Abstract
Background
Antibodies to human neutrophil antigens (HNA) can cause transfusion reactions, as well as autoimmune and neonatal neutropenia. This study is the first to report the frequencies of human neutrophil antigen genotypes in the Thai population.
Materials and methods
Three hundred unrelated, healthy Thai blood donors at the National Blood Centre, Thai Red Cross Society, Bangkok, Thailand were typed for HNA-1a, -1b, -1c, -3a, -3b and -4a using polymerase chain reaction with sequence-specific primers. Moreover, HNA-5a genotyping was performed using polymerase chain reaction-restriction fragment length polymorphism.
Results
The gene frequencies of HNA-1a, -1b and -1c were 0.470, 0.530 and 0.005, respectively. The frequencies of HNA-3a and -3b were 0.490 and 0.510, respectively. Additionally, the HNA-4a+/+ and HNA-4a+/− genotype frequencies were 0.947 and 0.053, respectively. The frequencies of HNA-5a+/+, HNA-5a+/− and HNA-5a−/− genotypes were 0.641, 0.297 and 0.062, respectively. Compared with other Asian populations, Thais have higher frequencies of HNA-1b (P <0.001). On the other hand, the frequency of HNA-5a observed in Thais is lower than that reported among Koreans (P <0.001).
Discussion
These findings suggest that Thais would be more susceptible to HNA-1b alloimmunisation. Furthermore, our results could establish a useful human neutrophil antigen donor file to provide more effective transfusion of blood and blood components.
Keywords: human neutrophil antigens, HNA, gene frequencies, Thais
Introduction
Human neutrophil antigens (HNA) are found on white blood cell membrane glycoproteins and are the target of HNA antibodies which are involved in various clinical conditions including neonatal immune neutropenia, transfusion-related acute lung injury (TRALI), refractoriness to granulocyte transfusions, febrile transfusion reaction, immune neutropenia after stem cell transplantation, autoimmune neutropenia and drug-induced immune neutropenia1. The probability of developing HNA alloantibodies in each patient depends on the individual’s existing antigens2,3. The Granulocyte Antigen Working Party of the International Society of Blood Transfusion (ISBT) agreed, in 1998, to establish a new nomenclature for well-defined neutrophil alloantigens based on the glycoprotein location of the antigens4. Currently, the HNA system includes eight antigens (HNA-1a, -1b, -1c, -2a, -3a, -3b, -4a and -5a) that are assigned to five antigen groups5–7. Commercial monoclonal antibodies specific to several HNA antigens are available and have been used to phenotype granulocytes; however, granulocyte viability is a test limitation8. Hence, to differentiate HNA alleles, polymerase chain reaction (PCR) assays such as PCR with sequence-specific primers (PCR-SSP) and PCR with restriction fragment length polymorphism (PCR-RFLP) have been implemented9–11. The frequencies of HNA have been characterized in different ethnic groups, and significant differences have been shown6,9,11–18. The frequency of HNA-1a is higher in most Asian populations, ranging between 0.300 and 0.680, whereas the frequency of HNA-1b is higher in Caucasians, ranging between 0.627 and 0.7186. The lower frequency of HNA-1c has also been reported19. The frequencies of HNA-2a, HNA-4a and HNA-5a in most populations are high1,13,20,21. The gene frequencies of HNA-3a and HNA-3b are 0.744 and 0.256, respectively, in the German population11. However, data about HNA gene frequencies in the Thai population are still unknown, and the commercial kit for HNA-1a, -1b, -1c, -3a, -3b, -4a, -4b, -5a and -5bw genotyping is expensive and not available in Thailand. The purpose of this study was to determine the genotype frequencies of the HNA-1, -3, -4 and -5 systems in the Thai population in order to estimate the potential risk for alloimmunisation.
Materials and methods
Subjects
Peripheral venous blood was collected into EDTA tubes from 300 unrelated, healthy Thai blood donors at the National Blood Centre, Thai Red Cross Society, Bangkok, Thailand. The donors were from central Thailand and their ages ranged from 19 to 58 years. Informed consent was obtained from each subject. This study was approved by the Committee on Human Rights Related to Research Involving Human Subjects, Thammasat University, Pathumtani, Thailand.
DNA standards
Samples with known HNA-1a, -1b, -1c, -3a, -3b, -4a and -5a DNA were provided by Dr. Núria Nogués, Laboratori d’Immunohematologia, Banc de Sang i Teixits, Passeig Taulat, Barcelona, Spain.
Primers
The primers used for HNA genotyping in this study are shown in Table I. The specific primers were similar to those previously described, with some modifications10,13,14,22.
Table I.
Sequence of the primers for HNA-1a, -1b, -1c, -3a, -4a, -5a and the HGH internal control.
| Primers | Sequence (5′→3′) | Product size (bp) |
|---|---|---|
| HNA-1a (F)14 | CAGTGGTTTCACAATGTGAA | 141 |
| HNA-1a (R)14 | ATGGACTTCTAGCTGCAC | |
| HNA-1b (F)14 | CAATGGTACAGCGTGCTT | 219 |
| HNA-1b (R)14 | ATGGACTTCTAGCTGCAC | |
| HNA-1c (F)14 | AAGATCTCCCAAAGGCTGTG | 191 |
| HNA-1c (R)14 | ACTGTCGTTGACTGTGTCAT | |
| HNA-3a (F)22 | AGTGGCTGAGGTGCTTCG | 601 |
| HNA-3b (F)22 | GAGTGGCTGAGGTGCTTCA | |
| HNA-3 (R)* | ATCGCCATGGCAATGACCA | |
| HNA-4a (F)10, 21 | CTCCCCACAGGGTGGTG | 124 |
| HNA-4a-positive (R)10, 13 | AGTGACTCACCCTGCATGC | |
| HNA-4a-negative (R)10, 13 | AGTGACTCACCCTGCATGT | |
| HNA-5a (F)10,13 | CTTCAGCATCTCCACCTTGC | 201 |
| HNA-5a (R)10** | TTCTGATATTCCCCACCCTGA | |
| HGH (F)9 | TGCCTTCCCAACCATTCCCTTA | 434 |
| HGH (R)9 | CCACTCACGGATTTCTGTTGTGTTTC |
Newly designed primer,
Primer with modification
HNA-1a, -1b and -1c genotyping by polymerase chain reaction - sequence-specific primers
HNA-1a, -1b and -1c genotyping was performed by a previously described PCR-SSP technique with some modifications14. Briefly, 2 μL of genomic DNA (100 ng/μL) were amplified in a total volume of 25 μL using 0.5 μM of HNA-1a forward and reverse primers for HNA-1a genotyping, 0.5 μM of HNA-1b forward and reverse primers for HNA-1b genotyping and 0.5 μM of HNA-1c forward and reverse primers for HNA-1c genotyping. Co-amplification of the human growth hormone gene (HGH) using 0.125 μM HGH (F) primer and 0.125 μM HGH (R) primer was run as an internal control. The PCR was performed with 12.5 μL of DreamTaqTM DNA polymerase (Thermo Fisher Scientific Inc., Glen Burnie, MD, USA) consisting of 2X Dream Taq green buffer, 0.4 mM of each dNTP, and 0.4 mM MgCl2 in a G-STORM GS1 thermal cycler (Gene Technologies Ltd., Essex, UK). The cycle parameters of the PCR programme began with a first step of one cycle of 300 s at 95 °C, followed by 30 cycles of 30 s at 95 °C, 60 s at 57 °C and 30 s at 72 °C. The final extension step lasted 5 min at 72 °C and then the sample was kept at 4 °C. After amplification, PCR products were analysed on a 1.5% agarose gel using 1X Tris borate ethylenediaminetetraacetate (TBE) buffer containing SYBR® Green I nucleic acid gel stain (Invitrogen, Grand Island, NY, USA) and were visualised under UV illumination. The PCR product sizes of HNA-1a, HNA-1b and HNA-1c were 141 bp, 219 bp and 191 bp, respectively, whereas the PCR product size of the HGH gene, used as an internal control, was 434 bp.
HNA-3 genotyping by polymerase chain reaction - sequence-specific primers
The PCR-SSP technique was used for HNA-3 genotyping with minor modifications as previously described22. Briefly, 2 μL of genomic DNA (100 ng/μL) were amplified in a total volume of 25 μL using 0.5 μM of HNA-3a (forward), HNA-3b (forward) and HNA-3 (reverse) primers. Co-amplification of the HGH gene using 0.125 μM HGH (F) primer and 0.125 μM HGH (R) primer was run as an internal control. PCR was performed with 12.5 μL of DreamTaqTM DNA polymerase (Thermo Fisher Scientific Inc.) consisting of 2X Dream Taq green buffer, 0.4 mM of each dNTP, and 0.4 mM MgCl2 in a G-STORM GS1 thermal cycler (Gene Technologies Ltd.). The cycle parameters of the PCR programme began with a first step of one cycle of 180 s at 95 °C, followed by 35 cycles of 30 s at 95 °C, 30 s at 57 °C and 60 s at 72 °C. The final extension step lasted for 10 min at 72 °C and then the sample was kept at 4 °C. After amplification, PCR products were analysed on a 1.5% agarose gel using 1X TBE buffer containing SYBR® Green I nucleic acid gel stain (Invitrogen) and were visualized under UV illumination. The PCR product size of both HNA-3a and -3b alleles was 601 bp, whereas that of the HGH gene, used as the internal control, was 434 bp.
HNA-4a genotyping by polymerase chain reaction - sequence-specific primers
HNA-4a genotyping was performed by a PCR-SSP technique, as previously described with some modifications10,13. Briefly, 2 μL of genomic DNA (100 ng/μL) were amplified in a total volume of 25 μL using 0.5 μM of HNA-4a (forward), HNA-4a-positive (reverse) and HNA-4a-negative (reverse) primers. Co-amplification of the HGH gene using 0.125 μM HGH (F) primer and 0.125 μM HGH (R) primer was run as an internal control. The PCR was performed with 12.5 μL of DreamTaqTM DNA polymerase (Thermo Fisher Scientific Inc.) consisting of 2X Dream Taq green buffer, 0.4 mM of each dNTP, and 0.4 mM MgCl2 in a G-STORM GS1 thermal cycler (Gene Technologies Ltd.). The cycle parameters of the PCR programme began with a first step of one cycle of 180 s at 95 °C, followed by 30 cycles of 60 s at 95 °C and 60 s at 58 °C and 60 s at 72 °C. The last step was final extension for 5 min at 72 °C and the sample was kept at 4 °C. After amplification, PCR products were analysed on a 1.5% agarose gel using 1X TBE buffer containing SYBR® Green I nucleic acid gel stain (Invitrogen) and were visualised under UV illumination. The PCR product size of both HNA-4a-positive and -4a-negative alleles was 124 bp, whereas that of the internal control, the HGH gene, was 434 bp.
HNA-5a genotyping by polymerase chain reaction - restriction fragment length polymorphism
HNA-5a genotyping was performed using a previously described PCR-RFLP technique with some modifications10,13. Briefly, the PCR was performed in a total volume of 25 μL made up of 0.5 μL of genomic DNA (50 ng); 12.5 μL of DreamTaqTM DNA polymerase (Thermo Fisher Scientific Inc.) consisting of 2X Dream Taq green buffer, 0.4 mM of each dNTP and 0.4 mM MgCl2; 0.5 μL of each HNA-5a primer (10 pmol/μL); and 11 μL of sterile-filtered H2O. The reaction was performed in a G-STORM GS1 thermal cycler (Gene Technologies Ltd.), under the following conditions: 95 °C for 180 s (initial denaturation); 35 cycles of 95 °C for 30 s (denaturation), 57 °C for 30 s (annealing), and 72 °C for 60 s (extension); and 72 °C for 5 min (final extension). The PCR product, without further purification, was digested with Bsp1286I (FastDigest®, Fermentas, CA, USA) for 15 min in at 37 °C water bath. PCR-RFLP products were electrophoresed at 100 V for 45 min with a 2% agarose-TBE gel containing SYBR® Green I nucleic acid gel stain (Invitrogen) and were visualised under UV illumination. The typical results of RFLP analysis for the HNA-5a-positive allele showed 136-bp and 65-bp fragments after Bsp1286I digestion, while the PCR products of the HNA-5a-negative allele were not split (201 bp) by the same treatment.
To validate the HNA genotyping in the Thai population, the PCR-SSP and PCR-RFLP were performed using known HNA-1a, -1b, -1c, -3a, -3b, -4a and -5a genotype panels.
Statistical analysis
Gene frequencies were calculated by gene counting as described by De La Vega et al. and Steffensen et al9,15. The chi-square test was used to test for Hardy-Weinberg equilibrium for the HNA-system and to compare the published HNA genotype frequencies among different populations. The analysis was performed using Excel software (Microsoft Office, 2007, version 12.0). P values of 0.001 or less were considered statistically significant, as previously described by Hauck et al11.
Results
In this study, the determined HNA genotypes are consistent with the Hardy-Weinberg equilibrium and the chi-square test results for observed and expected frequencies in the Thai population showed no significant difference for any HNA allele.
HNA-1a, 1b and -1c genotyping
HNA-1a, -1b and -1c gene frequencies were determined in 300 Thai blood donors. The HNA-1b allele was most frequent, 0.53 (273/300), followed by HNA-1a, 0.47 (255/300), with HNA-1c being the least frequent, 0.005 (3/300), in the Thai population.
HNA-3a and -3b genotyping
The gene frequencies of HNA-3a and HNA-3b were also determined in the 300 Thai blood donors. The gene frequency of HNA-3a was 0.493 (282/300) while that of HNA-3b was 0.507 (286/300). HNA-3ab heterozygous individuals were the most common (268/300).
HNA-4a and HNA-5a genotyping
The HNA-4a and HNA-5a genotype frequencies and gene frequencies are shown in Table II.
Table II.
HNA-4a and HNA-5a genotype frequencies and gene frequencies in Thais.
| HNA-4a | HNA-5a | ||
|---|---|---|---|
| Genotype frequencies | +/+ | 0.947 (284/300) | 0.641 (186/300) |
| +/− | 0.053 (16/300) | 0.297 (86/300) | |
| −/− | 0.000 (0/300) | 0.062 (18/300) | |
|
| |||
| Gene frequencies | + | 0.973 | 0.790 |
| − | 0.027 | 0.210 | |
HNA gene frequencies among different populations
A comparison of HNA gene frequencies reported from studies among Asian and Caucasians11,13,16–18, is presented in Table III. The frequencies of HNA-1a and -1b were similar to those found in a previous study in German and Turkish populations11, but were significantly different (P <0.001) from those in studies in Asian populations13,16–18. HNA-1c is the genotype with the lowest incidence among different populations. The frequency of HNA-3a was lower than that of HNA-3b in Thais, whereas the frequency of HNA-3a was higher in other populations11,13,16–18. The frequency of HNA-4a in Thais was similar to that in Koreans13,16, but was significantly higher than in German and Turkish populations11. In addition, the frequency of HNA-5a in the Thai population was significantly lower than in Korean populations13,16, while the frequency was similar to those in German and Turkish populations11.
Table III.
HNA gene frequencies in different populations.
| Populations | HNA gene frequencies | ||||||
|---|---|---|---|---|---|---|---|
|
|
|||||||
| HNA-1a | HNA-1b | HNA-1c | HNA-3a | HNA-3b | HNA-4a | HNA-5a | |
| Thai: this report* (n=300) | 0.47 | 0.53 | 0.005 | 0.49 | 0.51 | 0.97 | 0.79 |
| Korean13,16 (n=200), (n=110) | 0.58∂ | 0.42∂ | NA | NA | NA | NA | NA |
| NA | NA | NA | NA | NA | 0.99 | 0.96∂ | |
| Chinese14, 17 (n=138), (n=119) | 0.68∂ | 0.32∂ | 0.00 | NA | NA | NA | NA |
| NA | NA | NA | 0.62∂ | 0.38∂ | NA | NA | |
| Japanese11, 18 (n=400) | 0.62∂ | 0.38∂ | 0.00 | NA | NA | NA | NA |
| German11 (n=119) | 0.39 | 0.60 | 0.025 | 0.74∂ | 0.26∂ | 0.91∂ | 0.73 |
| Turkish11 (n=118) | 0.42 | 0.56 | 0.03 | 0.74∂ | 0.26∂ | 0.88∂ | 0.75 |
Thai blood donors,
P <0.001.
Discussion
Because the molecular basis of HNA has been previously elucidated, HNA genotyping using different PCR techniques was preferred over serological techniques. A disadvantage of serological techniques for HNA phenotyping is that it is time-consuming to isolate granulocytes, which may affect test results due to low viability. Generally, HNA-1, -3, -4 and -5 genotyping can be performed by PCR-SSP and PCR-RFLP; however, HNA-2a is only defined by serological techniques1,5,8.
This study is the first to report the gene frequencies of HNA-1a, 1b, -1c, -3a, -3b, -4a and -5a in the Thai population. Three hundred unrelated Thai blood donors were genotyped for HNA-1a, -1b, -1c, -3a, -3b and -4a by PCR-SSP and HNA-5a was genotyped by PCR-RFLP. For the HNA-1 system, the frequency of HNA-1a in most Asian populations ranges between 0.300 and 0.6806; however, a lower frequency of HNA-1a and a higher frequency of HNA-1b were found in Thais compared with other Asian populations, suggesting that Thais would be more susceptible to HNA-1b alloimmunisation13,16–18. Moreover, the lower frequency of HNA-1c was observed in Thais, as in previous studies10,19,21.
Unlike in Han Chinese and German populations, it was found that HNA-3a and HNA-3b frequencies in Thais were significantly different11,18, which may be due to the high frequency of heterozygous HNA-3ab in this study. Moreover, regarding a previous Thai Stem Cell Donor Registry study, it is proven that the population of central Thailand is a mixture of original Thais and Southern Chinese23. The results of this present study indicate that alloimmunisation against HNA-3 antibodies could be less frequent in Thais. Concerning the technique used for HNA-3a genotyping, a previous study showed that the PCR-SSP technique can produce misleading results in HNA-3ab heterozygotes with the additional CTL2-537T variation of the HNA-3a antigen24, suggesting the need for further study.
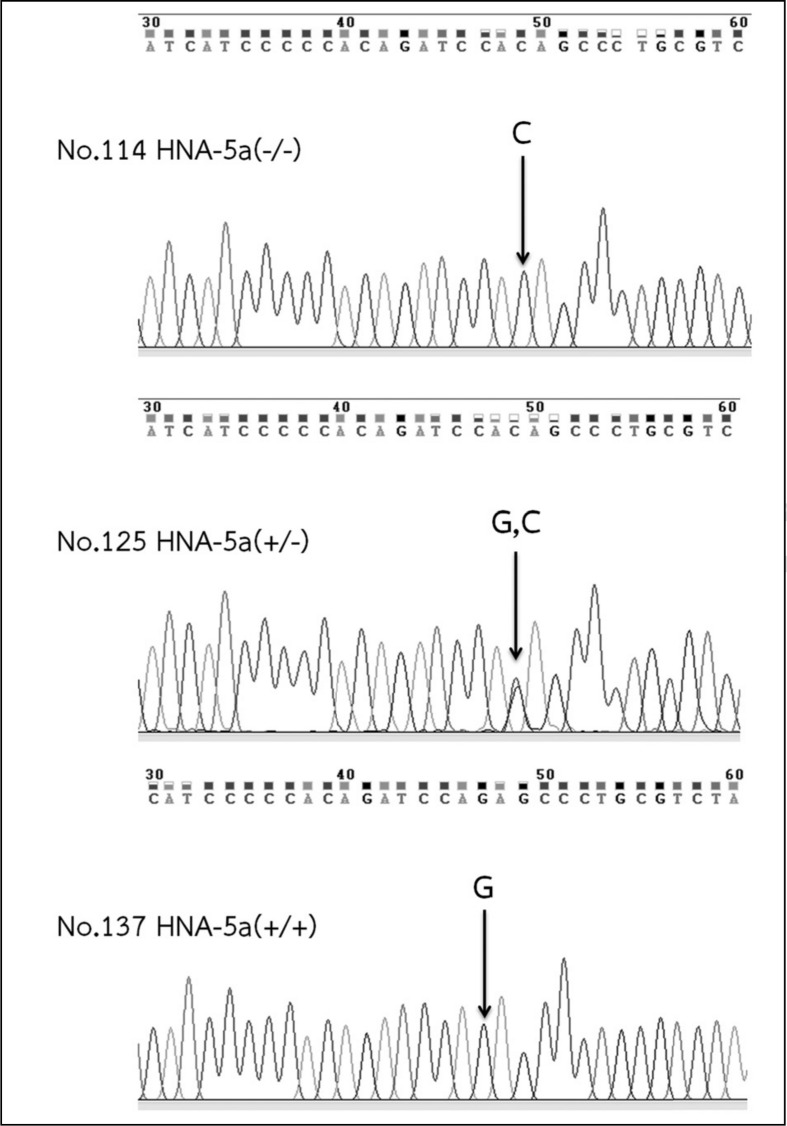
The gene frequency of HNA-4a in the Thai population is similar to that in Korean populations13,16; therefore, antibodies to HNA-4a may be involved in neonatal alloimmune neutropenia and autoimmune neutropenia as previously found in other studies20,25. Interestingly, the frequency of the HNA-5a gene in Thais is comparable to the frequencies reported in German and Turkish populations11. However, HNA-5a is significantly more frequent in Koreans than in Thais and other populations11,13. Hence, additional genotyping for HNA-5bw, using a commercially available kit, is needed in the Thai population. The selected DNA sequencing results of some HNA-5a negative donors by PCR-RFLP confirmed a single nucleotide polymorphism at position G2466C, as shown in Figure 1.
Figure 1.
Chromatogram of HNA-5a sequencing results in samples of 5a(−/−), 5a(+/−) and 5a(+/+).
In conclusion, the implementation of HNA genotyping in blood donors and patients is beneficial not only to provide more effective blood transfusions but also to increase the capability for HNA alloantibody investigations.
Acknowledgements
This work was supported by the National Research University Project of Thailand, Office of Higher Education Commission. We thank Dr. Núria Nogués, Laboratory of Immunohaematology, Blood and Tissue Bank, Barcelona, Spain for providing DNA controls. We also thank Ms. Siriporn Nathalang and the staff at the Apheresis Unit, National Blood Centre, Thai Red Cross Society for their support with specimen collection and Mr. Jesada Kittikul for his advice concerning statistics.
Footnotes
The Authors declare no conflicts of interest.
References
- 1.Moritz E, Norcia AM, Cardone JD, et al. Human neutrophil alloantigens systems. An Acad Bras Cienc. 2009;81:559–69. doi: 10.1590/s0001-37652009000300019. [DOI] [PubMed] [Google Scholar]
- 2.Fromont P, Pri’e N, Simon P, et al. Granulocyte antibody screening: evaluation of a bead-based assay in comparison with classical methods. Transfusion. 2010;50:2643–8. doi: 10.1111/j.1537-2995.2010.02762.x. [DOI] [PubMed] [Google Scholar]
- 3.Bux J, Behrens G, Jaeger G, Welte K. Diagnosis and clinical course of autoimmune neutropenia in infancy: analysis of 240 cases. Blood. 1998;91:181–6. [PubMed] [Google Scholar]
- 4.ISBT Working Party on Platelet and Granulocyte Serology. Nomenclature of granulocyte alloantigens. Vox Sang. 1999;77:251. [Google Scholar]
- 5.Bux J. Human neutrophil alloantigens. Vox Sang. 2008;94:277–85. doi: 10.1111/j.1423-0410.2007.01031.x. [DOI] [PubMed] [Google Scholar]
- 6.Xia W, Sachs U, Chen Y, et al. The frequencies of human neutrophil alloantigens in the Chinese Han population of Guangzhou. Transfusion. 2011;51:1271–7. doi: 10.1111/j.1537-2995.2010.02979.x. [DOI] [PubMed] [Google Scholar]
- 7.Fung YL, Minchinton RM. The fundamental of neutrophil antigen and antibody investigations. ISBT Science Series. 2011;6:381–6. [Google Scholar]
- 8.Clay ME, Schuller RM, Bachowski GJ. Granulocyte serology: current concepts and clinical significance. Immunohematol. 2010;26:11–21. [PubMed] [Google Scholar]
- 9.De La Vega Elena CD, Nogues N, Fernandes Montoya A, et al. HNA-1a, HNA-1b and HNA-1c gene frequencies in Argentineans. Tissue Antigens. 2008;71:475–7. doi: 10.1111/j.1399-0039.2008.01034.x. [DOI] [PubMed] [Google Scholar]
- 10.Cardone JDB, Bordin JO, Chiba AK, et al. Gene frequencies of the HNA-4a and -5a neutrophil antigens in Brazilian persons and a new polymerase chain reaction-restriction fragment length polymorphism method for HNA-5a genotyping. Transfusion. 2006;46:1515–20. doi: 10.1111/j.1537-2995.2006.00943.x. [DOI] [PubMed] [Google Scholar]
- 11.Hauck B, Philipp A, Eckstein R, et al. Human neutrophil alloantigen genotype frequencies among blood donors with Turkish and German descent. Tissue Antigens. 2011;78:416–20. doi: 10.1111/j.1399-0039.2011.01779.x. [DOI] [PubMed] [Google Scholar]
- 12.Norcia AMMI, Sugano EYK, Chiba AK, et al. Human neutrophil alloantigen -1a, -1b, -2, -3a and -4a frequencies in Brazilians. Tissue Antigens. 2009;74:404–7. doi: 10.1111/j.1399-0039.2009.01357.x. [DOI] [PubMed] [Google Scholar]
- 13.Han TH, Han KS. Gene frequencies of human neutrophil antigens 4a and 5a in the Korean population. Korean J Lab Med. 2006;26:114–8. doi: 10.3343/kjlm.2006.26.2.114. [DOI] [PubMed] [Google Scholar]
- 14.Kissel K, Hofmann C, Gittinger FS, et al. HNA-1a, HNA-1b, and HNA-1c (NA1, NA2, SH) frequencies in African and American Blacks and in Chinese. Tissue Antigens. 2000;56:143–8. doi: 10.1034/j.1399-0039.2000.560205.x. [DOI] [PubMed] [Google Scholar]
- 15.Steffensen R, Gulen T, Varming K, Jersild C. FcgammaRIIIB polymorphism: evidence that NA1/NA2 and SH are located in two closely linked loci and that the SH allele is linked to the NA1 allele in the Danish population. Transfusion. 1999;39:593–8. doi: 10.1046/j.1537-2995.1999.39060593.x. [DOI] [PubMed] [Google Scholar]
- 16.Seo DH, Park SS, Han KS. Genotype analysis of granulocyte-specific antigens in Koreans. Korean J Clin Patholol. 1997;17:1144–9. [Google Scholar]
- 17.Bowens KL, Sullivan MJ, Curtis BR. Determination of neutrophil antigen HNA-3a and HNA-3b genotype frequencies in six racial groups by high throughput 5′ exonuclease assay. Transfusion. 2012;52:2368–74. doi: 10.1111/j.1537-2995.2012.03600.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujiwara K, Watanabe Y, Mitsunaga S, et al. Determination of granulocyte-specific antigens on neutrophil FcA receptor IIIb by PCR-preferential homoduplex formation assay, and gene frequencies in the Japanese population. Vox Sang. 1999;77:218–22. doi: 10.1159/000031130. [DOI] [PubMed] [Google Scholar]
- 19.Bux J, Stein EL, Bierling P, et al. Characterization of a new alloantigen (SH) on the human neutrophil Fc gamma receptor IIIb. Blood. 1997;34:608–11. [PubMed] [Google Scholar]
- 20.Fung YL, Pitcher LA, Willett JE, et al. Alloimmune neonatal neutropenia linked to anti-HNA-4a. Transfus Med. 2003;13:49–52. doi: 10.1046/j.1365-3148.2003.00413.x. [DOI] [PubMed] [Google Scholar]
- 21.Simsek S, van der Schoot CE, Daams M, et al. Molecular characterization of antigenic polymorphisms (Ond(a) and Mart(a)) of the beta 2 family recognized by human leukocyte alloantisera. Blood. 1996;88:1350–8. [PubMed] [Google Scholar]
- 22.Reil A, Wesche J, Greinacher A, Bux J. Geno- and phenotyping and immunogenicity of HNA-3. Transfusion. 2011;51:18–24. doi: 10.1111/j.1537-2995.2010.02751.x. [DOI] [PubMed] [Google Scholar]
- 23.Kupatawintu P, Pheancharoen S, Srisuddee A, et al. HLA-A, -B, -DR haplotype frequencies in the Thai Stem Cell Donor Registry. Tissue Antigens. 2010;75:730–6. doi: 10.1111/j.1399-0039.2010.01450.x. [DOI] [PubMed] [Google Scholar]
- 24.Flesch BK, Reil A, Bux J. Genetic variation of the HNA-3a encoding gene. Transfusion. 2011;51:2391–7. doi: 10.1111/j.1537-2995.2011.03155.x. [DOI] [PubMed] [Google Scholar]
- 25.Hartman KR, Wright DG. Identification of autoantibodies specific for the neutrophil adhesion glycoproteins CD11b/CD18 in patient with autoimmune neutropenia. Blood. 1991;78:1096–104. [PubMed] [Google Scholar]