Summary
Since the cloning of the gene that encodes von Willebrand factor (VWF), 27 years ago, significant progress has been made in our understanding of the molecular basis of the most common inherited bleeding disorder, von Willebrand disease (VWD). The molecular pathology of this condition represents a range of genetic mechanisms, some of which are now very well characterized, and others that are still under investigation. In general, our knowledge of the molecular basis of type 2 and 3 VWD is now well advanced, and in some instances this information is being used to enhance clinical management. In contrast, our understanding of the molecular pathogenesis of the most common form of VWD, type 1 disease, is still at an early stage, with preliminary evidence that this phenotype involves a complex interplay between environmental factors and the influence of genetic variability both within and outside of the VWF locus.
Keywords: von Willebrand disease, von Willebrand factor, Factor VIII
In 1926, on the Island of Föglö in the Åland archipelago in the Baltic Sea, a Finnish physician, Erik von Willebrand, described the first cases of an inherited bleeding disease that now bears the eponymous name, von Willebrand disease (VWD) (Von Willebrand, 1926). His investigation of Family “S” on Föglö revealed a severe inherited bleeding condition that had resulted in the death of 3 children before the age of 4 years from gastrointestinal bleeds in two cases and a tongue bleed in the third sibling. His studies of the VWD index case, Hjördis, also documented evidence of significant bleeding from an early age, with a major lip and several ankle bleeds before the age of 5 years. Hjördis went on to die from her fourth menstrual bleed at the age of 14 years. At the time, Dr. von Willebrand thought that the underlying cause of this bleeding was severe platelet dysfunction combined with a vessel wall abnormality. The condition was given the name of hereditary pseudo-haemophilia (Von Willebrand, 1926).
We now know that Family “S” was affected by type 3 VWD and that the severe bleeding phenotype was the consequence of an absence of the pro-coagulant adhesive protein, von Willebrand factor (VWF).
Brief Recent History of von Willebrand Disease Advances
Following Erik von Willebrand’s observations on the original VWD families, the next period of investigation began in 1957, with the involvement of Drs. Inge Marie Nilsson and Margareta Blombäck from the Karolinska Institute in Stockholm (Nilsson et al, 1957). They documented prolonged bleeding times and a reduction in the level of plasma anti-haemophilic globulin (now recognized as factor VIII [FVIII]) in affected patients and showed that the infusion of the plasma fraction 1-0 (containing several large proteins including VWF, FVIII and fibrinogen) corrected the haemostatic defect. However, it was not until 1971, with the development of an immunological test to differentiate VWF and FVIII, that the characteristics of VWD and haemophilia A were clarified (Zimmerman et al, 1971). Very soon, it became apparent that variant forms of VWF were present in some patients with VWD (Peake et al, 1974), and by 1987 the detailed phenotypic testing available had resulted in the sub-classification of 11 variants of type 1 VWD and 13 type 2 variants. However, it was the simultaneous cloning of the VWF cDNA by four laboratories in 1985 that initiated the advances we have witnessed in the molecular characterization of VWD over the past 27 years (Ginsburg et al, 1985; Sadler et al, 1985; Lynch et al, 1985; Verweij et al, 1985).
Brief Summary of the von WiIlebrand Factor Gene and Protein
The gene that encodes VWF is located on the short arm of chromosome 12. The VWF gene comprises 178 kb of genomic DNA arranged into 52 exons ranging in size from 41 bp to 1.3 kb (exon 28). Complicating the genetic analysis of VWF is the fact that chromosome 22 carries a partial VWF pseudogene that replicates the chromosome 12 sequence between exons 23 and 34, with a 3% variance (Mancuso et al, 1991).
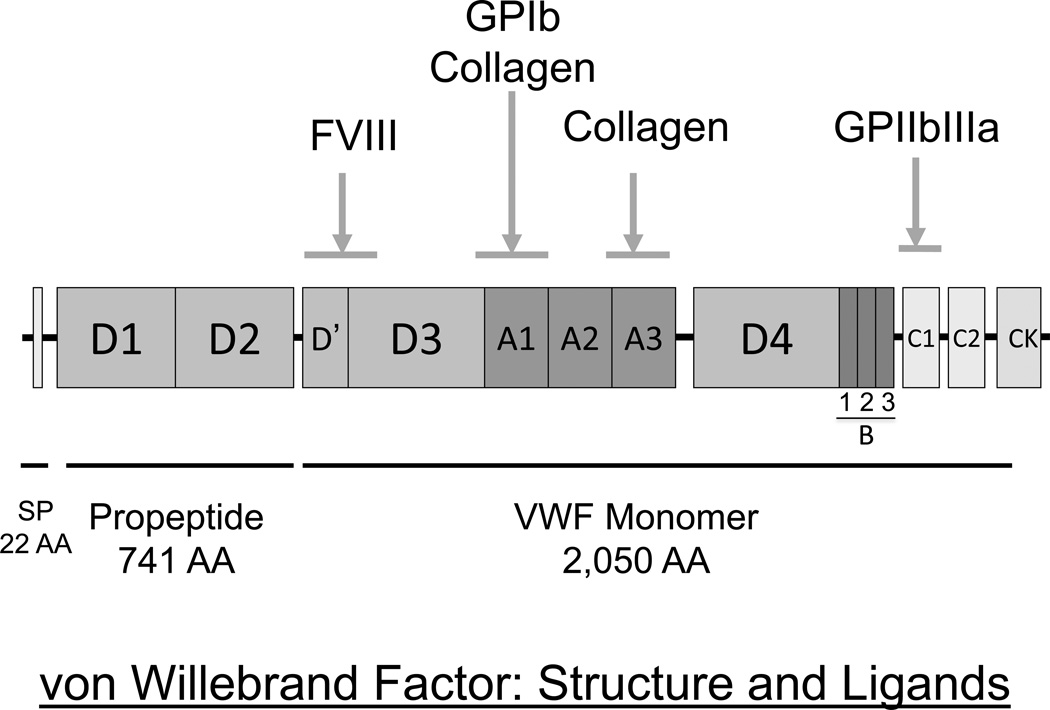
The VWF gene encodes a 9 kb transcript that is translated into a 2,813 amino acid pre-pro-polypeptide sequence (Figure 1). The 22 amino acid pre-sequence enables entry into the endoplasmic reticulum, and the 741 residue propeptide mediates alignment of VWF dimers into N-linked multimeric forms (Sadler, 1998). The VWF propeptide is cleaved from the mature subunit in the golgi and is secreted into the plasma as disulfide-linked dimers. The biological function of the VWF propeptide in the circulation, if any exists, is unknown. The 2,050 residue mature VWF subunit possesses binding domains for FVIII (D’D3 domains), platelet glycoprotein Ib (A1 domain), collagen (A1 and A3 domains) and platelet glycoprotein IIb/IIIa (C1 domain). These multiple binding functions enable VWF to mediate platelet adhesion and aggregation, and to protect FVIII from premature proteolysis by activated protein C (Koppelman et al, 1996).
Figure 1.
Primary structure of the von WiIlebrand factor (VWF) pre-pro-polypeptide and its ligands. There is a typical 22 amino acid (AA) hydrophobic signal peptide, a large 741 AA propeptide responsible for facilitating N-terminally aligned multimer formation, and a 2050 AA secreted mature VWF subunit containing the binding sites for FVIII, platelets and the subendothelium. CK=cysteine knot domain.
Following secretion from its cell of synthesis (either endothelial cells, ~85% of plasma VWF, or platelets – derived from megakaryocytic expression) (Wagner & Marder, 1983; Sporn et al, 1985), VWF undergoes limited proteolytic digestion by the metalloprotease ADAMTS13 through cleavage at between Tyr1605-Met1606 in the VWF A2 domain (Tsai, 1996). Access to this cleavage site requires partial unfolding of the A2 domain, an alteration mediated by circulatory shear stress (Zhang et al, 2009).
von Willebrand Disease Classification
The von Willebrand factor Scientific and Standardization Subcommittee (SSC) of the International Society on Thrombosis and Haemostasis (ISTH) has overseen the recent classification schemes for VWD, with the latest recommendation being published in 2006 (Sadler et al, 2006). There have been only minor changes in our thinking about VWD categorization since then, including recognition of the clinical impact of accelerated VWF clearance in a subset of Type1 VWD patients.
Type 1 VWD represents 70–80% of VWD cases and is characterized by a mild to moderate reduction in qualitatively normal VWF. The precise level of reduction below which a diagnosis of type 1 VWD can be made has been the subject of much recent debate and continues to be problematic (Sadler, 2003; Nichols et al, 2008).
Type 2 VWD represents a variety of traits in which qualitatively abnormal VWF molecules result in defective haemostasis. There are four variants of type 2 VWD: 2A, 2B, 2M and 2N that constitute approximately 20% of all cases of VWD. In most cases of types 2A, 2B and 2M disease, the qualitative VWF abnormality is accompanied by a reduction in plasma VWF antigen (VWF:Ag) and/or ristocetin cofactor activity (VWF:RCo) levels but there is a disproportionate reduction of VWF activity (VWF:RCo) compared to the VWF:Ag level (VWF:RCo to VWF:Ag ratio <0.6) (James et al, 2007a). Type 2A and 2M variants show a reduction in the platelet-dependent function of VWF. In type 2A disease this is due to the absence of high (and sometimes intermediate) molecular weight VWF multimers, while in type 2M VWD, there is a qualitative abnormality of VWF that is unrelated to the loss of high molecular weight (HMW) VWF and the defect in platelet-dependent function is due to missense substitutions in the A1 domain of VWF. In type 2B VWD, the missense substitutions result in a gain-of-function phenotype where the most haemostatically active forms of VWF bind to the platelet surface causing a thrombocytopenia in some cases and a plasma VWF deficit that results in a bleeding phenotype (Ruggeri, 2004). Finally, type 2N VWD presents with low plasma FVIII levels (between 0.05 and 0.40 u/ml) usually in the absence of any quantitative abnormalities of VWF (Mazurier et al, 2001).
It should be emphasized that the performance and interpretation of phenotypic tests for VWD is challenging (Chandler et al 2011; Meijer & Haverkate 2006).
In addition to the fact that there is significant temporal variability of VWF levels, the functional analysis (VWF:RCo and VWF:CB) and multimer profiling of VWF are tests that are usually confined to specialist laboratories in which the diagnosis of VWD is routinely investigated.
The most infrequent variant of VWD, type 3 disease, is due to a severe deficiency of VWF (<0.02 u/ml) and an accompanying major reduction in plasma FVIII, with levels only rarely being >0.10 u/ml (Eikenboom, 2001). The prevalence of type 3 VWD varies with geographical location, from levels of approximately 0.1 to 5 per million, depending upon factors such as rates of consanguineous marriages.
In this review, we will highlight advances in the molecular characterization of VWD associated with each of the sub-types described above. Suffice it to say that progress in understanding the molecular basis of types 2 and 3 VWD has been substantial, whereas, the molecular pathogenesis of type 1 disease continues to be complex and only partially explained.
We will discuss the details of current knowledge in the order, type 3, 2 and finally type 1 VWD.
Type 3 von Willebrand Disease Molecular Pathology
This condition has been thought of as a classical recessive trait, but further inspection of even the index Åland Island family with type 3 disease, shows that varying manifestations of a haemostatic defect and bleeding phenotype are not uncommon in type 3 VWD heterozygotes. Thus, while ~50% of Type 3 families will exhibit a typical recessive inheritance pattern, the remaining cases appear to demonstrate features of co-dominant inheritance, with either one or both parents showing laboratory and clinical evidence of type 1 VWD (Montgomery, 2006). Whatever the pattern of inheritance, two mutant alleles are required to generate this phenotype.
The first cases of type 3 VWD to be characterized by molecular genetic approaches showed complete and partial gene deletions and were associated with the development of anti-VWF immune responses following exposure of the patients to VWF replacement therapy (Ngo et al, 1988; Shelton-Inloes et al, 1987). The frequency of these immune reactions remains unclear, as no prospective, systematic study of this treatment complication has ever been undertaken. However, the few case series that have been reported indicate that this is a rare phenomenon with a prevalence of <5% (Mannucci et al, 1981).
Since these initial reports, more than 100 different type 3 VWF mutations have been described. Details of these mutations can be found at the ISTH-sponsored VWF Online Database (http://www.vwf.group.shef.ac.uk/ accessed on September 4th 2012). The mutations represent a miscellany of mutation types that are spread over the entire VWF gene sequence and involve regions of VWF from the extreme N-terminus of the pre-propolypeptide to the C-terminus of the mature VWF subunit. Most of the mutations result in clear null phenotypes (deletions, nonsense, frameshift and consensus splice site mutations) but a substantial number of missense variants have also been documented. In many instances, the pathogenetic mechanism underlying these missense substitutions has not been clarified. There are several recurrent type 3 mutations that have been identified which are probably the result of founder effects: the original Åland island small deletion mutation (c.2435delC) (Zhang et al, 1993), a partial deletion mutation involving loss of exons 1–3 (Mohl et al, 2008), and a deletion of exons 4 and 5 leading to an in-frame mutant transcript (Sutherland et al, 2009). Interestingly, neither of the two recurrent partial deletion mutations appears to be associated with the development of VWF alloantibodies.
In the small number of series of type 3 VWD patients that have been subjected to genetic analysis, causative mutations have been found in 80–90% of cases. The explanation for the missing mutations is unclear but most probably reflects the fact that all studies to date have only analysed the coding regions, splice sites and proximal promoter of the VWF gene. Further investigation of these cases may well reveal additional pathogenic variants deep in intron sequences and possibly also in more distant regulatory sequences. Certainly, to date, there is no evidence of genetic locus heterogeneity for type 3 VWD.
While the molecular pathology for type 3 VWD is now increasingly being characterized, and much more is being learnt about VWF structure/function details from these studies, this new information has relatively limited clinical utility. The diagnosis of type 3 VWD is still readily made with clinical findings and routine haemostasis testing for VWF and FVIII. However, there are two situations where molecular genetics does provide additional clinical benefit. The first is in the context of prenatal testing, where chorionic villus or amniocyte analysis can provide a definite fetal diagnosis of type 3 VWD, and the second is perhaps in facilitating the identification of patients with an increased propensity for anti-VWF antibody responses. Patients with partial or total VWF deletion mutations may be at greater risk of anti-VWF immune responses, although this proposal has not been substantiated with any prospective study, and, as noted above, there are recurrent partial VWF deletion mutations that are not associated with inhibitor development. At least part of the explanation for this discrepancy probably relates to the potential for the synthesis of small amounts of in-frame truncated mutant protein, sufficient to mediate central tolerance to VWF.
Type 2 von Willebrand Disease Molecular Pathology
In all of the four subtypes of type 2 VWD, a mutant VWF protein is produced that has an abnormal function. Three of the type 2 VWD subtypes (types 2A, 2B and 2M) show a dominant inheritance pattern with complete penetrance of the phenotype in heterozygotes. In contrast, type 2N VWD is transmitted as a recessive trait requiring the inheritance of either homozygous of compound heterozygous 2N mutations (either two different type 2N missense mutations or a combination of a 2N missense mutation and a VWF null mutation) to manifest the low plasma FVIII levels that result from reduced binding to FVIII. In the vast majority of cases, type 2 VWD is the result of missense substitutions affecting regions of the VWF protein involved in binding to either platelets (types 2A, 2B and 2M), collagen (type 2M) or FVIII (type 2N) (Meyer et al, 1997) (Figure 2). Recent studies employing both detailed phenotypic testing and molecular genetics have highlighted that, in some instances, the distinction between type 1 VWD and types 2A and 2M VWD continues to be challenging (Goodeve et al, 2007; James et al, 2007a)
Figure 2.
Location of mutations resulting in Type 2 von Willebrand disease. Almost all of these mutations are missense variants. Type 2A substitutions affect predominantly the A2 domain, with some additional substitutions in the propeptide, D’ domain and the cysteine knot region. Almost all type 2B and 2M substitutions are located in the A1 domain, and the loss-of-function FVIII binding mutations in type 2N disease are in the D’/D3 domains.
Type 2A von Willebrand Disease Molecular Details
In this loss-of-platelet dependent function phenotype, patients show reduced VWF:RCo/VWF:Ag ratios (<0.6) and a loss of high molecular weight VWF multimers.
More than 70 different mutations have been described to result in type 2A VWD. Greater than 95% of the mutations are missense substitutions and approximately 80% of the substitutions are located in exon 28. Most of these substitutions change amino acids in the A2 domain.
As described soon after the delineation of type 2A VWD, there are two pathogenetic mechanisms that contribute to this phenotype (Lyons et al, 1992): the inability to synthesize and secrete high molecular weight VWF multimers and an enhanced propensity for ADAMTS13-mediated proteolysis. More recent in vitro studies have shown that many type 2A mutations produce a complex phenotype with abnormalities affecting several stages of VWF biosynthesis, storage, release and proteolytic processing (Jacobi et al, 2012).
The large number of missense type 2A mutations located in exon 28 produce their effect through varying contributions from all of the above detailed pathogenetic mechanisms. Some of these mutations are recurrent (ie. R1597W, S1506L) and are generated through either CpG dinucleotide hypermutability (at arginine codons) and/or through founder effects.
Outside of exon 28 and the large number of A2 domain mutants, there are small numbers of type 2A mutations in the propeptide and at the extreme C-terminus of the mature subunit that interfere with the formation of VWF multimers and dimers, respectively.
Detailed characterization of the molecular pathology underlying type 2A VWD has added important knowledge to our understanding of the biosynthesis and proteolytic processing of VWF. However, the translational advances derived from this information are minimal. The diagnosis of type 2A disease is readily achievable with standardized testing of VWF:Ag, VWF:RCo, the VWF multimer profile and a test for ristocetin-induced platelet agglutination (RIPA). Molecular testing does not provide any enhancements over good standardized haemostasis testing, and in particular, with the exception of a few intensively investigated variants (ie. R1597W, associated with enhanced ADAMTS13-mediated proteolysis), does not predict the predominant pathogenetic mechanism responsible for the clinical phenotype. This latter shortcoming indicates that all type 2A VWD patients should probably undergo a trial of desmopressin responsiveness with inclusion of a 4-h time point to evaluate the possible occurrence of accelerated proteolysis.
Type 2B von Willebrand Disease Molecular Details
The mutations responsible for type 2B VWD produce a classical gain-of-function phenotype in which binding of the mutant A1 domain of VWF to the glycoprotein (Gp) Ibα receptor is enhanced (Ruggeri et al, 1980). The effect of these mutations mimics the influence of shear, and probably results in a change in conformation of the A1 domain from a ‘resting’ state to a hyperactive configuration.
There are >20 different substitutions that result in type 2B VWD, all within exon 28 of the VWF gene and most affecting residues at the base of the A1 domain that interact with the β-hairpin region of GpIbα (Huizinga et al, 2002). Some of these mutations are recurrent, most often affecting hypermutable CpG dinucleotide sequences in arginine codons – ie. R1341Q, R1308C and R1306W. Interestingly, the recurrent V1316M mutation is the cause of the phenotype in the previously described Montreal Platelet Disorder, a condition that is now recognized as one form of type 2B VWD (Jackson et al, 2009).
One of the phenotypic characteristics of type 2B disease is the persistent thrombocytopenia that worsens with increased mutant VWF levels (such as during pregnancy) in some cases. There is a good genotype/phenotype correlation with this phenomenon, with some mutations consistently being associated with more severe thrombocytopenic states that are accentuated by stress and predict for more bleeding (Federici et al, 2008).
In terms of translational advances pertaining to type 2B disease, the invariable localization of these mutations to exon 28 makes this VWD subtype a feasible target for molecular genetic diagnosis. Thus, after initial haemostasis studies suggesting this variant (ie. reduced VWF:RCo/VWF:Ag ratio, loss of high molecular weight multimers and enhanced RIPA), sequencing of exon 28 can be used to confirm the diagnosis. In cases where the VWF exon 28 sequence analysis is negative, the phenocopy, platelet-type VWD, should be considered and sequencing of the GP1BA gene undertaken (Othman, 2007).
Type 2M von Willebrand Disease Molecular Details
Type 2M VWD represents the loss-of-function equivalent to type 2B disease. In this subtype, missense mutations in the A1 domain adversely affect binding to platelet GpIbα. Notably, these substitutions occur throughout the apex of the VWF A1 domain and very likely interrupt a productive interaction with the GpIbα β-switch region (Huizinga et al, 2002). More than 25 different missense mutants have been described to result in this phenotype. There is recent evidence that type 2M VWD is associated with a milder bleeding phenotype than most other types of VWD (Castaman et al, 2012).
Type 2M VWD is differentiated from the other loss of platelet function variant, type 2A VWD, through the analysis of the VWF multimer profile, with type 2M plasma showing a normal complement of high molecular weight multimers. Some cases of type 2M VWD may be difficult to differentiate from type 1 disease, but the lower the VWF:RCo/VWF:Ag ratio (or VWF collagen binding activity [VWF:CB]/VWF:Ag ratio) below 0.6 the more likely that missense mutations located in the A1 domain will be identified characteristic of a 2M diagnosis (James et al, 2007a). This differentiation is of therapeutic importance as the administration of desmopressin will be of benefit in most type 1 cases but will not be able to rescue the loss-of-function phenotype in type 2M patients.
While the designation of type 2M disease was originally intended for mutants with defective binding to platelet GpIb, there are now several dysfunctional variants that show reduced binding to collagen, and these variants are also currently assigned the 2M categorization (Ribba et al, 2001; Riddell et al, 2009; Flood et al, 2010). VWF binds to three types of collagen: types I, III and VI, with VWF residues in the A3 domain interacting with types I and III collagen, and the A1 domain being involved in binding to all three types of collagen. In the routine clinical haemostasis laboratory, when VWF:CB is evaluated, the collagen substrates that are used are either collagen I or III, or a mix of I and III (Flood et al, 2012a). Binding to collagen type VI is rarely, if ever, tested.
To date, four collagen binding missense substitutions have been identified in the A3 domain: S1731T, W1745C, S1783A and H1786D (Ribba et al, 2001; Riddell et al, 2009; Flood et al, 2010). These mutants show a pattern of dominant inheritance with reduced binding in heterozygotes to collagens I and III. The bleeding symptoms in patients affected by these mutations are mild.
Very recently, the first mutations adversely affecting binding to collagen VI have been described: R1399H, S1387I and Q1402P (Flood et al, 2012b). These missense variants are located in the A1 domain. However, to complicate matters, these variants have also been found in some healthy individuals without bleeding symptoms. More studies are needed to determine the impact of abnormal collagen binding on the clinical bleeding phenotype. In some instances, it appears that these variants are sufficient alone to result in bleeding, whereas in other instances, these changes may act as secondary genetic modifiers of the phenotype.
Evaluation of the VWF:CB should be included in the diagnostic investigation of patients with mild bleeding phenotypes. In some of these patients, reduced collagen binding may be the only test abnormality found (Flood et al, 2012a). Whether this testing should be performed with only type I and III collagen, or should also include type VI collagen is yet to be resolved. While molecular genetic analysis of the A1 and A3 domain coding regions may identify missense variants in these cases, unless they are identical to previously reported cases, the significance of these changes may be unclear.
Type 2N von Willebrand Disease Molecular Details
Type 2N disease, for which there are now more than 25 different mutant alleles, presents as an autosomal recessive form of mild/moderate FVIII deficiency (Mazurier, 1992). VWF levels are often normal and FVIII levels can range from ~0.05 to 0.40 u/ml. In those type 2N patients with reduced VWF:Ag levels, the molecular basis of the phenotype probably represents the coinheritance of a 2N and null allele (see below).
The location of type 2N missense mutations is predictably in the D’D3 domains of VWF, the region of the protein to which FVIII binds. In addition, rare mutants have also been described at the extreme C-terminus of the VWF propeptide sequence that interfere with cleavage and release of the mature VWF subunit (Casonato et al, 2003). In these cases of ‘hereditary persistence of proVWF’ it is assumed that the attached propeptide sequence sterically interferes with FVIII binding to the adjacent D’D3 domains.
Type 2N disease is the only prevalent recessive form of type 2 VWD. While in many instances the two mutant alleles represent homozygous inheritance of a missense mutation or compound heterozygosity for two different missense alleles, there is a significant prevalence of compound heterozygosity for a 2N missense mutation in combination with a VWF null allele.
The other clinically relevant aspect of type 2N mutations is their correlation with the plasma level of FVIII and the potential to predict the therapeutic response to desmopressin (Mazurier et al, 2001). Thus, the most prevalent type 2N mutation, R854Q, is associated with FVIII levels of ~25% and a reasonable likelihood that desmopressin administration will provide a satisfactory therapeutic benefit for at least minor procedures. In contrast, the recurrent missense substitutions, R816W and T791M, result in levels of FVIII <10% and poor desmopressin responsiveness.
The significant majority of type 2N mutations are located in exons 18, 19 and 20, with infrequent mutations in exons 17, and 21–27. Thus, a targeted molecular genetic approach can be used to confirm haemostasis test studies that suggest this diagnosis. Whether this genetic approach to type 2N confirmation is preferable to a strategy involving a FVIII binding assay depends very much upon the local availability of resources.
Type 1 von Willebrand Disease Molecular Pathology
In most populations, type 1 VWD represents the most prevalent form of the disease, accounting for 70–80% of cases. In type 1 VWD patients, the mild/moderate mucocutaneous bleeding phenotype is associated with mild/moderate reductions (0.05–0.50 u/ml) in the plasma levels of qualitatively normal VWF. The recent debate about what VWF level should be chosen for defining the diagnosis of type 1 VWD continues (Sadler, 2003; Nichols et al, 2008), and while a positive bleeding history will often help resolve this dilemma, this may not be the case for young children and males.
In marked contrast to the genetic pathology associated with type 2 and 3 VWD, the pathogenetic details of type 1 VWD are complex, and have only recently begun to be characterized. While type 3 VWD is inherited as either a monogenic recessive or co-dominant trait and most forms of type 2 VWD as monogenic dominant traits (apart from type 2N, which is a recessively inherited), type 1 disease is a complex oligo- or multigenic dominant trait that demonstrates significant variability of penetrance and expressivity. Estimates of the heritability of plasma VWF levels range from ~30–75% depending upon the experimental approach undertaken to derive this figure (De Lange et al, 2001; Orstavik, 1990).
Prior to the recent, initial studies of type 1 VWD genetics, there was already evidence that various ‘environmental’ factors contributed to the variability in plasma VWF levels. Thus, increased levels of oestrogens and thyroid hormones are associated with increased VWF levels (Liu et al, 1993), while hypothyroidism results in low levels of VWF (Dalton et al, 1987). In addition, VWF is an acute phase protein that demonstrates increased levels with a variety of stress situations (Pottinger et al, 1989). Finally, and of significant importance in patients with mild VWF deficiency states, levels of VWF increase by 1–2% with each year of age (Conlan et al, 1993; Coppola et al, 2003). Thus, many type 1 VWD patients with initial levels of VWF of around 0.30 u/ml will show a progressive increase in their VWF concentration until, by middle age, they have normalized their levels. Whether the bleeding phenotype in these patients also disappears has yet to be determined. This may depend, at least in part, on whether levels of platelet VWF also show an increase with age, a fact that is currently unknown. Finally, it is noteworthy that the mechanistic basis for neither the hormone responsiveness nor the age-related increases in VWF protein levels has been characterized.
Until the report of the recent type 1 VWD studies described below, details of the genetic basis of this VWD subtype were limited. There had been occasional reports of VWF missense mutations, mostly affecting cysteine codons and interfering with VWF biosynthesis, but the most obvious genetic contributor to low VWF levels was the ABO (H) blood group locus (Orstavik, 1990; Gill et al, 1987). The ABO (H) effect on VWF levels represents approximately 30% of the genetic influence on this phenotype. The observation that levels of VWF are approximately 25% lower in individuals with blood group O compared to subjects with non-O blood groups has been recognized for several decades (Gill et al, 1987). The mechanistic basis for this effect seems most likely to be due to accelerated clearance of group O VWF (Gallinaro et al, 2008) and presumably relates to the complex glycan modifications experienced by VWF. Approximately 20% of the mass of VWF comprises carbohydrate, with 13 N-linked and 10 O-linked glycan sites on the VWF mature subunit (Canis et al, 2012)(Canis et al, 2010). ABO (H) antigen additions are made to almost all of the N-linked glycans, and are attached in a non-uniform distribution to ~13% of the glycan structures at these sites. Whether additional genetic modifiers of plasma VWF levels exist that relate to glycan variances remains to be determined.
Results from Initial Type 1 von Willebrand Disease Studies
Over the past five years, four cohort studies have undertaken the initial characterization of VWF gene variation in patients with type 1 VWD (Goodeve et al, 2007; James et al, 2007b; Cumming et al, 2006; Montgomery et al, 2011). In total, approximately 500 index cases of type 1 VWD have now been studied in these series and the genetic analysis has initially evaluated the VWF coding region, splice sites and proximal promoter regions. The results from all four studies have consistently reported that candidate VWF mutations have been documented in approximately 65% of index cases. These results are also consistent with previous genetic linkage and association studies showing that the type 1 phenotype segregates with the VWF gene in only approximately 50% of families (James et al, 2006; Eikenboom et al, 2006). Thus, both sets of data suggest that genetic loci outside of, and in addition to, the VWF gene are very likely to contribute to the determination of low plasma VWF levels in this quantitative trait.
In the four recent type 1 VWD cohort studies, the pattern of candidate VWF mutations was consistent and can be summarized as follows: more than 100 different candidate mutations have been documented, located throughout the coding region of the VWF sequence. Candidate missense mutations comprise the largest group of variants, accounting for approximately 70% of the changes. In approximately 15% of index cases, there is more than a single variant present. Finally, the likelihood of finding candidate mutations increased with lower VWF levels, such that in the Canadian study, VWF variants were found in 75% of index cases with VWF:Ag levels <0.30 u/ml, but only in 50% of cases with VWF:Ag levels >0.30 u/ml (James et al, 2007b).
Some of the candidate mutations identified in these studies have been found repeatedly (ie. Y1584C, R1205H, R924Q and R1315C), and at least some of these variants appear to derive from founder effects (O’Brien et al, 2003).
The major issue that currently remains unanswered for many of these candidate mutations is the lack of definitive evidence for their role in type 1 VWD pathogenesis. While in vitro studies addressing this issue have been performed for some of the variants (Eikenboom et al, 2009), combined in rare instances with results from in vivo mouse model studies (Pruss et al, 2011), the potential pathogenic nature of most of the variants remains unresolved. There is additional recent evidence to suggest that this dilemma will only become more problematic as the role of sequence variation in different ethnic groups is determined. Initial analysis in a healthy African American population has shown that >60% of this population had evidence of VWF sequence variation and that despite not reporting symptoms of a bleeding disorder, several of the previously reported VWF candidate mutations were found in these subjects (Bellissimo et al, 2012). These findings highlight the complexity of assigning primary pathogenicity to these variations, and we might expect over the next few years, as data from studies such as the 1,000 Genomes project emerge, that further challenges to this area of discovery will become apparent.
One particular group of candidate mutations that merit special mention is that associated with accelerated VWF clearance (Table I). There is evidence that in approximately 15% of type 1 VWD cases, accelerated clearance of VWF is the primary pathogenic mechanism (Type 1C VWD). This estimate derives from studies utilizing quantification of the plasma VWF propeptide/VWF:Ag ratio as an indirect measure of VWF clearance (Haberichter et al, 2006; Haberichter et al, 2008). The prototypic clearance mutant is the VWF Vicenza, R1205H, variant (Castaman et al, 2009). The phenotype associated with this variant shows VWF levels of ~0.15 u/ml, a markedly elevated VWFpp/VWF:Ag ratio (often >10), an exaggerated (4- to 10-fold increase in VWF levels) but short lived (VWF half- life <3 h) response to desmopressin, and sometimes subtly abnormal VWF multimer patterns (occasionally, supranormal high molecular weight bands) (Mannucci et al, 1988). Other accelerated clearance mutants have also been localized to the D3 region of VWF, suggesting that this region of the protein may play a key role in regulating VWF clearance. Recognition of this variant is important clinically because of the short-lived desmopressin response which may necessitate the use of VWF concentrates in some settings.
Table I.
Summary of the phenotypic features of the accelerated clearance mutants of type 1 von Willebrand disease (type 1C VWD). A predominant accelerated clearance phenotype can be documented in approximately 15% of type 1 VWD cases.
| Accelerated Clearance Type 1 von Willebrand Disease: (Type 1C) |
|---|
| • VWF:Ag <30% often <15% |
| • VWF:pp to VWF:Ag ratio >2 up to >10 |
| • Exaggerated DDAVP response >4-fold up to 10-fold |
| • Short term benefit from DDAVP (2–4 h) |
| • Plasma VWF half-lives <3 h |
| • Subtly abnormal VWF multimer profiles |
VWF:Ag, von Willebrand antigen; VWFpp, von WiIlebrand factor propeptide; DDAVP, desmopressin.
In terms of clinical utility, with the exception of the Type 1C variants, the recent discoveries pertaining to type 1 VWD may all be some distance from delivering clear benefits. While the issue has been debated, it seems increasingly improbable that molecular diagnostic testing for type 1 VWD will see widespread use in the foreseeable future (Peake & Goodeve, 2010; Favaloro, 2010), and even some of the genotype/phenotype associations that have been described, such as those attempting to predict desmopressin responsiveness (Castaman et al, 2008), are unlikely to change clinical practice.
Future Prospects for Type 1 von Willebrand Disease Genetic Characterization
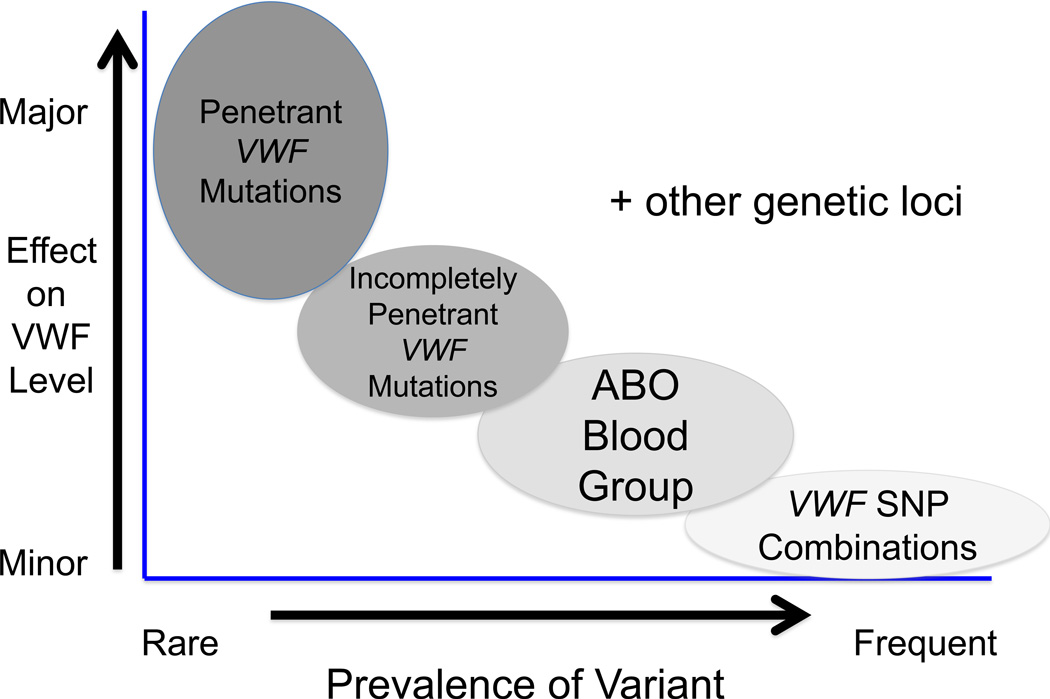
With the publication of the initial reports of the genetic landscape of type 1 VWD, it is rapidly becoming apparent that this clinically variable condition will be matched by significant genetic heterogeneity. It is already evident that there is major allelic heterogeneity and that at least some of the candidate mutant alleles have questionable primary pathogenetic relevance. In the ~35% of index cases where candidate mutations are not present in the VWF coding region, splice sites and promoter, further studies will first be aimed at evaluating novel pathogenic variation in the VWF introns and at more distant transcriptional regulatory regions. Furthermore, with increasing evidence of significant polymorphic variability of the VWF locus, the possibility exists that quantitative influences could derive not from single site variants, but from distinct polymorphic haplotypes within VWF (Campos et al, 2011) (Figure 3).
Figure 3.
Proposal for the molecular pathogenesis of the phenotype in type 1 VWD. There is growing evidence that many (most) cases of mild/moderately severe type 1 VWD are due to a combination of genetic variables both within and outside the VWF locus. Some of these sequence variances may involve distinct polymorphic haplotypes within VWF, some may be located adjacent to, but outside of the VWF gene, and additional modifiers may involve other genes involved in VWF biosynthesis, secretion and clearance. SNP, single nucleotide polymorphism.
In addition to considerable more work being required to characterize the role of the VWF locus in determining VWF levels, there is also a growing appreciation that genetic loci in addition to the VWF and ABO genes are contributing to the type 1 VWD phenotype.
One recent piece of supporting evidence for the role of genetic modifiers at other loci comes from the large CHARGE (Cohorts for Heart and Aging Research in Genome Epidemiology) genome-wide association study (GWAS) meta-analysis (Smith et al, 2010). In this analysis of genetic associations with plasma levels of FVII, FVIII and VWF in normal subjects, several important observations were made with regards to the genetic regulation of VWF. First, the important role for the VWF and ABO genes in determining VWF levels was robustly confirmed. Furthermore, a number of novel loci were also identified in both the discovery and replication GWAS cohorts as contributors to VWF plasma levels (Table II). An evaluation of the functional role of the proteins encoded by these loci suggests that most of them are plausible candidates for modifiers of VWF biosynthesis, storage and release, or clearance (Figure 4). In this GWAS meta-analysis involving normal subjects, the contribution of each of these individual loci to plasma VWF levels is very small (<5%). Thus, in type 1 VWD patients one might envisage at least two ways in which variations at these loci could influence VWF levels: common variants at these loci may act as minor genetic modifiers of levels, while rare variants could play a more significant role in determining the plasma VWF concentration.
Table II.
Novel genetic associations with plasma von Willebrand factor levels as determined by the CHARGE genome-wide association meta-analysis (Smith et al, 2010). Of the five genes identified by this strategy in the initial discovery populations and in an independent replication cohort, two encode proteins involved in vesicular trafficking, storage and secretion, and three encode receptor proteins that might contribute to VWF clearance.
| CHARGE GWAS-identified Novel Genetic Loci associated with plasma VWF levels | |
|---|---|
| Vesicular trafficking and exocytosis proteins | |
| a) STXBP5 | Syntaxin binding protein 5 |
| b) STX2 | Syntaxin 2 |
| Receptor proteins | |
| a) SCARA5 | Scavenger receptor class A member 5 |
| b) STAB2 | Stabilin 2 |
| c) CLEC4M | C-type Lectin Type 4 Member M |
Figure 4.
Potential sites of pathogenic variants resulting in quantitative von Willebrand factor (VWF) traits (type 1 and 3 von Willebrand disease). Genetic variables anywhere along the VWF life cycle from the regulation of gene expression to protein clearance may result in quantitative phenotypes. In some instances, single variables may produce a dominant effect but in mild/moderate quantitative phenotypes combinations of variables may be more prevalent.
Finally, in addition to the application of GWAS to identify novel loci influencing plasma VWF levels there are also ongoing studies employing linkage analysis as the initial strategy to localize novel modifier genes. These studies are showing preliminary evidence of additional modifying loci on chromosomes 2 (K Desch, University of Michigan, Ann Arbor, MI, USA), 6 and 16 (J DiPaola, University of Colorado, Denver, CO, USA). The magnitude of the influence of these loci remains to be seen.
Conclusions and Summary Statement
Overall, VWD represents an excellent paradigm for the range of pathogenetic possibilities for a human genetic disease, with clear examples of monogenic recessive, co-dominant, and dominant inheritance and now, with type 1 disease, growing evidence of a complex oligo (multi)genic disease model. In some instances, these advances in our understanding of the molecular pathology of VWD have clear clinical utility (ie. prenatal testing for type 3 VWD and genetic confirmation of type 2N disease (Table III). In other cases, such as the genetic basis of type 1 disease, our basic knowledge of the genetic factors that determine plasma VWF levels needs substantial enhancement before any clear translational benefit could be envisaged.
Table III.
Summary of the prevalence, mutation detection success rates and potential clinical utility of molecular genetic testing for the various subtypes of von Willebrand disease (VWD). Note that the prevalence figures for the various type 2 forms of VWD vary significantly between different geographical locations.
| Type of VWD |
Prevalence (%) |
Molecular Test Success (%) |
Utility of Diagnostic Molecular Analysis |
|---|---|---|---|
| 1 | 75% | ~65% | Currently not beneficial |
| 2A | ~8% | ~85% | Confirmatory of phenotypic analysis |
| 2B | ~8% | ~85% | Confirmatory of phenotypic analysis |
| 2M | ~2% | ~85% | Confirmatory of phenotypic analysis |
| 2N | ~8% | ~85% | Confirmatory of phenotypic analysis |
| 3 | <1% | ~85% | Prenatal testing |
Acknowledgements
DL is the recipient of a Canada Research Chair in Molecular Hemostasis. The authors’ research on von Willebrand disease is funded by the Canadian Institutes for Health Research (MOP 97849), the Heart and Stroke Foundation of Ontario (Grant # 000293), the US National Institutes of Health (PPG HL081588) and the Canadian Hemophilia Society.
References
- Bellissimo DB, Christopherson PA, Flood VH, Gill JC, Friedman KD, Haberichter SL, Shapiro AD, Abshire TC, Leissinger C, Hoots WK, Lusher JM, Ragni MV, Montgomery RR. Von Willebrand factor mutations and new sequence variations identified in healthy controls are more frequent in the African-American population. Blood. 2012;119:2135–2140. doi: 10.1182/blood-2011-10-384610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos M, Sun W, Yu F, Barbalic M, Tang W, Chambless LE, Wu KK, Ballantyne C, Folsom AR, Boerwinkle E, Dong J-F. Genetic determinants of plasma von Willebrand factor antigen levels: a target gene SNP and haplotype analysis of ARIC cohort. Blood. 2011;117:5224–5230. doi: 10.1182/blood-2010-08-300152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canis K, McKinnon T, Nowak A, Haslam SM, Panico M, Morris HR, Laffan MA, Dell A. Mapping the N-glycome of Human Von Willebrand Factor. The Biochemical journal. 2012;447:217–228. doi: 10.1042/BJ20120810. [DOI] [PubMed] [Google Scholar]
- Canis K, McKinnon TA, Nowak A, Panico M, Morris HR, Laffan M, Dell A. The plasma von Willebrand factor O-glycome comprises a surprising variety of structures including ABH antigens and disialosyl motifs. J Thromb Haemost. 2010;8:137–145. doi: 10.1111/j.1538-7836.2009.03665.x. [DOI] [PubMed] [Google Scholar]
- Casonato A, Sartorello F, Cattini MG, Pontara E, Soldera C, Bertomoro A, Girolami A. An Arg760Cys mutation in the consensus sequence of the von Willebrand factor propeptide cleavage site is responsible for a new von Willebrand disease variant. Blood. 2003;101:151–156. doi: 10.1182/blood-2002-04-1046. [DOI] [PubMed] [Google Scholar]
- Castaman G, Lethagen S, Federici AB, Tosetto A, Goodeve A, Budde U, Batlle J, Meyer D, Mazurier C, Fressinaud E, Goudemand J, Eikenboom J, Schneppenheim R, Ingerslev J, Vorlova Z, Habart D, Holmberg L, Pasi J, Hill F, Peake I, Rodeghiero F. Response to desmopressin is influenced by the genotype and phenotype in type 1 von Willebrand disease (VWD): results from the European Study MCMDM-1VWD. Blood. 2008;111:3531–3539. doi: 10.1182/blood-2007-08-109231. [DOI] [PubMed] [Google Scholar]
- Castaman G, Tosetto A, Rodeghiero F. Reduced von Willebrand factor survival in von Willebrand disease: pathophysiologic and clinical relevance. J Thromb Haemost. 2009;7(Suppl 1):71–74. doi: 10.1111/j.1538-7836.2009.03381.x. [DOI] [PubMed] [Google Scholar]
- Castaman G, Castaman G, Federici AB, Tosetto A, La Marca S, Stufano F, Mannucci PM, Rodeghiero F. Different bleeding risk in type 2A and 2M von Willebrand disease: a 2-year prospective study in 107 patients. J Thromb Haemost. 2012;10:632–638. doi: 10.1111/j.1538-7836.2012.04661.x. [DOI] [PubMed] [Google Scholar]
- Chandler WL, Peerschke EIB, Castellone DD, Meijer P. Von Willebrand factor assay proficiency testing. The North American Specialized Coagulation Laboratory Association experience. Am J Clin Path. 2011;135:62–869. doi: 10.1309/AJCPH5JK4ONENPAE. [DOI] [PubMed] [Google Scholar]
- Conlan MG, Folsom AR, Finch A, Davis CE, Sorlie P, Marcucci G, Wu KK. Associations of factor VIII and von Willebrand factor with age, race, sex, and risk factors for atherosclerosis. The Atherosclerosis Risk in Communities (ARIC) Study. Thromb.Haemost. 1993;70:380–385. [PubMed] [Google Scholar]
- Coppola R, Mari D, Lattuada A, Franceschi C. Von Willebrand factor in Italian centenarians. Haematologica. 2003;88:39–43. [PubMed] [Google Scholar]
- Cumming A, Grundy P, Keeney S, Lester W, Enayat S, Guilliatt A, Bowen D, Pasi J, Keeling D, Hill F, Bolton-Maggs PH, Hay C, Collins P. An investigation of the von Willebrand factor genotype in UK patients diagnosed to have type 1 von Willebrand disease. Thromb Haemost. 2006;96:630–641. [PubMed] [Google Scholar]
- Dalton RG, Dewar MS, Savidge GF, Kernoff PB, Matthews KB, Greaves M, Preston FE. Hypothyroidism as a cause of acquired von Willebrand’s disease. Lancet. 1987;1:1007–1009. doi: 10.1016/s0140-6736(87)92272-0. [DOI] [PubMed] [Google Scholar]
- De Lange M, Snieder H, Ariëns RA, Spector TD, Grant PJ. The genetics of haemostasis: a twin study. Lancet. 2001;357:101–105. doi: 10.1016/S0140-6736(00)03541-8. [DOI] [PubMed] [Google Scholar]
- Eikenboom JC. Congenital von Willebrand disease type 3: clinical manifestations, pathophysiology and molecular biology. Best.Pract.Res.Clin.Haematol. 2001;14:365–379. doi: 10.1053/beha.2001.0139. [DOI] [PubMed] [Google Scholar]
- Eikenboom J, Van V M, Putter H, Goodeve A, Rodeghiero F, Castaman G, Federici AB, Batlle J, Meyer D, Mazurier C, Goudemand J, Schneppenheim R, Budde U, Ingerslev J, Vorlova Z, Habart D, Holmberg L, Lethagen S, Pasi J, Hill F, Hashemi Soteh M, Baronciani L, Hallden C, Guilliatt A, Lester W, Peake I. Linkage analysis in families diagnosed with type 1 von Willebrand disease in the European study, molecular and clinical markers for the diagnosis and management of type 1 VWD. J Thromb Haemost. 2006;4:774–782. doi: 10.1111/j.1538-7836.2006.01823.x. [DOI] [PubMed] [Google Scholar]
- Eikenboom J, Hilbert L, Ribba AS, Hommais A, Habart D, Messenger S, Al-Buhairan A, Guilliatt A, Lester W, Mazurier C, Meyer D, Fressinaud E, Budde U, Will K, Schneppenheim R, Obser T, Marggraf O, Eckert E, Castaman G, Rodeghiero F, Federici AB, Batlle J, Goudemand J, Ingerslev J, Lethagen S, Hill F, Peake I, Goodeve A. Expression of 14 von Willebrand factor mutations identified in patients with type 1 von Willebrand disease from the MCMDM-1VWD study. Journal of thrombosis and haemostasis: JTH. 2009;7:1304–1312. doi: 10.1111/j.1538-7836.2009.03486.x. [DOI] [PubMed] [Google Scholar]
- Favaloro EJ. Genetic testing for von Willebrand disease: the case against. J Thromb Haemost. 2010;8:6–12. doi: 10.1111/j.1538-7836.2009.03482.x. [DOI] [PubMed] [Google Scholar]
- Federici AB, Mannucci PM, Castaman G, Baronciani L, Bucciarelli P, Canciani MT, Pecci A, Lenting PJ, De Groot PG. Clinical and molecular predictors of thrombocytopenia and risk of bleeding in patients with von Willebrand disease type 2B: A cohort study of 67 patients. Blood. 2008;113:526–534. doi: 10.1182/blood-2008-04-152280. [DOI] [PubMed] [Google Scholar]
- Flood VH, Lederman CA, Wren JS, Christopherson PA, Friedman KD, Hoffmann RG, Montgomery RR. Absent collagen binding in a VWF A3 domain mutant: utility of the VWF:CB in diagnosis of VWD. Journal of thrombosis and haemostasis: JTH. 2010;8:1431–1433. doi: 10.1111/j.1538-7836.2010.03869.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flood VH, Gill JC, Christopherson PA, Wren JS, Friedman KD, Haberichter SL, Hoffmann RG, Montgomery RR. Comparison of type I, type III and type VI collagen binding assays in diagnosis of von Willebrand disease. Journal of thrombosis and haemostasis: JTH. 2012a;10:1425–1432. doi: 10.1111/j.1538-7836.2012.04747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flood VH, Gill JC, Christopherson PA, Bellissimo DB, Friedman KD, Haberichter SL, Lentz SR, Montgomery RR. Critical von Willebrand factor A1 domain residues influence type VI collagen binding. Journal of thrombosis and haemostasis: JTH. 2012b;10:1417–1424. doi: 10.1111/j.1538-7836.2012.04746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallinaro L, Cattini MG, Sztukowska M, Padrini R, Sartorello F, Pontara E, Bertomoro A, Daidone V, Pagnan A, Casonato A. A shorter von Willebrand factor survival in O blood group subjects explains how ABO determinants influence plasma von Willebrand factor. Blood. 2008;111:3540–3545. doi: 10.1182/blood-2007-11-122945. [DOI] [PubMed] [Google Scholar]
- Gill JC, Endres-Brooks J, Bauer PJ, Marks WJ, Jr., Montgomery RR. The effect of ABO blood group on the diagnosis of von Willebrand disease. Blood. 1987;69:1691–1695. [PubMed] [Google Scholar]
- Ginsburg D, Handin RI, Bonthron DT, Donlon TA, Bruns GA, Latt SA, Orkin SH. Human von Willebrand factor (vWF): isolation of complementary DNA (cDNA) clones and chromosomal localization. Science. 1985;228:1401–1406. doi: 10.1126/science.3874428. [DOI] [PubMed] [Google Scholar]
- Goodeve A, Eikenboom J, Castaman G, Rodeghiero F, Federici AB, Batlle J, Meyer D, Mazurier C, Goudemand J, Schneppenheim R, Budde U, Ingerslev J, Habart D, Vorlova Z, Holmberg L, Lethagen S, Pasi J, Hill F, Hashemi SM, Baronciani L, et al. Phenotype and genotype of a cohort of families historically diagnosed with type 1 von Willebrand disease in the European study, Molecular and Clinical Markers for the Diagnosis and Management of Type 1 von Willebrand Disease (MCMDM-1VWD) Blood. 2007;109:112–121. doi: 10.1182/blood-2006-05-020784. [DOI] [PubMed] [Google Scholar]
- Haberichter SL, Balistreri M, Christopherson P, Morateck P, Gavazova S, Bellissimo DB, Manco-Johnson MJ, Gill JC, Montgomery RR. Assay of the von Willebrand factor (VWF) propeptide to identify patients with type 1 von Willebrand disease with decreased VWF survival. Blood. 2006;108:3344–3351. doi: 10.1182/blood-2006-04-015065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberichter SL, Castaman G, Budde U, Peake I, Goodeve A, Rodeghiero F, Federici AB, Batlle J, Meyer D, Mazurier C, Goudemand J, Eikenboom J, Schneppenheim R, Ingerslev J, Vorlova Z, Habart D, Holmberg L, Lethagen S, Pasi J, Hill FG, Montgomery RR. Identification of type 1 von Willebrand disease patients with reduced von Willebrand factor survival by assay of the VWF propeptide in the European study: molecular and clinical markers for the diagnosis and management of type 1 VWD (MCMDM-1VWD) Blood. 2008;111:4979–4985. doi: 10.1182/blood-2007-09-110940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huizinga EG, Tsuji S, Romijn RA, Schiphorst ME, De Groot PG, Sixma JJ, Gros P. Structures of glycoprotein Ibalpha and its complex with von Willebrand factor A1 domain. Science. 2002;297:1176–1179. doi: 10.1126/science.107355. [DOI] [PubMed] [Google Scholar]
- Jackson SC, Sinclair GD, Cloutier S, Duan Z, Rand ML, Poon M-C. The Montreal platelet syndrome kindred has type 2B von Willebrand disease with the VWF V1316M mutation. Blood. 2009;113:3348–3351. doi: 10.1182/blood-2008-06-165233. [DOI] [PubMed] [Google Scholar]
- Jacobi PM, Gill JC, Flood VH, Jakab DA, Friedman KD, Haberichter SL. Intersection of mechanisms of type 2A VWD through defects in VWF multimerization, secretion, ADAMTS-13 susceptibility, and regulated storage. Blood. 2012;119:4543–4553. doi: 10.1182/blood-2011-06-360875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James PD, Paterson AD, Notley C, Cameron C, Hegadorn C, Tinlin S, Brown C, O’Brien L, Leggo J, Lillicrap D. Genetic linkage and association analysis in type 1 von Willebrand disease: results from the Canadian type 1 VWD study. J Thromb Haemost. 2006;4:783–792. doi: 10.1111/j.1538-7836.2006.01860.x. [DOI] [PubMed] [Google Scholar]
- James PD, Notley C, Hegadorn C, Poon MC, Walker I, Rapson D, Lillicrap D. Challenges in defining type 2M von Willebrand disease: results from a Canadian cohort study. J Thromb Haemost. 2007a;5:1914–1922. doi: 10.1111/j.1538-7836.2007.02666.x. [DOI] [PubMed] [Google Scholar]
- James PD, Notley C, Hegadorn C, Leggo J, Tuttle A, Tinlin S, Brown C, Andrews C, Labelle A, Chirinian Y, O’Brien L, Othman M, Rivard G, Rapson D, Hough C, Lillicrap D. The mutational spectrum of type 1 von Willebrand disease: Results from a Canadian cohort study. Blood. 2007b;109:145–154. doi: 10.1182/blood-2006-05-021105.. [DOI] [PubMed] [Google Scholar]
- Koppelman SJ, van HM, Vink T, Lankhof H, Schiphorst ME, Damas C, Vlot AJ, Wise R, Bouma BN, Sixma JJ. Requirements of von Willebrand factor to protect factor VIII from inactivation by activated protein C. Blood. 1996;87:2292–2300. [PubMed] [Google Scholar]
- Liu L, Wang X, Lin Z, Wu H. Elevated plasma levels of VWF:Ag in hyperthyroidism are mediated through beta-adrenergic receptors. Endocrine research. 1993;19:123–133. doi: 10.3109/07435809309033019. [DOI] [PubMed] [Google Scholar]
- Lynch DC, Zimmerman TS, Collins CJ, Brown M, Morin MJ, Ling EH, Livingston DM. Molecular cloning of cDNA for human von Willebrand factor: authentication by a new method. Cell. 1985;41:49–56. doi: 10.1016/0092-8674(85)90060-1. [DOI] [PubMed] [Google Scholar]
- Lyons SE, Bruck ME, Bowie EJ, Ginsburg D. Impaired intracellular transport produced by a subset of type IIA von Willebrand disease mutations. J Biol Chem. 1992;267:4424–4430. [PubMed] [Google Scholar]
- Mancuso DJ, Tuley EA, Westfield LA, Lester-Mancuso TL, Le Beau MM, Sorace JM, Sadler JE. Human von Willebrand factor gene and pseudogene: structural analysis and differentiation by polymerase chain reaction. Biochemistry. 1991;30:253–269. doi: 10.1021/bi00215a036. [DOI] [PubMed] [Google Scholar]
- Mannucci PM, Ruggeri ZM, Ciavarella N, Kazatchkine MD, Mowbray JF. Precipitating antibodies to factor VIII/von Willebrand factor in von Willebrand’s disease: effects on replacement therapy. Blood. 1981;57:25–31. [PubMed] [Google Scholar]
- Mannucci PM, Lombardi R, Castaman G, Dent JA, Lattuada A, Rodeghiero F, Zimmerman TS. von Willebrand disease “Vicenza” with larger-than-normal (supranormal) von Willebrand factor multimers. Blood. 1988;71:65–70. [PubMed] [Google Scholar]
- Mazurier C. von Willebrand disease masquerading as haemophilia A. Thromb Haemost. 1992;67:391–396. [PubMed] [Google Scholar]
- Mazurier C, Goudemand J, Hilbert L, Caron C, Fressinaud E, Meyer D. Type 2N von Willebrand disease: clinical manifestations, pathophysiology, laboratory diagnosis and molecular biology. Best.Pract.Res.Clin.Haematol. 2001;14:337–347. doi: 10.1053/beha.2001.0138. [DOI] [PubMed] [Google Scholar]
- Meijer P, Haverkate K. An external quality assessment program for von Willebrand factor laboratory analysis: an overview from the European concerted action on thrombosis and disabilities foundation. Sem Thromb Hemost. 2006;32:485–491. doi: 10.1055/s-2006-947862. [DOI] [PubMed] [Google Scholar]
- Meyer D, Fressinaud E, Gaucher C, Lavergne JM, Hilbert L, Ribba AS, Jorieux S, Mazurier C. Gene defects in 150 unrelated French cases with type 2 von Willebrand disease: from the patient to the gene. INSERM Network on Molecular Abnormalities in von Willebrand Disease. Thrombosis & Haemostasis. 1997;78:451–456. [PubMed] [Google Scholar]
- Mohl A, Marschalek R, Masszi T, Nagy E, Obser T, Oyen F, Sallai K, Bodo I, Schneppenheim R. An Alu-mediated novel large deletion is the most frequent cause of type 3 von Willebrand disease in Hungary. J.Thromb.Haemost. 2008;6:1729–1735. doi: 10.1111/j.1538-7836.2008.03107.x. [DOI] [PubMed] [Google Scholar]
- Montgomery RR. When it comes to von Willebrand disease, does 1 + 1 = 3? Journal of thrombosis and haemostasis: JTH. 2006;4:2162–2163. doi: 10.1111/j.1538-7836.2006.02158.x. [DOI] [PubMed] [Google Scholar]
- Montgomery RR, Christopherson PA, Haberichter SL, Flood VH, Gill JC, The Zimmerman Program Investigators Diagnostic Fidelity of Historically Diagnosed Patients with VWD Enrolled from 27 Centers in the ZPMCB-VWD. ASH Annual Meeting Abstracts. 2011;118:381. [Google Scholar]
- Ngo KY, Glotz VT, Koziol JA, Lynch DC, Gitschier J, Ranieri P, Ciavarella N, Ruggeri ZM, Zimmerman TS. Homozygous and heterozygous deletions of the von Willebrand factor gene in patients and carriers of severe von Willebrand disease. Proc.Natl.Acad.Sci.U.S.A. 1988;85:2753–2757. doi: 10.1073/pnas.85.8.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols WL, Hultin MB, James AH, Manco-Johnson MJ, Montgomery RR, Ortel TL, Rick ME, Sadler JE, Weinstein M, Yawn BP. von Willebrand disease (VWD): evidence-based diagnosis and management guidelines, the National Heart, Lung, and Blood Institute (NHLBI) Expert Panel report (USA) Haemophilia. 2008;14:171–232. doi: 10.1111/j.1365-2516.2007.01643.x. [DOI] [PubMed] [Google Scholar]
- Nilsson IM, Blomback M, Jorpes E, Blomback B, Johansson SA. Von Willebrand’s disease and its correction with human plasma fraction 1-0. Acta medica Scandinavica. 1957;159:179–88. doi: 10.1111/j.0954-6820.1957.tb00123.x. [DOI] [PubMed] [Google Scholar]
- O’Brien LA, James PD, Othman M, Berber E, Cameron C, Notley CR, Hegadorn CA, Sutherland JJ, Hough C, Rivard GE, O’Shaunessey D, Lillicrap D. Founder von Willebrand factor haplotype associated with type 1 von Willebrand disease. Blood. 2003;102:549–557. doi: 10.1182/blood-2002-12-3693. [DOI] [PubMed] [Google Scholar]
- Orstavik KH. Genetics of plasma concentration of von Willebrand factor. Folia haematologica (Leipzig, Germany: 1928) 1990;117:527–531. [PubMed] [Google Scholar]
- Othman M. Platelet-type von Willebrand disease and type 2B von Willebrand disease: a story of nonidentical twins when two different genetic abnormalities evolve into similar phenotypes. Semin Thromb Hemost. 2007;33:780–786. doi: 10.1055/s-2007-1000368. [DOI] [PubMed] [Google Scholar]
- Peake IR, Bloom AL, Giddings JC. Inherited variants of factor-VIII-related protein in von Willebrand’s disease. The New England journal of medicine. 1974;291:113–117. doi: 10.1056/NEJM197407182910301. [DOI] [PubMed] [Google Scholar]
- Peake IR, Goodeve AC. Genetic testing for von Willebrand disease: the case for. J Thromb Haemost. 2010;8:13–16. doi: 10.1111/j.1538-7836.2009.03670.x. [DOI] [PubMed] [Google Scholar]
- Pottinger BE, Read RC, Paleolog EM, Higgins PG, Pearson JD. von Willebrand factor is an acute phase reactant in man. Thromb Res. 1989;53:387–394. doi: 10.1016/0049-3848(89)90317-4. [DOI] [PubMed] [Google Scholar]
- Pruss CM, Golder M, Bryant A, Hegadorn CA, Burnett E, Laverty K, Sponagle K, Dhala A, Notley C, Haberichter S, Lillicrap D. Pathologic mechanisms of type 1 VWD mutations R1205H and Y1584C through in vitro and in vivo mouse models. Blood. 2011;117:4358–4366. doi: 10.1182/blood-2010-08-303727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribba AS, Loisel I, Lavergne JM, Juhan-Vague I, Obert B, Cherel G, Meyer D, Girma JP. Ser968Thr mutation within the A3 domain of von Willebrand factor (VWF) in two related patients leads to a defective binding of VWF to collagen. Thromb.Haemost. 2001;86:848–854. [PubMed] [Google Scholar]
- Riddell A, Gomez K, Millar C, Mellars G, Brown S, Gill S, Laffan M, McKinnon TAJ. Characterisation of W1745C and S1783A, two novel collagen binding defects in the A3 domain of von Willebrand factor. Blood. 2009;114:3489–3496. doi: 10.1182/blood-2008-10-184317. [DOI] [PubMed] [Google Scholar]
- Ruggeri ZM. Type IIB von Willebrand disease: a paradox explains how von Willebrand factor works. J Thromb Haemost. 2004;2:2–6. doi: 10.1111/j.1538-7836.2003.00523.x. [DOI] [PubMed] [Google Scholar]
- Ruggeri ZM, Pareti FI, Mannucci PM, Ciavarella N, Zimmerman TS. Heightened interaction between platelets and factor VIII/von Willebrand factor in a new subtype of von Willebrand’s disease. The New England journal of medicine. 1980;302:1047–1051. doi: 10.1056/NEJM198005083021902. [DOI] [PubMed] [Google Scholar]
- Sadler JE. Biochemistry and genetics of von Willebrand factor. Annual review of biochemistry. 1998;67:395–424. doi: 10.1146/annurev.biochem.67.1.395. [DOI] [PubMed] [Google Scholar]
- Sadler JE. Von Willebrand disease type 1: a diagnosis in search of a disease. Blood. 2003;101:2089–2093. doi: 10.1182/blood-2002-09-2892. [DOI] [PubMed] [Google Scholar]
- Sadler JE, Shelton-Inloes BB, Sorace JM, Harlan JM, Titani K, Davie EW. Cloning and characterization of two cDNAs coding for human von Willebrand factor. Proc.Natl.Acad.Sci.U.S.A. 1985;82:6394–6398. doi: 10.1073/pnas.82.19.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler JE, Budde U, Eikenboom JC, Favaloro EJ, Hill FG, Holmberg L, Ingerslev J, Lee CA, Lillicrap D, Mannucci PM, Mazurier C, Meyer D, Nichols WL, Nishino M, Peake IR, Rodeghiero F, Schneppenheim R, Ruggeri ZM, Srivastava A, Montgomery RR, Federici AB, for the Working Party on von Willebrand Disease Classification Update on the pathophysiology and classification of von Willebrand disease: a report of the Subcommittee on von Willebrand Factor. J Thromb Haemost. 2006;4:2103–2114. doi: 10.1111/j.1538-7836.2006.02146.x. [DOI] [PubMed] [Google Scholar]
- Shelton-Inloes BB, Chehab FF, Mannucci PM, Federici AB, Sadler JE. Gene deletions correlate with the development of alloantibodies in von Willebrand disease. J Clin.Invest. 1987;79:1459–1465. doi: 10.1172/JCI112974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith NL, Chen MH, Dehghan A, Strachan DP, Basu S, Soranzo N, Hayward C, Rudan I, Sabater-Lleal M, Bis JC, De Maat MP, Rumley A, Kong X, Yang Q, Williams FM, Vitart V, Campbell H, Malarstig A, Wiggins KL, Van Duijn CM, et al. Novel associations of multiple genetic loci with plasma levels of factor VII, factor VIII, and von Willebrand factor: The CHARGE (Cohorts for Heart and Aging Research in Genome Epidemiology) Consortium. Circulation. 2010;121:1382–1392. doi: 10.1161/CIRCULATIONAHA.109.869156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporn LA, Chavin SI, Marder VJ, Wagner DD. Biosynthesis of von Willebrand protein by human megakaryocytes. J.Clin.Invest. 1985;76:1102–1106. doi: 10.1172/JCI112064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland MS, Cumming AM, Bowman M, Bolton-Maggs PHB, Bowen DJ, Collins PW, Hay CRM, Will AM, Keeney S. A novel deletion mutation is recurrent in von Willebrand disease types 1 and 3. Blood. 2009;114:1091–1098. doi: 10.1182/blood-2008-08-173278. [DOI] [PubMed] [Google Scholar]
- Tsai HM. Physiologic cleavage of von Willebrand factor by a plasma protease is dependent on its conformation and requires calcium ion. Blood. 1996;87:4235–4244. [PubMed] [Google Scholar]
- Verweij CL, De Vries CJ, Distel B, Van Zonneveld AJ, Van Kessel AG, Van Mourik JA, Pannekoek H. Construction of cDNA coding for human von Willebrand factor using antibody probes for colony-screening and mapping of the chromosomal gene. Nucleic Acids Res. 1985;13:4699–4717. doi: 10.1093/nar/13.13.4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner DD, Marder VJ. Biosynthesis of von Willebrand protein by human endothelial cells: identification of a large precursor polypeptide chain. J.Biol.Chem. 1983;258:2065–2067. [PubMed] [Google Scholar]
- Von Willebrand EA. Hereditar pseudohemofili. Finska Lakaresallskapets Handlingar. 1926;LXVII:87–112. [Google Scholar]
- Zhang X, Halvorsen K, Zhang C-Z, Wong WP, Springer TA. Mechanoenzymatic cleavage of the ultralarge vascular protein von Willebrand factor. Science (New York, N.Y.) 2009;324:1330–1334. doi: 10.1126/science.1170905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZP, Blomback M, Nyman D, Anvret M. Mutations of von Willebrand factor gene in families with von Willebrand disease in the Aland Islands. Proc.Natl.Acad.Sci.U.S.A. 1993;90:7937–7940. doi: 10.1073/pnas.90.17.7937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman TS, Ratnoff OD, Powell AE. Immunologic differentiation of classic hemophilia (factor 8 deficiency) and von Willebrand’s dissase, with observations on combined deficiencies of antihemophilic factor and proaccelerin (factor V) and on an acquired circulating anticoagulant against anti. The Journal of clinical investigation. 1971;50:244–254. doi: 10.1172/JCI106480. [DOI] [PMC free article] [PubMed] [Google Scholar]