Abstract
Amyotrophic lateral sclerosis (ALS) is a fatal, late-onset neurodegenerative disease primarily impacting motor neurons. A unifying feature of many proteins associated with ALS, including TDP-43 and Ataxin-2, is that they localize to stress granules. Unexpectedly, we found that genes that modulate stress granules are striking modifiers of TDP-43 toxicity in Saccharomyces cerevisiae and Drosophila melanogaster, eIF2α phosphorylation is upregulated by TDP-43 toxicity in flies, and TDP-43 interacts with a central stress granule component polyA binding protein (PABP). In human ALS spinal cord neurons, PABP accumulates abnormally, suggesting that prolonged stress granule dysfunction may contribute to pathogenesis. We investigated the efficacy of a small molecule inhibitor of eIF2α-phosphorylation in ALS models. This treatment mitigated TDP-43 toxicity in flies and mammalian neurons. These findings indicate that dysfunction induced by prolonged stress granule formation may contribute directly to ALS and that compounds that mitigate this process may represent a novel therapeutic approach.
INTRODUCTION
Amyotrophic Lateral Sclerosis (ALS) is an intractable neurodegenerative disease that affects upper and lower motor neurons, resulting in debilitating and lethal paralysis 1,2. There is no cure and only one effective treatment, riluzole, which extends life by only 3 months on average. Thus, novel therapeutic strategies are desperately needed. The genetic underpinnings of sporadic and familial ALS are heterogeneous and incompletely characterized, but almost all cases of ALS share a pathological endpoint: ubiquitin-positive cytoplasmic inclusions that contain the RNA binding protein TDP-43 3. Focus on TDP-43 and other RNA-binding proteins has intensified as mutations in members of an expanding family of RNA recognition motif-containing proteins with prion-like domains, FUS/TLS, TAF15, EWSR1, hnRNPA2B1, and hnRNPA1, have been found in ALS patients 4–7. Efforts to identify the molecular functions of TDP-43 that may be relevant to its role in proteinopathy have elucidated several roles in RNA processing 8, including a possible function in the formation and regulation of RNA granules that form in conditions of stress 9–11.
When primary neurons and cultured cells are stressed, TDP-43 translocates from the nucleus to cytoplasmic stress granules 11,12. Stress granules are thought to be protective during cellular stress, harboring translationally arrested poly(A)+ mRNA, polyA-binding protein (PABP), 40S ribosomal subunits, and eukaryotic initiation factors 13. The importance of nuclear-to-cytosolic trafficking of TDP-43 in stressed cells is not yet clear. The parallels between these observations in cells and the cytoplasmic mis-localization of TDP-43 and inclusion formation in patient spinal cord tissue has raised the possibility that stress granule formation may represent sites of early protein aggregation in ALS 14–17. Prolonged stress granule activity is predicted to lead to a prolonged stressed state and prolonged translational repression, which would be deleterious 15,18–20. In prion disease models, sustained translational repression by eIF2α phosphorylation mediates neurodegeneration 21,22.
To define toxic features of TDP-43 that may be relevant to human disease, we have used yeast and fly models coupled with human patient tissue analysis. Yeast and fly have proven to be powerful model systems for revealing mechanistic insight into several neurodegenerative diseases, including ALS, Parkinson, Huntington, and Alzheimer diseases 23–28. High throughput screens in yeast, coupled with functional testing in the Drosophila nervous system, have been highly successful at identifying modifiers of disease genes with findings that extend to the human condition 5,25. Here we find that a genome-wide yeast screen revealed genes in RNA metabolism, including several RNA-binding proteins with connections to stress granules, as critical to TDP-43 toxicity. Pursuing this, we unexpectedly found that eiF2α-phosphorylation, indicative of stress granules and translational repression, becomes abnormally upregulated upon TDP-43-associated neurodegeneration in Drosophila, and key genes that modulate eiF2α–phosphorylation dramatically affect the ability of TDP-43 to be neurotoxic. We pursued these studies to reveal a role of polyA binding protein, then extended these studies to show that a small molecule that mitigates eiF2α-phosphorylation may provide a promising avenue for therapeutic intervention in disease.
RESULTS
Yeast reveals a role for stress granule components in ALS
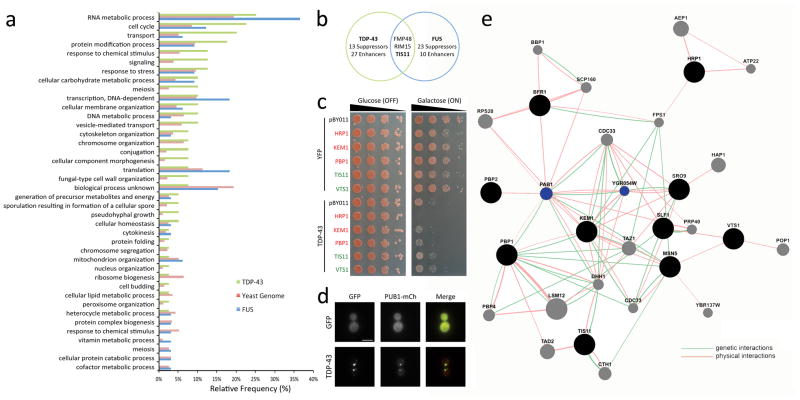
We previously developed a TDP-43 yeast model that recapitulates key features of TDP-43 associated with disease including toxicity and aggregation 29,30. Using this model, we performed a screen for genes that when upregulated would modify TDP-43 toxicity 25,29–31. A yeast query strain carrying an integrated copy of TDP-43 under the control of a galactose inducible promoter was transformed with the yeast FLEX gene library, which contains an arrayed set of 5500 genes that are overexpressed upon galactose induction 24,32. Transformed yeast cells were subsequently plated on glucose (TDP-43 expression OFF) and galactose plates (TDP-43 expression ON), and screened for genes that suppressed or enhanced TDP-43 toxicity. We identified 40 hits, which included 13 suppressors and 27 enhancers of TDP-43 toxicity (Table 1). GO term analysis on the entire set of screen hits showed a striking enrichment in RNA metabolic process, cell cycle, transport and protein modification process genes (Fig. 1a). While there was little overlap between the hits found in this TDP-43 screen and in a previous FUS overexpression screen 33, both screens were enriched for RNA metabolic processes, and to a lesser extent cell cycle genes (Fig. 1a,b). We performed the screen in triplicate, and hits confirmed all three times were independently verified with more sensitive spotting assays (Fig. 1c).
Table 1.
Genes that suppress or enhance TDP-43 toxicity in yeast when overexpressed
| Effect | Gene | Human Homolog | Description |
|---|---|---|---|
| Suppressor | ADY3 | CENPE | Protein wall formation |
| Suppressor | BFR1 | Component of mRNP complexes associated with polyribosomes | |
| Suppressor | CYC8 | Transcription co-repressor; part of complex that recruits SWI/SNF and SAGA complexes to promoters | |
| Suppressor | FMP48 | STK36 | Protein of unknown function |
| Suppressor | HSP104 | Heat shock protein chaperon | |
| Suppressor | ICS2 | Protein of unknown function | |
| Suppressor | NNK1 | DCLK1 | Protein kinase; interacts with TORC1, Ure2p and Gdh2p |
| Suppressor | PGM1 | PGM1 | Phosphoglucomutase |
| Suppressor | RDR1 | Transcriptional repressor | |
| Suppressor | RIM15 | STK38 | Glucose-repressible protein kinase |
| Suppressor | TIS1 | ZNF36/TTP | mRNA-binding protein; component of stress granules |
| Suppressor | VTS1 | SAM4B/Smaug | RNA-binding protein containing a SAM domain; component of P granules |
| Suppressor | XRS2 | Protein required for DNA repair | |
| Enhancer | CDC6 | CDC6 | Essential ATP-binding protein required for DNA replication |
| Enhancer | DIP5 | SLC7A7 | Dicarboxylic amino acid permease |
| Enhancer | HRP1 | Musashi 1 & 2 | RNA binding protein; component of stress granules |
| Enhancer | KEL1 | RAB9 | Protein required for proper cell fusion and cell morphology |
| Enhancer | KEM1 | XRN1 | 5′-3′ exonuclease component of P bodies |
| Enhancer | KIN3 | NEK2 | Serine/threonine protein kinase |
| Enhancer | MEC1 | ATR | Genome integrity checkpoint protein |
| Enhancer | MSA1 | Mucin 17 | Involved in regulation of timing of G1-specific gene transcription and cell cycle initiation |
| Enhancer | MSN5 | XPO5 | Karyopherin involved in nuclear import and export |
| Enhancer | MTH1 | Negative regulator of the glucose-sensing signal transduction pathway | |
| Enhancer | PBP1 | Ataxin 2 | Interacts with Pab1p; component of stress granules |
| Enhancer | PBP2 | PCBP1, 2, 3, & 4 | RNA binding protein |
| Enhancer | PCL6 | Pho85p cyclin of the Pho80p subfamily | |
| Enhancer | PIB2 | WDFY3 | Protein binding phosphatidylinositol 3-phosphate |
| Enhancer | RGA2 | ARHGAP15 | GTPase-activating protein for Cdc42p |
| Enhancer | ROM2 | NET1 | GDP/GTP exchange protein (GEP) for Rho1p and Rho2p |
| Enhancer | SAK1 | CAMKK1 | Upstream kinase for the SNF1 complex |
| Enhancer | SFG1 | Putative transcription factor | |
| Enhancer | SLF1 | LARP1 | RNA binding protein that associates with polysome |
| Enhancer | SLG1 | Sensor-transducer of the stress-activated PKC-MPK1 kinase pathway | |
| Enhancer | SOL1 | PGLS | Protein with possible role in tRNA transport |
| Enhancer | SRO9 | LARP2 | RNA-binding protein that associates with translating ribosomes |
| Enhancer | TSC11 | RICTOR | Subunit of TORC2 |
| Enhancer | UBP7 | USP21 | Ubiquitin-specific protease |
| Enhancer | VHS1 | MARK2 | Cytoplasmic serine/threonine kinase |
| Enhancer | YCK2 | CSNK1G2 | Casein kinase 1 isoform |
| Enhancer | YHR131C | Putative protein of unknown function |
Descriptive information from the Saccharomyces Gene Database, www.yeastgenome.org
Figure 1. Yeast plasmid overexpression screen highlights the role of stress granules in TDP-43 toxicity.
a) Histogram showing the functional categories of genes (GO term process) with the relative frequency of genes from the TDP-43 overexpression screen compared to the yeast genome and to the FUS overexpression screen 33. Both the TDP-43 and FUS screens were enriched for RNA metabolic process genes, while the TDP-43 screen was enriched for cell cycle, transport, and protein modification process genes.
b) While the TDP-43 and FUS screens were both enriched for RNA metabolic process genes, most of the hits did not overlap with the exception of three genes.
c) Stress granule genes identified in the screen enhance or suppress TDP-43 toxicity. Spotting assay showing that on the galactose plate (expression ON) the co-expression of HRP1, KEM1, or PBP1 with TDP-43 leads to enhanced toxicity (reduced growth) while the co-expression of TIS11 or VTS1 with TDP-43 leads to suppression of toxicity (increased growth).
d) RNA-binding protein focused yeast interaction network for TDP-43 screen hits reveal connections to PAB1 and EIF2A homolog. Ten out of forty yeast genes that modified TDP-43 toxicity when overexpressed are annotated as RNA-binding proteins. These are displayed as black circles. GeneMANIA 56 was used to search for interacting genes based physical and genetic interactions. Interacting genes are displayed as grey circles and network edges are colored based on the type of interaction (red=physical interaction and green=genetic interaction). This analysis revealed strong connections to PAB1 and YGR054W, which are highlighted in blue. YGR054W is the yeast homolog of a human translation initiation factor EIF2A.
e) GFP-tagged ALS-linked disease gene TDP-43 forms aggregates when expressed in yeast that colocalize with the stress granule marker PUB1-mCherry. Scale bar, 5 μm.
Analysis of the TDP-43 screen hits revealed that stress granule and P-body components (involved in RNA triage and degradation 34) were significantly enriched (Table 1; 5 out of 40 hits, 12.5%, versus 50 out of 6311 genes in the yeast genome, 0.8%; P=1x10−4). These included both enhancers and suppressors of toxicity (Fig. 1b,c). Among the genes was PBP1, the yeast homolog of Ataxin-2, which we previously defined as a protein that synergizes with TDP-43 in promoting toxicity, and whose polyQ repeat expansions are a genetic contributor to ALS 25,35–37. Because of the importance of RNA metabolic processes in ALS pathogenesis 36 and the enrichment of RNA-binding proteins as modifiers, we performed a network analysis of the identified RNA-binding proteins, defining genetic and physical interactions between hits and additional genes that interacted with the hits (Fig. 1d). Strikingly, this analysis revealed multiple interactions between genes from the screen and PAB1, which encodes polyA-binding protein. Additionally, a previously uncharacterized yeast gene, YGR054W, was identified as interacting physically and genetically with several of the TDP-43 modifier genes (Fig. 1d); YGR054W encodes the yeast homolog of a translation initiation factor EIF2A 38. This network analysis and the additional interactors predicted by it helped us to focus our further analyses on PAB1 and translation initiation factors (see below).
Given these findings, we then asked whether TDP-43 colocalized with the stress granule marker PUB1. These studies revealed that TDP-43 accumulations in yeast co-localize with PUB1 (Fig. 1e). Stress granule and P-body markers have been found to colocalize with TDP-43 aggregates in patient samples and multiple models of disease 10,39, the results of this unbiased genetic screen indicate that stress granule components, and the stressed state induced by such components, may play a direct role in TDP-43 toxicity, as modifying their expression impacts TDP-43 toxicity. Thus, not only are RNA granules markers of TDP-43 pathology in disease, but they may play a more direct role in neurodegenerative disease pathogenesis.
Stress granule genes modulate TDP-43 toxicity in the fly
To test the significance of the interaction between stress granules and ALS-associated RNA binding protein toxicity in the nervous system, we used Drosophila. Although it is challenging to image stress granules in vivo in flies, eIF2α-phosphorylation induces the accumulation of non-functional translation initiation complexes that concentrate in stress granules, thus levels of eIF2α-phosphorylation are directly correlated with the levels of stress granules 13. We extracted protein from heads of control and TDP-43-expressing animals, and immunoblotted extracts with a phospho-specific eIF2α antibody (Ser51). This approach revealed a progressive increase in eIF2α-phosphorylation upon expression of TDP-43 in the brain: there was no difference between control and TDP-43-expressing flies in the levels of eIF2α-phosphorylation at 5d, but by 8d and 14d, the levels of eIF2α-phosphorylation were significantly increased to 1.4±0.1-fold (s.e.m.), and 1.6±0.1-fold (s.e.m.), respectively (Fig. 2a). There was no observed increase in levels of eIF2α protein, showing that the change in phospho-eIF2α levels represented a change in the stress granule specific form (Supplementary Fig. 1). These data suggest that TDP-43 expression in the fly brain induces chronic eIF2α-phosphorylation. Moreover, the increase in eIF2α-phosphorylation indicates a state of prolonged translational repression 22.
Figure 2. Genes that impact stress granule formation modulate TDP-43 toxicity.
a) TDP-43 expression increases eIF2α-phosphorylation levels. Genes were expressed in the nervous system in a drug-inducible manner with the elavGS driver. eIF2α- phosphorylation level of elavGS/UAS-YFP and elavGS, TDP-43/UAS-YFP flies fed as adults on RU486 (40μg/ml), and assessed at the indicated time-points. Genotypes: Control is elavGS/UAS-YFP. TDP-43 is elavGS, UAS-TDP-43(S)/UAS-YFP. Mean ± s.e.m., n=3 independent experiments. *p<0.05, *** p<0.001, n.s., not significant. (Student’s t-test).
b) Altering the levels of genes that reduce stress granule formation mitigates TDP-43 toxicity, and that promote stress granule formation enhances TDP-43 toxicity. Mean ± 95% CI of four experiments. Genotypes: elavGS/UAS-YFP is elavGS/UAS-YFP. elavGS, TDP-43/UAS-YFP is elavGS, UAS-TDP-43(S)/UAS-YFP. elavGS, TDP-43/Gadd34 RNAi is elavGS, UAS-TDP-43(S)/UAS-Gadd34. RNAiHMS00811. elavGS, TDP-43/Rox8 RNAi is elavGS, UAS-TDP-43(S)/UAS-Rox8. RNAiHMS00472. elavGS, TDP-43/PEK RNAi is elavGS, UAS-TDP-43(S)/UAS-PEK. RNAiGL00030. All flies raised with RU486 (40μg/ml) (125 flies per genotype). ANOVA for significance, followed by Tukey’s multiple comparison test, **p<0.01, *** p<0.001, # p<0.0001, n.s., not significant.
c) eIF2α-phosphorylation level of elavGS, TDP-43/UAS-YFP, elavGS, TDP-43/Rox8 RNAi, elavGS, TDP-43/Gadd34 RNAi and elavGS, TDP-43/PEK RNAi. Mean ± s.e.m., n=3 independent experiments. *p<0.05, n.s., not significant (Student’s t-test).
d) Total, nuclear and cytosolic TDP-43 protein level in 10d fly heads. Genes predicted to increase stress granules formation increase cytoplasmic TDP-43 protein levels. Mean ± s.e.m., n=3 independent experiments. *** p<0.001, n.s., not significant (Student’s t-test).
To determine whether modulation of this could have an effect on TDP-43-associated neurodegeneration, we examined whether altering the levels of key genes that impact eIF2α-phosphorylation could modulate TDP-43 toxicity in vivo. We knocked down the levels of PEK, Rox8, and Gadd34 by RNAi in the presence of TDP-43 and examined the progressive effects on TDP-43-induced climbing dysfunction. PEK is the homolog of mammalian PERK and is a kinase that phosphorylates eIF2α, Rox8 is the Drosophila homologue of TIA1 which facilitates the physical aggregation of stress granules, and Gadd34 is a phosphatase for eIF2α 40–43. In mammalian cell lines, loss of PERK blocks calcium-induced stress granule formation, stimulation of eIF2α-phosphorylation is necessary and sufficient for stress granule induction 44, expression of dominant negative TIA1 prevents stress granule formation 45, and GADD34 inhibitor treatment induces massive stress granule formation 46. Thus, reduction of PEK and Rox8 are predicted to inhibit, whereas knockdown of Gadd34 should enhance, stress granule formation in Drosophila.
Flies expressing TDP-43 show a markedly reduced climbing ability compared to normal animals aged to 14d (28±5% (95% CI) compared to 82±5% in controls, Fig. 2b). This climbing deficit was greatly suppressed by knockdown of either Rox8 or PEK, with 62±6% (95%CI) of flies retaining climbing ability; conversely, knockdown of Gadd34 led TDP-43-expressing flies to lose their motility almost entirely (Fig. 2b). Importantly, we confirmed that downregulation of PEK decreased eIF2α-phosphorylation, and that Gadd34 increased eIF2α-phosphorylation (Fig. 2c). Knockdown of Gadd34, PEK or Rox8 on their own had no effect on climbing abillity (data not shown). Intriguingly, Gadd34 reduction, which enhanced TDP-43 toxicity in our locomotion assay, also led to accumulation of TDP-43 protein in the cytoplasm (Fig. 2d). Taken together, these data indicate that TDP-43-induced neural toxicity can be dramatically enhanced and suppressed by altering genes that converge on eIF2α phosphorylation.
The TDP-43/Ataxin-2 interaction requires the PAM2 motif
To provide more insight into this potentially critical role of stress granules in TDP-43 toxicity, we investigated in greater detail the interaction between Ataxin-2 and TDP-43. Ataxin-2, like TDP-43, localizes to stress granules and regulates their assembly and functions 47,48. PolyQ expansions in Ataxin-2 are a risk for ALS, and the Ataxin-2 protein has several functional motifs including one for the key stress granule protein, PolyA Binding Protein (PABP) 49. We generated transgenic fly lines that express human Ataxin-2 with a normal length polyQ repeat (ATXN2-22Q), or an expanded repeat within the risk range for ALS (ATXN2-32Q). As with fly Ataxin-2 25, co-expression of either protein with TDP-43 dramatically enhanced toxicity, with faster loss of lifespan and climbing ability (Supplementary Fig. 2). Intriguingly, ATXN2-32Q enhanced the toxicity of TDP-43 measured by lifespan more significantly than ATXN2-22Q, demonstrating that an expansion of only ten glutamine repeats from normal into the ALS-associated range has a significant effect in vivo.
Ataxin-2 has two key domains implicated in RNA binding function (Fig. 3a): the Lsm (like Sm) domain is important for RNA binding affinity 50, whereas the PAM2 motif mediates interaction with PABP 51. We generated domain deletion forms of ATXN2-32Q and selected transgenic lines that express these proteins at the same level as intact ATXN2-32Q. Expression of the deletion-bearing ATXN2-32Q proteins alone showed effects comparable to the intact ATXN2-32Q protein: a mild disruption of retinal structure and slightly reduced lifespan (Supplementary Fig. 3). When we then assessed the ability of these forms to enhance toxicity of TDP-43. Strikingly, whereas the ATXN2-Q32-ΔLSM protein acted the same as ATXN2-Q32, the ATXN2-32Q-ΔPAM2 protein completely lost the ability to interact with TDP-43 in retinal toxicity (Fig. 3b-c). These results were similar for lifespan and climbing ability, with the ATXN2-32Q-ΔPAM2 form showing almost complete loss of interaction with TDP-43 (Fig. 3). TDP-43 protein accumulation was also unaffected by ATXN2-32Q-ΔPAM2 (Fig. 3e), consistent with functional loss of the interaction. This finding raised the possibility that PABP, which binds the PAM2 motif, may have a role in TDP-43-induced neurodegeneration and that the shared interaction of TDP-43 and Ataxin-2 with PABP may represent a critical axis in stress granule formation and the biological effects of that stress.
Figure 3. The interaction between TDP-43 and Ataxin-2 is mediated through the polyA binding protein motif of Ataxin-2.
a) Schematic representation of the versions of Ataxin-2 generated and tested for interactions with TDP-43. Colored boxes indicate the putative RNA-binding Lsm domain, the Lsm-associated domain LsmAD, the PABP-interacting motif PAM2 and the polyQ stretch. Deletion region of ΔLSM is aa277-346, and ΔPAM2 is aa920-932 of ATXN2-32Q. Both domains are well conserved from yeast to mammal.
b) TDP-43 toxicity is not affected by co-expression of ATXN2 lacking the PAM2 motif. Whereas TDP-43 toxicity is enhanced with co-expression of ATXN2-32Q or ATXN2-32Q-ΔLSM (TDP-43+ATXN2-32Q or TDP-43+ΔLSM), there is no effect with co-expression of ATXN2-32Q-ΔPAM2 (TDP-43+ΔPAM2). Genotypes: YFP is gmr-GAL4(YH3)/UAS-YFP. TDP-43 is UAS-TDP-43(M)/+; gmr-GAL4(YH3)/+. TDP-43+ATXN2-32Q is UAS-TDP-43(M)/+; gmr-GAL4(YH3)/UAS-ATXN2-32Q. TDP-43+ΔPAM2 is UAS-TDP-43(M)/+; gmr-GAL4(YH3)/UAS-ATXN2-32Q.ΔPAM2. TDP-43+ΔLSM is UAS-TDP-43(M)/+; gmr-GAL4(YH3)/UAS- ATXN2-32Q.ΔLSM. Scale bar for eyes, 100 μm; for sections, 10 μm.
c) Expression of TDP-43 in the nervous system reduces lifespan (green, compared with normal in brown). Expression of ATXN2-32Q greatly enhanced TDP-43 toxicity (blue), but the ATXN2-32Q.ΔPAM2 deletion mutant form did not (red). Genotypes: Elav/+ is elav3A-GAL4/+ (n=177 flies). Elav, TDP-43/+ is elav3A-GAL4, UAS-TDP-43(S)/+ (n=156). Elav, TDP-43/ATXN2-32Q is elav3A-GAL4, UAS-TDP-43(S)/UAS-ATXN2-32Q (n=152). Elav, TDP-43/ΔPAM2 is elav3A-GAL4, UAS-TDP-43(S)/UAS-ATXN2-32Q. ΔPAM2 (n=138).
d) TDP-43 caused progressive loss of climbing ability when expressed in the adult nervous system (green) which is dramatically enhanced by co-expression of ATXN2-32Q (blue). However, the ΔPAM2 form of ATXN2-Q32 has little effect (red) compared to TDP-43 alone. Mean ± 95% CI of four experiments (n=125 flies per genotype). # p<0.0001, n.s., not significant (ANOVA for significance, followed by Tukey’s multiple comparison test).
e) Total, nuclear and cytosolic TDP-43 protein levels in 10d fly heads. Cytosolic TDP-43 protein level is greatly increased by ATXN2-32Q; ΔPAM2 expression does not affect TDP-43 localization or level. Mean ± s.e.m., n=3-4. # p<0.0001, n.s., not significant (ANOVA for significance, followed by Tukey’s multiple comparison test).
d–e Genotypes: elavGS, TDP-43/+ is elavGS, UAS-TDP-43(S)/+.elavGS, TDP-43/ATXN2-32Q is elavGS, UAS-TDP-43(S)/ UAS-ATXN2-32Q. elavGS, TDP-43/ΔPAM2 is elavGS, UAS-TDP-43(S)/ UAS-ATXN2-32Q.ΔPAM2. All flies raised with RU486 (20 μg/ml). *p<0.05, **p< 0.01, n.s. not significant. (ANOVA for significance, followed by Student’s t-test).
Poly-A binding protein modulates TDP-43 toxicity in Drosophila
To investigate the potential role of PABP in ALS-associated gene toxicity, we examined the effects of the fly gene dPABP (the Drosophila homologue of the cytoplasmic protein PABPC1) on toxicity of TDP-43 and ATXN2. Whereas upregulation of dPABP on its own had minimal effects, co-expression with either TDP-43 or ATXN2-32Q enhanced toxicity, resulting in more severe retinal degeneration (Fig. 4a). Consistent with a role for dPABP in conveying toxicity due to the interaction of TDP-43 with ATXN2, dPABP did not modulate ATXN2-32Q-ΔPAM2 (Fig. 4a). The effect of dPABP on TDP-43 was exquisitely dose-dependent: reduction of endogenous levels using RNAi significantly delayed the progressive loss of motility that occurs upon expression of TDP-43 in the nervous system (Fig. 4b). dPABP downregulation did not impact total TDP-43 protein levels in the brain, but did cause a reduction to 0.7±0.1 (s.e.m.) fold of the cytoplasmic levels of TDP-43, compared to TDP-43 with normal dPABP levels (Fig. 4c). Consistent with the essential function of the RNA binding activity of TDP-43, expression of TDP-43 with point mutations in the aromatic residues of the RNA-recognition motifs (RRMs) 25,52 failed to induce neurodegeneration or synergize with ATXN2 or dPABP (Supplementary Fig. 4). We confirmed that dPABP had no effect on the efficiency of the GAL4/UAS driver system (Supplementary Fig. 5). These data indicate that interactions with PABP modulate TDP-43 toxicity, suggesting that PABP function may be critical in human disease.
Figure 4. Poly(A) binding protein is required for TDP-43 toxicity.
a) dPABP upregulation enhances TDP-43 and ATXN2-32Q toxicity. However, dPABP does not interact with the ΔPAM2 form of ATXN2-32Q. Genotype: TDP-43 is UAS-TDP-43(M)/+; gmr-GAL4(YH3)/+. TDP-43+dPABP is UAS-TDP-43(M)/+; gmr-GAL4(YH3)/UAS-dPABP. PABP is gmr-GAL4(YH3)/UAS-dPABP. ATXN2-32Q+dPABP is gmr-GAL4(YH3), UAS-dPABP/ UAS-ATXN2-32Q.ΔPAM2+dPABP is gmr-GAL4(YH3), UAS-dPABP/ UAS-ATXN2-32Q.ΔPAM2. Scale bar for eyes, 100 μm; for sections, 10 μm.
b) TDP-43 loss of climbing ability is suppressed by downregulation of dPABP. Genotypes: elavGS, TDP-43/+ is elavGS, UAS-TDP-43(S)/+. elavGS, TDP-43/dPABP RNAi is elavGS, UAS-TDP-43(S)/UAS-dPABP.RNAiJF03104 (125 flies/genotype). Mean ± 95% CI of four experiments. *** p<0.001, n.s. not significant (Student’s t-test).
c) Total, nuclear and cytosolic TDP-43 protein levels in 10d fly heads. PABP downregulation decreases cytosolic TDP-43 protein level. Genotype: elavGS, TDP-43/+ is elavGS, UAS-TDP-43(S)/+. elavGS, TDP-43/dPABP. RNAi is elavGS, UAS-TDP-43(S)/UAS-dPABP.RNAiJF03104. Mean ± s.e.m., n=3 independent experiments *** p<0.001, n.s. not significant (Student’s t-test).
PABPC1 is mislocalized in spinal cord motor neurons in ALS
To determine if PABP function may be altered in human disease, we immunostained control and ALS patient spinal cord tissue for PABPC1 protein, the cytoplasmic PABP, to examine whether there were pathological associations between PABPC1 localization and ALS disease. In control spinal cord neurons, PABPC1 shows a diffuse cytoplasmic localization pattern in most neurons (Fig. 5a), occasionally being present in denser accumulations (2±0.7% (s.e.m.) motor neurons, Fig. 5b, 5e). In ALS patient spinal cord tissue, robust cytoplasmic inclusions of PABPC1 were observed in 13±4% (s.e.m.) of motor neurons (Fig. 5c-e; Supplementary Table 1). Recent analysis of co-localization pathological phosphorylated TDP-43 with PABPC1 indicated ~70% overlap in ALS human tissues 10. These data suggest that interactions with or altered function of PABPC1 may occur in ALS disease, and that these punctate accumulations containing PABPC1 in ALS spinal cords may represent structures with a functional role similar to that of stress granules, and be indicative of a prolonged stress state and associated altered signaling pathways. Taken together, these findings raised the possibility that treatment approaches that alter activities that impact stress granules, and their contingent effects on translation and signaling pathways, may impact ALS disease progression.
Figure 5. PABPC1 is mislocalized in motor neurons from ALS patient tissue.

a–d, Immunostaining for PABPC1 in the cervical spinal cord of control and ALS patients.
a) PABPC1 is expressed in spinal cord motor neurons, and is localized throughout the cytoplasm in a diffuse pattern.
b) Occasionally PABPC1 cytoplasmic accumulations were observed in normal motorneurons (arrow).
c) In ALS patient motor neurons, PABPC1 is present in distinct cytoplasmic and dense accumulations (arrow).
d) PABPC1 cytoplasmic accumulations were also observed in the ALS motorneurons where there was a clearing in the middle. Scale bars, 30 μm.
e) The number of motor neurons containing cytoplasmic accumulation of PABPC1 in ALS patients was significantly greater (p=0.01, one-tailed Mann-Whitney test) than in motor neurons from controls. For quantification of accumulations of PABPC1 in individual normal versus ALS cases, see Supplementary Table 1. n=5 controls, 4 ALS patients, mean ± s.e.m.
Pharmacologic rescue of TDP-43 toxicity
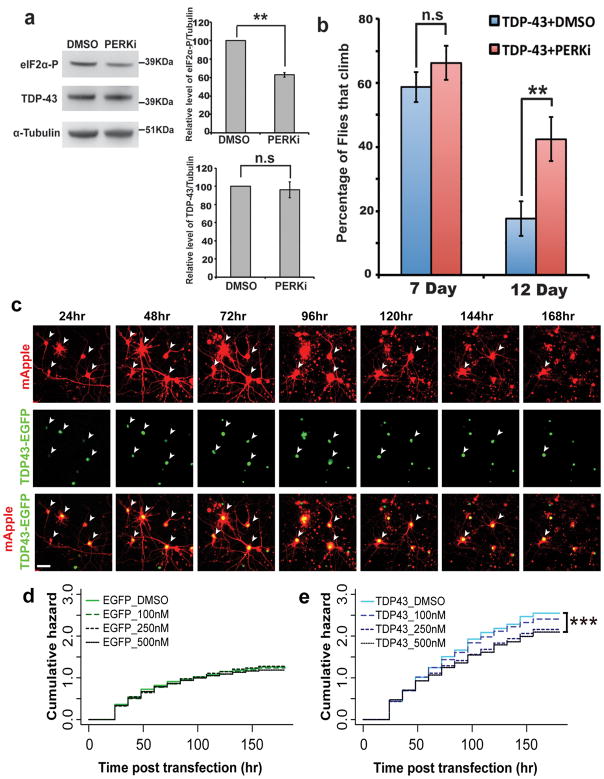
To test the idea that targeting eIF2α-phosphorylation in a therapeutic manner may modulate TDP-43 toxicity in the nervous system, we turned back to the fly. Our data with gene manipulation indicated that inhibition of PEK activity might be effective to mitigate TDP-43 toxicity. Compounds that inhibit PERK, the human homologue of PEK, have been developed. We therefore tested whether treating flies with the PERK inhibitor compound GSK2606414 53 could impact TDP-43-associated neurodegeneration. Flies fed 10uM GSK2606414 showed a markedly reduced level of eIF2a-phosphorylation, to 63±3% (s.e.m.) of that in heads of TDP-43-expressing animals (Fig. 6a). Remarkably, at this level of treatment, there was a dramatic mitigation of TDP-43-induced climbing dysfunction: whereas normally at 12d, TDP-43-expressing flies had only 18±5% (95% CI) climbing ability, with inhibitor treatment they now retained 42±7% (95% CI) climbing ability (Fig. 6b).
Figure 6. PERK inhibitor treatment rescues TDP-43 toxicity in Drosophila and primary rat neurons.
a,b, PERK inhibitor reduces eIF2α-phosphorylation levels and mitigates TDP-43 toxicity to flies.
a) Immunoblot of flies expressing TDP-43 in the nervous system. PERK inhibitor decreases eIF2α-phosphorylation levels, but not TDP-43 protein levels. Quantification performed on 3 independent experiments, normalized to tubulin. Mean ± s.e.m. **p<0.01, n.s., not significant (Student’s t-test).
b) Flies expressing TDP-43 by elav were treated with DMSO or PERK inhibitor (GSK2606414,10μM). Genotypes: elav3A-GAL4, UAS-TDP-43(S)/UAS-YFP. Mean ± s.e.m. n=4 experiments. **p<0.01, n.s., not significant (Student’s t-test).
c-f, PERK inhibitor (PERKi) reduces TDP43 toxicity in primary rat cortical neurons.
c) Representative micrographs showing longitudinal imaging of primary cortical neurons transfected with mApple as morphology marker and TDP43-EGFP. White arrows show cells still counted as alive in survival analysis. Cells were transfected at 4 DIV; hr indicate imaging time post transfection. Scale bar, 20μM.
d-e) Cumulative hazard plots for survival analysis performed on primary cortical neurons treated with PERKi and transfected with mApple and d) EGFP, e) TDP43-EGFP. PERKi treatment (500nM) of neurons expressing TDP43-EGFP decreased the risk of death by 14%, compared to neurons expressing TDP43-EGFP with vehicle (DMSO) only treatment. Three independent experiments were analyzed to determine statistical significance and cumulative hazard plots. Statistics was performed by Cox proportional hazards analysis; see Supplementary Table 2 for cells per condition and hazard ratios. *, p<0.05. **, p<0.01. ***, p<0.001. #, p<0.0001. n.s., not significant.
To extend these studies to the mammalian neurons, we assessed the ability of the PERK inhibitor to mitigate toxicity of TDP-43 to primary neurons. Rat primary cortical neurons were transfected with two constructs: one expressing mApple, and the other expressing either enhanced green florescent protein (EGFP) alone or EGFP tagged TDP-43. We imaged ~800–1000 individual neurons from each condition at 24-hr intervals for 7d using automated microscopy 54. Death of neurons was scored by the loss of detectable cell body mApple fluorescence. Previous studies have shown that upregulation of TDP-43 is toxic to neurons, inducing degeneration, cytoplasmic mislocalization of TDP-43 and aggregation of the protein 55, thus reflecting fundamental features of pathological TDP-43 associated with disease. To then assess the ability of GSK2606414 to mitigate TDP-43 toxicity, we first performed a dose-response to the compound, testing a range of compound concentrations (100, 250, 500nM, 1, 2, 5 uM). Concentrations of 1 uM or higher were deleterious on their own, but the nM concentrations were tolerated (Fig. 6d-e; Supplementary Table 2). We thus assessed the effect of compound on TDP-43 toxicity with concentrations of 100, 250 and 500 nM. These studies showed that treatment of primary neurons with PERKi GSK2606414 mitigated TDP-43 toxicity at 250 and 500 nM (p<0.01, Fig. 6e, Supplementary Table 2c). Taken together, these data indicate that prolonged activity of eIF2α-phosphorylation functionally impacts TDP-43 toxicity, and suggest the possibility that compounds that mitigate this, such as GSK2606414, could be developed as a therapeutic strategy for ALS and related TDP-43 proteinopathies.
DISCUSSION
A common biological feature of several RNA-binding proteins connected to neurodegenerative diseases is their localization to cytoplasmic stress granules, and a working hypothesis for disease is that stress granule formation facilitates or promotes TDP-43 aggregation in ALS 11,15,17–20,39 Moreover, stress granule markers such as TIA1 and eIF3 co-localize with TDP-43 inclusions in ALS and the clinicopathologically-related disorder, FTLD-U 16. However, the relationship between these observations and neurotoxicity is not clear. These findings do suggest, however, that ALS may be associated with a prolonged stress state and the outcome of such a state. Thus, it has been unclear whether genes that impact stress granule formation could directly affect TDP-43 neurotoxicity. The impact of prolonged stress granules would long-term translational repression and altered signalling pathways; sustained translational repression is thought to be causal in prion-mediated neurodegeneration and mitigating this has an effect to mitigate disease 21,22. Here, we show that eIF2α-phosphorylation is increased upon TDP-43-induced neural dysfunction in Drosophila, that genes that are implicated in stress granule formation impact TDP-43 toxicity in yeast, and that TDP-43 induced climbing deficits are mitigated by knockdown of genes whose activity normally modulates eIF2α-phosphorylation. Moreover, the interaction between Ataxin-2 and TDP-43 is mediated through PABP, which is mislocalized in ALS. As a proof of principle that modulating eIF2α-phosphorylation could be beneficial, a small molecule inhibitor of PERK activity significantly suppressed TDP-43 toxicity in vivo in the fly, and mitigated TDP-43 toxicity to mammalian primary cortical neurons. Thus, inhibitory activity of molecules on stress granule formation, and thus on a potentially prolonged stress state in disease, may accurately gauge potential therapeutic activity in vivo. Molecules that decrease stress granule formation or stability could be a promising treatment avenue for ALS and other TDP-43 proteinopathies. Such compounds may affect TDP-43 accumulation, and also mitigate the outcome of prolonged stress on cells that contributes to their degeneration.
Online Methods
Yeast Plasmid Overexpression Screen
The query strain was generated by integrating TDP-43 into the HIS3 locus using a p303-GAL-TDP-43 vector 25. Plasmids from the yeast FLEXgene collection (5500 full-length yeast ORFs) were transformed into the query strain. The screen was performed similarly to previous screens 33 and as described 25,31. The screen was replicated three times and only those hits that consistently reproduced were considered to be modifiers of TDP-43. A control strain with YFP (p303-GAL-YFP) integrated into the HIS3 locus was used in experiments to confirm that the overexpression strains did not lead to toxicity or a growth advantage when expressed in the absence of TDP-43. GO-term analysis of the hits was performed using the GO-slim mapper tool available at the SGD website, www.yeastgenome.org. GeneMANIA 56 was used to search for interacting genes based physical and genetic interactions.
Yeast Transformation and Spotting Assays
PEG/Lithium acetate yeast transformations and spotting assays were performed according to standard protocols.
Visualizing Stress Granules in Yeast
A plasmid expressing PUB1-mCherry, a stress granule marker 57, was transformed into the wild type yeast strain BY4741. Subsequently, p415-GAL-GFP or p415-GAL-TDP-43-GFP plasmids were transformed into the strain. The TDP-43 expression plasmid was created using an LR reaction with the destination vector p415-GAL-ccdB-GFP and the donor vector pDONR221-TDP-43 (CCSB Human ORFeome Collection).
Transformants were grown overnight in raffinose-containing selective media (CSM-Ura-Leu) and then added to galactose-containing selective media and allowed to grow for 6 hours to induce the expression of the galactose inducible genes. Yeasts were mounted in the media they were growing in on coverslips coated with concanavalin A (Sigma #L7647) 58. Images were captured on a Leica DM6000B with a ProEM EMCCD camera (Princeton Instruments).
Fly strains
Drosophila stocks were maintained on standard cornmeal agar media at 25°C unless otherwise noted. elav3A–GAL4 was a gift from Mark A. Tanouye 59. UAS-TDP-43 is described previously 25. The Bloomington stock center provided other fly stocks.
DNA constructs for germ line transformation
For generating random insertion transgenic lines, we used the pUAST vector 60. Human ATXN2 cDNA that has 32 CAG repeats was cloned into pUAST vector as BglII-XbaI fragments. The pUAST-ATXN2-Q32-ΔPAM2 construct was generated by deleting the region encoding amino acids 910-922 of the PAM2 domain. The pUAST-ATXN2-32Q construct containing encoding the full length ATXN2 protein with 32Q repeats was used as a template for two PCR reactions. The region 5′ of the PAM2 domain was amplified using a forward primer containing an attached BglII cut site primer PAM2-N1 (primer sequences listed in Supplementary Table 3) and a reverse primer complimentary to the beginning of the PAM2 domain and containing an attached HindIII cut site primer PAM2-N2. The region 3′ of the PAM2 domain was amplified using a forward primer complimentary to the end of the PAM2 domain and containing an attached HindIII cut site primer PAM2-C1 and a reverse primer containing an attached XbaI cut site primer PAM2-C2. Each PCR product was subcloned into TOPO vectors, cloned into pUAST vectors, cut out of the pUAST vector, ligated together, and reinserted into a pUAST vector.
Similarly, the pUAST-ATXN2-Q32-ΔLSM construct was generated by deleting the region encoding amino acids 267-325 of the PAM2 domain. The pUAST-ATXN2-Q32 construct was used as a template for two PCR reactions. The region 5′ of the LSM domain was amplified using a forward primer containing an attached BglII cut site primer LSM-N1 and a reverse primer complimentary to the beginning of the LSM domain and containing an attached HindIII cut site primer LSM-N2. The region 3′ of the LSM domain was amplified using a forward primer complimentary to the end of the LSM domain and containing an attached HindIII cut site primer LSM-C1, and a reverse primer containing an attached XbaI cut site primer LSM-C2. Each PCR product was subcloned into TOPO vectors, cloned into pUAST vectors, cut out of the pUAST vector, ligated together, and reinserted into a pUAST vector.
For generation of site-specific integration transgenic lines, we used pJFRC5 and pJFRC5-myc vector 61. ATXN2 cDNAs that with 32 CAG or 22 CAG repeats were cloned into pJFRC5-myc vector as SwaI-XbaI fragment. These DNA constructs were integrated into the attP2 landing site.
External eye and retinal tissue microscopy
For fly eye pictures, we used a Leica Z16-Apo A motorized zoom microscope system with DFC420 digital camera and Leica Application Suite Montage module software (Leica Microsystems). Retinal tissue was visualized by autofluorescence in horizontal paraffin sections. 0-1d males were used for experiments.
Lifespan and adult climbing assays
0-1d males were separated and transferred onto experimental vials containing fly media mixed with or without RU486 (20μg/ml or 40μg/ml) at a density of 20 (for life span) or 25 (for climbing assay) flies per vial. Dead flies were scored everyday and flies were transferred to fresh media every other day. All flies for climbing assays or life span analysis raised at 29°C. Adult locomotor function was assessed by a previously described method, with 125 flies per genotype per time point in all experiments, except for SFig. 2d where 150 flies per genotype per time point were used 62. Experiments were repeated twice to assure consistent results.
Western immunoblotting
For total protein extraction, 10 male fly heads were homogenized in 1X LDS sample buffer (Invitrogen). For subcellular fractionation, 20 male fly heads were lysed in NE-PER extraction reagent (Pierce) according to the manufacturer’s protocol. The extracts (equivalent to 2 heads/lane) were then run on 3–8% Tris Acetate gel or 4–12% Bis-Tris gel (Invitrogen). Western blotting was performed according to standard protocols. The following primary antibodies were used: anti-TDP-43 (1:800, Proteintech, catalog # 10782-2-AP); anti-β-Actin (1:2000, Abcam, catalog # ab16039); anti-α-Tubulin (1:2000, conjugated with HRP, Cell Signaling, catalog # 9099); anti-Lamin C (1:1000, Deveopmental Studies Hybridoma Bank, catalog # LC28.26); anti-phospho-eIF2α (1:1000, Cell signaling tech, Catalog #9722); anti-mouse-Ataxin-2 (1:300, BD biosciences, catalog # 6113378), anti-rabbit-eIF2α (1:1000, Abcam, catalog # ab26197) and anti-mouse β-galactosidase (Promega, catalog # Z3781). After incubation with HRP-coupled secondary antibodies (goat anti-rabbit diluted 1:2000 and goat anti-mouse diluted 1:2000; Pierce), blots were visualized using ECL plus or prime kit (Amersham Biosciences). Quantification of Western blots was performed using ImageGauge4.22 (GE Healthcare).
Immunohistochemistry of human post-mortem spinal cord
Human spinal cord tissue fixed in 10% neutral buffered formalin was processed into paraffin wax. The tissue was cut into 7 μm sections and PABPC1 was immunolocalized with antigen retrieval (Antigen unmasking solution, citric acid based, Vector Laboratories, catalog # H-3300) and the avidin-biotin complex detection method with 3,3-diaminobenzidine as the chromagen (Vectastain ABC detection kit, Vector Labs, catalog # PK-6100). PABPC1 rabbit polyclonal antibody was used at 1:800 (Cell Signaling, catalog # 4992S). Biotinylated anti-rabbit IgG (Vector Labs, catalogue #: BA-1000) was used at 1:1000. Stained sections were counterstained with hematoxylin and eosin.
Drug treatment
GSK2606414 (PERK inhibitor, EMD Millipore, catalog #: 516535) was added directly to media from a 1mg/ml stock (dissolved in DMSO) to give final concentration 10μM. The same amount of DMSO was added for vehicle control.
Statistical analyses
For climbing and immunoblot quantifications, data were analyzed by Student’s t-test (Vassar Stats, www.vassarstats.net) or first analyzed using one-way ANOVA followed by Tukey’s multiple comparisons test (GraphPad Prism Software, La Jolla, CA). Human immunohistochemistry data was analyzed by one-tailed Mann-Whitney test (VassarStats). Differences were considered significant when p<0.05, and are indicated as follows: * p<0.05; ** p<0.01; *** p<0.001; # p<0.0001; n.s., not significant.
Plasmids
For primary cell culture, human TDP43 was cloned into pGW1-CMV plasmid, C-terminally fused to EGFP as described 55. EGFP and mApple were cloned into pGW1-CMV as described 55,63.
Cell culture
Cortical neurons were isolated from E20-21 Long Evans rat embryos (Charles River) and cultured at 100,000 cells/well of 96 well plate in serum-free Neurobasal medium (Invitrogen) supplemented with B27, GlutaMax, and pen/strep (Invitrogen). At 4d in vitro (DIV), neurons were transfected with Lipofectamine 2000 (Invitrogen) per manufacturers protocol. Neurons were cotransfected with plasmids encoding survival markers (mApple) or proteins of interest (TDP43-EGFP or EGFP control) in a 1:1 molar ratio for a total of 0.2 μg total DNA per well. After transfection cells were returned to Neurobasal media mixed 1:1 with conditioned media. For cells treated with PERK inhibitor (GSK2606414, CalBiochem), drug was added once immediately after transfection.
Robotic microscopy
For neuronal survival analysis, we use a robotic imaging system as described 63. Briefly, images are obtained with an inverted Nikon microscope (Ti-E) equipped with PerfectFocus, an extra-long working distance (ELWD) 20x objective lens, and a back-illuminated Andor iXON 888 14-bit, cooled, electron multiplying charge coupled device (EMCCD). Illumination is provided a xenon lamp and liquid light guide. All movements of the stage are controlled with electrical stepper motors. Coordination of fluorescence excitation and emission filters, stage movements, focusing, and imaging acquisition are accomplished with custom-designed and commercially available programs.
Image analysis and statistics
Digitized images were assembled into montages in Pipeline Pilot and ImageJ using original programs. Background fluorescence from neighboring regions of interest was subtracted, montages from each time point are assembled into stacks in chronological order and aligned with one another. Cell bodies of transfected neurons, identified by morphology marker fluorescence (mApple), were automatically segmented and followed over time. Cell death was determined by an abrupt loss of fluorescence, indicating shrinkage or disappearance of the cell body. The time of death for each neuron was considered the last time that the neuron was present. Kaplan-Meier and cumulative risk of death curves were generated in R. Statistical significance of survival differences between cohorts of neurons is determined by the log-rank test, and Cox proportional hazards analysis used to measure the relative change in the risk of death attributed to various experimental conditions.
Supplementary Material
Acknowledgments
We thank Xiuyin Teng and Dr. Yongqing Zhu for technical assistance, Dr. William Motley, Dr. Amit Berson and other laboratory members for insightful comments. This work was funded by grants from the Howard Hughes Medical Institute (N.M.B.), R01NS073660 (A.D.G. and N.M.B.), NIH Director’s New Innovator Award DP2OD004417 (A.D.G.) and R01NS065317 (A.D.G.), the Robert Packard Center for ALS and the Williams H. Adams Foundation (S.F.), and AG10124, AG32953, AG17586 and NS53488 (J.Q.T. and V.M.-Y.L.). A.D.G. and S.F. are supported by a grant from Target ALS. A.R.R. is supported by a BrightFocus Alzheimer’s disease research grant.
Footnotes
Author Contributions
H.-J.K., A.R.R., E.S.L., L.M., A.D.G. conceived, designed and performed experiments, performed statistical analysis, analyzed data; R.W. performed experiments; J.Q.T, V. M.-Y.L. contributed reagents and materials, and experimental input; S.F., A.D.G., N.M.B. conceived and designed experiments, analyzed data, and supervised the research; H.-J.K., and N.M.B., with input from A.R.R. and A.D.G., wrote the paper.
Financial Disclosures:
A.D.G. is an inventor on patents and patent applications that have been licensed to FoldRx.
References
- 1.Boillee S, Vande Velde C, Cleveland DW. ALS: a disease of motor neurons and their nonneuronal neighbors. Neuron. 2006;52:39–59. doi: 10.1016/j.neuron.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 2.Cleveland DW, Rothstein JD. From Charcot to Lou Gehrig: deciphering selective motor neuron death in ALS. Nat Rev Neurosci. 2001;2:806–19. doi: 10.1038/35097565. [DOI] [PubMed] [Google Scholar]
- 3.Al-Chalabi A, et al. The genetics and neuropathology of amyotrophic lateral sclerosis. Acta Neuropathol. 2012;124:339–52. doi: 10.1007/s00401-012-1022-4. [DOI] [PubMed] [Google Scholar]
- 4.Couthouis J, et al. Evaluating the role of the FUS/TLS-related gene EWSR1 in amyotrophic lateral sclerosis. Hum Mol Genet. 2012;21:2899–911. doi: 10.1093/hmg/dds116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Couthouis J, et al. A yeast functional screen predicts new candidate ALS disease genes. Proc Natl Acad Sci U S A. 2011;108:20881–90. doi: 10.1073/pnas.1109434108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim HJ, et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013 doi: 10.1038/nature11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.King OD, Gitler AD, Shorter J. The tip of the iceberg: RNA-binding proteins with prion-like domains in neurodegenerative disease. Brain Res. 2012;1462:61–80. doi: 10.1016/j.brainres.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Warraich ST, Yang S, Nicholson GA, Blair IP. TDP-43: a DNA and RNA binding protein with roles in neurodegenerative diseases. Int J Biochem Cell Biol. 2010;42:1606–9. doi: 10.1016/j.biocel.2010.06.016. [DOI] [PubMed] [Google Scholar]
- 9.Aulas A, Stabile S, Vande Velde C. Endogenous TDP-43, but not FUS, contributes to stress granule assembly via G3BP. Mol Neurodegener. 2012;7:54. doi: 10.1186/1750-1326-7-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bentmann E, et al. Requirements for stress granule recruitment of fused in sarcoma (FUS) and TAR DNA-binding protein of 43 kDa (TDP-43) J Biol Chem. 2012;287:23079–94. doi: 10.1074/jbc.M111.328757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parker SJ, et al. Endogenous TDP-43 localized to stress granules can subsequently form protein aggregates. Neurochem Int. 2012;60:415–24. doi: 10.1016/j.neuint.2012.01.019. [DOI] [PubMed] [Google Scholar]
- 12.Dewey CM, et al. TDP-43 is directed to stress granules by sorbitol, a novel physiological osmotic and oxidative stressor. Mol Cell Biol. 2011;31:1098–108. doi: 10.1128/MCB.01279-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thomas MG, Loschi M, Desbats MA, Boccaccio GL. RNA granules: the good, the bad and the ugly. Cell Signal. 2011;23:324–34. doi: 10.1016/j.cellsig.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hart MP, Gitler AD. ALS-associated ataxin 2 polyQ expansions enhance stress-induced caspase 3 activation and increase TDP-43 pathological modifications. J Neurosci. 2012;32:9133–42. doi: 10.1523/JNEUROSCI.0996-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li YR, King OD, Shorter J, Gitler AD. Stress granules as crucibles of ALS pathogenesis. J Cell Biol. 2013;201:361–72. doi: 10.1083/jcb.201302044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu-Yesucevitz L, et al. Tar DNA binding protein-43 (TDP-43) associates with stress granules: analysis of cultured cells and pathological brain tissue. PLoS One. 2010;5:e13250. doi: 10.1371/journal.pone.0013250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolozin B. Regulated protein aggregation: stress granules and neurodegeneration. Mol Neurodegener. 2012;7:56. doi: 10.1186/1750-1326-7-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bentmann E, Haass C, Dormann D. Stress granules in neurodegeneration--lessons learnt from TAR DNA binding protein of 43 kDa and fused in sarcoma. FEBS J. 2013;280:4348–70. doi: 10.1111/febs.12287. [DOI] [PubMed] [Google Scholar]
- 19.Ramaswami M, Taylor JP, Parker R. Altered Ribostasis: RNA-Protein Granules in Degenerative Disorders. Cell. 2013;154:727–36. doi: 10.1016/j.cell.2013.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thomas M, Alegre-Abarrategui J, Wade-Martins R. RNA dysfunction and aggrephagy at the centre of an amyotrophic lateral sclerosis/frontotemporal dementia disease continuum. Brain. 2013;136:1345–60. doi: 10.1093/brain/awt030. [DOI] [PubMed] [Google Scholar]
- 21.Moreno JA, et al. Oral Treatment Targeting the Unfolded Protein Response Prevents Neurodegeneration and Clinical Disease in Prion-Infected Mice. Sci Transl Med. 2013;5:206ra138. doi: 10.1126/scitranslmed.3006767. [DOI] [PubMed] [Google Scholar]
- 22.Moreno JA, et al. Sustained translational repression by eIF2alpha-P mediates prion neurodegeneration. Nature. 2012;485:507–11. doi: 10.1038/nature11058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Auluck PK, Chan HY, Trojanowski JQ, Lee VM, Bonini NM. Chaperone suppression of alpha-synuclein toxicity in a Drosophila model for Parkinson’s disease. Science. 2002;295:865–8. doi: 10.1126/science.1067389. [DOI] [PubMed] [Google Scholar]
- 24.Cooper AA, et al. Alpha-synuclein blocks ER-Golgi traffic and Rab1 rescues neuron loss in Parkinson’s models. Science. 2006;313:324–8. doi: 10.1126/science.1129462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elden AC, et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature. 2010;466:1069–75. doi: 10.1038/nature09320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gitler AD, et al. Alpha-synuclein is part of a diverse and highly conserved interaction network that includes PARK9 and manganese toxicity. Nat Genet. 2009;41:308–15. doi: 10.1038/ng.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Outeiro TF, Lindquist S. Yeast cells provide insight into alpha-synuclein biology and pathobiology. Science. 2003;302:1772–5. doi: 10.1126/science.1090439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Treusch S, et al. Functional links between Abeta toxicity, endocytic trafficking, and Alzheimer’s disease risk factors in yeast. Science. 2011;334:1241–5. doi: 10.1126/science.1213210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson BS, McCaffery JM, Lindquist S, Gitler AD. A yeast TDP-43 proteinopathy model: Exploring the molecular determinants of TDP-43 aggregation and cellular toxicity. Proc Natl Acad Sci U S A. 2008;105:6439–44. doi: 10.1073/pnas.0802082105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson BS, et al. TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity. J Biol Chem. 2009;284:20329–39. doi: 10.1074/jbc.M109.010264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Armakola M, Hart MP, Gitler AD. TDP-43 toxicity in yeast. Methods. 2011;53:238–45. doi: 10.1016/j.ymeth.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu XH, et al. Genetic dissection of ethanol tolerance in the budding yeast Saccharomyces cerevisiae. Genetics. 2007;175:1479–87. doi: 10.1534/genetics.106.065292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun Z, et al. Molecular determinants and genetic modifiers of aggregation and toxicity for the ALS disease protein FUS/TLS. PLoS Biol. 2011;9:e1000614. doi: 10.1371/journal.pbio.1000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jain S, Parker R. The discovery and analysis of P Bodies. Adv Exp Med Biol. 2013;768:23–43. doi: 10.1007/978-1-4614-5107-5_3. [DOI] [PubMed] [Google Scholar]
- 35.Lagier-Tourenne C, Polymenidou M, Cleveland DW. TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Hum Mol Genet. 2010;19:R46–64. doi: 10.1093/hmg/ddq137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ling SC, Polymenidou M, Cleveland DW. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron. 2013;79:416–38. doi: 10.1016/j.neuron.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tazen S, et al. Amyotrophic Lateral Sclerosis and Spinocerebellar Ataxia Type 2 in a Family With Full CAG Repeat Expansions of ATXN2. JAMA Neurol. 2013 doi: 10.1001/jamaneurol.2013.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zoll WL, Horton LE, Komar AA, Hensold JO, Merrick WC. Characterization of mammalian eIF2A and identification of the yeast homolog. J Biol Chem. 2002;277:37079–87. doi: 10.1074/jbc.M207109200. [DOI] [PubMed] [Google Scholar]
- 39.Dewey CM, et al. TDP-43 aggregation in neurodegeneration: are stress granules the key? Brain Res. 2012;1462:16–25. doi: 10.1016/j.brainres.2012.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anderson P, Kedersha N. Visibly stressed: the role of eIF2, TIA-1, and stress granules in protein translation. Cell Stress Chaperones. 2002;7:213–21. doi: 10.1379/1466-1268(2002)007<0213:vstroe>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brand S, Bourbon HM. The developmentally-regulated Drosophila gene rox8 encodes an RRM-type RNA binding protein structurally related to human TIA-1-type nucleolysins. Nucleic Acids Res. 1993;21:3699–704. doi: 10.1093/nar/21.16.3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dalton LE, Healey E, Irving J, Marciniak SJ. Phosphoproteins in stress-induced disease. Prog Mol Biol Transl Sci. 2012;106:189–221. doi: 10.1016/B978-0-12-396456-4.00003-1. [DOI] [PubMed] [Google Scholar]
- 43.Khong A, Jan E. Modulation of stress granules and P bodies during dicistrovirus infection. J Virol. 2011;85:1439–51. doi: 10.1128/JVI.02220-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kimball SR, Horetsky RL, Ron D, Jefferson LS, Harding HP. Mammalian stress granules represent sites of accumulation of stalled translation initiation complexes. Am J Physiol Cell Physiol. 2003;284:C273–84. doi: 10.1152/ajpcell.00314.2002. [DOI] [PubMed] [Google Scholar]
- 45.Kedersha N, et al. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J Cell Biol. 2000;151:1257–68. doi: 10.1083/jcb.151.6.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ruggieri A, et al. Dynamic oscillation of translation and stress granule formation mark the cellular response to virus infection. Cell Host Microbe. 2012;12:71–85. doi: 10.1016/j.chom.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nonhoff U, et al. Ataxin-2 interacts with the DEAD/H-box RNA helicase DDX6 and interferes with P-bodies and stress granules. Mol Biol Cell. 2007;18:1385–96. doi: 10.1091/mbc.E06-12-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Swisher KD, Parker R. Localization to, and effects of Pbp1, Pbp4, Lsm12, Dhh1, and Pab1 on stress granules in Saccharomyces cerevisiae. PLoS One. 2010;5:e10006. doi: 10.1371/journal.pone.0010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kozlov G, Menade M, Rosenauer A, Nguyen L, Gehring K. Molecular determinants of PAM2 recognition by the MLLE domain of poly(A)-binding protein. J Mol Biol. 2010;397:397–407. doi: 10.1016/j.jmb.2010.01.032. [DOI] [PubMed] [Google Scholar]
- 50.Chowdhury A, Raju KK, Kalurupalle S, Tharun S. Both Sm-domain and C-terminal extension of Lsm1 are important for the RNA-binding activity of the Lsm1–7-Pat1 complex. RNA. 2012;18:936–44. doi: 10.1261/rna.029876.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Satterfield TF, Pallanck LJ. Ataxin-2 and its Drosophila homolog, ATX2, physically assemble with polyribosomes. Hum Mol Genet. 2006;15:2523–32. doi: 10.1093/hmg/ddl173. [DOI] [PubMed] [Google Scholar]
- 52.Buratti E, Baralle FE. Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9. J Biol Chem. 2001;276:36337–43. doi: 10.1074/jbc.M104236200. [DOI] [PubMed] [Google Scholar]
- 53.Axten JM, et al. Discovery of 7-methyl-5-(1-{[3-(trifluoromethyl)phenyl]acetyl}-2,3-dihydro-1H-indol-5-yl)-7H-p yrrolo[2,3-d]pyrimidin-4-amine (GSK2606414), a potent and selective first-in-class inhibitor of protein kinase R (PKR)-like endoplasmic reticulum kinase (PERK) J Med Chem. 2012;55:7193–207. doi: 10.1021/jm300713s. [DOI] [PubMed] [Google Scholar]
- 54.Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–10. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- 55.Barmada SJ, et al. Cytoplasmic mislocalization of TDP-43 is toxic to neurons and enhanced by a mutation associated with familial amyotrophic lateral sclerosis. J Neurosci. 2010;30:639–49. doi: 10.1523/JNEUROSCI.4988-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zuberi K, et al. GeneMANIA prediction server 2013 update. Nucleic Acids Res. 2013;41:W115–22. doi: 10.1093/nar/gkt533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hofmann S, Cherkasova V, Bankhead P, Bukau B, Stoecklin G. Translation suppression promotes stress granule formation and cell survival in response to cold shock. Mol Biol Cell. 2012;23:3786–800. doi: 10.1091/mbc.E12-04-0296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Buchan JR, Yoon JH, Parker R. Stress-specific composition, assembly and kinetics of stress granules in Saccharomyces cerevisiae. J Cell Sci. 2011;124:228–39. doi: 10.1242/jcs.078444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hekmat-Scafe DS, Dang KN, Tanouye MA. Seizure suppression by gain-of-function escargot mutations. Genetics. 2005;169:1477–93. doi: 10.1534/genetics.104.036558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–15. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 61.Pfeiffer BD, et al. Refinement of tools for targeted gene expression in Drosophila. Genetics. 2010;186:735–55. doi: 10.1534/genetics.110.119917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Feany MB, Bender WW. A Drosophila model of Parkinson’s disease. Nature. 2000;404:394–8. doi: 10.1038/35006074. [DOI] [PubMed] [Google Scholar]
- 63.Arrasate M, Finkbeiner S. Automated microscope system for determining factors that predict neuronal fate. Proc Natl Acad Sci U S A. 2005;102:3840–5. doi: 10.1073/pnas.0409777102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.