Abstract
The ubiquitin ligase CUL4A has been implicated in tumorigenesis but its contributions to progression and metastasis have not been evaluated. Here we show that CUL4A is elevated in breast cancer as well as ovarian, gastric and colorectal tumors where its expression level correlates positively with distant metastasis. CUL4A overexpression in normal or malignant human mammary epithelial cells increased their neoplastic properties in vitro and in vivo, markedly increasing epithelial-mesenchymal transition (EMT) and the metastatic capacity of malignant cells. In contrast, silencing CUL4A in aggressive breast cancer cells inhibited these processes. Mechanistically, we found CUL4A modulated histone H3K4me3 at the promoter of the EMT regulatory gene ZEB1 in a manner associated with its transcription. ZEB1 silencing blocked CUL4A-driven proliferation, EMT, tumorigenesis and metastasis. Further, in human breast cancers ZEB1 expression correlated positively with CUL4A expression and distant metastasis. Taken together, our findings reveal a pivotal role of CUL4A in regulating the metastatic behavior of breast cancer cells.
Keywords: CUL4A, proliferation, epithelial-mesenchymal transition, invasion, metastasis
INTRODUCTION
Breast cancer (BC), the most frequent malignancy in women worldwide (1, 2), is a complex and intrinsically heterogeneous disease. It is believed that BC develops through the accumulation of a wide spectrum of genomic aberrations (3). Amplification of 13q34 is found in 5% of all human BCs and as high as 20% of basal-type BCs (4), the subtype of BCs most often associated with aggressive growth and poor prognosis (5), and in other malignant tumors (6, 7). Several candidate genes including CUL4A have been proposed for this region (4, 8, 9). CUL4A mRNA and protein levels are highly correlated with 13q34 amplification and has been hypothesized to be the candidate driver of this amplicon based on its amplification and significant elevation in BCs (4, 10), as well as in other cancers (9, 11–15). The fact that overexpression of CUL4A is associated with poor prognosis in node-negative BCs and other malignancies such as ovarian carcinoma (16, 17) further supports its possible role in the aggressive behavior of certain cancers.
CUL4A, a member of the cullin family of proteins that composes the multifunctional ubiquitin ligase E3 complex, is essential for the ubiquitination of several well-defined tumor suppressor genes such as p21 (18), p27 (19) and p53 (20). Changes in CUL4A potentially exert pleiotropic effects that alter cellular functions including proliferation, differentiation, and apoptosis. Thus, CUL4A may act as an oncogene, but whether CUL4A plays a role in BC metastasis remains unknown.
The vast majority of BC patients succumb to their disease as a result of metastasis (21, 22). Currently, treatment options for metastatic BCs are limited and ineffective. Therefore, tremendous effort has been focused on the understanding of the mechanisms by which metastasis occurs in order to provide a more rational approach in the development of future metastatic breast cancer treatments. However, how metastases are formed remains less understood. Mounting evidence shows that in epithelial cancers, including BCs, induction of epithelial-mesenchymal transition (EMT) is a major event that provides mobility to cancer cells in order to generate metastases (23). EMT is characterized by the loss of epithelial characteristics and acquisition of a mesenchymal phenotype, which confers the ability for cancer cells to invade adjacent tissue and migrate to distant sites (24), where these cancer cells proliferate to generate new tumors. Hence, clarifying the regulation of proliferation and EMT will greatly benefit our understanding of tumor metastasis.
In this study, we found that CUL4A overexpression induced proliferation and EMT in normal and malignant human mammary epithelial cells, resulting in enhancement of growth, migration and invasion in vitro and metastasis in vivo. Depletion of CUL4A inhibited proliferation, led to mesenchymal-to-epithelial transition (MET) and blocked distant metastasis. These functional effects of CUL4A were exerted through control of ZEB1 transcriptional expression via trimethylation of H3K4 (H3K4me3). Our findings provide a novel mechanistic role of CUL4A in BC metastasis and a reasonable explanation of our clinical observation that CUL4A expression is correlated with BC metastasis, suggesting that CUL4A may serve as a potential therapeutic target for advanced BCs.
MATERIALS AND METHODS
Chemicals and antibodies
Lipofectamine 2000 transfection and TRIZOL LS Reagents were purchased from Invitrogen (Grand Island, NY, USA). Antibodies against CUL4A, fibronectin, Histone H3, H3K4me1, H3K4me2, H3K4me3, H3K9me3, H3K27me3 were purchased from Abcam (Cambridge, MA, USA). E-cadherin, N-cadherin, vimentin, ZEB1, p21, p27, Ki67 and β-actin antibodies were from Cell Signaling technology (Danvers, MA, USA). α-catenin antibody was from BD (Franklin Lakes, NJ, USA). Unless otherwise noted, all other chemicals were from Sigma (St. Louis, MO, USA).
Histological and immunohistochemical analyses
The tumors, lungs and livers dissected from mice were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) overnight and subsequently embedded in paraffin wax. Sections cut at a thickness of 4 µm were stained with hematoxylin and eosin for histological analysis. Tissue microarrays for breast (BR954), colon (C0812) and ovary (OV806, OV8010) cancers were from Alenabio (Xian, China), and for gastric cancer (HStm-Ade080CD-01) was from Shanghai Outdo Biotech (Shanghai, China). Clinical and pathologic information was provided by the manufacturers. Immunohistochemical analysis was performed for different markers in these arrays as described previously (25). The proportion of stained cells (lower, <30% staining; higher, ≥30% staining) was semiquantitatively determined following published protocols (26).
Cell culture
The human BC cell lines, MDA-MB-468, MDA-MB-231, BT549, MCF7, HCC1569, normal human breast epithelial cell line, MCF10A, and HEK 293 Phoenix ampho packaging cells were purchased from the American Type Culture Collection (Manassas, VA, USA), where they were characterized by DNA-fingerprinting and isozyme detection. Cell culture was according to manufacturer’s protocol. All the cell lines were grown at 37°C in a 5% CO2/95% air atmosphere and were revived every 3 to 4 months.
Chromatin immunoprecipitation (ChIP)-qPCR
ChIP kit was purchased from Millipore and ChIP experiments were carried out essentially as described (27). Immnuoprecipitated DNA was analyzed on the ABI PRISM 7900HT sequence detection system. The primers used for detection of promoters after ChIP are available upon request.
In vivo tumor growth and metastasis
Nude mice were purchased from Shanghai Slac Laboratory Animal Co. Ltd and maintained in microisolator cages. All animals were used in accordance with institutional guidelines and the current experiments were approved by the Use Committee for Animal Care. For subcutaneous inoculation, different numbers of tumor cells were resuspended in PBS medium with 50% Matrigel and inoculated subcutaneously into 8-week-old nude mice. The tumors were measured every 3 days after appearance of tumors and the tumor volume was calculated by the formula length × width2/2. The mice were killed 40 days after the inoculation. For metastasis assays, cells were resuspended in PBS at a concentration of 1×107 cells ml−1. Cell suspension (0.1ml) was injected into tail veins of nude mice. All of the mice were killed by CO2 60 days after inoculation.
Statistical analysis
Data was described as the mean ± s.d.. Association between ZEB1 and CUL4A expression in breast tissue microarray was assessed using Spearman’s rank correlation test. Comparisons between different groups were undertaken using the Student’s two-tailed t-test. The criteria of statistical significance was P<0.05. Statistical analysis was done with SPSS/Win11.0 software (SPSS, Inc., Chicago, Illinois, USA).
RESULTS
CUL4A is highly expressed in malignant cancers
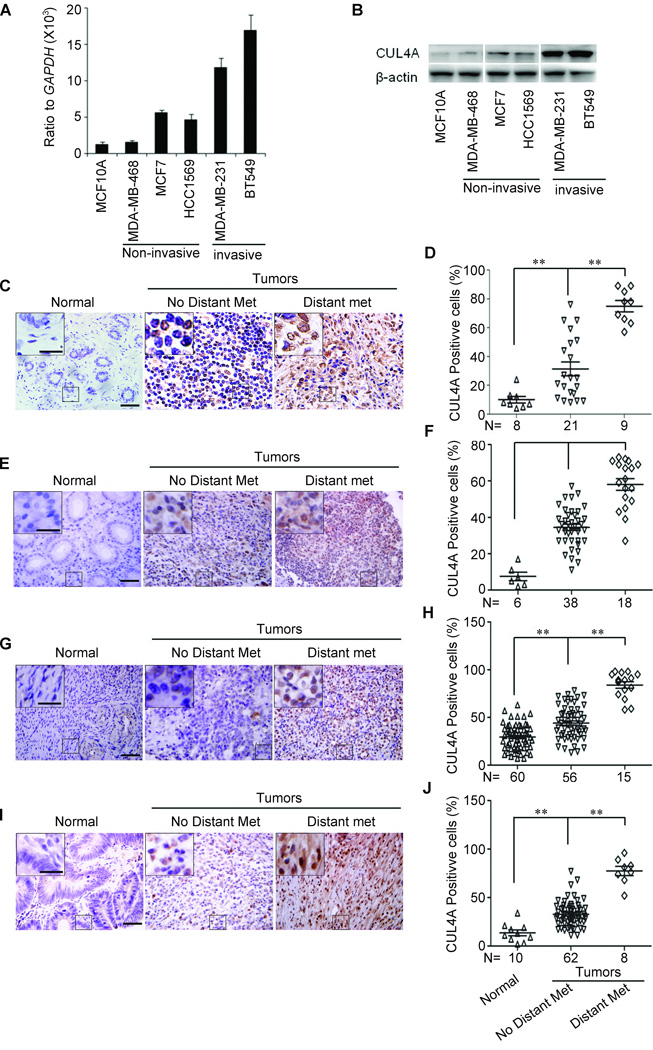
CUL4A was highly expressed in BC cells, especially in invasive cancer cells compared with normal mammary epithelial cells (Fig. 1A and B). We then analyzed CUL4A expression in normal breast tissues and BCs without or with distant metastasis using BC tissue microarray. Consistent with previous report (10), CUL4A was highly expressed in BC tissues compared with adjacent normal breast tissues (Fig. 1C and D). Most importantly, we found that CUL4A overexpression was significantly correlated with distant metastasis (Fig. 1D). In our samples, all BC patients with distant metastasis but only 8 of 21 (38.1%) BC patients without distant metastasis had high CUL4A expression (Fig. 1D).
Figure 1.
CUL4A expression is correlated with metastasis of human cancers. A and B, CUL4A expression was analyzed by qRT-PCR (A) and Western blotting (B) in human BC cell lines. C–J, Immunohistochemical analysis of CUL4A expression in human breast (C and D), gastric (E and F), ovarian (G and H), and colorectal (I and J) cancers using tissue microarrays. (C, E, G and I) From left to right are representative images showing CUL4A expression, and (D, F, H and J) semi-quantification of CUL4A expression in normal tissues, primary cancer tissues without or with distant metastasis. Normal: normal tissues; No Distant Met: primary cancers without distant metastasis (in situ and lymph node metastasis); Distant Met: primary cancers with distant metastasis. Scale bars indicate 50 µm (C, E, G and I) and 20 µm in the inserts (C, E, G and I), **P < 0.01 is based on Student’s t-test (D, F, H and J). Error bars indicate standard deviation.
Since CUL4A overexpression or amplification has been reported in other cancers, we examined CUL4A expression in other types of carcinomas. CUL4A exhibited high expression in ovarian (Fig. 1E and F), colorectal (Fig. 1G and H), and gastric (Fig. 1I and J) cancer tissues compared with adjacent normal tissues and CUL4A overexpression was consistently significantly correlated to distant metastasis in those types of carcinomas. These results collectively indicate a functional role of CUL4A in aggressive behaviors of cancers. We then used BC as a model to verify the function and underlying mechanisms of CUL4A in promoting cancer metastasis.
CUL4A promotes proliferative capacity of BC cells
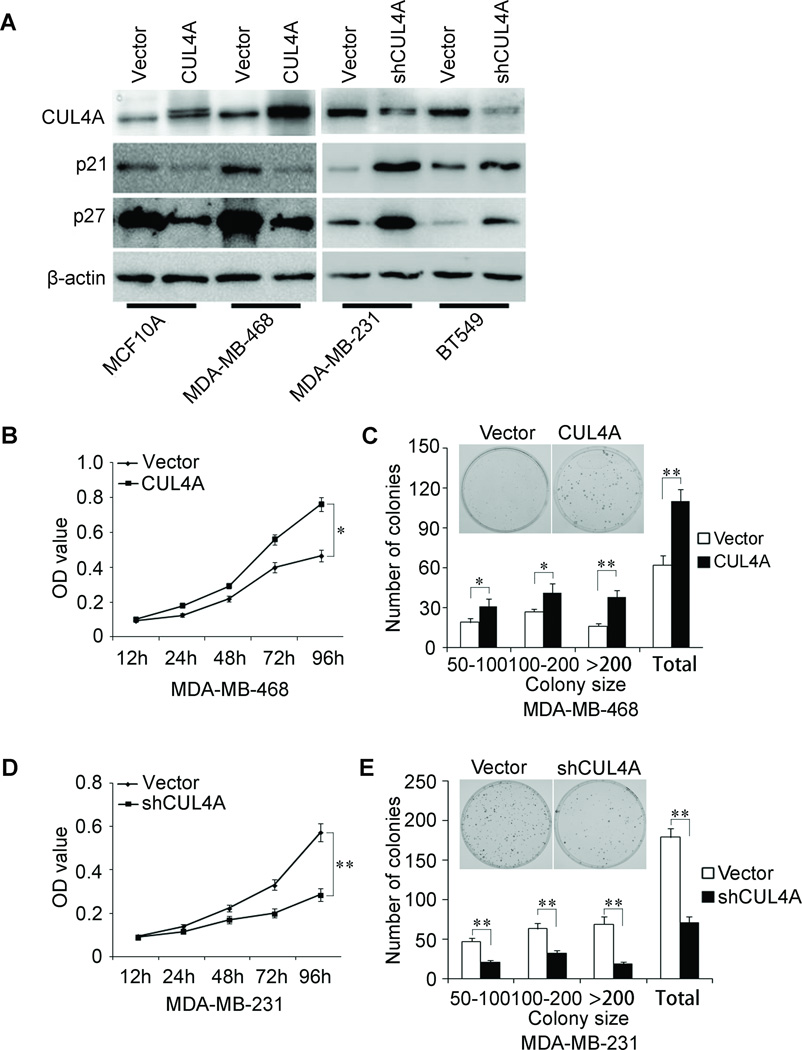
In order to test the oncogenic activity of CUL4A in BCs, we retrovirally established stable overexpression of CUL4A in MCF10A and MDA-MB-468 cells (designated as MCF10A-CUL4A and MDA-MB-468-CUL4A), and silencing CUL4A in MDA-MB-231 and BT549 cells (designated as MDA-MB-231-shCUL4A and BT549-shCUL4A). The levels of CUL4A in these resultant cell lines were verified by western blotting (Fig. 2A). Compared to vector-only controls, both MCF10A-CUL4A and MDA-MB-468-CUL4A cells had significant increases in cell proliferation by MTT assay (Supplemental Fig. 1A; Fig. 2B) and generated more numbers and larger colonies (Supplemental Fig. 1B; Fig. 2C). In contrast, silencing CUL4A in MDA-MB-231 and BT549 cells significantly reduced cell proliferation (Fig. 2D; Supplemental Fig. 1C) and clonogenicity (Fig. 2E; Supplemental Fig. 1D). Consistent with these observations, the expression of two major proliferation related protein, p21 and p27, was modulated upon CUL4A expression, CUL4A overexpression significantly decreased the expression levels of both p21 and p27 while silencing CUL4A dramatically increased their expression levels (Fig. 2A). Taking together, these results suggest that CUL4A is an important regulator of proliferation in BC cells.
Figure 2.
CUL4A promotes proliferation of human mammary epithelial and BC cells. A, Expression levels of CUL4A, p21 and p27 by Western blotting Analysis. B–E, Cell proliferation was examined by MTT (B and D) and colony formation (C and E) assays. For colony formation assay, colonies containing more than 50 cells were counted and plotted; representative plates are shown in inserts. * P < 0.05 and ** P < 0.01 are based on Student's t-test. All results are from three independent experiments. Error bars indicate standard deviation.
CUL4A regulates the transition between epithelial and mesenchymal phenotypes in BC cells
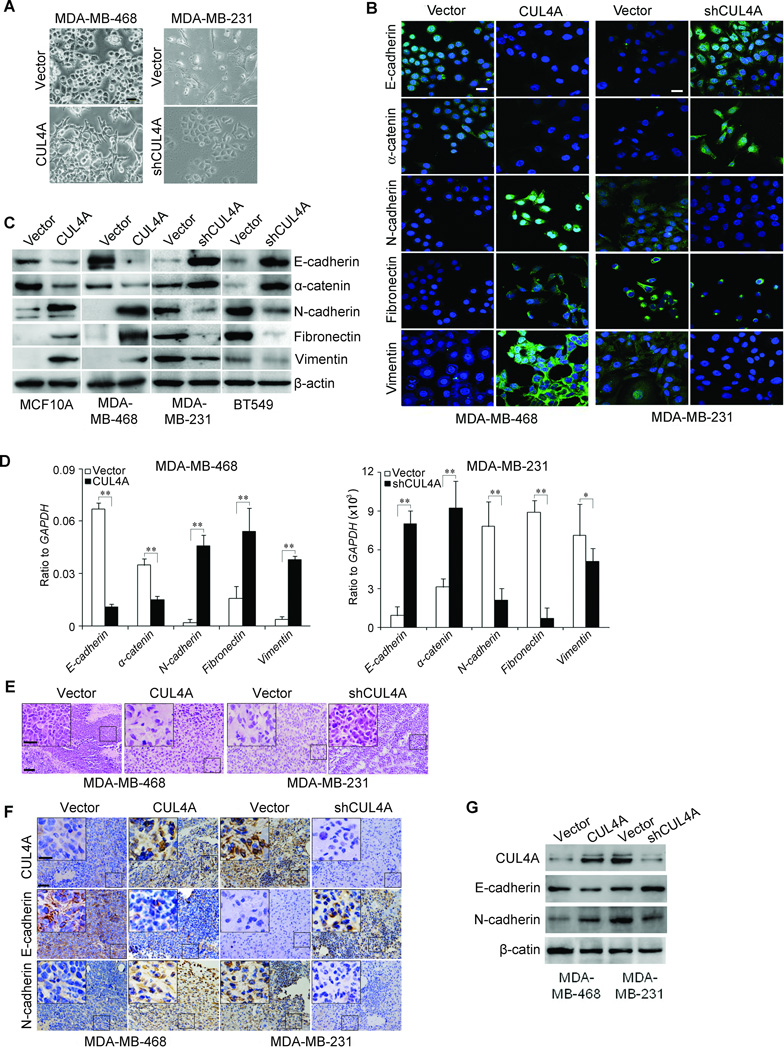
To investigate whether CUL4A positively regulates cell migration and invasion, we first observed the morphological changes and found that both MCF10A-CUL4A and MDA-MB-468-CUL4A cells exhibited fibroblastic morphology (Supplemental Fig. 2A; Fig. 3A). This observation was further confirmed by expression analysis of epithelial and mesenchymal markers. We showed that CUL4A overexpression decreased the levels of epithelial markers (E-cadherin and α-catenin) and increased the levels of mesenchymal markers (N-cadherin, fibronectin and vimentin) in both cell lines (Supplemental Fig. 2B; Fig. 3B and C). Moreover, mRNA levels correlated with the corresponding protein levels (Supplemental Fig. 2C; Fig. 3D), suggesting that CUL4A affected the expression of epithelial and mesenchymal markers at the transcript level.
Figure 3.
CUL4A regulates the transition between epithelial and mesenchymal phenotypes in human BC cells. A, Representative phase-contrast images of MDA-MD-468 and MDA-MD-231 cells showed CUL4A-modulateded morphological changes. B-D, Expression of epithelial and mesenchymal marker was analyzed by Immunofluorescence stains (B), Western blotting (C) and qRT-PCR (D). E, Sections of xenograft tumors were stained with H&E. F and G, Expression of CUL4A, epithelial and mesenchymal marker in xenograft tumors was analyzed by immunohistochemical staining (F) and Western blotting (G). Scale bars indicate 20 µm (A), 20 µm (B), 50 µm (E and F) and 20 µm in the insert in (E, F). Each experiment in (A to D) is repeated at least three times. n = 5 for subcutaneous transplantation. **P < 0.01 is based on Student’s t-test. Error bars indicate standard deviation.
Conversely, both MDA-MB-231-shCUL4A and BT549-shCUL4A cells reverted to an epithelial phenotype as compared to their respective control cells (Fig. 3A; Supplemental Fig. 2A). Consistent with this, silencing CUL4A increased levels of epithelial markers, and decreased levels of mesenchymal markers (Fig. 3B–D; Supplemental Fig. 2B and C).
MDA-MB-468-CUL4A, MDA-MB-231-shCUL4A and their corresponding control cells were subcutaneously injected into nude mice. Xenograft tumors from MDA-MB-468-CUL4A cells also showed a mesenchymal morphology, indicating that CUL4A overexpression maintained EMT in vivo (Fig. 3E). In contrast, changes consistent with MET were observed in xenograft tumors from MDA-MD-231-shCUL4A cells (Fig. 3E). These observations were further confirmed by expression analysis of epithelial and mesenchymal markers using both immunochemical staining (Fig. 3F) and Western blotting (Fig. 3G) in these tumors. Taken together, these findings suggest that CUL4A plays an important role in regulating EMT-MET plasticity of BC cells.
CUL4A promotes migratory and invasive capacities of BC cells in vitro
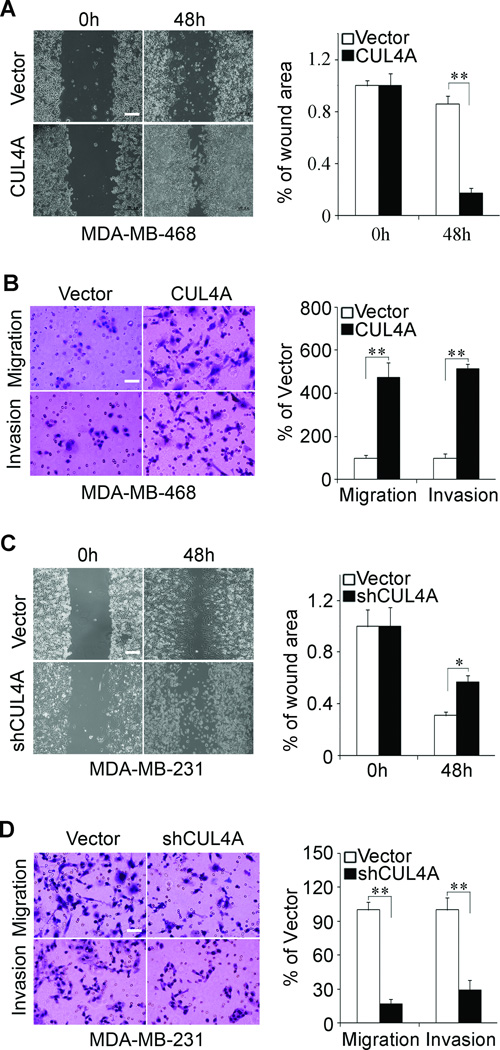
The effect of CUL4A on cell migration was first assessed by wound healing assay. Both MCF10A-CUL4A and MDA-MB-468-CUL4A cells had significantly faster closure of the wound area compared to their control cells (Supplemental Fig. 3A; Fig. 4A). This result was confirmed by Boyden's chamber assay (Supplemental Fig. 3C; Fig. 4B). Moreover, MCF10A-CUL4A and MDA-MB-468-CUL4A cells showed a greater degree of invasion through Matrigel (Supplemental Fig. 3C; Fig. 4B). In contrast, silencing CUL4A dramatically reduced the migratory and invasive capacity of MDA-MB-231 and BT549 cells (Fig. 4C and D; Supplemental Fig. 3B and D), suggesting that restoration of an epithelial phenotype through MET may dampen or inhibit their mobility potential. These results indicate that CUL4A promotes migratory and invasive behaviors in BC cells.
Figure 4.
CUL4A promotes migration and invasion of BC cells. MDA-MB-468-CUL4A and MDA-MB-231-shCUL4A cells or control vector cells were subjected to wound healing assay (A and C), transwell migration (B and D, upper panels) and Matrigel invasion assays (B and D, lower panels). The uncovered areas in the wound healing assays were quantified as a percentage of the original wound area (A and C). Quantification of migrated cells through the membrane and invaded cells through Matrigel of each cell line are shown as proportions of their vector controls (B and D). Scale bars indicate 500 µm (A and C) and 50 µm (B and D). ** P < 0.01 is based on Student's t-test. All results are from three or four independent experiments. Error bars indicate standard deviation.
CUL4A promotes tumorigenesis and metastasis in vivo
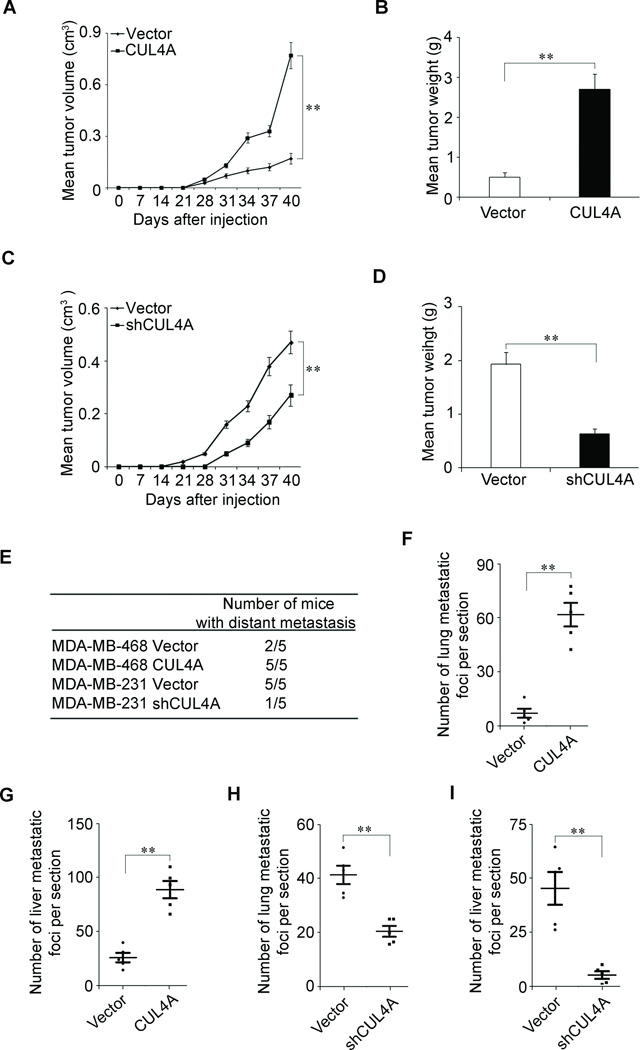
To extend our in vitro observations, we investigated whether CUL4A could regulate tumorigenic and metastatic capacity of BC cells in vivo. MDA-MB-468-CUL4A, MDA-MB-231-shCUL4A and their corresponding control cells were subcutaneously injected into nude mice. As expected, the tumors from MDA-MB-468-CUL4A cells grew more rapidly at the implantation site than their cells (Fig. 5A and B). Increased cell proliferation in MDA-MB-468-CUL4A-derived tumors was further confirmed by ki67 level (Supplemental Fig. 4A). In contrast, silencing CUL4A in the typically aggressive MDA-MD-231 cells led to a dramatic decrease in cell proliferation (Supplemental Fig. 4B), tumor volume and weight (Fig. 5C and D).
Figure 5.
CUL4A promotes tumorigenesis and metastasis of human breast cancer. A and C, Growth curve of tumors formed by MDA-MB-468-CUL4A cells (A), MDA-MB-231-shCUL4A (C) or their control cells by subcutaneous injection. B and D, The weight of tumors formed by MDA-MB-468-CUL4A cells (B), MDA-MB-231-shCUL4A (D) or their control cells at harvest time. E, The total numbers of mice with distant metastasis at 60 days after injection of MDA-MB-468-CUL4A, MDA-MB-231-shCUL4A, or their respective control cells into tail vein. F and G, The numbers of metastatic foci per section in lung (F) and liver (G) of individual mouse with injection of MDA-MB-468-CUL4A or its control cells. H and I, The numbers of metastatic foci per section in lung (H) and liver (I) of individual mouse with injection of MDA-MB-231-shCUL4A or its control cells. n = 5 for subcutaneous transplantation and n = 5 for tail-veil injection. **P < 0.01 is based on Student’s t-test. Error bars indicate standard deviation.
We then investigated the functional relevance of CUL4A for metastasis in vivo. MDA-MB-468-CUL4A, MDA-MD-231-shCUL4A and their corresponding control cells were injected into nude mice through the tail vein. CUL4A overexpression not only significantly increased the number of mice with distant metastasis (Fig. 5E), but also dramatically increased the number of metastatic tumors in both lung and liver of each mouse (Fig. 5F and G; Supplemental Fig. 4C). Silencing CUL4A in MDA-MB-231 cells inhibited metastatic behavior, both in terms of the number of mice with distant metastasis (Fig. 5E) and the number of metastatic tumors in the lung and liver of each mouse (Fig. 5H and I; Supplemental Fig. 4D). Therefore, the in vivo results further demonstrate the critical role of CUL4A in BC metastasis.
CUL4A regulates ZEB1 expression through H3K4 trimethylation
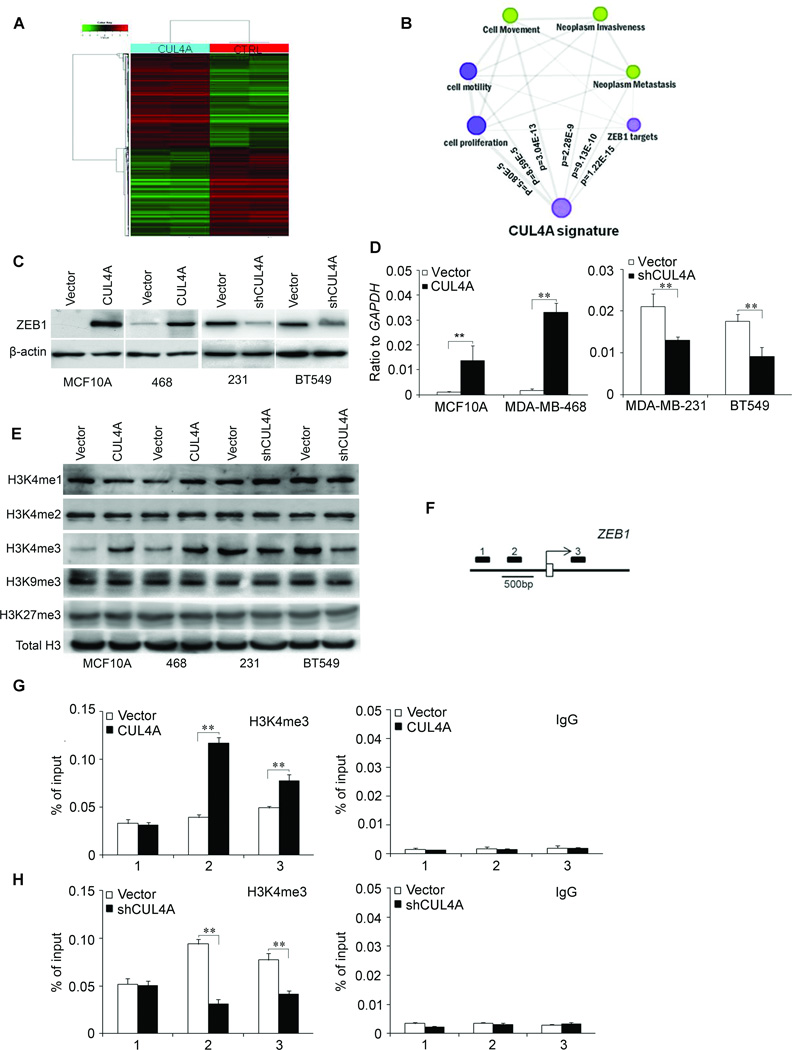
To better understand the mechanisms by which CUL4A engaged in BC development and progression, we performed gene expression profiling on MDA-MB-468-CUL4A and its control cells. Microarray analysis identified a list of genes significantly differentially expressed after CUL4A overexpression including upregulation of ZEB1 (Fig. 6A; Supplemental Table 1). Furthermore, gene set enrichment analysis indicated that proliferation, neoplasm metastasis and invasion, cell movement and motility, and ZEB1 related gene signatures (28) were significantly enriched in CUL4A overexpression cells (Fig. 6B), supporting the idea that CUL4A regulates proliferation, EMT and cancer invasion and metastasis. These data also led us to hypothesize that CUL4A exerts these functions possibly via ZEB1. To test this, we first determined whether ZEB1 is a downstream target of CUL4A in BC cells. Expression of ZEB1 in the cells with altered CUL4A expression was further evaluated by Western blotting and qRT-PCR. MCF10A-CUL4A and MDA-MB-468-CUL4A cells exhibited greatly increased both ZEB1 protein and mRNA levels, whereas silencing CUL4A in MDA-MB-231 and BT549 cells dramatically decreased its protein and mRNA levels (Fig. 6C and D). Similar observations were found in xenograft tumors from MDA-MB-468-CUL4A and MDA-MB-231-shCUL4A cells (Supplemental Fig. 5B and C), suggesting the regulation of ZEB1 expression by CUL4A is at transcriptional level. This was confirmed by ZEB1 gene promoter luciferase assay (Supplemental Fig. 5A).
Figure 6.
CUL4A regulates ZEB1 transcriptional expression through H3K4 trimethylation. A, Supervised hierarchical clustering of the genes differentially expressed after CUL4A overexpression. B, Gene set enrichment analysis was carried out using ConceptGen. Edge indicates significant overlap between two gene sets. The P values for enrichment between CUL4A signature and others determined with ConceptGen are shown. C and D, Protein and mRNA levels of ZEB1 were measured in BC cells with CUL4A overexpression or silencing by Western blotting (C) and qRT-PCR (D) assay. E, The abundance of H3 lysine methylation was assessed by Western blotting using whole cell lysate; total H3 was used as a loading control. F, Schematic presentation of three regions relative to the ZEB1 transcriptional start site used as primers to test histone occupied abundance. G and H, qChIP was performed to assess H3K4me3 occupancy in MDA-MB-468-CUL4A (G) and MDA-MB-231-shCUL4A (H) cells. IgG was used as negative control (G and H, left panels). “Percentage of input” indicates the ratio of DNA fragment of each promoter region bound by H3K4me3 to the total amount of input DNA fragment without H3K4me3 antibody pull-down. ** P < 0.01 in (D, G and H) is based on Student's t-test. All experiments except expression array are repeated three or four times. Error bars indicate standard deviation.
We then explored how CUL4A regulates ZEB1 expression at the transcriptional level. Cullin-RING ligase complexes are frequently involved in chromatin regulation (29, 30). To determine whether CUL4A regulates specific histone modifications in BC cells, histone modification patterns were measured after modulation of CUL4A expression. Among histone H3K4, H3K9 and H3K27, we found that only H3K4me3 was affected by CUL4A expression (Fig. 6E). Ectopic expression of CUL4A increased H3K4me3 while silencing CUL4A decreased this modification.
Because H3K4me3 is associated with active transcription, we tested whether CUL4A expression was correlated with the H3K4me3 modification at the ZEB1 gene promoter in BC cells. Quantitative ChIP (qChIP) assay was performed in MDA-MB-468-CUL4A and MDA-MB-231-shCUL4A cells. We found that CUL4A expression was associated with increased H3K4me3 levels at region −616 to −371bp and +239 to +464 bp of the ZEB1 promoter in MDA-MB-468-CUL4A cells (Fig. 6F and G). Less occupancy of those ZEB1 gene promoter regions by H3K4me3 was detected in MDA-MB-231-shCUL4A cells (Fig. 6H). The occupancy of chromatin repressors such as methylated H3K9 and H3K27 at the ZEB1 gene promoter was not changed by altered CUL4A expression (Supplemental Fig. 5D). These results clearly indicate that CUL4A induces transcriptional activation of ZEB1 through regulating H3K4me3 and enriching H3K4me3 to the ZEB1 gene promoter.
ZEB1 is a mediator for CUL4A-induced EMT, migration, invasion, and tumor metastasis
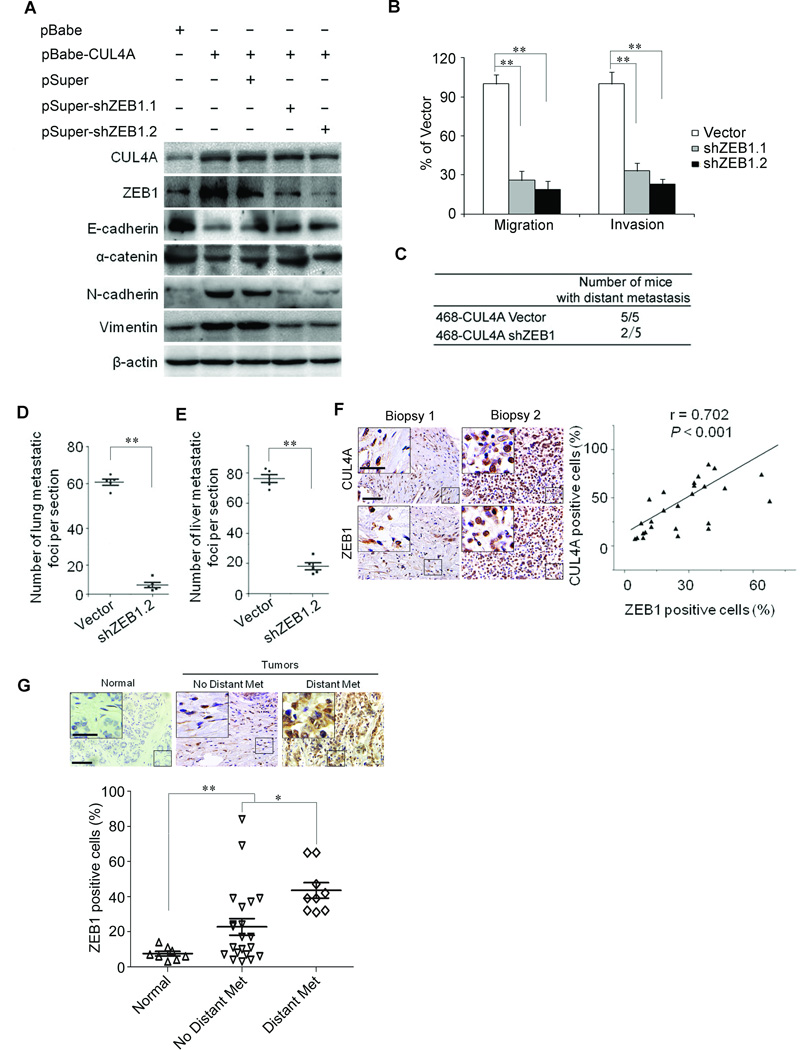
To test whether CUL4A-induced metastatic capacity was mediated by ZEB1, shRNAs were used to silence ZEB1 gene expression by virally transfecting MDA-MD-468-CUL4A cells with two distinct ZEB1 shRNAs (Fig. 7A). Knockdown of ZEB1 in MDA-MB-468-CUL4A cells resulted in increase in epithelial marker expression and decrease in mesenchymal marker expression at protein (Fig. 7A) and transcriptional levels (Supplemental Fig. 6A), and was accompanied with the reduction of migratory and invasive capacities (Fig. 7B; Supplemental Fig. 6B). Taken together, these results show that ZEB1 mediates CUL4A-induced EMT, migration and invasion in breast cancer cells.
Figure 7.
ZEB1 mediates CUL4A-induced EMT and metastasis. A, Silencing ZEB1 restored the epithelial marker expression and decreased mesenchymal markers in MDA-MB-468-CUL4A cells. B, Silencing ZEB1 inhibited CUL4A-driven transwell migration and Matrigel invasion in MDA-MB-468-CUL4A cells. C, The total numbers of mice with distant metastasis at 60 days after injection of MDA-MB-468-CUL4A cells with or without silencing ZEB1 into tail vein. 468 in (C) indicates MDA-MB-468 cell line. D and E, The numbers of metastatic foci per section in lung (D) and liver (E) of individual mouse with injection of MDA-MB-468-CUL4A cells with or without silencing ZEB1. F, ZEB1 expression was positively correlated with CUL4A expression in BC tissue arrays. Left panel showed the representive stainings of CUL4A and ZEB1. G, Correlation of ZEB1 expression in primary BC tissues with metastasis was analyzed using BC tissue array as describe in Fig. 1. Upper panel showed the representive staining of ZEB1. Scale bars indicate 50 µm (F and G) and 20 µm in the inserts (F and G). * P < 0.05 and ** P < 0.01 are based on Student's t-test. P < 0.001 in (F) was obtained from Pearson correlation test. All experiments except tissue array analysis are repeated at least three times. Error bars indicate standard deviation.
To verify if ZEB1 eventually mediates CUL4A-induced metastasis in vivo, MDA-MB-468-CUL4A cells with or without silencing ZEB1 were injected into nude mice through the tail vein. Silencing ZEB1 not only significantly decreased the number of mice with distant metastasis (Fig. 7C), but also dramatically decreased the number of metastatic tumors in both lung and liver of each mouse (Fig. 7D and E; Supplemental Fig. 6C). Therefore, the in vivo results further demonstrated the critical role of ZEB1 in mediating CUL4A-promoted metastatic behavior in BC cells.
To recognize any clinical correlation of CUL4A and ZEB1, we analyzed ZEB1 expression in the same human BC tissue microarray. Highly positive correlation between CUL4A and ZEB1 expression was drawn (Fig. 7F). Similar to CUL4A, ZEB1 was highly expressed in BC tissues compared with their adjacent normal breast tissues, and high level of ZEB1 was significantly correlated to distant metastasis (Fig. 7G). This result is consistent with and further supports our above in vitro and in vivo analyses.
DISCUSSION
To our knowledge, this is the first study to show that CUL4A plays a functional role in metastasis. CUL4A overexpression in BC cells induced proliferation, EMT, migration, invasion in vitro and enhanced tumorigenic and metastatic capacities in vivo. In contrast, silencing CUL4A reversed these events in otherwise aggressive and invasive BC cells. We also showed a mechanistic link between CUL4A and ZEB1 through CUL4A-mediated regulation of H3K4m3, which subsequently leads to transcriptional upregulation of ZEB1 expression. Knockdown of ZEB1 attenuated CUL4A function and had effects similar to those elicited by direct silencing CUL4A.
The putative role of CUL4A as an oncogene in cancer development is supported by the observations that CUL4A is highly expressed in BCs and other malignant tumors relative to normal tissues (4, 9, 10). The fact that CUL4A ubiquitinates and degrades several well-known tumor suppressor genes (18–20, 31) lends further support to, and potential mechanistic insight into, the possibility of CUL4A as an oncogene. Perhaps the most convincing evidence is the genetic study showing that deletion of Cul4a in mouse resulted in dramatically increased resistance to UV-induced skin carcinogenesis (18). Consistent with these reports, we showed that CUL4A overexpression promoted BC cell proliferation and enhanced tumor formation in vivo. Interestingly, our study points to a novel function of CUL4A in BC metastasis through regulating EMT.
First, BC cells with ectopic expression of CUL4A displayed an EMT phenotype, including the associated stimulatory effects on migration and invasion in vitro. Interestingly, our results indicate that CUL4A not only promotes EMT, but silencing CUL4A also leads to MET. This observation suggests that EMT-MET is a fluid process. Consistent with the notion that EMT is essential for tumor cells to disseminate from adjacent tissues and seed new tumors in distant sites, all of these characteristics induced by CUL4A in vitro culminated to increased numbers of distant metastases in vivo. These experimental findings provide a mechanistic framework to explain the clinical observations that BC patients with high levels of CUL4A in tissues have higher probabilities of distant metastasis and significant shorter overall and disease-free survivals (16), and that amplification of CUL4A-containing 13q34 region is associated with aggressive basal subtype of BCs (4).
The roles of several transcription factors as EMT regulators have been extensively reported. In our effort to elucidate the mechanism how CUL4A modulates EMT in BC cells, we identified ZEB1 as an effective mediator of these CUL4A-induced phenomena. The mechanistic connection between CUL4A and ZEB1 was previously unknown, indeed, regulation of ZEB1 is not well understood. In this study, we showed that modulation of CUL4A expression altered the methylation status of H3K4 at the ZEB1 gene promoter, which in turn transcriptionally controlled ZEB1expression. However, we did not detect any influence of CUL4A expression on the methylation status of H3K9 and H3K27, nor did we find recruitment of H3K9me3 and H3K27me3 on ZEB1 gene promoter. Thus, we conclude that CUL4A transcriptionally activates ZEB1 expression through regulation of H3K4 trimethylation and recruitment of H3K4me3 to ZEB1 gene promoter, and consequently promotes EMT in vitro and metastasis in vivo. How CUL4A modulates H3K4me3 remains further clarification, a few reports suggest CUL4A regulates H3K4me3 possibly through a substrate-specific adaptor WDR5 (30), which is an essential component of the MLL histone methylation complexes that catalyze the critical trimethylation at H3K4 (32, 33).
Our observations that silencing CUL4A in aggressive BC cells with high level of CUL4A dramatically blocked tumor growth and metastasis in vivo provide us a therapeutic option by targeting CUL4A in clinical practice. Considering that CUL4A is highly expressed in all breast, ovarian, colorectal, and gastric cancers with distant metastasis we analyzed, and that the vast majority of BC patients succumb to their disease as a result of distant metastasis (21, 22) together with our previous report that tumor cells with high CUL4A levels are sensitive to thalidomide treatment (12), our studies suggest a promising therapeutic target in certain subtypes of BCs and probably in other types of metastatic malignant tumors.
Supplementary Material
Acknowledgments
Financial support:
This work was supported by National Natural Science Foundation of China No. 81172528, 31271461, Doctoral Fund of Ministry of Education of China No. 20110131110035, Natural Science Foundation of Shandong Province No. ZR2011HM034 and Taishan Scholar Program of Shandong Province (GW); by National Institutes of Health, National Cancer Institute grant R01 CA116481, and Low Dose Scientific Focus Area, Office of Biological & Environmental Research, US Department of Energy (DE-AC02-05CH11231) (JHM).
Footnotes
Disclosure of Potential Conflicts of Interest:
The authors disclose no potential conflicts of interest.
REFERENCES
- 1.Forouzanfar MH, Foreman KJ, Delossantos AM, Lozano R, Lopez AD, Murray CJ, et al. Breast and cervical cancer in 187 countries between 1980 and 2010: a systematic analysis. Lancet. 2011;378:1461–1484. doi: 10.1016/S0140-6736(11)61351-2. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 3.Di Cosimo S, Baselga J. Management of breast cancer with targeted agents: importance of heterogeneity. [corrected] Nat Rev Clin Oncol. 2010;7:139–147. doi: 10.1038/nrclinonc.2009.234. [DOI] [PubMed] [Google Scholar]
- 4.Melchor L, Saucedo-Cuevas LP, Munoz-Repeto I, Rodriguez-Pinilla SM, Honrado E, Campoverde A, et al. Comprehensive characterization of the DNA amplification at 13q34 in human breast cancer reveals TFDP1 and CUL4A as likely candidate target genes. Breast Cancer Res. 2009;11:R86. doi: 10.1186/bcr2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carey L, Winer E, Viale G, Cameron D, Gianni L. Triple-negative breast cancer: disease entity or title of convenience? Nat Rev Clin Oncol. 2010;7:683–692. doi: 10.1038/nrclinonc.2010.154. [DOI] [PubMed] [Google Scholar]
- 6.Knosel T, Petersen S, Schwabe H, Schluns K, Stein U, Schlag PM, et al. Incidence of chromosomal imbalances in advanced colorectal carcinomas and their metastases. Virchows Arch. 2002;440:187–194. doi: 10.1007/s004280100493. [DOI] [PubMed] [Google Scholar]
- 7.Isinger-Ekstrand A, Johansson J, Ohlsson M, Francis P, Staaf J, Jonsson M, et al. Genetic profiles of gastroesophageal cancer: combined analysis using expression array and tiling array--comparative genomic hybridization. Cancer Genet Cytogenet. 2010;200:120–126. doi: 10.1016/j.cancergencyto.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 8.Abba MC, Fabris VT, Hu Y, Kittrell FS, Cai WW, Donehower LA, et al. Identification of novel amplification gene targets in mouse and human breast cancer at a syntenic cluster mapping to mouse ch8A1 and human ch13q34. Cancer Res. 2007;67:4104–4112. doi: 10.1158/0008-5472.CAN-06-4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yasui K, Arii S, Zhao C, Imoto I, Ueda M, Nagai H, et al. TFDP1, CUL4A, and CDC16 identified as targets for amplification at 13q34 in hepatocellular carcinomas. Hepatology. 2002;35:1476–1484. doi: 10.1053/jhep.2002.33683. [DOI] [PubMed] [Google Scholar]
- 10.Chen LC, Manjeshwar S, Lu Y, Moore D, Ljung BM, Kuo WL, et al. The human homologue for the Caenorhabditis elegans cul-4 gene is amplified and overexpressed in primary breast cancers. Cancer Res. 1998;58:3677–3683. [PubMed] [Google Scholar]
- 11.Shinomiya T, Mori T, Ariyama Y, Sakabe T, Fukuda Y, Murakami Y, et al. Comparative genomic hybridization of squamous cell carcinoma of the esophagus: the possible involvement of the DPI gene in the 13q34 amplicon. Genes Chromosomes Cancer. 1999;24:337–344. [PubMed] [Google Scholar]
- 12.Ren S, Xu C, Cui Z, Yu Y, Xu W, Wang F, et al. Oncogenic CUL4A determines the response to thalidomide treatment in prostate cancer. J Mol Med (Berl) 2012;90:1121–1132. doi: 10.1007/s00109-012-0885-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hung MS, Mao JH, Xu Z, Yang CT, Yu JS, Harvard C, et al. Cul4A is an oncogene in malignant pleural mesothelioma. J Cell Mol Med. 2011;15:350–358. doi: 10.1111/j.1582-4934.2009.00971.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Michiels EMC, Weiss MM, Hoovers JMN, Baak JPA, Voute PA, Baas F, et al. Genetic alterations in childhood medulloblastoma analyzed by comparative genomic hybridization. J Pediat Hematol Onc. 2002;24:205–210. doi: 10.1097/00043426-200203000-00009. [DOI] [PubMed] [Google Scholar]
- 15.Dohna M, Reincke M, Mincheva A, Allolio B, Solinas-Toldo S, Lichter P. Adrenocortical carcinoma is characterized by a high frequency of chromosomal gains and high-level amplifications. Gene Chromosome Canc. 2000;28:145–152. [PubMed] [Google Scholar]
- 16.Schindl M, Gnant M, Schoppmann SF, Horvat R, Birner P. Overexpression of the human homologue for Caenorhabditis elegans cul-4 gene is associated with poor outcome in node-negative breast cancer. Anticancer Res. 2007;27:949–952. [PubMed] [Google Scholar]
- 17.Birner P, Schoppmann A, Schindl M, Dinhof C, Jesch B, Berghoff AS, et al. Human homologue for Caenorhabditis elegans CUL-4 protein overexpression is associated with malignant potential of epithelial ovarian tumours and poor outcome in carcinoma. J Clin Pathol. 2012;65:507–511. doi: 10.1136/jclinpath-2011-200463. [DOI] [PubMed] [Google Scholar]
- 18.Liu L, Lee S, Zhang J, Peters SB, Hannah J, Zhang Y, et al. CUL4A abrogation augments DNA damage response and protection against skin carcinogenesis. Mol Cell. 2009;34:451–460. doi: 10.1016/j.molcel.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bondar T, Kalinina A, Khair L, Kopanja D, Nag A, Bagchi S, et al. Cul4A and DDB1 associate with Skp2 to target p27Kip1 for proteolysis involving the COP9 signalosome. Mol Cell Biol. 2006;26:2531–2539. doi: 10.1128/MCB.26.7.2531-2539.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gupta A, Yang LX, Chen L. Study of the G2/M cell cycle checkpoint in irradiated mammary epithelial cells overexpressing Cul-4A gene. Int J Radiat Oncol Biol Phys. 2002;52:822–830. doi: 10.1016/s0360-3016(01)02739-0. [DOI] [PubMed] [Google Scholar]
- 21.Grayson M. Breast cancer. Nature. 2012;485:S49. doi: 10.1038/485S49a. [DOI] [PubMed] [Google Scholar]
- 22.Rice J. Metastasis: The rude awakening. Nature. 2012;485:S55–S57. doi: 10.1038/485S55a. [DOI] [PubMed] [Google Scholar]
- 23.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nature reviews. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 24.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 25.Singh AP, Moniaux N, Chauhan SC, Meza JL, Batra SK. Inhibition of MUC4 expression suppresses pancreatic tumor cell growth and metastasis. Cancer Res. 2004;64:622–630. doi: 10.1158/0008-5472.can-03-2636. [DOI] [PubMed] [Google Scholar]
- 26.Castillo SD, Angulo B, Suarez-Gauthier A, Melchor L, Medina PP, Sanchez-Verde L, et al. Gene amplification of the transcription factor DP1 and CTNND1 in human lung cancer. J Pathol. 2010;222:89–98. doi: 10.1002/path.2732. [DOI] [PubMed] [Google Scholar]
- 27.Ang YS, Tsai SY, Lee DF, Monk J, Su J, Ratnakumar K, et al. Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell. 2011;145:183–197. doi: 10.1016/j.cell.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aigner K, Dampier B, Descovich L, Mikula M, Sultan A, Schreiber M, et al. The transcription factor ZEB1 (deltaEF1) promotes tumour cell dedifferentiation by repressing master regulators of epithelial polarity. Oncogene. 2007;26:6979–6988. doi: 10.1038/sj.onc.1210508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jackson S, Xiong Y. CRL4s: the CUL4-RING E3 ubiquitin ligases. Trends Biochem Sci. 2009;34:562–570. doi: 10.1016/j.tibs.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Higa LA, Wu M, Ye T, Kobayashi R, Sun H, Zhang H. CUL4-DDB1 ubiquitin ligase interacts with multiple WD40-repeat proteins and regulates histone methylation. Nat Cell Biol. 2006;8:1277–1283. doi: 10.1038/ncb1490. [DOI] [PubMed] [Google Scholar]
- 31.Kopanja D, Stoyanova T, Okur MN, Huang E, Bagchi S, Raychaudhuri P. Proliferation defects and genome instability in cells lacking Cul4A. Oncogene. 2009;28:2456–2465. doi: 10.1038/onc.2009.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Couture JF, Collazo E, Trievel RC. Molecular recognition of histone H3 by the WD40 protein WDR5. Nat Struct Mol Biol. 2006;13:698–703. doi: 10.1038/nsmb1116. [DOI] [PubMed] [Google Scholar]
- 33.Schuetz A, Allali-Hassani A, Martin F, Loppnau P, Vedadi M, Bochkarev A, et al. Structural basis for molecular recognition and presentation of histone H3 by WDR5. Embo J. 2006;25:4245–4252. doi: 10.1038/sj.emboj.7601316. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.