SUMMARY
Histone deacetylases (HDACs) are a family of proteins that play an important role in regulating transcription as well as the function of a variety of cellular proteins. While these proteins are expressed abundantly in the brain, little is known about their roles in brain function. A growing body of evidence suggests that HDACs regulate neuronal survival. Results from studies conducted in vertebrate and mammalian experimental systems indicate that while some of these proteins are involved in promoting neuronal death, a majority of the HDACs studied thus far protect against neurodegeneration. Here we review the research performed on the role played by individual members of the HDAC family in the regulation of neuronal death. Chemical inhibitors of HDACs have been used in a variety of models of neurodegenerative disorders. We summarize the results from these studies, which indicate that HDAC inhibitors show great promise as therapeutic agents for human neurodegenerative disorders.
Neurodegenerative diseases constitute a set of pathological conditions characterized by persistent loss of neurons within specific regions of the brain or spinal cord, resulting in progressive mental and physical dysfunction. Current medications alleviate only the symptoms associated with the disorder and are generally only modestly effective. Because neuronal loss continues unabated, such palliative treatments have no effect on disease progression. The development of a cure or treatment for neurodegenerative diseases thus represents an urgent and most significant medical challenge.
A strategy for treating neurodegenerative diseases that has generated considerable recent enthusiasm is the use of small-molecule inhibitors of histone deacetylases (HDACs). HDACs are a family of enzymes that were initially identified by their ability to remove an acetyl group from lysine residues within histone tails. The effects of HDACs are reversed by another family of enzymes called histone acetyl transferases (HATs), which acetylate histones. Acetylation of histone tails neutralizes their positive charge, thereby promoting the formation of a relaxed chromatin structure that is more accessible to transcription factors, and thus promoting transcriptional activation. Conversely, histone deacetylation favors transcriptional repression by causing chromatin compactation. The balance between the actions of HATs and HDACs serves as a pivotal regulatory mechanism for gene expression, controlling diverse physiological processes. It is now known that HATs and HDACs also act on a large number of nonhistone substrates both in the nucleus and in the cytoplasm. These include transcription factors, hormone receptors, chaperones and cytoskeletal proteins. Acetylation/deacetylation of these proteins can affect their functional activity, stability, intracellular localization and associations with other proteins, resulting in effects on cell growth, survival and differentiation as well as on cytoskeleton dynamics, endocytosis and energy metabolism.
Perturbation of the balance between HAT and HDAC activities is emerging as an important event in the pathogenesis of a number of disorders. This was first observed in cancer, many forms of which are associated with increased expression and activity of HDACs.1–5 Elevated deacetylase activity has been found to result in the transcriptional repression of a variety of genes, mainly involved in promoting differentiation or cell death. Treatment with pharmacological HDAC inhibitors reverses epigenetic silencing and exerts antineoplastic effects in tissue cultures and animal models of tumorigenesis. Consequently, a variety of HDAC inhibitors are currently being tested in clinical trials for the treatment of cancer. It was later found that these inhibitors may have therapeutic utility in other human disorders as well, leading to an explosion in interest in their development and testing (reviewed in6, 7).
The focus of this review is not on HDAC inhibitors themselves, but on their primary targets. Specifically, it covers much of what is known about the role of individual HDAC proteins in the regulation of neurodegeneration. Although results from studies utilizing small-molecule HDAC inhibitors in experimental models of neurodegenerative disease have been summarized, the reader is referred to other recent reviews that describe the literature on this subject in more detail.8, 9
THE HDAC PROTEIN FAMILY IN MAMMALS
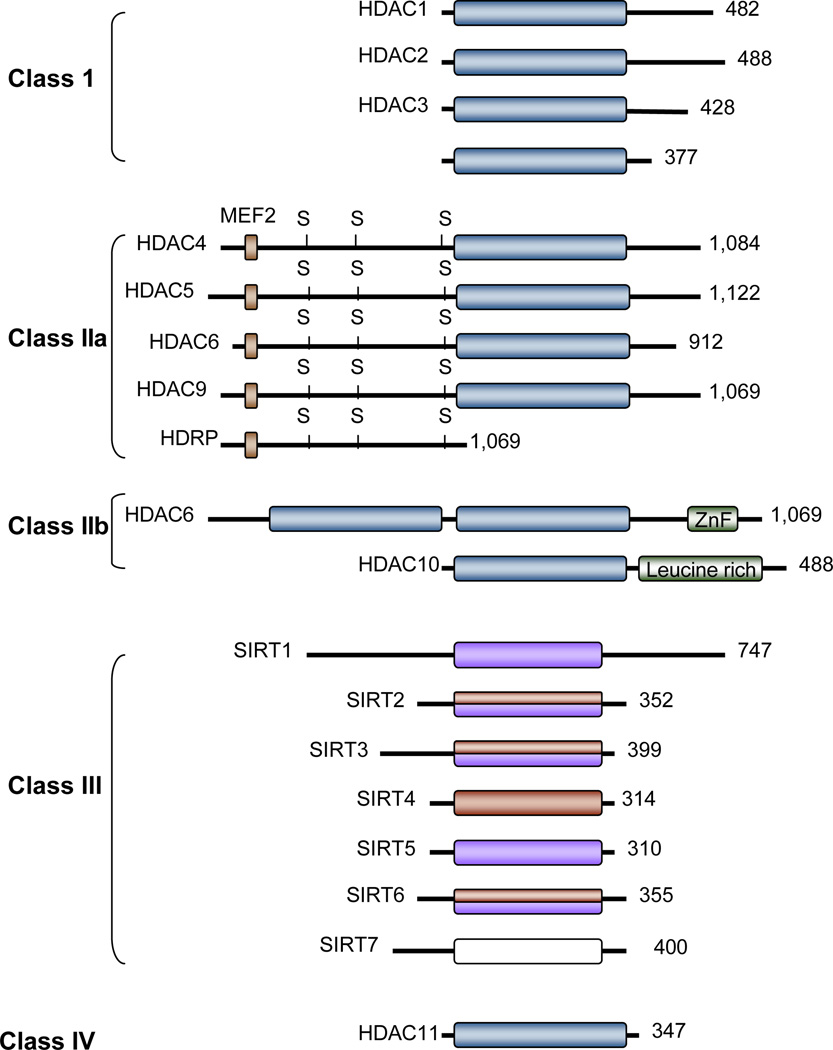
Mammals express 18 HDAC proteins, which have been grouped into four classes based on their homology to yeast deacetylase proteins (reviewed in10, 11). Class I HDACs (HDAC1, HDAC2, HDAC3 and HDAC8) are homologues of the yeast HDAC RPD3 protein. These HDACs are expressed ubiquitously, localized predominantly in the nucleus (with the exception of HDAC3, which can also be found in the cytoplasm) and possess high enzymatic activity. HDAC1 and HDAC2 are structurally very similar and within cells, are found complexed with corepressors such as the mammalian paired amphipathic helix protein Sin3 and the protein CoREST, as well as with the polycomb-repressive complex 2 (PRC2) and the nucleosome remodeling and histone deacetylation (NuRD) complex. HDAC3 associates with distinct complexes such as the N-CoR-SMRT complex. Finally, HDAC8 does not appear to function as part of a protein complex.10, 11
Class II HDACs are homologous to the yeast HDAC HDA1 and are further divided into class IIa (HDAC4, -5, -7 and -9) and class IIb (HDAC6 and -10) HDACs. Class IIa HDACs are characterized by large N-terminal extensions with conserved binding sites for the transcription factor myocyte-specific enhancer factor 2A (MEF2) and 14-3-3 protein eta, and can shuttle between the nucleus and cytoplasm in a phosphorylation-dependent manner (Fig. 1). Phosphorylation is mediated by kinases such as calcium/calmodulin-dependent protein kinase (CaMK) and protein kinase D at conserved serine residues in the N-terminal region of these proteins. Class IIa HDACs also display a restricted expression pattern. For example, HDAC4, -5 and -9 are highly expressed in the brain, heart and skeletal muscle, while HDAC7 is abundant in endothelial cells and thymocytes. Class IIb HDACs lack the N-terminal extension but possess two nonidentical catalytic domains in tandem. HDAC6 is localized exclusively in the cytoplasm where it is physically associated with tubulin. Deacetylation of tubulin as well as other cytoskeletal and transmembrane proteins by HDAC6 has been reported. Relatively little is known about the intracellular localization, expression pattern or functions of HDAC10.
Figure 1.
The histone deacetylase (HDAC) family of proteins. The conserved deacetylase domain of the classical HDACs (blue rectangles), deacetylase domain of the sirtuins (purple rectangles) and the ADP-ribosyl transferase catalytic domain (red rectangles) are indicated. Histone deacetylase-related protein (HDRP), also referred to as MITR, is a truncated form of HDAC9, generated by alternative splicing. SIRT2, SIRT3 and SIRT6 have both deacetylase and ADP-ribosyl transferase activities. No catalytic activity has been reported for SIRT7. The myocyte-specific enhancer factor 2A (MEF2)-binding site is marked by a blue square and the location of key serine residues that are phosphorylated, is indicated with “S”. The leucine-rich region in HDAC10 and the C-terminal zinc finger (ZnF) are also shown.
HDAC11 shares sequence conservation with both RPD3 and HDA1 and is therefore placed in class IV. Its expression is highest in the brain, heart, testis and kidney. Little is known about the protein’s enzymatic activity, its substrates or the proteins with which it associates.
Classes I, II, and IV HDACs are all zinc-dependent enzymes, sometimes referred to as “classical HDACs”. In contrast, class III HDACs require a nicotinamide adenine dinucleotide ion (NAD+) for their catalytic activity and are homologous to the NAD-dependent HDAC SIR2 yeast protein.12 These proteins, called sirtuins, share no sequence or structural similarity with the classical HDACs and deacetylate proteins by a mechanism that is distinct from the acetyl-lysine hyrolysis mechanism utilized by classical HDACs.13 The sirtuin subclass is composed of 7 members (SIRT1–7), which share a conserved catalytic core domain of approximately 275 amino acids but differ in their N- and C-terminal protein sequences flanking this core (reviewed in12, 14, 15); Fig. 1). While SIRT1 and SIRT5 are deacetylases, SIRT2 and SIRT3 have both deacetylase and mono-ADP-ribosyl transferase activity. SIRT4 is believed to have only ADP-ribosyl transferase activity. Although initially reported to act as an ADP-ribosyltransferase, SIRT6 was recently demonstrated to also possess histone deacetylase activity.16 An enzymatic activity for SIRT7 has yet to be established. The sirtuins also show differences in their subcellular localization. SIRT1, which has the highest sequence similarity to yeast SIR2, is generally nuclear where it deacetylates histones H3 and H4 as well as transcription factors such as nuclear factor NF-kappa-B, tumor suppressor p53, forkhead box protein O (FOXO), Ku70 and PGC-1-alpha.15 A few recent studies have described nucleo–cytoplasmic shuttling of SIRT1 in response to oxidative stress.17 SIRT2 is predominantly cytoplasmic where it associates with microtubules and deacetylates alpha-tubulin.18, 19 SIRT2 can also deacetylate histone H4 when the nuclear membrane disassembles during mitosis.20 SIRT3, SIRT4 and SIRT5 have been reported to localize to the mitochondria,21 although one recent study described that SIRT5 localizes to the nucleus and cytoplasm in neurons.22 Within the mitochondria, these SIRTs appear to localize to different subcompartments, suggesting distinct functions.23 A recent study reported that SIRT5 localizes to the mitochondrial matrix where it deacetylates and activates carbamoyl-phosphate synthetase I, the enzyme catalyzing the first step of the urea cycle for ammonia detoxification.24 These authors found that under fasting conditions, SIRT5 knock-out mice fail to upregulate carbamoyl-phosphate synthetase I activity, resulting in elevated blood ammonia. SIRT5-deficient hepatocytes also show reduced viability in nutrient-depleted medium, which mimics starvation in animals.24 Like SIRT1, SIRT6 and SIRT7 are nuclear proteins although the three proteins display distinct subnuclear localization patterns; SIRT6 associates with heterochromatin, SIRT7 localizes to nucleoli and SIRT1 is largely associated with euchromatin within the nucleus.21
ROLE OF CLASSICAL HDACs IN THE REGULATION OF NEURONAL SURVIVAL AND DEATH
A large number of studies have shown that small-molecule HDAC inhibitors, particularly of the classical HDACs, have impressive protective effects in a variety of tissue culture and in vivo models of neurodegeneration (reviewed in8, 9, 25). A shortcoming of these inhibitors however, is that they inhibit the catalytic activity of all 11 classical HDAC proteins effectively. As a result, the members of the HDAC family that are relevant to the neuroprotective action of these inhibitors remain largely unidentified. Uncovering their identity would be important in the design and development of inhibitors targeting only those HDAC proteins responsible for the promotion of neurodegeneration. A number of recent studies have focused on individual HDAC proteins with the goal of determining whether or not they regulate neuronal survival or death. Somewhat unexpectedly, the results that have been reported so far indicate that a majority of HDAC proteins does not promote neurodegeneration even when the proteins are overexpressed. On the contrary, most of the classical HDAC proteins studied thus far have neuroprotective effects.
HDAC1, a class I HDAC, was shown to play a protective role in a p25/cyclin-dependent kinase 5-inducible transgenic mouse model. Following induction of p25 expression, these mice display postnatal neurodegeneration found to be mediated by a direct interaction between p25 and a region within the catalytic domain of HDAC1. This results in reduced HDAC1 activity, which in turn was reported to trigger aberrant expression of cell cycle proteins and DNA damage, leading to neuronal death. The in vivo findings were recapitulated in primary cortical neurons.26 In these neurons, the overexpression of HDAC1, but not a catalytically inactive mutant, protected cultured neurons against p25-induced neurotoxicity.26 Pharmacological inhibition of HAT activity was also protective against p25 neurotoxicity and the extent of protection was not increased by HDAC1 coexpression. These findings suggest that the neuroprotective effect of HDAC1 is mediated by histone deacetylation. Excitingly, these scientists showed that virally delivered HDAC1 protects against ischemia-induced neuronal death in vivo.
Most studies pertaining to the regulation of neurodegeneration have focused on on the class II HDACs (reviewed in27). Morrison et al. (2006), demonstrated that histone deacetylase-related protein (HDRP), a naturally expressed truncated form of HDAC9, is a neuroprotective protein. These authors reported that HDRP expression is downregulated in cultured cerebellar granule neurons induced to die by low potassium treatment. Forced expression completely prevented neuronal death whereas knock-down of endogenous HDRP reduced the survival of healthy neurons. Similarly, neurons cultured from HDRP- and HDAC9-deficient mice were more vulnerable to apoptosis. Neuroprotection by HDRP was suggested to involve repressed transcription of JUN, the gene encoding the proto-oncogene c-jun, through histone deacetylation at the JUN promoter.28 Although HDRP lacks a catalytic domain, it was found to recruit deacetylase activity by direct interaction with HDAC1. Treatment with HDAC inhibitors antagonizes the survival-promoting effect of HDRP consistent with the requirement of deacetylase activity in the neuroprotection.28 In addition to suppressing JUN transcription, HDRP inhibits the phosphorylation and activation of the proto-oncogene c-jun through direct interaction with c-Jun N-terminal kinase (JNK).28
Disagreement exists over the role of HDAC4 in regulating neurodegeneration. Yao and Bolger (2006) initially reported that the forced expression of HDAC4 in cultured cerebellar granule neurons causes cell death. A subsequent study, also performed in cerebellar granule neurons by Majdzadeh (2008), reported that HDAC4 expression protects neurons against low-potassium-induced cell death. In this study, HDAC4 was also was shown to protect cultured cortical neurons against homocysteic acid and 6-hydroxy dopamine induced toxicity. Arguing strongly for a survival-promoting role for HDAC4 in neurons is the finding that the cerebellums of HDAC4 knock-out mice, which die within 2 weeks of birth, display postnatal degeneration of Purkinje neurons.29 Using HDAC inhibitors and mutant HDAC4 constructs lacking the deactylase domain, it was found that HDAC4-mediated neuroprotection does not require deacetylase activity. Rather, neuroprotection by HDAC4 involves an inhibition of cell cycle proteins such as cyclin-dependent kinase-1. A recently published report by Chen and Cepko (2009) confirms a neuroprotective role for HDAC4. These authors demionstrated that HDAC4 reduced naturally ocurring neuronal death in the retina, and that HDAC4 overexpression in a mouse model of retinal degeneration prolonged photoreceptor survival.30
The role of HDAC5 has been examined in cultured cerebellar granule neurons. Linseman et al. (2003) reported that low-potassium-induced death of these neurons leads to a dephosphorylation of HDAC5 and its translocation from the cytoplasm to the nucleus, where it induced apoptosis. Although depolarizing stimuli were shown to inactivate HDAC5’s proapoptotic action by CaMK-mediated phosphorylation,31 our group has failed to reproduce this finding (Majdzadeh and D’Mello, unpublished). Furthermore, we found that overexpression of HDAC5 has little effect on the survival of cerebellar granule neurons either in depolarizing or nondepolarizing medium.
Compelling evidence has been described for HDAC6 involvement in the protection of neurons against the toxic effects of abnormal protein aggregates. Through interaction with the microtubule-associated dynein motor and aggregated protein, HDAC6 transports ubiquitinated protein aggregates to the aggrosome for subsequent clearance by autophagy; cells deficient in HDAC6 fail to clear misfolded protein aggregates.321 HDAC6 was shown to be required for the autophagic clearance of aggregated huntingtin protein.33 HDAC6 also interacts with polyubiquitinated DJ-1, a protein mutated in early-onset form of PD, facilitating the transport of mutant DJ-1 into aggresomes.34 HDAC6 localizes to Lewy bodies in sporadic PD patients,33 suggesting that it may also play a similar role in the formation of these inclusions characterizing PD, now believed to serve as a means of sequestering otherwise harmful protein aggregates. Ectopically expressed HDAC6 rescues neurons from degeneration in a Drosophila model of spinobulbar muscular atrophy in which the androgen receptor containing a polyglutamine expansion (polyQ-AR) is overexpressed.35 Once more, protection by HDAC6 involved the activation of the autophagic degradation pathway that compensated for the impairment of the ubiquitin-proteosome system caused by polyQ-AR aggregation. Besides stimulating autophagic degradation, HDAC6 protects cells from the cytotoxic effects of protein aggregates by deacetylating and activating major cellular chaperones such as HSP 90, which then helps protein refolding or directs their delivery to the ubiquitin-proteosome system.36, 37 Taken together, these findings indicate that HDAC6 is a key player in dealing with the accumulation of protein aggregates in neurons and perhaps other cell types as well. It is not clear whether HDAC6 is protective against neurodegenerative stimuli besides protein aggregation.
The contribution of the different HDAC proteins to neurodegeneration has also been studied in a Caenorhabditis elegans model of Huntington’s disease, generated by expression of polyglutamine-expanded human huntingtin protein (polyQ-Htt). Loss of function alleles and RNA interference was used to systematically reduce the function of each of the eight classical HDACs expressed in the worm.38 While knock-down of Hda-3 (the C. elegans orthologue of human HDAC3) reduced polyQ-Htt neurotoxicity, suppression of expression of all other classical HDACs enhanced toxicity.38 Restoration of Hda-3 expression restored polyQ-Htt toxicity, confirming that Had-3 promotes neurodegeneration in C. elegans. The enhanced neurodegeneration observed with suppression of function of the other HDACs suggests that they play a protective role against neurodegeneration at least in response to polyglutamine toxicity. Taken together with the results emerging from mammalian paradigms, this study’s data strengthen the contention that many of the HDAC proteins are neuroprotective. Mice lacking most of the classical HDACs have been generated (Table 1).
Table I.
Phenotypes of HDAC knock-out mice
| HDAC | Time of lethality |
Phenotype | Reference |
|---|---|---|---|
| HDAC1 | E10.5 | Proliferation defects in embryonic stem cells | 111, 112 |
| HDAC2 | P1 | Cardiac malformation | 112, 113 |
| HDAC3 | E9.5 | Vascular defects | 114 |
| HDAC4 | P7–P14 | Small size, bone and skeletal defects, neurodegeneration | 29, 115 |
| HDAC5 | Viable | Cardiac hypertrophy after stress | 116 |
| HDAC6 | Viable | Highly increased alpha-tubulin acetylation | 117 |
| HDAC7 | E11 | Cardiovascular defects | 118 |
| HDAC8 | P1 | Craniofacial defects | 119 |
| HDAC9 | Viable | Cardiac hypertrophy after stress | 120 |
| HDAC10 | ND | ND | |
| HDAC11 | ND | ND | |
| SIRT1 | P1–P7 | Small size, genomic instability, severe developmental defects | 121, 122 |
| SIRT2 | ND | ND | |
| SIRT3 | Viable | Hyperacetylated mitochondrial proteins, no overt phenotype | 123 |
| SIRT4 | Viable | Increased glutamate dehydrogenase activity, no overt phenotypic abnormalities | 125 |
| SIRT5 | Viable | No overt phenotypic abnormalities | 24 |
| SIRT6 | 4 weeks | Small size, genomic instability, premature aging | 124 |
| SIRT7 | 1 year | Degenerative heart hypertrophy | 125 |
E, embryonic day; P, postnatal day; ND, not determined.
With the exception of the HDAC4 knock-out mice, no overt effect on survival or death of neurons has been observed. It deserves mention that all knock-out mice of class I HDACs die at or before birth, precluding detailed analysis of the role of these proteins on the regulation of neuronal viability. It is possible that compensatory effects of other HDACs in the mutant mice could explain the absence of any neurodegeneration in mice lacking HDACs 1, 6 and 9, all of which have been found to have neuroprotective effects as described above.
SIRTUINS AND THE REGULATION OF NEURODEGENERATION
Sirtuins also play an important role in the regulation of neurodegeneration (for reviews,9, 39, 40). Emerging evidence suggests both protective and degenerative actions for members of the sirtuin family, as described for the classical HDACs. The first direct evidence of neuroprotective effects of sirtuins came from a study by Araki et al. (2004) showing that the delay in axonal degeneration displayed by slow Wallerian degeneration (WldS) mice is due to an increase in nicotinamide mononucleotide adenylyltransferase 1 activity. The protection afforded by that enzyme against axonal degeneration is blocked by small interfering RNA (siRNA)-mediated suppression of SIRT1 expression or exposure to sirtinol, a pharmacological SIRT1 inhibitor.41 It was thus concluded that the neuroprotective effect of nicotinamide mononucleotide adenylyltransferase 1 was mediated by SIRT1 activation.41 This conclusion was challenged, however, by another study, which found no differences in the extent of axonal degeneration between neurons from wild-type and SIRT1 knock-out mice.42 Furthermore, sirtinol did not inhibit the protective effect of nicotinamide mononucleotide adenylyltransferase 1 on axonal degeneration in this second study.42 Despite these differences, a rapidly growing body of evidence indicates that SIRT1 does exert impressive neuroprotective effects. In a mixed cortical neuronal/glia culture paradigm, overexpression of the sirtuin protected against microglia-mediated amyloid beta neurotoxicity by inhibiting the activation of NF-kappaB in microglia.43 Neuroprotection by SIRT1 has also been observed in the Tg2576 transgenic mouse model of Alzheimer’s disease (AD).44 This study found that caloric restriction caused an elevation of SIRT1 deacetylase activity in the brains of Tg2576 mice, which coincided with significantly decreased amyloid beta peptide levels.
Encouragingly, the reduced amyloid beta production was accompanied by a dramatic reduction of AD-type amyloid neuropathology.44 The neuroprotective effect of SIRT1 also correlated with decreased expression of the rho-associated protein kinase 1, which has previously been implicated in promoting nonamyloidogenic processing of amyloid beta A4 protein (APP). Using neuronal cultures from Tg2576 mice, these authors confirmed that SIRT1 overexpression stimulates alpha-secretase activity through an inhibition of rho-associated protein kinase 1 activity, resulting in dramatically reduced amyloid beta generation.44
SIRT1 expression is upregulated in the p25-inducible mouse model of AD and the Sod1G37R transgenic mouse model of amyotrophic lateral sclerosis.45 This increase was proposed to represent a protective response to neurodegenerative conditions. Injection of SIRT1 lentivirus in the hippocampus of these mice conferred significant protection against p25-mediated neurodegeneration. Deacetylation and inactivation of cellular tumor antigen p53 was proposed to be the mechanism by which SIRT1 protected against p25 neurotoxicity.45 Parker et al. (2005) described that elevated levels of protein sir-2.1, the nematode homologue of SIRT1, increased the life span of C. elegans and reduced neuronal dysfunction in a model of Huntington’s disease. The protective effect of sir-2.1 was attributed to the activation of abnormal dauer formation protein 16, the nematode homologue of the mammalian FOXO transcription factor.46
Much evidence of a role for SIRT1 in neuroprotection has come from the use of resveratrol, a polyphenol with strong antioxidant effects.47 In the p25-inducible transgenic mouse model, administration of resveratrol reduced hippocampal neurodegeneration and prevented cognitive decline. This was accompanied by reduced acetylation of known SIRT1 substrates.45 Two separate studies have shown that resveratrol reduces amyloid beta plaque formation in mice. In one of these studies, this was due to an alteration in the pattern of APP processing, resulting in lowered amyloid beta production.44 This effect was proposed to be mediated by activation of SIRT1. The other study reported no activation of SIRT1 and found no difference in the pattern of APP processing.48 Resveratrol protects rats against ischemic brain damage and worms from polyQ-induced neurotoxicity.46, 49 The protective effect of resveratrol in both of these studies was inhibited by pharmacological sirtuin inhibitors, suggesting a SIRT1-mediated mechanism. Although resveratrol also provided robust protection against degeneration of the substantia nigral induced by the dopaminergic neurotoxin 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP), and against kainite-induced hippocampal neurodegeneration, evidence for the involvement of SIRT1 was not presented in these reports.50, 51 In a study in which resveratrol was found to be protective in the organotypic slice model of Parkinson’s disease (PD) involving exposure to 1-methyl-4-phenylpyridinium (MPP+), the neurotoxic metabolite of MPTP, the protection was not reduced by sirtuin inhibitors.52 Instead, resveratrol was found to prevent accumulation of reactive oxygen species, depletion of cellular glutathione and cellular oxidative damage induced by MPP+, suggesting involvement of an antioxidative rather than SIRT1-activating mechanism in the neuroprotection.52 In another study, resveratrol was shown to protect rats against 3-nitropropionic acid-induced motor and cognitive impairment. Again, the beneficial effect of resveratrol was attributed to its antioxidant activity.53 A SIRT1-independent mechanism for the neuroprotective effect of resveratrol has also been suggested by others.52, 54 It is noteworthy that in addition to SIRT1, resveratrol modulates the activity of other important signaling proteins including enzymes such as 5'-AMP-activated protein kinase, COX-1 and PI3-kinase, many transcription factors, as well as other proteins that regulate cell death (reviewed in55–58). Also noteworthy are reports from two different laboratories that resveratrol is incapable of activating SIRT1 in intact cells. These scientists discovered that the stimulatory effects of resveratrol on SIRT1 measured in vitro are dependent on the fluorescently modified substrate used in the assay.59, 60 Resveratrol had no effect when the same acetylated substrates were used without the fluorophore. It is possible that some of the neuroprotective effects of resveratrol attributed to SIRT1 activation may be mediated by other signaling molecules and mechanisms instead.
In sharp contrast to the neuroprotective activity of SIRT1, SIRT2 is involved in promoting the toxic effects of a PD-associated mutant form of alpha-synuclein (alpha-SynA53T) in rat midbrain neurons. Suppressing SIRT2 expression by siRNA or pharmacological inhibition of its activity protected against alpha-synuclein-induced toxicity.61 These findings were confirmed in a Drosophila model of PD in which alpha-synuclein was overexpressed specifically in neurons. In addition to reducing toxicity, the inhibition of SIRT2 reduced the number of intracellular alpha-synuclein aggregates, converting them into fewer but larger aggregates.61 The mechanism by which SIRT2 inhibition affects alpha-synuclein aggregation is not clear but it was proposed that SIRT2 inhibition might increase acetylation of tubulin, a target of SIRT2.61 Since alpha-synuclein is associated with tubulin, tubulin acetylation could stimulate aggregation of alpha-synuclein by mechanisms that have yet to be uncovered. Intriguingly, SIRT2 is highly expressed in the brain. The relative level of SIRT2 expression in the postnatal hippocampus was found to be at least ten fold higher than that of any other sirtuin protein.62 It was also the most highly expressed of all the 18 HDACs in the hippoacampus.62 This suggests that SIRT2’s proapoptotic activity must be inhibited by other proteins under normal circumstances. Downregulation or inactivation of this protein would then permit SIRT2 to promote cell death.
Reduced SIRT2 activity has also been suggested to contribute to delayed Wallerian degeneration in the WldS mice, and the resistance of these mice to axonal degeneration caused by microtubule-depolymerizing drugs.63 Using cerebellar granule neurons cultured from wild-type and WldS mice, Suzuki and Koike (2007) found that SIRT2 expression was lower in neurons from WldS mice, and that this correlates with increased tubulin acetylation and stabilization of microtubules in response to treatment with the microtubule-destabilizing drug, colchicine. Overexpression of SIRT2 in neurons from WldS mice abrogated microtubule hyperacetylation and resistance to axonal degeneration, whereas siRNA-mediated SIRT2 knock-down enhanced microtubule acetylation and resistance to axonal degeneration in wild-type neurons.63
Relatively little is known about the effects of the other sirtuin proteins on the regulation of neurodegeneration. In a recent study in which each of the seven sirtuins was overexpressed in cultured cerebellar granule neurons, SIRT1 protected against low potassium-induced apoptosis while SIRT2 was proapoptotic.22 These results are consistent with the previously reported effects of SIRT1 and SIRT2 in other experimental paradigms of neurodegeneration. In this study, SIRT3 and SIRT6 induced apoptosis in otherwise healthy neurons.22 Intriguingly, SIRT5 was described to exhibit both protective and apoptotic effects, depending on its subcellular localization. When localized in the nucleus and cytoplasm, SIRT5 was found to exert a protective effect whereas it promoted neuronal death when localized in the mitochondria. As observed by other investigators, Pfister et al. (2008) found that pharamacological inhibition of SIRT1 did not reduce the protein’s protective effect, and neuronal death was not inhibited by SIRT1 activators. Interestingly, complete neuroprotection was observed with two separate mutant forms of SIRT1 lacking deacetylase activity, indicating that SIRT1 can protect neurons by a mechanism that does not require its catalytic activity. Pfister et al. also analyzed the effect of sirtuin expression in HT-22 neuroblastoma cells, which can be induced to die by homocysteic acid treatment. While the effects of most SIRTs were similar to those observed in primary neurons, SIRT6 was modestly protective against homocysteic acid toxicity in HT-22 cells. SIRT5 was localized predominantly in the mitochondria of these cells and was apoptotic.22
Knock-out mice have been produced of every sirtuin, except for SIRT2. These mice all survive until birth (Table 1). While SIRT1−/− mice die perinatally, SIRT6−/− mice are viable for about a month, by which time the development of the nervous system is complete. Mice lacking SIRT3, SIRT4 or SIRT5 have a normal life span. As described for the classical HDACs, none of the SIRT knock-out mice display overt effects on the nervous system (Table 1). Compensatory effects by other sirtuins or HDACs could explain the lack of effect on the regulation of neuronal survival.
EFFECT OF CLASSICAL HDAC INHIBITORS ON NEURODEGENERATION
HDAC inhibitors that are currently being used can be classified into four structurally distinct groups: hydroxamic acids such as vorinostat and trichostatin A, cyclic tetrapeptides such as depsipeptide and apicidin, benzamides such as entinostat, and short-chain fatty acids such as sodium butyrate and valproic acid (for recent reviews,64–66). Of these, hydroxamic acids are the largest class and represent the most potent HDAC inhibitors, functioning at nanomolar concentrations by entering the HDAC’s active site where they chelate zinc.
Since the deacetylation of histones by HDACs has a repressive activity on gene transcription, treatment with HDAC inhibitors could be expected to stimulate global transcriptional activity within cells. Analysis of transcriptional effects resulting from HDAC inhibition has revealed, however, both stimulatory and inhibitory effects on gene expression. While previous studies have demonstrated that expression of less than 10% of the genes are regulated by HDAC inhibitors,67–69 a more recent microarray study utilizing two different HDAC inhibitors (vorinostat and depsipeptide) found that the expression of at least 22% was altered, with slightly more genes being repressed than activated.70 It is possible that the inhibitory effects of HDAC inhibitors on transcription might be secondary or downstream effects resulting from the activation of genes encoding transcriptional repressors and corepressors.
Gene profiling studies conducted using RNA isolated from the brains of patients with Huntington’s disease and from different Huntington’s disease mouse models have revealed alterations in the expression of a large number of genes.71 In the mouse models, many of these alterations occur before symptoms become obvious, suggesting that they play a causal role in disease pathogenesis. In contrast to Huntington’s disease and other triplet-repeat expansion disorders in which transcriptional dysregulation and alterations in acetylation patterns are well described, evidence for a causal role of transcriptional dysfunction in the pathogenesis of the more prevalent neurodegenerative disorders (e.g., AD, PD, amyotrophic lateral sclerosis and stroke-induced brain injury) is somewhat limited. Despite the lack of mechanistic rationale, however, HDAC inhibitors have been used in experimental models of a large number of neurodegenerative disorders, and with great success, as is described below. Indeed, their effectiveness might well be the best evidence for transcriptional dysfunction in most neurodegenerative diseases.
The utility of HDAC inhibitors as potential therapeutic agents for neurodegenerative disorders was first described in a Drosophila model of polyglutamine expansion disease.72 The best studied polyglutamine disorder is Huntington’s disease, which is caused by glutamine expansion within the coding region of the huntingtin gene (HTT). Mutant Htt has been shown to disrupt transcription through different mechanisms. For example, it inhibits the activity of CREB-binding protein, a histone acetyl transferase with global effects on transcription, through direct interaction and sequestration. Mutant Htt also interacts with transcription factors such as Sp1, which regulates the transcription of a large number of genes. Indeed, known targets of Sp1 display decreased expression in human Huntington’s disease and in mouse models thereof. In addition to these mechanisms involving protein–protein interaction, polyQ-expanded Htt can alter post-transcriptional modifications of histones resulting in the condensation of chromatin to a more repressed conformation. Vorinostat and butyrate were shown to arrest ongoing progressive neuronal degeneration and reduce lethality induced by the polyglutamine domain of Htt (polyQ-Htt) or just a polyglutamine peptide (Q48). Even when administered to flies already exhibiting neurodegeneration, HDAC inhibitors reduced further neurodegeneration.72 A similar reduction in the extent and rate of neurodegeneration was observed when a partial loss of function mutant form of Drosophila Sin3A, a corepressor protein that is part of active HDAC complexes, was overexpressed in the flies.72 Because this genetic manipulation could be expected to reduce HDAC activity, it suggested that the HDAC inhibitors act by inhibiting the activity of HDACs as opposed to affecting cellular processes other than the deacetylase pathways. Drosophila expresses five different classical HDACs (Rpd3, Hdac3, HDAC4, HDAC6 and histone deacetylase X, an orthologue of HDAC11) and it was not known which HDACs were responsible for the neuroprotective effects of vorinostat and butyrate. In a more recent study, the same group reported that polyQ-Htt-overexpressing flies with a reduced level of Rpd3 suffered less neurodegeneration and lived longer than those with normal Rpd3 expression.73 Reduced expression of each of the other four HDACs using multiple heterozygous mutations and/or short hairpin-type RNA constructs had no beneficial effect on neurodegeneration or life span.73 These observations suggest that pharmacological inhibitors that selectively target human orthologues of Rpd3 would be excellent therapeutic agents for neurodegenerative disorders. Rpd3 is most homologous to human HDAC1, HDAC2 and HDAC3. Intriguingly, and as described above, HDAC1 can also be neuroprotective in C. elegans and mammalian models of neurodegeneration.38, 74 In addition to flies, HDAC inhibitors are protective in C. elegans and mouse models of polyglutamine-induced neurodegeneration.38, 75–78 In the case of a vorinostat-related HDAC inhibitor called HDACi 4b, improved behavioral performance in the R6/2 transgenic mouse model of Huntington’s disease was observed even when the treatment was initiated after the onset of motor deficits.78 Interestingly, microarray analysis revealed that HDACi 4b administration reversed only a relatively small proportion of gene expression alterations caused by mutant Htt in the transgenic mice.
In a transgenic mouse model of spinal and bulbar muscular atrophy, caused by a polyglutamine repeat in the androgen gene, the HDAC inhibitor sodium butyrate partially alleviated disease symptoms. Benefit was observed within a narrow dose range, with higher doses actually aggravating motor dysfunction.76
HDAC inhibitors may be useful in the treatment of Friedrich’s ataxia, a neurodegenerative disorder caused by a triplet-repeat expansion within the frataxin gene (FXN), which leads to its silencing (reviewed in79). Increased trimethylation of histone H3 and hypoacetylation of histones H3 and H4 is associated with silencing of FXN.80 Administration of novel HDAC inhibitors of the benzamide class to lymphocytes cultured from patients with Friedrich’s ataxia was found to increase H3 and H4 acetylation and reverse FXN gene silencing.81 Increased FXN expression and histone acetylation in the brain after administration of HDAC inhibitors has also been observed in a mouse knock-in model of Friedrich’s ataxia.82
HDAC inhibitors have been shown to prevent neuronal death in experimental models of ischemic stroke. Administration of voronistat after induction of ischemic stroke by middle cerebral artery occlusion reduced brain injury.83 Protection was accompanied by the inhibition of histone H3 deactylation and the stimulation of expression of neuroprotective proteins such as Bcl-2 and HSP70. Similar protection against brain injury and neurological deficits caused by ischemic stroke was described following administration of valproic acid administration.84 In this study, valproic acid also stimulated HSP70 expression and histone acetylation. It also delays disease onset, reduces neurological deficits and prolongs survival in the Sod1-1 G93A transgenic mouse model of amyotrophic lateral sclerosis. The beneficial effect of valproic acid in this case was found to involve the inhibition of GSK-3 activity.85
HDAC inhibitors might be useful in treating spinal muscular atrophy (SMA), a neuromuscular disease caused by the homozygous loss of the SMN1 gene (encoding the survival motor neuron protein), which is characterized by motor neuron degeneration in the spinal cord’s anterior horn cells. SMA disease severity depends on the splicing pattern of a gene that is nearly identical to SMN1, called SMN2. The production of a protein from a full-length form of SMN2 mRNA compensates for the loss of function the protein encoded by SMN1 and impedes the onset and progression of the disease. However, the predominantly produced SMN2 transcript is an alternatively spliced form lacking an exon and generates a truncated and unstable protein, which is unable to compensate effectively. Treatment of lymphoid cells obtained from SMA patients with sodium butyrate was found to increase production of the full-length SMN2 mRNA and protein.86 Another study using valproic acid confirmed these results using fibroblast cell lines derived from SMA patients.87 In these and other studies, HDAC inhibitors increased transcription of SMN2 and altered the splicing pattern to produce more full-length protein encoded by SMN2.86–89 HDAC inhibitors have been found to ameliorate neurological symptoms and increase life span in a mouse model of SMA.86 In a more recent study performed in humans, valproic acid was administered to SMA patients and the levels of SMN2 transcripts and proteins were measured in blood collected from these subjects.90 While full-length SMN2 mRNA and protein were elevated in a third of the patients, one third of them showed no change and a third had reduced levels after valproic acid treatment.90 Whether the treatment improved motor function in the patients was not reported. The possibility that the alteration in SMN2 expression observed in blood, does not occur in spinal cord motorneurons cannot be excluded.
In addition to neurodegenerative disorders, HDAC inhibitors may have potential to treat neurodevelopmental disorders such as Rubinstein-Taybi syndrome and Rett’s syndrome that are caused by mutations affecting transcriptional and epigenetic mechanisms as previously suggested.8, 9, 91 The potential utility of HDAC inhibitors in the treatment of mood and anxiety disorders as well as schizophrenia has also been proposed.6, 8, 9
A limitation of the existing HDAC inhibitors as potential therapeutic drugs is their nonselectivity against different members of the HDAC family. This is of particular significance to diseases of the central nervous system because all HDACs are abundantly expressed in the brain. Moreover, and as described in this review, it is becoming increasingly evident that some HDACs have neuroprotective roles. Indeed, HDAC inhibitors have been found to induce apoptosis in cultures of cerebellar granule neurons, even when administered to the cells for only a few hours.28, 92, 93 Extended exposure to HDAC inhibitors has also been reported to be toxic to cultured cortical neurons.94 Intensive efforts are underway to develop class- and isoform-specific HDAC inhibitors that are also potent, stable and brain-permeating.
SIRTUIN INHIBITORS AND NEURODEGENERATION
One of the most widely used sirtuin inhibitors in experimental systems is nicotinamide, an end product inhibitor of the deacetylase reaction that suppresses sirtuin activity by directly binding within a conserved pocket of the enzyme and inhibiting NAD+ hydrolysis. Not unexpectedly, given its mechanism of action, it inhibits the activities of all sirtuins. Moreover, nicotinamide inhibits other NAD+-dependent enzymes. One of these is poly (ADP-ribose) polymerase-1 (PARP-1), a molecule that is often implicated in promoting neurodegeneration (reviewed in95–97).
The neuroprotective effects of nicotinamide were first demonstrated in Drosophila models of polyglutamine neurotoxicity.98 Nicotinamide administration is also protective in rodent models of MPTP-induced neurodegeneration,99 focal cerebral ischemic stroke100 and traumatic brain injury.101, 102 Furthermore, nicotinamide reduces ethanol-induced neurodegeneration in the developing mouse brain and prevents hyperactivity and memory impairment in adult mice caused by perinatal exposure to alcohol.103 These studies did not assess the status of sirtuins or whether other sirtuin inhibitors had similar effects. Hence, the involvement of mechanisms other than sirtuin inhibition, such as PARP inhibition, in the neuroprotective effects of nicotinamide cannot be ruled out. In some studies, however, evidence for sirtuin inhibition as the possible mechanism for nicotinamide’s beneficial effects has been described. For example, nicotinamide was reported to prevent cognitive deficits in a transgenic mouse model of Alzheimer’s disease. In this study, inhibition of the effects of nicotinamide were similar to those obtained with SIRT1 deficiency. Nicotinamide also dramatically increased acetylated alpha-tubulin in the brain of the transgenic mice. Because alpha-tubulin is a primary SIRT2 substrate, the authors suggested that SIRT2 inhibition was also involved.104 Similarly, inhibition of SIRT1 activity was demonstrated in a study in which nicotinamide protected cultured cortical neurons against excitotoxicity. Sirtinol, an inhibitor of both SIRT1 and SIRT2, was also protective in this study.100 Neuroprotection by nicotinamide has also been found to be mediated through inhibition of molecules other than sirtuins. For example, protection against N-methyl-N-nitrosourea-induced retinal degeneration in rats was proposed to be mediated through inhibition of PARP-1 and JNK signaling.105, 106
Several chemical sirtuin inhibitors have been synthesized recently. Among these, the most commonly used are hydroxynapthaldehyde derivatives such as sitinol and cambinol, splitomicin and its derivatives, indoles and suramins.107, 108 Most of these compounds, however, are not particularly selective against individual sirtuin proteins. Given the beneficial effects of SIRT1 on neuronal survival, SIRT1-specific inhibitors are unlikely to be useful for treatment of neurodegenerative disorders. On the other hand, pharmacologic inhibition of SIRT2 activity could represent a therapeutic approach for degenerative disorders of the central nervous system. Outiero and colleagues designed a chemical library of 200 analogues using an alpha-cyanopropenamide scaffold and identified AGK-2, a compound that selectively inhibited SIRT2 over SIRT1 and SIRT3. AGK-2 protected cultured neurons and flies from alpha-synuclein-induced neurotoxicity.61 A number of other selective SIRT2 inhibitors have been synthesized including para-sirtinol109 and EX-527,110 although these compounds have not been tested in paradigms of neurodegeneration.
CONCLUSIONS
A growing body of evidence suggests that HDAC proteins regulate the survival of neurons. A majority of the HDACs studied thus far in mammalian systems, including HDAC1, HDAC4 and HDAC6, have neuroprotective effects. Similarly evidence from C. elegans also indicates that many HDACs protect against neurodegeneration. This is somewhat unexpected given the impressive protection afforded by HDAC inhibitors in a variety of experimental models of neurodegenerative disease. Existing HDAC inhibitors, and particularly ones used in the studies described in this review, do not discriminate between the different HDAC proteins. Such inhibitors can be expected to affect diverse cellular processes that depend on protein deacetylation and acetylation, most of which may not be involved in neurodegeneration. Determining the most relevant HDACs driving neurodegeneration and developing inhibitors specifically targeting each of them, would be an ideal approach. Given that several of the HDACs have neuroprotective effects, pharmacological activators of these proteins could also be of value in treating neurodegenerative disorders. Indeed, resveratrol, a compound that can activate SIRT1, has been shown to be highly neuroprotective in a variety of experimental models of neurodegenerative disease. Alternatively, the overexpression of neuroprotective HDACs via viral vectors in vulnerable neuronal polulations can be considered.
An unanswered question is whether the neuroprotective effects of HDAC inhibitors are epigenetic (via histone acetylation), non-epigenetic, or both. A related matter is whether HDAC inhibitors target common pathways in different neurodegenerative conditions or whether neuroprotection by the same HDAC inhibitor in different experimental models of neurodegenerative disorders (e.g., Alzheimer’s, Huntington’s and Parkinson’s disease) are mediated by affecting distinct molecules and mechanisms. The answers to these questions notwithstanding, HDAC inhibitors represent an exciting therapeutic tool for the treatment of neurodegenerative disorders.
ACKNOWLEDGEMENTS
This work was supported by grants from the National Institute of Neurological Diseases and Stroke (NS40408 and NS058462) to SRD
Footnotes
DISCLOSURE
The author has nothing to disclose.
REFERENCES
- 1.Halkidou K, Gaughan L, Cook S, Leung HY, Neal DE, Robson CN. Upregulation and nuclear recruitment of HDAC1 in hormone refractory prostate cancer. Prostate. 2004;2(59):177–189. doi: 10.1002/pros.20022. [DOI] [PubMed] [Google Scholar]
- 2.Wilson AJ, Byun DS, Popova N, Murray LB, L'Italien K, Sowa Y, Arango D, Velcich A, Augenlicht LH, Mariadason JM. Histone deacetylase 3 (HDAC3) and other class I HDACs regulate colon cell maturation and p21 expression and are deregulated in human colon cancer. J. Biol. Chem. 2006;19(281):13548–13558. doi: 10.1074/jbc.M510023200. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Z, Yamashita H, Toyama T, Sugiura H, Ando Y, Mita K, Hamaguchi M, Hara Y, Kobayashi S, Iwase H. Quantitation of HDAC1 mRNA expression in invasive carcinoma of the breast*. Breast Cancer Res. Treat. 2005;1(94):11–16. doi: 10.1007/s10549-005-6001-1. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Z, Yamashita H, Toyama T, Sugiura H, Omoto Y, Ando Y, Mita K, Hamaguchi M, Hayashi S, Iwase H. HDAC6 expression is correlated with better survival in breast cancer. Clin. Cancer Res. 2004;20(10):6962–6968. doi: 10.1158/1078-0432.CCR-04-0455. [DOI] [PubMed] [Google Scholar]
- 5.Weichert W. HDAC expression and clinical prognosis in human malignancies. Cancer Lett. 2009;2(280):168–176. doi: 10.1016/j.canlet.2008.10.047. [DOI] [PubMed] [Google Scholar]
- 6.Abel T, Zukin RS. Epigenetic targets of HDAC inhibition in neurodegenerative and psychiatric disorders. Curr. Opin. Pharmacol. 2008;1(8):57–64. doi: 10.1016/j.coph.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carew JS, Giles FJ, Nawrocki ST. Histone deacetylase inhibitors: mechanisms of cell death and promise in combination cancer therapy. Cancer Lett. 2008;1(269):7–17. doi: 10.1016/j.canlet.2008.03.037. [DOI] [PubMed] [Google Scholar]
- 8.Sleiman SF, Basso M, Mahishi L, Kozikowski AP, Donohoe ME, Langley B, Ratan RR. Putting the 'HAT' back on survival signalling: the promises and challenges of HDAC inhibition in the treatment of neurological conditions. Expert Opin. Investig. Drugs. 2009;5(18):573–584. doi: 10.1517/13543780902810345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kazantsev AG, Thompson LM. Therapeutic application of histone deacetylase inhibitors for central nervous system disorders. Nat. Rev. Drug Discov. 2008;10(7):854–868. doi: 10.1038/nrd2681. [DOI] [PubMed] [Google Scholar]
- 10.Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat. Rev. Genet. 2009;1(10):32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang XJ, Seto E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat. Rev. Mol. Cell Biol. 2008;3(9):206–218. doi: 10.1038/nrm2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gan L, Mucke L. Paths of convergence: sirtuins in aging and neurodegeneration. Neuron. 2008;1(58):10–14. doi: 10.1016/j.neuron.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith BC, Hallows WC, Denu JM. Mechanisms and molecular probes of sirtuins. Chem. Biol. 2008;10(15):1002–1013. doi: 10.1016/j.chembiol.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taylor DM, Maxwell MM, Luthi-Carter R, Kazantsev AG. Biological and potential therapeutic roles of sirtuin deacetylases. Cell Mol. Life Sci. 2008;24(65):4000–4018. doi: 10.1007/s00018-008-8357-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saunders LR, Verdin E. Sirtuins: critical regulators at the crossroads between cancer and aging. Oncogene. 2007;37(26):5489–5504. doi: 10.1038/sj.onc.1210616. [DOI] [PubMed] [Google Scholar]
- 16.Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, Damian M, Cheung P, Kusumoto R, Kawahara TL, Barrett JC, Chang HY, Bohr VA, Ried T, Gozani O, Chua KF. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;7186(452):492–496. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tanno M, Sakamoto J, Miura T, Shimamoto K, Horio Y. Nucleocytoplasmic shuttling of the NAD+-dependent histone deacetylase SIRT1. J. Biol. Chem. 2007;9(282):6823–6832. doi: 10.1074/jbc.M609554200. [DOI] [PubMed] [Google Scholar]
- 18.North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol. Cell. 2003;2(11):437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- 19.Hisahara S, Chiba S, Matsumoto H, Tanno M, Yagi H, Shimohama S, Sato M, Horio Y. Histone deacetylase SIRT1 modulates neuronal differentiation by its nuclear translocation. Proc. Natl. Acad. Sci. U. S. A. 2008;40(105):15599–15604. doi: 10.1073/pnas.0800612105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vaquero A, Scher MB, Lee DH, Sutton A, Cheng HL, Alt FW, Serrano L, Sternglanz R, Reinberg D. SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes Dev. 2006;10(20):1256–1261. doi: 10.1101/gad.1412706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol. Biol. Cell. 2005;10(16):4623–4635. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfister JA, Ma C, Morrison BE, D'Mello SR. Opposing effects of sirtuins on neuronal survival: SIRT1-mediated neuroprotection is independent of its deacetylase activity. PLoS ONE. 2008;12(3):e4090. doi: 10.1371/journal.pone.0004090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakamura Y, Ogura M, Tanaka D, Inagaki N. Localization of mouse mitochondrial SIRT proteins: shift of SIRT3 to nucleus by co-expression with SIRT5. Biochem. Biophys. Res. Commun. 2008;1(366):174–179. doi: 10.1016/j.bbrc.2007.11.122. [DOI] [PubMed] [Google Scholar]
- 24.Nakagawa T, Lomb DJ, Haigis MC, Guarente L. SIRT5 Deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell. 2009;3(137):560–570. doi: 10.1016/j.cell.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morrison BE, Majdzadeh N, D'Mello SR. Histone deacetylases: focus on the nervous system. Cell Mol. Life Sci. 2007;17(64):2258–2269. doi: 10.1007/s00018-007-7035-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim D, Frank CL, Dobbin MM, Tsunemoto RK, Tu W, Peng PL, Guan JS, Lee BH, Moy LY, Giusti P, Broodie N, Mazitschek R, Delalle I, Haggarty SJ, Neve RL, Lu Y, Tsai LH. Deregulation of HDAC1 by p25/Cdk5 in neurotoxicity. Neuron. 2008;5(60):803–817. doi: 10.1016/j.neuron.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Majdzadeh N, Morrison BE, D'Mello SR. Class IIA HDACs in the regulation of neurodegeneration. Front. Biosci. 2008;(13):1072–1082. doi: 10.2741/2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morrison BE, Majdzadeh N, Zhang X, Lyles A, Bassel-Duby R, Olson EN, D'Mello SR. Neuroprotection by histone deacetylase-related protein. Mol. Cell. Biol. 2006;9(26):3550–3564. doi: 10.1128/MCB.26.9.3550-3564.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Majdzadeh N, Wang L, Morrison BE, Bassel-Duby R, Olson EN, D'Mello SR. HDAC4 inhibits cell-cycle progression and protects neurons from cell death. Dev. Neurobiol. 2008;8(68):1076–1092. doi: 10.1002/dneu.20637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen B, Cepko CL. HDAC4 regulates neuronal survival in normal and diseased retinas. Science. 2009;5911(323):256–259. doi: 10.1126/science.1166226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Linseman DA, Bartley CM, Le SS, Laessig TA, Bouchard RJ, Meintzer MK, Li M, Heidenreich KA. Inactivation of the myocyte enhancer factor-2 repressor histone deacetylase-5 by endogenous Ca(2+) //calmodulin-dependent kinase II promotes depolarization-mediated cerebellar granule neuron survival. J. Biol. Chem. 2003;42(278):41472–41481. doi: 10.1074/jbc.M307245200. [DOI] [PubMed] [Google Scholar]
- 32.Kawaguchi Y, Kovacs JJ, McLaurin A, Vance JM, Ito A, Yao TP. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell. 2003;6(115):727–738. doi: 10.1016/s0092-8674(03)00939-5. [DOI] [PubMed] [Google Scholar]
- 33.Iwata A, Riley BE, Johnston JA, Kopito RR. HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J. Biol. Chem. 2005;48(280):40282–40292. doi: 10.1074/jbc.M508786200. [DOI] [PubMed] [Google Scholar]
- 34.Olzmann JA, Li L, Chudaev MV, Chen J, Perez FA, Palmiter RD, Chin LS. Parkin-mediated K63-linked polyubiquitination targets misfolded DJ-1 to aggresomes via binding to HDAC6. J. Cell Biol. 2007;6(178):1025–1038. doi: 10.1083/jcb.200611128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pandey UB, Nie Z, Batlevi Y, McCray BA, Ritson GP, Nedelsky NB, Schwartz SL, DiProspero NA, Knight MA, Schuldiner O, Padmanabhan R, Hild M, Berry DL, Garza D, Hubbert CC, Yao TP, Baehrecke EH, Taylor JP. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;7146(447):859–863. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- 36.Boyault C, Zhang Y, Fritah S, Caron C, Gilquin B, Kwon SH, Garrido C, Yao TP, Vourc'h C, Matthias P, Khochbin S. HDAC6 controls major cell response pathways to cytotoxic accumulation of protein aggregates. Genes Dev. 2007;17(21):2172–2181. doi: 10.1101/gad.436407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kovacs JJ, Murphy PJ, Gaillard S, Zhao X, Wu JT, Nicchitta CV, Yoshida M, Toft DO, Pratt WB, Yao TP. HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Mol. Cell. 2005;5(18):601–607. doi: 10.1016/j.molcel.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 38.Bates EA, Victor M, Jones AK, Shi Y, Hart AC. Differential contributions of Caenorhabditis elegans histone deacetylases to huntingtin polyglutamine toxicity. J. Neurosci. 2006;10(26):2830–2838. doi: 10.1523/JNEUROSCI.3344-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Outeiro TF, Marques O, Kazantsev A. Therapeutic role of sirtuins in neurodegenerative disease. Biochim. Biophys. Acta. 2008;6(1782):363–369. doi: 10.1016/j.bbadis.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 40.Tang BL, Chua CE. SIRT1 and neuronal diseases. Mol. Aspects Med. 2008;3(29):187–200. doi: 10.1016/j.mam.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 41.Araki T, Sasaki Y, Milbrandt J. Increased nuclear NAD biosynthesis and SIRT1 activation prevent axonal degeneration. Science. 2004;5686(305):1010–1013. doi: 10.1126/science.1098014. [DOI] [PubMed] [Google Scholar]
- 42.Wang J, Zhai Q, Chen Y, Lin E, Gu W, McBurney MW, He Z. A local mechanism mediates NAD-dependent protection of axon degeneration. J. Cell Biol. 2005;3(170):349–355. doi: 10.1083/jcb.200504028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen J, Zhou Y, Mueller-Steiner S, Chen LF, Kwon H, Yi S, Mucke L, Gan L. SIRT1 protects against microglia-dependent amyloid-beta toxicity through inhibiting NF-kappaB signaling. J. Biol. Chem. 2005;48(280):40364–40374. doi: 10.1074/jbc.M509329200. [DOI] [PubMed] [Google Scholar]
- 44.Qin W, Yang T, Ho L, Zhao Z, Wang J, Chen L, Zhao W, Thiyagarajan M, MacGrogan D, Rodgers JT, Puigserver P, Sadoshima J, Deng H, Pedrini S, Gandy S, Sauve AA, Pasinetti GM. Neuronal SIRT1 activation as a novel mechanism underlying the prevention of Alzheimer disease amyloid neuropathology by calorie restriction. J. Biol. Chem. 2006;31(281):21745–21754. doi: 10.1074/jbc.M602909200. [DOI] [PubMed] [Google Scholar]
- 45.Kim D, Nguyen MD, Dobbin MM, Fischer A, Sananbenesi F, Rodgers JT, Delalle I, Baur JA, Sui G, Armour SM, Puigserver P, Sinclair DA, Tsai LH. SIRT1 deacetylase protects against neurodegeneration in models for Alzheimer's disease and amyotrophic lateral sclerosis. EMBO J. 2007;13(26):3169–3179. doi: 10.1038/sj.emboj.7601758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parker JA, Arango M, Abderrahmane S, Lambert E, Tourette C, Catoire H, Neri C. Resveratrol rescues mutant polyglutamine cytotoxicity in nematode and mammalian neurons. Nat. Genet. 2005;4(37):349–350. doi: 10.1038/ng1534. [DOI] [PubMed] [Google Scholar]
- 47.Anekonda TS. Resveratrol--a boon for treating Alzheimer's disease? Brain Res. Rev. 2006;2(52):316–326. doi: 10.1016/j.brainresrev.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 48.Karuppagounder SS, Pinto JT, Xu H, Chen HL, Beal MF, Gibson GE. Dietary supplementation with resveratrol reduces plaque pathology in a transgenic model of Alzheimer's disease. Neurochem. Int. 2009;2(54):111–118. doi: 10.1016/j.neuint.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Della-Morte D, Dave KR, DeFazio RA, Bao YC, Raval AP, Perez-Pinzon MA. Resveratrol pretreatment protects rat brain from cerebral ischemic damage via a sirtuin 1-uncoupling protein 2 pathway. Neuroscience. 2009;3(159):993–1002. doi: 10.1016/j.neuroscience.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Q, Yu S, Simonyi A, Rottinghaus G, Sun GY, Sun AY. Resveratrol protects against neurotoxicity induced by kainic acid. Neurochem. Res. 2004;11(29):2105–2112. doi: 10.1007/s11064-004-6883-z. [DOI] [PubMed] [Google Scholar]
- 51.Blanchet J, Longpre F, Bureau G, Morissette M, DiPaolo T, Bronchti G, Martinoli MG. Resveratrol, a red wine polyphenol, protects dopaminergic neurons in MPTP-treated mice. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2008;5(32):1243–1250. doi: 10.1016/j.pnpbp.2008.03.024. [DOI] [PubMed] [Google Scholar]
- 52.Okawara M, Katsuki H, Kurimoto E, Shibata H, Kume T, Akaike A. Resveratrol protects dopaminergic neurons in midbrain slice culture from multiple insults. Biochem. Pharmacol. 2007;4(73):550–560. doi: 10.1016/j.bcp.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 53.Kumar P, Padi SS, Naidu PS, Kumar A. Effect of resveratrol on 3-nitropropionic acid-induced biochemical and behavioural changes: possible neuroprotective mechanisms. Behav. Pharmacol. 2006;5–6(17):485–492. doi: 10.1097/00008877-200609000-00014. [DOI] [PubMed] [Google Scholar]
- 54.Alvira D, Yeste-Velasco M, Folch J, Verdaguer E, Canudas AM, Pallas M, Camins A. Comparative analysis of the effects of resveratrol in two apoptotic models: inhibition of complex I and potassium deprivation in cerebellar neurons. Neuroscience. 2007;3(147):746–756. doi: 10.1016/j.neuroscience.2007.04.029. [DOI] [PubMed] [Google Scholar]
- 55.Harikumar KB, Aggarwal BB. Resveratrol: a multitargeted agent for age-associated chronic diseases. Cell. Cycle. 2008;8(7):1020–1035. doi: 10.4161/cc.7.8.5740. [DOI] [PubMed] [Google Scholar]
- 56.Pirola L, Frojdo S. Resveratrol: one molecule, many targets. IUBMB Life. 2008;5(60):323–332. doi: 10.1002/iub.47. [DOI] [PubMed] [Google Scholar]
- 57.Pallas M, Casadesus G, Smith MA, Coto-Montes A, Pelegri C, Vilaplana J, Camins A. Resveratrol and neurodegenerative diseases: activation of SIRT1 as the potential pathway towards neuroprotection. Curr. Neurovasc Res. 2009;1(6):70–81. doi: 10.2174/156720209787466019. [DOI] [PubMed] [Google Scholar]
- 58.Athar M, Back JH, Kopelovich L, Bickers DR, Kim AL. Multiple molecular targets of resveratrol: Anti-carcinogenic mechanisms. Arch. Biochem. Biophys. 2009;2(486):95–102. doi: 10.1016/j.abb.2009.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kaeberlein M, McDonagh T, Heltweg B, Hixon J, Westman EA, Caldwell SD, Napper A, Curtis R, DiStefano PS, Fields S, Bedalov A, Kennedy BK. Substrate-specific activation of sirtuins by resveratrol. J. Biol. Chem. 2005;17(280):17038–17045. doi: 10.1074/jbc.M500655200. [DOI] [PubMed] [Google Scholar]
- 60.Borra MT, Smith BC, Denu JM. Mechanism of human SIRT1 activation by resveratrol. J. Biol. Chem. 2005;17(280):17187–17195. doi: 10.1074/jbc.M501250200. [DOI] [PubMed] [Google Scholar]
- 61.Outeiro TF, Kontopoulos E, Altmann SM, Kufareva I, Strathearn KE, Amore AM, Volk CB, Maxwell MM, Rochet JC, McLean PJ, Young AB, Abagyan R, Feany MB, Hyman BT, Kazantsev AG. Sirtuin 2 inhibitors rescue alpha-synuclein-mediated toxicity in models of Parkinson's disease. Science. 2007;5837(317):516–519. doi: 10.1126/science.1143780. [DOI] [PubMed] [Google Scholar]
- 62.Pandithage R, Lilischkis R, Harting K, Wolf A, Jedamzik B, Luscher-Firzlaff J, Vervoorts J, Lasonder E, Kremmer E, Knoll B, Luscher B. The regulation of SIRT2 function by cyclin-dependent kinases affects cell motility. J. Cell Biol. 2008;5(180):915–929. doi: 10.1083/jcb.200707126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Suzuki K, Koike T. Mammalian Sir2-related protein (SIRT) 2-mediated modulation of resistance to axonal degeneration in slow Wallerian degeneration mice: a crucial role of tubulin deacetylation. Neuroscience. 2007;3(147):599–612. doi: 10.1016/j.neuroscience.2007.04.059. [DOI] [PubMed] [Google Scholar]
- 64.Balasubramanian S, Verner E, Buggy JJ. Isoform-specific histone deacetylase inhibitors: the next step? Cancer Lett. 2009;2(280):211–221. doi: 10.1016/j.canlet.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 65.Buchwald M, Kramer OH, Heinzel T. HDACi--targets beyond chromatin. Cancer Lett. 2009;2(280):160–167. doi: 10.1016/j.canlet.2009.02.028. [DOI] [PubMed] [Google Scholar]
- 66.Frew AJ, Johnstone RW, Bolden JE. Enhancing the apoptotic and therapeutic effects of HDAC inhibitors. Cancer Lett. 2009;2(280):125–133. doi: 10.1016/j.canlet.2009.02.042. [DOI] [PubMed] [Google Scholar]
- 67.Glaser KB, Staver MJ, Waring JF, Stender J, Ulrich RG, Davidsen SK. Gene expression profiling of multiple histone deacetylase (HDAC) inhibitors: defining a common gene set produced by HDAC inhibition in T24 and MDA carcinoma cell lines. Mol. Cancer. Ther. 2003;2(2):151–163. [PubMed] [Google Scholar]
- 68.Mitsiades CS, Mitsiades NS, McMullan CJ, Poulaki V, Shringarpure R, Hideshima T, Akiyama M, Chauhan D, Munshi N, Gu X, Bailey C, Joseph M, Libermann TA, Richon VM, Marks PA, Anderson KC. Transcriptional signature of histone deacetylase inhibition in multiple myeloma: biological and clinical implications. Proc. Natl. Acad. Sci. U. S. A. 2004;2(101):540–545. doi: 10.1073/pnas.2536759100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mariadason JM, Corner GA, Augenlicht LH. Genetic reprogramming in pathways of colonic cell maturation induced by short chain fatty acids: comparison with trichostatin A, sulindac, and curcumin and implications for chemoprevention of colon cancer. Cancer Res. 2000;16(60):4561–4572. [PubMed] [Google Scholar]
- 70.Peart MJ, Smyth GK, van Laar RK, Bowtell DD, Richon VM, Marks PA, Holloway AJ, Johnstone RW. Identification and functional significance of genes regulated by structurally different histone deacetylase inhibitors. Proc. Natl. Acad. Sci. U. S. A. 2005;10(102):3697–3702. doi: 10.1073/pnas.0500369102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brochier C, Gaillard MC, Diguet E, Caudy N, Dossat C, Segurens B, Wincker P, Roze E, Caboche J, Hantraye P, Brouillet E, Elalouf JM, de Chaldee M. Quantitative gene expression profiling of mouse brain regions reveals differential transcripts conserved in human and affected in disease models. Physiol. Genomics. 2008;2(33):170–179. doi: 10.1152/physiolgenomics.00125.2007. [DOI] [PubMed] [Google Scholar]
- 72.Steffan JS, Bodai L, Pallos J, Poelman M, McCampbell A, Apostol BL, Kazantsev A, Schmidt E, Zhu YZ, Greenwald M, Kurokawa R, Housman DE, Jackson GR, Marsh JL, Thompson LM. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature. 2001;6857(413):739–743. doi: 10.1038/35099568. [DOI] [PubMed] [Google Scholar]
- 73.Pallos J, Bodai L, Lukacsovich T, Purcell JM, Steffan JS, Thompson LM, Marsh JL. Inhibition of specific HDACs and sirtuins suppresses pathogenesis in a Drosophila model of Huntington's disease. Hum. Mol. Genet. 2008;23(17):3767–3775. doi: 10.1093/hmg/ddn273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim D, Nguyen MD, Dobbin MM, Fischer A, Sananbenesi F, Rodgers JT, Delalle I, Baur JA, Sui G, Armour SM, Puigserver P, Sinclair DA, Tsai LH. SIRT1 deacetylase protects against neurodegeneration in models for Alzheimer's disease and amyotrophic lateral sclerosis. EMBO J. 2007;13(26):3169–3179. doi: 10.1038/sj.emboj.7601758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hockly E, Richon VM, Woodman B, Smith DL, Zhou X, Rosa E, Sathasivam K, Ghazi-Noori S, Mahal A, Lowden PA, Steffan JS, Marsh JL, Thompson LM, Lewis CM, Marks PA, Bates GP. Suberoylanilide hydroxamic acid, a histone deacetylase inhibitor, ameliorates motor deficits in a mouse model of Huntington's disease. Proc. Natl. Acad. Sci. U. S. A. 2003;4(100):2041–2046. doi: 10.1073/pnas.0437870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Minamiyama M, Katsuno M, Adachi H, Waza M, Sang C, Kobayashi Y, Tanaka F, Doyu M, Inukai A, Sobue G. Sodium butyrate ameliorates phenotypic expression in a transgenic mouse model of spinal and bulbar muscular atrophy. Hum. Mol. Genet. 2004;11(13):1183–1192. doi: 10.1093/hmg/ddh131. [DOI] [PubMed] [Google Scholar]
- 77.Ferrante RJ, Kubilus JK, Lee J, Ryu H, Beesen A, Zucker B, Smith K, Kowall NW, Ratan RR, Luthi-Carter R, Hersch SM. Histone deacetylase inhibition by sodium butyrate chemotherapy ameliorates the neurodegenerative phenotype in Huntington's disease mice. J. Neurosci. 2003;28(23):9418–9427. doi: 10.1523/JNEUROSCI.23-28-09418.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Thomas EA, Coppola G, Desplats PA, Tang B, Soragni E, Burnett R, Gao F, Fitzgerald KM, Borok JF, Herman D, Geschwind DH, Gottesfeld JM. The HDAC inhibitor 4b ameliorates the disease phenotype and transcriptional abnormalities in Huntington's disease transgenic mice. Proc. Natl. Acad. Sci. U. S. A. 2008;40(105):15564–15569. doi: 10.1073/pnas.0804249105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pandolfo M, Pastore A. The pathogenesis of Friedreich ataxia and the structure and function of frataxin. J. Neurol. 2009;(256) Suppl 1:9–17. doi: 10.1007/s00415-009-1003-2. [DOI] [PubMed] [Google Scholar]
- 80.Soragni E, Herman D, Dent SY, Gottesfeld JM, Wells RD, Napierala M. Long intronic GAA*TTC repeats induce epigenetic changes and reporter gene silencing in a molecular model of Friedreich ataxia. Nucleic Acids Res. 2008;19(36):6056–6065. doi: 10.1093/nar/gkn604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rai M, Soragni E, Jenssen K, Burnett R, Herman D, Coppola G, Geschwind DH, Gottesfeld JM, Pandolfo M. HDAC inhibitors correct frataxin deficiency in a Friedreich ataxia mouse model. PLoS One. 2008;4(3):e1958. doi: 10.1371/journal.pone.0001958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Herman D, Jenssen K, Burnett R, Soragni E, Perlman SL, Gottesfeld JM. Histone deacetylase inhibitors reverse gene silencing in Friedreich's ataxia. Nat. Chem. Biol. 2006;10(2):551–558. doi: 10.1038/nchembio815. [DOI] [PubMed] [Google Scholar]
- 83.Faraco G, Pancani T, Formentini L, Mascagni P, Fossati G, Leoni F, Moroni F, Chiarugi A. Pharmacological inhibition of histone deacetylases by suberoylanilide hydroxamic acid specifically alters gene expression and reduces ischemic injury in the mouse brain. Mol. Pharmacol. 2006;6(70):1876–1884. doi: 10.1124/mol.106.027912. [DOI] [PubMed] [Google Scholar]
- 84.Ren M, Leng Y, Jeong M, Leeds PR, Chuang DM. Valproic acid reduces brain damage induced by transient focal cerebral ischemia in rats: potential roles of histone deacetylase inhibition and heat shock protein induction. J. Neurochem. 2004;6(89):1358–1367. doi: 10.1111/j.1471-4159.2004.02406.x. [DOI] [PubMed] [Google Scholar]
- 85.Feng HL, Leng Y, Ma CH, Zhang J, Ren M, Chuang DM. Combined lithium and valproate treatment delays disease onset, reduces neurological deficits and prolongs survival in an amyotrophic lateral sclerosis mouse model. Neuroscience. 2008;3(155):567–572. doi: 10.1016/j.neuroscience.2008.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chang JG, Hsieh-Li HM, Jong YJ, Wang NM, Tsai CH, Li H. Treatment of spinal muscular atrophy by sodium butyrate. Proc. Natl. Acad. Sci. U. S. A. 2001;17(98):9808–9813. doi: 10.1073/pnas.171105098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Brichta L, Hofmann Y, Hahnen E, Siebzehnrubl FA, Raschke H, Blumcke I, Eyupoglu IY, Wirth B. Valproic acid increases the SMN2 protein level: a well-known drug as a potential therapy for spinal muscular atrophy. Hum. Mol. Genet. 2003;19(12):2481–2489. doi: 10.1093/hmg/ddg256. [DOI] [PubMed] [Google Scholar]
- 88.Sumner CJ, Huynh TN, Markowitz JA, Perhac JS, Hill B, Coovert DD, Schussler K, Chen X, Jarecki J, Burghes AH, Taylor JP, Fischbeck KH. Valproic acid increases SMN levels in spinal muscular atrophy patient cells. Ann. Neurol. 2003;5(54):647–654. doi: 10.1002/ana.10743. [DOI] [PubMed] [Google Scholar]
- 89.Andreassi C, Angelozzi C, Tiziano FD, Vitali T, De Vincenzi E, Boninsegna A, Villanova M, Bertini E, Pini A, Neri G, Brahe C. Phenylbutyrate increases SMN expression in vitro: relevance for treatment of spinal muscular atrophy. Eur. J. Hum. Genet. 2004;1(12):59–65. doi: 10.1038/sj.ejhg.5201102. [DOI] [PubMed] [Google Scholar]
- 90.Brichta L, Holker I, Haug K, Klockgether T, Wirth B. In vivo activation of SMN in spinal muscular atrophy carriers and patients treated with valproate. Ann. Neurol. 2006;6(59):970–975. doi: 10.1002/ana.20836. [DOI] [PubMed] [Google Scholar]
- 91.Alarcon JM, Malleret G, Touzani K, Vronskaya S, Ishii S, Kandel ER, Barco A. Chromatin acetylation, memory, and LTP are impaired in CBP+/− mice: a model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron. 2004;6(42):947–959. doi: 10.1016/j.neuron.2004.05.021. [DOI] [PubMed] [Google Scholar]
- 92.Boutillier AL, Trinh E, Loeffler JP. Constitutive repression of E2F1 transcriptional activity through HDAC proteins is essential for neuronal survival. Ann. N. Y. Acad. Sci. 2002;(973):438–442. doi: 10.1111/j.1749-6632.2002.tb04679.x. [DOI] [PubMed] [Google Scholar]
- 93.Salminen A, Tapiola T, Korhonen P, Suuronen T. Neuronal apoptosis induced by histone deacetylase inhibitors. Brain Res. Mol. Brain Res. 1998;1–2(61):203–206. doi: 10.1016/s0169-328x(98)00210-1. [DOI] [PubMed] [Google Scholar]
- 94.Langley B, D'Annibale MA, Suh K, Ayoub I, Tolhurst A, Bastan B, Yang L, Ko B, Fisher M, Cho S, Beal MF, Ratan RR. Pulse inhibition of histone deacetylases induces complete resistance to oxidative death in cortical neurons without toxicity and reveals a role for cytoplasmic p21(waf1/cip1) in cell cycle-independent neuroprotection. J. Neurosci. 2008;1(28):163–176. doi: 10.1523/JNEUROSCI.3200-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.David KK, Andrabi SA, Dawson TM, Dawson VL. Parthanatos, a messenger of death. Front. Biosci. 2009;(14):1116–1128. doi: 10.2741/3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dawson VL. Inhibition of poly(adenosine diphosphate-ribose) polymerase (PARP) in experimental models of neurologic diseases: cell death prevention. Retina. 2005;8(25) Suppl:S31–S32. doi: 10.1097/00006982-200512001-00012. [DOI] [PubMed] [Google Scholar]
- 97.Kauppinen TM, Swanson RA. The role of poly(ADP-ribose) polymerase-1 in CNS disease. Neuroscience. 2007;4(145):1267–1272. doi: 10.1016/j.neuroscience.2006.09.034. [DOI] [PubMed] [Google Scholar]
- 98.Ghosh S, Feany MB. Comparison of pathways controlling toxicity in the eye and brain in Drosophila models of human neurodegenerative diseases. Hum. Mol. Genet. 2004;18(13):2011–2018. doi: 10.1093/hmg/ddh214. [DOI] [PubMed] [Google Scholar]
- 99.Anderson DW, Bradbury KA, Schneider JS. Broad neuroprotective profile of nicotinamide in different mouse models of MPTP-induced parkinsonism. Eur. J. Neurosci. 2008;3(28):610–617. doi: 10.1111/j.1460-9568.2008.06356.x. [DOI] [PubMed] [Google Scholar]
- 100.Liu D, Gharavi R, Pitta M, Gleichmann M, Mattson MP. Nicotinamide prevents NAD+ depletion and protects neurons against excitotoxicity and cerebral ischemia: NAD+ consumption by SIRT1 may endanger energetically compromised neurons. Neuromolecular Med. 2009;1(11):28–42. doi: 10.1007/s12017-009-8058-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hoane MR, Pierce JL, Holland MA, Anderson GD. Nicotinamide treatment induces behavioral recovery when administered up to 4 hours following cortical contusion injury in the rat. Neuroscience. 2008;3(154):861–868. doi: 10.1016/j.neuroscience.2008.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hoane MR, Kaplan SA, Ellis AL. The effects of nicotinamide on apoptosis and blood-brain barrier breakdown following traumatic brain injury. Brain Res. 2006;1(1125):185–193. doi: 10.1016/j.brainres.2006.10.019. [DOI] [PubMed] [Google Scholar]
- 103.Ieraci A, Herrera DG. Nicotinamide protects against ethanol-induced apoptotic neurodegeneration in the developing mouse brain. PLoS Med. 2006;4(3):e101. doi: 10.1371/journal.pmed.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Green KN, Steffan JS, Martinez-Coria H, Sun X, Schreiber SS, Thompson LM, LaFerla FM. Nicotinamide restores cognition in Alzheimer's disease transgenic mice via a mechanism involving sirtuin inhibition and selective reduction of Thr231-phosphotau. J. Neurosci. 2008;45(28):11500–11510. doi: 10.1523/JNEUROSCI.3203-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Uehara N, Miki K, Tsukamoto R, Matsuoka Y, Tsubura A. Nicotinamide blocks N-methyl-N-nitrosourea-induced photoreceptor cell apoptosis in rats through poly (ADP-ribose) polymerase activity and Jun N-terminal kinase/activator protein-1 pathway inhibition. Exp. Eye Res. 2006;3(82):488–495. doi: 10.1016/j.exer.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 106.Kiuchi K, Yoshizawa K, Shikata N, Matsumura M, Tsubura A. Nicotinamide prevents N-methyl-N-nitrosourea-induced photoreceptor cell apoptosis in Sprague-Dawley rats and C57BL mice. Exp. Eye Res. 2002;3(74):383–392. doi: 10.1006/exer.2001.1127. [DOI] [PubMed] [Google Scholar]
- 107.Schemies J, Sippl W, Jung M. Histone deacetylase inhibitors that target tubulin. Cancer Lett. 2009;2(280):222–232. doi: 10.1016/j.canlet.2009.01.040. [DOI] [PubMed] [Google Scholar]
- 108.Neugebauer RC, Sippl W, Jung M. Inhibitors of NAD+ dependent histone deacetylases (sirtuins) Curr. Pharm. Des. 2008;6(14):562–573. doi: 10.2174/138161208783885380. [DOI] [PubMed] [Google Scholar]
- 109.Mai A, Massa S, Lavu S, Pezzi R, Simeoni S, Ragno R, Mariotti FR, Chiani F, Camilloni G, Sinclair DA. Design, synthesis, and biological evaluation of sirtinol analogues as class III histone/protein deacetylase (Sirtuin) inhibitors. J. Med. Chem. 2005;24(48):7789–7795. doi: 10.1021/jm050100l. [DOI] [PubMed] [Google Scholar]
- 110.Napper AD, Hixon J, McDonagh T, Keavey K, Pons JF, Barker J, Yau WT, Amouzegh P, Flegg A, Hamelin E, Thomas RJ, Kates M, Jones S, Navia MA, Saunders JO, DiStefano PS, Curtis R. Discovery of indoles as potent and selective inhibitors of the deacetylase SIRT1. J. Med. Chem. 2005;25(48):8045–8054. doi: 10.1021/jm050522v. [DOI] [PubMed] [Google Scholar]
- 111.Lagger G, O'Carroll D, Rembold M, Khier H, Tischler J, Weitzer G, Schuettengruber B, Hauser C, Brunmeir R, Jenuwein T, Seiser C. Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression. EMBO J. 2002;11(21):2672–2681. doi: 10.1093/emboj/21.11.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Montgomery RL, Davis CA, Potthoff MJ, Haberland M, Fielitz J, Qi X, Hill JA, Richardson JA, Olson EN. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes Dev. 2007;14(21):1790–1802. doi: 10.1101/gad.1563807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Trivedi CM, Luo Y, Yin Z, Zhang M, Zhu W, Wang T, Floss T, Goettlicher M, Noppinger PR, Wurst W, Ferrari VA, Abrams CS, Gruber PJ, Epstein JA. Hdac2 regulates the cardiac hypertrophic response by modulating Gsk3 beta activity. Nat. Med. 2007;3(13):324–331. doi: 10.1038/nm1552. [DOI] [PubMed] [Google Scholar]
- 114.Montgomery RL, Potthoff MJ, Haberland M, Qi X, Matsuzaki S, Humphries KM, Richardson JA, Bassel-Duby R, Olson EN. Maintenance of cardiac energy metabolism by histone deacetylase 3 in mice. J. Clin. Invest. 2008;11(118):3588–3597. doi: 10.1172/JCI35847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Vega RB, Matsuda K, Oh J, Barbosa AC, Yang X, Meadows E, McAnally J, Pomajzl C, Shelton JM, Richardson JA, Karsenty G, Olson EN. Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell. 2004;4(119):555–566. doi: 10.1016/j.cell.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 116.Chang S, McKinsey TA, Zhang CL, Richardson JA, Hill JA, Olson EN. Histone deacetylases 5 and 9 govern responsiveness of the heart to a subset of stress signals and play redundant roles in heart development. Mol. Cell. Biol. 2004;19(24):8467–8476. doi: 10.1128/MCB.24.19.8467-8476.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhang Y, Kwon S, Yamaguchi T, Cubizolles F, Rousseaux S, Kneissel M, Cao C, Li N, Cheng HL, Chua K, Lombard D, Mizeracki A, Matthias G, Alt FW, Khochbin S, Matthias P. Mice lacking histone deacetylase 6 have hyperacetylated tubulin but are viable and develop normally. Mol. Cell. Biol. 2008;5(28):1688–1701. doi: 10.1128/MCB.01154-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chang S, Young BD, Li S, Qi X, Richardson JA, Olson EN. Histone deacetylase 7 maintains vascular integrity by repressing matrix metalloproteinase 10. Cell. 2006;2(126):321–334. doi: 10.1016/j.cell.2006.05.040. [DOI] [PubMed] [Google Scholar]
- 119.Haberland M, Mokalled MH, Montgomery RL, Olson EN. Epigenetic control of skull morphogenesis by histone deacetylase 8. Genes Dev. 2009;14(23):1625–1630. doi: 10.1101/gad.1809209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhang CL, McKinsey TA, Chang S, Antos CL, Hill JA, Olson EN. Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell. 2002;4(110):479–488. doi: 10.1016/s0092-8674(02)00861-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Cheng HL, Mostoslavsky R, Saito S, Manis JP, Gu Y, Patel P, Bronson R, Appella E, Alt FW, Chua KF. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc. Natl. Acad. Sci. U. S. A. 2003;19(100):10794–10799. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.McBurney MW, Yang X, Jardine K, Hixon M, Boekelheide K, Webb JR, Lansdorp PM, Lemieux M. The mammalian SIR2alpha protein has a role in embryogenesis and gametogenesis. Mol. Cell. Biol. 2003;1(23):38–54. doi: 10.1128/MCB.23.1.38-54.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, Mostoslavsky R, Kim J, Yancopoulos G, Valenzuela D, Murphy A, Yang Y, Chen Y, Hirschey MD, Bronson RT, Haigis M, Guarente LP, Farese RV, Jr, Weissman S, Verdin E, Schwer B. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol. Cell. Biol. 2007;24(27):8807–8814. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, Mills KD, Patel P, Hsu JT, Hong AL, Ford E, Cheng HL, Kennedy C, Nunez N, Bronson R, Frendewey D, Auerbach W, Valenzuela D, Karow M, Hottiger MO, Hursting S, Barrett JC, Guarente L, Mulligan R, Demple B, Yancopoulos GD, Alt FW. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;2(124):315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 125.Vakhrusheva O, Smolka C, Gajawada P, Kostin S, Boettger T, Kubin T, Braun T, Bober E. Sirt7 increases stress resistance of cardiomyocytes and prevents apoptosis and inflammatory cardiomyopathy in mice. Circ. Res. 2008;6(102):703–710. doi: 10.1161/CIRCRESAHA.107.164558. [DOI] [PubMed] [Google Scholar]