Abstract
Longitudinal observational clinical data on pediatric patients in electronic format is becoming widely available. A new era of multi-institutional data networks that study pediatric diseases and outcomes across disparate health delivery models and care settings are also enabling an innovative collaborative rapid improvement paradigm called the Learning Health System. However, the potential alignment of routine clinical care, observational clinical research, pragmatic clinical trials, and health systems improvement requires a data infrastructure capable of combining information from systems and workflows that historically have been isolated from each other. Removing barriers to integrating and reusing data collected in different settings will permit new opportunities to develop a more complete picture of a patient’s care and to leverage data from related research studies. One key barrier is the lack of a common terminology that provides uniform definitions and descriptions of clinical observations and data. A well-characterized terminology ensures a common meaning and supports data reuse and integration. A common terminology allows studies to build upon previous findings and to reuse data collection tools and data management processes. We present the current state of terminology harmonization and describe a governance structure and mechanism for coordinating the development of a common pediatric research terminology that links to clinical terminologies and can be used to align existing terminologies. By reducing the barriers between clinical care and clinical research, a Learning Health System can leverage and reuse not only its own data resources but also broader extant data resources.
Keywords: terminology harmonization, clinical research, learning health system, governance, pediatric research, ontology
Longitudinal patient-level pediatric data are captured in unprecedented quantities through a growing array of electronic systems, including electronic health records (EHRs), automated physiologic instruments, digital images, patient portals, and research data collection systems. Pediatric research programs that historically developed customized and unique data definitions, data collection forms, and data management systems are yielding to reusable and sharable informatics infrastructures that support a wide range of clinical research programs.1–3 Similar approaches for collecting and reusing detailed clinical data also support national performance improvement collaboratives and disease-specific registries.4–7 These new informatics tools can expand the size, scope, and reach of child health research by connecting to clinical settings that previously were unable to participate. A reusable research data infrastructure can reduce costs, accelerate study start-up and patient recruitment, and ensure broader participation and generalizability in patient populations and practice characteristics.8–10
The Health Information Technology for Economic and Clinical Health (HITECH) Act, part of the 2009 American Recovery and Reinvestment Act, provides a significant financial investment for deploying EHRs, which has resulted in a substantial increase in EHR implementations across a variety of practice settings. As of 2012, nearly 3 in every 4 office-based physicians in the United States used an EHR system.11,12 Data collected during routine clinical care can be extracted and reused to support patient-centered and population-based clinical care management, quality reporting, and clinical research.6,13,14 The concept of “enter once, use many times” has enormous appeal and potential, especially given the high cost of dedicated single-purpose data collection or chart abstraction.15,16
A clinical research project that develops its own data definitions, data collection forms, and protocols specific to the unique population, interventions, and outcomes has effectively developed its own independent “language” to convey the meaning of concepts being studied. Precision in terms and definitions is critical to ensuring data are accurate, consistent, and represent the intended meaning. Even a simple term such as “current medications” can have different definitions, such as the list of medications a patient is taking at the start of a visit or the medications prescribed at the end of a visit.
Reuse of data definitions from previous studies or across studies has historically been limited. Unfortunately, across the pediatric clinical research landscape, a Tower of Babel has been erected with numerous nearly identical measures being defined and collected for the same clinical concepts. Barriers to data reuse are further exacerbated by project-specific terms and definitions across groups studying the same disease and across programs studying potentially related diseases. Variability in clinical terms results in “data silos” that constrain the return on what is often a substantial investment. Because most pediatric diseases involve patient populations with limited numbers, data silos weaken the ability to perform meta-analyses and therefore threaten the generalizability and statistical power that can be gained by combining data across studies. Custom definitions also prevent integrating data across multiple data sources to develop a more comprehensive picture of patients than is possible by using any 1 data source alone. As resources for large-scale, long-term sustainable child health research become more limited, efforts that can leverage previous investments are both timely and necessary.13,14
Study-specific terminology may reflect evolving understanding of conditions being studied. But these one-off terminologies also arise in significant part from the limited adoption of standardized controlled terminologies in pediatric research, a technology that can reduce or eliminate data silos and enhance data reuse and integration.17 Because of mandates in the American Recovery and Reinvestment Act HITECH legislation, standard terminologies must be deployed in EHR systems for use in clinical care processes and quality measures reporting, but linkage to research is uncertain. This article describes how controlled terminologies and common data definitions are defined, maintained, and used in clinical research, reviews current efforts across a range of pediatric clinical networks, and presents a model of coordinated terminology development and maintenance integrated with clinical terminologies to establish a common pediatric research terminology.
The Need for a Comprehensive Pediatric Research Terminology
A case scenario illustrates how a lack of a common research terminology can impede pediatric research:
Two neonatologists at separate institutions seek to investigate new therapeutic interventions for necrotizing enterocolitis (NEC). Based on their own expertise, each develops highly specific but different diagnostic definitions for NEC and for assigning levels of severity. The 2 NEC definitions are overlapping but neither definition includes all of the patients identified by the other. There is no direct mapping between the levels of severity defined by each investigator. In addition, abdominal radiology findings use different terms and values. Neither investigator uses terms contained in Systematized Nomenclature of Medicine–Clinical Terms (SNOMED–CT), an extensive terminology being incorporated into EHRs to describe clinical findings nor terms contained in RadLex, a comprehensive lexicon of radiology findings endorsed by the Radiological Society of North Americas.
If the 2 studies have different findings, it is not possible to combine the 2 populations for meta-analysis or reanalysis of a combined data set. Future studies will need to select data definitions from 1 or the other study if they wish to incorporate previous data elements. In addition, 2 independent parallel research activities may ensue, with neither group able to leverage the infrastructure developed by the other.
Several solutions can be applied. Perhaps least likely is that 1 group convinces the other group to adopt their terms and definitions and to discard their previous work. This approach wastes previous resources and results in 2 data silos being replaced by 1 data silo. Another solution is to harmonize the 2 terminologies, identifying terms in each set that are sufficiently similar to be combined into a common term while leaving incompatible terms as separate entities. This solution allows data to be shared where there is agreement while allowing for clinically meaningful differences. An important variation to the previous solution is to also map harmonized terms to 1 or more existing terminologies, which would enable integration with other data sets that also link to the same terminologies. All of the above solutions enable reuse of existing data but do not result in leveraging data infrastructures. A more efficient solution would be for both research groups, along with relevant clinical, administrative, governmental, and funding stakeholders, to participate in developing and using a common pediatric terminology with standardized definitions that can be maintained and expanded as community needs dictate. When this common terminology is integrated into other terminologies, significant leverage and scale can be achieved.
The negative impact of data silos on pediatric research findings is not hypothetical. For example, after years of off-label use of inhaled nitric oxide in premature infants, the National Institutes of Health (NIH) funded 3 concurrent randomized trials to evaluate its efficacy to prevent bronchopulmonary dysplasia.18–20 Each study employed different definitions of bronchopulmonary dysplasia and yielded conflicting results. The NIH subsequently convened a multidisciplinary panel to interpret the disparate findings.21 An independent systematic review of the trials revealed that “the effect of inhaled nitric oxide on the severity of bronchopulmonary dysplasia in the randomized controlled trials was compromised by the wide variation in bronchopulmonary dysplasia definitions” and concluded that “insufficient data are available to perform a meta-analysis for any measure of severity due to the lack of uniformity in definitions and study measures used.”22
Employing well-characterized terminology from the earliest stages of research design ensures that data will be generated according to accepted standards. By providing definitions and synonyms, and by codifying the relationships between terms and concepts, terminologies enable automated data collection; accelerate data analysis; reduce human-introduced errors; and support data sharing and integration. A standardized terminology involves an agreement among investigators to use specific definitions for research terms, and agreement when distinct terms signify an equivalent (interchangeable) concept. Standardization enables data collected from different studies, centers, or hospitals to be comparable, understandable, and exchangeable (interoperable). By standardizing terms, clinicians and researchers can combine results from their research efforts with those of other studies and networks, as well as with clinical care records, or link to subject-level data with population-based environmental23 or public health data,24 leading to broader scientific understanding and extending the return on research investments. Moreover, shared terminologies can improve the translation of research to patient care by eliminating the differences between research findings and clinical decision-making, thereby facilitating the development of evidence-based decision support and quality measures.
Controlled Terminologies: A Brief Primer
Medical science consists of basic concepts that represent unique entities such as diseases, diagnostic tests, treatments, outcomes, and their associated processes and relationships. New knowledge can generate new concepts, alter the meaning of existing concepts, or determine that a concept is incorrect or obsolete. Whereas concepts are intended to represent a single unique idea, there can be many expressions that describe the same concept. For example, adult onset diabetes mellitus and type 2 diabetes are medical terms that represent the same concept. Multiple terms assigned to the same concept are called synonyms. Standard terminologies assign a “preferred term” as determined by consensus or rules of a particular system. Preferred terms support consistency, indexing, and searching. Concepts and terms are often assigned codes to support analysis and maintain uniqueness between different uses of the same term. The term “cold” could represent a relative temperature or a state of illness associated with upper respiratory infection or a finding on a nuclear scan. The different uses of the same term would be assigned different codes.
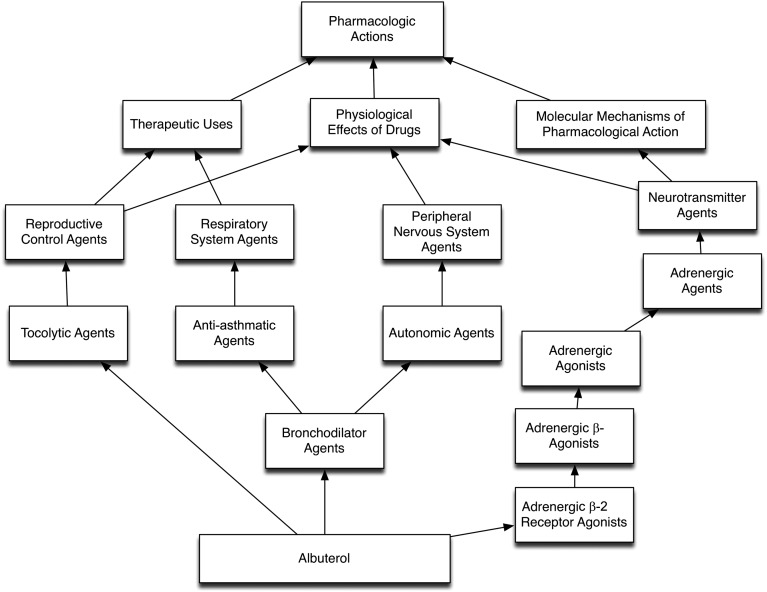
Controlled terminologies are more than a collection of concepts and terms. An ontology is a specification of all terms and their relationships in a knowledge domain that is understood by humans and machines. Hierarchies represent connections between codes organized according to a classification or navigation system. Figure 1 reveals a partial hierarchy from the National Library of Medicine’s (NLM) Unified Medical Language System (UMLS) related to pharmacologic actions for the drug albuterol, using the RxNav graphical browser25 (see http://rxnav.nlm.nih.gov). Three hierarchies are shown in this view: therapeutic uses, physiologic effects, and molecular mechanisms of pharmacological action. Intermediate concepts link to other medications that have the same therapeutic use or physiologic effect. As one moves up the hierarchy, concepts become broader and encompass a wider spectrum of medications.
FIGURE 1.
A partial display of an “is-a” hierarchy for the concept “albuterol.” Notice that albuterol is a tocolytic, anti-asthmatic, autonomic, and adrenergic agent. Additional hierarchies reveal relationships such as physiologic effects and other perspectives.
Figure 1 illustrates an “is-a” hierarchy (albuterol is a bronchodilator agent). Another common structure is a “part-of” hierarchy, which is often used to organize anatomic concepts. The aortic valve is part of the heart, whereas the hypothalamus is part of the brain. The UMLS Semantic Network defines 54 hierarchy link types organized in 5 major categories: physically related to, spatially related to, temporally related to, functionally related to, and conceptually related to.26 A visual depiction of the differences in the various concepts, terminologies, ontologies, and other vocabulary structures can be found at the Marine Metadata Interoperability Web site.27
Hierarchies are also important for harmonizing disparate terms by placing terms that are similar, but not identical, in the same location within a hierarchy. Hierarchies allow distinctions between terms to be retained yet also allow investigators to use a less specific term to combine data when appropriate. Hierarchies also help organize large data sets. For example, although a study may record specific drugs prescribed to patients, a drug classification hierarchy allows investigators to view all drugs that are adrenergic agonists or tocolytic drugs (Fig 1).
Terminologies are developed by numerous organizations to meet diverse information and data processing needs (Table 1). Typically, terminologies address general clinical topics, medicinal/medical products, or biomedical/life sciences. In the clinical domain, existing terminologies primarily cover clinical care, services, or research; findings; diagnoses; statistical reporting; health care billing; clinical images; procedures; patient outcomes; medical laboratory observations; function; disability; or medical specialties, such as oncology. Medicinal/medical product terminologies describe drugs, biopharmaceuticals, or vaccines; devices; adverse events; ingredients; chemical structure; dose; and physiologic effect, mechanism of action, or pharmacokinetics. Biomedical and life sciences terminologies describe microorganisms; research model organisms; substances; physiologic components, functions, interactions, regulation, or pathways; phenotype; exposure; development; phylogeny and taxonomy; research and experimental procedures; or units of measurement. Numerous existing terminologies are available for use by research investigators to meet particular needs.
TABLE 1.
Examples of Biomedical Terminologies
| Content | Terminology | Organization | Web Site |
|---|---|---|---|
| Preclinical and clinical representations | BioPortal: Clinically Relevant Terminologies39 | National Center for Biomedical Ontology40 | http://bioportal.bioontology.org/ |
| Clinical research | CDISC Terminology41 | CDISC | http://www.cdisc.org/terminology |
| Biomedical sciences | Controlled Biomedical Vocabularies42 | UMLS, NLM | http://www.nlm.nih.gov/research/umls/ |
| Medical, surgical, and diagnostic procedures and services | Current Procedural Terminology43 | American Medical Association | http://www.ama-assn.org/ama/pub/physician-resources/solutions-managing-your-practice/coding-billing-insurance/cpt.page |
| Findings, diagnoses, disease, statistics, health care billing | International Classification of Diseases, 10th Revision44 | WHO | http://www.who.int/classifications/icd/en/ |
| Health conditions, diagnoses, procedures, statistics, health care billing | International Classification of Diseases, Ninth Revision, Clinical Modification and Procedure Code System | WHO | http://www.who.int/classifications/icd/en/ |
| Classification of function and disability | International Classification of Functioning, Disability, and Health45 | WHO | http://www.who.int/classifications/icf/en/ |
| Medical laboratory observations, clinical results/care | Logical Observation Identifiers Names and Codes46 | Regenstrief Institute | http://loinc.org/ |
| Adverse event, biopharmaceuticals, medical products | Medical Dictionary for Regulatory Affairs47 | International Conference on Harmonization | http://www.meddramsso.com/index.asp |
| Clinical trial related adverse events reporting | Common Terminology Criteria for Adverse Events28 | Cancer Therapy Evaluation Program, US NCI | http://ctep.cancer.gov/protocolDevelopment/electronic_applications/ctc.htm |
| Life sciences indexing controlled vocabulary | Medical Subject Headings48 | NLM | http://www.nlm.nih.gov/mesh/ |
| Ingredients, chemical structure, dose, physiologic effect, mechanism of action, pharmacokinetics, diseases | NDF–RT49 | US Department of Veterans Affairs, Veterans Health Administration | http://www.nlm.nih.gov/research/umls/sourcereleasedocs/current/NDFRT/ |
| Vocabulary for clinical care, translational and basic research | NCI Thesaurus34 | NCI | http://ncit.nci.nih.gov/ |
| Clinical drugs | RxNorm50 | NLM | http://www.nlm.nih.gov/research/umls/rxnorm/overview.html |
| Nursing activity classification | Nursing Interventions Classification51 | Center for Nursing Classification, University of Iowa, College of Nursing | http://www.ncvhs.hhs.gov/970416w4.htm |
| Patient outcomes classification | Nursing Outcomes Classification52 | Center for Nursing Classification, University of Iowa, College of Nursing | http://www.nursing.uiowa.edu/cncce/nursing-outcomes-classification-overview |
| Diseases, findings, procedures, microorganisms, substances | SNOMED–CT53 | International Health Terminology Standards Development Organization | http://www.ihtsdo.org/ |
| Units of measurement in science, business, engineering | UCUM54 | The UCUM Organization | http://unitsofmeasure.org/trac/ |
| Vaccines | Vaccine Administered from Centers for Disease Control and Prevention | Centers for Disease Control and Prevention | http://www2a.cdc.gov/vaccines/iis/iisstandards/vaccines.asp?rpt=cvx |
CDISC, Clinical Data Interchange Standards Consortium; UCUM, Unified Code for Units of Measure; WHO, World Health Organization.
Using Controlled Terminologies to Accelerate Child Health Research
We previously included a vignette that highlights the barriers to data reuse and sharing that can occur when controlled terminologies and other tools are not used. Using the same example, we envision what would be different if common terminology and definitions, linked to external controlled terminologies, had been used.
Two neonatologists at separate institutions seek to investigate the impact of new therapeutic interventions for NEC. Because a common terminology for neonatology exists, the 2 investigators reuse data collection forms and database tools developed by others that have incorporated these definitions. In addition, linkages to clinical terminologies, such as SNOMED–CT for clinical observations, Logical Observation Identifiers Names and Codes for laboratory results, and RadLex for radiology findings, allow both investigators to incorporate data contained in electronic systems including clinical, laboratory, medication, image, and pathology data from local clinical systems. In addition, mortality data from local and state death registries can be linked and incorporated. In both studies, not all concepts were present in the existing terminologies. Recognizing that important concepts were missing, the investigators provided detailed definitions to a pediatric research terminology-coordinating center, which curated the proposed definitions and incorporated the missing information into an expanded version of the controlled terminology. Missing terms were also submitted to national and international standards bodies and were incorporated for future use in EHRs for clinical care documentation. At the conclusion of each study, the 2 investigators combined their data sets to conduct a secondary analysis that would not have had sufficient power if either of the 2 data sets were used alone. Using connections through other controlled terminologies that linked phenotype descriptions in anonymized genomic databases, an unanticipated potential association between specific NEC outcomes and 2 loci associated with systemic inflammatory response was discovered.
A key feature of the previous vignette is the integration of clinical research and clinical care terminologies to facilitate the use of EHR data to support research studies. Similarly, the integration with genomic databases facilitates cross-sectional, population-based genome-wide association studies that could reveal novel biological processes that predict outcomes and suggest new interventions. In both cases, the effectiveness of terminologies depends on their ability to accurately describe concepts of interest, and the relationships between them. But it also depends on establishing common ground for use across research efforts that may never interact directly but are able to use information from each other’s work to extend their own scope. Often these uses are not planned as part of the original research but take advantage of the generality of terminologies to develop linkages to new resources. Leveraging these opportunities requires investment in establishing and maintaining the terminologies, and in choosing to adopt them rather than developing incompatible vocabularies for each area of research. As research produces new concepts and changes our understanding, common terminologies must also adapt through periodic review and curation. The curation process not only incorporates new understanding, but by codifying evolving versions of terms, also improves validity by allowing research that incorporates previously captured data to account for this evolution. Done correctly, this ongoing evolution supports reuse of existing information in new contexts for new analyses. This capability to retain the intended meanings of concepts is particularly important to child health research, where outcomes of interest may not occur for extended times, and will likely be evaluated in settings other than the original research sites as subjects grow into adulthood.
Standard terminologies are established in a limited number of research contexts. Perhaps the most widely adopted research terminology is the Common Terminology Criteria for Adverse Events that is used to ensure consistent and comparable toxicity assessments in multisite clinical trials sponsored by the Cancer Therapy Evaluation Program within the US National Cancer Institute (NCI).28 The promise of more effective collaboration across research programs has led to recent efforts in several domains to develop common terminologies. The Neonatal Research Networks Terminology retrospectively mapped terms used by existing research networks to a common hierarchy that extends concepts present in SNOMED–CT.29 This effort identified a large number of terms critical to neonatal research that are not present in the existing version of SNOMED–CT. The Global Rare Diseases Registry (GRDR; http://www.grdr.info) project, led by the NIH Office of Rare Diseases Research, represents one of the first attempts in pediatric research to incorporate standard terminologies both prospectively and retrospectively into a new platform designed specifically to support long-term studies and data sharing. Based on a set of consensus concepts developed with Office of Rare Diseases Research guidance, the GRDR common data elements (CDEs) provide detailed specifications for rare disease registries to contribute core information from their patient population to the GRDR to enable cross-disease research.30 As illustrated in Table 2, each CDE specification is linked to concepts in an appropriate reference terminology (Normalized Naming System for Drugs [RxNorm], National Drug File–Reference Terminology [NDF–RT], SNOMED–CT), ensuring that data in the GRDR have consistent representation and can be linked to other data sources, including EHRs. New registries developed by using GRDR CDEs incorporate these standardized terms from the point of patient contact, providing consistency through the process of data definition, collection, and transfer. Participating registries that antedate the GRDR CDEs create mappings from their existing data elements to CDEs where possible and contribute this subset as core data that are consistent and can be combined with prospective data collection. As CDEs are reviewed, updated, and expanded, increasing amounts of existing data move from siloed registry-specific representations into a sharable common terminology.
TABLE 2.
Three CDE Definitions From the GRDR
| GRDR No. | Item Concept | Question Text | Reference Categories | Reference Categories Link |
|---|---|---|---|---|
| GRDR038 | Current medications | What are the medications that this participant is currently taking? | Registry responses are mapped to RxNorm identifiers specifying the active ingredient(s) and route of administration. | RxNorm |
| GRDR039 | Medical foods/special diet | Does the participant take any medical foods or follow a special diet for treatment of his/her rare disease? | Registry responses are mapped to NDF–RT identifiers. | NDF–RT |
| GRDR040 | Previous surgeries | Has the participant had any of the following surgeries to treat his/her rare disease? | Registry responses are mapped to SNOMED–CT surgery concepts, and the GRDR stores the SNOMED–CT identifiers. | SNOMED–CT |
Not all columns are shown. Three standardized terminologies (RxNorm, NDF–RT, and SNOMED–CT) are used to ensure common values across registries that can be linked to other data sources and to EHRs.
Figure 2 illustrates how CDEs and controlled terminologies are combined to enable data integration, sharing, and reuse. CDEs use controlled terminologies to represent data. Without the use of controlled terminologies in CDEs, each data element would be a stand-alone concept not connected to any terminology or hierarchy and therefore unable to be combined without an extensive manual concept-to-concept mapping effort that is resource intensive and can be prone to technical errors, bias, and personal judgment. CDEs also contain concept definitions, which are text descriptions for standardizing data collection and evaluation of subjective clinical findings. Although these definitions do not completely eliminate variation in clinical observations, CDE definitions can provide some guidance to reduce interobserver variation.
FIGURE 2.
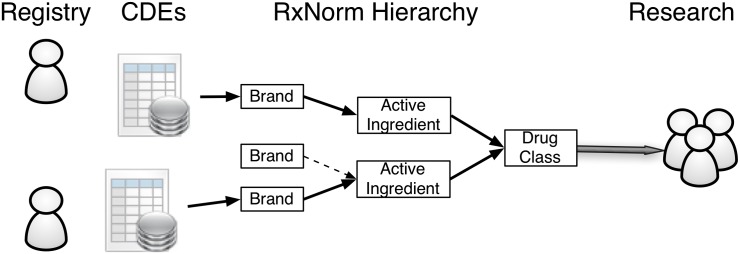
Linking CDEs to controlled terminologies to enable data integration, sharing, and reuse.
Our second vignette also highlights the integration of research terminology into pediatric practices via terms within the EHR. Under the HITECH legislation, the Office of the National Coordinator issued new EHR certification requirements that require vendors to incorporate a growing list of national and international terminology standards.31 Ensuring that critical pediatric research concepts are included in these mandated terminologies will enable new opportunities to perform observational and comparative effectiveness studies by using existing EHR systems. Although significant clinical information currently resides in free-text documents, meaningful use reporting requirements and technical advances in natural language processing are making more of these data available as structured terms.
Pediatric Research Terminologies for Child Health Research: A Proposed Path Forward
The development and use of well-described terminologies and CDEs to support clinical research is a focus within the NIH through the Trans-NIH BioMedical Informatics Coordinating Committee. A growing number of funding opportunities require the use of CDEs to ensure broad data integration, reuse, and sharing. An extensive set of resources on CDEs, including databases of standardized data elements, case report forms, and other resources can be found at the NIH CDE Resource Portal hosted by the NLM.32 For researchers interested in using standard terminologies, the NLM’s Unified Terminology Services,33 the NCI’s Enterprise Vocabulary Services,34 and the NIH CDE Resource Portal provide Web-based access to a broad set of terminologies covering many biomedical subject areas.
Currently, there is no unified semantic framework for integrating terminology efforts across clinical research disciplines. There is no unified semantic authority to promote the development, use, and maintenance of pediatric research terminologies. And there is no coherent training curriculum to help investigators navigate the technical details of terminology tools and systems. These challenges need to be addressed broadly across all research domains as promoted by the NIH CDE Resource Portal.
Developing high quality pediatric research terminologies to address unmet needs should be a collaborative, community effort, requiring the expertise of clinicians and researchers from multiple subdisciplines, the terminology engineering expertise of content curators, and an extensive array of computer-based tools. Identifying, harmonizing, and sustaining pediatric research terminologies also require coordination, prioritization, and sustainable development and support structures. Without some level of oversight, independent terminology efforts are likely to diverge into inconsistent and incompatible terms and definitions. In September 2012, the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) and the Pediatric Electronic Health Record Data Sharing Network jointly sponsored an all-day symposium entitled “Bridging the terminology gap in pediatrics: developing an action plan to support the continuum from clinic to research” (http://www.nichd.nih.gov/about/meetings/2012/Pages/092112.aspx). Over 100 participants across the pediatric research community reviewed current requirements for pediatric terminology, and discussed a proposed governance structure and prioritization mechanism to support coordination and communication across pediatric terminology efforts.
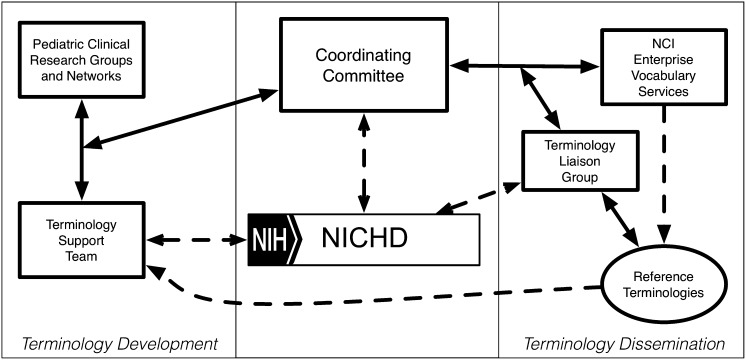
The governance model illustrated in Fig 3 provides a framework that coordinates terminology development activities between multiple clinical research groups and guides the development, use, and maintenance of pediatric research terminologies. Pediatric clinical research groups and networks are paired with a dedicated terminology support team that has access to a rich array of terminology tools housed at the NCI, the NLM, and the National Center for Biomedical Ontology. A Governance Committee, in collaboration with the NICHD, provides oversight and coordination to minimize duplication of effort and standardization of outputs across multiple simultaneous terminology efforts. The Governance Committee also ensures that representatives from relevant clinical domains are present within each terminology team.
FIGURE 3.
Proposed governance structure for coordinating pediatric research terminology development.
In 2010, an initial pediatric terminology project led by the NICHD successfully harmonized child life stages from 6 primary sources that had overlapping definitions. A second harmonization project integrated over 2000 unique terms used by 3 neonatal research networks. The NICHD Pediatric Terminologies currently include newborn screening, pregnancy and childbirth, childhood immunization, neurologic development, and the neonatal research network terminologies. These terminologies are developed in partnership with and maintained by and available from the NCI Enterprise Vocabulary Services (http://www.nichd.nih.gov/health/clinicalresearch/clinical-researchers/terminology/Pages/current.aspx). In April 2013, the Governance Committee, using the proposed governance model in Fig 3, identified 3 additional pediatric research domains for terminology harmonization. Leaders of national research efforts in pediatric rheumatology, pediatric adverse events, and perinatology were identified. A core group of clinical investigators are teamed with terminology experts from the NCI’s Enterprise Vocabulary Services to develop harmonized terminologies within their domains. The governance committee meets twice monthly to evaluate progress and to identify future opportunities and potential clinical domain leaders for future terminology harmonization efforts. Steady progress with these initial harmonization efforts indicates both the governance and execution mechanisms appear effective.
Key components of the Learning Health System model include opportunities for rapid transfer of information from a variety of clinical settings to support clinical research at a scale previously not feasible, and for rapid translation of validated conclusions to the point of care.35–38 This concept is grounded in the principle that the physical and conceptual distance between patient care and research must be reduced to make effective use of the opportunity to learn from each patient, and to use the resulting knowledge to improve the health of every patient. Because the Learning Health System vision is inherently distributed, it is dependent on the existence of broadly expressive, well-curated terminologies to ensure efficient use of limited health care resources, fidelity of data capture across different times and locations, and accurate translation of results to future patients. In addition, it allows more effective use of limited health care resources, intellectual as well as financial, as information collected in 1 context can be analyzed in others. Careful stewardship of data reflects another core principle of the Learning Health System: that health information is part of the trust patients’ grant to the health care system; making the best use of this information is not only an opportunity, but an ethical imperative. A comprehensive, well-curated, sustainable common pediatric research terminology is a necessary component of achieving this imperative.
Acknowledgments
Anjali Kastorf, Daniela Smith, Ranjana Srivastava, Daniel Lyman, and Michael Keller from Booz Allen Hamilton and Elizabeth Humphries and Clem McDonald of the NLM provided subject matter expertise in terminology harmonization and background materials on terminology standards organizations used to generate Table 1. Anjali Kastorf from Booz Allen Hamilton and Elizabeth Hines from Children’s Hospital of Philadelphia provided critical project management.
Glossary
- CDE
common data element
- EHR
electronic health record
- GRDR
Global Rare Diseases Registry
- HITECH
Health Information Technology for Economic and Clinical Health
- NCI
National Cancer Institute
- NDF–RT
National Drug File–Reference Terminology
- NEC
necrotizing enterocolitis
- NICHD
Eunice Kennedy Shriver National Institute of Child Health and Human Development
- NIH
National Institutes of Health
- NLM
National Library of Medicine
- RxNorm
Normalized Naming System for Drugs
- SNOMED–CT
Systematized Nomenclature of Medicine–Clinical Terms
- UMLS
Unified Medical Language System
Footnotes
Dr Kahn was an organizer of and presenter at the 2012 Pediatric Terminology Harmonization Workshop that developed the proposed governance model, and drafted the initial manuscript; Drs Bailey, Forrest, Padula, and Hirschfeld were organizers of and presenters at the 2012 Pediatric Terminology Harmonization Workshop that developed the proposed governance model, and critically reviewed and revised the manuscript; and all authors approved the final manuscript as submitted.
The views and comments expressed in this document do not necessarily reflect the views and policies of the Eunice Kennedy Shriver National Institute of Child Health and Human Development.
FINANCIAL DISCLOSURE: The authors have indicated they have no financial relationships relevant to this article to disclose.
FUNDING: Supported by National Children’s Study contracts HHSN275200800018C (to Dr Kahn) and HHSN267200700020C (to Drs Bailey, Forrest, and Padula); National Institutes of Health, National Center for Advancing Translational Sciences, Colorado Clinical and Translational Sciences Institute grant UL1 TR000154 to Dr Kahn and Agency for Healthcare Research & Quality R01HS020024-03 (to Drs Bailey and Forrest). Funded by the National Institutes of Health (NIH).
POTENTIAL CONFLICT OF INTEREST: The authors have indicated they have no potential conflicts of interest to disclose.
References
- 1.Pace WD, Cifuentes M, Valuck RJ, Staton EW, Brandt EC, West DR. An electronic practice-based network for observational comparative effectiveness research. Ann Intern Med. 2009;151(5):338–340 [DOI] [PubMed] [Google Scholar]
- 2.Maro JC, Platt R, Holmes JH, et al. Design of a national distributed health data network. Ann Intern Med. 2009;151(5):341–344 [DOI] [PubMed] [Google Scholar]
- 3.Brown JS, Holmes JH, Shah K, Hall K, Lazarus R, Platt R. Distributed health data networks: a practical and preferred approach to multi-institutional evaluations of comparative effectiveness, safety, and quality of care. Med Care. 2010;48(suppl 6):S45–S51 [DOI] [PubMed] [Google Scholar]
- 4.Kantak AD, Grow JL, Ohlinger J, Adams HJ, Knupp AM, Lavin JP, Jr. Management of high-order multiple births: application of lessons learned because of participation in Vermont Oxford Network collaboratives. Pediatrics. 2006;118(suppl 2):S159–S168 [DOI] [PubMed] [Google Scholar]
- 5.Payne NR, Finkelstein MJ, Liu M, Kaempf JW, Sharek PJ, Olsen S. NICU practices and outcomes associated with 9 years of quality improvement collaboratives. Pediatrics. 2010;125(3):437–446 [DOI] [PubMed] [Google Scholar]
- 6.Slora EJ, Harris DL, Bocian AB, Wasserman RC. Pediatric clinical research networks: current status, common challenges, and potential solutions. Pediatrics. 2010;126(4):740–745 [DOI] [PubMed] [Google Scholar]
- 7.Crandall WV, Margolis PA, Kappelman MD, et al. ImproveCareNow Collaborative . Improved outcomes in a quality improvement collaborative for pediatric inflammatory bowel disease. Pediatrics. 2012;129(4). Available at: http://www.pediatrics.org/cgi/content/full/129/4/e1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holve E, Segal C, Lopez MH, Rein A, Johnson BH. The Electronic Data Methods (EDM) forum for comparative effectiveness research (CER). Med Care. 2012;50(suppl):S7–S10 [DOI] [PubMed] [Google Scholar]
- 9.Lopez MH, Holve E, Sarkar IN, Segal C. Building the informatics infrastructure for comparative effectiveness research (CER): a review of the literature. Med Care. 2012;50(suppl):S38–S48 [DOI] [PubMed] [Google Scholar]
- 10.Holve E, Segal C, Hamilton Lopez M. Opportunities and challenges for comparative effectiveness research (CER) with Electronic Clinical Data: a perspective from the EDM forum. Med Care. 2012;50(suppl):S11–S18 [DOI] [PubMed] [Google Scholar]
- 11.Hing E, Hsiao CJ. Electronic medical record use by office-based physicians and their practices: United States, 2007. Natl Health Stat Report. 2010;(23):1–11 [PubMed]
- 12.Hsiao CJ, Hing E. Use and characteristics of electronic health record systems among office-based physician practices: United States, 2001–2012. NCHS Data Brief. 2012;(111):1–8 [PubMed]
- 13.Spooner SA, Classen DC. Data standards and improvement of quality and safety in child health care. Pediatrics. 2009;123(suppl 2):S74–S79 [DOI] [PubMed] [Google Scholar]
- 14.Wasserman RC. Electronic medical records (EMRs), epidemiology, and epistemology: reflections on EMRs and future pediatric clinical research. Academic Pediatr. 2011;11(4):280–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cimino JJ. Collect once, use many. Enabling the reuse of clinical data through controlled terminologies. J AHIMA. 2007;78(2):24–29; quiz 31–32 [PubMed] [Google Scholar]
- 16.Marsolo K. In search of a data-in-once, electronic health record-linked, multicenter registry: how far we have come and how far we still have to go. eGEMs. 2012;1(1):3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kahn MG, Weng C. Clinical research informatics: a conceptual perspective. J Am Med Inform Assoc. 2012;19(e1):e36–e42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Meurs KP, Wright LL, Ehrenkranz RA, et al. Preemie Inhaled Nitric Oxide Study . Inhaled nitric oxide for premature infants with severe respiratory failure. N Engl J Med. 2005;353(1):13–22 [DOI] [PubMed] [Google Scholar]
- 19.Ballard RA, Truog WE, Cnaan A, et al. NO CLD Study Group . Inhaled nitric oxide in preterm infants undergoing mechanical ventilation. N Engl J Med. 2006;355(4):343–353 [DOI] [PubMed] [Google Scholar]
- 20.Kinsella JP, Cutter GR, Walsh WF, et al. Early inhaled nitric oxide therapy in premature newborns with respiratory failure. N Engl J Med. 2006;355(4):354–364 [DOI] [PubMed] [Google Scholar]
- 21.Cole FS, Alleyne C, Barks JD, et al. NIH consensus development conference: inhaled nitric oxide therapy for premature infants. NIH Consens State Sci Statements. 2010;27(5):1–34 [PubMed] [Google Scholar]
- 22.Donohue PK, Gilmore MM, Cristofalo E, et al. Inhaled nitric oxide in preterm infants: a systematic review. Pediatrics. 2011;127(2). Available at: www.pediatrics.org/cgi/content/full/127/2/e414 [DOI] [PubMed] [Google Scholar]
- 23.Padula AM, Mortimer K, Hubbard A, Lurmann F, Jerrett M, Tager IB. Exposure to traffic-related air pollution during pregnancy and term low birth weight: estimation of causal associations in a semiparametric model. Am J Epidemiol. 2012;176(9):815–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bradley CJ, Penberthy L, Devers KJ, Holden DJ. Health services research and data linkages: issues, methods, and directions for the future. Health Serv Res. 2010;45(5 pt 2):1468–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zeng K, Bodenreider O, Kilbourne J, Nelson S. RxNav: a web service for standard drug information. AMIA Annu Symp Proc. 2006:1156 [PMC free article] [PubMed] [Google Scholar]
- 26.McCray AT. An upper-level ontology for the biomedical domain. Comp Funct Genomics. 2003;4(1):80–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marine Metadata Initiative. Marine metadata interoperability categories of controlled vocabularies. Available at: https://marinemetadata.org/guides/vocabs/voctypes/voccat. Accessed March 1, 2013
- 28.National Cancer Institute. NCI Wiki common terminology criteria for adverse events. Available at: https://wiki.nci.nih.gov/display/VKC/Common+Terminology+Criteria+for+Adverse+Events. Accessed April 15, 2013
- 29.Pallotto EK, Hunt PG, Reber K, Evans JR, Padula MA. Topics in neonatal informatics: standardizing diagnoses in neonatology: bronchopulmonary dysplasia and beyond. NeoReviews. 2012;13:e577–e582 [Google Scholar]
- 30.NIH Office of Rare Diseases Research. CDE overview. Available at: www.grdr.info/index.php?option=com_content&view=article&id=3&Itemid=5. Accessed April 15, 2013
- 31.Office of the National Coordinator. 45 CFR Part 170 Health Information Technology: Standards, Implementation Specifications, and Certification Criteria for Electronic Health Record Technology, 2014 Edition; Revisions to the Permanent Certification Program for Health Information Technology. In: Services DoHaH, ed. Federal Register; 2012;77:54163–54292 [PubMed] [Google Scholar]
- 32.National Library of Medicine. NIH Common Data Element (CDE) resource portal. Available at: www.nlm.nih.gov/cde/index.html. Accessed April 15, 2013
- 33.National Library of Medicine. UMLS Reference Manual 2009. Available at: www.ncbi.nlm.nih.gov/books/NBK9676/. Accessed August 3, 2013
- 34.Sioutos N, de Coronado S, Haber MW, Hartel FW, Shaiu WL, Wright LW. NCI Thesaurus: a semantic model integrating cancer-related clinical and molecular information. J Biomed Inform. 2007;40(1):30–43 [DOI] [PubMed] [Google Scholar]
- 35.Greene SM, Reid RJ, Larson EB. Implementing the learning health system: from concept to action. Ann Intern Med. 2012;157(3):207–210 [DOI] [PubMed] [Google Scholar]
- 36.Friedman C, Rigby M. Conceptualising and creating a global learning health system. Int J Med Inform. 2013;82(4):e63–e71 [DOI] [PubMed] [Google Scholar]
- 37.Friedman CP, Wong AK, Blumenthal D. Achieving a nationwide learning health system. Sci Transl Med. 2010;2(57):57cm29. [DOI] [PubMed] [Google Scholar]
- 38.Etheredge LM. A rapid-learning health system. Health Aff (Millwood). 2007;26(2):w107–w118 [DOI] [PubMed] [Google Scholar]
- 39.Whetzel PL, Noy NF, Shah NH, et al. BioPortal: enhanced functionality via new Web services from the National Center for Biomedical Ontology to access and use ontologies in software applications. Nucleic Acids Res. 2011;39(Web Server Issue):W541–W545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Musen MA, Noy NF, Shah NH, et al. NCBO team . The National Center for Biomedical Ontology. J Am Med Inform Assoc. 2012;19(2):190–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Richesson RL, Krischer J. Data standards in clinical research: gaps, overlaps, challenges and future directions. J Am Med Inform Assoc. 2007;14(6):687–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res. 2004;32(Database Issue):D267–D270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abraham M, Ahlman JT, Boudreau AJ, Connelly JK, Evans DD, Glenn RL. CPT 2011 Standard Edition. Chicago, IL: American Medical Association Press; 2010 [Google Scholar]
- 44.Israel RA. The International Classification of Disease. Two hundred years of development. Public Health Rep. 1978;93(2):150–152 [PMC free article] [PubMed] [Google Scholar]
- 45.Stucki G, Cieza A, Melvin J. The International Classification of Functioning, Disability and Health (ICF): a unifying model for the conceptual description of the rehabilitation strategy. J Rehabil Med. 2007;39(4):279–285 [DOI] [PubMed] [Google Scholar]
- 46.Huff SM, Rocha RA, McDonald CJ, et al. Development of the Logical Observation Identifier Names and Codes (LOINC) vocabulary. J Am Med Inform Assoc. 1998;5(3):276–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brown EG, Wood L, Wood S. The medical dictionary for regulatory activities (MedDRA). Drug Saf. 1999;20(2):109–117 [DOI] [PubMed] [Google Scholar]
- 48.Rogers FB. Medical subject headings. Bull Med Libr Assoc. 1963;51:114–116 [PMC free article] [PubMed] [Google Scholar]
- 49.Brown SH, Elkin PL, Rosenbloom ST, et al. VA National Drug File Reference Terminology: a cross-institutional content coverage study. Stud Health Technol Inform. 2004;107(pt 1):477–481 [PubMed] [Google Scholar]
- 50.Nelson SJ, Zeng K, Kilbourne J, Powell T, Moore R. Normalized names for clinical drugs: RxNorm at 6 years. J Am Med Inform Assoc. 2011;18(4):441–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bulechek GM, Butcher HK, Dochterman JM. Nursing Interventions Classification (NIC). 5th ed. St Louis, MO: Mosby/Elsevier; 2008 [Google Scholar]
- 52.Moorhead S. Nursing Outcomes Classification (NOC). 4th ed. St Louis, MO: Mosby/Elsevier; 2008 [Google Scholar]
- 53.Spackman KA, Campbell KE, Cote RA. SNOMED RT: a reference terminology for health care. Proc AMIA Annu Fall Symp. 1997:640–644 [PMC free article] [PubMed] [Google Scholar]
- 54.Schadow G, McDonald CJ, Suico JG, Föhring U, Tolxdorff T. Units of measure in clinical information systems. J Am Med Inform Assoc. 1999;6(2):151–162 [DOI] [PMC free article] [PubMed] [Google Scholar]