Abstract
The bromodomain is a highly conserved motif of 110 amino acids that is bundled into four anti-parallel α-helices and found in proteins that interact with chromatin, such as transcription factors, histone acetylases and nucleosome remodelling complexes. Bromodomain proteins are chromatin ‘readers’; they recruit chromatin-regulating enzymes, including ‘writers’ and ‘erasers’ of histone modification, to target promoters and to regulate gene expression. Conventional wisdom held that complexes involved in chromatin dynamics are not ‘druggable’ targets. However, small molecules that inhibit bromodomain and extraterminal (BET) proteins have been described. We examine these developments and discuss the implications for small molecule epigenetic targeting of chromatin networks in cancer.
This Review begins with the recollection of a 1991 report of an unusual chromosome translocation. Investigators identified a novel t(15;19)(q15;p13) translocation that was associated with a case of poorly differentiated thymic carcinoma (known as NUT midline carcinoma (NMC)) that proved fatal for a young Japanese woman1. This type of carcinoma is rare (affecting only 20–40 patients annually in the United States), is refractory to all treatment, uniquely aggressive and is almost uniformly lethal. The tumours involve balanced translocations of the nuclear protein in testis (NUT) gene on chromosome 15q14 and the translocation of the bromodomain-containing protein 4 (BRD4) gene2 on chromosome 19p13.1, or sometimes the closely related3 BRD3 at 9q34. These genes encode transcriptional regulators that contain a double, mutually related motif that comprises 110 amino acids called a bromodomain in the amino-terminal region (FIG. 1) and an extraterminal (ET) protein–protein interaction domain in the carboxy-terminal region. The BRD2, BRD3 and BRD4 proteins share these structural features4,5 and hence are known as BET family proteins6. Studies to define the structure and function of the bromodomain motif have been the object of considerable research interest over the past few years, and data from recent noteworthy studies of BET protein phenotypes that have relevance to cancer are the focus of this Review.
Figure 1. Structure and relationships among bromodomain-containing proteins.
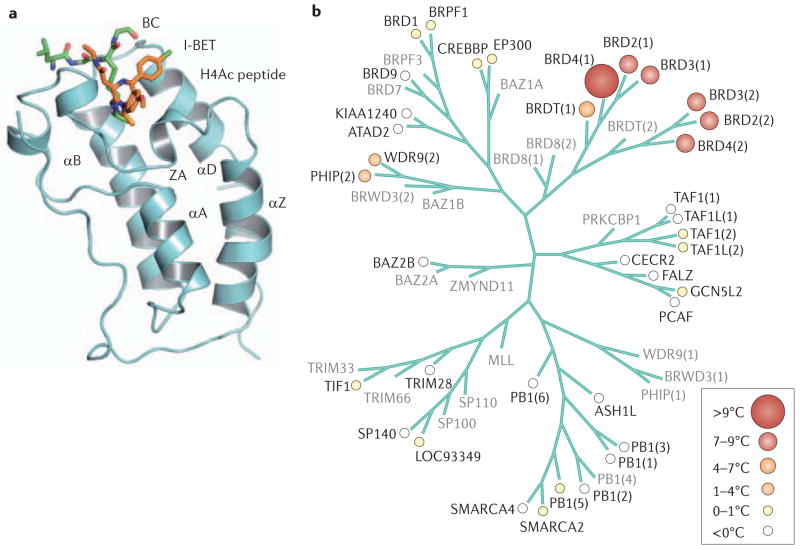
a. The anti-parallel α-helices of the bromodomain bundle are shown in association with the small-molecule inhibitor I-BET and a histone H4 lysine peptide acetylated at position 12 (REF. 97). The BC and ZA loops form the binding pocket for the ε-acetyl-lysine groups of nucleosomal histones in the structure, which the Zhou group first described in detail13. b. Relatedness among bromodomain families, as defined by selectivity for JQ1, is measured by differential scanning fluorimetry79. The BET proteins BRD2, BRD3 and BRD4 are shown to be closely related, with respect to both the first and second bromodomains, as well as the first bromodomain of BRDT. The second bromodomain of BRDT was not tested (shown in grey). Part a is reproduced, with permission, from REF. 97 © (2010) Macmillan Publishers Ltd. All rights reserved. Part b reproduced, with permission, from REF. 79 © (2010) Macmillan Publishers Ltd. All rights reserved.
The characteristics of BET proteins
Bromodomains6–9 were first identified in connection with components of the SWI/SNF nucleosome remodelling complex and the Mediator transcription complex10–12. The bromodomain comprises a highly conserved, four-helix, left-twisted bundle with a characteristic hydrophobic cleft between two conserved loops. The so-called ZA and BC loops in the bromodomain bind to the ε-aminoacetyl groups of nucleosomal histone lysines13 (FIG. 1a). In humans, there are estimated to be 56 bromodomains encoded in 42 proteins14. Bromodomains are found in the closely related DNA helicase superfamily members SWI/SNF-related matrix-associated, actin-dependent, regulator of chromatin, subfamily A, member 2 (SMARCA2; also known as brahma, BRM and SNF2α) and SMARCA4 (also known as BRG1 and SNF2β)15. SMARCA2 and SMARCA4 are the mutually exclusive core catalytic ATPase subunits of a SWI/SNF complex16 that regulates chromatin status9,17. The brahma subunit (its name conferred by Drosophila melanogaster biologists who were searching the Hindu pantheon for evocative descriptors) gives bromodomain its etymology. The bromodomain is also found in many transcriptional and developmental regulators that function through histone modification and nucleosome remodelling6–8 (FIG. 1b). These regulators include authentic histone acetyltransferases (HATs), such as CREB-binding protein (CBP)18,19 and TBP-associated factor 1 (TAF1; also known as CCG1 and TAFII250)6,20.
In proteins that contain two bromodomains, such as BET proteins, these domains are mutually related and arranged in tandem. However, in homologous proteins, first bromodomains are more closely related to each other than they are to second bromodomains in the same protein. In yeast and plants, double bromodomains do not exist and instead are found on separate genes that encode single bromodomain proteins4,5. Such proteins combine to give the functionality seen in a single protein in humans. For example, yeast Bdf1 and Bdf2 proteins are encoded by different genes but are closely related to each other, as well as to TAF1. Bdf1 and Bdf2 combine with yeast TAFII130/145 to execute the analogous functions of human TAF1 (REF. 21).
The BRD2 gene, which Trowsdale and colleagues6,20 originally named RING3, is situated within the human class II major histocompatibility complex (MHC) on chromosome 6 at p21.3, and in syntenic regions of other organisms20,22–24. The RING etymology was unfortunate because BRD2 is completely unrelated to the RING domain family of zinc finger proteins that are involved in protein ubiquitylation and degradation25, a confusion that persists to this day. The BRD2 gene was also identified as part of a major effort by the Sanger Centre to sequence and publish all the open reading frames on human chromosome 6. BRD2 is flanked by genes involved in antigen presentation, inflammation and other immune functions, although structurally BRD2 is highly dissimilar to these nearby genes26. BRD2 was the first mammalian BET protein to be functionally characterized27 as a nuclear-localized28 non-canonical protein kinase29,30 and effector of mitogenic signal transduction. BRD2 binds to ε-aminoacetyl groups of nucleosomal histone lysines31, particularly acetyl-histone H4 (REFS 32,33), recruits transcription factors, transcriptional co-activators and transcriptional co-repressors29,34 and regulates transcription35. BRD2 and related bromodomain proteins provide a scaffold on chromatin36 to recruit E2F proteins29,37, histone deacetylases (HDACs)34, histone H4-specific acetyltransferase (HAT)38 and proteins involved in chromatin remodelling, including SWI/SNF subunits and elements of the Mediator complex10–12,34, thereby coupling histone acetylation to transcription35,38. The ATP-dependence of association of these complex components34 suggests that other BET proteins such as BRD4 also participate in multiprotein complexes that are conserved in composition or that may be shared among BET proteins.
The close similarity between the bromodomains of BRD2 and BRD4 (about 80% identity at the amino acid level in humans and mice) implies that the substantial functional divergence between BRD2- and BRD4-manipulated phenotypes in vivo probably does not lie in the relative specificity of the dual bromodomains for their target promoters per se, so much as in the enzymes recruited through interaction with the ET protein–protein interaction domain4,5 and C-terminal domain (CTD) (FIG. 2). All mammalian BET family members possess the ET domain39 in some form, along with putative nuclear localization signals28. The ET domain is also about 80% identical among BET family members4,5, thus, factors that are recruited through this domain might be expected to be shared among the family and to possibly contribute to functional redundancy across certain promoters. In addition, the ET domain of BRD2, BRD3 and BRD4 independently recruits transcription-modifying factors, including glioma tumour suppressor candidate region gene 1 (GLTSCR1); NSD3, a SET domain-containing histone methyltransferase; JMJD6, a histone arginine demethylase; and CHD4, a catalytic component of the NuRD nucleosome remodelling complex40. Depletion of GLTSCR1, JMJD6 and NSD3 through small interfering RNA (siRNA) establishes the importance of these factors for BRD4 transactivation of a model viral promoter, the bovine papilloma virus 1 locus control region40, as well as important cellular genes, including cyclin D1 (CCND1). These results suggest potential new targets to treat cancers for which cyclin D1 is particularly important, such as breast cancer41–43. Small-molecule inhibitors of BET proteins would also be expected to displace these activities from chromatin, by virtue of their association with the ET domain; the functional consequences of such a loss should be resolved from the loss of the CTD-associated activities and further investigated.
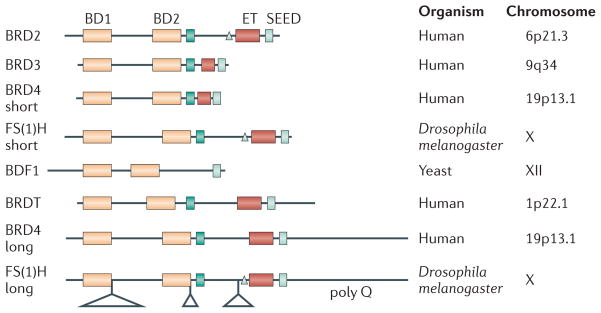
Figure 2. Motif alignment of double bromodomain-containing proteins.
The dual, tandem bromodomains (BDs) are mutually related and always positioned at the amino terminus, where anchoring to nucleosomal histones is encoded, and the carboxy-terminal end of each polypeptide is available for interaction with chromatin-modifying factors, transcription factors, histone-modification enzymes and other proteins. This recruitment takes place either through poorly understood protein–protein interaction ET domains or through SEED domains rich in acidic, phosphorylatable amino acids that resemble the C-terminal domain of RNA polymerase II. The human, fruitfly and yeast proteins contain putative or verified nuclear localization signals (shown in blue) or ATP binding, kinase catalytic sites (shown by grey triangles). Additional features of the Drosophila melanogaster protein include large insertions (represented by triangles) and stretches of polyglutamine (poly Q), a motif that is frequently associated with transcriptional activation. The C-terminal domain of the long isoform of BRD4 is unstructured and does not contain well-established protein–protein interaction or transcriptional-activation motifs, but is nevertheless partly responsible for functional differences between this isoform of BRD4 and other, shorter BET proteins. The chromosome on which each gene is located is identified.
Several isoforms of each BET protein have been reported, but most fall into long or short isoform categories owing to alternative splicing (FIG. 2), an organizational pattern first described in D. melanogaster (BOX 1). It is not understood how short and long forms, if co-expressed in the same cell, compete for binding to chromatin at the same promoter, or whether BRD2, BRD3 and BRD4 exhibit unique or partially overlapping functions on crucial promoters. Different protein–protein interaction motifs in each isoform or related BET proteins are likely to define important differences in the functional interactions. The CTD of BRD4 is several hundred amino acids longer than the CTD of BRD2 and includes polyserine stretches interspersed with glutamate and aspartate (SEED) motifs and a proline- and glutamine-rich unstructured region of about 500 amino acids in length that is not found in other BET proteins4,5. This unstructured region is similar to the highly phosphorylatable CTD of RNA polymerase II and, like this region in RNA polymerase II, the BRD4 CTD interacts with the positive transcription elongation factor b (P-TEFb; a heterodimer of cyclin-dependent kinase 9 (CDK9) and cyclin T)44,45. P-TEFb has a crucial role in transcription elongation, and phosphorylates the CTD of RNA polymerase46. The M phase to G1 phase transition is likely to be partly controlled by BRD4 recruitment of P-TEFb to target postmitotic genes for transcription immediately on the resumption of G1 phase47.
Box 1. BET proteins and Drosophila melanogaster homeosis.
An interesting insight into BET protein function in cancer originated in studies on the Drosophila melanogaster homologue of human BET genes, called female sterile homeotic (fs(1)h), which was the first BET gene to be functionally characterized. This developmental gene activates Ultrabithorax and exhibits maternal effects: mutational analysis has shown that maternally supplied mRNA controls developmental programming before a zygotic mRNA takes over. Maternal effect has also been demonstrated in zebrafish133, raising the possibility that BET genes might have maternal effects in humans, but no reports are yet available to address this question. The FS(1)H protein exerts chromatin-modification functions during fly development121,122. Both FS(1)H and BRD2 are highly homologous to TAF1 (REF. 20). Like TAF1 and BRD2, FS(1)H exhibits protein kinase activity121, a property that has also recently been reported to be shared with BRD4 (REF. 147), although the physiological importance of phosphorylation events that BET proteins catalyse directly has not been explored mechanistically. Mutation of fs(1)h causes severe defects in differentiation and cell fate; fs(1)h-null is lethal148–150. The fs(1)h locus is an upstream activator of trithorax in D. melanogaster151,152, an important, homeotic control gene that in mice positively regulates Hox-controlled differentiation, countering repression by the Polycomb group (PcG) proteins. The trithorax gene (formerly called ALL1, HRX or HTRX1 and now known as MLL) encodes a bromodomain-containing transcription factor that is disrupted in human 11q23 mixed lineage leukaemias153–156. Several recurring chromosomal translocations associated with human acute leukaemias are characterized by breakpoints that interrupt genes that encode transcription factors of importance for D. melanogaster development138, suggesting that the multiprotein interactions that regulate D. melanogaster development may be conserved in human haematopoiesis. Fly developmental systems remain an underexploited resource for the formulation of mechanistic hypothesis to investigate human malignant transformation. The above facts led to a proposal in 1996 that these functional relationships were conserved among D. melanogaster and humans, linking BET proteins to MLL27, a functional connection that has since been supported by studies of small-molecule BET protein inhibitors that exhibit potent antineoplastic activity in human and murine MLL cell lines92.
BET proteins in cell cycle control
BRD4 (REFS 48,49) and BRD2 (REFS 28,29,38) have crucial roles in cell cycle control of normal mammalian cells32. Initial reports showed that E2F1 and E2F2 (REFS 34,37), which are key transcriptional regulators of S phase genes, are associated with BRD2 multiprotein complexes29,34,38. BRD3-dependent functional relationships with the cell cycle control machinery in normal cells are poorly understood, although there is some evidence that forced expression of Brd3 downregulates the RB–E2F pathway in nasopharyngeal carcinoma cells50. BRD4 seems to be required for the G2 to M phase transition of the cell cycle because microinjection of BRD4-specific antibodies leads to cell cycle arrest51. Interestingly, forced expression of BRD4 opposes the function of replication factor C and also results in G1 to S phase arrest48. More recent evidence shows that, unlike non-BET bromodomain proteins, BRD2 and BRD4 remain bound to mitotic chromatin35,51,52, a property that has been postulated to be important for the maintenance of epigenetic memory47,53– 55. This mechanism primes a set of M phase to G1 phase postmitotic genes for transcription at the outset of the next cell cycle after cytokinesis is complete56, which is consistent with the BRD4 dependence of P-TEFb recruitment. BRD2 also associates with postmitotic chromatin, but does not recruit P-TEFb — the functional importance of which is still unclear. Association with mitotic chromatin is a highly conserved property of BET proteins in yeast57, thale cress (Arabidopsis thaliana) 58,59, roundworm60, zebrafish55, murine49,51,53,56 and human cells61,62.
In view of the elemental role of BET proteins in the normal cell cycle and their ubiquitous expression, it is not surprising that the study of BET protein function through genetic deletion experiments has been difficult: Brd2 and Brd4 are essential for cell growth (TABLE 1). Knockout of Brd2 is lethal in mice63–65, Brd4+/− mice have severe defects in differentiation and organogenesis, and Brd4-null animals die in utero66 owing to mitotic defects48,51. Such defects may occur owing to the failure of the Aurora B spindle checkpoint during mitosis, as Aurora B expression is BRD4 dependent67. Similarly, yeast requires a functional copy of either of the transcription factors BDF1 or BDF2, and double mutants are lethal57,68. The BDF1-null phenotype is also lethal if combined with a block in histone H4 acetylation69. There are no Brd3-knockout model systems available.
Table 1.
Selected reported BET protein phenotypes*
| Organism | Gene | Mutant or forced expression phenotype | Null | Refs |
|---|---|---|---|---|
| Saccharomyces cerevisiae | BDF1 | Sporulation defect and chromatin | Lethal | 57,68,69,120 |
| Arabidopsis thaliana | GTE4 and GTE6 | Mitosis, cell cycle, cell fate pattern formation and histone acetylation | Unknown | 58,59,181 |
| Drosophila melanogaster | fs(1)h | Chromatin, maternal effect and homeotic transformation | Lethal | 121,122,148–150 |
| Danio rerio | brd2a, brd2b and brd4 | Pattern formation, mitosis maternal effect and mitotic chromatin | Unknown | 55,133 |
| Caenorhabditis elegans | bet-1 | Unstable cell fate | Lethal | 60 |
| Mus musculus | Brd2 | Cancer, mitosis and cell cycle | Lethal | 63–65,93 |
| M. musculus | Brd4 | Organ defects, mitosis and cell cycle | Lethal | 48,49,51,66 |
| Homo sapiens | BRD2 | Cancer, virus transcription and inflammation | Unknown | 27,92,96,101,166–168 |
| H. sapien | BRD4 | Cancer, episomal maintenance, virus transcription and higher order chromatin structure | Unknown | 1–3,40,45,47,61, 159–165,182 |
Published reports of phenotypes arising from mutation or expression of BET proteins under the control of a heterologous promoter in different species: yeast (S. cerevisiae), plants (Arabidopsis), flies (D. melanogaster), zebrafish (D. rerio), roundworms (C. elegans), mice (M. musculus) and humans (H. sapiens). Genes are indicated along with phenotypes. Comprehensive information for null phenotypes is not available for all species.
It is interesting that these studies indicate that, although each of the BET proteins is closely related to the others, whatever functional redundancy might exist among them is insufficient to rescue the null phenotypes. Moreover, the TAF1 transcription factor — which also has a double bromodomain, binds acetylated histone H4 (REF. 70) and is crucial for cell cycle progression71,72 — also does not compensate for BET protein loss, despite the fact that it shares some sequence homology and functional similarities with the BET proteins. Indeed, small-molecule inhibitors of BET proteins bind relatively poorly to other bromodomain-containing factors (FIG. 1b).
BET protein interactions, inhibitors and cancer
The data discussed above indicate that BET proteins have a crucial role in regulating gene transcription through the recruitment of proteins to form complexes that modify chromatin. Histone-binding, chromatin-regulatory proteins have long been implicated in cancer; a considerable amount of literature concerning their carcinogenic mechanisms has grown up alongside literature that addresses how dysfunctions of sequence-specific DNA-binding transcription factors are linked to cancer. Indeed, epigenetic deregulation of transcription is now appreciated to be as important for carcinogenesis as genetic mutation73. For example, chromosomal translocations can mistarget bromodomain-containing histone-modification enzymes, such as CBP and p300, or chromatin remodelling machines to incorrect promoters, accounting for a substantial number of haematological malignancies74. Specifically, the t(8;16)(p11;p13) translocation that is associated with the M4/M5 subtype of acute myeloid leukaemias (AMLs) was the first report of a translocation involving CBP75. The t(11:16) (q23;p13.3) translocation, which arises in treatment-related myelodysplasias and AML, fuses mixed lineage leukaemia (MLL) to CBP76. Full oncogenicity of MLL–CBP is retained only if both the histone acetylase activity and the bromodomain of CBP are present in the transforming fusion gene77. In acute promyelocytic leukaemia, a histone acetylase complex replaces the NCoR–SIN3–HDAC repression complex, resulting in inappropriate transactivation of target genes78. Similarly, the recruitment of a transcriptional co-activator instead of a co-repressor is the mechanism by which the oncoprotein AML1–ETO alters gene expression and accounts for >10% of AML74. Mistargeted BET protein fusions probably work in a similar manner, inappropriately recruiting chromatin-modifying enzymes to the promoters of cell cycle control genes to corrupt proliferation programmes. Recent work with BET protein inhibitors supports this proposition.
The NMC tumours that arise from the reciprocally translocated BRD4 or BRD3 genes came to the attention of James Bradner, a medical chemist who had been developing targeted, small-molecule inhibitors of histone deacetylases. Bradner led a group that built on the known interaction of thienodiazepines with BRD4 (for which these compounds had been patented by the Mitsubishi Tanabe Pharma Corporation) to develop a novel, small-molecule inhibitor of the binding interface between acetylated histone H4 in chromatin and a bromodomain. The resulting first-generation BET-specific inhibitor, JQ1, contains a bulky hydrophobic substituent at a chiral centre in the molecule that prevents its binding to the central benzodiazepine receptor (FIG. 3); thus, such compounds should have no psychotropic activity. JQ1 proved to be highly effective against NMC xenografts in mice and promoted both growth arrest and differentiation of NMC cells in vitro79. No drug regimen had been shown to be effective against these tumours before, much less from the surprising angle of inhibiting the binding of proteins to acetylated histones80, rather than inhibiting the HAT or HDAC enzymes themselves. Thus, the discovery received substantial publicity. There is now a small but growing number of structural variations on the JQ1 theme (FIG. 3) that might have clinical value for orphan cancers such as NMC, as well as for other applications.
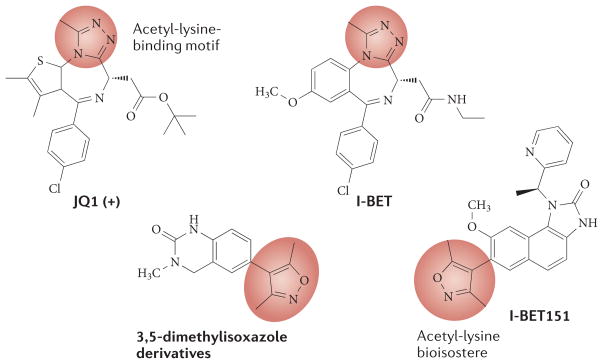
Figure 3. Small-molecule inhibitors of BET proteins.
Recently reported chemical structures and measurements of inhibition constants (IC50) or dissociation constants (Kd) for JQ1 (REF. 79), I-BET (REF. 97), I-BET151 (REF. 92) and other structures that incorporate acetyl-lysine bioisoteres99 are shown. The parts of each molecule that displace the ε-acetyl-lysine group of the histone are circled in red.
Other investigations of the consequences of BET protein inhibition have shown that the transcription of the proto-oncogene MYC is downregulated81. This could account for much of the anti-proliferative effect of JQ1 in human cancer cell lines. BRD3 and BRD4 seem to be involved in chromatin control at the MYC promoter, and MYC in turn represses CDKN2A transcription (which encodes the cell cycle regulator p21) in response to DNA damage and p53 activation82. Thus, ablation of MYC would be expected to promote cell cycle arrest in part through the derepression of p21 (REF. 81). Therefore, there is an emerging rationale to test whether BET protein inhibitors might be clinically useful to target specific human cancers that are strongly dependent on MYC-regulated transcriptional networks, such as Burkitt’s lymphoma83 and certain types of AML81,84. Coincidentally, the single bromodomain protein ATAD2 co-activates MYC and is implicated in breast and prostate cancer85,86. However, the bromodomain of ATAD2 is not closely related to BET bromodomains and none of the small molecules reported to date significantly inhibits its binding to acetylated histones79. Epigenetic inhibitors for non-BET bromodomain proteins await additional design and screening in cancer model systems, particularly breast and prostate cancer. It will be interesting to learn whether any of these small molecules also ablate MYC function.
Independent studies have shown somewhat surprisingly that mice can be treated with chemotherapeutic doses of JQ1 (50 mg per kg per day daily for 1 month) apparently without catastrophic weight loss, metabolic collapse, lethal bone marrow suppression or other systemic toxicity79,81,84. These findings are not in agreement with the Brd2- and Brd4-haploinsufficient or null phenotypes described above (TABLE 1), nor do they indicate that MYC is the primary target of JQ1, because inhibition of MYC activity has substantial effects on constantly renewing tissues, such as intestinal epithelium and skin87,88. Further investigation is clearly needed to understand the mechanisms of JQ1 activity in cancers that express MYC and to study potential, as yet unappreciated or longer-term, toxicities of BET inhibitors in general.
The full range of involvement of BET proteins in cancer is still being established. The interaction of BRD4 with P-TEFb and of TAF1 with the E2 protein of human papillomaviruses (HPVs) that are associated with cervical cancer risk89 (BOX 2) focus attention on relationships between host BRD4 and virus-encoded proteins that are important for transcriptional control and cancer. Importantly, in cells infected with HIV, the HIV transcription factor Tat competes with host BRD4 for binding to host P-TEFb that is present at the HIV long terminal repeat90 to control transcription together with recruited HAT and SWI/SNF activities (BOX 3). Thus, small molecule inhibition of BRD4 might be predicted to release BRD4 from chromatin and remove it from the P-TEFb–Tat equilibrium, which could potentially promote HIV transcription and virus reactivation from latency. This process could also redistribute BRD4 to other multiprotein complexes that associate with chromatin through interactions other than BRD4 bromodomains, or that are unbound to chromatin, with unpredictable results. However, because mice seem to tolerate JQ1 well79,81,84, targeting BRD4 could be preferable to targeting P-TEFb in HIV therapeutics or in malignancies, such as MLL, in which P-TEFb is important91. The potential efficacy of inhibiting BET proteins in MLL has recently been published92. The BET protein inhibitor GSK1210151A (I-BET151) induced apoptosis in both mouse and human leukaemic cell lines with different MLL-fusion genes. Displacement of BRD3 and BRD4 from the chromatin and the polymerase association factor complex (PAFC) and super elongation complex (SEC) that interact with BET proteins resulted in reduced transcription of BCL2, MYC and cyclin-dependent kinase 6 (CDK6)92. These data indicate that the displacement of BET proteins from chromatin, and the factors that associate with them, may have therapeutic efficacy. Incidentally, a number of investigators continue to refer to JQ1 or I-BET as BRD4 inhibitors. This usage is incorrect: JQ1 and I-BET are not highly selective for BRD4 over BRD2, BRD3 or BRDT. Until more specific compounds are available, mechanistic studies that use genetic ablation or overexpression of each BET protein member are required to rule out the participation of the other BET proteins in each case before conclusions can confidently be drawn.
Box 2. Virus replication, latency and transcription.
In certain virus infections, host-encoded BET proteins have been shown to be crucial for both transcriptional activation and transcriptional repression of virus promoters. Certain animal and human papilloma viruses (HPVs) use BET proteins as cellular adaptors to anchor viral genomes to mitotic chromosomes157. The E2 protein of HPV, which is required for virus episome maintenance and virus transcription158, interacts with the carboxy-terminal domain (CTD) of BRD4 (REF. 159) to enable both E2 transcriptional activation of E2-target genes160–162 and E2 repression of the oncogenic E6 and E7 genes163,164. Recombinant truncated CTD of BRD4 exerts dominant-negative effects on E2 transcriptional activation165, possibly through redistribution of multiprotein complexes. Similarly, BRD2 (REFS 166,167), BRD3 (REF. 168) and BRD4 (REFS 61,168) interact with Kaposi’s sarcoma-associated herpesvirus (KSHV; also known as HHV8)-encoded latent nuclear antigen 1 (LANA1), a functional analogue of HPV E2 protein. These BRD proteins contribute to LANA1-regulated transcription61, promoting cell cycle progression. KSHV is a common co-infection among patients infected with HIV and has additionally been implicated in two lymphoid tumours: primary effusion lymphoma and multicentric Castleman’s disease. Reinforcing the theme of context-dependent co-activation or co-repression by the same factor, LANA1 transcriptionally activates some genes, such as E2F-dependent cell cycle genes169, whereas it represses others, such as p53-dependent pro-apoptotic genes170, to promote the proliferation of KSHV-infected cells.
It has been proposed that in Epstein–Barr virus (EBV) episomal maintenance, transformation and latency, the EBV nuclear antigen (EBNA) proteins, particularly EBNA2, provide a functional analogue of LANA1. EBV is the causative agent of lymphoproliferative diseases, lymphomas and certain other malignancies that develop on EBV-driven immortalization. BRD4 recruits the transcription elongation complex P-TEFb to the viral C promoter, and JQ1 (50 nM for 48 hours in vitro) reduces BRD4 association with the promoter, as demonstrated by chromatin immunoprepitation171. This result introduces the possibility that JQ1 or newer small molecules that are more specific for BRD4 might have therapeutic value for EBV-associated malignancies.
A serious safety concern arises on consideration of these small-molecule inhibitors that alleviate BET protein co-repression: human genomes carry diverse, asymptomatic, cell type-specific DNA viruses in a latent form, including HIV. In the case of HIV, this reactivation might offer a solution for virus eradication, but only in the context of intensive anti-retroviral therapy (M. Montano, personal communication). Inadvertent re-activation of some of these latent viruses, of uncertain provenance, could promote viraemia with highly undesirable oncogenic or immunotoxic effects.
Unlike for EBV, inhibition of BRD4 function with JQ1 treatment has not yet been reported for HPV- or KSHV-associated tumours, but represents an obvious, immediately available approach for additional experiments. Targeted delivery of JQ1 or next-generation small molecules is likely to perturb viral transcription and latency in possibly useful therapeutic ways. In each case, the net outcome will probably be determined primarily by re-association and redistribution of the enzymatic and nucleosome remodelling activities that are separately recruited by the BRD4 CTD and ET domains40, which depend on promoter context and respond to signal transduction demands, not by ablation of BRD4 itself.
Box 3. Parallels with the SWI/SNF nucleosome remodelling complex.
Bromodomain-containing transcription complexes functionally resemble SWI/SNF chromatin remodelling complexes (with which BRD2 proteins associate) and exert opposing effects in cell cycle control172,173. Chromatin remodelling machines, such as the SWI/SNF complex, alter the structure of the nucleosome in an ATP-dependent manner by modifying the histone–DNA interface and causing nucleosome sliding174; they are capable of activating or repressing genes. Mammalian SWI/SNF comprises a 2 MDa subunit complex that possess SMARCA4 (also known as BRG1) or SMARCA2 (also known as BRM), and an additional 9–12 proteins known as BRM/BRG1-associated factors (BAFs)16. The majority of genes frequently classified as targets of SWI/SNF enzymes are dependent on either SMARCA4 or SMARCA2, but not on both. Although the different SWI/SNF complexes share many if not most of the same subunits, they are distinguished by the presence of either SMARCA4 or SMARCA2, and unique subunits or tissue-specific variants16,175,176. Smarca4−/− mice are embryonic lethal177, whereas Smarca2−/− mice show a relatively mild phenotype178. The involvement of bromodomain-containing factors in both modes of chromatin remodelling, establishment of transcriptionally active euchromatin or transcriptionally silent heterochromatin, was first appreciated in yeast, in which swi/snf mutations were observed to turn on as many genes as were turned off179,180. Thus, SWI/SNF and BET complexes both co-activate and co-repress genes, depending on the context.
BRD2 activity is also increased in some human leukaemias27, but there is only limited genetic evidence linking BRD2 disruption to human cancer. The Mitelman database of recurrent chromosomal abnormalities associated with cancer (see the Mitelman database; see Further information) identifies >500 patient haematological malignancies involving breakpoints at 6p21 that could potentially affect BRD2, but only a small minority have been mapped with high resolution. The extraordinary amount of polymorphism in this region makes accurate mapping onerous and has slowed progress on this investigation. However, data from mice indicate that BRD2 could potentially be oncogenic in humans. Constitutive expression of a Brd2 transgene in the lymphoid lineage of mice (under Eμ promoter or enhancer control, resulting in B cell-restricted expression), transcriptionally co-activates cyclin A (Ccna2)38 in resting B cells, eventually causing B cell malignancy93. Transcriptional profiling of this aggressive tumour reveals a transcriptional signature that is most similar to the activated B cell (ABC) type of diffuse large B cell lymphoma, in which several genes with developmental functions are re-activated93,94. ABC lymphomas are characterized by the activation of nuclear factor-κB (NF-κB)-regulated genes, which is consistent with the phenotype of Brd2-hypomorphic mice described below and the effect of I-BET on NF-κB-regulated cytokines. Evidence for the involvement of BRD3 in cancer remains preliminary. BRD3 is involved in certain NMC translocations3,95, and it potentially associates with MLL fusion oncoproteins in leukemogenesis92 and with MYC in multiple myeloma96. Brd3-transgenic or knockout animals have not been reported, nor have Brd4-transgenic animals been developed. New mouse models of BET protein-driven malignancy are clearly needed; these models could be of great translational importance for the mechanistic study of haematological malignancy, as well as for the testing of next-generation, small-molecule BET protein inhibitors as cancer chemotherapeutic agents.
BET proteins, inflammation and obesity
Inhibitors of BET proteins also have anti-inflammatory properties. An I-BET compound that is structurally similar97 to JQ1 demonstrates anti-inflammatory properties; however, I-BET does not discriminate among BET family members (FIG. 1b). Nicodeme and colleagues have shown, in a well-established model system of pro-inflammatory cytokine production from bone marrowderived macrophages challenged by bacterial endotoxin, that I-BET suppresses the expression of several crucial pro-inflammatory cytokines and chemokines (such as interleukin-1β (IL-1β), IL-6, IL-12α, CXCL9 and CCL12). Chromatin immunoprecipitation experiments show that I-BET displaces BRD2, BRD3 and BRD4 from the IL6 promoter, which was used as a model97. Most dramatically, I-BET injections (30 mg per kg by the retro-orbital or the tail-vein route) rescued mice from endotoxin-induced death, and caecal ligation and puncture-induced death. An important observation noted in that report97 is that I-BET upregulates hexamethylene bis-acetamide inducible protein 1 (HEXIM1), and this has also been reported for JQ1 in other model systems81,96. HEXIM1 inhibits P-TEFb function, suggesting that I-BET and JQ1 can also block proliferation through MYC-independent mechanisms. HEXIM1 also inhibits NF-κB-dependent target genes and so inhibits the transcription of pro-inflammatory cytokine genes98. The investigators report that the expression of key inflammatory molecules, for example, tumour necrosis factor (TNF), monocyte chemotactic protein 1 and a number of other chemokines, was not ablated by I-BET treatment, which does not make sense in view of the coordinate regulation of NF-κB-dependent transcriptional programmes that are known to be mobilized in macrophages in response to endotoxin challenge. In addition, a new class of dimethyl isoxazole derivatives that inhibit BET bromodomains99,100 has also been shown to have anti-inflammatory properties in cellular assays, in which they block IL-6 and TNF production from bacterial endotoxin-challenged human peripheral blood mononuclear cells101.
Consistent with the ability of small-molecule BET protein inhibitors to ablate inflammation, gene disruption of Brd2 in mice ablates a broad range of inflammatory responses and protects animals from the inflammatory complications of obesity-induced insulin resistance65. Gene disruption was accomplished by lacZ insertion into a region of the gene that is 5′ to the translational start site. This insertion only reduces rather than eliminates whole-body expression of BRD2 (Brd2 lo) to produce a hypomorphic phenotype, thus making these mice viable. The resultant low-inflammatory characteristics of Brd2 lo mice include uncoupling of Toll-like receptor (TLR) and TNF signalling65,102 from NF-κB-directed transcription of diverse pro-inflammatory cytokine genes103. The mouse phenotype provides a model for a population of obese humans for whom obesity is not entirely bad104; population studies have not unfailingly identified an inviolable association between obesity and cardiometabolic risk. These metabolically healthy obese (MHO) subjects105 comprise about 25% of the adult obese population in the United States. Importantly, along with preserved insulin sensitivity and excellent glucose tolerance106, MHO subjects exhibit a reduced inflammatory profile, including less severe elevations of C-reactive protein, TNF and other pro-inflammatory cytokines in the context of their obesity107,108. Compared with insulin-resistant obese subjects, MHO subjects have fewer metabolic complications associated with obesity, including insulin resistance, metabolic syndrome, hypertension, type 2 diabetes and cardiovascular disease, a protection that has been attributed in no small part to their low-inflammatory responses109,110. These subjects are also protected from all-cause cancer mortality compared with insulin-resistant obese subjects111. It is intriguing to speculate that MHO individuals may harbour alleles of BET genes, particularly BRD2, that confer protection from metabolic dysfunction or cancer in obesity.
Beyond haematological malignancy, it remains to be tested whether single nucleotide polymorphisms of human BET genes or alternatively spliced forms of BET mRNAs influence body composition of adults, including adiposity and fat distribution, insulin resistance, inflammatory risks and associated co-morbidities, such as the obesity-associated cancers103. There is recent evidence that, in addition to BRD2, BRD4 can also co-activate pro-inflammatory genes that depend on NF-κB transcription, through interaction with acetylated RELA112. A full account of the interplay of BRD2, BRD3 and BRD4, and how they co-activate NF-κB and cooperate with SWI/SNF complexes113 to regulate the transcription responses of genes that encode important pro-inflammatory cytokines, such as TNF and IL-6, awaits exposition. These data are potentially relevant to the links between unresolved chronic inflammation or irritation and increased cancer risk114, a long-established association115. For example, the bowel inflammation that is characteristic of Crohn’s disease and related conditions116 is strongly linked to colorectal cancer117. It is possible that inflammation promotes certain obesity-associated cancers that are resident in or near to inflamed white adipose tissue in insulin-resistant obese subjects103. The role of unresolved, chronic inflammation and metabolic dysfunction in obesity-associated cancers is a considerable public health problem, and new epigenetically acting drugs such as the BET protein inhibitors might provide a novel pathway for treating or preventing obesity-associated cancer. Additional preclinical studies are required to more firmly establish the mechanisms underlying hypotheses that the anticancer and anti-inflammatory properties of BET protein inhibitors usefully combine in a chemopreventive strategy for the obesity-associated cancers.
BET proteins and transcriptional repression
The obese phenotype of Brd2-hypomorphic mice is partly due to the activation of peroxisome proliferator-activated receptor-γ (PPARγ)-directed transcription in adipocytes65,103. Independent support for this mechanism comes from experiments in which short hairpin RNA (shRNA) knockdown of Brd2 in 3T3-L1 pre-adipocytes strongly promotes adipogenic differentiation. BRD2 directly interacts with PPARγ and opposes its transcriptional function65,118. The discovery that a BET protein can also function to repress transcription in differentiated adult cells raises some interesting questions. This mechanism suggests that a balance exists on PPARγ-controlled promoters between BRD2-associated repressive factors, including N-CoR, SMRT and repressive SWI/SNF complexes, and co-activating factors, such as PPARγ–RXR-ligand complexes, HATs and activating SWI/SNF complexes (FIG. 4). Interestingly, the Nicodeme group119 has described a new class of benzodiazepine-based small-molecule inhibitors with anti-inflammatory effects. These molecules were identified in a screen for compounds that upregulate the transcription of the atheroprotective gene apolipoprotein A1 (REF. 119). Although these authors do not explicitly define the molecular mechanism for this upregulation, the screen almost certainly depends on the alleviation of BET protein repression complex that is present at the human apolipoprotein A1 promoter in hepatocyte HepG2 cells.
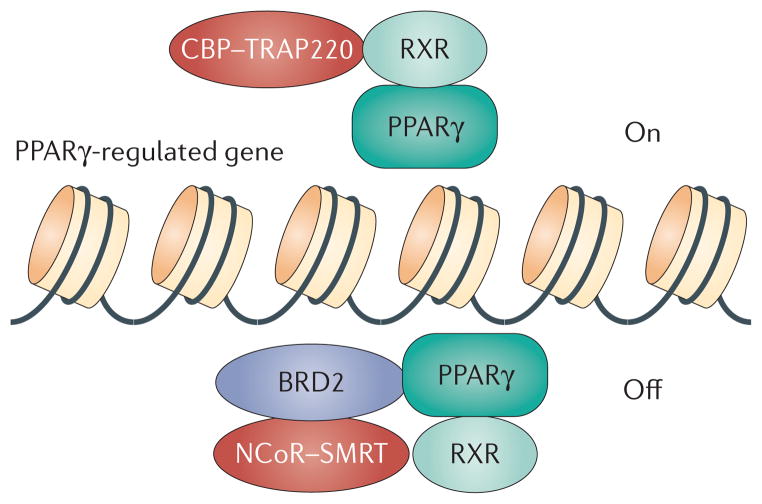
Figure 4. Model for BET protein co-repression of PPARγ-responsive genes.
Transcriptional co-repression of specific loci is an active process that requires the recruitment of repressor complexes. In the case of peroxisome proliferator-activated receptor-γ (PPARγ), co-repression is enabled through BRD2 association with RXR, which is known to heterodimerize with PPARγ. Removal of BRD2 by genetic ablation promotes the transcription of adipogenic networks118, analogous to thiazolidinedione drug treatment. Small-molecule inhibitors of BET proteins would be expected to produce a similar result.
In retrospect, it is not surprising that BET protein knockdown or inhibition should derepress the transcription of certain genes, as this was first suggested for BRD2 more than 10 years ago29, and transcriptional repression mechanisms have long been apparent in BET homologues in other model organisms. The gene product of BDF1 (REF. 68), is a ‘reader’ of histone acetylation120 in Saccharomyces cerevisiae and is important for chromatin restructuring69. In D. melanogaster, the BET homologue female sterile (1) homeotic (fs(1)h) has important transcriptional repression functions that are essential for proper differentiation in the early embryo121,122. Like the SWI/SNF complexes, BET protein complexes can function as both co-activators and co-repressors103,123. They recruit either HATs or HDACs depending on the requirements of signal transduction and the promoter to which they are bound36. This behaviour is enacted with other bromodomain proteins, such as SMARCA2, which binds to the tumour suppressor protein RB124,125 and facilitates co-repression of cell cycle genes through the recruitment of histone deacetylases126–129 (BOX 3).
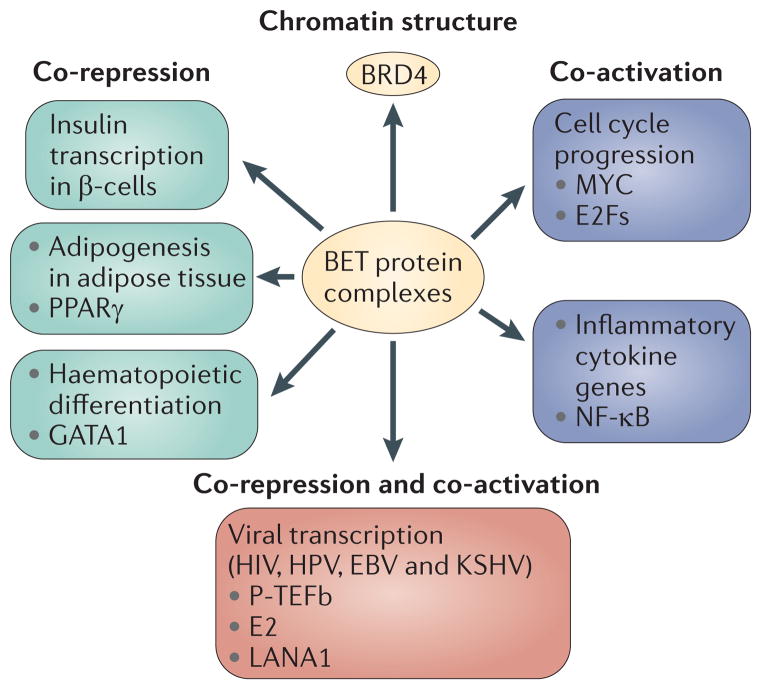
It has recently been suggested that small-molecule inhibitors of BET proteins, such as JQ1, exert their anti-proliferative effects in AML simply as anti-proliferative agents, with more rapidly growing cells being more sensitive130, much the same way that antimetabolite therapies work in diverse malignancies by targeting cells with high mitotic indices. Although downregulation of MYC in several haematological malignancies undoubtedly contributes to the anti-proliferative phenotype, because of the dual nature of bromodomain-containing multiprotein complexes and their cousins the SWI/SNF complexes, this view is incomplete. Rather, we propose that inhibition of BET proteins works through transcriptional reprogramming of a network of target genes, ablating co-activation of cell cycle genes while simultaneously ablating co-repression of other specific, differentiation-associated genes (FIG. 5). Thus, the altered balance of co-activation and co-repression reprogrammes cell fate. The long-appreciated antagonism between haematopoietic proliferation and differentiation131,132 might be explained in part by these dual, opposing functions123 of BET protein-containing co-regulator complexes.
Figure 5. BET proteins co-regulate transcriptional networks of transcriptional activation and repression.
Several functional networks are co-regulated by BET protein interactions. Some interactions involve transcriptional co-repression, such as insulin transcription65, peroxisome proliferator-activated receptor-γ (PPARγ)-controlled adipogenic differentiation in adipose tissue65,118 and GATA1-controlled haematopoietic differentiation143,144. Other interactions involve transcriptional co-activation, such as the activation of genes that promote cell cycle progression controlled by MYC81,84,92,96 and E2F proteins28,29,34,37,38; nuclear factor-κB (NF-κB)-controlled synthesis of pro-inflammatory cytokines65,97,101,102; and cellular genes regulated by P-TEFb. The transcription and replication of latent viruses seem to exploit BET protein capacity for either co-repression or co-activation, depending on the demands of the virus, through P-TEFb40,44,45,47,171, human papilloma virus (HPV) E2 protein157–165 or Kaposi’s sarcoma-associated herpesvirus (KSHV) LANA1 protein61,166–170. Recent data also implicate BRD4 in the maintenance of higher order chromatin structure. EBV, Epstein–Barr virus.
The requirement for BET protein homologues in proper development, pattern formation and stable cell fate in Metazoans, particularly BET-1 in the roundworm60, FS(1)H in the fruitfly121 and Brd2a and Brd2b in the zebrafish133 indicates that this co-regulator system is functionally well conserved. The high degree of relatedness at the amino acid level among bromodomains of diverse species4,5 attests to evolutionary conservation. It follows from this observation and their trithorax-like functions (BOX 1) that BET proteins are also likely to engage an antagonistic relationship with the Polycomb (PcG) group of co-repressors. The balance of trithorax group and PcG genes in the silencing and derepression of transcriptional networks of genes during the development of model organisms is well studied, but BET gene interactions in different tissues during human development are totally unexplored. Interestingly, the single bromodomain protein human BRD7 (REF. 134) tightly interacts with protein arginine methyltransferase 5-containing SWI/SNF complexes, as well as with three core subunits of the PcG repressor complex (PRC)2, that differentially regulate transcriptional silencing and derepression135. In yeast, there is evidence that Bdf1 opposes silencing by restricting the heterochromatic spreading of silent information regulator (SIR) sirtuin proteins136. In addition, BDF1, but not BDF2, was recently identified as a suppressor of mutation in the variant histone H2A.Z pathway, which regulates heterochromatin silencing in S. cerevisiae137, which is consistent with a crucial, fundamental role for BET proteins in chromatin states. It seems likely that additional bromodomain proteins will be found to interact as co-regulators with different families of trithorax and PcG proteins to direct transcriptional programmes that influence development and pattern formation.
It is not coincidental that when developmentally crucial circuits of homeotic genes are disrupted by reciprocal chromosomal translocations in humans, malignancies, particularly acute leukaemias, are a common outcome138,139. For example, perturbation of BET protein expression can lead not only to increased proliferation in malignancy, but also to the reactivation of developmentally silenced genes94, supporting a long-held view that a class of malignancies derived from corrupted transcription factors bear the morphological and transcriptional signatures of poorly differentiated tissues2,3,95 or progenitors of the cell lineage in which the tumour arises. Similar patterns of poorly differentiated sarcomas have been reported for certain tumours arising from mutations in SWI/SNF subunits140. Mutations in SWI/SNF subunits likewise reveal crucial roles for chromatin regulation in adipogenesis, osteogenesis and haematopoiesis. Thus, both SWI/SNF complexes and BET protein complexes are likely to contribute to cell fate in the adult by associating with lineage-specific transcription factors to regulate exit from the highly proliferative state of progenitors and entry into the terminally differentiated state of specialized tissues.
Conclusions and implications
Consideration of BET protein functions discussed above informs the hypothesis that networks of apparently orthogonal transcriptional programmes — adipogenesis, inflammatory cytokine production, cell cycle control and developmental programmes — are actually deeply interrelated because they share this limited set of epigenetic actors. It is true that dedicated adipogenic transcription factors (such as PPARγ), inflammatory transcription factors (such as NF-κB), cell cycle transcription factors (such as E2Fs) and lineage-specific transcription factors (such as PU.1 and TBET) are required for cell-specific responses to signal transduction in each case. However, the phenotypes observed in Brd2-manipulated mice or cells cannot be explained by the alteration of a single pathway, nor do they lead to genome-wide transcriptional confusion, senescence, apoptosis or other catastrophic outcomes. Thus, the extent of BET protein-dependent transcriptional programmes is limited. A possible complication to small-molecule inhibition of BET family bromodomain interactions is that other histone-binding proteins might become available to assume transcriptional co-regulator functions once the BET protein is displaced from chromatin. Similarly, changes in histone acetylation patterns can redistribute important catalytic activities such as DNA topoisomerase II (REF. 141) and mutation of chromatin-regulatory enzymes can redistribute histone variant proteins142 with major consequences for DNA replication and the cell cycle. Redistribution of co-activator and co-repressor complexes on ablation of a BET protein might in some contexts cause unexpected transcriptional activation or repression of transcriptional networks. However, small-molecule BET protein inhibitors have now been shown to be well-tolerated, potent, epigenetically acting, potential anti-neoplastic agents for BRD4-driven NUT midline carcinoma79, multiple myeloma96, AML84 and MLL92. Mice seem to tolerate high doses of JQ1 without major, acute systemic side effects79,81,84. The discovery that BRD3 associates with acetylated GATA1 (REF. 143), a transcription factor that is crucial for normal erythroid and megakaryocytic development, raises concerns that BET protein inhibition might be associated with haematopoietic toxicity. Both JQ1 and an I-BET-like derivative GW841819X inhibit this interaction144, but apparently do not show obvious myelotoxic side effects in mice. Animal models chronically treated with these inhibitors should be more fully evaluated for the suppression of specific haematopoietic lineages. Alternatively, the apparently minimal side effect profile may indicate that BET protein complexes are mobilized in response to specific signal transduction in specialized, differentiated cells of the adult and do not constitutively regulate ordinary metabolic processes or housekeeping functions.
It follows from the above discussion that ablation of BET co-regulator complexes is likely to be simultaneously anti-neoplastic and anti-inflammatory. Pro-adipogenic transcriptional networks that are controlled by PPARγ are also expected to be activated by BET protein inhibition, based on the phenotype of Brd2-hypomorphic mice. It will be interesting to determine whether MHO humans have a lower risk for obesity-associated cancers, attributable to reduced inflammatory responses103. The mechanistic links between inflammation and cancer114, and between inflammation and insulin-resistant obesity106–110, ground an overarching hypothesis that, at the transcriptional level, chronic inflammation in obesity exacerbates risk for both metabolic complications and cancer. The ongoing and anticipated dire consequences of the world wide epidemic of obesity145 highlight the translational importance of this discovery, because about 90% of type 2 diabetes is attributable to obesity146. Chemoprevention of obesity-associated cancers by uncoupling NF-κB-driven cytokine gene expression with small-molecule BET protein inhibitors would represent an innovative, epigenetically based approach to protect obese subjects who are at risk of both diabetes and cancer. Targeting one set of processes with a BET protein inhibitor might confer benefit by targeting other, apparently orthogonal transcriptional networks that are actually fundamentally related. An extraordinary interconnectedness of chromatin-dependent transcription programmes is thus revealed.
At a glance.
Mammalian BET proteins, a class of transcriptional co-regulators that contain dual, mutually related bromodomain motifs and an extraterminal domain, are important in the control of networks of genes; these proteins bind to acetylated lysines in the histones of nucleosomal chromatin, recruit chromatin-modification enzymes to target promoters and function as co-activators or co-repressors of gene expression, depending on the context.
New small-molecule inhibitors have recently been developed that disrupt the binding interface between the bromodomain and the acetylated lysine groups; the inhibitors have remarkable potency, selectivity and are well tolerated. They have recently been used as anticancer and anti-inflammatory agents.
These developments are important because chromatin was not considered to be a druggable target; as a result of these new drugs, a whole field of new epigenetically targeted therapeutics has become available for investigation.
As this field of therapeutics rapidly expands, several features of BET protein function will need to be considered, including possible redundancy among the closely related family members, the selectivity of next-generation agents for specific BET proteins, and possible undesirable consequences of systemic administration without cellular targeting. These side effects might include uncontrolled transcriptional derepression of genes, altered haematopoiesis, immunosuppression or reactivation of latent viruses.
Acknowledgments
This work is supported by grants from the American Cancer Society, Leukaemia and Lymphoma Society and National Institutes of Health. The authors thank J. Bradner, B. Corkey, T. Gilmore, B. Nikolajczyk and M. Obin for useful discussions, and they thank their virology colleagues P. Howley, K. Kaye, D. Margolis, M. Montano and G. Viglianti for generously sharing reagents and ideas.
Glossary
- Mediator transcription complex
Mediator is a large (1.2 MDa) multiprotein complex of up to 30 subunits that responds to signal transduction and that regulates transcription from a diverse set of RNA polymerase II-controlled promoters
- Thienodiazepines
A chemical structural class built on a seven-membered 1,4-diazepine ring fused to a thiophene ring that provides a scaffold for derivatives of great current interest to medicinal chemistry because they target several pharmacologically important proteins, such as certain benzodiazepine derivatives that have neurological activity
- Insulin resistance
A complex phenotype arising from reduced insulin responsiveness in tissues that transport glucose in response to insulin action, such as adipose tissue and skeletal muscle
Biographies
Anna C. Belkina is a doctoral student in the Program in Molecular Medicine at Boston University School of Medicine, USA. Along with Fangnian Wang and others, she contributed the first report of functional involvement of a BET protein in mammalian inflammation, adipogenesis and energy balance.
Gerald V. Denis is a faculty member in the Cancer Research Center and Department of Pharmacology and Experimental Therapeutics of Boston University School of Medicine. He discovered mammalian BET protein function. Building on the knowledge base of BET protein mechanisms in Drosophila melanogaster homeosis, he was the first to show that constitutive expression of a BET protein in mice is oncogenic and that reduced expression reveals a co-repression mechanism of major importance for obesity and inflammation. These observations underpin the new hypothesis that reduced inflammation protects against obesity-associated cancers, in part through BET protein co-regulation of transcriptional networks.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
References
- 1.Kubonishi I, et al. Novel t(15;19)(q15;p13) chromosome abnormality in a thymic carcinoma. Cancer Res. 1991;51:3327–3328. [PubMed] [Google Scholar]
- 2.French CA, et al. BRD4-NUT fusion oncogene: a novel mechanism in aggressive carcinoma. Cancer Res. 2003;63:304–307. [PubMed] [Google Scholar]
- 3.French CA, et al. BRD-NUT oncoproteins: a family of closely related nuclear proteins that block epithelial differentiation and maintain the growth of carcinoma cells. Oncogene. 2008;27:2237–2242. doi: 10.1038/sj.onc.1210852. [DOI] [PubMed] [Google Scholar]
- 4.Florence B, Faller DV. You bet-cha: a novel family of transcriptional regulators. Front Biosci. 2001;6:D1008–D1018. doi: 10.2741/florence. [DOI] [PubMed] [Google Scholar]
- 5.Wu SY, Chiang CM. The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J Biol Chem. 2007;282:13141–13145. doi: 10.1074/jbc.R700001200. [DOI] [PubMed] [Google Scholar]
- 6.Haynes SR, et al. The bromodomain: a conserved sequence found in human, Drosophila and yeast proteins. Nucl Acids Res. 1992;20:2603. doi: 10.1093/nar/20.10.2603. This report offered the first suggestion that the bromodomain module is widely conserved and is likely to have a role in chromatin status and transcriptional regulation in diverse organisms. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jeanmougin F, Wurtz JM, Le Douarin B, Chambon P, Losson R. The bromodomain revisited. Trends Biochem Sci. 1997;22:151–153. doi: 10.1016/s0968-0004(97)01042-6. [DOI] [PubMed] [Google Scholar]
- 8.Winston F, Allis CD. The bromodomain: a chromatin-targeting module? Nature Struct Biol. 1999;6:601–604. doi: 10.1038/10640. [DOI] [PubMed] [Google Scholar]
- 9.Horn PJ, Peterson CL. The bromodomain: a regulator of ATP-dependent chromatin remodeling? Front Biosci. 2001;6:D1019–1023. doi: 10.2741/horn. [DOI] [PubMed] [Google Scholar]
- 10.Jiang YW, et al. Mammalian mediator of transcriptional regulation and its possible role as an end-point of signal transduction pathways. Proc Natl Acad Sci USA. 1998;95:8538–8543. doi: 10.1073/pnas.95.15.8538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuras L, Borggrefe T, Kornberg RD. Association of the Mediator complex with enhancers of active genes. Proc Natl Acad Sci USA. 2003;100:13887–13891. doi: 10.1073/pnas.2036346100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kornberg RD. Mediator and the mechanism of transcriptional activation. Trends Biochem Sci. 2005;30:235–239. doi: 10.1016/j.tibs.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 13.Dhalluin C, et al. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 14.Sanchez R, Zhou MM. The role of human bromodomains in chromatin biology and gene transcription. Curr Opin Drug Discov Dev. 2009;12:659–665. [PMC free article] [PubMed] [Google Scholar]
- 15.Tamkun JW, et al. brahma – a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SWI2/SNF2. Cell. 1992;68:561–572. doi: 10.1016/0092-8674(92)90191-e. [DOI] [PubMed] [Google Scholar]
- 16.Wang W, et al. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 1996;15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- 17.Pazin MJ, Kadonaga JT. SWI2/SNF2 and related proteins: ATP-driven motors that disrupt protein-DNA interactions? Cell. 1997;88:737–740. doi: 10.1016/s0092-8674(00)81918-2. [DOI] [PubMed] [Google Scholar]
- 18.Ogryzko VV, Schiltz OL, Russanova V, Howard BH, Nakatani Y. The transcriptional co-activators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 19.Bannister AJ, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 20.Beck S, Hanson I, Kelly A, Pappin DJC, Trowsdale J. A homologue of the Drosophila female sterile homeotic (fsh) gene in the class II region of the human MHC. DNA Seq. 1992;2:203–210. doi: 10.3109/10425179209020804. [DOI] [PubMed] [Google Scholar]
- 21.Matangkasombut O, Buratowski RM, Swilling NW, Buratowski S. Bromodomain factor 1 corresponds to the missing piece of yeast TFIID. Genes Dev. 2000;14:951–962. [PMC free article] [PubMed] [Google Scholar]
- 22.Thorpe KL, Abdulla S, Kaufman J, Trowsdale J, Beck S. Phylogeny and structure of the RING3 gene. Immunogenet. 1996;44:391–396. doi: 10.1007/BF02602785. [DOI] [PubMed] [Google Scholar]
- 23.Salter-Cid L, Pasquier L, Flajnik M. RING3 is linked to the Xenopus major histocompatibility complex. Immunogenet. 1996;44:397–399. [PubMed] [Google Scholar]
- 24.Takami K, Zaleska-Rutczynska Z, Figueroa F, Klein J. Linkage of LMP, TAP, and RING3 with Mhc class I rather than class II genes in the zebrafish. J Immunol. 1997;159:6052–6060. [PubMed] [Google Scholar]
- 25.Deshaies RJ, Joazeiro CAP. RING domain E3 ubiquitin ligases. Ann Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 26.Ting JP, Trowsdale J. Genetic control of MHC class II expression. Cell. 2002;109:S21–33. doi: 10.1016/s0092-8674(02)00696-7. [DOI] [PubMed] [Google Scholar]
- 27.Denis GV, Green MR. A novel, mitogen-activated nuclear kinase is related to a Drosophila developmental regulator. Genes Dev. 1996;10:261–271. doi: 10.1101/gad.10.3.261. This was the first report of a BET protein phenotype in mammalian cells. [DOI] [PubMed] [Google Scholar]
- 28.Guo N, Faller DV, Denis GV. Activation-induced nuclear translocation of RING3. J Cell Sci. 2000;113:3085–3091. doi: 10.1242/jcs.113.17.3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Denis GV, Vaziri C, Guo N, Faller DV. RING3 kinase transactivates promoters of cell cycle regulatory genes through E2F. Cell Growth Diff. 2000;11:417–424. [PMC free article] [PubMed] [Google Scholar]
- 30.Ostrowski J, Florio SK, Denis GV, Suzuki H, Bomsztyk K. Stimulation of p85/RING3 kinase in multiple organs after systemic administration of mitogens into mice. Oncogene. 1998;16:1223–1227. doi: 10.1038/sj.onc.1201624. [DOI] [PubMed] [Google Scholar]
- 31.Crowley TE, Kaine EM, Yoshida M, Nandi A, Wolgemuth DJ. Reproductive cycle regulation of nuclear import, euchromatic localization, and association with components of Pol. II mediator of a mammalian double-bromodomain protein. Mol Endocrinol. 2002;16:1727–1737. doi: 10.1210/me.2001-0353. [DOI] [PubMed] [Google Scholar]
- 32.Kanno T, et al. Selective recognition of acetylated histones by bromodomain proteins visualized in living cells. Mol Cell. 2004;13:33–43. doi: 10.1016/s1097-2765(03)00482-9. [DOI] [PubMed] [Google Scholar]
- 33.Nakamura Y, et al. Crystal structure of the human BRD2 bromodomain: insights into dimerization and recognition of acetylated histone H4. J Biol Chem. 2007;282:4193–4201. doi: 10.1074/jbc.M605971200. [DOI] [PubMed] [Google Scholar]
- 34.Denis GV, et al. Identification of transcription complexes that contain the double bromodomain protein Brd2 and chromatin remodeling machines. J Proteome Res. 2006;5:502–511. doi: 10.1021/pr050430u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.LeRoy G, Rickards B, Flint SJ. The double bromodomain proteins Brd2 and Brd3 couple histone acetylation to transcription. Mol Cell. 2008;30:51–60. doi: 10.1016/j.molcel.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Denis GV. Bromodomain motifs and “scaffolding”? Front Biosci. 2001;6:D1066–D1068. doi: 10.2741/a668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peng J, et al. Brd2 is a TBP-associated protein and recruits TBP into E2F-1 transcriptional complex in response to serum stimulation. Mol Cell Biochem. 2007;294:45–54. doi: 10.1007/s11010-006-9223-6. [DOI] [PubMed] [Google Scholar]
- 38.Sinha A, Faller DV, Denis GV. Bromodomain analysis of Brd2-dependent transcriptional activation of cyclin A. Biochem J. 2005;387:257–269. doi: 10.1042/BJ20041793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin YJ, et al. Solution structure of the extraterminal domain of the bromodomain-containing protein BRD4. Protein Sci. 2008;17:2174–2179. doi: 10.1110/ps.037580.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rahman S, et al. The Brd4 extraterminal domain confers transcription activation independent of pTEFb by recruiting multiple proteins, including NSD3. Mol Cell Biol. 2011;31:2641–2652. doi: 10.1128/MCB.01341-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bartkova J, et al. Cyclin D1 protein expression and function in human breast cancer. Int J Cancer. 1994;57:353–361. doi: 10.1002/ijc.2910570311. [DOI] [PubMed] [Google Scholar]
- 42.Courjal F, et al. Mapping of DNA amplifications at 15 chromosomal localizations in 1875 breast tumors: Definition of phenotypic groups. Cancer Res. 1997;57:4360–4367. [PubMed] [Google Scholar]
- 43.Ormandy CJ, et al. Cyclin D1, EMS1 and 11q13 amplification in breast cancer. Breast Cancer Res Treat. 2003;78:323–335. doi: 10.1023/a:1023033708204. [DOI] [PubMed] [Google Scholar]
- 44.Jang MK, et al. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 45.Yang Z, et al. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 46.Marshall NF, Peng J, Xie Z, Price DH. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J Biol Chem. 1996;271:27176–27183. doi: 10.1074/jbc.271.43.27176. [DOI] [PubMed] [Google Scholar]
- 47.Yang Z, He N, Zhou Q. Brd4 recruits P-TEFb to chromosomes at late mitosis to promote G1 gene expression and cell cycle progression. Mol Cell Biol. 2008;28:967–976. doi: 10.1128/MCB.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maruyama T, et al. A Mammalian bromodomain protein, Brd4, interacts with replication factor C and inhibits progression to S phase. Mol Cell Biol. 2002;22:6509–6520. doi: 10.1128/MCB.22.18.6509-6520.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dey A, Chitsaz F, Abbasi A, Misteli T, Ozato K. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc Natl Acad Sci USA. 2003;100:8758–8763. doi: 10.1073/pnas.1433065100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang L, et al. microRNA-141 is involved in a nasopharyngeal carcinoma-related genes network. Carcinogenesis. 2010;31:559–566. doi: 10.1093/carcin/bgp335. [DOI] [PubMed] [Google Scholar]
- 51.Dey A, et al. A bromodomain protein, MCAP, associates with mitotic chromosomes and affects G(2)-to-M transition. Mol Cell Biol. 2000;20:6537–6549. doi: 10.1128/mcb.20.17.6537-6549.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Umehara T, et al. Structural basis for acetylated histone H4 recognition by the human BRD2 bromodomain. J Biol Chem. 2010;285:7610–7618. doi: 10.1074/jbc.M109.062422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nishiyama A, Dey A, Miyazaki J, Ozato K. Brd4 is required for recovery from antimicrotubule drug-induced mitotic arrest: preservation of acetylated chromatin. Mol Biol Cell. 2006;17:814–823. doi: 10.1091/mbc.E05-08-0729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Laue K, et al. The multidomain protein Brpf1 binds histones and is required for Hox gene expression and segmental identity. Development. 2008;135:1935–1946. doi: 10.1242/dev.017160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Toyama R, Rebbert ML, Dey A, Ozato K, Dawid IB. Brd4 associates with mitotic chromosomes throughout early zebrafish embryogenesis. Dev Dyn. 2008;237:1636–1644. doi: 10.1002/dvdy.21576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dey A, Nishiyama A, Karpova T, McNally J, Ozato K. Brd4 marks select genes on mitotic chromatin and directs postmitotic transcription. Mol Biol Cell. 2009;20:4899–4909. doi: 10.1091/mbc.E09-05-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chua P, Roeder GS. Bdf1, a yeast chromosomal protein required for sporulation. Mol Cell Biol. 1995;15:3685–3696. doi: 10.1128/mcb.15.7.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Della Rovere F, et al. The Arabidopsis BET bromodomain factor GTE4 regulates the mitotic cell cycle. Plant Signal Behav. 2010;5:677–680. doi: 10.4161/psb.5.6.11571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Airoldi CA, et al. The Arabidopsis BET bromodomain factor GTE4 is involved in maintenance of the mitotic cell cycle during plant development. Plant Physiol. 2010;152:1320–1334. doi: 10.1104/pp.109.150631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shibata Y, Takeshita H, Sasakawa N, Sawa H. Double bromodomain protein BET-1 and MYST HATs establish and maintain stable cell fates in C. elegans. Development. 2010;137:1045–1053. doi: 10.1242/dev.042812. [DOI] [PubMed] [Google Scholar]
- 61.You J, et al. Kaposi’s Sarcoma-associated herpesvirus latency-associated nuclear antigen interacts with bromodomain protein Brd4 on host mitotic chromosomes. J Virol. 2006;80:8909–8919. doi: 10.1128/JVI.00502-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhao R, Nakamura T, Fu Y, Lazar Z, Spector DL. Gene bookmarking accelerates the kinetics of post-mitotic transcriptional re-activation. Nature Cell Biol. 2011;13:1295–1304. doi: 10.1038/ncb2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gyuris A, et al. The chromatin-targeting protein Brd2 is required for neural tube closure and embryogenesis. Biochim Biophys Acta. 2009;1789:413–421. doi: 10.1016/j.bbagrm.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shang E, Wang X, Wen D, Greenberg DA, Wolgemuth DJ. Double bromodomain-containing gene Brd2 is essential for embryonic development in mouse. Dev Dyn. 2009;238:908–917. doi: 10.1002/dvdy.21911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang F, et al. Brd2 disruption in mice causes severe obesity without type 2 diabetes. Biochem J. 2009;425:71–83. doi: 10.1042/BJ20090928. References 63–65 established that Brd2-null mice are not viable. Reference 65 reports the surprising finding of low-inflammatory, metabolically protected obesity in Brd2-hypomorphic mice. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Houzelstein D, et al. Growth and early postimplantation defects in mice deficient for the bromodomain-containing protein Brd4. Mol Cell Biol. 2002;22:3794–3802. doi: 10.1128/MCB.22.11.3794-3802.2002. Along with references 48 and 49, this report established that Brd4-null mice are not viable owing to cell cycle defects. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.You J, et al. Regulation of aurora B expression by the bromodomain protein Brd4. Mol Cell Biol. 2009;29:5094–5103. doi: 10.1128/MCB.00299-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lygerou Z, et al. The yeast BDF1 gene encodes a transcription factor involved in the expression of a broad class of genes including snRNAs. Nucl Acids Res. 1994;22:5332–5340. doi: 10.1093/nar/22.24.5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Matangkasombut O, Buratowski S. Different sensitivities of bromodomain factors 1 and 2 to histone H4 acetylation. Mol Cell. 2003;11:353–363. doi: 10.1016/s1097-2765(03)00033-9. [DOI] [PubMed] [Google Scholar]
- 70.Jacobson RH, Ladurner AG, King DS, Tjian R. Structure and function of a human TAFII250 double bromodomain module. Science. 2000;288:1422–1425. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- 71.Dunphy EL, Johnson T, Auerbach SS, Wang EH. Requirement for TAF(II)250 acetyltransferase activity in cell cycle progression. Mol Cell Biol. 2000;20:1134–1139. doi: 10.1128/mcb.20.4.1134-1139.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang EH, Zou S, Tjian R. TAFII250-dependent transcription of cyclin A is directed by ATF activator proteins. Genes Dev. 1997;11:2658–2669. doi: 10.1101/gad.11.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Iacobuzio-Donahue CA. Epigenetic changes in cancer. Ann Rev Pathol. 2009;4:229–249. doi: 10.1146/annurev.pathol.3.121806.151442. [DOI] [PubMed] [Google Scholar]
- 74.Redner RL, Wang J, Liu JM. Chromatin remodeling and leukaemia: new therapeutic paradigms. Blood. 1999;94:417–428. [PubMed] [Google Scholar]
- 75.Borrow J, et al. The translocation t(8;16)(p11;p13) of acute myeloid leukaemia fuses a putative acetyltransferase to the CREB-binding protein. Nature Genet. 1996;14:33–41. doi: 10.1038/ng0996-33. [DOI] [PubMed] [Google Scholar]
- 76.Sobulo OM, et al. MLL is fused to CBP, a histone acetyltransferase, in therapy-related acute myeloid leukaemia with a t(11;16)(q23;p13.3) Proc Natl Acad Sci USA. 1997;94:8732–8737. doi: 10.1073/pnas.94.16.8732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lavau C, Du C, Thirman M, Zeleznik-Le N. Chromatin-related properties of CBP fused to MLL generate a myelodysplastic-like syndrome that evolves into myeloid leukaemia. EMBO J. 2000;19:4655–4664. doi: 10.1093/emboj/19.17.4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Martens JH, et al. PML-RARalpha/RXR alters the epigenetic landscape in Acute Promyelocytic Leukaemia. Cancer Cell. 2010;17:173–185. doi: 10.1016/j.ccr.2009.12.042. [DOI] [PubMed] [Google Scholar]
- 79.Filippakopoulos P, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. First report of small-molecule inhibition of bromodomain proteins that demonstrated that epigenetically acting therapeutics that disrupt chromatin–co-activator interactions are feasible and have therapeutic benefit. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Muller S, Filippakopoulos P, Knapp S. Bromodomains as therapeutic targets. Expert Rev Mol Med. 2011;13:e29. doi: 10.1017/S1462399411001992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mertz JA, et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci USA. 2011;108:16669–16674. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Seoane J, Le HV, Massagué J. Myc suppression of the p21(Cip1) Cdk inhibitor influences the outcome of the p53 response to DNA damage. Nature. 2002;419:729–734. doi: 10.1038/nature01119. [DOI] [PubMed] [Google Scholar]
- 83.Boxer LM, Dang CV. Translocations involving c-myc and c-myc function. Oncogene. 2001;20:5595–5610. doi: 10.1038/sj.onc.1204595. [DOI] [PubMed] [Google Scholar]
- 84.Zuber J, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ciro M, et al. ATAD2 is a novel cofactor for MYC, overexpressed and amplified in aggressive tumors. Cancer Res. 2009;69:8491–8498. doi: 10.1158/0008-5472.CAN-09-2131. [DOI] [PubMed] [Google Scholar]
- 86.Caron C, et al. Functional characterization of ATAD2 as a new cancer/testis factor and a predictor of poor prognosis in breast and lung cancers. Oncogene. 2010;29:5171–5181. doi: 10.1038/onc.2010.259. [DOI] [PubMed] [Google Scholar]
- 87.Soucek L, et al. Modelling Myc inhibition as a cancer therapy. Nature. 2008;455:679–683. doi: 10.1038/nature07260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Soucek L, Nasi S, Evan GI. Omomyc expression in skin prevents Myc-induced papillomatosis. Cell Death Differ. 2004;11:1038–1045. doi: 10.1038/sj.cdd.4401443. [DOI] [PubMed] [Google Scholar]
- 89.Centeno F, Ramírez-Salazar E, García-Villa E, Gariglio P, Garrido E. TAF1 interacts with and modulates human papillomavirus 16 E2-dependent transcriptional regulation. Intervirology. 2008;51:137–143. doi: 10.1159/000141706. [DOI] [PubMed] [Google Scholar]
- 90.Zhou M, et al. Bromodomain protein Brd4 regulates human immunodeficiency virus transcription through phosphorylation of CDK9 at threonine 29. J Virol. 2009;83:1036–1044. doi: 10.1128/JVI.01316-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mueller D, et al. A role for the MLL fusion partner ENL in transcriptional elongation and chromatin modification. Blood. 2007;110:4445–4454. doi: 10.1182/blood-2007-05-090514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dawson MA, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478:529–533. doi: 10.1038/nature10509. This article describes BET protein inhibitors in MLL, and confirms a hypothesis first proposed in reference 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Greenwald RJ, et al. Eμ-BRD2 transgenic mice develop B cell lymphoma and leukaemia. Blood. 2004;103:1475–1484. doi: 10.1182/blood-2003-06-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lenburg ME, Sinha A, Faller DV, Denis GV. Tumor-specific and proliferation-specific gene expression typifies murine transgenic B cell lymphomagenesis. J Biol Chem. 2007;282:4803–4811. doi: 10.1074/jbc.M605870200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.French CA. Pathogenesis of NUT midline carcinoma. Ann Rev Pathol Mech Dis. 2012;7:247–265. doi: 10.1146/annurev-pathol-011811-132438. [DOI] [PubMed] [Google Scholar]
- 96.Delmore JE, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. This article describes MYC as a co-activator of BET proteins in human multiple myeloma and responsiveness to JQ1 therapeutics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nicodeme E, et al. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468:1119–1123. doi: 10.1038/nature09589. First demonstration that small-molecule BET protein inhibitors have potent and broad-spectrum anti-inflammatory properties. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ouchida R, et al. Suppression of NF-κB-dependent gene expression by a hexamethylene bisacetamide-inducible protein HEXIM1 in human vascular smooth muscle cells. Genes Cells. 2003;8:95–107. doi: 10.1046/j.1365-2443.2003.00618.x. [DOI] [PubMed] [Google Scholar]
- 99.Hewings DS, et al. 3,5-dimethylisoxazoles act as acetyl-lysine-mimetic bromodomain ligands. J Med Chem. 2011;54:6761–6770. doi: 10.1021/jm200640v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chung CW, Dean AW, Woolven JM, Bamborough P. Fragment-based discovery of bromodomain inhibitors part 1: inhibitor binding modes and implications for lead discovery. J Med Chem. 2012;55:576–586. doi: 10.1021/jm201320w. [DOI] [PubMed] [Google Scholar]
- 101.Bamborough P, et al. Fragment-based discovery of bromodomain inhibitors part 2: optimization of phenylisoxazole sulfonamides. J Med Chem. 2012;55:587–596. doi: 10.1021/jm201283q. [DOI] [PubMed] [Google Scholar]
- 102.Belkina AC, Blanton W, Wang F, Liu H, Denis GV. Whole body Brd2 deficiency protects obese mice from insulin resistance by creating a low inflammatory environment. Obesity. 2010;18:S58. [Google Scholar]
- 103.Denis GV. Bromodomain coactivators in cancer, obesity, type 2 diabetes and inflammation. Discov Med. 2010;10:489–499. [PMC free article] [PubMed] [Google Scholar]
- 104.Andres R. Effect of obesity on total mortality. Int J Obes. 1980;4:381–386. [PubMed] [Google Scholar]
- 105.Sims EAH. Are there persons who are obese, but metabolically healthy? Metabolism. 2001;50:1499–1504. doi: 10.1053/meta.2001.27213. [DOI] [PubMed] [Google Scholar]
- 106.Klöting N, et al. Insulin-sensitive obesity. Am J Physiol Endocrinol Metab. 2010;299:E506–E515. doi: 10.1152/ajpendo.00586.2009. [DOI] [PubMed] [Google Scholar]
- 107.Bonora E, et al. U-shaped and J-shaped relationships between serum insulin and coronary heart disease in the general population. The Bruneck Study. Diabetes Care. 1991;21:221–230. doi: 10.2337/diacare.21.2.221. [DOI] [PubMed] [Google Scholar]
- 108.Wildman RP, et al. The obese without cardiometabolic risk factor clustering and the normal weight with cardiometabolic risk factor clustering: prevalence and correlates of 2 phenotypes among the US population. Arch Intern Med. 2008;168:1617–1624. doi: 10.1001/archinte.168.15.1617. [DOI] [PubMed] [Google Scholar]
- 109.Karelis AD, et al. The metabolically healthy but obese individual presents a favorable inflammation profile. J Clin Endocrinol Metab. 2005;90:4145–4150. doi: 10.1210/jc.2005-0482. [DOI] [PubMed] [Google Scholar]
- 110.Succurro E, et al. Insulin secretion in metabolically obese, but normal weight, and in metabolically healthy but obese individuals. Obesity (Silver Spring) 2008;16:1881–1886. doi: 10.1038/oby.2008.308. [DOI] [PubMed] [Google Scholar]
- 111.Calori G, et al. Prevalence, metabolic features, and prognosis of metabolically healthy obese Italian individuals: the Cremona Study. Diabetes Care. 2011;34:210–215. doi: 10.2337/dc10-0665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Huang B, Yang XD, Zhou MM, Ozato K, Chen LF. Brd4 coactivates transcriptional activation of NF-kappaB via specific binding to acetylated RelA. Mol Cell Biol. 2009;29:1375–1387. doi: 10.1128/MCB.01365-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Cullen SJ, Ponnappan S, Ponnappan U. Catalytic activity of the proteasome finetunes Brg1-mediated chromatin remodeling to regulate the expression of inflammatory genes. Mol Immunol. 2009;47:600–605. doi: 10.1016/j.molimm.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Karin M. NF-κB as a critical link between inflammation and cancer. Cold Spring Harb Perspect Biol. 2009;1:a000141. doi: 10.1101/cshperspect.a000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Virchow R. In: Cellular Pathology, as Based on Physiological and Pathological Histology. Chance F, editor. J. Churchill; 1860. [DOI] [PubMed] [Google Scholar]
- 116.Goldgraber MB, Humphreys EM, Kirsner JB, Palmer WL. Carcinoma and ulcerative colitis, a clinical-pathologic study. II. Statistical analysis. Gastroenterology. 1958;34:840–846. [PubMed] [Google Scholar]
- 117.Kraus S, Arber N. Inflammation and colorectal cancer. Curr Opin Pharmacol. 2009;9:405–410. doi: 10.1016/j.coph.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 118.Denis GV, Nikolajczyk BN, Schnitzler GR. An emerging role for bromodomain-containing proteins in chromatin regulation and transcriptional control of adipogenesis. FEBS Lett. 2010;584:3260–3268. doi: 10.1016/j.febslet.2010.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chung CW, et al. Discovery and characterization of small molecule inhibitors of the BET family bromodomains. J Med Chem. 2011;54:3827–3838. doi: 10.1021/jm200108t. [DOI] [PubMed] [Google Scholar]
- 120.Pamblanco M, et al. Bromodomain factor 1 (Bdf1) protein interacts with histones. FEBS Lett. 2001;496:31–35. doi: 10.1016/s0014-5793(01)02397-3. [DOI] [PubMed] [Google Scholar]
- 121.Chang YL, King B, Lin SC, Kennison JA, Huang DH. A double-bromodomain protein, FSH-S, activates the homeotic gene ultrabithorax through a critical promoter-proximal region. Mol Cell Biol. 2007;27:5486–5498. doi: 10.1128/MCB.00692-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Florence BL, Faller DV. Drosophila female sterile (1) homeotic is a multifunctional transcriptional regulator that is modulated by Ras signaling. Dev Dyn. 2008;237:554–564. doi: 10.1002/dvdy.21432. [DOI] [PubMed] [Google Scholar]
- 123.Denis GV. Duality in bromodomain-containing protein complexes. Front Biosci. 2001;6:D849–D852. doi: 10.2741/denis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Trouche D, Le Chalony C, Muchardt C, Yaniv M, Kouzarides T. RB and hbrm cooperate to repress the activation functions of E2F1. Proc Natl Acad Sci USA. 1997;94:11268–11273. doi: 10.1073/pnas.94.21.11268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhang HS, et al. Exit from G1 and S phase of the cell cycle is regulated by repressor complexes containing HDAC-Rb-hSWI/SNF and Rb-hSWI/SNF. Cell. 2000;101:79–89. doi: 10.1016/S0092-8674(00)80625-X. [DOI] [PubMed] [Google Scholar]
- 126.Brehm A, et al. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 127.Ferreira R, Magnaghi-Jaulin L, Robin P, Harel-Bellan A, Trouche D. The three members of the pocket proteins family share the ability to repress E2F activity through recruitment of a histone deacetylase. Proc Natl Acad Sci USA. 1998;95:10493–10498. doi: 10.1073/pnas.95.18.10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Magnaghi-Jaulin L, et al. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature. 1998;391:601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- 129.Stiegler P, De Luca A, Bagella L, Giordano A. The COOH-terminal region of pRb2/p130 binds to histone deacetylase 1 (HDAC1), enhancing transcriptional repression of the E2F-dependent cyclin A promoter. Cancer Res. 1998;58:5049–5052. [PubMed] [Google Scholar]
- 130.Blobel GA, Kalota A, Sanchez PV, Carroll M. Short hairpin RNA screen reveals bromodomain proteins as novel targets in acute myeloid leukaemia. Cancer Cell. 2011;20:287–288. doi: 10.1016/j.ccr.2011.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sachs L. Control of normal cell differentiation in leukemic cells. Haematologica. 1976;61:93–97. [PubMed] [Google Scholar]
- 132.Sachs L. Leukemogenesis and differentiation. Ann Rev Med. 1985;36:177–184. doi: 10.1146/annurev.me.36.020185.001141. [DOI] [PubMed] [Google Scholar]
- 133.Dibenedetto AJ, et al. Zebrafish brd2a and brd2b are paralogous members of the bromodomain-ET (BET) family of transcriptional coregulators that show structural and expression divergence. BMC Dev Biol. 2008;8:39. doi: 10.1186/1471-213X-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sun H, et al. Solution structure of BRD7 bromodomain and its interaction with acetylated peptides from histone H3 and H4. Biochem Biophys Res Commun. 2007;358:435–441. doi: 10.1016/j.bbrc.2007.04.139. [DOI] [PubMed] [Google Scholar]
- 135.Tae S, et al. Bromodomain protein 7 interacts with PRMT5 and PRC2, and is involved in transcriptional repression of their target genes. Nucleic Acids Res. 2011;39:5424–5438. doi: 10.1093/nar/gkr170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ladurner AG, Inouye C, Jain R, Tjian R. Bromodomains mediate an acetyl-histone encoded antisilencing function at heterochromatin boundaries. Mol Cell. 2003;11:365–376. doi: 10.1016/s1097-2765(03)00035-2. [DOI] [PubMed] [Google Scholar]
- 137.Hang M, Smith MM. Genetic analysis implicates the Set3/Hos2 histone deacetylase in the deposition and remodeling of nucleosomes containing H2AZ. Genetics. 2011;187:1053–1066. doi: 10.1534/genetics.110.125419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Look AT. Oncogenic transcription factors in the human acute leukaemias. Science. 1997;278:1059–1064. doi: 10.1126/science.278.5340.1059. [DOI] [PubMed] [Google Scholar]
- 139.Rowley JD. The critical role of chromosome translocations in human leukaemias. Ann Rev Genet. 1998;32:495–519. doi: 10.1146/annurev.genet.32.1.495. [DOI] [PubMed] [Google Scholar]
- 140.Wilson BG, Roberts CWM. SWI/SNF nucleosome remodelers and cancer. Nature Rev Cancer. 2011;11:481–492. doi: 10.1038/nrc3068. [DOI] [PubMed] [Google Scholar]
- 141.Cowell IG, Papageorgiou N, Padget K, Watters GP, Austin CA. Histone deacetylase inhibition redistributes topoisomerase IIβ from heterochromatin to euchromatin. Nucleus. 2011;2:61–71. doi: 10.4161/nucl.2.1.14194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gkikopoulos T, et al. The SWI/SNF complex acts to constrain distribution of the centromeric histone variant Cse4. EMBO J. 2011;30:1919–1927. doi: 10.1038/emboj.2011.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lamonica JM, et al. Bromodomain protein Brd3 associates with acetylated GATA1 to promote its chromatin occupancy at erythroid target genes. Proc Natl Acad Sci USA. 2011;108:E159–E168. doi: 10.1073/pnas.1102140108. This report is one of the few that describes a function for the BET protein BRD3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Gamsjaeger R, et al. Structural basis and specificity of acetylated transcription factor GATA1 recognition by BET family bromodomain protein Brd3. Mol Cell Biol. 2011;31:2632–2640. doi: 10.1128/MCB.05413-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Haslam DW, James WP. Obesity. Lancet. 2009;366:1197–1209. doi: 10.1016/S0140-6736(05)67483-1. [DOI] [PubMed] [Google Scholar]
- 146.Hossain P, Kawar B, El Nahas M. Obesity and diabetes in the developing world--a growing challenge. N Engl J Med. 2007;356:213–215. doi: 10.1056/NEJMp068177. [DOI] [PubMed] [Google Scholar]
- 147.Devaiah BN, et al. BRD4 is an atypical kinase that phosphorylates serine 2 of the RNA Polymerase II carboxy-terminal domain. Proc Natl Acad Sci USA. 2012;109:6927–6932. doi: 10.1073/pnas.1120422109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Gans M, Audit C, Masson M. Isolation and characterization of sex-linked female-sterile mutants in Drosophila melanogaster. Genetics. 1975;81:683–704. doi: 10.1093/genetics/81.4.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Gans M, Forquignon F, Masson M. The role of dosage of the region 7D1-7D5-6 of the X chromosome in the production of homeotic transformations in Drosophila melanogaster. Genetics. 1980;96:887–902. doi: 10.1093/genetics/96.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Digan ME, et al. Genetic and molecular analysis of fs(1)h, a maternal effect homeotic gene in Drosophila. Dev Biol. 1986;114:161–169. doi: 10.1016/0012-1606(86)90392-1. [DOI] [PubMed] [Google Scholar]
- 151.Mozer BA, Dawid IB. Cloning and molecular characterization of the trithorax locus of Drosophila melanogaster. Proc Natl Acad Sci USA. 1989;86:3738–3742. doi: 10.1073/pnas.86.10.3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Mazo AM, Huang DH, Mozer BA, Dawid IB. The trithorax gene, a trans-acting regulator of the bithorax complex in Drosophila, encodes a protein with zinc-binding domains. Proc Natl Acad Sci USA. 1990;87:2112–2116. doi: 10.1073/pnas.87.6.2112. This report establishes that D. melanogaster BET proteins are activators of the Trithorax locus, and, with references 6 and 150, was the first described to describe functions for Metazoan bromodomain proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Cimino G, et al. Cloning of ALL-1, the locus involved in leukemias with the t(4;11)(q21;q23), t(9;11) (p22;q23), and t(11;19)(q23;p13) chromosome translocations. Cancer Res. 1991;51:6712–6714. [PubMed] [Google Scholar]
- 154.Djabali M, et al. A trithorax-like gene is interrupted by chromosome 11q23 translocations in acute leukaemias. Nature Genet. 1992;2:113–118. doi: 10.1038/ng1092-113. [DOI] [PubMed] [Google Scholar]
- 155.Gu Y, et al. The t(4;11) chromosome translocation of human acute leukaemias fuses the ALL-1 gene, related to Drosophila trithorax, to the AF-4 gene. Cell. 1992;71:701–708. doi: 10.1016/0092-8674(92)90603-a. [DOI] [PubMed] [Google Scholar]
- 156.Tkachuk DC, Kohler S, Cleary ML. Involvement of a homolog of Drosophila trithorax by 11q23 chromosomal translocations in acute leukaemias. Cell. 1992;71:691–700. doi: 10.1016/0092-8674(92)90602-9. [DOI] [PubMed] [Google Scholar]
- 157.McPhillips MG, Ozato K, McBride AA. Interaction of bovine papillomavirus E2 protein with Brd4 stabilizes its association with chromatin. J Virol. 2005;79:8920–8932. doi: 10.1128/JVI.79.14.8920-8932.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Hebner CM, Laimins LA. Human papillomaviruses: basic mechanisms of pathogenesis and oncogenicity. Rev Med Virol. 2006;16:83–97. doi: 10.1002/rmv.488. [DOI] [PubMed] [Google Scholar]
- 159.You J, Croyle JL, Nishimura A, Ozato K, Howley PM. Interaction of the bovine papillomavirus E2 protein with Brd4 tethers the viral DNA to host mitotic chromosomes. Cell. 2004;117:349–360. doi: 10.1016/s0092-8674(04)00402-7. [DOI] [PubMed] [Google Scholar]
- 160.Schweiger MR, You J, Howley PM. Bromodomain protein 4 mediates the papillomavirus E2 transcriptional activation function. J Virol. 2006;80:4276–4285. doi: 10.1128/JVI.80.9.4276-4285.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.McPhillips MG, Oliveira JG, Spindler JE, Mitra R, McBride AA. Brd4 is required for e2-mediated transcriptional activation but not genome partitioning of all papillomaviruses. J Virol. 2006;80:9530–9543. doi: 10.1128/JVI.01105-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Senechal H, Poirier GG, Coulombe B, Laimins LA, Archambault J. Amino acid substitutions that specifically impair the transcriptional activity of papillomavirus E2 affect binding to the long isoform of Brd4. Virology. 2007;358:10–17. doi: 10.1016/j.virol.2006.08.035. [DOI] [PubMed] [Google Scholar]
- 163.Wu SY, et al. Brd4 links chromatin targeting to HPV transcriptional silencing. Genes Dev. 2006;20:2383–2396. doi: 10.1101/gad.1448206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Smith JA, et al. Genome-wide siRNA screen identifies SMCX, EP400, and Brd4 as E2-dependent regulators of human papillomavirus oncogene expression. Proc Natl Acad Sci USA. 2010;107:3752–3757. doi: 10.1073/pnas.0914818107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Ilves I, Maemets K, Silla T, Janikson K, Ustav M. Brd4 is involved in multiple processes of the bovine papillomavirus type 1 life cycle. J Virol. 2006;80:3660–3665. doi: 10.1128/JVI.80.7.3660-3665.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Platt GM, Simpson GR, Mittnacht S, Schulz TF. Latent nuclear antigen of Kaposi’s sarcoma-associated herpesvirus interacts with RING3, a homolog of the Drosophila female sterile homeotic (fsh) gene. J Virol. 1999;73:9789–9795. doi: 10.1128/jvi.73.12.9789-9795.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Viejo-Borbolla A, et al. Brd2/RING3 interacts with a chromatin-binding domain in the Kaposi’s Sarcoma-associated herpesvirus latency-associated nuclear antigen 1 (LANA-1) that is required for multiple functions of LANA-1. J Virol. 2005;79:13618–13629. doi: 10.1128/JVI.79.21.13618-13629.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Ottinger M, et al. Kaposi’s sarcoma-associated herpesvirus LANA-1 interacts with the short variant of BRD4 and releases cells from a BRD4- and BRD2/RING3-induced G1 cell cycle arrest. J Virol. 2006;80:10772–10786. doi: 10.1128/JVI.00804-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Radkov SA, Kellam P, Boshoff C. The latent nuclear antigen of Kaposi sarcoma-associated herpesvirus targets the retinoblastoma-E2F pathway and with the oncogene Hras transforms primary rat cells. Nature Med. 2000;6:1121–1127. doi: 10.1038/80459. [DOI] [PubMed] [Google Scholar]
- 170.Friborg J, Jr, Kong W, Hottiger MO, Nabel GJ. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature. 1999;402:889–894. doi: 10.1038/47266. [DOI] [PubMed] [Google Scholar]
- 171.Palermo RD, Webb HM, West MJRNA. Polymerase II stalling promotes nucleosome occlusion and pTEFb recruitment to drive immortalization by Epstein-Barr Virus. PLoS Pathog. 2011;7:e1002334. doi: 10.1371/journal.ppat.1002334. This was the first study to suggest that small-molecule BET protein inhibitors could have value in the treatment of latent virus infections by disrupting transcriptional co-regulator interactions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Nagl NG, Jr, Zweitzig DR, Thimmapaya B, Beck GR, Jr, Moran E. The c-myc gene is a direct target of mammalian SWI/SNF-related complexes during differentiation associated cell cycle arrest. Cancer Res. 2006;66:1289–1293. doi: 10.1158/0008-5472.CAN-05-3427. [DOI] [PubMed] [Google Scholar]
- 173.Nagl NG, Jr, Wang X, Patsialou A, Van Scoy M, Moran E. Distinct mammalian SWI/SNF chromatin remodeling complexes with opposing roles in cell-cycle control. EMBO J. 2007;26:752–763. doi: 10.1038/sj.emboj.7601541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Peterson CL. Chromatin remodeling: nucleosomes bulging at the seams. Curr Biol. 2002;12:R245–R247. doi: 10.1016/s0960-9822(02)00782-0. [DOI] [PubMed] [Google Scholar]
- 175.Sif S, Saurin AJ, Imbalzano AN, Kingston RE. Purification and characterization of mSin3A-containing Brg1 and hBrm chromatin remodeling complexes. Genes Dev. 2001;15:603–618. doi: 10.1101/gad.872801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Olave I, Wang W, Xue Y, Kuo A, Crabtree GR. Identification of a polymorphic, neuron-specific chromatin remodeling complex. Genes Dev. 2002;16:2509–2517. doi: 10.1101/gad.992102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Bultman S, et al. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol Cell. 2000;6:1287–1295. doi: 10.1016/s1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- 178.Reyes JC, et al. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2alpha) EMBO J. 1998;17:6979–6991. doi: 10.1093/emboj/17.23.6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Burns LG, Peterson CL. The yeast SWI-SNF complex facilitates binding of a transcriptional activator to nucleosomal sites in vivo. Mol Cell Biol. 1997;17:4811–4819. doi: 10.1128/mcb.17.8.4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Holstege FC, et al. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. This report reveals an important principle that chromatin regulatory factors such as the Swi/Snf complex usually have dual functions as co-repressors and co-activators, implying that mutation or inhibition of their function will have pleiotropic effects. Combined with references 65 and 123, this principle suggests that JQ1, I-BET and related small-molecule inhibitors should be evaluated for the alleviation of co-repressor functions of BET proteins. [DOI] [PubMed] [Google Scholar]
- 181.Chua YL, Channeliere S, Mott E, Gray JC. The bromodomain protein GTE6 controls leaf development in Arabidopsis by histone acetylation at ASYMMETRIC LEAVES1. Genes Dev. 2005;19:2245–2254. doi: 10.1101/gad.352005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Wang R, Li Q, Helfer CM, Jiao J, You J. Bromodomain protein Brd4 associated with acetylated chromatin is important for maintenance of higher-order chromatin structure. J Biol Chem. 2012;287:10738–10752. doi: 10.1074/jbc.M111.323493. [DOI] [PMC free article] [PubMed] [Google Scholar]