PMN influx but can be rescued by PMN depletion (27). It remains elusive, however, whether PMN influx is the cause or consequence of increased TB susceptibility.
Recruitment of PMNs to sites of inflammation is partly driven by the CXCR2 receptor and its ligands (28, 29). CXCR2 binds chemokines with the ELR motif, including, as most prominent cognates, CXCL1 (or keratinocyte-derived chemokine), CXCL2 (or macrophage inflammatory protein 2), CXCL5 (or LPS-induced CXC chemokine), and CXCL15 (or lungkine). Chemokine cognates, which, like the CXCR2 ligands, target a shared receptor, are generally considered redundant. Yet, distinct cellular sources of these ligands point toward a spatial regulation. Using BM chimeric mice and Klebsiella pneumoniae infection, Cai et al. showed that CXCL1 was expressed by both radioresistant and radiosensitive cells (30). CXCL1 and CXCL2 are secreted by myeloid cells, such as macrophages and PMNs (31), and their importance in PMN recruitment to inflamed lungs is well appreciated in various infectious diseases (30, 32–36). CXCL15, which is produced by bronchial epithelial cells (29), contributes to pulmonary defense against K. pneumoniae infection (37). CXCL5, which plays a role in LPS-induced lung inflammation, is secreted by type I and type II alveolar epithelial cells (AECI and AECII, respectively) (31, 38–41). During M. tuberculosis infection, CXCR2 and its ligand CXCL5 are strongly upregulated in lungs (9, 42, 43). While the immunomodulatory functions of chemokines are becoming increasingly appreciated in TB research (43), most studies have focused on lymphocyte recruitment and granuloma formation (44–47). The role of chemokines and of lung-resident cells in governing innate immune cell recruitment remains ill defined.
Here, we describe for the first time an essential and nonredundant role of lung epithelial cell–derived CXCL5 in experimental TB pathogenesis of mice. Our studies pinpoint a critical role of CXCL5 in PMN recruitment to the infected lung. Despite shared use of the receptor CXCR2 by several chemokine cognates, TB was significantly ameliorated in the absence of CXCL5.
Results
M. tuberculosis infection induces expression of CXCR2 and its ligand chemokines.
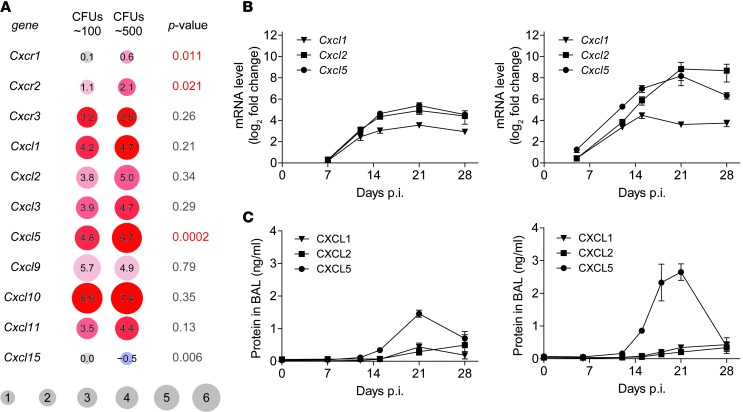
As a first step toward characterizing the relationship between susceptibility and PMN-driven inflammation in TB, we compared the expression of different CC and CXC chemokine receptors and ligands by cross-analyzing microarray data from C57BL/6 (WT) mice after aerosol infection with either a low or a high dose of M. tuberculosis (Figure 1, A and B, and Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI72030DS1). Both CXCR2 and CCR1 function as neutrophil chemokine receptors (28, 29, 48, 49). Several murine CCR1 ligands, including Ccl3, Ccl5, and Ccl7, were expressed upon M. tuberculosis infection; Ccl4 was expressed in a dose-dependent manner (Supplemental Figure 1). The ELR+ chemokines Cxcl1, Cxcl2, Cxcl3, and Cxcl5 as well as their shared receptors Cxcr1 and Cxcr2 were all induced in lung tissue upon M. tuberculosis infection (Figure 1, A and B). The receptors Cxcr1/Cxcr2 and their ligand Cxcl5 formed the sole chemokine receptor/ligand pair that showed a significant increase in expression between low- and high-dose infections (Figure 1A). Consistent with this finding, protein levels of PMN chemoattractants were elevated in BAL fluid of low- and high-dose M. tuberculosis–infected mice (Figure 1C). Thus, compared with other PMN-attracting chemokines, CXCL5 was expressed earlier (starting day 12 after infection [p.i.]) and more abundantly, confirming previous results (43). Furthermore, we observed that CXCL5 expression correlates with the M. tuberculosis infection dose.
Figure 1. Expression of CXC chemokine receptors and ligands in lungs of M. tuberculosis–infected mice.
(A) Effect of M. tuberculosis infection dose on chemokine and chemokine receptor gene expression. Comparison of low-dose infection (∼100 CFUs) naive and day 23 p.i. microarray gene expression data and high-dose infection (∼500 CFUs) naive and day 21 p.i. microarray gene expression data. Lungs from 5 WT mice were pooled for microarray analysis. Experiment was performed in duplicate with color swaps. Adjusted P values correspond to interaction between dose and infection. The range of red colors represents the degree of upregulation, with dark red indicating increased upregulation; blue (Cxcl15) represents downregulation. Numbers in circles are log2 fold change. (B) Expression of chemokine mRNA at different time points p.i. in WT lungs following low-dose (left, ∼150 CFUs) and high-dose (right, ∼550 CFUs) infection relative to naive lungs (mean ± SEM; npooled ≥ 8). Data are pooled from 2 independent experiments. (C) Chemokine protein levels in BAL fluid collected at different time points p.i. from WT mice following low-dose (left, ∼150 CFUs) and high-dose (right, ∼550 CFUs) infection (mean ± SEM; npooled = 10). Data are pooled from 2 independent experiments.
To characterize the biological role of CXCL5-dependent PMN recruitment in TB in more depth, we generated CXCL5-deficient mutant mice. To this end, we deleted all Cxcl5-coding regions by replacing them with the β-galactosidase gene (Supplemental Figure 2, A–C). Cxcl5–/– mice were viable, fertile, and showed no apparent abnormalities or growth retardation. Consistent with results from a Cxcl5–/– mouse generated by Mei et al., PMN numbers were elevated in naive BM (Supplemental Figure 2, D and E, and ref. 39).
Impaired PMN response in Cxcl5–/– mice after low- and high-dose infection.
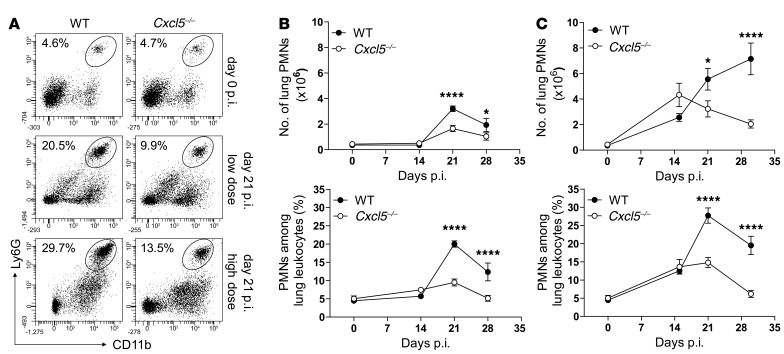
CXCL5 is well known to attract neutrophils (39). To evaluate the role of CXCL5 in pulmonary PMN recruitment during TB, we infected WT and Cxcl5–/– mice with low and high doses of M. tuberculosis and analyzed their innate immune cell responses in lungs. Overall, the kinetics of neutrophil accumulation were similar between mouse strains and infection doses (Figure 2). However, the PMN responses differed in magnitude and were lower in Cxcl5–/– mice compared with their WT controls (Cxcl5–/– low dose < WT low dose ≤ Cxcl5–/– high dose < WT high dose) (Figure 2B). Notably, after day 21 p.i., lung PMN numbers declined in WT and Cxcl5–/– mice after low-dose infection and in Cxcl5–/– mice, but not in WT mice, after high-dose infection (Figure 2B). Accordingly, we observed fewer myeloperoxidase+ (MPO+) cells in lung parenchyma from Cxcl5–/– mutants compared with WT mice after high-dose infection (Supplemental Figure 3A).
Figure 2. Reduced pulmonary PMN recruitment in Cxcl5–/– mice.
(A) Dot plots showing representative frequencies of lung PMNs following low-dose and high-dose M. tuberculosis infection. (B and C) Numbers and frequencies of PMNs among lung leukocytes in (B) naive and low-dose (∼150 CFUs) and (C) high-dose (∼500 CFUs) M. tuberculosis–infected WT and Cxcl5–/– mice determined by flow cytometry (mean ± SEM; npooled = 10 per time point). Each time point shows pooled data from at least 2 independent experiments. Curve data were pooled from a total of 7 independent experiments (2-way ANOVA/Bonferroni post-test). *P < 0.05; ****P < 0.0001.
Blood PMNs represent a systemic cell pool that can be rapidly recruited to the lung. In both Cxcl5–/– and WT mice, the frequencies of segmented PMNs among blood leukocytes increased upon high-dose M. tuberculosis infection, reaching a peak at day 21 p.i. (Supplemental Figure 3B). The number of alveolar macrophages, lung macrophages, and lung monocytes, on the other hand, did not differ substantially between mouse strains and infection dose (Supplemental Figure 4). Taken together these results demonstrate that following low- and high-dose M. tuberculosis infection, PMNs were the main cell type among myeloid innate immune cells affected by the lack of CXCL5.
Abrogation of the CXCL5/CXCR2 pathway renders mice resistant to fatal TB.
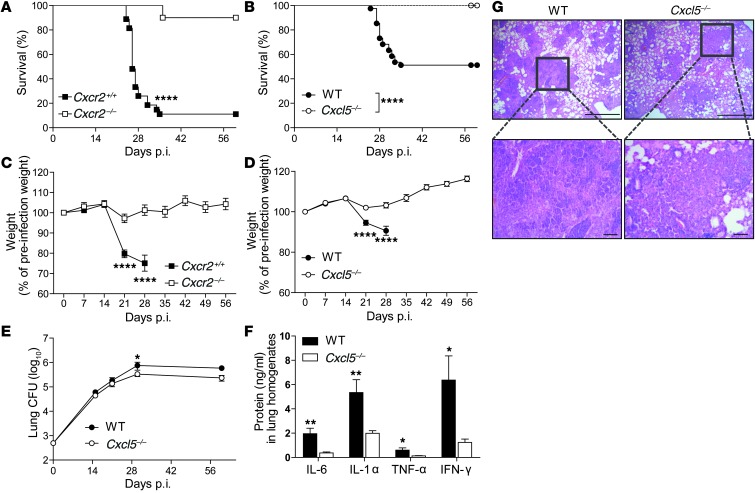
We identified CXCL5-dependent pulmonary PMN recruitment as a general mechanism during murine TB, independent of infection dose. However, a high-dose M. tuberculosis infection caused higher expression of Cxcl5 and Cxcr2 and increased pulmonary PMN recruitment compared with a low-dose M. tuberculosis infection. To further corroborate the relevance of CXCL5-mediated PMN recruitment in TB, we continued our analysis using high-dose infection. After backcrossing for over 9 generations on the C57BL/6 background, Cxcl5–/– mice, like WT controls, survived low-dose M. tuberculosis infection (G. Nouailles, unpublished observations). Next, Cxcr2–/–, Cxcl5–/–, and WT control mice were infected with a high dose of M. tuberculosis, and different disease parameters were determined, including survival, bacterial burden, inflammatory status, and lung pathology. A total of 90% of Cxcr2–/– mice and 100% of Cxcl5–/– mice survived acute and chronic TB for more than 60 days p.i., while 50% to 80% of corresponding WT controls died between days 25 and 35 p.i. (Figure 3, A and B). When compared with Cxcr2–/– and Cxcl5–/– mice, WT controls showed increased weight loss starting on day 21 p.i. (Figure 3, C and D). Surviving WT mice showed a moderate increase in M. tuberculosis burden by days 30 and 60 p.i. compared with Cxcl5–/– mice (Figure 3E). Proinflammatory cytokines in M. tuberculosis–infected lungs, including IL-6, IL-1α, TNF-α, and IFN-γ, were elevated in WT mice compared with those in Cxcl5–/– mice at day 30 p.i. but not day 21 p.i. (Figure 3F and Supplemental Figure 3C). At day 21 p.i., numerous inflammatory foci were observed in lung parenchyma of both WT and Cxcl5–/– mice (Figure 3G). The granulomatous infiltrates were surrounded by regions of functional and dilated alveoli. WT specimens presented abundant polymorphonuclear cells disposed in clusters within the inflammatory lesions. Granulocytes had accumulated in the airways in these samples. Immunostaining for MPO, a marker for neutrophils and inflammatory macrophages, indicated more abundant MPO+ cells in lung tissue from WT mice (Supplemental Figure 3A). Based on cellular morphology and staining pattern, we concluded that CXCL5 likely limits neutrophilic pneumonia (Figure 3G and Supplemental Figure 3A). Thus, the absence of CXCL5 or its receptor, CXCR2, heightened host resistance to TB, and CXCL5 was critically involved in lung inflammation.
Figure 3. Absence of CXCL5 or its receptor CXCR2 protects from TB-associated wasting disease and death.
(A) Kaplan-Meier curves showing survival of Cxcr2+/+ (n = 27) and Cxcr2–/– (n = 20) mice after M. tuberculosis infection (∼570 CFUs). Results are pooled from 2 independent experiments (log-rank test). (B) Kaplan-Meier curves showing survival of WT (n = 41) and Cxcl5–/– (n = 36) mice after M. tuberculosis infection (∼500 CFUs). Results are pooled from 4 independent experiments (log-rank test). (C) Relative weight curves of Cxcr2+/+ and Cxcr2–/– mice during M. tuberculosis infection (∼570 CFUs, n ≥ 20 [nWT day 28 p.i. = 9]). Data are pooled from 2 independent experiments. (D) Relative weight curves of WT and Cxcl5–/– mice during M. tuberculosis infection (∼500 CFUs; n ≥ 36 [nWT day 28 p.i. = 35]). Data are pooled from 4 independent experiments. (E) Pulmonary M. tuberculosis burden in WT and Cxcl5–/– mice (∼500 CFUs; npooled = 9–10 per time point). Data are pooled from 5 independent experiments. (C–E) Mean ± SEM, 2-way ANOVA/Bonferroni post-test. (F) Proinflammatory cytokine levels in lung homogenates of WT and Cxcl5–/– mice at day 30 p.i. (mean ± SEM; npooled = 10). Data are pooled from 2 independent experiments (unpaired t test). (G) Lung histopathology at 21 days p.i. as shown by hematoxylin and eosin staining. Scale bar: 1,000 μm (top row); 100 μm (bottom row). Data are representative of 3 independent experiments (n = 3–5). *P < 0.05; **P < 0.01; ****P < 0.0001.
Unaltered type 1 T cell responses in absence of CXCL5.
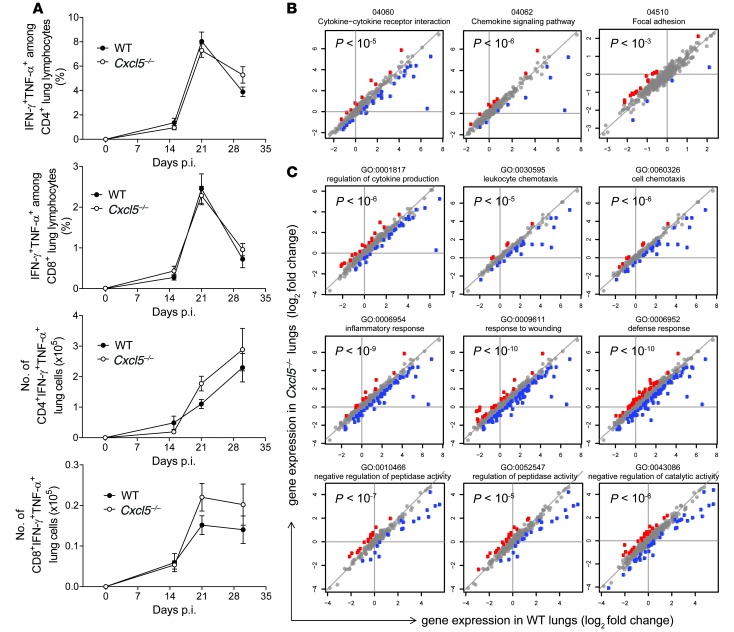
Because T cells are crucial for TB control (3), we also investigated the consequences of CXCL5 deficiency on adaptive immunity. M. tuberculosis–specific type 1 CD4 and CD8 T cell responses, as measured by IFN-γ and TNF-α secretion, were comparable in WT and Cxcl5–/– mice (Figure 4A and Supplemental Figure 5A). Likewise, total CD4 and CD8 T cell numbers did not differ between WT and Cxcl5–/– mice (G. Nouailles, unpublished observations). Consistently, among chemokines and cytokines associated with T cell responses, only the chemokine CCL5 was more abundant in lung homogenates of WT mice on day 30 p.i., likely reflecting increased inflammation in WT mice (Supplemental Figure 5B). In BAL fluid, none of the analyzed chemokines and cytokines was measurable above background level (data not shown). Hence, we conclude that type 1 T cell responses were not affected and therefore were not critically involved in increased mortality of WT mice compared with that of Cxcl5–/– mice.
Figure 4. M. tuberculosis–specific type 1 T cell response and pathway analysis in Cxcl5–/– mice.
(A) Frequencies of IFN-γ+TNF-α+ cells among lung CD4 and CD8 T cells and numbers of IFN-γ+TNF-α+ lung CD4 and CD8 T cells after short-term in vitro restimulation with M. tuberculosis–derived peptides determined by flow cytometry (mean ± SEM; npooled ≥ 14 per time point except on day 0, when n = 10). Mice were infected with high-dose (∼500 CFU) M. tuberculosis. Each time point represents pooled data from at least 2 independent experiments. Curve data pooled from a total of 4 independent experiments (2-way ANOVA/Bonferroni post-test) did not reveal significant differences between groups at analyzed time points. (B) Significant selected pathways by SPIA analysis. P values correspond to pGFWER composite and multiple testing–corrected P values from SPIA analysis. Plots show log2 fold changes in gene expression of given pathways between naive and day 21 p.i. WT lungs (horizontal axis) and Cxcl5–/– lungs (vertical axis). Mice were infected with high-dose (∼500 CFUs) M. tuberculosis. Red denotes genes for which log fold change is significantly higher in the KO, and blue denotes genes for which log fold change is significantly higher in the WT. (C) Selected GO terms significant in GO enrichment analysis. P values correspond to significance of GO enrichment, corrected for multiple testing. Colors and axes are as in B.
To complement the specific analysis of innate and adaptive cell populations, we pursued a broader approach, analyzing the general pulmonary host response by microarray-based comparison of mRNA expression in whole lungs of WT and Cxcl5–/– mice (Figure 4, B and C). Genes were analyzed for differential expression upon infection (day 21 p.i. vs. naive lungs) in both mouse strains (WT and Cxcl5–/–), and the regulation of gene expression upon infection was compared between the two strains. We examined genes showing significant difference in regulation between the two strains for enrichment in gene ontology (GO) terms and pathways. Genes belonging to selected significant GO terms and pathways are visualized as dots in Figure 4, B and C. Genes corresponding to GO terms and pathways related to inflammation and chemotaxis were significantly less regulated in Cxcl5–/– mice compared with those in WT mice. Collectively, these results suggest that genetic ablation of CXCL5 resulted in impaired PMN recruitment without affecting type 1 T cell responses and, consequently, reduced lung inflammation.
CXCL5/CXCR2 pathway drives PMN recruitment into the bronchoalveolar space.
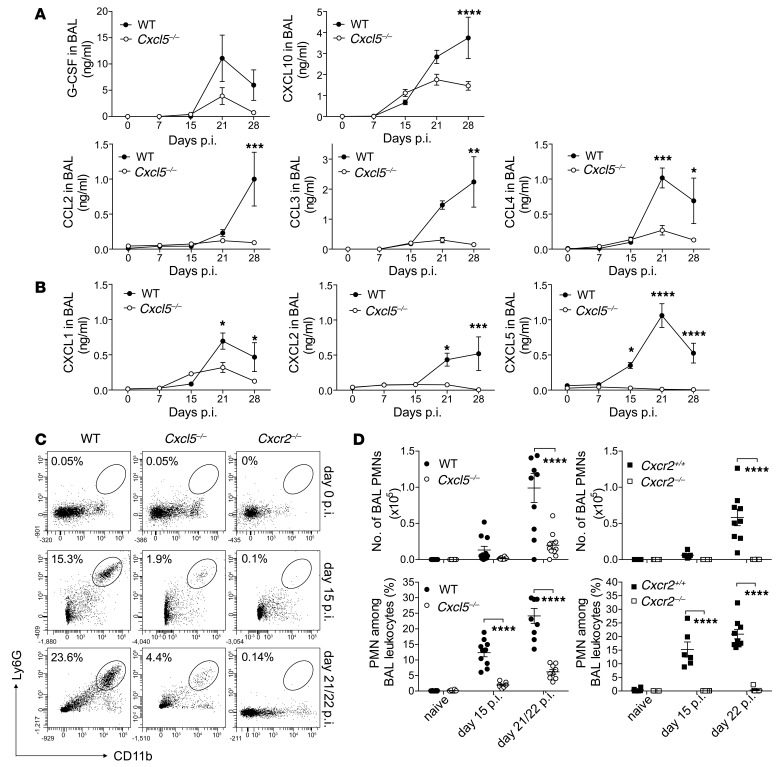
So far, our analysis revealed increased resistance and decreased inflammation in Cxcr2–/– and Cxcl5–/– mice compared with that in WT controls. To identify underlying mechanisms, we measured different leukocyte attractants in lung homogenates and BAL. Numbers of alveolar macrophages, lung macrophages, and lung monocytes remained similar between WT and Cxcl5–/– mice throughout the course of infection (Supplemental Figure 4). Consistently, similar concentrations of CXCL10, CCL3, and CCL4 (monocyte/macrophage chemoattractants) were detected in WT and Cxcl5–/– mice before day 30 p.i. in lung homogenates and before day 21 p.i. in BAL fluid (Figure 5A and Supplemental Figure 6). Although we could not quantify CXCL5 protein in lung homogenates due to nonspecific binding (data not shown), CXCL5 was not detected above background levels in BAL fluid of Cxcl5–/– mice. Levels of CXCL1 and CXCL2 were significantly reduced on days 21 and 30 p.i. in Cxcl5–/– mice compared with WT mice (Figure 5B). Interestingly, differences in chemokine concentrations became apparent in BAL fluid prior to lung homogenates. Accordingly, flow cytometric analysis of BAL cells revealed defective PMN recruitment in Cxcl5–/– mice and absence of PMN recruitment in Cxcr2–/– mice as early as day 15 p.i. (Figure 5, C and D). Thus, early PMN accumulation in the bronchoalveolar spaces depended exclusively on CXCR2 and largely on CXCL5.
Figure 5. Reduced chemokine expression and BAL PMN recruitment in Cxcl5–/– mice.
(A and B) Chemokine measurements by (A) multiplex analysis and (B) ELISA in BAL fluid of naive and high-dose (∼500 CFUs) M. tuberculosis–infected WT and Cxcl5–/– mice (mean ± SEM; npooled = 10). Data are pooled from 2 independent experiments (2-way ANOVA/Bonferroni post-test). (C) Dot plots showing representative frequencies of BAL PMNs following high-dose M. tuberculosis infection. (D) Numbers and frequencies of BAL PMNs in naive and high-dose (∼500 CFUs) M. tuberculosis–infected WT control (WT and Cxcr2+/+), Cxcl5–/–, and Cxcr2–/– mice determined by flow cytometry (mean ± SEM). Data are pooled from 6 independent experiments (2-way ANOVA/Bonferroni post-test). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Lung epithelial-derived CXCL5 recruits PMNs to M. tuberculosis–infected lungs.
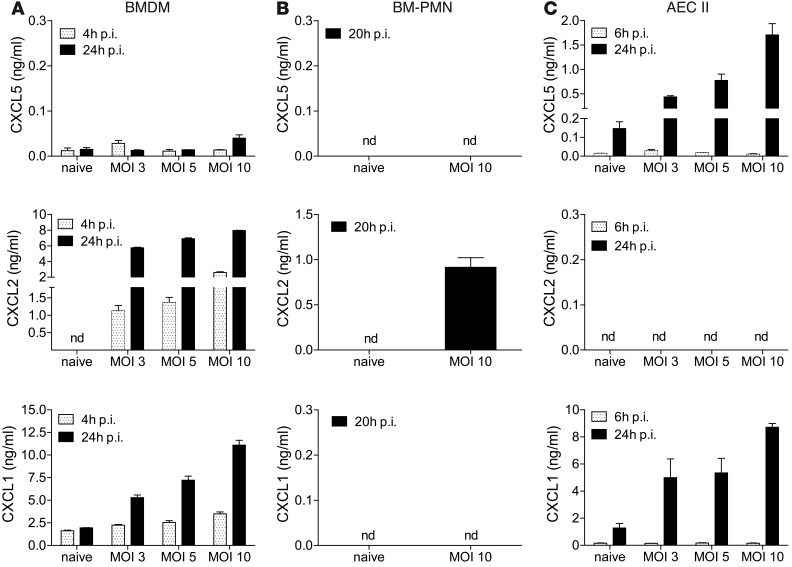
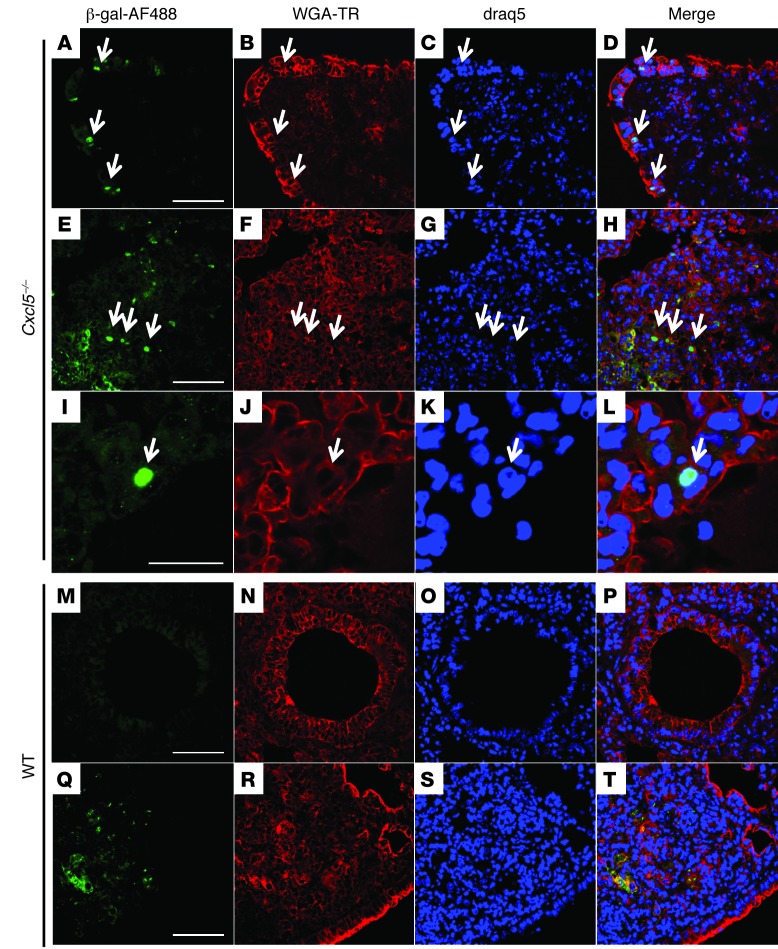
In order to identify the cellular source of CXCL5 in TB, we infected candidate cell populations with M. tuberculosis in vitro and measured their capacity to secrete CXCL5, CXCL1, and CXCL2 by ELISA. We generated BM-derived macrophages (BMDMs) and isolated PMNs from BM. Neither of these cell types secreted CXCL5 upon M. tuberculosis infection (Figure 6, A and B). In contrast, M. tuberculosis–infected T7 cells (an AECII line) secreted CXCL5 and CXCL1 but not CXCL2 (Figure 6C), suggesting that only epithelial cells were capable of secreting CXCL5. In contrast, CXCL1 was produced by both epithelial and myeloid cells and CXCL2 was produced only by myeloid cells. Replacement of the Cxcl5 gene with a β-galactosidase (LacZ) gene with a nuclear localization signal in Cxcl5–/– mutant mice allowed us to harness immunohistochemistry to monitor in situ induction of Cxcl5 during TB. Nuclear β-galactosidase staining was observed exclusively in tissues collected from Cxcl5–/– mice upon M. tuberculosis infection (Figure 7 and Supplemental Figure 7). The β-galactosidase+ cells were detected in the bronchial (Figure 7, A–D) and alveolar epithelium (Figure 7, I–L) and scattered within granulomatous cellular infiltrates (Figure 7, E–H). A nonspecific granular staining pattern with cytosolic localization was caused by crossreactivity of the polyclonal anti–β-galactosidase antibody with mycobacterial antigens, due to the use of CFA for the original immunization. This was observed in M. tuberculosis–infected mice but not in naive WT and Cxcl5–/– mice (Figure 7, E–H and Q–T, and Supplemental Figure 7). Collectively, our data indicate that the respiratory epithelium in the lungs was the main source of CXCL5 upon M. tuberculosis infection.
Figure 6. CXCL5 release by M. tuberculosis–infected epithelial cells.
CXCL5, CXCL1, and CXCL2 levels measured by ELISA in supernatants of M. tuberculosis–infected (A) BMDMs, (B) PMNs isolated from BM (BM-PMNs), and (C) T7 cells, an AECII cell line (mean ± SEM; n = 3). Data are representative of 3 independent experiments with 3 replicates each. nd, not detected.
Figure 7. In situ expression of CXCL5 by lung epithelial cells during TB.
Nuclear β-galactosidase (AF488, green) staining with specific antiserum reveals Cxcl5 induction in lung tissue at 21 days p.i. (∼500 CFUs). (A–D) β-Galactosidase+ nuclei in bronchial epithelial cells (arrows), (E–H) granulomatous infiltrates (arrows), and (I–L) alveolar epithelial cells (arrows) from Cxcl5–/– mice. (M–P) Absence of β-galactosidase+ nuclei in WT epithelia and (Q–T) granular staining pattern with cytosolic location in WT control mice. Tissue was counterstained with wheat germ agglutinin labeled with Texas Red (WGA-TR; red) and draq5 (blue). Scale bar: 50 μm (A–H and M–T); 20 μm (I–L). Representative confocal images are from 1 experiment (n = 3).
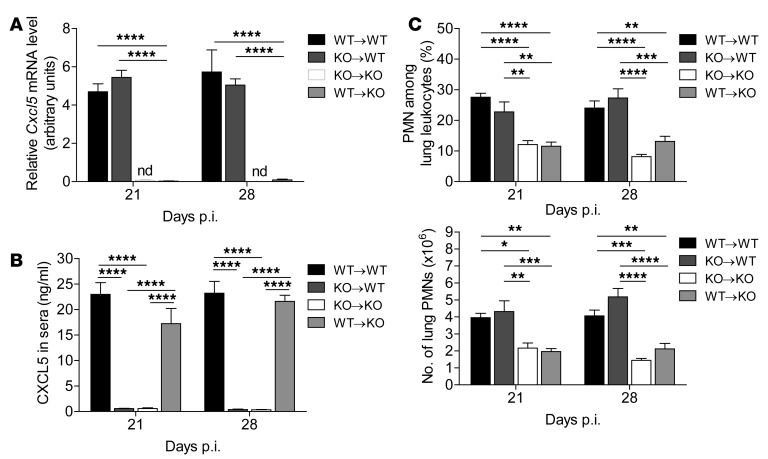
To verify that CXCL5-dependent neutrophilic inflammation was driven by epithelial and not by hematopoietic cells in vivo, we infected BM chimeric mice with M. tuberculosis. In irradiated WT mice that had received Cxcl5–/– BM (KO→WT chimera) only radioresistant cells (including the lung epithelium) are capable of secreting CXCL5. In irradiated Cxcl5–/– mice that had received WT BM (WT→KO chimera) only radiosensitive (hematopoietic cells, including platelets) are capable of secreting CXCL5. Analysis of chimeric mice allowed us to examine which compartment is responsible for CXCL5 secretion and CXCL5-dependent pulmonary PMN recruitment. In lungs, we detected Cxcl5 mRNA expression in chimeras with radioresistant WT cells and Cxcl5–/– BM cells (KO→WT) but not in chimeras with radioresistant Cxcl5–/– cells and WT BM cells (Figure 8A). In sera, we measured high abundance of CXCL5 in chimeras with radioresistant Cxcl5–/– cells and WT BM cells (WT→KO) but not in chimeras with radioresistant WT cells and Cxcl5–/– BM cells (KO→WT) (Figure 8B). In an exploratory experiment, we MACS-sorted lung cells from high-dose M. tuberculosis–infected WT mice (Supplemental Figure 8A). Cxcl5 mRNA expression was significantly increased (∼14-fold) in the epithelial and residual cell fraction (LSEC–CD45–) compared with that in unsorted lung cells. In contrast, leukocytes (LSEC–CD45+) showed a decreased expression of Cxcl5 mRNA (∼12-fold) compared with that in unsorted lung cells. Cxcl5 mRNA expression measured in the endothelial fraction (LSEC+) was similar to expression found in unsorted lung cells and likely resulted from contamination with LSEC–CD45– cells (Supplemental Figure 8, B and C), which argues against LSEC+ endothelial cells as major source of Cxcl5.
Figure 8. Radioresistant cells are responsible for defective PMN recruitment in Cxcl5–/– mice.
Chimeric mice were generated as follows: (a) WT BM in WT recipients (WT→WT); (b) Cxcl5–/– BM in WT recipients (KO→WT); (c) Cxcl5–/– BM in Cxcl5–/– recipients (KO→KO); and (d) WT BM in Cxcl5–/– recipients (WT→KO). Mice were infected with high-dose (∼400 CFUs) M. tuberculosis. (A) Relative Cxcl5 mRNA levels in lungs of M. tuberculosis–infected chimeric mice measured by quantitative RT-PCR of Cxcl5 and Gapdh mRNA expression with the formula 1.8(ct Gapdh - ct Cxcl5) × 100. nd, Cxcl5 mRNA not detected, Ct >40. (B) CXCL5 in sera of M. tuberculosis–infected chimeric mice as measured by ELISA. (C) Frequencies and numbers of lung PMNs in M. tuberculosis–infected chimeric mice as measured by flow cytometry. Data are representative of 2 independent experiments (mean ± SEM; n = 5; 2-way ANOVA/Bonferroni post-test). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Thus, analysis of BM chimeric mice and MACS-sorted lung cells confirmed in vitro and in situ data showing that the CD45– and radioresistant compartment, including epithelial cells, was the major cellular source of pulmonary CXCL5. In contrast, all CXCL5 measured in sera was derived from hematopoietic cells (e.g., platelets).
Since PMN migration to the lung follows chemokine gradients (29), absence of CXCL5 — not only in lungs, but also in blood — could influence PMN migration and hence also influence the observed phenotype in Cxcl5–/– mice. To address this, we determined the recruitment of lung PMNs in BM chimeric mice. Absolute numbers and frequencies of lung PMNs differed significantly between mice with different radioresistant genotypes (KO→WT and WT→KO) but not between mice with the same radioresistant genotype (WT→WT and KO→WT as well as KO→KO and WT→KO) and were independent of a radiosensitive genotype (Figure 8C). Thus, presence or absence of hematopoietic cell–derived CXCL5 in blood did not influence pulmonary PMN recruitment. Moreover, CXCL5-dependent PMN recruitment in pulmonary TB was exclusively driven by radioresistant epithelial cells.
M. tuberculosis induces epithelial CXCL5 expression via TLR2.
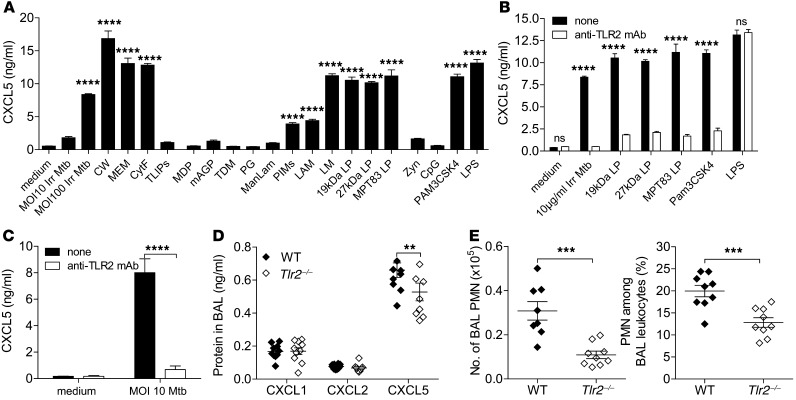
Finally, to understand how M. tuberculosis induces epithelial cell CXCL5 expression, we stimulated the AECII cell line T7 with different M. tuberculosis–derived and control PRR agonists (Figure 9A). γ-Irradiated whole M. tuberculosis stimulated CXCL5 expression in AECII in a dose-dependent fashion. Likewise, the M. tuberculosis cell wall and cell membrane as well as cytosolic fraction induced CXCL5 secretion. Subsequently, using a panel of M. tuberculosis–derived PRR agonists, we identified phosphatidylinositol mannosides 1 and 2, lipoarabinomannan, lipomannan; 19-kDA lipoprotein, 27-kDa lipoprotein, and MPT83 — all TLR1/TLR2 agonists — as potent inducers of CXCL5 (Figure 9A). In contrast, CXCL5 was not induced by M. tuberculosis–derived total lipids, nucleotide-binding oligomerization domain-containing protein 2 (NOD2) agonists muramyl dipeptide and mycolylarabinogalactan-peptidoglycan, macrophage-inducible C-type lectin (Mincle) agonist trehalose-6,6′-dimycolate, TLR2 and NOD2 agonist peptidoglycan, and DC-SIGN agonist mannose-capped lipoarabinomannan (Figure 9A). The control TLR agonists PAM3CSK4 (TLR1/2) and LPS (TLR4) induced chemokine secretion, whereas zymosan, an agonist of dectin-1 and TLR2, as well as CpG, a ligand for TLR9, failed to do so.
Figure 9. CXCL5 expression in epithelial cells is induced by M. tuberculosis TLR2 agonists.
(A) CXCL5 abundance measured by ELISA in supernatants of the AECII cell line (T7) stimulated with M. tuberculosis components and TLR agonists: γ-irradiated M. tuberculosis (Irr Mtb; MOI 10 and 100), 10 μg/ml cell wall (CW), 10 μg/ml membrane (MEM), 10 μg/ml cytosolic fraction (CytF), 10 μg/ml total lipids (TLIPs), 5 μg/ml muramyl dipeptide (MDP), 5 μg/ml mycolylarabinogalactan-peptidoglycan (mAGP), 5 μg/ml trehalose-6,6′-dimycolate (TDM), 5 μg/ml peptidoglycan (PG), 5 μg/ml mannose-capped lipoarabinomannan (ManLAM), 5 μg/ml phosphatidyl-myo-inositol mannosides 1 and 2 (PIMs), 5 μg/ml lipoarabinomannan (LAM), 1 μg/ml lipomannan (LM), 1 μg/ml 19-kDa lipopeptide (19kDa LP), 1 μg/ml 27-kDa lipopeptide (27kDa LP), 1 μg/ml MPT83 lipopeptide (MPT83 LP), 10 μg/ml zymosan (Zyn), 1 μM CpG, 1 μg/ml PAM3CSK4, 1 μg/ml LPS (1-way ANOVA/Bonferroni post-test). (B and C) CXCL5 concentration measured by ELISA in supernatants of AECII (T7) treated with and without anti-TLR2 mAb and (B) stimulated for 24 hours with γ-irradiated M. tuberculosis, 19-kDa and 27-kDa lipopeptides, MPT83, PAM3SCK4, or LPS and (C) infected with M. tuberculosis (Mtb) (2-way ANOVA/Bonferroni post-test). (A–C) Data are representative of 2 independent experiments (mean ± SEM; n = 3). (D) Chemokine measurements by ELISA and (E) PMN numbers and frequencies determined by flow cytometry in BAL fluid of high-dose (∼400 CFUs) M. tuberculosis–infected WT and Tlr2–/– mice (mean ± SEM; npooled = 10). Data are pooled from 2 independent experiments (2-way ANOVA/Bonferroni post-test). **P < 0.01; ***P < 0.001; ****P < 0.0001.
To understand the role of TLR signaling in CXCL5 secretion in more depth, we neutralized TLR2 signaling with anti-TLR2 blocking antibody (T2.5) and subsequently stimulated the AECII with M. tuberculosis, irradiated M. tuberculosis, M. tuberculosis lipopeptides (19 kDa, 27 kDA, and MPT83), or classical TLR agonists PAM3CSK4 and LPS (Figure 9, B and C). CXCL5 induction in AECII by M. tuberculosis and M. tuberculosis lipopeptides was completely abolished by blocking TLR2. To investigate further the in vivo relevance of M. tuberculosis– and TLR2-dependent CXCL5 expression, we subsequently infected WT and Tlr2–/– mice with a high dose of M. tuberculosis. At day 15 p.i., CXCL5 protein expression (Figure 9D) and PMN recruitment (Figure 9E) into BAL fluid were reduced in Tlr2–/– mice compared with those in WT controls. Thus, signaling via TLR2 contributed to the overall abundance of CXCL5 in BAL fluid from M. tuberculosis–infected mice, resulting in reduced PMN recruitment.
Based on the results presented here, we conclude that epithelial cells orchestrate the recruitment of PMNs via chemokines, notably CXCL5, which can be directly induced by M. tuberculosis–derived molecular pattern molecules that are sensed by TLR2.
Discussion
Studies aimed at deciphering the pathophysiological role of the chemokine network have largely focused on chemokine receptors, rather than ligands, due to the high redundancy of the latter. In line with this, the critical role of CXCR2 expressed on PMNs in host defense against acute infections has been well documented (34, 35, 50, 51).
Thus far, in the context of mycobacterial infections, only one report is available. Upon intratracheal infection with Mycobacterium avium, Cxcr2–/– mice do not show reduced lung PMN recruitment, whereas after intraperitoneal infection, PMN recruitment was impaired in Cxcr2–/– mice, albeit without effect on bacterial burdens (52). Cxcr2–/– mice suffer from numerous abnormalities, including reduced fertility and reduced body weight (53), rendering this mouse strain less suitable for chronic infection and inflammation studies. To circumvent these deficiencies and to gain deeper understanding of a PMN-targeting chemokine in TB, we constructed a “single-chemokine” KO mouse. We chose CXCL5 due to its earlier and more abundant expression compared with CXCL1 and CXCL2 as well as its dose-dependent coexpression with Cxcr2 after M. tuberculosis infection, indicating their coregulation in TB.
Using the Cxcl5–/– mouse strain, we identified an essential role for a single PMN-attracting ELR+ CXC chemokine, CXCL5, in pulmonary TB. We demonstrated that (a) CXCL5 is largely responsible for pulmonary PMN attraction; (b) TLR2 sensing of M. tuberculosis molecular patterns induces expression of CXCL5 by alveolar epithelial cells; and (c) CXCL5-dependent PMN recruitment via CXCR2 causes exacerbated inflammation, resulting in wasting disease and eventually death of the M. tuberculosis–infected host following high-dose infection. These data show conclusively that a single chemokine ligand of CXCR2 was essential for PMN-driven detrimental inflammation in TB, which could not be compensated for by cognates for the same receptor.
Deletion of Cxcl5 allowed us to investigate the role of PMNs in chronic infection and inflammation, which remains controversial in TB (11). We demonstrate that PMN recruitment to the lung following a high-dose M. tuberculosis infection caused exaggerated inflammation with fatal consequences due to tissue inflammation rather than increased bacterial load. The detrimental effects of CXCL5 and CXCR2 in TB stand in marked contrast to several reports on the protective role of CXCR2-dependent PMN recruitment in acute bacterial infections (30, 34, 35). This discrepancy might best be explained by differential dependencies on PMNs for bacterial clearance.
PMNs have been described to promote T cell responses in TB. Following low-dose M. tuberculosis aerosol infection, Kang et al. observed decreased numbers of IFN-γ+ CD4 T cells in lungs after depletion of Gr-1+ PMNs, leaving bacterial burdens unaffected (42). Additionally, Ernst and colleagues described a contribution of PMNs to early T cell priming in the draining lymph nodes (54, 55). We did not detect changes in pulmonary M. tuberculosis–specific type 1 CD4 and CD8 T cell responses following high-dose infection of Cxcl5–/– mice despite reduced PMN recruitment. Similarly, Cxcr2–/– mice devoid of PMNs in BAL fluid showed improved resistance to high-dose M. tuberculosis infection, which argues against defective type 1 T cell responses (56). Differential experimental setups, notably low- versus high-dose M. tuberculosis inocula, appear to influence the outcome of PMN-governed host defense. After high-dose M. tuberculosis infection, the beneficial role of PMNs in early T cell priming could be overcome by their detrimental impact on tissue inflammation and destruction.
CXCL5 is not only temporally, but also spatially, differentially regulated compared with other ELR+ CXC chemokines (42, 43). Platelets and endothelial and epithelial cells can secrete CXCL5 (31, 39, 41). Our experiments revealed that upon in vitro M. tuberculosis infection, only AECII produced CXCL5, while CXCL2 was secreted by PMNs and BMDMs, and CXCL1 was secreted by BMDMs and AECII. In situ, we detected CXCL5+ alveolar cells as well as bronchial epithelial cells. Experiments with BM chimeric mice and MACS-sorted lung cells confirmed and extended our in vitro and in situ findings that CXCL5-dependent PMN recruitment was exclusively driven by radioresistant lung-resident cells and not by radiosensitive hematopoietic cells. CXCL5 expression is neither cell type nor organ specific but, rather, is expressed by diverse tissue-resident cells, including cardiac myocytes (57), alveolar epithelial cells (31, 41), enterocytes (38), aortic endothelial cells (58), and kidney tubular cells (E. Disteldorf and C. Krebs et al., unpublished observations). This broad expression emphasizes the general role of CXCL5-secreting resident tissue cells in orchestrating PMNs in response to microbial and inflammatory danger signals (6). Thus, epithelial cells do not only serve as a barrier for invading M. tuberculosis but also participate in the coordination of canonical leukocyte-driven immune defense and pathology. Pulmonary CXCL5 failed to enter the blood circulation, as evidenced by the absence of CXCL5 in sera in KO→WT chimeras. Moreover, despite high concentrations in sera, hematopoietic cell–derived CXCL5 had no effect on PMN recruitment to the lung. This observation contrasts findings for severe Escherichia coli pneumonia (39) and, at first sight, appears unexpected, since PMNs follow chemokine gradients to reach the lung. We assume that CXCL5 measured in sera was released during sample preparation by activated platelets (39). Alternatively, it is possible that CXCL5 is present in sera but in an inactive form. CXCL5 binding to scavenger receptors, such as Duffy antigen/receptor for chemokines on erythrocytes, has been described as an underlying decoy mechanism (39).
We identified TLR2-dependent M. tuberculosis sensing by alveolar epithelial cells as an inducer of CXCL5 secretion in TB. In vitro, M. tuberculosis–triggered CXCL5 secretion by AECII was abolished by blocking TLR2 signaling. In vivo, however, it was not surprising that Tlr2–/– mice had reduced, though not abolished, levels of CXCL5 in BAL fluid compared with those of WT mice. In contrast to in vitro cell culture consisting solely of AECII, other cell types likewise responding to M. tuberculosis challenge, e.g., by cytokine secretion, are present in the bronchoalveolar spaces in vivo. In fact, various inflammatory mediators secreted by cells other than pneumocytes are reportedly capable of inducing CXCL5 release by epithelia (49, 59), among which at least IL-1β and TNF-α were detected at day 15 p.i. in WT and Tlr2–/– mice, albeit at low concentrations (G. Nouailles, unpublished observations). Despite the well-described importance of TLR2 in M. tuberculosis sensing (60), TLR2-deficient mice are able to control low-dose M. tuberculosis infection and succumb only to higher doses (61–63). In fact, TLR2 plays a dual role in TB, including both activation of antimycobacterial effector mechanisms in macrophages and downregulation of MHCII expression on antigen-presenting cells, thereby impairing CD4 T cell responses (64). Our data add to these findings a role of TLR2 expressed on the surface of epithelial cells in regulation of the inflammatory host response in the lung.
The spatial and temporal regulation of chemokine expression, as observed for CXCL5 in TB, could serve as general mechanism to regulate delicate immune responses, such as PMN recruitment into tissue compartments. In a murine model of noninfectious inflammatory arthritis, multiple sequential events are required for PMN recruitment into the joints, including BLT1, CCR1, and CXCR2 expression (49). In the case of TB, pathogen-derived TLR2 ligands directly induced PMN-attractant chemokines in resident pulmonary cells. Similarly, in bacterial pneumonia, genetic deletion or antibody-mediated depletion of single CXC chemokines lead to reduced pulmonary PMN recruitment. Cxcl15–/– mice and Cxcl1–/– mice are more susceptible to K. pneumoniae infection and show impaired PMN recruitment into airspaces (30, 37). Depletions of CXCL2 or CXCL1 during pulmonary Legionella pneumoniae and Pseudomonas aeruginosa infection reduce PMN recruitment modestly, but blockage of CXCR2 causes pronounced reduction in PMNs followed by increased mortality (35, 36). Likewise, in M. tuberculosis infection, CXCR2 was absolutely required for PMN recruitment into the bronchoalveolar spaces, but CXCL5 alone accounted for about 60% of the recruited PMNs in the airspaces. Thus, Cxcl5–/– mice are not a phenocopy of Cxcr2–/– mice. CXCR2 ligands, rather than acting redundantly, have additive effects on PMN recruitment. CXCL1 and possibly CXCL2 and CXCL3 contributed to PMN recruitment in TB but to a lesser extent than CXCL5 because of low abundance in BAL fluid. In addition to the type of activated cell — tissue-resident versus hematopoietic — the mode of chemokine activation, through inflammatory mediators, such as leukotriene B4 and IL-17, or through bacterial TLR ligands, determines the strength of PMN attraction.
Murine CXCL5 is most closely related to human neutrophil attractants ENA-78 (human CXCL5) and GCP-2 (human CXCL6) (65, 66). Reportedly, ENA-78 and CXCL8 are made by human pulmonary epithelial cell lines (67, 68). It is tempting to hypothesize that ENA-78 is involved in PMN recruitment to airspaces in human TB. However, it remains to be investigated whether ENA-78 like murine CXCL5 is the dominant chemokine in this regard.
Our results demonstrate an unexpected and nonredundant role of epithelial-derived CXCL5 in PMN-driven pathology in TB. Our characterization of a unique role of CXCL5 in PMN-driven pathology is not restricted to TB. Similar findings were made in a model of noninfectious chronic inflammation, crescentic glomerulonephritis, in which this chemokine contributes to renal tissue injury mediated by pathogenic PMNs (E. Disteldorf and C. Krebs et al., unpublished observations). The essentiality of CXCL5 calls for a more detailed analysis of single chemokine cognates that target shared receptors. Moreover, our experiments strengthen the growing appreciation of epithelial cells as counterparts of leukocytes in the orchestration of immune defense (25).
Methods
Mice
Generation of Cxcl5–/– mice.
The gene-targeting strategy is depicted in Supplemental Figure 1A. In brief, following homologous recombination, coding exons of the Cxcl5 gene were replaced by a β-galactosidase gene (LacZ) using the endogenous start codon (ATG) of the Cxcl5 coding frame in exon 1; a loxP site-flanked self-excising ACN cassette, consisting of a neomycin resistance gene (Neo); and a testes-specific promoter from the angiotensin-converting enzyme (tACE) gene promoter-driven cre gene. Thymidine kinase from herpes simplex virus was used for negative selection (Supplemental Figure 1, A–C). Primers used were LIXLO_FW, TTATGGCGCGCCGGTTATTCCTCTTCTGAGAGCCCAG, and LIXLO_RV, ATATAGTCGACTGTGGACAAGATCCCGGACCAGG, for amplification of the upstream homologous region and LIXSH_FW, TATAGTCGACCATCACATCCATTATTTTAAGCCTGTAC, and LIXSH_RV, AAATTGCGGCCGCTTAGTGTTTCCTACAACCCAGGGGAT, for amplification of the downstream homologous region of the Cxcl5 gene. Cxcl5–/– mice were backcrossed on C57BL/6 background for at least 9 generations.
Breeding and housing.
C57BL/6 (WT) and Cxcr2–/– mice were originally purchased from The Jackson Laboratory. WT, Cxcl5–/–, Cxcr2–/–, and Cxcr2+/+ littermates were bred in our own facilities. Mice were 8 to 9 weeks old and sex- and age-matched for experiments. Infected mice were kept in a biosafety level 3 facility under specific pathogen–free conditions.
Generation of chimeric mice
Recipient mice were lethally γ-irradiated (9 Gy) 24 hours before transplantation. BM from donor mice was harvested from both the tibiae and femora. After lysis of red cells, approximately 5 × 106 cells were intravenously injected into recipient mice. BM transplantation was performed in 4 groups of mice: (a) WT BM in WT recipients (WT→WT); (b) Cxcl5–/– BM in WT recipients (KO→WT); (c) Cxcl5–/– BM in Cxcl5–/– recipients (KO→KO); and (d) WT BM in Cxcl5–/– recipients (WT→KO). Approximately 10 weeks after reconstitution, chimeric mice were infected with M. tuberculosis.
Bacteria and infection of mice
M. tuberculosis strain H37Rv (ATCC, no. 27294) was grown in Middlebrook 7H9 broth (BD Biosciences) supplemented with 0.2% glycerol, 0.05% Tween 80, and 10% ADC enrichment (BD Biosciences). Midlogarithmic cultures were harvested and stored at –80°C. Stocks were tested for virulence and titrated prior to use. Animals were aerosol infected with M. tuberculosis, using a Glas-Col inhalation exposure system at a low dose of approximately 100 CFUs per mouse or a high dose of approximately 500 CFUs per mouse. At designated time points, serial dilutions of tissue homogenates were plated onto Middlebrook 7H11 agar supplemented with 10% OADC Enrichment (BD Biosciences) and ampicillin (25 μg/ml). CFUs were determined after 3 to 4 weeks at 37°C.
Histology
Lung lobes were collected at 21 days p.i., fixed overnight with 4% w/v paraformaldehyde and embedded in paraffin. Hematoxylin and eosin–stained tissues cut at 2 μm were assessed for pathology. A commercially available antibody (rabbit polyclonal anti-MPO, Abcam) was used to detect MPO+ cells. Nuclear β-galactosidase was revealed after protease-mediated antigen retrieval using a polyclonal antibody (Europa Bioproducts), which was preadsorbed on Mycobacterium bovis bacille Calmette-Guérin. Secondary antibodies were from Jackson ImmunoResearch Laboratories. Slides were counterstained either with hematoxylin (Sigma-Aldrich) or with wheat germ agglutinin (Vector) and draq5 (Molecular Probes). Pictures were acquired using a Leica DMLB microscope or a LSM 510 meta microscope and the LSM software (Zeiss) for confocal microscopy analysis.
BAL
Airways were washed twice with 1 ml 1×PBS supplemented with a protease inhibitor cocktail (Roche). The BAL suspension was centrifuged, and the supernatant (BAL fluid) was frozen at –20°C until analysis. Cell pellets were resuspended in media. Viable cells were counted by trypan blue exclusion.
Isolation of lung leukocytes
Lungs were perfused with 1×PBS to reduce numbers of blood leukocytes. The left lung lobe was used for leukocyte purification. Lungs were mechanically digested and incubated for 30 minutes with RPMI 1640 medium supplemented with glutamine, Na-pyruvate, 2-ME, penicillin, streptomycin, 10% heat-inactivated FCS, collagenase D (Roche), and collagenase type VIII (Sigma-Aldrich). Single-cell suspensions of lungs were then prepared using an iron mesh sieve and red blood cell lysis. Viable cells were counted by trypan blue exclusion.
Flow cytometry
For intracellular cytokine staining, 2 × 106 to 4 × 106 cells were cultured for 6 hours in a total volume of 200 μl medium containing 5 μg/ml Brefeldin A and the following M. tuberculosis–derived peptides at 10–4 M concentrations: MHC-II, Ag85AR241–260R (QDAYNAGGGHNGVFDFPDSG), Ag85BR240–260R (FQDAYNAAGGHNAVFNFPPNG), and ESAT6R1–20R (MTEQQWNFAGIEAAASAIQG); MHC-I, PepA (GAPINSATAM; derived from M. tuberculosis 32A protein). Cells were blocked with rat serum and anti-CD16/anti-CD32, surface stained with anti-CD4 (RM4-5; BD) and anti-CD8 (53-6.7, BD) mAbs, fixed with 2% paraformaldehyde, permeabilized with saponin buffer, and stained intracellularly with anti–IFN-γ (XMG1.2, BD) and anti–TNF-α (XT-22, ATCC) mAbs. For flow cytometry of innate immune cells, isolated leukocytes were stained with anti-Ly6G (1A8, BD), anti-CD11b (M1/70, BD), anti-F4/80 (BM8, eBioscience), and anti-CD11c (HL3, ATCC) mAbs. The FL-1 channel was used to determine autofluorescence of cells. All stained cells were acquired using a BD FACS Canto II and analyzed with BD FACSDiva and FACS Analyzer software. Cell numbers were calculated from frequencies among total leukocytes.
MACS cell separation
Selection of LSEC+, LSEC–CD45–, and LSEC–CD45+ cell fractions from isolated lung cells was performed with magnetic beads according to the manufacturer’s instructions (Miltenyi Biotech). Cell fractions were processed for RNA extraction using TRIzol reagent.
Lung homogenates
Lungs were perfused with 1×PBS to reduce numbers of blood leukocytes. Lung lobes were immediately dispersed in 1×PBS supplemented with a protease inhibitor cocktail (Roche) and frozen at –20°C until analysis.
Blood smears and serum samples
Blood samples for serum analysis and blood smears were taken postmortem from the aorta abdominalis. For serum analysis, blood was collected in Serum Separator Tubes (BD) and left for 30 minutes at room temperature, followed by centrifugation at 12,000 g for 3 minutes. Sera were stored at –20°C until analysis. For blood smears, one drop of blood was dispersed on a glass slide, left to dry, and stained with Diff-Quick (Behring). Frequencies of segmented PMNs were calculated from 100 counted cells.
Multiplex analysis and ELISA on cytokines/chemokines
Samples from lung homogenates, BAL fluid, sera, and cell culture supernatants were thawed and appropriately diluted. Measurements were performed using Multiplex Bead-Based Immunoassay Kits (Millipore) or ELISA (R&D) according to the manufacturers’ instructions. Samples were acquired on a Bio-Plex 200 System (Bio-Rad) or SpectraMax ELISA Reader (Molecular Devices).
RNA extraction, cDNA transcription, and quantitative RT-PCR
Lung lobes were immediately homogenized in TRIzol (Invitrogen) or cells were resuspended in TRIzol, and total RNA extraction was carried out according to the manufacturer’s instructions. RNA concentrations and quality were determined using NanoDrop 2000c equipment (Thermo Scientific). cDNA synthesis was carried out using SuperScript III Reverse Transcriptase (Invitrogen) according to the manufacturer’s instructions. Quantitative RT-PCR was performed using Power SYBR Green PCR Master Mix (Applied Biosystems). Amplification was performed with an ABI PRISM 7900HT thermocycler. Expression levels of Cxcl5, Cxcl2, Cxcl1, and housekeeping gene Gapdh (internal control) in lung tissue were analyzed using QuantiTect primer probes (Qiagen). Fold changes in gene expression were calculated with Pfaffl’s method (69) using naive tissue as calibrator or normalized for the expression of the housekeeping gene Gapdh with the formula 1.8(ct Gapdh – ct Cxcl5) × 100 to evaluate Cxcl5 expression in tissues from BM chimeric mice and MACS-sorted lung cells.
Microarray design and hybridization
Total RNA was isolated by the TRIzol (Invitrogen) method following the manufacturer’s protocol. Microarray experiments were performed as dual-color hybridizations. To compensate for dye-specific effects, an independent dye-reversal color-swap was applied (70). Quality control and quantification of total RNA amount were assessed using an Agilent 2100 bioanalyzer (Agilent Technologies) and a NanoDrop 1000 spectrophotometer (Kisker). RNA labeling was performed with the QuickAmp Labeling Kit (Agilent Technologies). In brief, mRNA was reverse transcribed and amplified using an oligo-dT-T7 promotor primer, and the resulting cRNA was labeled either with cyanine 3-CTP or cyanine 5-CTP. After precipitation, purification, and quantification, 1.25 μg of each labeled cRNA was fragmented and hybridized to whole mouse genome 44 k microarrays (AMADID 014868) according to the supplier’s protocol (Agilent Technologies). Scanning of microarrays was performed with 5 μm resolution and extended range using a G2565CA high-resolution laser microarray scanner (Agilent Technologies). Raw microarray image data were analyzed with the Image Analysis/Feature Extraction software G2567AA (version A.10.5.1.1, Agilent Technologies) using default settings and the GE2_105_Jan09 protocol. The extracted MAGE-ML files were further analyzed with the Rosetta Resolver Biosoftware, build 7.2.2 SP1.31 (Rosetta Biosoftware). Data have been deposited in NCBI’s Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo) and are accessible with GEO series accession number GSE43337 ( http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=vzcfbwioesmsefk&acc=GSE43337).
Microarray analysis
Limma version 3.12.1 (71) was used: normalization of raw microarray data was done using normexp background correction, loess within-array normalization, and quantile between-array normalization. Spots were annotated using the Agilent mouse annotation data (mgug4121a), and duplicate array probes were averaged. The differences in the reaction of the mouse strains in response to M. tuberculosis infection were asserted by testing for significant interaction between infection and strain using the moderated eBayes statistic in Limma. Calculated P values were corrected for multiple testing using Benjamini and Hochberg method (72), and the resulting q values were used throughout the subsequent analyses.
Functional analysis
To test for enrichment in sets of differentially expressed genes, we applied the Gorilla algorithm (73) first in a single-list mode, in which genes were sorted by log fold change of the interaction contrast, and second by comparing differentially expressed sets of genes defined by absolute log2 fold change > 0.5 and q < 0.005, with the set of remaining genes as background. Resulting P values were corrected for multiple testing using the Benjamini and Hochberg method (72). Pathway analysis was done using the SPIA package for R (74), which allows testing of both pathway enrichment and pathway perturbation. The resulting composite P value is corrected for multiple testing using the Bonferroni method.
Cell cultures of primary cells and cell lines
BMDMs were obtained and generated as described previously (27). PMNs were purified from BM with anti-Ly6G magnetic beads according to the manufacturer’s instructions (Miltenyi Biotech). T7 cells, a murine AECII cell line (ECACC 07021402), were grown in T7 media (DMEM supplemented with 10% FCS [heat inactivated], 0.2 mM l-glutamine, 10 mM HEPES, and insulin-transferrin-selenium A [GIBCO]). Prior to infection/stimulation cells were harvested, seeded in OPTI-MEM (GIBCO), and allowed to rest for 3.5 hours; anti-TLR2–blocking antibody (T2.5; eBioscience) was added to the cells 0.5 hours before stimulations. Cells were kept at 37°C in a 5% CO2 atmosphere.
In vitro M. tuberculosis infections
Primary cells and cell lines were infected with M. tuberculosis obtained from early log phase cultures. Bacteria were grown in Middlebrook 7H9 broth (BD Biosciences) supplemented with 10% ADC enrichment (BD Biosciences), 0.05% glycerol, and Tween 80. Single bacteria were resuspended in culture media at various MOIs. Cell culture supernatants were harvested at indicated time points p.i. and stored at –20°C until further analysis.
In vitro stimulation
γ-Irradiated M. tuberculosis, cell wall, membrane, cytosolic fraction, total lipids, mycolylarabinogalactan-peptidoglycan, trehalose-6,6′-dimycolate, peptidoglycan, mannose-capped lipoarabinomannan, phosphatidyl-myo-inositol mannosides 1 and 2, lipoarabinomannan, and lipomannan were derived from M. tuberculosis and obtained through BEI Resources, National Institute of Allergy and Infectious Diseases, NIH (M. tuberculosis strain H37Rv). Muramyl dipeptide was from Calbiochem. LPS, CpG ODN 1826, and zymosan were obtained from InvivoGen. Synthetic PAM3CSK4, MPT3, and 19-kDA and 27-kDA lipopeptides were obtained from EMC Microcollections. For all treatments, AECII were prepared as described for the M. tuberculosis infection.
Statistics
PRISM GraphPad software was used for the following statistical analyses. Survival curves were analyzed using the Kaplan-Meier method and log-rank test. Two groups were compared using 2-tailed, unpaired t test; 3 or more groups were compared by 1-way ANOVA/Bonferroni post-test; and grouped analyses were performed using 2-way ANOVA/Bonferroni post-test. P v–alues of less than 0.05 were considered statistically significant.
Study approval
All animal experiments were conducted according to German animal protection law (T0070/08, T0223/11, T0087/13, G0119/06, G0266/11).
Supplementary Material
Acknowledgments
The authors thank U. Zedler and D. Oberbeck-Mueller for immunohistochemical stainings, K. Hahnke for assistance with microarrays, C. Koeberle for developing a program to assist analysis of flow cytometric data, M.L. Grossman for editorial help, and M. Kursar for scientific advice. This work was supported by the European Union’s Seventh Framework Programme (EU FP7) projects “NewTBVAC” (Health-F3-2009-241745) and “ADITEC” (HEALTH-F4-2011-280873).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2014;124(3):1268–1282. doi:10.1172/JCI72030.
Markus Koch’s present address is: Bayer Pharma AG, Berlin, Germany.
Geraldine Nouailles’s present address is: Charité — Universitätsmedizin Berlin, Berlin, Germany.
Jens Zerrahn’s present address is: Parexel, Berlin, Germany.
References
- 1.World Health Organization. Global Tuberculosis Report 2012. Geneva, Switzerland: World Health Organization; 2012. [Google Scholar]
- 2.Gengenbacher M, Kaufmann SH. Mycobacterium tuberculosis: success through dormancy. FEMS Microbiol Rev. 2012;36(3):514–532. doi: 10.1111/j.1574-6976.2012.00331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cooper AM. T cells in mycobacterial infection and disease. Curr Opin Immunol. 2009;21(4):378–384. doi: 10.1016/j.coi.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kleinnijenhuis J, Oosting M, Joosten LA, Netea MG, Van Crevel R. Innate immune recognition of Mycobacterium tuberculosis. Clin Dev Immunol. 2011;2011:405310. doi: 10.1155/2011/405310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thoma-Uszynski S, et al. Induction of direct antimicrobial activity through mammalian toll-like receptors. Science. 2001;291(5508):1544–1547. doi: 10.1126/science.291.5508.1544. [DOI] [PubMed] [Google Scholar]
- 6.Li Y, Wang Y, Liu X. The role of airway epithelial cells in response to mycobacteria infection. Clin Dev Immunol. 2012;2012:791392. doi: 10.1155/2012/791392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ernst JD. The immunological life cycle of tuberculosis. Nat Rev Immunol. 2012;12(8):581–591. doi: 10.1038/nri3259. [DOI] [PubMed] [Google Scholar]
- 8.Philips JA, Ernst JD. Tuberculosis pathogenesis and immunity. Annu Rev Pathol. 2012;7:353–384. doi: 10.1146/annurev-pathol-011811-132458. [DOI] [PubMed] [Google Scholar]
- 9.Keller C, Hoffmann R, Lang R, Brandau S, Hermann C, Ehlers S. Genetically determined susceptibility to tuberculosis in mice causally involves accelerated and enhanced recruitment of granulocytes. Infect Immun. 2006;74(7):4295–4309. doi: 10.1128/IAI.00057-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eum SY, et al. Neutrophils are the predominant infected phagocytic cells in the airways of patients with active pulmonary TB. Chest. 2010;137(1):122–128. doi: 10.1378/chest.09-0903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lowe DM, Redford PS, Wilkinson RJ, O’Garra A, Martineau AR. Neutrophils in tuberculosis: friend or foe? Trends Immunol. 2012;33(1):14–25. doi: 10.1016/j.it.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 12.Jones GS, Amirault HJ, Andersen BR. Killing of Mycobacterium tuberculosis by neutrophils: a nonoxidative process. J Infect Dis. 1990;162(3):700–704. doi: 10.1093/infdis/162.3.700. [DOI] [PubMed] [Google Scholar]
- 13.Brown AE, Holzer TJ, Andersen BR. Capacity of human neutrophils to kill Mycobacterium tuberculosis. J Infect Dis. 1987;156(6):985–989. doi: 10.1093/infdis/156.6.985. [DOI] [PubMed] [Google Scholar]
- 14.Kisich KO, Higgins M, Diamond G, Heifets L. Tumor necrosis factor alpha stimulates killing of Mycobacterium tuberculosis by human neutrophils. Infect Immun. 2002;70(8):4591–4599. doi: 10.1128/IAI.70.8.4591-4599.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martineau AR, et al. Neutrophil-mediated innate immune resistance to mycobacteria. J Clin Invest. 2007;117(7):1988–1994. doi: 10.1172/JCI31097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corleis B, Korbel D, Wilson R, Bylund J, Chee R, Schaible UE. Escape of Mycobacterium tuberculosis from oxidative killing by neutrophils. Cell Microbiol. 2012;14(7):1109–1121. doi: 10.1111/j.1462-5822.2012.01783.x. [DOI] [PubMed] [Google Scholar]
- 17.Denis M. Human neutrophils, activated with cytokines or not, do not kill virulent Mycobacterium tuberculosis. J Infect Dis. 1991;163(4):919–920. doi: 10.1093/infdis/163.4.919. [DOI] [PubMed] [Google Scholar]
- 18.Aston C, Rom WN, Talbot AT, Reibman J. Early inhibition of mycobacterial growth by human alveolar macrophages is not due to nitric oxide. Am J Respir Crit Care Med. 1998;157(6 pt 1):1943–1950. doi: 10.1164/ajrccm.157.6.9705028. [DOI] [PubMed] [Google Scholar]
- 19.Reyes-Ruvalcaba D, Gonzalez-Cortes C, Rivero-Lezcano OM. Human phagocytes lack the ability to kill Mycobacterium gordonae, a non-pathogenic mycobacteria. Immunol Lett. 2008;116(1):72–78. doi: 10.1016/j.imlet.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 20.Ramos-Kichik V, et al. Neutrophil extracellular traps are induced by Mycobacterium tuberculosis. Tuberculosis (Edinb). 2009;89(1):29–37. doi: 10.1016/j.tube.2008.09.009. [DOI] [PubMed] [Google Scholar]
- 21.Yang CT, Cambier CJ, Davis JM, Hall CJ, Crosier PS, Ramakrishnan L. Neutrophils exert protection in the early tuberculous granuloma by oxidative killing of mycobacteria phagocytosed from infected macrophages. Cell Host Microbe. 2012;12(3):301–312. doi: 10.1016/j.chom.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pedrosa J, Saunders BM, Appelberg R, Orme IM, Silva MT, Cooper AM. Neutrophils play a protective nonphagocytic role in systemic Mycobacterium tuberculosis infection of mice. Infect Immun. 2000;68(2):577–583. doi: 10.1128/IAI.68.2.577-583.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fulton SA, Reba SM, Martin TD, Boom WH. Neutrophil-mediated mycobacteriocidal immunity in the lung during Mycobacterium bovis BCG infection in C57BL/6 mice. Infect Immun. 2002;70(9):5322–5327. doi: 10.1128/IAI.70.9.5322-5327.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eruslanov EB, et al. Neutrophil responses to Mycobacterium tuberculosis infection in genetically susceptible and resistant mice. Infect Immun. 2005;73(3):1744–1753. doi: 10.1128/IAI.73.3.1744-1753.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Desvignes L, Ernst JD. Interferon-gamma-responsive nonhematopoietic cells regulate the immune response to Mycobacterium tuberculosis. Immunity. 2009;31(6):974–985. doi: 10.1016/j.immuni.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nandi B, Behar SM. Regulation of neutrophils by interferon-gamma limits lung inflammation during tuberculosis infection. J Exp Med. 2011;208(11):2251–2262. doi: 10.1084/jem.20110919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dorhoi A, et al. The adaptor molecule CARD9 is essential for tuberculosis control. J Exp Med. 2010;207(4):777–792. doi: 10.1084/jem.20090067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iyoda T, Kobayashi Y. Involvement of MIP-2 and CXCR2 in neutrophil infiltration following injection of late apoptotic cells into the peritoneal cavity. Apoptosis. 2004;9(4):485–493. doi: 10.1023/B:APPT.0000031450.95188.e7. [DOI] [PubMed] [Google Scholar]
- 29.Bhatia M, Zemans RL, Jeyaseelan S. Role of chemokines in the pathogenesis of acute lung injury. Am J Respir Cell Mol Biol. 2012;46(5):566–572. doi: 10.1165/rcmb.2011-0392TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cai S, Batra S, Lira SA, Kolls JK, Jeyaseelan S. CXCL1 regulates pulmonary host defense to Klebsiella infection via CXCL2, CXCL5, NF-κB, and MAPKs. J Immunol. 2010;185(10):6214–6225. doi: 10.4049/jimmunol.0903843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jeyaseelan S, et al. Induction of CXCL5 during inflammation in the rodent lung involves activation of alveolar epithelium. Am J Respir Cell Mol Biol. 2005;32(6):531–539. doi: 10.1165/rcmb.2005-0063OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Batra S, Cai S, Balamayooran G, Jeyaseelan S. Intrapulmonary administration of leukotriene B(4) augments neutrophil accumulation and responses in the lung to Klebsiella infection in CXCL1 knockout mice. J Immunol. 2012;188(7):3458–3468. doi: 10.4049/jimmunol.1101985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ritzman AM, Hughes-Hanks JM, Blaho VA, Wax LE, Mitchell WJ, Brown CR. The chemokine receptor CXCR2 ligand KC (CXCL1) mediates neutrophil recruitment and is critical for development of experimental Lyme arthritis and carditis. Infect Immun. 2010;78(11):4593–4600. doi: 10.1128/IAI.00798-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moore TA, Newstead MW, Strieter RM, Mehrad B, Beaman BL, Standiford TJ. Bacterial clearance and survival are dependent on CXC chemokine receptor-2 ligands in a murine model of pulmonary Nocardia asteroides infection. J Immunol. 2000;164(2):908–915. doi: 10.4049/jimmunol.164.2.908. [DOI] [PubMed] [Google Scholar]
- 35.Tsai WC, Strieter RM, Mehrad B, Newstead MW, Zeng X, Standiford TJ. CXC chemokine receptor CXCR2 is essential for protective innate host response in murine Pseudomonas aeruginosa pneumonia. Infect Immun. 2000;68(7):4289–4296. doi: 10.1128/IAI.68.7.4289-4296.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tateda K, et al. Chemokine-dependent neutrophil recruitment in a murine model of Legionella pneumonia: potential role of neutrophils as immunoregulatory cells. Infect Immun. 2001;69(4):2017–2024. doi: 10.1128/IAI.69.4.2017-2024.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen SC, et al. Impaired pulmonary host defense in mice lacking expression of the CXC chemokine lungkine. J Immunol. 2001;166(5):3362–3368. doi: 10.4049/jimmunol.166.5.3362. [DOI] [PubMed] [Google Scholar]
- 38.Mei J, et al. Cxcr2 and Cxcl5 regulate the IL-17/G-CSF axis and neutrophil homeostasis in mice. J Clin Invest. 2012;122(3):974–986. doi: 10.1172/JCI60588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mei J, et al. CXCL5 regulates chemokine scavenging and pulmonary host defense to bacterial infection. Immunity. 2010;33(1):106–117. doi: 10.1016/j.immuni.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jeyaseelan S, Chu HW, Young SK, Worthen GS. Transcriptional profiling of lipopolysaccharide-induced acute lung injury. Infect Immun. 2004;72(12):7247–7256. doi: 10.1128/IAI.72.12.7247-7256.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamamoto K, et al. Type I alveolar epithelial cells mount innate immune responses during pneumococcal pneumonia. J Immunol. 2012;189(5):2450–2459. doi: 10.4049/jimmunol.1200634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kang DD, Lin Y, Moreno JR, Randall TD, Khader SA. Profiling early lung immune responses in the mouse model of tuberculosis. PLoS One. 2011;6(1):e16161. doi: 10.1371/journal.pone.0016161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Slight SR, Khader SA. Chemokines shape the immune responses to tuberculosis. Cytokine Growth Factor Rev. 2013;24(2):105–113. doi: 10.1016/j.cytogfr.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Khader SA, et al. In a murine tuberculosis model, the absence of homeostatic chemokines delays granuloma formation and protective immunity. J Immunol. 2009;183(12):8004–8014. doi: 10.4049/jimmunol.0901937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Slight SR, et al. CXCR5+ T helper cells mediate protective immunity against tuberculosis. . J Clin Invest. 2013;123(2):712–726. doi: 10.1172/JCI65728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gopal R, et al. Interleukin-17-dependent CXCL13 mediates mucosal vaccine-induced immunity against tuberculosis. Mucosal Immunol. 2013;6(5):972–984. doi: 10.1038/mi.2012.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seiler P, et al. Early granuloma formation after aerosol Mycobacterium tuberculosis infection is regulated by neutrophils via CXCR3-signaling chemokines. Eur J Immunol. 2003;33(10):2676–2686. doi: 10.1002/eji.200323956. [DOI] [PubMed] [Google Scholar]
- 48.Lionakis MS, et al. Chemokine receptor Ccr1 drives neutrophil-mediated kidney immunopathology and mortality in invasive candidiasis. PLoS Pathog. 2012;8(8):e1002865. doi: 10.1371/journal.ppat.1002865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chou RC, et al. Lipid-cytokine-chemokine cascade drives neutrophil recruitment in a murine model of inflammatory arthritis. Immunity. 2010;33(2):266–278. doi: 10.1016/j.immuni.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Herbold W, et al. Importance of CXC chemokine receptor 2 in alveolar neutrophil and exudate macrophage recruitment in response to pneumococcal lung infection. Infect Immun. 2010;78(6):2620–2630. doi: 10.1128/IAI.01169-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Del Rio L, Bennouna S, Salinas J, Denkers EY. CXCR2 deficiency confers impaired neutrophil recruitment and increased susceptibility during Toxoplasma gondii infection. J Immunol. 2001;167(11):6503–6509. doi: 10.4049/jimmunol.167.11.6503. [DOI] [PubMed] [Google Scholar]
- 52.Goncalves AS, Appelberg R. The involvement of the chemokine receptor CXCR2 in neutrophil recruitment in LPS-induced inflammation and in Mycobacterium avium infection. Scand J Immunol. 2002;55(6):585–591. doi: 10.1046/j.1365-3083.2002.01097.x. [DOI] [PubMed] [Google Scholar]
- 53.Cacalano G, et al. Neutrophil and B cell expansion in mice that lack the murine IL-8 receptor homolog. Science. 1994;265(5172):682–684. doi: 10.1126/science.8036519. [DOI] [PubMed] [Google Scholar]
- 54.Blomgran R, Desvignes L, Briken V, Ernst JD. Mycobacterium tuberculosis inhibits neutrophil apoptosis, leading to delayed activation of naive CD4 T cells. Cell Host Microbe. 2012;11(1):81–90. doi: 10.1016/j.chom.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Blomgran R, Ernst JD. Lung neutrophils facilitate activation of naive antigen-specific CD4+ T cells during Mycobacterium tuberculosis infection. J Immunol. 2011;186(12):7110–7119. doi: 10.4049/jimmunol.1100001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Green AM, Difazio R, Flynn JL. IFN-γ from CD4 T cells is essential for host survival and enhances CD8 T cell function during Mycobacterium tuberculosis infection. J Immunol. 2013;190(1):270–277. doi: 10.4049/jimmunol.1200061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Madorin WS, Rui T, Sugimoto N, Handa O, Cepinskas G, Kvietys PR. Cardiac myocytes activated by septic plasma promote neutrophil transendothelial migration: role of platelet-activating factor and the chemokines LIX and KC. Circ Res. 2004;94(7):944–951. doi: 10.1161/01.RES.0000124395.20249.AE. [DOI] [PubMed] [Google Scholar]
- 58.Rousselle A, et al. CXCL5 limits macrophage foam cell formation in atherosclerosis. J Clin Invest. 2013;123(3):1343–1347. doi: 10.1172/JCI66580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu Y, et al. IL-17A and TNF-α exert synergistic effects on expression of CXCL5 by alveolar type II cells in vivo and in vitro. J Immunol. 2011;186(5):3197–3205. doi: 10.4049/jimmunol.1002016. [DOI] [PubMed] [Google Scholar]
- 60.Drage MG, et al. TLR2 and its co-receptors determine responses of macrophages and dendritic cells to lipoproteins of Mycobacterium tuberculosis. Cell Immunol. 2009;258(1):29–37. doi: 10.1016/j.cellimm.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bafica A, Scanga CA, Feng CG, Leifer C, Cheever A, Sher A. TLR9 regulates Th1 responses and cooperates with TLR2 in mediating optimal resistance to Mycobacterium tuberculosis. J Exp Med. 2005;202(12):1715–1724. doi: 10.1084/jem.20051782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reiling N, et al. Cutting edge: Toll-like receptor (TLR)2- and TLR4-mediated pathogen recognition in resistance to airborne infection with Mycobacterium tuberculosis. J Immunol. 2002;169(7):3480–3484. doi: 10.4049/jimmunol.169.7.3480. [DOI] [PubMed] [Google Scholar]
- 63.Drennan MB, et al. Toll-like receptor 2-deficient mice succumb to Mycobacterium tuberculosis infection. Am J Pathol. 2004;164(1):49–57. doi: 10.1016/S0002-9440(10)63095-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stenger S, Modlin RL. Control of Mycobacterium tuberculosis through mammalian Toll-like receptors. Curr Opin Immunol. 2002;14(4):452–457. doi: 10.1016/S0952-7915(02)00355-2. [DOI] [PubMed] [Google Scholar]
- 65.Zlotnik A, Yoshie O. The chemokine superfamily revisited. Immunity. 2012;36(5):705–716. doi: 10.1016/j.immuni.2012.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zlotnik A, Yoshie O. Chemokines: a new classification system and their role in immunity. Immunity. 2000;12(2):121–127. doi: 10.1016/S1074-7613(00)80165-X. [DOI] [PubMed] [Google Scholar]
- 67.Murdoch C, Read RC, Zhang Q, Finn A. Choline-binding protein A of Streptococcus pneumoniae elicits chemokine production and expression of intercellular adhesion molecule 1 (CD54) by human alveolar epithelial cells. J Infect Dis. 2002;186(9):1253–1260. doi: 10.1086/344527. [DOI] [PubMed] [Google Scholar]
- 68.Lin Y, Zhang M, Barnes PF. Chemokine production by a human alveolar epithelial cell line in response to Mycobacterium tuberculosis. Infect Immun. 1998;66(3):1121–1126. doi: 10.1128/iai.66.3.1121-1126.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Churchill GA. Fundamentals of experimental design for cDNA microarrays. Nat Genet. 2002;32(suppl):490–495. doi: 10.1038/ng1031. [DOI] [PubMed] [Google Scholar]
- 71.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 72.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc Ser B (Methodol). 1995;57(1):289–300. [Google Scholar]
- 73.Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tarca AL, et al. A novel signaling pathway impact analysis. Bioinformatics. 2009;25(1):75–82. doi: 10.1093/bioinformatics/btn577. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.