Abstract
During the past 25 years, our knowledge concerning the pathogenesis, diagnostic strategies and treatment of von Willebrand disease (VWD) has increased significantly. Following the immunological differentiation of factor VIII (FVIII) and von Willebrand factor (VWF) in the 1970s and the cloning of the FVIII and VWF genes in the mid-1980s, substantial progress has been made in our understanding of this, the most common inherited bleeding disorder. We now recognize that VWD represents a range of genetic diseases all with the clinical endpoint of increased mucocutaneous bleeding. The molecular pathology of Type 2 and Type 3 VWD is now comprehensively documented and involves rare sequence variants at the VWF locus. In contrast, the genetic causation of Type 1 disease remains incompletely defined and in many cases appears to involve genetic determinants in addition to or instead of VWF. The diagnostic triad of a personal history of excessive mucocutaneous bleeding, laboratory tests for VWF that are consistent with VWD, and a family history of the condition remain the keystone to VWD identification. In the laboratory, measurement of VWF antigen and function continue to be the most important diagnostic studies, and while our understanding of the molecular genetic pathology of VWD has advanced considerably in the past decade, genetic testing as a component of diagnosis is limited to certain distinct subtypes of the disorder. Treatment of VWD has been relatively unchanged for the past decade and continues to involve either stimulation of the release of intrinsic VWF with desmopressin or the infusion of VWF concentrates.
Keywords: von WIllebrand factor, von Willebrand disease
Introduction
In 1926, the Finnish physician, Erik von Willebrand, described his seminal study of a large family (Family S) in the Ǻland Islands, in whom excessive bleeding had resulted in the early hemorrhagic deaths of several children [1]. Most famously, the index case in this study, Hjördis, one of seven sisters of whom five had bleeding symptoms, presented at the age of five years with recurrent epistaxis, bleeding from her lips and following tooth extractions. At the age of three years, she had bled for three days from an injury to her lip. Then, at age 14, Hjördis bled to death at the time of her fourth menstrual period.
During these investigations, von Willebrand recognized that the pattern of bleeding and the fact that both men and women were affected by the condition, differentiated this condition from classical hemophilia. Subsequent infusion studies of Cohn fraction I from human plasma in the 1950s confirmed that a novel plasma coagulation factor was missing in the Ǻland Island kindred [2], and in 1971 von Willebrand factor (VWF) was first detected immunologically [3].
Recent Historical Perspectives of von Willebrand Disease
Following the immunological characterization of VWF in the early 1970s, it soon became apparent that the clinical features of VWD were associated with a miscellany of laboratory abnormalities reflecting a significant heterogeneity at the level of VWF structure/function [4] . Then, in 1985, four groups simultaneously cloned the VWF gene and set in motion the next 25 years of investigation into the molecular pathology of VWD [5–8]. These studies have included several updates to the classification of VWD [9] and a progressive improvement in our understanding of the pathogenetic mechanisms underlying the disorder. Finally, following the production of several generations of plasma-derived VWF concentrates and the discovery that the synthetic vasopressin analogue, desmopressin, can stimulate the release of stored VWF [10], we are now witnessing the development of recombinant forms of VWF for treatment in VWD.
With this brief historical background, the aim of this review will be to highlight new information concerning basic, laboratory and clinical aspects of VWD that have derived from large VWD population studies performed in the last few years. The two specific areas that will be discussed in detail in this review will be VWD pathogenesis and diagnosis.
von Willebrand Disease Pathogenesis
von Willebrand disease is a genetic disorder and thus, the major contributor to the phenotype in VWD concerns variations in the genome. With our current understanding of genetic causation, one can regard VWD as exhibiting a diverse array of genetic pathologies ranging from clear examples of monogenic dominant and recessive traits to the more common incompletely penetrant and variably expressed complex trait that is type 1 VWD. In terms of pathogenetic mechanisms, VWD can be thought of as the hemostatic equivalent of the globin pathologies responsible for the quantitative diseases seen in the thalassemias (ie. Types 1 and 3 VWD) and the qualitative hemoglobinopathies (type 2 VWD).
Acquired/Environmental Determinants of Plasma VWF Levels
Before describing details of the recent advances in understanding the genetic pathology of VWD, it is worthwhile to summarize knowledge pertaining to acquired, environmental factors that influence VWF. von Willebrand factor is one of several hemostatic proteins (FVIII, fibrinogen and plasminogen activator inhibitor-1 being the others) that respond as acute phase reactants. Increases of approximately 3-fold in VWF plasma levels are routinely seen in subjects suffering from bacterial and viral infectious diseases with elevated levels persisting for 1–2 weeks during the period of infection [11]. Long-term elevations of VWF can also accompany disorders in which chronic endothelial damage occurs such as diabetes. In contrast, acute transient elevations of VWF (between 2 to 4-fold) can be seen following episodes of increased physical activity and following epinephrine infusion, a phenomenon that reflects a secretion/release response mediated through beta adrenergic receptor signaling as opposed to increased de novo synthesis [12,13]. Chronic elevations of VWF (2 to 3-fold) are also seen in patients with hyperthyroidism, where again a beta adrenergic receptor-mediated mechanism has been implicated [14]. In contrast, hypothyroidism is associated with a reduction of VWF (15–45% levels) that reverts to normal with thyroid hormone replacement [15].
Von Willebrand factor levels also increase in response to increasing levels of estragens as seen with the use of the oral contraceptive and hormone replacement therapy and during pregnancy, in which plasma levels of VWF can often reach 300–400% at term [16,17]. Details of the molecular mechanistic basis for these significant changes in VWF synthesis remains unexplained.
Another significant factor that influences the plasma level of VWF is age. Throughout life, VWF levels show a gradual increase of approximately 1–2% per year. This increase has been well documented in large populations of normal subjects [18,19] and has also been observed in patients with type 1 VWD in whom an age-related rise in VWF results in the eventual resolution of the quantitative deficiency state that characterizes this disorder. Once again, the mechanisms underlying these age-related changes are unknown.
Genetic Determinants of von Willebrand Factor Levels and Function
In the past decade, the assessment of genetic factors that influence both quantitative and qualitative characteristics of VWF has progressed significantly. These studies have involved the investigation of two types of population: patients with von Willebrand disease and healthy individuals. While the genetic basis for VWD has been a productive focus of activity for the past two decades [20], the incorporation of genetic information from large populations of healthy subjects is a relatively new experimental approach. These latter studies have been made possible through the development of new genomic methodologies such as genomewide association studies (GWAS) and next generation sequencing strategies. Twin studies have indicated that plasma VWF levels are strongly influenced by genetic factors, with heritability values between 60–75% [21–23]. Recent evidence derived from GWAS and more focused genetic association studies in healthy subjects suggest that there may be many genetic variances that in combination determine the plasma VWF level in healthy individuals. By extension, these same genetic variants may act as either modifiers of the phenotype in VWD or alternatively, rare variants at the same loci may sometimes be the primary pathogenetic changes in VWD.
This discussion of genetic determination of VWF pathobiology will be divided into two sections: the first dealing with the VWF gene and the second section relating to genetic variants at other non-VWF loci.
Variation at the VWF Locus Resulting in Qualitative VWD Subtypes
Qualitative subtypes of VWD encompass types 2A, 2B, 2M and 2N disease. The genetic pathology underlying these traits has been very well characterized during the past two decades by work from many groups, but especially by the French INSERM Network on Molecular Abnormalities in VWD [24]. In aggregate, these VWD subtypes constitute approximately 25% of all VWD cases.
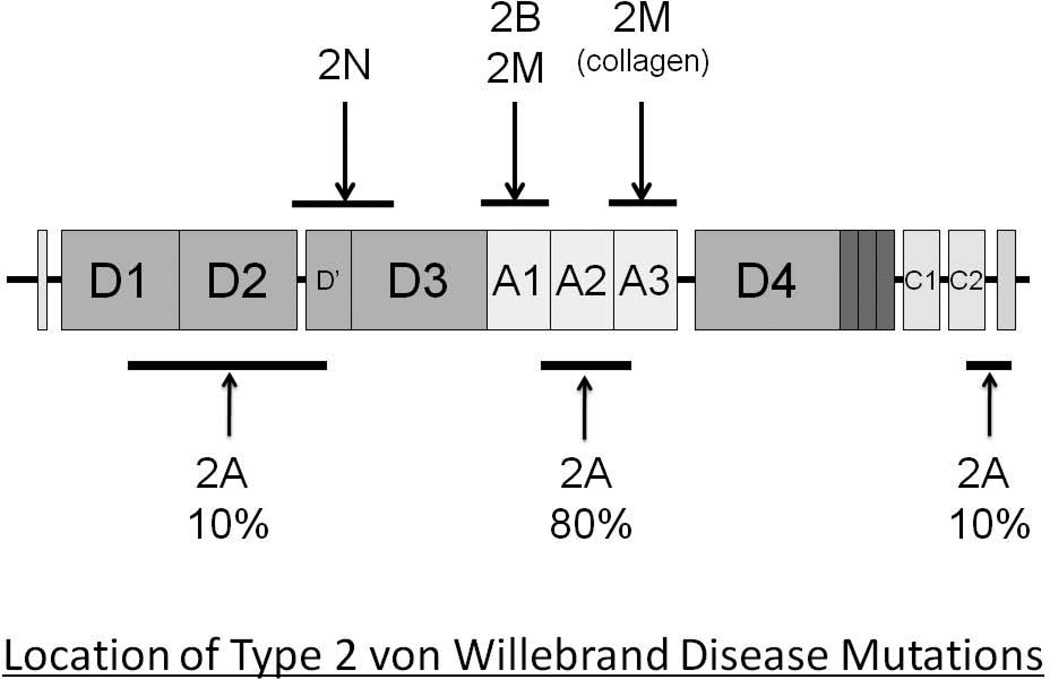
To date, all cases of VWF functional pathology have demonstrated genetic changes at the VWF locus, with one interesting exception, the case of platelet-type VWD where the causative mutations are located in the GPIBA gene [25] – (Figure 1).
Figure 1.
Location of the missense mutations resulting in type 2 VWD.
Von Willebrand Factor Mutations Resulting in Reduced Platelet Binding Capability: Types 2A and 2M VWD
von Willebrand factor plays a key role in mediating platelet adhesion to the exposed subendothelium of a damaged vessel wall. To perform this function effectively, the circulating VWF must possess high molecular weight multimers (HMWMs) and the structure and configuration of the glycoprotein Ib binding region in the A1 domain must be normal.
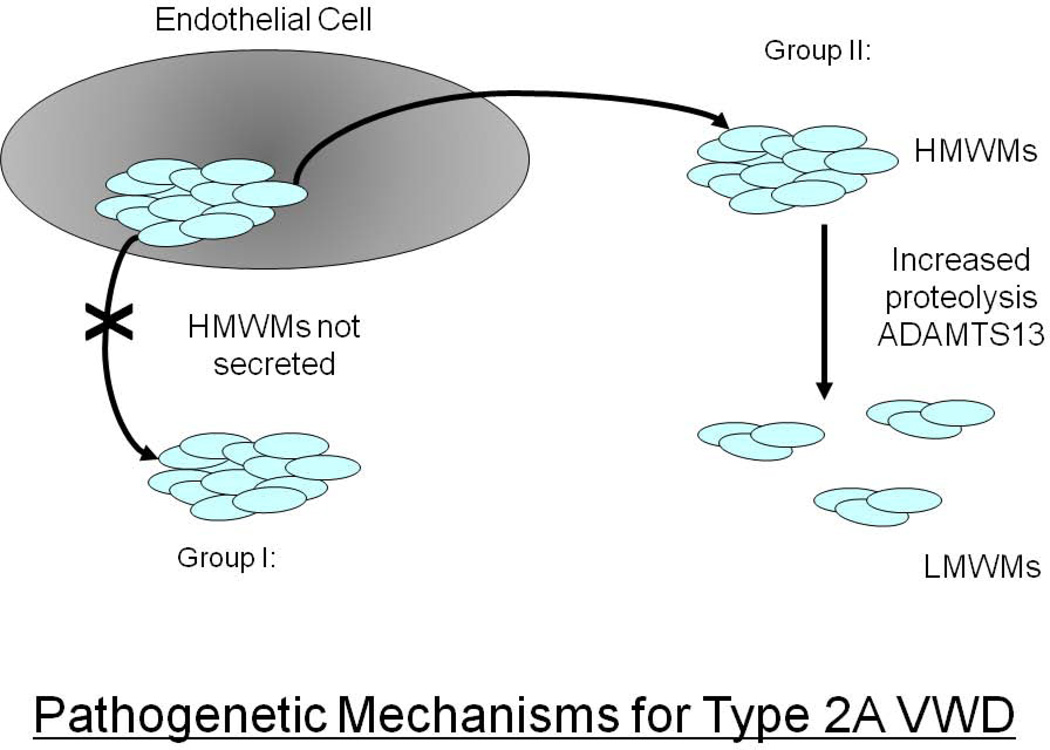
The absence of HMWMs can result from either a failure in the VWF biosynthetic process (failure to form VWF dimers and/or multimers), so called Group I Type 2A VWD mutations [26] or the synthesis of a VWF molecule that is more susceptible to ADAMTS13-mediated cleavage (Group II, Type 2A VWD mutations) [27] (Figure 2). Approximately 75% of the 73 different type 2A VWD mutations are located in exon 28 of the VWF gene and most often involve missense substitutions in the A2 domain of the protein (see ISTH-SSC VWF Online Database at http://www.vwf.group.shef.ac.uk/. Accessed on January 3rd 2012). Several of these mutations are frequent and recurrent (ie. R1597W and S1506L). These mutations in the A1 domain result in either an abnormal protein structure that interferes with VWF multimer assembly (Group I mutations) or in an A2 domain conformation that enables enhanced ADAMTS13-mediated proteolysis (Group II mutations). In addition to the predominant A2 domain Type 2A mutations, ∼10% of mutations are located in the VWF propeptide or in the N terminus of the mature subunit and interfere with multimer formation, while a further ∼10% of Type 2A variants are localized in the C-terminal dimerization domains.
Figure 2.
Pathogenetic mechanisms resulting in Type 2A VWD.
In Type 2M VWD, the VWF has a normal complement of HMWMs, but a reduced capability for binding to platelets due to changes in A1 domain structure and configuration [28]. Approximately 90% of Type 2M mutations involve missense substitutions in the A1 domain- encoding region of exon 28. Indeed, the few non-A1 domain variants listed in the VWF mutation database may well be misclassified [29]. Detailed structural analysis of the location of Type 2M missense substitutions indicates that they are located at or very close to the site of contact with the beta switch region of the glycoprotein Ib alpha receptor [28].
Mutations Resulting in VWF with Enhanced Platelet Binding Capability: Type 2B VWD
von Willebrand factor binds to the platelet GPIb alpha receptor through its central A1 domain. As discussed above, loss-of-function variants in this region result in Type 2M VWD, whereas gain-of-function mutants produce a Type 2B phenotype in which HMWMs of VWF are missing from the plasma and the mutant protein binds platelets in the absence of any facilitating factor [30].
To date, approximately 26 different Type 2B VWD mutations have been described, all in exon 28 of the VWF gene and all affecting the structure of the VWF A1 domain in such a way as to mimic the physiologically activated state of this GPIb alpha binding domain. Several of the Type 2B missense mutations are frequent and recurrent (ie. R1306W, V1316M and R1341Q), and there is a good correlation between specific 2B variants and phenotypic characteristics such as thrombocytopenia and clinical bleeding [31]. While the origin of most of these mutations has not been established, and most are found in families known to transmit the mutations, at least one case of germinal mosaicism for a Type 2B mutation has been described [32].
Platelet-Type von Willebrand Disease Mutations
To date, the only evidence of locus heterogeneity for a qualitative VWF trait concerns the mutations described at the GPIBA locus that result in a glycoprotein Ib alpha receptor with enhanced binding capability for the VWF A1 domain. This phenocopy appears to be significantly less frequent than Type 2B mutants and only four mutations have been documented to date to result in a gain-of-function GPIb receptor. These mutations involve three missense substitutions in the glycoprotein Ib alpha, beta switch region (two different mutations at codon 233 and one at codon 239) and a 9 amino acid deletion mutant in the receptor’s macroglycopeptide domain [25, 33–36]. Presumably all of these changes stabilize the beta switch region of the receptor in the activated, VWF binding conformation [37,38].
Mutations Resulting in Reduced Collagen Binding
Rather surprisingly, given the central hemostatic role of VWF binding to subendothelial collagen, there have only been four missense variants (S1731T, W1745C, S1783A and H1786D) that adversely affect this interaction and result in a mild bleeding phenotype [39–41]. The index cases are all heterozygous for these variations.
All four of these mutations are located in the collagen binding A3 domain of VWF and laboratory analysis shows that they exhibit variable binding deficits to collagens I and III. Currently, these mutations are classified as Type 2M VWD, although pleas have been made to assign these cases to their own distinct subtype.
Mutations Resulting in Reduced Factor VIII Binding: Type 2N VWD
The second important hemostatic function of VWF is as a carrier protein for factor VIII (FVIII). The region of VWF involved in this function, encompasses the N-terminal D’/D3 domains of the mature VWF monomer.
Approximately 25 different Type 2N VWD mutations have now been described, all affecting the structure of the FVIII binding region of the protein [42,43]. Rarely, these mutations interfere with cleavage of the VWF propeptide, which then sterically hinders FVIII binding. However, most of the 2N mutations are missense substitutions in the VWF D’ and D3 domains and the phenotype is recessive in nature. Significant numbers of homozygous and compound heterozygous Type 2N cases have been documented, with, in particular, an important contribution from individuals in whom there has been co-inheritance of a 2N and null VWF allele. As with most of the qualitative VWF variants, Type 2N VWD demonstrates good genotype/phenotype correlations. Thus, the common R854Q mutation is associated with a mild phenotype and FVIII levels of approximately 25%, while both the recurrent T791M and R816W mutations result in FVIII levels of <10%.
Inheritance Patterns of Qualitative VWF Variants
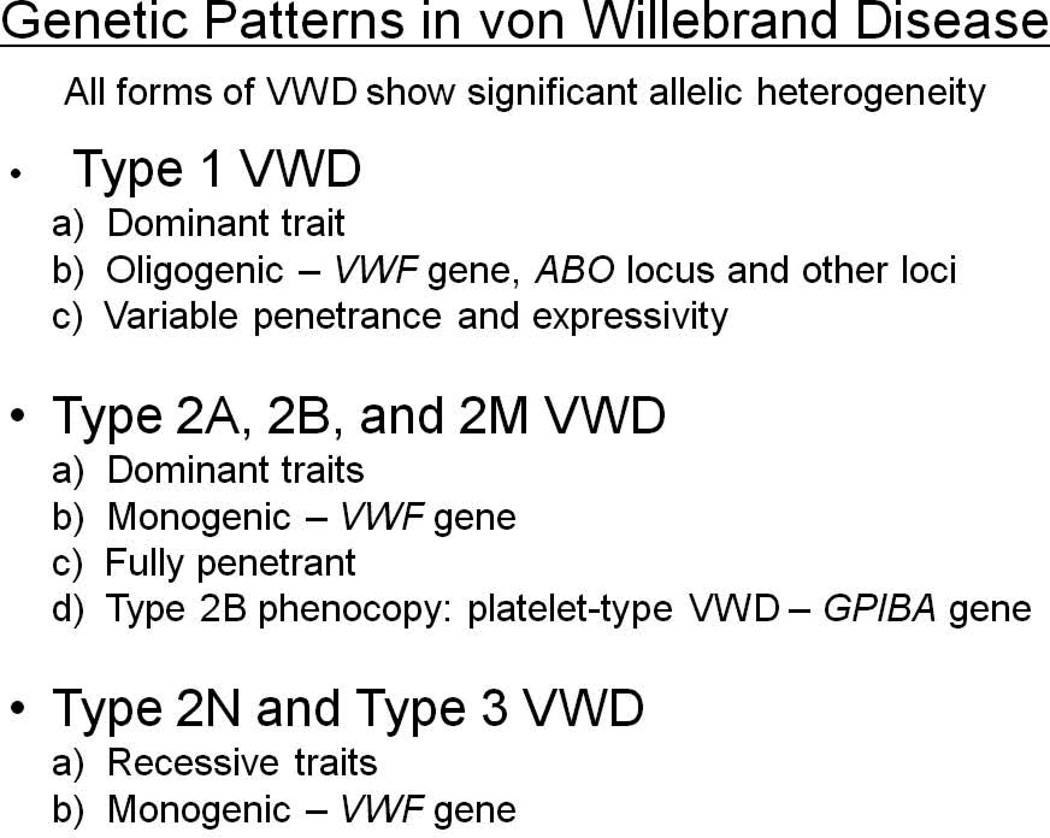
Almost all of the mutations resulting in Types 2A, 2M, 2B and platelet-type VWD show classical dominant inheritance patterns (there are rare cases of recessive 2A disease), with complete penetrance and minimally variable expressivity (Figure 3). Given the rarity of these conditions there is no documentation of homozygous cases of any of these phenotypes, although mouse models of 2A and 2B VWD suggest that, at least for these traits, the mutations are indeed dominant and not co-dominant variants [44,45]. In contrast, all cases of Type 2N disease are recessive in nature, as described in detail above.
Figure 3.
The genetic characteristics of von Willebrand disease subtypes.
Severe Quantitative von Willebrand Factor Mutations: Type 3 VWD
Type 3 VWD is the most severe and infrequent form of the disease. The prevalence of Type 3 disease ranges from 0.5 to 4 per million, and varies significantly depending upon factors such as the rates of consanguinity in different geographies.
The Type 3 VWD phenotype is characterized by undetectable levels of VWF, and FVIII levels that are usually <10% [46]. In accord with these laboratory findings, patients with Type 3 VWD usually demonstrate a more severe clinical phenotype than those with other VWD subtypes.
Type 3 VWD is a recessive trait with all affected subjects being either homozygous or compound heterozygous for mutations at the VWF locus. To date, there is no evidence of locus heterogeneity for type 3 disease.
The first group of mutations described to result in Type 3 VWD comprised of large total and partial VWF deletions [47,48]. In addition to demonstrating the typical severe Type 3 clinical phenotype, some of these patients had also developed anti-VWF antibodies following replacement therapy with VWF concentrates, a treatment-associated complication that occurs in approximately 5% of patients with this disorder.
The ISTH-SSC VWF online database currently lists 131 VWF mutations associated with Type 3 disease. This listing includes a wide array of mutation types including 26 nonsense (20% of total) and 27 different missense substitutions (20% of total) [49,50]. These latter variants likely result in the Type 3 phenotype through a variety of mechanisms involving abnormal biosynthetic processing, packaging and protein secretion [51]. However, it should be emphasized that details of the pathogenetic mechanisms for the majority of Type 3 missense alleles remain unresolved.
In addition to the large number of discrete mutations causing Type 3 disease, approximately 10% of the causative variants represent partial and total VWF gene deletions. In some instances, such as the recently described exon 4–5 deletion, where the mutational mechanism involves an Alu repeat-mediated unequal homologous recombination event, the deletions are recurrent in nature and represent a significant proportion of the Type 3 alleles in specific populations [52,53]. The prevalence of partial deletion alleles in Type 3 disease signifies that it is important for any mutation screening approach to include a methodology, such as ligation-mediated probe amplification, that will detect heterozygous deletion events.
In the Type 3 VWD populations that have been tested for mutations to date, >90% of the causative variants have been identified. Nevertheless, in all of these populations, there are still cases in which there are missing pathogenic alleles. With the advent of re-sequencing of the VWF locus, it will be interesting to see if these cases harbor intronic mutations or distant regulatory alleles that contribute to the severe phenotype.
Mild/Moderate Quantitative VWF Mutations: Type 1 VWD
The least well characterized, and easily most complex form of VWD is also the most common subtype, Type 1 VWD [54]. Approximately 75% of VWD cases fall into the category of Type 1 disease and thus most of the 1 per 1,000 prevalence of VWD relates to this condition.
While Type 1 VWD is classified in older text books as a monogenic dominant trait, there is a growing appreciation of the fact that the disorder represents a complex trait in which several (and possibly many) genetic influences interact with acquired factors to result in a low plasma level of functionally normal VWF. As VWF levels fall from the lower end of the normal range of 50% to less than 10% there is evidence from recent studies that there is a gradient of increasing influence of pathogenic VWF mutations [55,56]. To further complicate the genetic characterization of Type 1 disease, the condition shows incomplete penetrance and variable expressivity of some disease-causing alleles. All of these issues have combined to generate significant controversy about the definition of Type 1 VWD and this issue will be further addressed in the next section on VWD diagnosis.
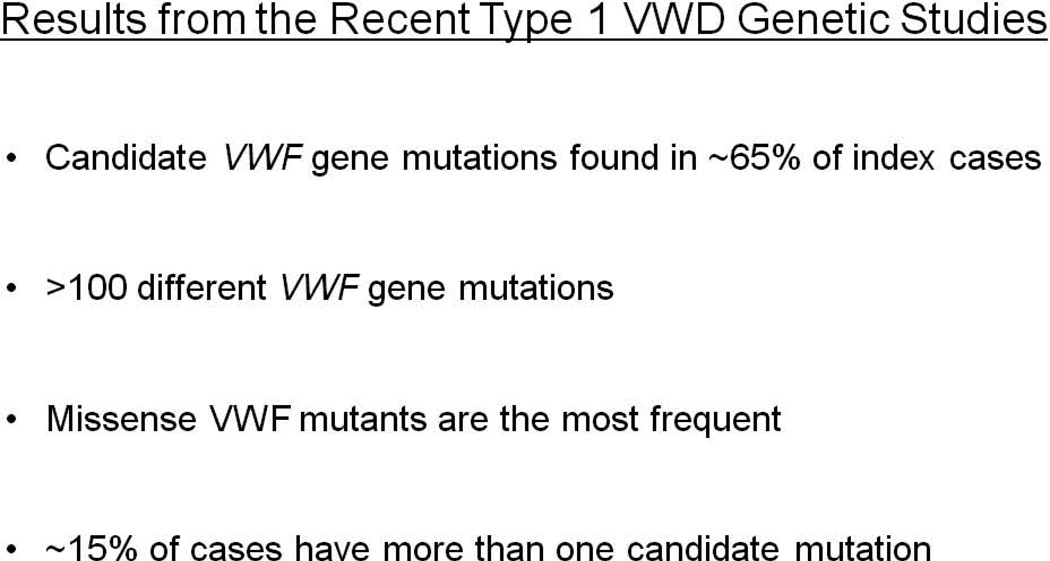
The recent large population studies of Type 1 VWD have all shown several common features concerning the pathogenetic details of the condition [55–58] (Figure 4): first, putative causative variants can be identified in approximately 65% of cases when the VWF proximal promoter, exons and splice boundaries are screened. The proportion of cases that demonstrate positive results increases as the Type 1 phenotype becomes more severe. Secondly, the disease demonstrates very significant allelic heterogeneity with >100 different mutations already described in the first four Type 1 VWD genetic studies. Thirdly, the most prevalent mutation type involves missense variants, although the precise pathogenic mechanisms involved in these cases, with rare exceptions, remains unresolved. Fourth, in approximately 15% of index cases, more than a single candidate VWF variant is present. Finally, as is becoming increasingly evident in many complex genetic traits, the distinction between pathogenic mutations and benign polymorphisms is often far from straightforward.
Figure 4.
Summary of key findings in the four recent Type 1 VWD genetic studies.
Two of the most frequently encountered Type 1 mutations, Y1784C and R1205H, illustrate the diversity of genetic characteristics and range of pathogenic mechanisms associated with this condition.
The Y1584C mutation has been identified in all Type 1 VWD populations examined to date and appears to be a frequent recurrent cause of this phenotype [59]. In the Canadian and UK studies this variant accounted for 14 and 25% of the Type 1 VWD alleles respectively [55,57]. In the common heterozygous state, Y1584C is associated with a VWF level of approximately 40%, whereas in rare homozygous subjects the VWF levels have been significantly lower at approximately 25%, suggesting a co-dominant mode of inheritance. This mutation, like many mild Type 1 VWD variants is often co-inherited with blood group “O”. This association will be discussed in more detail below. The phenotype associated with Y1584C is penetrant in approximately 70% of subjects who inherit the mutation. In terms of pathogenic mechanisms, in vitro analysis and evaluation of a mouse model of the mutation suggest that a combination of impaired biosynthesis and enhanced ADAMTS13 proteolysis contribute to the phenotype, although the latter effect is insufficient to justify re-classification of this variant as Type 2A VWD [59,60].
In contrast to the Y1584C mutant, R1205H, another recurrent missense mutation found in approximately 3% of Type 1 populations, is fully penetrant and always associated with a fairly severe phenotype, with VWF levels of 10–15% in heterozygotes [61]. Once again, the pathogenic mechanisms associated with this variant appear complex and involve evidence of accelerated clearance [62] and, in some instances, an abnormal multimer profile that may be secondary to the rapid clearance of the protein [63].
Other missense variants found in the Type 1 VWD population pose additional questions. For example the prototypic Type 2N mutation, R854Q, has been found repeatedly in the heterozygous state in type 1 VWD populations, but in these Type 1 patients it is associated with a quantitative VWF phenotype and not with disproportionately low FVIII levels. This finding remains unexplained. The R924Q variant is another missense substitution found with a prevalence of approximately 5% of Type 1 patients [55]. However, in vitro analysis of this mutant VWF has shown no evidence of biosynthetic changes and no changes to the multimer profile or rate of clearance. Thus, to date, it seems that R924Q may be a polymorphism that marks the causative variant that is located in an adjacent region of the VWF locus [64].
Another group of candidate mutations identified during the initial search of Type 1 VWD populations has involved a series of variants in the proximal regulatory region of the VWF locus. However, as with most of the candidate missense mutations identified in these studies, the mechanistic proof that these variants adversely affect transcription of the VWF gene is lacking. This proof has however been generated for a 13 bp deletion mutation that disrupts binding of an Ets transcription factor to the proximal promoter and significantly reduces transcriptional activity of a reporter transgene [65].
While the pathogenic mechanisms of most Type 1 VWD variants remain unresolved, recent evidence suggests that, at least for those that adversely affect biosynthesis, the examination of patient-derived endothelial cells (circulating progenitor cells that can be derived as blood outgrowth endothelial cells) may shed some light onto abnormalities in VWF trafficking, sorting into storage organelles and regulated secretion. Preliminary studies using this approach have already shown abnormal Weibel-Palade body formation and intrinsic VWF secretion in a few Type 1 missense variants [66]. These abnormalities are also pertinent to recent observations that additional non-VWF loci that play a role in vesicular trafficking and exocytosis may be important contributors to the Type 1 VWF phenotype. Finally, evidence of an accelerated clearance phenotype has also now been demonstrated for approximately 15% of Type 1 VWD cases [67,68] a finding that again pertains to some of the newly identified non-VWF loci associated with VWF levels (Figure 5).
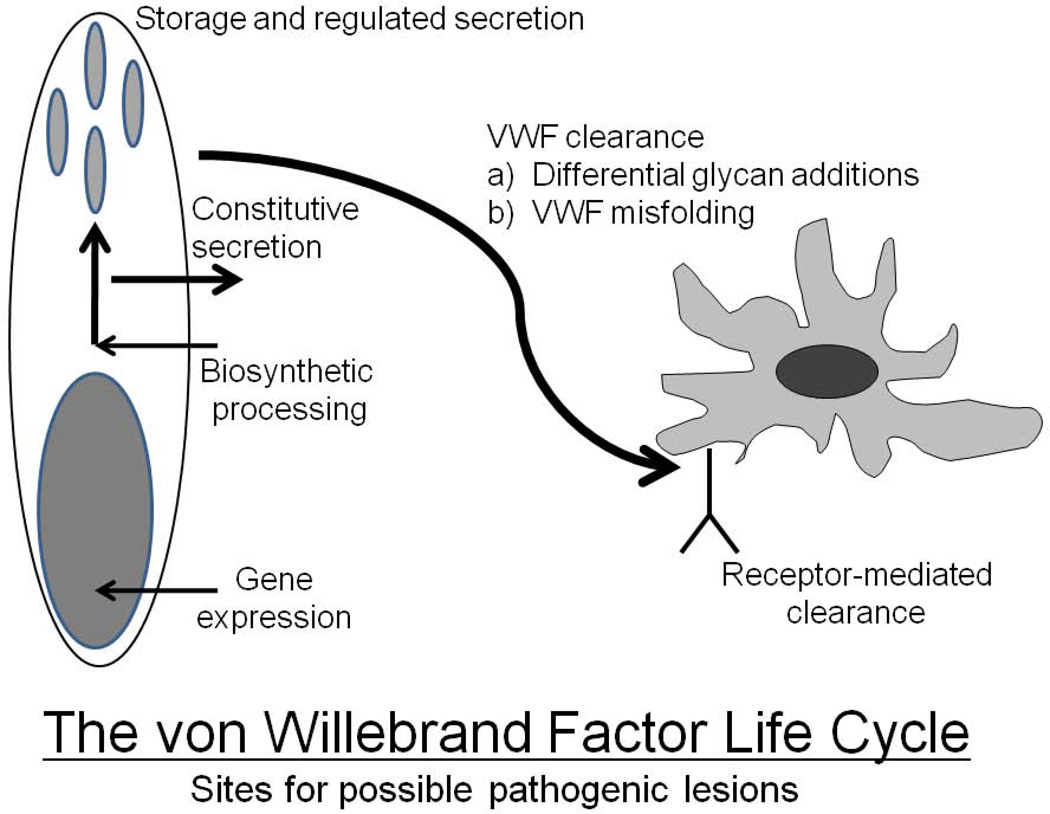
Figure 5.
The life-cycle of von Willebrand factor from gene expression to receptor-mediated clearance. Pathogenic mechanisms localized to any of these processes can result in a reduction in plasma VWF levels.
While candidate mutations have not been found in approximately 35% of all Type 1 VWD cases, of particular note, a few cases of relatively severe Type 1 disease (with VWF levels <25%) have demonstrated no mutations. In these cases, sequencing of the full VWF gene may identify causative intronic variants or more distant regulatory sequence mutations. Finally, and of direct relevance to the next section of this review, these cases may harbor mutations at alternative genetic loci that contribute to VWF synthesis, secretion and clearance.
Non-VWF Genetic Loci Involved in Quantitative VWF Trait Determination
While the phenotypes of Type 2 and 3 VWD are essentially monogenic traits with all of the influence derived from variation at the VWF locus, the quantitative trait represented by Type 1 VWD is increasingly recognized to be affected by changes at other genes and also by acquired environmental influences.
The best documented and most influential genetic modifier of VWF levels is the ABO blood group system. It has been known for several decades that subjects with blood group “O” have VWF levels that are approximately 25% lower than subjects with non-O blood groups [69]. This effect is also seen as a major contributor to mild cases of type 1 VWD where the co-inheritance of blood group “O” appears to play an important pathogenic role. Thus, in the Canadian Type 1 VWD study, the prevalence of blood group “O” in Type 1 index cases with VWF levels of >30% was 66%, whereas in cases where the VWF levels were <30%, blood group “O” was found in 50% of patients, a prevalence that reflects that found in the general Canadian population [55]. Studies of the influence of the ABO system on VWF levels indicate that this is the single most important genetic modifying factor for this trait, accounting for approximately 30% of the variability. Subsequent recent confirmation of the importance of the ABO system has come from genome-wide association studies (GWAS) where the statistical significance of association has been calculated to be as high as p< 5 × 10−342 [70]. The mechanism underlying the influence of the ABO glycan effect relates to the fact that approximately 13% of the N-linked glycan structures on VWF possess ABO antigens [71]. This post-translational modification, in turn influences the differential clearance of VWF, with group “O” VWF being cleared significantly faster than non-group “O” protein [72].
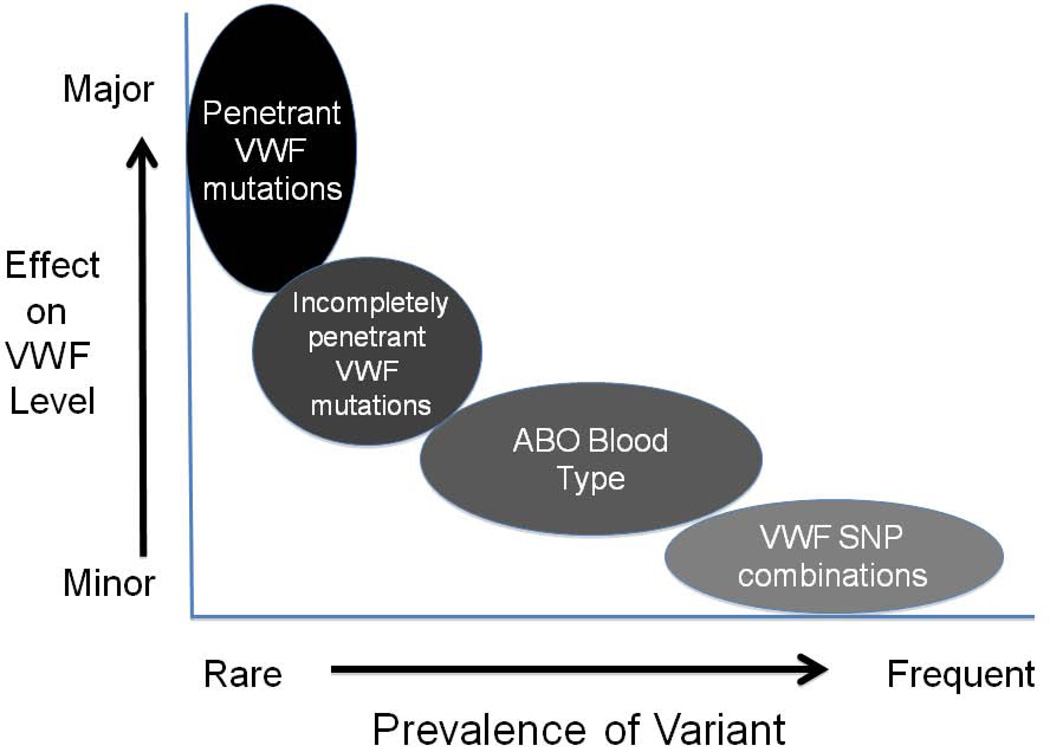
The role of other genetic loci as contributors to plasma VWF levels is now a subject of ongoing studies. Recent genome-wide and more focused association studies have identified several candidate loci that demonstrate both strong statistical association and have biologic plausibility for regulating VWF levels. In particular, the large CHARGE GWAS meta-analysis has identified several novel loci that show strong statistical associations with VWF levels that were reproduced in independent discovery and replication populations (Figure 6). Two of these novel loci, syntaxin 2 and syntaxin binding protein 5, are involved in vesicular trafficking and exocytosis, while three clearance receptor genes (Stabilin-2, C-type lectin domain family 4, member M and Scavenger receptor class A, member 5) were also found to be strongly associated with plasma VWF levels [70]. These initial studies therefore suggest that a number of novel genetic loci may contribute to both the biosynthetic and clearance behavior of VWF, although much remains to be learnt about the details and (patho)physiological significance of these effects. Of note, in interpreting the results from these large genetic association studies, it is important to keep in mind that all of these analyses have been performed in populations of healthy subjects. In these study groups, the individual contributions of each one of the loci to the variance in VWF levels is very small (∼1–3%), but the distinct possibility exists that rare variants at these same loci may act as either more significant genetic modifiers of the Type 1 VWD phenotype or even possibly as the primary pathogenic mutations (Figure 7).
Figure 6.
Novel non-VWF genetic loci characterized in the recent CHARGE genome-wide association study meta-analysis as being associated with plasma levels of VWF.
Figure 7.
A model for the influence of rare and frequent genetic variants on the levels of VWF.
The final and most recent piece of information relating to genetic modifiers of VWF levels also derives from genetic association studies but, in this instance, reverts to the VWF gene itself. In two recent association studies of VWF levels in healthy subjects, SNPs within the VWF gene again showed a strong association with VWF levels but both studies documented that this effect was only seen with SNPs located in the region of the gene that encodes the amino-terminal domains of the VWF monomer – ie. The D’ and D3 domains [73,74]. The significance of these observations requires further study, but may indicate that variants in this region of the protein which in addition to regulating FVIII binding is also crucial for multimer generation may play a role in determining VWF plasma levels through an influence on biosynthesis and/or clearance.
The Diagnosis of von Willebrand Disease: Conclusions from Recent VWD Population Studies
It is not the intent of this review to revisit details of the diagnostic process for VWD. This has been very well discussed in other recent reviews and consensus documents and has not changed appreciably in the last five years [9,75]. However, the results of the recent large population studies have highlighted several diagnostic points that merit brief presentation. Overall, a conclusion that is hard to avoid is that while most of the Type 2 variants and Type 3 VWD are relatively easy to diagnose, the diagnostic certainty for Type 1 disease often remains problematic and may be inherently impossible to simplify.
First to consider is the assessment of the clinical bleeding phenotype. The recent population studies employed different strategies to determine the bleeding phenotype in the study subjects. However, over the past 5–10 years there has been a progressive realization that objective, quantifiable scoring systems for mucocutaneous bleeding may facilitate the identification of clinical bleeders and also provide an estimate of the severity of the phenotype [76,77]. While these tools were initially devised for the assessment of bleeding in adults [78], recent evidence suggests that, with minimal revision, they can also prove useful in children [79]. Whether these bleeding assessment tools will have a significant impact upon the diagnostic definition of VWD remains to be seen, but at the very least, they provide a more objective measure of a bleeding tendency that can be easily communicated between health care professionals.
The second issue that warrants attention is the differentiation between Type 1 and certain forms (most often Type 2A) of qualitative VWD variants. These distinctions have usually involved demonstration of subtly abnormal VWF multimer patterns that have been generated by expert laboratories but are quite likely to be missed by most routine hemostasis laboratories [80]. Interestingly, when evaluated for VWF mutations as a group, the rate of positivity in these cases is close to 100%. Whether the re-classification of these cases as Type 2A variants is clinically useful is debatable, as in most cases, the therapeutic approach will still involve an initial trial of desmopressin.
The final issue that has been highlighted by the recent population studies is the uncertainty of a historical diagnosis of Type 1 VWD. The issue of when to assign this diagnostic label was initially raised in a Blood “Perspectives” article several years ago [81], and the controversy still resonates in the hemostasis community, despite comprehensive attempts to provide a definitive resolution [75]. The problem is unfortunately likely to be irreconcilably complex, involving, as it does, a continual quantitative variable (VWF plasma level), and both transient and progressive permanent variances to this level. The progressive influence of age on VWF levels (approximately 1% increase/year) makes the diagnosis of Type 1 VWD especially challenging in children, in whom exposure to bleeding events may also limit the potential for evaluation of the bleeding phenotype. These issues explain, at least in part, the 1–2% prevalence of VWD described in two earlier studies of children [82,83]. Recent guidelines have been proposed to facilitate the diagnosis but there will always be individuals in whom the diagnosis is tenuous and may change over time. An additional inevitable conclusion from the recent large genetic studies of this trait is that the extreme allelic heterogeneity and lack of identifiable candidate mutations in approximately 35% of Type 1 VWD cases makes the incorporation of genetic testing to assist in making this diagnosis very difficult to justify.
Concluding Summary
As the most common inherited bleeding disease, VWD has attracted significant interest from the basic science and clinical hemostasis communities over the past several decades. In the past five years, advances have been made in characterizing the genetic determinants that influence the complex quantitative trait, Type 1 VWD. Prior to then, the molecular pathology of the Type 2 VWD variants and of Type 3 VWD had already been comprehensively described. While some advance in our pathogenetic understanding of Type 1 VWD has clearly been achieved, the diagnostic challenges presented by this trait persist. Finally, the therapeutic options that are already safe and usually very effective at preventing or treating bleeding in VWD have not changed appreciably in recent years, apart from the arrival of a recombinant VWF concentrate in the clinic [84]. There is no doubt that much more needs to be learnt about the natural and pathological life cycles of VWF, and the recent large scale genetic studies have provided excellent starting points for these future investigations.
Acknowledgements
The authors’ studies of VWD are funded by grants from the Canadian Institutes for Health Research (MOP97849), the Heart and Stroke Foundation of Canada (NA 6386), The National Institutes of Health (HL081588) and the Canadian Hemophilia Society. DL is the recipient of a Canada Research Chair in Molecular Hemostasis.
References
- 1.von Willebrand EA. Hereditar pseudohemofili. Finska Lakaresallskapets Handlingar. 1926;LXVII:87–112. [Google Scholar]
- 2.Nilsson IM, Blomback M, Jorpes E, Blomback B, Johansson SA. Von Willebrand’s disease and its correction with human plasma fraction 1-0. Acta medica Scandinavica. 1957;159(3):179–188. doi: 10.1111/j.0954-6820.1957.tb00123.x. [DOI] [PubMed] [Google Scholar]
- 3.Zimmerman TS, Ratnoff OD, Powell AE. Immunologic differentiation of classic hemophilia (factor VIII deficiency) and von Willebrand’s dissase, with observations on combined deficiencies of antihemophilic factor and proaccelerin (factor v) and on an acquired circulating anticoagulant against antihemophilic factor. The Journal of clinical investigation. 1971;50(1):244–254. doi: 10.1172/JCI106480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ruggeri ZM, Zimmerman TS. Classification of variant von Willebrand’s disease subtypes by analysis of functional characteristics and multimeric composition of factor viii/von willebrand factor. Annals of the New York Academy of Sciences. 1981;370:205–209. doi: 10.1111/j.1749-6632.1981.tb29733.x. [DOI] [PubMed] [Google Scholar]
- 5.Ginsburg D, Handin RI, Bonthron DT, et al. Human von Willebrand factor (VWF): isolation of complementary DNA (cDNA) clones and chromosomal localization. Science. 1985;228(4706):1401–1406. doi: 10.1126/science.3874428. [DOI] [PubMed] [Google Scholar]
- 6.Sadler JE, Shelton-Inloes BB, Sorace JM, et al. Cloning and characterization of two cDNAs coding for human von Willebrand factor. Proc.Natl.Acad.Sci.U.S.A. 1985;82(19):6394–6398. doi: 10.1073/pnas.82.19.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lynch DC, Zimmerman TS, Collins CJ, et al. Molecular cloning of cDNA for human von Willebrand factor: authentication by a new method. Cell. 1985;41(1):49–56. doi: 10.1016/0092-8674(85)90060-1. [DOI] [PubMed] [Google Scholar]
- 8.Verweij CL, de Vries CJ, Distel B, et al. Construction of cDNA coding for human von Willebrand factor using antibody probes for colony-screening and mapping of the chromosomal gene. Nucleic Acids Res. 1985;13(13):4699–4717. doi: 10.1093/nar/13.13.4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sadler JE, Budde U, Eikenboom JC, et al. Update on the pathophysiology and classification of von Willebrand disease: a report of the subcommittee on von Willebrand factor. J Thromb Haemost. 2006;4(10):2103–2114. doi: 10.1111/j.1538-7836.2006.02146.x. [DOI] [PubMed] [Google Scholar]
- 10.Mannucci PM. Desmopressin (DDAVP) and factor VIII: the tale as viewed from Milan (and Malmo) J.Thromb.Haemost. 2003;1:1538–7933. doi: 10.1046/j.1538-7836.2003.00221.x. [DOI] [PubMed] [Google Scholar]
- 11.Pottinger BE, Read RC, Paleolog EM, Higgins PG, Pearson JD. Von Willebrand factor is an acute phase reactant in man. Thromb Res. 1989;53(4):387–394. doi: 10.1016/0049-3848(89)90317-4. [DOI] [PubMed] [Google Scholar]
- 12.Prentice CRM, Forbes CDSS. Rise of factor FVIII after exercise and adrenaline infusion measured by immunological and biological techniques. Thromb Res. 1972;1:493–506. [Google Scholar]
- 13.Vischer UM, Wollheim CB. Epinephrine induces von Willebrand factor release from cultured endothelial cells: involvement of cyclic AMP-dependent signalling in exocytosis. Thromb Haemost. 1997;77(6):1182–1188. [PubMed] [Google Scholar]
- 14.Liu L, Wang X, Lin Z, Wu H. Elevated plasma levels of VWF:Ag in hyperthyroidism are mediated through beta-adrenergic receptors. Endocrine Research. 1993;19(2–3):123–133. doi: 10.3109/07435809309033019. [DOI] [PubMed] [Google Scholar]
- 15.Dalton RG, Dewar MS, Savidge GF, et al. Hypothyroidism as a cause of acquired von Willebrand’s disease. Lancet. 1987;1(8540):1007–1009. doi: 10.1016/s0140-6736(87)92272-0. [DOI] [PubMed] [Google Scholar]
- 16.Clark P, Brennand J, Conkie JA, et al. Activated protein C sensitivity, protein C, protein S and coagulation in normal pregnancy. Thromb Haemost. 1998;79(6):1166–1170. [PubMed] [Google Scholar]
- 17.Stirling Y, Woolf L, North WR, Seghatchian MJ, Meade TW. Haemostasis in normal pregnancy. Thromb Haemost. 1984;52(2):176–182. [PubMed] [Google Scholar]
- 18.May M, Lawlor DA, Patel R, et al. Associations of von Willebrand factor, fibrin d-dimer and tissue plasminogen activator with incident coronary heart disease: British women’s heart and health cohort study. European journal of cardiovascular prevention and rehabilitation. 2007;14(5):638–645. doi: 10.1097/HJR.0b013e3280e129d0. [DOI] [PubMed] [Google Scholar]
- 19.Kumari M, Marmot M, Brunner E. Social determinants of von Willebrand factor: the Whitehall II study. Arteriosclerosis, Thrombosis, and Vascular Biology. 2000;20(7):1842–1847. doi: 10.1161/01.atv.20.7.1842. [DOI] [PubMed] [Google Scholar]
- 20.Ginsburg D. Molecular genetics of von Willebrand disease. Thromb Haemost. 1999;82(2):585–591. [PubMed] [Google Scholar]
- 21.Orstavik KH. Genetics of plasma concentration of von willebrand factor. Folia haematologica (Leipzig, Germany : 1928) 1990;117(4):527–531. [PubMed] [Google Scholar]
- 22.de Lange M, Snieder H, Ariëns RA, Spector TD, Grant PJ. The genetics of haemostasis: a twin study. Lancet. 2001;357(9250):101–105. doi: 10.1016/S0140-6736(00)03541-8. [DOI] [PubMed] [Google Scholar]
- 23.de Lange M, de Geus EJC, Kluft C, et al. Genetic influences on fibrinogen, tissue plasminogen activator-antigen and von Willebrand factor in males and females. Thrombosis and haemostasis. 2006;95(3):414–419. doi: 10.1160/TH05-09-0596. [DOI] [PubMed] [Google Scholar]
- 24.Meyer D, Fressinaud E, Gaucher C, et al. Gene defects in 150 unrelated French cases with type 2 von Willebrand disease: from the patient to the geneinserm network on molecular abnormalities in von willebrand disease. Thromb Haemost. 1997;78(1):451–456. [PubMed] [Google Scholar]
- 25.Miller JL, Cunningham D, Lyle VA, Finch CN. Mutation in the gene encoding the alpha chain of platelet glycoprotein Ib in platelet-type von willebrand disease. Proceedings of the National Academy of Sciences of the United States of America. 1991;88(11):4761–4765. doi: 10.1073/pnas.88.11.4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lyons SE, Bruck ME, Bowie EJ, Ginsburg D. Impaired intracellular transport produced by a subset of type IIA von Willebrand disease mutations. J Biol Chem. 1992;267(7):4424–4430. [PubMed] [Google Scholar]
- 27.O’Brien LA, Sutherland JJ, Weaver DF, Lillicrap D. Theoretical structural explanation for group I and group II, type 2A von Willebrand disease mutations. J Thromb Haemost. 2005;3(4):796–797. doi: 10.1111/j.1538-7836.2005.01219.x. [DOI] [PubMed] [Google Scholar]
- 28.Hillery CA, Mancuso DJ, Evan SJ, et al. Type 2M von Willebrand disease: F606I and I662F mutations in the glycoprotein Ib binding domain selectively impair ristocetin- but not botrocetin-mediated binding of von Willebrand factor to platelets. Blood. 1998;91(5):1572–1581. [PubMed] [Google Scholar]
- 29.James PD, Notley C, Hegadorn C, et al. Challenges in defining type 2M von Willebrand disease: results from a Canadian cohort study. J Thromb Haemost. 2007;5(9):1914–1922. doi: 10.1111/j.1538-7836.2007.02666.x. [DOI] [PubMed] [Google Scholar]
- 30.Ruggeri ZM. Type IIb von willebrand disease: a paradox explains how von Willebrand factor works. J Thromb Haemost. 2004;2(1):2–6. doi: 10.1111/j.1538-7836.2003.00523.x. [DOI] [PubMed] [Google Scholar]
- 31.Federici AB, Mannucci PM, Cast+aman G, et al. Clinical and molecular predictors of thrombocytopenia and risk of bleeding in patients with von Willebrand disease type 2B: a cohort study of 67 patients. Blood. 2008;113:526–534. doi: 10.1182/blood-2008-04-152280. [DOI] [PubMed] [Google Scholar]
- 32.Murray EW, Giles AR, Lillicrap D. Germ-line mosaicism for a valine-to-methionine substitution at residue 553 in the glycoprotein Ib-binding domain of von Willebrand factor, causing type IIB von Willebrand disease. Am.J.Hum Genet. 1992;50:199–207. [PMC free article] [PubMed] [Google Scholar]
- 33.Russell SD, Roth GJ. Pseudo-von Willebrand disease: a mutation in the platelet glycoprotein Ib alpha gene associated with a hyperactive surface receptor. Blood. 1993;81(7):1787–1791. [PubMed] [Google Scholar]
- 34.Takahashi H, Murata M, Moriki T, et al. Substitution of Val for Met at residue 239 of platelet glycoprotein Ib alpha in Japanese patients with platelet-type von Willebrand disease. Blood. 1995;85(3):727–733. [PubMed] [Google Scholar]
- 35.Matsubara Y, Murata M, Sugita K, Ikeda Y. Identification of a novel point mutation in platelet glycoprotein Ib alpha, Gly to Ser at residue 233, in a Japanese family with platelet-type von Willebrand disease. Journal of Thromb Haemost. 2003;1(10):2198–2205. doi: 10.1046/j.1538-7836.2003.00369.x. [DOI] [PubMed] [Google Scholar]
- 36.Othman M, Notley C, Lavender FL, et al. Identification and functional characterization of a novel 27-bp deletion in the macroglycopeptide-coding region of the GPIba gene resulting in platelet-type von Willebrand disease. Blood. 2005;105(11):4330–4336. doi: 10.1182/blood-2002-09-2942. [DOI] [PubMed] [Google Scholar]
- 37.Huizinga EG, Tsuji S, Romijn RA, et al. Structures of glycoprotein Ib alpha and its complex with von Willebrand factor A1 domain. Science. 2002;297(5584):1176–1179. doi: 10.1126/science.107355. [DOI] [PubMed] [Google Scholar]
- 38.Sadler JE. Biomedicine. contact--how platelets touch von Willebrand factor. Science. 2002;297(5584):1128–1129. doi: 10.1126/science.1075452. [DOI] [PubMed] [Google Scholar]
- 39.Ribba AS, Loisel I, Lavergne JM, et al. Ser968Thr mutation within the A3 domain of von Willebrand factor (VWF) in two related patients leads to a defective binding of VWF to collagen. Thromb.Haemost. 2001;86:848–854. [PubMed] [Google Scholar]
- 40.Riddell A, Gomez K, Millar C, et al. Characterisation of W1745C and S1783A, two novel collagen binding defects in the A3 domain of von Willebrand factor. Blood. 2008;112 doi: 10.1182/blood-2008-10-184317. [DOI] [PubMed] [Google Scholar]
- 41.Flood VH, Lederman CA, Wren JS, et al. Absent collagen binding in a VWF A3 domain mutant: utility of the VWF:CB in diagnosis of VWD. Journal of Thromb Haemost. 2010;8(6):1431–1433. doi: 10.1111/j.1538-7836.2010.03869.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mazurier C. Von Willebrand disease masquerading as haemophilia A. Thromb Haemost. 1992;67(4):391–396. [PubMed] [Google Scholar]
- 43.Mazurier C, Hilbert L. Type 2N von Willebrand disease. Curr.Hematol Rep. 2005;4(5):350–358. [PubMed] [Google Scholar]
- 44.Golder M, Pruss CM, Hegadorn C, et al. Mutation-specific hemostatic variability in mice expressing common type 2B von Willebrand disease substitutions. Blood. 2010;115(23):4862–4869. doi: 10.1182/blood-2009-11-253120. [DOI] [PubMed] [Google Scholar]
- 45.Pruss CM, Golder M, Hegadorn C, et al. The impact of von Willebrand factor sequence changes that affect ADAMTS13-mediated cleavage in the von Willebrand factor knock-out mouse model. J Thromb Haemost. 2009;7:11. s2. [Google Scholar]
- 46.Eikenboom JC. Congenital von Willebrand disease type 3: clinical manifestations, pathophysiology and molecular biology. Best.Pract.Res.Clin.Haematol. 2001;14:365–379. doi: 10.1053/beha.2001.0139. [DOI] [PubMed] [Google Scholar]
- 47.Ngo KY, Glotz VT, Koziol JA, et al. Homozygous and heterozygous deletions of the von Willebrand factor gene in patients and carriers of severe von willebrand disease. Proc.Natl.Acad.Sci.U.S.A. 1988;85(8):2753–2757. doi: 10.1073/pnas.85.8.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shelton-Inloes BB, Chehab FF, Mannucci PM, Federici AB, Sadler JE. Gene deletions correlate with the development of alloantibodies in von willebrand disease. J Clin.Invest. 1987;79(5):1459–1465. doi: 10.1172/JCI112974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baronciani L, Cozzi G, Canciani MT, et al. Molecular defects in type 3 von willebrand disease: updated results from 40 multiethnic patients. Blood Cells Mol.Dis. 2003;30(3):264–270. doi: 10.1016/s1079-9796(03)00033-0. [DOI] [PubMed] [Google Scholar]
- 50.Sutherland MS, Keeney S, Bolton-Maggs PHB, et al. The mutation spectrum associated with type 3 von Willebrand disease in a cohort of patients from the North West of England. Haemophilia. 2009;15(5):1048–1057. doi: 10.1111/j.1365-2516.2009.02059.x. [DOI] [PubMed] [Google Scholar]
- 51.Baronciani L, Federici AB, Cozzi G, et al. Expression studies of missense mutations p.D141Y, p.C275S located in the propeptide of von willebrand factor in patients with type 3 von Willebrand disease. Haemophilia. 2008;14:549–555. doi: 10.1111/j.1365-2516.2008.01682.x. [DOI] [PubMed] [Google Scholar]
- 52.Sutherland MS, Cumming AM, Bowman M, et al. A novel deletion mutation is recurrent in von Willebrand disease types 1 and 3. Blood. 2009;114(5):1091–8. doi: 10.1182/blood-2008-08-173278. [DOI] [PubMed] [Google Scholar]
- 53.Schneppenheim R, Castaman G, Federici AB, et al. A common 253-kb deletion involving VWF and TMEM16B in German and Italian patients with severe von Willebrand disease type 3. J Thromb Haemost. 2007;5(4):722–728. doi: 10.1111/j.1538-7836.2007.02460.x. [DOI] [PubMed] [Google Scholar]
- 54.Mohlke KL, Ginsburg D. Von Willebrand disease and quantitative variation in von Willebrand factor. J Lab Clin.Med. 1997;130(3):252–261. doi: 10.1016/s0022-2143(97)90019-6. [DOI] [PubMed] [Google Scholar]
- 55.James PD, Notley C, Hegadorn C, et al. The mutational spectrum of type 1 von Willebrand disease: results from a Canadian cohort study. Blood. 2007;109(1):145–154. doi: 10.1182/blood-2006-05-021105.. [DOI] [PubMed] [Google Scholar]
- 56.Goodeve A, Eikenboom J, Castaman G, et al. Phenotype and genotype of a cohort of families historically diagnosed with type 1 von Willebrand disease in the European study, molecular and clinical markers for the diagnosis and management of type 1 von Willebrand disease (MCMDM-1VWD) Blood. 2007;109(1):112–121. doi: 10.1182/blood-2006-05-020784. [DOI] [PubMed] [Google Scholar]
- 57.Cumming A, Grundy P, Keeney S, et al. An investigation of the von Willebrand factor genotype in UK patients diagnosed to have type 1 von Willebrand disease. Thromb Haemost. 2006;96(5):630–641. [PubMed] [Google Scholar]
- 58.Bellissimo DB, Christopherson PA, Flood VH, et al. Von Willebrand factor mutations and new sequence variations identified in healthy controls are more frequent in the African-American population. Blood. 2011 doi: 10.1182/blood-2011-10-384610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.O’Brien LA, James PD, Othman M, et al. Founder von Willebrand factor haplotype associated with type 1 von Willebrand disease. Blood. 2003;102(2):549–557. doi: 10.1182/blood-2002-12-3693. [DOI] [PubMed] [Google Scholar]
- 60.Pruss CM, Golder M, Bryant A, et al. Pathologic mechanisms of type 1 VWD mutations R1205H and Y1584C through in vitro and in vivo mouse models. Blood. 2011;117(16):4358–4366. doi: 10.1182/blood-2010-08-303727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schneppenheim R, Federici AB, Budde U, et al. Von Willebrand disease type 2M “Vicenza” in Italian and German patients: identification of the first candidate mutation (G3864A; R1205H) in 8 families. Thromb Haemost. 2000;83(1):136–140. [PubMed] [Google Scholar]
- 62.Casonato A, Pontara E, Sartorello F, et al. Reduced von Willebrand factor survival in type Vicenza von Willebrand disease. Blood. 2002;99(1):180–184. doi: 10.1182/blood.v99.1.180. [DOI] [PubMed] [Google Scholar]
- 63.Gézsi A, Budde U, Deák I, et al. Accelerated clearance alone explains ultra-large multimers in von Willebrand disease Vicenza. Journal of thromb and Haemost. 2010;8(6):1273–180. doi: 10.1111/j.1538-7836.2010.03753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hickson N, Hampshire D, Winship P, et al. Von Willebrand factor variant p.Arg924Gln marks an allele associated with reduced von Willebrand factor and factor FVIII levels. J Thromb Haemost. 2010 doi: 10.1111/j.1538-7836.2010.03927.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Othman M, Chirinian Y, Brown C, et al. Functional characterization of a 13 bp deletion (c.-1522_-1510del13) in the promoter of the von Willebrand factor gene in type 1 von Willebrand disease. Blood. 2010;116(18):3645–1252. doi: 10.1182/blood-2009-12-261131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang J-W, Groeneveld DJ, Cosemans G, et al. Biogenesis of Weibel-Palade bodies in von Willebrand disease variants with impaired von Willebrand factor intrachain or interchain disulfide bond formation. Haematologica. 2011 doi: 10.3324/haematol.2011.057216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Haberichter SL, Castaman G, Budde U, et al. Identification of type 1 von willebrand disease patients with reduced von Willebrand factor survival by assay of the VWF propeptide in the European study: molecular and clinical markers for the diagnosis and management of type 1 VWD (MCMDM-1VWD) Blood. 2008;111(10):4979–4985. doi: 10.1182/blood-2007-09-110940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Haberichter SL, Balistreri M, Christopherson P, et al. Assay of the von Willebrand factor (VWF) propeptide to identify patients with type 1 von Willebrand disease with decreased vwf survival. Blood. 2006;108(10):3344–3351. doi: 10.1182/blood-2006-04-015065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gill JC, Endres-Brooks J, Bauer PJ, Marks WJ, Jr, Montgomery RR. The effect of ABO blood group on the diagnosis of von Willebrand disease. Blood. 1987;69(6):1691–1695. [PubMed] [Google Scholar]
- 70.Smith NL, Chen MH, Dehghan A, et al. Novel associations of multiple genetic loci with plasma levels of factor VII, factor VIII, and von Willebrand factor: the charge (cohorts for heart and aging research in genome epidemiology) consortium. Circulation. 2010;121(12):1382–1392. doi: 10.1161/CIRCULATIONAHA.109.869156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Matsui T, Titani K, Mizuochi T. Structures of the asparagine-linked oligosaccharide chains of human von Willebrand factor occurrence of blood group A, B, and H (O) structures. J Biol.Chem. 1992;267(13):8723–8731. [PubMed] [Google Scholar]
- 72.Gallinaro L, Cattini MG, Sztukowska M, et al. A shorter von Willebrand factor survival in O blood group subjects explains how ABO determinants influence plasma von Willebrand factor. Blood. 2008;111:3540–3545. doi: 10.1182/blood-2007-11-122945. [DOI] [PubMed] [Google Scholar]
- 73.Campos M, Sun W, Yu F, et al. Genetic determinants of plasma von Willebrand factor antigen levels: a target gene SNP and haplotype analysis of ARIC cohort. Blood. 2011;117(19):5224–5230. doi: 10.1182/blood-2010-08-300152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zabaneh D, Gaunt TR, Kumari M, et al. Genetic variants associated with von Willebrand factor levels in healthy men and women identified using the HumanCVD beadchip. Annals of human genetics. 2011;75(4):456–467. doi: 10.1111/j.1469-1809.2011.00654.x. [DOI] [PubMed] [Google Scholar]
- 75.Nichols WL, Hultin MB, James AH, et al. Von Willebrand disease (VWD): evidence-based diagnosis and management guidelines, the National Heart, Lung, and Blood Institute (NHLBI) expert panel report (USA) Haemophilia. 2008;14:171–232. doi: 10.1111/j.1365-2516.2007.01643.x. [DOI] [PubMed] [Google Scholar]
- 76.Tosetto A, Castaman G, Rodeghiero F. Bleeding scores in inherited bleeding disorders: clinical or research tools? Haemophilia. 2008;14:415–422. doi: 10.1111/j.1365-2516.2007.01648.x. [DOI] [PubMed] [Google Scholar]
- 77.Rodeghiero F, Castaman G, Tosetto A, et al. The discriminant power of bleeding history for the diagnosis of type 1 von willebrand disease: an international, multicenter study. J.Thromb.Haemost. 2005;3(12):2619–2626. doi: 10.1111/j.1538-7836.2005.01663.x. [DOI] [PubMed] [Google Scholar]
- 78.Rodeghiero F, Tosetto A, Castaman G. How to estimate bleeding risk in mild bleeding disorders. J Thromb Haemost. 2007;5:157–166. doi: 10.1111/j.1538-7836.2007.02520.x. [DOI] [PubMed] [Google Scholar]
- 79.Biss TT, Blanchette VS, Clark DS. Quantitation of bleeding symptoms in children with von Willebrand disease: use of a standardized pediatric bleeding questionnaire. J Thromb Haemost. 2010;8:950–956. doi: 10.1111/j.1538-7836.2010.03796.x. [DOI] [PubMed] [Google Scholar]
- 80.Budde U, Schneppenheim R, Eikenboom J, et al. Detailed von Willebrand factor multimer analysis in patients with von Willebrand disease in the European study, molecular and clinical markers for the diagnosis and management of type 1 von Willebrand disease (MCMDM-1VWD) Journal of Thromb Haemost. 2008;6:762–771. doi: 10.1111/j.1538-7836.2008.02945.x. [DOI] [PubMed] [Google Scholar]
- 81.Sadler JE. Von Willebrand disease type 1: a diagnosis in search of a disease. Blood. 2003;101(6):2089–2093. doi: 10.1182/blood-2002-09-2892. [DOI] [PubMed] [Google Scholar]
- 82.Rodeghiero F, Castaman G, Dini E. Epidemiological investigation of the prevalence of von Willebrand's disease. Blood. 1987;69(2):454–459. [PubMed] [Google Scholar]
- 83.Werner EJ, Broxson EH, Tucker EL. Prevalence of von Willebrand disease in children: a multiethnic study. J Pediatr. 1993;123(6):893–898. doi: 10.1016/s0022-3476(05)80384-1. [DOI] [PubMed] [Google Scholar]
- 84.Turecek PL, Schrenk G, Rottensteiner H, et al. Structure and function of a recombinant von Willebrand factor drug candidate. Seminars in Thrombosis and Hemostasis. 2010;36(5):510–21. doi: 10.1055/s-0030-1255445. [DOI] [PubMed] [Google Scholar]