Abstract
Age-associated deterioration in both the quality and quantity of mitochondria occurs in older women. The main aim of this study was to examine the effect of age on mitochondrial DNA copy number (mtDNA number) in early developmental stage bovine embryos as well as the dynamics of mtDNA number during early embryo development. Real-time PCR was used to determine mtDNA number. In vitro-produced embryos 48 h after insemination derived from Japanese black cows, ranging in age from 25 to 209 months were categorized based on their cleavage status. There was an overall negative relationship between the age of the cow and cleavage status, to the extent that the ratio of embryos cleaved over the 4-cell stage was greater in younger cows. The mtDNA number did not differ among the cleaved status of embryos. In the next experiment, oocytes collected from each donor cow were divided into 2 groups containing 10 oocytes each, in order to compare the mtDNA number of mature oocytes and early developmental stage embryos within individuals. Upon comparing the mtDNA number between oocytes at the M2 stage and early developmental stage 48 h post insemination, mtDNA number was found to decrease in most cows, but was found to increase in some cows. In conclusion, age affects the cleaving ability of oocytes, and very old cows (> 180 months) tend to have lower mtDNA numbers in their oocytes. The change in mtDNA number during early development varied among individual cows, although overall, it showed a tendency to decrease.
Keywords: Aging, Cow, Embryo, Mitochondrial DNA copy number
Mitochondria play various cellular roles, including roles in calcium homeostasis, oxidative phosphorylation, signal transduction, steroid and heme synthesis, and apoptosis [1]. In oocytes, mitochondria have a small, spherical, undifferentiated shape [2], and their numbers increase during oocyte growth [3].
Mitochondrial DNA copy number (mtDNA number) has generally been examined based on the premise that the mitochondria in oocytes contain only 1 or 2 genomes [4]. It has long been believed that mtDNA number increases during oocyte growth and then decreases from fertilization to the early developmental stage embryos in cows, although it remains constant from the oocyte to the blastocyst stage in mice [5,6,7]. Conversely, when the mtDNA number is artificially reduced, the number then increases during embryo development [8], indicating that in some cases, the mtDNA number can increase in zygotes. There is, however, no definite consensus as to whether oocytes decrease or increase their mtDNA number during early embryo development, because it is not possible to measure the mtDNA number noninvasively.
Age-associated subfertility is a common feature of mammals. Oocytes collected from aged females have low fertilization rates and developmental abilities [9,10,11]. There is accumulating evidence demonstrating that aging affects both the quality and quantity of mitochondria in oocytes [12,13,14]. Age-associated reproductive deterioration occurs gradually in humans (>35 years) [15], whereas in mice, a common animal model, it occurs within 1 year [16]. In light of the fact that it is difficult to collect a sufficient number of mouse oocytes due to their diminishing reproductive performance, how maternal aging affects the mtDNA number in early developmental stage embryos remains unclear. Therefore, other animal models with long reproductive lives and dozens of available oocytes are required for fertility research. Cows have a similar oocyte selection system and reproductive life length to humans [17,18,19,20,21], and dozens of oocytes can be easily collected from their ovaries. Moreover, cows in Japan are readily tractable, as we are able to identify breeds and age in months at a slaughterhouse.
Recently, we showed that oocytes derived from aged cows have a low mtDNA number and a reduced ability for in vitro maturation and fertilization [22]. In this study, we compared the mtDNA number between both in vitro matured oocytes and early developmental stage embryos 48 h post insemination (p.i.) within a donor using a novel comparison method. We examined the effect of aging on cleavage and mitochondrial number in early developmental stage embryos. In addition, we examined the dynamics in mitochondrial number between in vitro matured oocytes and early developmental stage embryos 48 h after insemination.
Materials and Methods
Chemicals
All reagents used in this study were purchased from Nacalai Tesque (Kyoto, Japan) unless stated otherwise. Synthetic oviductal fluid (SOF) was the medium used for fertilization and culture [23]. We used TCM-199 medium (Sigma-Aldrich, St Louis, MO, USA), containing 5 mM taurine and 10% fetal calf serum (FCS; 5703H; ICN Pharmaceuticals, Costa Mesa, CA, USA), SOF supplemented with 5 mg/ml BSA (fatty acid free) and 10 U/ml heparin (Sigma-Aldrich) as the in vitro fertilization (IVF) medium. SOF supplemented with amino acids, 1% FCS, 5 mM taurine and 1.5 mM glucose was used as the in vitro culture (IVC) medium. Cultures were maintained at 38.5 C with 5% CO2 in air at maximum humidity for in vitro maturation (IVM) and IVF and at 38.5 C with 5% CO2, 5% O2, and 90% N2 in air at maximum humidity for IVC.
Ovary and oocyte collection
Japanese Black cows aged 20–204 months were used as donors. The breed and age (in months) of each cow were confirmed at the slaughterhouse. Ovaries with 1 dominant follicle and a functional luteum were collected and stored at 25 C in phosphate-buffered saline containing antibiotics and were transported to the laboratory within 4 h. This ovary selection was intended to minimize the effect of the estrus cycle and to avoid collecting ovaries from aged cows that have already lost their reproductive ability. The preservation period of the ovaries used for all experiments was 3–4 h. Cumulus-oocyte complexes (COCs) were collected from both ovaries of each cow by using a syringe with an 18-G needle.
In vitro maturation and fertilization
COCs were matured in the IVM medium for 21 h (10 COCs/100 μl drop). After maturation, the complexes were washed in the IVF medium and co-incubated with frozen-thawed semen. The semen was obtained from a Japanese Black bull and was washed with a 45 to 60% Percoll solution (Amersham Biosciences, Uppsala, Sweden) to create a discontinuous gradient for centrifugation (800 × g for 10 min). The final sperm concentration in the IVF medium was 1 × 106 cells/ml. After 7 h of co-incubation, the oocytes were denuded from the surrounding cumulus cells by vortexing. Forty hours after insemination, oocytes and embryos were divided into 4 categories: >7-cell stage embryos, 4- to 7-cell stage embryos, 2- to 3-cell stage embryos, and noncleaved oocytes.
Assessment of mitochondrial DNA number
The average mtDNA number in both mature (after 21 h of culture) oocytes and embryos was determined by examining a group of oocytes or embryos from each donor. The oocytes or embryos from each donor were lysed in 6 μl lysis buffer (20 mM Tris, 0.4 mg/ml pronase K, 0.9% Nonidet P-40 and 0.9% Tween 20) at 55 C for 30 min followed by 95 C for 5 min. Mitochondrial DNA number was then determined by real-time PCR as described previously [22]. In brief, the PCR reaction mixture (20 µl) was prepared with 6 µl of extracted DNA, 0.5 µM of each primer set (5′-ATTTACAGCAATATGCGCCC-3′ and 5′-AAAAGGCGTGGGTACAGATG-3′) and MESA Blue (qPCR MasterMix Plus for SYBR Assay; Eurogentec, Seraing, Belgium). PCR was performed with initial denaturation at 95 C for 5 min followed by 40 cycles at 95 C, 58 C and 72 C each for 30 sec. SYBR green fluorescence was measured at the end of each extension step (72 C). A standard curve was generated for each run using 10-fold serial dilutions representing copies of the external standard. The external standard we used was the PCR product of the corresponding gene cloned into a vector by using a Zero Blunt TOPO PCR Cloning Kit (Invitrogen, Carlsbad, CA, USA). Data from 2 trials in which the amplification efficiency was less than 1.9 were discarded.
Experimental designs
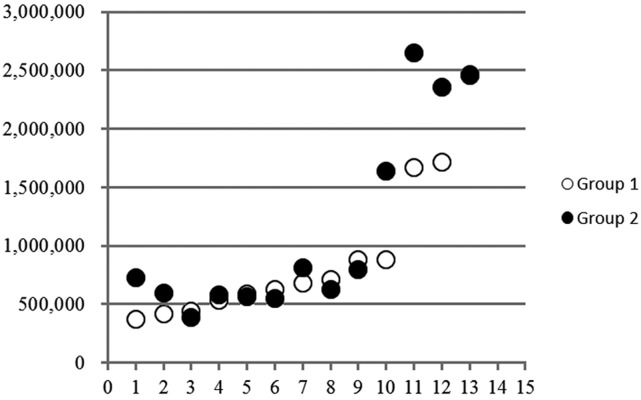
To compare the mtDNA number between oocytes and early developmental stage embryos, we conducted the following pilot experiment. Twenty oocytes were collected from each donor, and the average mtDNA number was calculated using 10 oocytes and compared with the average mtDNA number obtained using the other 10 oocytes. As shown in Fig. 1, there was a significantly high correlation between the mtDNA numbers in the 2 groups (r = 0.91, P < 0.01). This result suggests that mtDNA numbers could be confidently compared between 2 different cell stages within the same donor.
Fig. 1.
Comparison of mtDNA numbers between 2 groups consisting of 10 oocytes each collected from the same donor cows. The 2 values were significantly correlated. Y-axis, mtDNA number; X-axis, donor number.
In experiment 1, we collected oocytes from each of 49 cows, ranging in age from 25 to 209 months. All oocytes collected from each donor were in vitro matured and fertilized as described above. Forty-eight hours after insemination, the embryos were categorized by cleavage stage as either >7-cell, 4- to 7-cell, 2- to 3-cell or noncleaved oocytes. The group of the embryos categorized to each cleavage stage were used to determine the mtDNA number for each embryo stage. Then, the mtDNA number was related to donor age and cleavage ratio.
In experiment 2, we examined whether the mtDNA number increased or decreased throughout early development. Over twenty oocytes were collected from each donor, and 10 in vitro-matured oocytes were used to determine the mtDNA number. The remaining oocytes were fertilized and categorized based on the number of blastomeres, as described above, and the mtDNA copy number was determined for each stage. In this experiment only young cows were used, and the average age in months was 28.1 ± 0.5.
Statistical analysis
Pearson's correlation coefficients between parameters and regression lines were calculated using SPSS 17.0 (SPSS, Chicago, IL, USA). The mtDNA number was compared using 2-tailed Student's t-tests. A P value < 0.05 was considered statistically significant.
Results
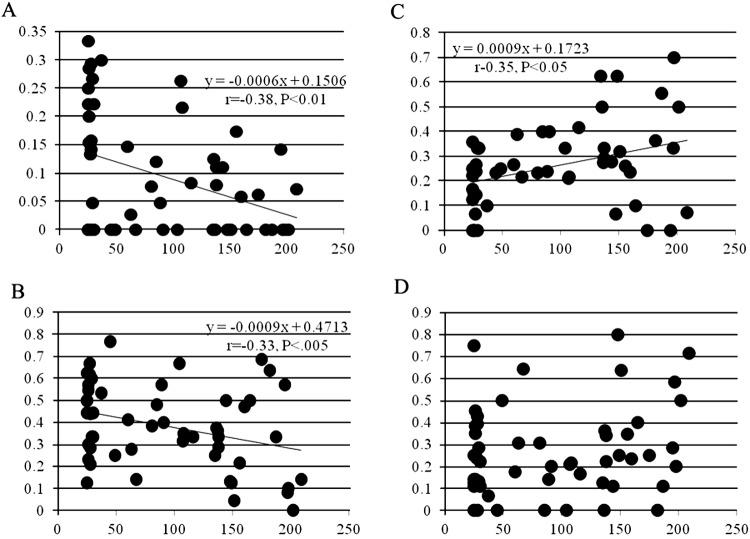
In experiment 1, the cleavage states of 882 zygotes in total were examined: 10.9, 37.3, 25.4, and 26.4% of the zygotes were categorized as >7-cell stage, 4- to 7-cell stage, 2- to 3-cell stage, and noncleaved oocytes, respectively. Figure 2 shows the relationship between donor age (in months) and the ratio of zygotes categorized into each developmental stage. There was a significant negative correlation between donor age and the ratio of both >7-stage embryos and 4- to 7-cell stage embryos (Fig. 2A and B), while the relationship between donor age and the ratio of 2- to 3-cell stage embryos was significantly positive (Fig. 2C). There was no relationship between age and the ratio of noncleaved oocytes (Fig. 2D).
Fig. 2.
Correlation between age and ratio of cleavage. X-axis, donor age in months; Y-axis, ratio of cleavage, determined 48 h after insemination. A: >7-cell stage embryos, B: 4- to 7-cell stage embryos, C: 2- to 3-cell stage embryos, and D: noncleaved oocytes.
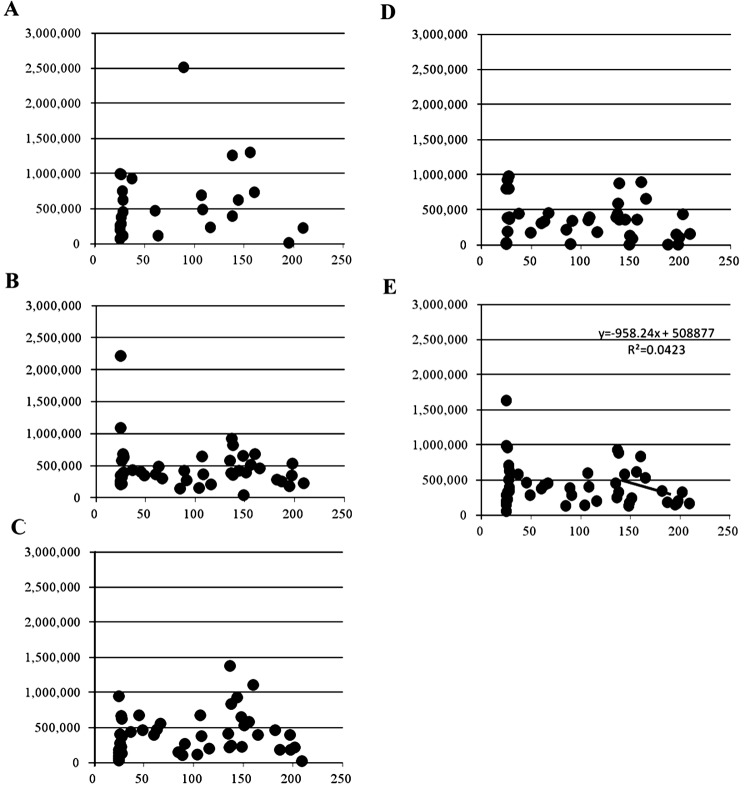
The mtDNA numbers of embryos for all ages, and in each of 3 age groups (>120, 45–120, and 25–45 months), are shown in Table 1. When we look at the data of all age groups (n = 49), the highest mtDNA number was observed in >7-cell stage embryos (587,536 ± 104,277), which was significantly greater than that observed in noncleaved oocytes (353,039 ± 43,345, P < 0.05). Similar to this result, the highest mtDNA number was observed in >7-cell stage embryos, and the lowest mtDNA number was observed in noncleaved oocytes in both the middle-aged (n = 12) and aged cows groups (n= 19), whereas in young cows (n = 18), embryos had high mtDNA numbers after the 4-cell stage, and the lowest mtDNA number was observed in 2–3-cell stage embryos (Table 1). Interestingly, when we looked at the mtDNA number of early developmental stage embryos derived from very old cows (>180 in months), it was found to be lower numbers than those of the other age groups, although the differences did not reach statistical significance. Figure 3 shows that overall, there was no significant correlation between the mtDNA number and donor age at any of the developmental stages of embryos. However, using only data obtained from the more aged donors (>120 months), a significantly negative correlation was evident between mtDNA number and donor age (Fig. 3E; r = –0.49, y = –2E-05x + 183.85, P < 0.05). Similarly, embryos derived from aged cows had lower mtDNA numbers in both 4- to 7-cell stage and >7-cell stage embryos (Figs. 3A and B).
Table 1. Comparison of mitochondrial DNA copy number of embryos 48 h after insemination among age groups.
| Developmental stage | mtDNA number (Mean ± SE) | ||||||||||||||
| All ages (n=49) | Young (n=18) | Middle (n=12) | Aged (n=19) | Aged (>180)* (n=7) | |||||||||||
| >7-cell | 587,536 | ± | 104,277a | 501,818 | ± | 163,051 | 674,356 | ± | 216,656 | 647,661 | ± | 203,372 | 118,408 | ± | 106,535 |
| 4-to 7-cell | 463,796 | ± | 49,815ab | 563,569 | ± | 52,664 | 357,376 | ± | 64,052 | 446,055 | ± | 81,851 | 301,181 | ± | 52,016 |
| 2-to 3-cell | 397,852 | ± | 43,731ab | 335,085 | ± | 74,154 | 336,218 | ± | 87,734 | 494,735 | ± | 69,936 | 240,436 | ± | 65,043 |
| Non cleaved | 353,039 | ± | 43,345b | 444,072 | ± | 78,958 | 286,554 | ± | 69,663 | 332,980 | ± | 63,891 | 141,590 | ± | 64,694 |
| All | 418,020 | ± | 42,183ab | 495,612 | ± | 91,018 | 342,556 | ± | 40,022 | 392,173 | ± | 60,445 | 216,472 | ± | 30,764 |
Embryos 48 h after insemination were divided into >7-cell stage, 4-to 7-cell stage and 2-to 3-cell stage embryos and noncleaved oocytes. mtDNA number per stage was measured. Cows were divided into age groups: Young 25–45 months, Middle, 45–120 months, Aged, >120 months, and Aged* >180 months. a–b: P<0.05.
Fig. 3.
Correlation between age and mtDNA number in each embryo stage. X-axis, donor age in months; Y-axis, mtDNA number. A: >7-cell stage embryo, B: 4- to 7-cell stage embryos, C: 2- to 3-cell stage embryos, D: noncleaved oocytes, and E: all stages.
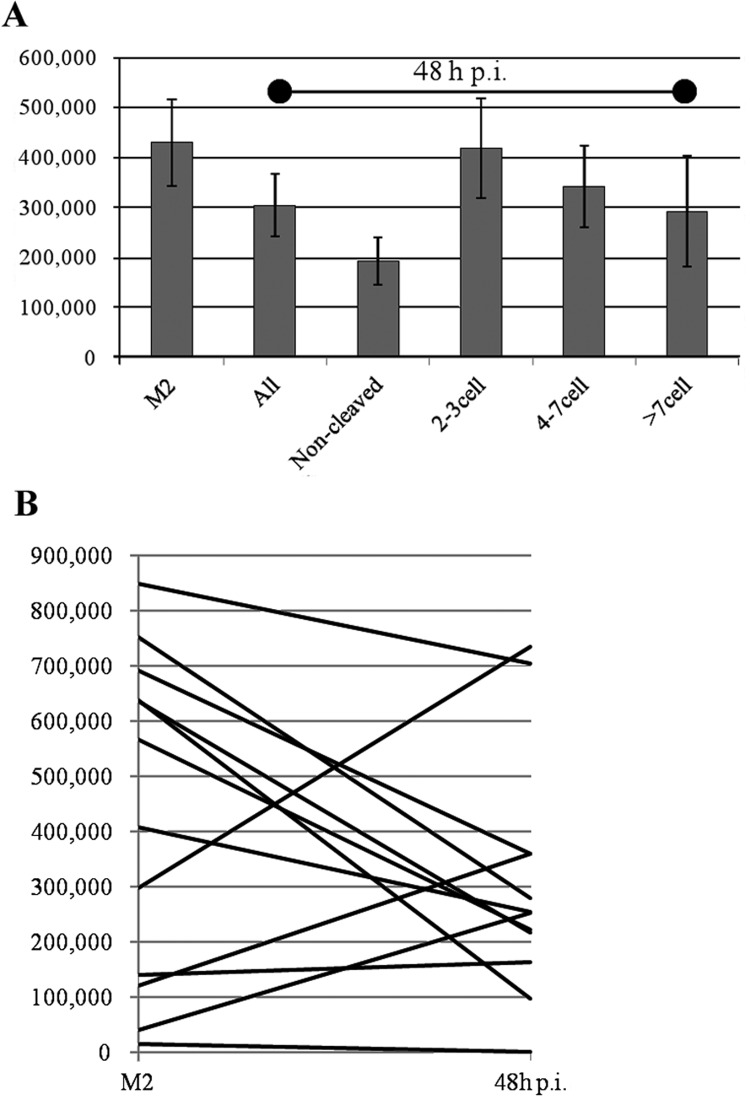
In experiment 2, the dynamics of mtDNA number over the first 48 h p.i. were examined in a total of 11 cows (Fig. 4A). Overall, the mtDNA number tended to decrease during the early developmental period (430,262 ± 85,897 in matured oocytes; 304,828 ± 63,077 in embryos 48 h after insemination, P= 0.25). However, when the mtDNA numbers were examined at the individual level, the mtDNA number was found to be increased in 4 of the 11 cows, with increase ratios as follows (48 h p.i./M2): 1.2, 2.5, 3.0 and 6.3 (Fig. 4B).
Fig. 4.
Comparison of mtDNA number between M2 stage and early developmental stage embryos 48 h after insemination. A: Comparison of mtDNA number among several developmental stage embryos. B: Comparison of mtDNA number between M2 stage oocytes and embryos 48 h after insemination.
Discussion
The present study showed that age affects the ability of oocytes to cleave. Although the mtDNA copy number in early cleavage stage embryos was not related to donor age, mtDNA number tended to decrease in embryos of cows older than 120 months. In addition, mtDNA number showed an overall tendency to decrease during the 3 early cell cycles, although it increased in some cows.
An age-associated decline in bovine reproductive performance was shown in a field-based study [24]. In addition, Melhi et al. [25] compared embryo production between 13- to 16-year-old cows and their younger daughters after superovulation and showed that higher ratios of noncleaved oocytes were recovered from aged cows. There have been no studies examining in vitro developmental competence of oocytes collected from aged cows. This study is the first to demonstrate a negative relationship between donor age and the ratio of embryos developed beyond the 4-cell stage, along with a positive relationship between donor age and the ratio of embryos developed to less than the 4-cell stage. In experiment 1, cleavage ratio was examined 48 h after insemination, a time point at which a competent bovine zygote develops to the 4- to 16-cell stage, because in our preliminary experiment, embryos as less than the 4-cell stage at this point rarely developed into blastocysts. We found that many parts of oocytes were more likely to arrest at early developmental stages when derived from aged cows. This result agrees with the result of a previous study showing that a greater number of noncleaved oocytes following superovulation was found in aged cows compared with their daughters [25]. Although the cause of the low cleavage rate in oocytes derived from aged cows remains unknown, our finding of a negative relationship between donor age and normal fertilization [22] indicates that a possible reason is a high ratio of abnormal fertilization.
Understanding the relationship between mtDNA number in oocytes and developmental ability has recently attracted the attention of many researchers. An elegant study using TFAMKO mice revealed that decreasing mitochondrial number in oocytes does not affect fertilization or early development [26]. In addition, centrifugation followed by the removal of the enriched mitochondrial fraction did not affect the developmental competence of embryos [8]. Conversely, reducing the mtDNA number using DCC during IVM resulted in a low development ratio of blastocysts [27]. In experiment 1, the mtDNA number in noncleaved oocytes was the lowest in the 2- to 3-cell, 4- to 7-cell, and >7-cell stage embryos, although the mtDNA number did not differ among the 3 stages. Possible reasons for this are as follows: (1) poor, incompetent oocytes have a low mitochondrial number or (2) poor, incompetent zygotes reduce their mitochondrial number during the first 2 days of development.
It is generally believed that mtDNA number decreases during early embryo development and then increases during the morula and blastocyst stages in cows [5, 28]. Conversely, Chiaratti et al. [8] demonstrated that artificially reduced mitochondrial numbers replenish during early development up to the blastocyst stage. In addition, Mitango et al. [29] showed that the expression of genes related to mitochondrial replication exhibited a hybridization pattern, indicating that the mitochondrial number represents a balance between biosynthesis and degradation. Using next-generation sequencing technology, we recently detected the expression of genes related to mitochondrial biosynthesis and mitochondrial quality control in the immature, mature, and 8- to 16-cell stages, including HTRA2, LONP1, LONP1, CLLP, TEM1L1, AFG3L2, PARL, PINK, MUL1, PARLK, POLG2, C26H10ORF1, SSBP1, TFAM, TFB1M and TFB2M (unpublished data). Thus, it is plausible that mtDNA number is controlled by both biosynthesis and degradation. In addition, our recent study shows that mtDNA number in oocytes increases during in vitro oocyte growth from the early antral follicle stage to the antral follicle stage and that the increase ratio varies to a great extent and depends on the individual donors [30]. In agreement with this result, in the present study, the increase ratio for mtDNA number also varied ranging from 0.15 to 6.26. In experiment 3, we found that the mtDNA number decreased overall between mature oocytes and early developmental stage embryos, which agrees with results of previous studies. Interestingly, when the mtDNA number was compared among individual cows, it was evident that it increased in some cows. This suggests that under certain conditions, mitochondrial biosynthesis may be upregulated. Although many studies have demonstrated a decrease in mtDNA number during early embryo development, this may actually be due to the fact that differences in the mtDNA number were not assessed within the same donor.
Mitochondrial DNA copy number is reduced as maternal age increases in both humans [31] and cows [22]. In the present study, the mitochondrial DNA number in each developmental stage was not related to maternal age. However, there was a trend for the mtDNA number to decline once donor cows reached the age of 120 months (Fig. 3E). In addition, the mtDNA number of embryos derived from very old cows (>180 months) was lower than in the other groups, although the difference did not reach significance (Table 1). Together, these results suggest that aging affects the mtDNA copy number in early developmental stage embryos as well as in oocytes.
In conclusion, age affects the cleaving ability of oocytes, and embryos derived from very old cows (> 180 months) tend to have lower mtDNA numbers. Although mtDNA number generally decreases during early development, its exact dynamics also appear to vary between individual cows.
Acknowledgment
This study was supported by the Strategic Research Foundation Grant-aided Project for Private Universities of the Ministry of Education, Culture, Sports, Science and Technology of Japan (S0801025).
References
- 1.Van Blerkom J. Mitochondria in human oogenesis and preimplantation embryogenesis: engines of metabolism, ionic regulation and developmental competence. Reproduction 2004; 128: 269–280 [DOI] [PubMed] [Google Scholar]
- 2.Van Blerkom J. Mitochondrial function in the human oocyte and embryo and their role in developmental competence. Mitochondrion 2011; 11: 797–813 [DOI] [PubMed] [Google Scholar]
- 3.Jansen RP, de Boer K. The bottleneck: mitochondrial imperatives in oogenesis and ovarian follicular fate. Mol Cell Endocrinol 1998; 145: 81–88 [DOI] [PubMed] [Google Scholar]
- 4.Pikó L, Taylor KD. Amounts of mitochondrial DNA and abundance of some mitochondrial gene transcripts in early mouse embryos. Dev Biol 1987; 123: 364–374 [DOI] [PubMed] [Google Scholar]
- 5.Smith LC, Thundathil J, Filion F. Role of the mitochondrial genome in preimplantation development and assisted reproductive technologies. Reprod Fertil Dev 2005; 17: 15–22 [DOI] [PubMed] [Google Scholar]
- 6.McConnell JM, Petrie L. Mitochondrial DNA turnover occurs during preimplantation development and can be modulated by environmental factors. Reprod Biomed Online 2004; 9: 418–424 [DOI] [PubMed] [Google Scholar]
- 7.Kameyama Y, Filion F, Yoo JG, Smith LC. Characterization of mitochondrial replication and transcription control during rat early development in vivo and in vitro. Reproduction 2007; 133: 423–432 [DOI] [PubMed] [Google Scholar]
- 8.Chiaratti MR, Bressan FF, Ferreira CR, Caetano AR, Smith LC, Vercesi AE, Meirelles FV. Embryo mitochondrial DNA depletion is reversed during early embryogenesis in cattle. Biol Reprod 2010; 82: 76–85 [DOI] [PubMed] [Google Scholar]
- 9.Pellestor F, Andréo B, Arnal F, Humeau C, Demaille J. Maternal aging and chromosomal abnormalities: new data drawn from in vitro unfertilized human oocytes. Hum Genet 2003; 112: 195–203 [DOI] [PubMed] [Google Scholar]
- 10.Miyara F, Aubriot FX, Glissant A, Nathan C, Douard S, Stanovici A, Herve F, Dumont-Hassan M, LeMeur A, Cohen-Bacrie P, Debey P. Multiparameter analysis of human oocytes at metaphase II stage after IVF failure in non-male infertility. Hum Reprod 2003; 18: 1494–1503 [DOI] [PubMed] [Google Scholar]
- 11.Lopes FL, Fortier AL, Darricarrère N, Chan D, Arnold DR, Trasler JM. Reproductive and epigenetic outcomes associated with aging mouse oocytes. Hum Mol Genet 2009; 18: 2032–2044 [DOI] [PubMed] [Google Scholar]
- 12.Wilding M, Dale B, Marino M, di Matteo L, Alviggi C, Pisaturo ML, Lombardi L, De Placido G. Mitochondrial aggregation patterns and activity in human oocytes and preimplantation embryos. Hum Reprod 2001; 16: 909–917 [DOI] [PubMed] [Google Scholar]
- 13.Jansen RP, Burton GJ. Mitochondrial dysfunction in reproduction. Mitochondrion 2004; 4: 577–600 [DOI] [PubMed] [Google Scholar]
- 14.Kushnir VA, Ludaway T, Russ RB, Fields EJ, Koczor C, Lewis W. Reproductive aging is associated with decreased mitochondrial abundance and altered structure in murine oocytes. J Assist Reprod Genet 2012; 29: 637–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spandorfer SD, Davis OK, Barmat LI, Chung PH, Rosenwaks Z. Relationship between maternal age and aneuploidy in in vitro fertilization pregnancy loss. Fertil Steril 2004; 81: 1265–1269 [DOI] [PubMed] [Google Scholar]
- 16.Tatone C, Di Emidio G, Barbaro R, Vento M, Ciriminna R, Artini PG. Effects of reproductive aging and postovulatory aging on the maintenance of biological competence after oocyte vitrification: insights from the mouse model. Theriogenology 2011; 76: 864–873 [DOI] [PubMed] [Google Scholar]
- 17.Adams GP. Comparative patterns of follicle development and selection in ruminants. J Reprod Fertil Suppl 1999; 54: 17–32 [PubMed] [Google Scholar]
- 18.Ireland JJ, Mihm M, Austin E, Diskin MG, Roche JF. Historical perspective of turnover of dominant follicles during the bovine estrous cycle: key concepts, studies, advancements, and terms. J Dairy Sci 2000; 83: 1648–1658 [DOI] [PubMed] [Google Scholar]
- 19.Baerwald AR, Adams GP, Pierson RA. A new model for ovarian follicular development during the human menstrual cycle. Fertil Steril 2003; 80: 116–122 [DOI] [PubMed] [Google Scholar]
- 20.Baerwald AR, Adams GP, Pierson RA. Characterization of ovarian follicular wave dynamics in women. Biol Reprod 2003; 69: 1023–1031 [DOI] [PubMed] [Google Scholar]
- 21.Adams GP, Jaiswal R, Singh J, Malhi P. Progress in understanding ovarian follicular dynamics in cattle. Theriogenology 2008; 69: 72–80 [DOI] [PubMed] [Google Scholar]
- 22.Iwata H, Goto H, Tanaka H, Sakaguchi Y, Kimura K, Kuwayama T, Monji Y. Effect of maternal age on mitochondrial DNA copy number, ATP content and IVF outcome of bovine oocytes. Reprod Fertil Dev 2011; 23: 424–432 [DOI] [PubMed] [Google Scholar]
- 23.Takahashi Y, First NL. In vitro development of bovine one-cell embryos: Influence of glucose, lactate, pyruvate, amino acids and vitamins. Theriogenology 1992; 37: 963–978 [DOI] [PubMed] [Google Scholar]
- 24.Erickson BH, Reynolds RA, Murphree RL. Ovarian characteristics and reproductive performance of the aged cow. Biol Reprod 1976; 15: 555–560 [DOI] [PubMed] [Google Scholar]
- 25.Malhi PS, Adams GP, Mapletoft RJ, Singh J. Oocyte developmental competence in a bovine model of reproductive aging. Reproduction 2007; 134: 233–239 [DOI] [PubMed] [Google Scholar]
- 26.Wai T, Ao A, Zhang X, Cyr D, Dufort D, Shoubridge EA. The role of mitochondrial DNA copy number in mammalian fertility. Biol Reprod 2010; 83: 52–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ge H, Tollner TL, Hu Z, Dai M, Li X, Guan H, Shan D, Zhang X, Lv J, Huang C, Dong Q. The importance of mitochondrial metabolic activity and mitochondrial DNA replication during oocyte maturation in vitro on oocyte quality and subsequent embryo developmental competence. Mol Reprod Dev 2012; 79: 392–401 [DOI] [PubMed] [Google Scholar]
- 28.May-Panloup P, Vignon X, Chrétien MF, Heyman Y, Tamassia M, Malthièry Y, Reynier P. Increase of mitochondrial DNA content and transcripts in early bovine embryogenesis associated with upregulation of mtTFA and NRF1 transcription factors. Reprod Biol Endocrinol 2005; 3: 65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitango NR, Harvey AJ, Latham KE, Brenner CA. Molecular control of mitochondrial function in developing rhesus monkey oocytes and preimplantation-stage embryos. Reprod Fertil Dev 2008; 20: 846–859 [DOI] [PubMed] [Google Scholar]
- 30.Endo M, Kimura K, Kuwayama T, Monji Y, Iwata H. Effect of estradiol in in vitro follicle culture medium on mitochondrial DNA copy number of oocytes and telomere length of the granulosa cells. Zygote 2012; accept. [DOI] [PubMed] [Google Scholar]
- 31.Chan CC, Liu VW, Lau EY, Yeung WS, Ng EH, Ho PC. Mitochondrial DNA content and 4977 bp deletion in unfertilized oocytes. Mol Hum Reprod 2005; 11: 843–846 [DOI] [PubMed] [Google Scholar]