Abstract
Gastric cancer is the second leading cause of cancer-related deaths worldwide. Conventional cytotoxic chemotherapy has limited efficacy for metastatic gastric cancer, with an overall survival of approximately ten months. Recent advances in high-throughput technologies have enabled the implementation of personalized cancer therapy for high-risk patients. The use of such high-throughput technologies, including microarray and next generation sequencing, have promoted the discovery of novel targets that offer new treatment strategies for patients lacking other therapeutic options. Many molecular pathways are currently under investigation as therapeutic targets in gastric cancer, including those related to the epidermal growth factor receptor family, the mesenchymal-epithelial transition factor axis, and the phosphatidylinositol 3-kinase-AKT-mammalian target of rapamycin factors. Advances in molecular diagnostic tools further support the discovery of new molecular targets. Limitations exist, however; not all patients can be tested for biomarkers, and numerous challenges hamper implementation of targeted therapy in clinical settings. Indeed, the scale of tumor genomic profiling is rapidly outpacing our ability to appropriately synthesize all the information in order to optimally refine patient care. Therefore, clinicians must continue to educate themselves regarding new tools and frameworks, and to utilize multidisciplinary team science, comprised of oncologists, geneticists, pathologists, biologists and bioinformaticians, to successfully implement this genomic approach therapeutically.
Keywords: Gastric cancer, Targeted therapy, Biomarker, Microarray, Sequencing
Core tip: Understanding the molecular mechanisms governing carcinogenesis, progression and prognosis of gastric cancer is a prerequisite for development of effective management strategies. Analysis of genomic and proteomic expression profiles of oncogenic signaling pathways have revealed different molecular subtypes of gastric cancer. Development of personalized cancer therapy regimens will specifically target aberrations that drive tumor growth and survival. Therefore, identifying and administering the appropriate drug based on genetic profiling will improve clinical outcomes and decrease toxicity. We anticipate that identification of novel cancer targets will further aid in understanding of cancer heterogeneity and in refinement of personalized therapeutic strategies.
INTRODUCTION
Advances in high-throughput technologies, such as microarray and next generation sequencing (NGS), have led to the discovery of novel therapeutic targets and revealed the power of predictive and prognostic markers in patient care. In addition, such comprehensive genomic approaches have increased our understanding of critical cellular and molecular mechanisms of cancer[1,2]. To date, hundreds of cancer-causing mutations have been discovered by genome-wide sequencing of the entire exome of more than 3000 tumors[3-5]. Most of the mutated cancer-causing genes may already have been identified, as the more recent NGS studies have reported previously identified mutants from different tumor types[5]. Because of this depth of knowledge regarding cancer genomes, we can now target specific aberrations that drive tumor growth and survival and customize drug combinations for individual patients. Despite the feasibility and clinical advantages of personalized cancer therapy, only a small minority of patients are being tested for biomarkers and treated accordingly. Moreover, most emerging drug candidates with no predictive biomarkers fail in clinical trials. Therefore, it is essential to determine which specific targeted therapies are most efficacious for particular sets of patients, and optimization of these treatment regimens should be a priority of future research.
Gastric cancer is one of the most common malignancies worldwide and is the most frequent cancer diagnosed in East Asian countries[6]. Since gastric cancer is a heterogeneous disease, both histologically and genetically, it is difficult to predict patient outcomes using classical histologic and molecular classifications[7]. Surgery is the only curative treatment strategy; yet, even when the primary tumor is resected, some early gastric cancer patients will ultimately succumb to the disease as a result of recurrence of local or distant tumors. Although not necessary in all patients, adjuvant chemotherapy has been shown to benefit some patients with early gastric cancer, whereas conventional chemotherapy has limited efficacy for advanced gastric cancer, with an overall survival of approximately ten mo. Therefore, to improve prognosis of these high-risk patients, it is important to identify predictive biomarkers and to develop refined treatment strategies.
MOLECULAR HETEROGENEITY OF GASTRIC CANCER
Human epidermal growth factor receptor 2 (HER2; also known as ERBB2) is a receptor associated with cell survival, proliferation, migration, adhesion, and differentiation. In the trastuzumab for gastric cancer trial, 594 patients with gastric cancer with overexpression of the HER2 protein were randomly assigned to two treatment groups, chemotherapy (capecitabine/fluorouracil plus cisplatin) or chemotherapy in combination with trastuzumab (a monoclonal antibody that inhibits HER2)[8]. Trastuzumab extended the median overall survival from 11.1 to 13.8 mo (HR = 0.74; P = 0.0046). This finding satisfied the primary objective of the trial and was later referenced in the National Comprehensive Cancer Network guideline.
In addition to the HER2 pathway, there are many other pathways abnormally regulated in gastric cancer. These include the fibroblast growth factor receptor (FGFR) family, the hepatocyte growth factor (HGF)-mesenchymal epithelial transition factor axis, the phosphatidylinositol 3-kinase (PI3K)-AKT-mammalian target of rapamycin (mTOR) factors, and the RAS/RAF/MEK/mitogen-activated protein kinase factors.
Genetic amplification, translocation or mutation of FGFR accelerates growth in a variety of cancers. More specifically, FGFR2 has been reported as amplified in 9% of gastric cancer specimens[9]. For the HGF/Met pathway, dysregulation can occur by aberrant paracrine and autocrine activation via inappropriate ligand production, activating mutations, genomic amplification, increased transcription, or Met receptor overexpression. Overexpression of Met has been detected in human gastric cancers and is associated with a more aggressive phenotype[10-12]. Notably, rilotumumab (a monoclonal antibody targeting the Met-HGF axis) yielded superior overall survival rates in a subgroup analysis of a phase II randomized study. Patients with high levels of Met expression were treated with either epirubicin, cisplatin, and capecitabine (ECX) in combination with rilotumumab or ECX alone in the first-line setting (11.1 mo vs 5.7 mo, HR = 0.29; 95%CI: 0.11-0.76, P = 0.012)[13]. Activation of the PI3K/Akt/mTOR signaling pathway is correlated with poor prognosis and has also been studied as a therapeutic target[14,15]. A phase III trial that evaluated supportive care in combination with either everolimus (an inhibitor of mTOR) or placebo for patients with advanced stage gastric cancer yielded negative results, where there was no significant difference in overall survival between the treatment arms (5.4 mo vs 4.3 mo for the everolimus and placebo groups, respectively)[16]. Histone deacetylase and poly (ADP-ribose) polymerase (PARP; a family of proteins involved in a number of cellular processes involving DNA repair and programmed cell death) have been investigated as treatment targets for gastric cancer[17,18]. Angiogenesis is essential in cancer development, growth, and proliferation, and the vascular endothelial growth factor (VEGF) and receptor (VEGFR) have been spotlighted as therapeutic targets. Recently, the REGARD Trial reported that ramucirumab (a VEGFR-2 monoclonal antibody) improved both progression-free survival and overall survival (vs placebo)[19]. Many clinical trials have evaluated molecular targeting agents that correspond to the various aforementioned signaling pathways (Figure 1), and many of them are ongoing (Table 1)[8,18-23].
Figure 1.
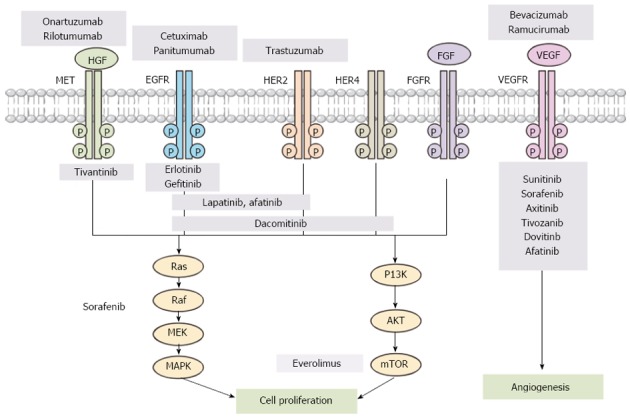
Molecular targeting agents for different signaling pathways in gastric cancer. PI3K: Phosphatidylinositol 3-kinase; HER: Human epidermal growth factor receptor; FGFR: Fibroblast growth factor (FGF) receptor; mTOR: Mammalian target of rapamycin; HGF: Hepatocyte growth factor; VEGFR: Vascular endothelial growth factor (VEGF) receptor; EGF: Epidermal growth factor; MAPK: Mitogen-activated protein kinase.
Table 1.
Recent phase III clinical trials investigating molecular targeting agents in gastric cancer
| Clinical trial | Biomarker | n | Results | Achievement of primary objective | Ref. |
| HER2 inhibitor | |||||
| Capecitabine/cisplatin ± trastuzumab (ToGA) | HER2 | 584 | PFS 6.7 mo vs 5.5 mo, P = 0.0002 | Positive | [8] |
| OS 13.8 mo vs 11.1 mo, P = 0.0046 | |||||
| Capecitabine/oxaliplatin ± lapatinib (LOGiC) | HER3 | 545 | Enrollment done | NP (NCT00680901) | |
| Paclitaxel ± lapatinib (TYTAN) | HER4 | 261 | OS 11.3 mo vs 8.8 mo, P = 0.2088 | Negative | [20] |
| EGFR inhibitor | |||||
| Capecitabine/cisplatin ± cetuximab (EXPAND) | NA | 904 | PFS 5.6 mo vs 4.4 mo, P = 0.3158 | Negative | [21] |
| OS 10.7 mo vs 9.4 mo, P = 0.9547 | |||||
| Increased toxicity | |||||
| Epirubicin/oxaliplatin/capecitabine ± panitumumab (REAL-3) | NA | 553 | PFS 6.0 mo vs 7.4 mo, P = 0.068 | Negative | [22] |
| OS 8.8 mo vs 11.3 mo, P = 0.013 | |||||
| Increased toxicity | |||||
| Angiogenesis inhibitor | |||||
| Capecitabine/cisplatin ± bevacizumab (AVAGAST) | NA | 774 | PFS 6.7 mo vs 5.3 mo, P = 0.0037 | Negative | [23] |
| OS 12.1 mo vs 10.1 mo, P = 0.1002 | |||||
| Ramucirumab vs placebo (REGARD) | NA | 355 | PFS 2.1 mo vs 1.3 mo, P < 0.0001 | Positive | [19] |
| OS 5.2 mo vs 3.8 mo, P = 0.0473 | |||||
| Paclitaxel ± ramucirumab (RAINBOW) | NA | 665 | Enrollment done | NP (NCT01170663) | |
| Afatinib vs placebo | NA | 270 | Enrolling | NP (NCT01512745) | |
| C-MET/HGF pathway inhibitor | |||||
| Epirubicin/cisplatin/capecitabine ± rilotumumab (RILOMET-1) | MET | 450 | Enrolling | NP (NCT01697072) | |
| Fluorouracil/folinic acid/oxaliplatin ± onartuzumab (MetGastric) | MET | 800 | Enrolling | NP (NCT01662869) | |
| HER2 | |||||
| PI3K/Akt/mTOR pathway inhibitor | |||||
| Everolimus vs placebo | NA | 648 | PFS 1.68 mo vs 1.41 mo, P < 0.00001 | Negative | [16] |
| OS 5.39 mo vs 4.34 mo, P = 0.1244 | |||||
| Paclitaxel ± everolimus (AIO-STO-0111) | NA | 480 | Enrolling | NP (NCT01248403) | |
NA: Not applicable; NCT: ClinicalTrials.gov identifier; NP: Not published; OS: Overall survival; PFS: Progression-free survival; PI3K: Phosphatidylinositol 3-kinase; HER: Human epidermal growth factor receptor; mTOR: Mammalian target of rapamycin.
Due to large-scale molecular techniques, our understanding of the molecular complexity underlying gastric cancer has increased, and the development of prognostic classifications based on gene expression profiles is rapidly evolving[24-28]. For example, two distinct gastric cancer subclasses that were strongly associated by prognosis were linked by analyzing gene expression profiles[29]. Interestingly, whole exome sequencing of a gastric adenocarcinoma revealed recurrent somatic mutations in both cell adhesion and chromatin remodeling genes[30]. The characterization of more molecular processes and interactions will yield effective tailored therapies to improve patient outcome and reduce drug toxicity.
GENE EXPRESSION MICROARRAY
A recent study by Ahn et al[29] identified the prognostic gene expression signatures of six genes significantly associated with survival and relapse and developed a scoring system based on these genes. Specifically, reverse transcriptase polymerase chain reaction was performed on paraffin-embedded tissues, and microarray was used to generate and analyze gene expression profiling data from 65 gastric cancer patients. Two distinct subgroups were strongly associated with prognosis (Figure 2). The C1 subgroup was linked with a poor prognosis, and six genes were identified whose expressions were unique to this subgroup (CTNNB1, EXOCS3, TOP2A, LBA1, CCL5, and LXTR1). Next, a scoring system based on the six genes was developed that independently predicted the likelihood of relapse after curative resection. This suggested that the distinct gene expression signature accurately reflected the clinical differences between patient subgroups and that such screens can provide information on disease trajectory. In the future, the efficacy of the risk score will be evaluated as a predictive marker for clinical response to adjuvant chemotherapy.
Figure 2.
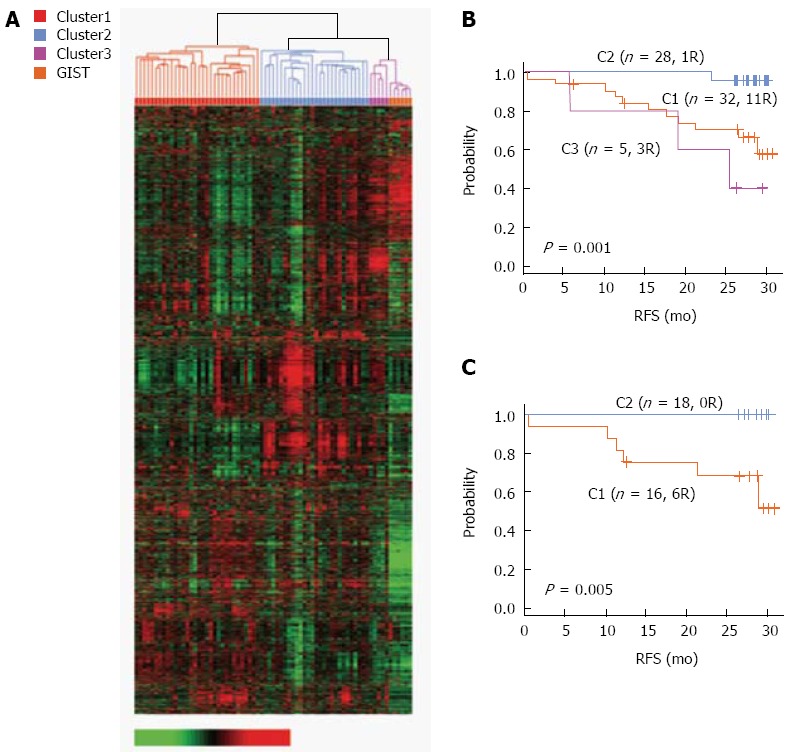
Prognostic expression signatures of genes associated with survival. A: Hierarchical clustering of gene expression data from 65 gastric cancer and 6 gastrointestinal stromal tumors patients in the Yonsei gastric cancer (YGC) cohort; B: Kaplan-Meier plots of 3 gastric cancer clusters in the YGC cohort; C: Kaplan-Meier plots of stage III patients in 2 clusters (C1 and C2) in the YGC cohort. RFS: Recurrence-free survival.
In another study using a gene expression microarray, the M2 isoform of pyruvate kinase (PKM2) was found to be overexpressed in gastric cancers at both the mRNA and protein levels relative to normal gastric tissues. Its expression was negatively correlated with survival in signet-ring cell gastric cancer patients. PKM2 expression may be an adverse prognostic factor for signet-ring cell carcinomas, and the biological role of PKM2 in gastric cancer development and its prognostic value need to be further elucidated[31].
MICRORNA MICROARRAY
MicroRNA (miRNA) play a role in the pathogenesis of various human cancers[32]. Although some miRNAs have been shown to function as oncogenes and others as tumor suppressors, their mechanisms remain to be elucidated[33,34]. The relationship between miRNA expression profile and gastric cancer prognosis[35,36] and pathogenesis[37,38] has been actively explored. For example, miR-196b may be a useful marker, as overexpression of miR-196b has been linked to leukemia and several solid cancers, including gastric cancer[39,40]. Whether miR-196b is an oncogene or tumor suppressor and whether it has a role in gastric carcinogenesis and progression have not yet been confirmed[41]. Recently, miR-21, miR-106b, miR-17, miR-18a and miR-20a were identified as the five most consistently identified miRNAs in screens of gastric cancer. The association between expression level of these miRNAs and clinicopathological features of gastric cancer was significant, and these miRNAs are potential diagnostic and/or prognostic markers that warrant further investigation[42].
RNA-sequencing
RNA-sequencing (RNA-seq) technology enables investigators to simultaneously quantify gene expression levels, assess alternative splicing and gene fusion events, and detect nucleotide variations in transcribed regions. In particular, whole-transcriptome RNA-seq provides a detailed view of the spectrum of expressed transcripts of both coding and noncoding mRNA[43]. Recently, a central metabolic regulator AMP-activated protein kinase (AMPK) was identified as a potential functional target in Asian gastric cancer. As seen in Figure 3, a simple scoring analysis found that only PRKAA2 (AMPKα2) was identified as a potential key modulator of gastric cancer progression. Importantly, the translational relevance of this gene as a target for early-stage gastric cancer was suggested by functional studies in gastric cancer cell lines[44]. Relative to previous RNA-seq studies, this whole-transcriptome RNA-seq approach has several advantages. First, two protocols were used that complementarily covered RNA fragments of different sizes. In this manner, quantification of mRNA, long noncoding RNA and miRNA expression could be conducted simultaneously. Second, ribosome-depleted RNA samples, rather than poly A-enriched RNA samples, were sequenced, thereby generating a less biased population of transcribed molecules. Third, strand-specific short reads were used, and this allowed for more accurate quantification of gene expression. In the future, further functional studies will be necessary to elucidate whether AMPKα2 is an effective therapeutic target.
Figure 3.
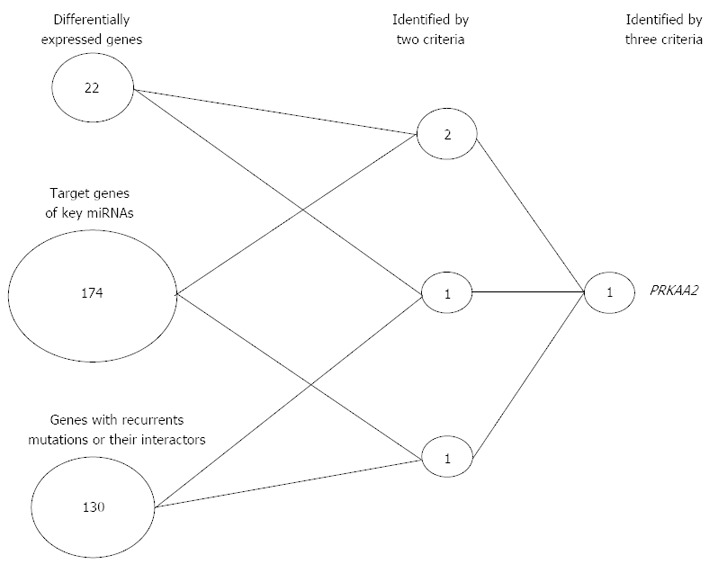
Simple scoring analysis of Asian gastric cancer. Three criteria were used to select key genes: (1) Genes that were identified in both 5-group and 4-stage differential expression analysis; (2) Target genes of six key differentially expressed miRNAs, where the target genes were predicted by TargetScan (Rs < -0.4, P < 0.05); and (3) Genes with recurrent somatic mutations or their interactors (Ingenuity Pathway Analysis program annotation). Only two genes met two criteria, and PRKAA2 was the only gene that met all three criteria. miRNA: MicroRNA.
CONNECTIVITY MAP
The Connectivity Map is a web-based interface (http://www.broadinstitue.org/cmap) that contains more than 7000 expression profiles representing effects of 1309 compounds on several cultured human cells[45]. Analyses using the Connectivity Map reveal functional connections between drugs, genes, and disease and provide a novel approach for cancer treatment. Candidate agents against a specific disease can be recognized by applying disease-specific gene expression profiles to Connectivity Map analysis[46]. Recently, a gastric cancer gene signature was applied to the Connectivity Map, and analysis revealed that histone deacetylase inhibitors, including vorinostat and trichostatin A, were potential drugs for the treatment of gastric cancer. These findings were validated in vitro using gastric cancer cell lines, wherein vorinostat significantly inhibited cell viability in a dose-dependent manner[17]. Therefore, application of unique gene expression profiles to the Connectivity Map may be a viable strategy for the discovery of novel therapeutic agents for gastric cancer.
CONCLUSION
Advances in molecular diagnostics rely more on classifying tumors based on the pathways that drive the oncogenic process, rather than by the tissue of origin. Such molecular tumor classification schemes are very useful in selecting the appropriate pathway inhibitor to apply to an individual patient. Genomic technologies enable robust tumor genomic profiling in the clinical arena (Figure 4) and make it possible to match plausible genetic alterations with rational therapeutic regimens. This means that data from cancer genomes may dictate rational treatment decisions that are tailored for a specific tumor. Importantly, the patients benefit from use of these biomarkers, as the most effective therapy would be selected upfront, sparing the patient from the considerable toxicity associated with conventional “trial and error” therapy. However, the genome era also poses clinical challenges. This unprecedented flow of tumor and germline genomic information needs to be supported by clinical-grade data interpretation. Oncologists will be required to conduct a new generation of evidence-based clinical trials to accommodate smaller numbers of patients with discrete genetic alterations. To aid in this discovery, platforms for repeat biopsy and tissue banking should be established. Moreover, the roles of personalized surgery and radiotherapy should not be underestimated. Defining a subgroup of patients who benefit from radiotherapy and the potential interactions between patient characteristics and the efficacy of radiotherapy should be further explored in the future. We believe this confluence of science, technology, drug discovery, and clinical trial will lead to successful implementation of informed personalized cancer medicine.
Figure 4.
Schematic for genomics-driven cancer medicine where the cancer treatment regimen is adjusted according to each patient’s genome.
Footnotes
P- Reviewers: Merrett ND, Sun LM, Yun S S- Editor: Gou SX L- Editor: A E- Editor: Liu XM
References
- 1.Ludwig JA, Weinstein JN. Biomarkers in cancer staging, prognosis and treatment selection. Nat Rev Cancer. 2005;5:845–856. doi: 10.1038/nrc1739. [DOI] [PubMed] [Google Scholar]
- 2.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M, Subramanian G, Thomas PD, Zhang J, Gabor Miklos GL, Nelson C, Broder S, Clark AG, Nadeau J, McKusick VA, Zinder N, Levine AJ, Roberts RJ, Simon M, Slayman C, Hunkapiller M, Bolanos R, Delcher A, Dew I, Fasulo D, Flanigan M, Florea L, Halpern A, Hannenhalli S, Kravitz S, Levy S, Mobarry C, Reinert K, Remington K, Abu-Threideh J, Beasley E, Biddick K, Bonazzi V, Brandon R, Cargill M, Chandramouliswaran I, Charlab R, Chaturvedi K, Deng Z, Di Francesco V, Dunn P, Eilbeck K, Evangelista C, Gabrielian AE, Gan W, Ge W, Gong F, Gu Z, Guan P, Heiman TJ, Higgins ME, Ji RR, Ke Z, Ketchum KA, Lai Z, Lei Y, Li Z, Li J, Liang Y, Lin X, Lu F, Merkulov GV, Milshina N, Moore HM, Naik AK, Narayan VA, Neelam B, Nusskern D, Rusch DB, Salzberg S, Shao W, Shue B, Sun J, Wang Z, Wang A, Wang X, Wang J, Wei M, Wides R, Xiao C, Yan C, Yao A, Ye J, Zhan M, Zhang W, Zhang H, Zhao Q, Zheng L, Zhong F, Zhong W, Zhu S, Zhao S, Gilbert D, Baumhueter S, Spier G, Carter C, Cravchik A, Woodage T, Ali F, An H, Awe A, Baldwin D, Baden H, Barnstead M, Barrow I, Beeson K, Busam D, Carver A, Center A, Cheng ML, Curry L, Danaher S, Davenport L, Desilets R, Dietz S, Dodson K, Doup L, Ferriera S, Garg N, Gluecksmann A, Hart B, Haynes J, Haynes C, Heiner C, Hladun S, Hostin D, Houck J, Howland T, Ibegwam C, Johnson J, Kalush F, Kline L, Koduru S, Love A, Mann F, May D, McCawley S, McIntosh T, McMullen I, Moy M, Moy L, Murphy B, Nelson K, Pfannkoch C, Pratts E, Puri V, Qureshi H, Reardon M, Rodriguez R, Rogers YH, Romblad D, Ruhfel B, Scott R, Sitter C, Smallwood M, Stewart E, Strong R, Suh E, Thomas R, Tint NN, Tse S, Vech C, Wang G, Wetter J, Williams S, Williams M, Windsor S, Winn-Deen E, Wolfe K, Zaveri J, Zaveri K, Abril JF, Guigó R, Campbell MJ, Sjolander KV, Karlak B, Kejariwal A, Mi H, Lazareva B, Hatton T, Narechania A, Diemer K, Muruganujan A, Guo N, Sato S, Bafna V, Istrail S, Lippert R, Schwartz R, Walenz B, Yooseph S, Allen D, Basu A, Baxendale J, Blick L, Caminha M, Carnes-Stine J, Caulk P, Chiang YH, Coyne M, Dahlke C, Mays A, Dombroski M, Donnelly M, Ely D, Esparham S, Fosler C, Gire H, Glanowski S, Glasser K, Glodek A, Gorokhov M, Graham K, Gropman B, Harris M, Heil J, Henderson S, Hoover J, Jennings D, Jordan C, Jordan J, Kasha J, Kagan L, Kraft C, Levitsky A, Lewis M, Liu X, Lopez J, Ma D, Majoros W, McDaniel J, Murphy S, Newman M, Nguyen T, Nguyen N, Nodell M, Pan S, Peck J, Peterson M, Rowe W, Sanders R, Scott J, Simpson M, Smith T, Sprague A, Stockwell T, Turner R, Venter E, Wang M, Wen M, Wu D, Wu M, Xia A, Zandieh A, Zhu X. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 3.Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, Jia M, Shepherd R, Leung K, Menzies A, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garraway LA, Lander ES. Lessons from the cancer genome. Cell. 2013;153:17–37. doi: 10.1016/j.cell.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 5.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 7.Shah MA, Ajani JA. Gastric cancer--an enigmatic and heterogeneous disease. JAMA. 2010;303:1753–1754. doi: 10.1001/jama.2010.553. [DOI] [PubMed] [Google Scholar]
- 8.Bang YJ, Van Cutsem E, Feyereislova A, Chung HC, Shen L, Sawaki A, Lordick F, Ohtsu A, Omuro Y, Satoh T, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet. 2010;376:687–697. doi: 10.1016/S0140-6736(10)61121-X. [DOI] [PubMed] [Google Scholar]
- 9.Deng N, Goh LK, Wang H, Das K, Tao J, Tan IB, Zhang S, Lee M, Wu J, Lim KH, et al. A comprehensive survey of genomic alterations in gastric cancer reveals systematic patterns of molecular exclusivity and co-occurrence among distinct therapeutic targets. Gut. 2012;61:673–684. doi: 10.1136/gutjnl-2011-301839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peters S, Adjei AA. MET: a promising anticancer therapeutic target. Nat Rev Clin Oncol. 2012;9:314–326. doi: 10.1038/nrclinonc.2012.71. [DOI] [PubMed] [Google Scholar]
- 11.Lennerz JK, Kwak EL, Ackerman A, Michael M, Fox SB, Bergethon K, Lauwers GY, Christensen JG, Wilner KD, Haber DA, et al. MET amplification identifies a small and aggressive subgroup of esophagogastric adenocarcinoma with evidence of responsiveness to crizotinib. J Clin Oncol. 2011;29:4803–4810. doi: 10.1200/JCO.2011.35.4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee J, Seo JW, Jun HJ, Ki CS, Park SH, Park YS, Lim HY, Choi MG, Bae JM, Sohn TS, et al. Impact of MET amplification on gastric cancer: possible roles as a novel prognostic marker and a potential therapeutic target. Oncol Rep. 2011;25:1517–1524. doi: 10.3892/or.2011.1219. [DOI] [PubMed] [Google Scholar]
- 13.Oliner KS, Tang R, Anderson A, Lan Y, Iveson T, Donehower RC, Jiang Y, Dubey S, Loh E. Evaluation of Met pathway biomarkers in a phase II study of rilotumumab or placebo in combination with epirubicin, cisplatin, and Capecitabine (ECX) in patients with locally advanced or metastatic gastric or esophagogastric junction cancer [abstract] J Clin Oncol. 2012;30(Suppl):4005. [Google Scholar]
- 14.Yu G, Wang J, Chen Y, Wang X, Pan J, Li G, Jia Z, Li Q, Yao JC, Xie K. Overexpression of phosphorylated mammalian target of rapamycin predicts lymph node metastasis and prognosis of chinese patients with gastric cancer. Clin Cancer Res. 2009;15:1821–1829. doi: 10.1158/1078-0432.CCR-08-2138. [DOI] [PubMed] [Google Scholar]
- 15.Tran TN, Brettingham-Moore K, Duong CP, Mitchell C, Clemons NJ, Phillips WA. Molecular changes in the phosphatidylinositide 3-kinase (PI3K) pathway are common in gastric cancer. J Surg Oncol. 2013;108:113–120. doi: 10.1002/jso.23357. [DOI] [PubMed] [Google Scholar]
- 16.Van Cutsem E, Yeh K, Bang Y, Shen L, Ajani JA, Bai YX, Chung HC, Pan HM, Chin K, Muro K, et al. Phase III trial of everolimus in previously treated patients with advanced gastric cancer (AGC): GRANITE-1. J Clin Oncol. 2012;30(Suppl 4):Abstr LBA3. [Google Scholar]
- 17.Claerhout S, Lim JY, Choi W, Park YY, Kim K, Kim SB, Lee JS, Mills GB, Cho JY. Gene expression signature analysis identifies vorinostat as a candidate therapy for gastric cancer. PLoS One. 2011;6:e24662. doi: 10.1371/journal.pone.0024662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Virág L, Szabó C. The therapeutic potential of poly(ADP-ribose) polymerase inhibitors. Pharmacol Rev. 2002;54:375–429. doi: 10.1124/pr.54.3.375. [DOI] [PubMed] [Google Scholar]
- 19.Fuchs CS, Tomasek J, Cho JY, Dumitru F, Passalacqua R, Goswami C, Safran H, dos Santos LV, Aprile G, Ferry DR, et al. REGARD: A phase III, randomized, double-blind trial of ramucirumab and best supportive care (BSC) versus placebo and BSC in the treatment of metastatic gastric or gastroesophageal junction (GEJ) adenocarcinoma following disease progression on first-line platinum- and/or fluoropyrimidine-containing combination therapy. J Clin Oncol. 2012;30(Suppl 34):Abstr LBA5. [Google Scholar]
- 20.Bang YJ. A randomized, open-label, phase III study of lapatinib in combination with weekly paclitaxel versus weekly paclitaxel alone in the second-line treatment of HER2 amplified advanced gastric cancer in Asian population: TyTAN study. J Clin Oncol. 2012;30(suppl 34):Abstr 11. doi: 10.1200/JCO.2013.53.6136. [DOI] [PubMed] [Google Scholar]
- 21.Lordick F, Kang YK, Chung HC, Salman P, Oh SC, Bodoky G, Kurteva G, Volovat C, Moiseyenko VM, Gorbunova V, et al. Capecitabine and cisplatin with or without cetuximab for patients with previously untreated advanced gastric cancer (EXPAND): a randomised, open-label phase 3 trial. Lancet Oncol. 2013;14:490–499. doi: 10.1016/S1470-2045(13)70102-5. [DOI] [PubMed] [Google Scholar]
- 22.Waddell TS, Chau I, Barbachano Y, de Castro DG, Wotherspoon A, Saffery C, Middleton GW, Wadsley J, Ferry DR, Mansoor W, et al. A randomized multicenter trial of epirubicin, oxaliplatin, and capecitabine (EOC) plus panitumumab in advanced esophagogastric cancer (REAL3) J Clin Oncol. 2012;30(Suppl 18):Abstr LBA4000. [Google Scholar]
- 23.Ohtsu A, Shah MA, Van Cutsem E, Rha SY, Sawaki A, Park SR, Lim HY, Yamada Y, Wu J, Langer B, et al. Bevacizumab in combination with chemotherapy as first-line therapy in advanced gastric cancer: a randomized, double-blind, placebo-controlled phase III study. J Clin Oncol. 2011;29:3968–3976. doi: 10.1200/JCO.2011.36.2236. [DOI] [PubMed] [Google Scholar]
- 24.Kim B, Bang S, Lee S, Kim S, Jung Y, Lee C, Choi K, Lee SG, Lee K, Lee Y, et al. Expression profiling and subtype-specific expression of stomach cancer. Cancer Res. 2003;63:8248–8255. [PubMed] [Google Scholar]
- 25.Chen CN, Lin JJ, Chen JJ, Lee PH, Yang CY, Kuo ML, Chang KJ, Hsieh FJ. Gene expression profile predicts patient survival of gastric cancer after surgical resection. J Clin Oncol. 2005;23:7286–7295. doi: 10.1200/JCO.2004.00.2253. [DOI] [PubMed] [Google Scholar]
- 26.Hippo Y, Taniguchi H, Tsutsumi S, Machida N, Chong JM, Fukayama M, Kodama T, Aburatani H. Global gene expression analysis of gastric cancer by oligonucleotide microarrays. Cancer Res. 2002;62:233–240. [PubMed] [Google Scholar]
- 27.Tay ST, Leong SH, Yu K, Aggarwal A, Tan SY, Lee CH, Wong K, Visvanathan J, Lim D, Wong WK, et al. A combined comparative genomic hybridization and expression microarray analysis of gastric cancer reveals novel molecular subtypes. Cancer Res. 2003;63:3309–3316. [PubMed] [Google Scholar]
- 28.Chen X, Leung SY, Yuen ST, Chu KM, Ji J, Li R, Chan AS, Law S, Troyanskaya OG, Wong J, et al. Variation in gene expression patterns in human gastric cancers. Mol Biol Cell. 2003;14:3208–3215. doi: 10.1091/mbc.E02-12-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ahn MJ, Park BB, Ahn JS, Kim SW, Kim HT, Lee JS, Kang JH, Cho JY, Song HS, Park SH, et al. Are there any ethnic differences in molecular predictors of erlotinib efficacy in advanced non-small cell lung cancer. Clin Cancer Res. 2008;14:3860–3866. doi: 10.1158/1078-0432.CCR-07-4608. [DOI] [PubMed] [Google Scholar]
- 30.Zang ZJ, Cutcutache I, Poon SL, Zhang SL, McPherson JR, Tao J, Rajasegaran V, Heng HL, Deng N, Gan A, et al. Exome sequencing of gastric adenocarcinoma identifies recurrent somatic mutations in cell adhesion and chromatin remodeling genes. Nat Genet. 2012;44:570–574. doi: 10.1038/ng.2246. [DOI] [PubMed] [Google Scholar]
- 31.Lim JY, Yoon SO, Seol SY, Hong SW, Kim JW, Choi SH, Cho JY. Overexpression of the M2 isoform of pyruvate kinase is an adverse prognostic factor for signet ring cell gastric cancer. World J Gastroenterol. 2012;18:4037–4043. doi: 10.3748/wjg.v18.i30.4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hammond SM. MicroRNAs as oncogenes. Curr Opin Genet Dev. 2006;16:4–9. doi: 10.1016/j.gde.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 35.Ueda T, Volinia S, Okumura H, Shimizu M, Taccioli C, Rossi S, Alder H, Liu CG, Oue N, Yasui W, et al. Relation between microRNA expression and progression and prognosis of gastric cancer: a microRNA expression analysis. Lancet Oncol. 2010;11:136–146. doi: 10.1016/S1470-2045(09)70343-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li X, Zhang Y, Zhang Y, Ding J, Wu K, Fan D. Survival prediction of gastric cancer by a seven-microRNA signature. Gut. 2010;59:579–585. doi: 10.1136/gut.2008.175497. [DOI] [PubMed] [Google Scholar]
- 37.Bandres E, Bitarte N, Arias F, Agorreta J, Fortes P, Agirre X, Zarate R, Diaz-Gonzalez JA, Ramirez N, Sola JJ, et al. microRNA-451 regulates macrophage migration inhibitory factor production and proliferation of gastrointestinal cancer cells. Clin Cancer Res. 2009;15:2281–2290. doi: 10.1158/1078-0432.CCR-08-1818. [DOI] [PubMed] [Google Scholar]
- 38.Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, de Martino I, Iliopoulos D, Pilozzi E, Liu CG, Negrini M, et al. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13:272–286. doi: 10.1016/j.ccr.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 39.Popovic R, Riesbeck LE, Velu CS, Chaubey A, Zhang J, Achille NJ, Erfurth FE, Eaton K, Lu J, Grimes HL, et al. Regulation of mir-196b by MLL and its overexpression by MLL fusions contributes to immortalization. Blood. 2009;113:3314–3322. doi: 10.1182/blood-2008-04-154310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsai KW, Hu LY, Wu CW, Li SC, Lai CH, Kao HW, Fang WL, Lin WC. Epigenetic regulation of miR-196b expression in gastric cancer. Genes Chromosomes Cancer. 2010;49:969–980. doi: 10.1002/gcc.20804. [DOI] [PubMed] [Google Scholar]
- 41.Bhatia S, Kaul D, Varma N. Potential tumor suppressive function of miR-196b in B-cell lineage acute lymphoblastic leukemia. Mol Cell Biochem. 2010;340:97–106. doi: 10.1007/s11010-010-0406-9. [DOI] [PubMed] [Google Scholar]
- 42.Wang JL, Hu Y, Kong X, Wang ZH, Chen HY, Xu J, Fang JY. Candidate microRNA biomarkers in human gastric cancer: a systematic review and validation study. PLoS One. 2013;8:e73683. doi: 10.1371/journal.pone.0073683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim YH, Liang H, Liu X, Lee JS, Cho JY, Cheong JH, Kim H, Li M, Downey TJ, Dyer MD, et al. AMPKα modulation in cancer progression: multilayer integrative analysis of the whole transcriptome in Asian gastric cancer. Cancer Res. 2012;72:2512–2521. doi: 10.1158/0008-5472.CAN-11-3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lamb J. The Connectivity Map: a new tool for biomedical research. Nat Rev Cancer. 2007;7:54–60. doi: 10.1038/nrc2044. [DOI] [PubMed] [Google Scholar]
- 46.Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 47.Pietrantonio F, De Braud F, Da Prat V, Perrone F, Pierotti MA, Gariboldi M, Fanetti G, Biondani P, Pellegrinelli A, Bossi I, et al. A review on biomarkers for prediction of treatment outcome in gastric cancer. Anticancer Res. 2013;33:1257–1266. [PubMed] [Google Scholar]


