Abstract
The ability to regulate and even target mutagenesis is an extremely valuable cellular asset. Enzyme-catalyzed DNA cytosine deamination is a molecular strategy employed by vertebrates to promote antibody diversity and defend against foreign nucleic acids. Ten years ago, a family of cellular enzymes was first described with several proving capable of deaminating DNA and inhibiting HIV-1 replication. Ensuing studies on the apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like 3 (APOBEC3) restriction factors have uncovered a broad-spectrum innate defense network that suppresses the replication of numerous endogenous and exogenous DNA-based parasites. Although many viruses possess equally elaborate counter-defense mechanisms, the APOBEC3 enzymes offer a tantalizing possibility of leveraging innate immunity to fend off viral infection. Here we focus on mechanisms of retroelement restriction by the APOBEC3 family of restriction enzymes and we consider the therapeutic benefits, as well as the possible pathological consequences, of arming cells with active DNA deaminases.
1. DNA Deaminase Evolution
Central to nucleic acid metabolism is the near-ubiquitous process of enzymatic deamination of adenine and cytosine bases, individually or in the context of larger nucleic acid constituents (Conticello et al. 2005; Grosjean 2009). For instance, in most species the wobble base in several of the tRNA anti-codons is frequently changed by deamination of adenosine to inosine (A-to-I), which can then base pair with cytosine, uracil, or adenosine, thereby increasing the flexibility and decoding capacity of a tRNA anti-codon (Gerber and Keller 1999). These enzymes belong to the adenosine deaminase acting on tRNA (ADAT) family. Related proteins in most metazoans from nematodes and flies to humans catalyze A-to-I editing of a variety of RNA targets (Kim et al. 1994; Nishikura 2010). These enzymes are called, appropriately, adenosine deaminases that act on RNA (ADAR). Editing events that occur in the coding region of an mRNA can result in amino acid substitutions in the resulting protein. However, the majority of editing events occur in non-coding regions of mRNA or in non-coding RNAs, and these A-to-I editing events can alter RNA secondary structure, stability, function, and/or capacity to be bound by regulatory RNAs such as siRNAs (Morse et al. 2002; Levanon et al. 2004; Agranat et al. 2008; Li et al. 2009).
Cytosine to uracil (C-to-U) deamination is almost as ancient as A-to-I editing (Conticello et al. 2007b; Grosjean 2009). The pyrimidine salvage pathways of most organisms use cytidine deaminase (CDA) to produce the essential RNA and DNA building blocks of uridine directly, and thymidine after additional enzymatic steps (Zrenner et al. 2006). However, at some point near the root of the vertebrate tree, polynucleotide cytosine deaminases emerged, with the lamprey CDA being a present day example (Rogozin et al. 2007). This enzyme is thought to underpin a unique form of adaptive immunity in which the DNA segments that encode arrays of highly diverse leucine rich repeats are assembled into mature variable lymphocyte receptor genes by a recombination-mediated process. It is thought that an ancestor of the present day lamprey enzyme served as the original substrate for expansion of the polynucleotide cytosine deaminase gene family during vertebrate evolution (Fig. 1a) (Rogozin et al. 2007; Conticello 2008). The result in most vertebrates alive today is a much larger repertoire of polynucleotide C-to-U editing enzymes that execute diverse biological functions from lipid metabolism to adaptive and innate immunity (Figs. 1 & 2).
Fig. 1.
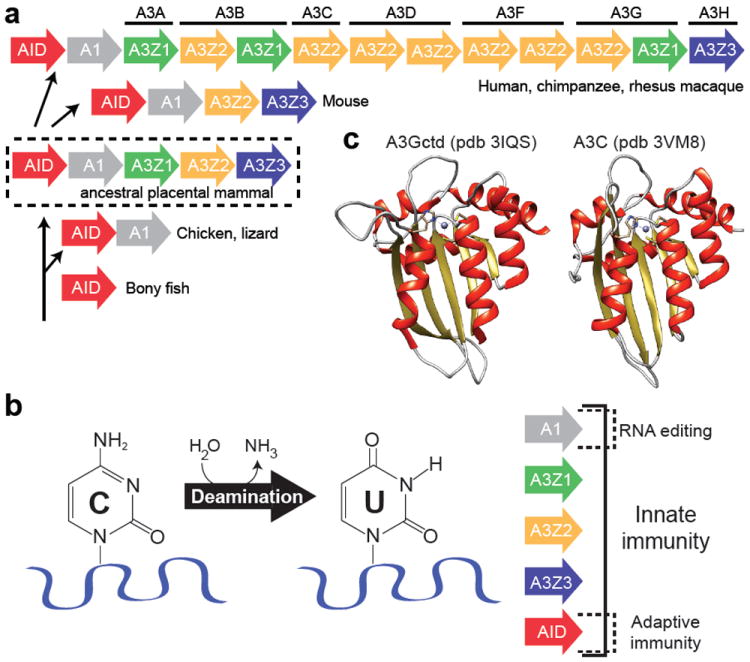
Evolution and structure-function of APOBEC cytosine deaminases. (a) Expansion of the modern primate APOBEC3 locus encoding seven APOBEC3 genes with eleven zinc-coordinating (Z) domains [reprinted with permission from (Lackey et al. 2012)]. (b) Deamination of cytosine to uracil plays a central role in innate immunity. (c) Three-dimensional structures of the APOBEC3G C-terminal domain and APOBEC3C. A zinc ion (purple) is shown coordinated in both proteins by one histidine and two cysteine residues in the α2-β3-α3 core.
Fig. 2.
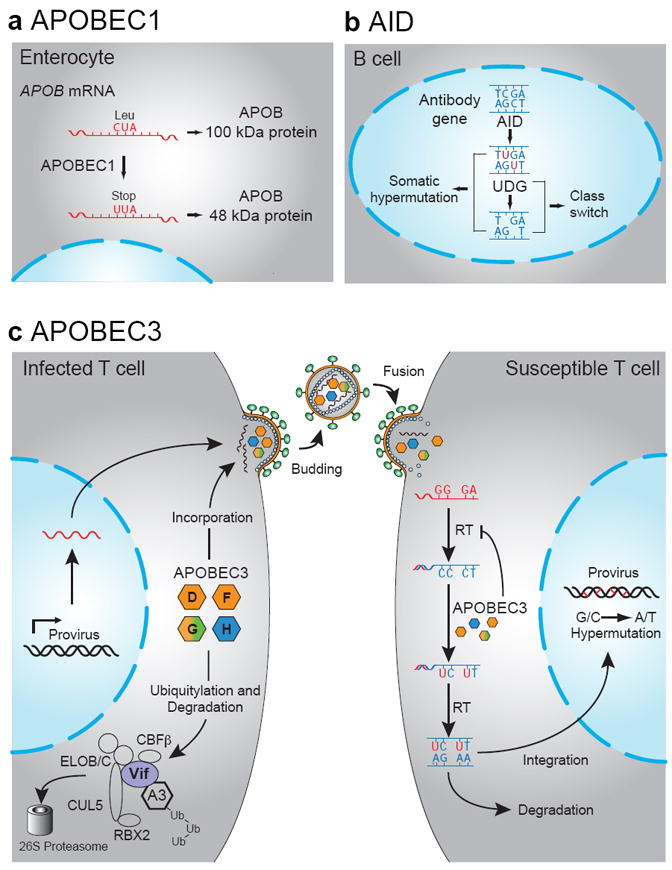
The physiological functions of the APOBEC family. (a) RNA editing by APOBEC1 generates a truncated APOB protein. (b) AID activity underpins the processes of somatic hypermutation and class switch recombination in B-cell germinal centers. (c) Four APOBEC3 proteins restrict HIV through cytosine deamination in the absence of Vif [reprinted with permission from (Hultquist et al. 2011)].
All vertebrate polynucleotide cytosine deaminases belong to the so-called ‘APOBEC’ family. The defining feature of this family is a conserved His-X-Glu-X25-31-Pro-Cys-X2-4-Cys zinc (Z)-coordinating motif, which is strictly required for deaminase activity (where X can be a variety of amino acids) (Wedekind et al. 2003; Harris and Liddament 2004; Conticello et al. 2005; LaRue et al. 2009). As described in more detail below, key residues within this motif position zinc at the active site of the enzyme (Fig. 1c). The protein sequences within these motifs enable phylogenetic groupings into three sub-families: APOBEC1, AID, and the APOBEC3s. The APOBEC3s can be further sub-divided into three sub-groups: Z1, Z2, and Z3 (Fig. 1a) (Conticello 2008; LaRue et al. 2009). Importantly, the number and organization of the A3 Z-domains can vary dramatically from branch to branch throughout the mammalian portion of the vertebrate phylogenetic tree (e.g., human versus mouse loci depicted in Fig. 1a).
Apolipoprotein B mRNA editing catalytic subunit 1 (APOBEC1) has provided the namesake to the larger family. It was discovered as an enzyme that catalyzes the deamination of a specific cytosine within the APOB mRNA (Figs. 1b & 2a) (Teng et al. 1993). This produces a premature translation stop codon and a smaller secondary gene product. These two APOB proteins (APOB100 and APOB48) differentially regulate the secretion of lipoproteins from the liver (Chan 1992). Many mammalian APOBEC1 enzymes also possess DNA C-to-U deaminase activity (Harris et al. 2002; Ikeda et al. 2008; Petit et al. 2009; Ikeda et al. 2011). Taken together with the fact that earlier vertebrate lineages, such as the one represented by birds and lizards, lack an APOB-like gene, it is probable that DNA editing function preceded involvement in RNA editing (Severi et al. 2011).
The most conserved DNA cytosine deaminase in vertebrates is activation-induced deaminase (AID; gene name AICDA) (Fig. 1a). AID has a central role in adaptive immunity by seeding somatic hypermutation, gene conversion, and class switch recombination with its DNA deaminase activity (Figs. 1b & 2b) [(Muramatsu et al. 1999; Muramatsu et al. 2000; Di Noia and Neuberger 2002; Petersen-Mahrt et al. 2002); reviewed by (Longerich et al. 2006; Di Noia and Neuberger 2007; Conticello 2008)]. Interestingly, the genes that encode APOBEC1 and AID are positioned adjacent to each other in the genomes of most vertebrates (an inversion has placed the human gene farther away on the same chromosome). This suggests that an ancestral AID gene duplicated and diverged to produce APOBEC1 (Fig. 1a). It is likely that duplication of an ancestral AID/APOBEC1 locus produced the genetic seeds for the mammal-exclusive APOBEC3 sub-family (Fig. 1a) (Jarmuz et al. 2002; Harris and Liddament 2004).
In humans, the seven APOBEC3 proteins are encoded by a tandemly arranged gene cluster (Fig. 1a) (Jarmuz et al. 2002). These present day genes are the products of continual evolution, in which an ancestral cluster of three Z domains is predicted to have undergone a minimum of eight duplication events over the past 100 million years to produce the locus found in most primates (LaRue et al. 2008; Münk et al. 2012). These domains are either expressed singly or one enzyme may consist of two Z domains (LaRue et al. 2009). In contrast, the ancestral APOBEC3 locus experienced a deletion in the rodent lineage of one of the ancestral Z-domains leading to the present day two domain locus, which encodes a single protein quite distinct from any of the primate enzymes (Fig. 1a).
One possible explanation for why some mammalian lineages, like primates, have many APOBEC3s, while other lineages, such as rodents have few is that these enzymes have overlapping innate immune functions to protect the host from a variety of parasitic elements (e.g., in HIV-1 restriction, Fig. 2c; mechanism elaborated in section 3, below). Because multiple distinct innate immune mechanisms serve to suppress the spread of such parasitic elements, it is reasonable to postulate that some mammals will be fortified at the APOBEC3 locus and weaker at other loci, with each mammalian lineage being distinct. For instance, primates encode a single TRIM5α protein, whereas mice have the capacity to encode a total of eight TRIM5α-like proteins [(Sawyer et al. 2007; Tareen et al. 2009); see also Chapter 13 in (Lever et al. 2010)]. It is likely that each species’ present day innate immune fortifications were independently shaped by past pathogenic pressures, which one can only speculate may have been the ancestors of present day viruses and transposable elements.
2. Biochemical and Structural Insights
Zinc-dependent deaminases, such as the APOBECs, catalyze the conversion of cytosine to uracil in polynucleotide substrates (Fig. 1b). This reaction requires the activation of water by a zinc ion coordinated by the enzyme (Fig. 1c). A glutamic acid in the active site of the enzyme protonates N3, priming the nucleophilic attack on the C4 position of the pyrimidine ring, followed by the removal and subsequent protonation of an amino group (NH2) that results in the release of ammonia (NH3) and uracil as products. This conversion can theoretically occur within both RNA and single-stranded DNA substrates. However, apart from APOBEC1, which has both RNA and DNA editing activities, AID and the APOBEC3s have proven specific to DNA substrates in vitro and in vivo.
The extent of amino acid homology to APOBEC1 originally suggested that the APOBEC3 enzymes might be a family of RNA editing proteins (Jarmuz et al. 2002). Three lines of evidence, however, demonstrated that this view was incorrect and established the APOBEC3 enzymes as single-stranded DNA cytosine deaminases. First, APOBEC3 has a high degree of homology with AID and experiments in E. coli demonstrated AID, APOBEC3C, and APOBEC3G are capable of inducing high levels of mutation in an antibiotic resistance gene (Harris et al. 2002; Petersen-Mahrt et al. 2002). This was clearly due to DNA editing because mutation levels rose synergistically in a bacterial strain deficient for uracil DNA glycosylase (UDG), an enzyme that initiates base excision repair by recognizing and removing uracil exclusively from DNA (Lindahl 2000; Di Noia and Neuberger 2002; Harris et al. 2002). Second, unambiguous evidence for DNA versus RNA editing comes from head-to-head biochemical studies using recombinant enzymes. AID and APOBEC3G, have a strong preference for single-stranded DNA substrates, with no detectable RNA editing activity (Bransteitter et al. 2003; Iwatani et al. 2006). Third, a strong preference for single-stranded DNA substrates is also evident in sequencing studies of retroviruses produced in the presence of a given APOBEC3 protein, such as APOBEC3G (Fig. 2c) (Harris et al. 2003; Lecossier et al. 2003; Mangeat et al. 2003; Zhang et al. 2003). In this experimental system, each APOBEC3 protein presumably has a chance to deaminate viral genomic RNA cytosines before the reverse transcription process converts it to a single-stranded cDNA intermediate and then to the double-stranded DNA required for integration. However, the most common APOBEC3-dependent mutations detected in integrated viral DNA that has survived this process are genomic strand G-to-A mutations, entirely attributable to cDNA strand C-to-U deamination events. Genomic strand C-to-T editing events possibly due to RNA editing are rarely detected. Importantly, APOBEC3 DNA editing activity is required to explain previously reported G-to-A mutation biases in HIV-1 substrates in vivo (Vartanian et al. 1994; Janini et al. 2001).
The solved structures of bacterial and yeast cytidine and cytosine deaminases were used to inform early functional and structural studies of various APOBEC3 family members (Betts et al. 1994; Ireton et al. 2003; Ko et al. 2003; Johansson et al. 2004; Xie et al. 2004). Each of these bacterial and yeast proteins, in monomeric form, is globular with a hydrophobic β-stranded core and several surrounding α-helices. The most conserved structural feature is the active site, which is defined by a histidine and two cysteines in the yeast enzyme and three cysteines in the bacterial enzymes (Xiang et al. 1997; Ireton et al. 2003; Ko et al. 2003). In both instances, these residues are positioned similarly by alpha helices and they serve to coordinate a zinc ion in the active site, which, as described above, is essential for the deamination reaction (Fig. 1b). Although these conserved features have been useful for generating models of APOBEC3 structures, they have also been misleading because the oligomeric state of each enzyme is variable. For instance, the E. coli CDA is homodimeric and the yeast enzyme is homotetrameric (Betts et al. 1994; Johansson et al. 2002). This has fuelled (likely incorrect) speculation that APOBEC3 family members must also function as oligomers.
Generating high-resolution structures of APOBEC3 family members has proved challenging in large part due to insolubility at higher protein concentrations [e.g., (Iwatani et al. 2006)]. However, several NMR and crystal structures have been achieved for the APOBEC3G catalytic domain (representing Z1-type deaminases) and crystal structures were obtained recently for APOBEC3C and the APOBEC3F catalytic domain (representing Z2-type deaminases) (e.g., Fig. 1c) (Chen et al. 2008; Holden et al. 2008; Furukawa et al. 2009; Shandilya et al. 2010; Kitamura et al. 2012b; Li et al. 2012; Bohn 2013).
These structures have several conserved features that provide insight into how these enzymes may function. First, these proteins are all globular with a hydrophobic core consisting of five beta strands surrounded by six alpha helices and the hallmark α2-β3-α3 zinc-coordinating motif that defines the larger cytosine deaminase superfamily. Second, β-strands 3, 4, and 5 are arranged in parallel, similar to the RNA editing enzyme TadA (an ADAT) but different from the antiparallel arrangement found in bacterial and yeast CDAs. This parallel β3-β4-β5 organization may be a key feature that distinguishes polynucleotide from non-polynucleotide deaminases. Third, although many potential oligomeric interfaces have been captured in the crystal lattices, none have proven critical for enzymatic activity and no common themes have emerged (Furukawa et al. 2009; Shandilya et al. 2010; Kitamura et al. 2012b; Bohn 2013). This is consistent a number of other studies indicating that oligomerization may not be essential for binding and deaminating single-stranded DNA substrates (Opi et al. 2006; Nowarski et al. 2008; Shlyakhtenko et al. 2011; Shlyakhtenko et al. 2012). However, more work on this topic is clearly needed to define the role of oligomerization in vivo, because several of the family members, including APOBEC3G, elicit such a property in living cells (Bransteitter et al. 2003; Chiu et al. 2006; Soros et al. 2007; Chen et al. 2013 in preparation). Finally, it is notable that the majority of structural and amino acid differences between APOBEC3 structures are confined to non-catalytic loop regions. Such differences likely relate to substrate targeting and possible co-factor binding, ultimately reflecting physiological function.
A significant remaining question in our understanding of APOBEC3 function is how these enzymes bind single-stranded DNA substrates. A current working model proposes a positively charged brim in the region surrounding the active site consisting of R213, R215, R313, and R320 in APOBEC3G (Chen et al. 2008; Shindo et al. 2012). These residues are predicted to position single-stranded DNA substrates in a manner that allows the target cytosine to enter the active site (Chen et al. 2008). This model also predicts that in order to access to the catalytic glutamic acid, the target C will be flipped-out with respect to the phosphodiester backbone. A base-flipping mechanism is in good agreement with the structure of the adenosine deaminase TadA complexed with its RNA substrate (Losey et al. 2006).
Finally, the brim-domain model and TadA structures suggest an explanation for the different local single-stranded DNA deamination preferences among APOBEC family members (Conticello et al. 2007a; Chen et al. 2008). Unlike bacterial restriction enzymes with 4, 6, or 8 base palindromic recognition sequences, APOBEC3 family members have a notable preference for the base immediately 5’ of the target C (Harris et al. 2002; Harris et al. 2003; Mangeat et al. 2003; Zhang et al. 2003; Bishop et al. 2004; Liddament et al. 2004; Wiegand et al. 2004; Yu et al. 2004a; Yu et al. 2004b; Zheng et al. 2004; Doehle et al. 2005a; Langlois et al. 2005; Dang et al. 2006; Aguiar et al. 2008; Harari et al. 2009; Stenglein et al. 2010). Specifically, AID prefers a 5’ purine base (5’-AC or GC), APOBEC3G a 5’ cytosine (5’-CC), and all other family members a 5’ thymine (5’-TC). Several studies have recently converged on a loop adjacent to the active site, positioned between β4 and α4 secondary structural elements (Conticello et al. 2007b; Chen et al. 2008; Holden et al. 2008; Kohli et al. 2009; Kohli et al. 2010; Rathore et al. 2013 in preparation). This is most dramatically evidenced by loop grafting experiments, in which this loop in AID can be replaced by the homologous loop from APOBEC3G or APOBEC3F resulting in a complete switch of the preferred base immediately 5’ of the target cytosine (Kohli et al. 2009; Carpenter et al. 2010; Kohli et al. 2010; Wang et al. 2010). Moreover, exchanging the same loop (or even a single amino acid) between APOBEC3A and APOBEC3G completely swaps the dinucleotide preference of these enzymes (Rathore et al. 2013 in preparation). Despite this progress, the field still anxiously awaits high-resolution structures of enzyme-substrate complexes that will more precisely define the substrate binding mechanism and advance our understanding of how these enzymes function in vivo.
3. Biological Functions
3.1 APOBEC3 Proteins in HIV Restriction
Pathogens including the retrovirus HIV-1 (hereafter HIV) must both engage and avoid numerous host factors to replicate and cause disease. Genome-wide knockdown and proteomic studies suggest that up to 10% of human proteins either directly or indirectly impact HIV replication (Brass et al. 2008; Konig et al. 2008; Zhou et al. 2008; Yeung et al. 2009; Jäger et al. 2012a; Jäger et al. 2012b). The majority of these proteins are required in some capacity for virus replication (i.e., dependency factors). In contrast, a small number of these cellular proteins are dominant proteins that directly suppress virus replication (i.e., restriction factors). Restriction factor hallmarks include the capacity to potently inhibit virus replication, signatures of rapid evolution (positive selection), responsiveness to interferon, and neutralization by at least one viral counter-restriction strategy (Malim and Emerman 2008; Harris et al. 2012; Malim and Bieniasz 2012). Here, we focus on the mechanism of HIV restriction by APOBEC3 DNA cytosine deaminases, and we encourage readers to see chapters on equally interesting restriction and counter-restriction mechanisms (TRIM5α in Chap. 2, Tetherin/BST-2 in Chap. 3, and SAMHD1 in Chap. 4).
Original studies showed that the viral infectivity factor (Vif) protein of HIV is required for virus replication in primary CD4+ lymphocytes and in several common laboratory T cell lines (e.g., CEM, H9, so-called non-permissive), but it was dispensable in several others (e.g., CEM-SS, SupT1, or permissive lines) (Fisher et al. 1987; Strebel et al. 1987; Gabuzda et al. 1992). This phenotypic difference led to the cloning of APOBEC3G as one cDNA sequence expressed differentially (of many) between CEM and CEM-SS (Sheehy et al. 2002). However, APOBEC3G proved remarkable as it could convert a permissive cell line to a non-permissive phenotype (Sheehy et al. 2002). Taken together with the independent and near-simultaneous discoveries of APOBEC3G as a putative RNA editing factor and as a DNA editing enzyme, an editing mechanism of restriction was predicted and shortly after demonstrated (Harris et al. 2002; Jarmuz et al. 2002; Harris et al. 2003; Lecossier et al. 2003; Mangeat et al. 2003; Zhang et al. 2003).
Over five hundred papers have now been published on APOBEC3G and the related APOBEC3 proteins that have culminated in a current Trojan horse-like model for HIV restriction as shown in Fig. 2c. To be effective as an HIV restriction factor, APOBEC3G must first be expressed in the cytoplasmic compartment of an infected virus-producing cell (Mangeat et al. 2003). Second, cytoplasmic APOBEC3G is thought to interact with a Gag-ribonucleoprotein complex, and this interaction is required for APOBEC3G packaging (Alce and Popik 2004; Luo et al. 2004; Schafer et al. 2004; Svarovskaia et al. 2004; Khan et al. 2005; Burnett and Spearman 2007; Bogerd and Cullen 2008). This interaction can be disrupted by RNase treatment and is therefore thought to involve a bridging RNA between APOBEC3G and the nucleocapsid region of Gag (Cen et al. 2004; Svarovskaia et al. 2004; Iwatani et al. 2007; Bogerd and Cullen 2008). The identity of the bridging RNA is still an active area of investigation; while several reports indicate a role for the Alu-like 7SL RNA, a role for viral genomic RNA has not been excluded (Khan et al. 2005; Khan et al. 2007; Tian et al. 2007; Wang et al. 2007; Bogerd and Cullen 2008). Third, by an ill-defined mechanism, packaged APOBEC3G must breach the nucleocapsid core of the viral particle. This is a genetically (but not mechanistically) defined step, as various chimeric constructs have the capacity to be packaged into viral particles, yet do not enter the core or restrict (Haché et al. 2005; Martin et al. 2011; Song et al. 2012).
Once an APOBEC3G-loaded viral core is deposited into a target cell, reverse transcription proceeds and a susceptible single-stranded cDNA intermediate is generated. Here, APOBEC3G deaminates C-to-U (Harris et al. 2003; Lecossier et al. 2003; Mangeat et al. 2003; Zhang et al. 2003; Yu et al. 2004b). The uracilated cDNA is either subjected to degradation, or templates the second-strand synthesis, preceding integration into the target cell’s genome (Kaiser and Emerman 2006; Mbisa et al. 2007; Yang et al. 2007). The uracils in the cDNA strand template the insertion of adenines in the nascent plus strand and thereby immortalize G-to-A mutations that limit subsequent rounds of viral replication (Harris et al. 2003; Lecossier et al. 2003; Mangeat et al. 2003; Zhang et al. 2003).
Human T cell lines that express near-physiological levels of an APOBEC3G catalytic mutant do not suppress the replication of a Vif-deficient virus, indicating that the predominant mechanism of HIV restriction depends upon deaminase activity (Bishop et al. 2004; Miyagi et al. 2007; Schumacher et al. 2008; Browne et al. 2009). However, a component of APOBEC3G’s capacity to restrict HIV may be deaminase-independent (Newman et al. 2005; Bishop et al. 2006; Iwatani et al. 2006; Opi et al. 2006; Iwatani et al. 2007; Mbisa et al. 2007; Bishop et al. 2008). The most convincing of these studies has shown that APOBEC3G is capable of binding viral genomic RNA and sterically hindering the processivity of reverse transcriptase (Bishop et al. 2006; Bishop et al. 2008). Nevertheless, the importance of deaminase-independent mechanisms, at least for APOBEC3G and HIV, is questionable given the aforementioned results at physiologic levels.
Soon after the initial discovery and functional characterization of APOBEC3G, attention turned towards its six, highly related family members. Proviral sequences isolated from HIV-infected individuals exhibit two distinct patterns of G-to-A mutation consistent with APOBEC3-mediated deamination, both 5’-GG-to-AG and 5’-GA-to-AA (Fitzgibbon et al. 1993; Janini et al. 2001; Caride et al. 2002; Kieffer et al. 2005; Pace et al. 2006; Gandhi et al. 2008; Land et al. 2008). As mentioned previously, APOBEC3G has a demonstrated preference for deaminating 5’-CC dinucleotides on the viral minus strand resulting in 5’-GG-to-AG mutations and is therefore the most likely source of this mutational pattern (Harris et al. 2003; Mangeat et al. 2003; Zhang et al. 2003; Liddament et al. 2004; Yu et al. 2004b). However, high levels of proviral 5’-GA-to-AA mutation in patients’ proviral sequences suggested the involvement of at least one additional family member. Unfortunately, all six other APOBEC3 family members prefer to deaminate a 5’-TC, therefore excluding the possibility of identifying an additional APOBEC3 source based solely on mutational preference [e.g., (Bishop et al. 2004); reviewed in (Albin and Harris 2010)].
In cell-based model systems with forced cDNA expression, all seven APOBEC3s have reported activity [e.g., (Bishop et al. 2004; Liddament et al. 2004; Wiegand et al. 2004; Yu et al. 2004a; Zheng et al. 2004; Doehle et al. 2005a; Rose et al. 2005; Dang et al. 2006; Goila-Gaur et al. 2007; Dang et al. 2008; OhAinle et al. 2008); reviewed in (Albin and Harris 2010)]. However, cellular expression patterns, ability to encapsidate into viral particles, and restriction in relevant T cell models indicate not all the APOBEC3s function in vivo as HIV restriction factors. For example, in primary CD4+ T lymphocytes, a major target of HIV in vivo, six APOBEC3s are expressed at appreciable levels, but APOBEC3A mRNA is undetectable (Koning et al. 2009; Refsland et al. 2010). In addition, only four: APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H (but not APOBEC3A, APOBEC3B, and APOBEC3C), package into viral-like particles and inhibit viral replication when stably expressed in human T cell lines (Hultquist et al. 2011). Finally, endogenous APOBEC3D and APOBEC3F combine to inflict the 5’-GA-to-AA mutation pattern observed in the non-permissive T cell line CEM2n (Refsland et al. 2012). Of note, CEM2n does not express appreciable levels of APOBEC3H mRNA; therefore the endogenous contribution of this protein could not be evaluated. Seven variants of APOBEC3H have been reported and cell-based studies indicate only a subset of these haplotypes are stable at the protein level and capable of HIV restriction (Dang et al. 2008; OhAinle et al. 2008; Harari et al. 2009; Wang et al. 2011). The significance of this natural variation in human populations and the relevance of it to HIV restriction require further investigation. Taken together, while all of the APOBEC3s can be compelled to deaminate a single-stranded viral DNA in vitro or in heterologous cell lines by gross overexpression, only four APOBEC3s — APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H — appear to have the capacity to function as HIV restriction factors in T lymphocytes.
All successful viruses have evolved sophisticated immune suppression and/or evasion mechanisms to neutralize the cellular defense systems they must face. One of the best-studied APOBEC3 counter-restriction mechanisms is orchestrated by the HIV viral infectivity factor, Vif. This protein is small (23 kDa), highly basic, and strictly required for pathogenesis in vivo as well as for virus replication ex vivo in monocytes, macrophages, primary CD4+ T lymphocytes, and non-permissive T cell lines (Kan et al. 1986; Lee et al. 1986; Sodroski et al. 1986; Fisher et al. 1987; Strebel et al. 1987; Gabuzda et al. 1992; von Schwedler et al. 1993). Vif recruits a multiprotein E3 ubiquitin ligase complex consisting of CUL5/NEDD8, ELOB, ELOC, RBX2 and CBFβ to mediate proteasomal degradation of the cellular APOBEC3 restriction factors (Fig 2c) (Conticello et al. 2003; Marin et al. 2003; Sheehy et al. 2003; Yu et al. 2003; Mehle et al. 2004; Jäger et al. 2012b). In addition to this primary role, it has been suggested that Vif can relieve APOBEC3G-mediated restriction by alternative mechanisms such as directly blocking deaminase activity, preventing encapsidation, sequestering the protein in catalytically inactive conformations, or impeding APOBEC3G translation (Mariani et al. 2003; Stopak et al. 2003; Kao et al. 2004; Santa-Marta et al. 2005; Opi et al. 2007; Goila-Gaur et al. 2008; Britan-Rosich et al. 2011). The extent to which these mechanisms function to counteract APOBEC3 during a productive infection warrants further investigation but is certain to be secondary to the proteasomal degradation mechanism.
3.2 APOBEC3 proteins in general innate immune defense
Exogenous retroviruses are rare in humans, consistent with the idea that the multifaceted APOBEC3 defense provides robust protection against this type of pathogen. HIV is one exception to this rule. It is successful, at least in part, due to Vif and degradation of the cellular APOBEC3s. However, evidence for a long history of positive selection acting on the human APOBEC3 locus suggests this family has been defending the genomic integrity of its host’s cells long before HIV was transmitted into the human population (Sawyer et al. 2004; Zhang and Webb 2004; Sanville et al. 2010). Given the significant presence of endogenous retroelements in the genomes of mammals (nearly 50%), suppression of these elements may represent the sub-family’s true raison d’être and the primary source of the positive selection that has shaped the complex present day locus and the diverse functions of these innate immunity factors.
Endogenous retroelements, including those containing long terminal repeats (LTR) like endogenous retroviruses, as well as non-LTR elements like long interspersed nuclear elements (LINEs) and short interspersed nuclear elements (SINEs), may have provided the evolutionary pressure necessary for the maintained expansion of the APOBEC3 locus in primates. Additionally, the differences in retrotransposition frequency between rodents and primates could be attributable to possessing an arsenal of seven APOBEC3 proteins versus only a single one (Maksakova et al. 2006; Stenglein and Harris 2006). In support of this hypothesis, human APOBEC3s have demonstrated activity on LTR retrotransposons from mice and yeast (Dutko et al. 2005; Esnault et al. 2005; Schumacher et al. 2005; Bogerd et al. 2006a; Chen et al. 2006; Esnault et al. 2006; Lee and Bieniasz 2007; Jern and Coffin 2008; Lee et al. 2008). Additionally, some endogenous retroviruses exhibit the characteristic scars from APOBEC3F and APOBEC3G activity in their genomes (Anwar et al. 2013 in press). Non-LTR elements, including LINE1 and Alu, are also restricted by human APOBEC3s but, in contrast to the HIV restriction mechanism described above, this mechanism appears entirely deamination-independent (Bogerd et al. 2006b; Chiu et al. 2006; Muckenfuss et al. 2006; Stenglein and Harris 2006; Carmi et al. 2011).
The generation of a single-stranded DNA intermediate in the life cycle of a parasitic element may render it susceptible to APOBEC3 restriction. In addition to HIV, other viruses including simian immunodeficiency virus, murine leukemia virus, foamy virus, porcine endogenous retrovirus, human T-cell leukemia virus, and hepatitis B virus have all been reported to be susceptible to APOBEC3-mediated editing of their genomes (Harris et al. 2003; Mangeat et al. 2003; Mariani et al. 2003; Kobayashi et al. 2004; Turelli et al. 2004; Yu et al. 2004a; Doehle et al. 2005b; Lochelt et al. 2005; Russell et al. 2005; Suspène et al. 2005; Abudu et al. 2006; Delebecque et al. 2006; Jonsson et al. 2007). Whether or not the APOBEC3-mediated restriction of all of these viral pathogens is part of a natural innate immune response or a by-product of the particular model systems used awaits further investigation.
Endogenous retroelements and their mobility are believed to have played a central role early in shaping the human genome during speciation (Kazazian 2004; Carmi et al. 2011). However, this process has its associated costs. Ultimately, cells have devised strategies to defend and preserve genomic integrity by curbing the movement of these genetic elements. The evolution of the APOBEC3 family has likely played a prominent role in this defense and in diversifying the retroelements to make them more useful for the host species [e.g., (Carmi et al. 2011)].
4. Pathological Consequences of DNA Deamination
Although DNA cytosine deamination has obvious benefits for individual cells and the organism as a whole, this process may have considerable pathological consequences, most notably cancer. For instance, overexpression of APOBEC1 in the liver of mice has been shown to cause hepatocellular carcinoma and liver dysplasia (Yamanaka et al. 1995). Likewise, AID transgenesis also causes cancer, and it has been implicated in initiating the chromosomal translocations responsible for some lymphocyte neoplasias including the hallmark c-myc/IgH rearrangement in Burkitt’s lymphoma [(Okazaki et al. 2003; Ramiro et al. 2004; Unniraman et al. 2004); reviewed by (Perez-Duran et al. 2007)]. AID may also promote resistance to the chemotherapeutic drug imatinib (Klemm et al. 2009). However, the overall impact of APOBEC1 and AID on human cancer is questionable because their expression is largely limited to tissues associated with their biological functions, APOBEC1 in the enterocytes of the small intestine and AID in B cells (Fig. 3a) (Powell et al. 1987; Teng et al. 1993; Muramatsu et al. 1999; Muramatsu et al. 2000).
Fig. 3.
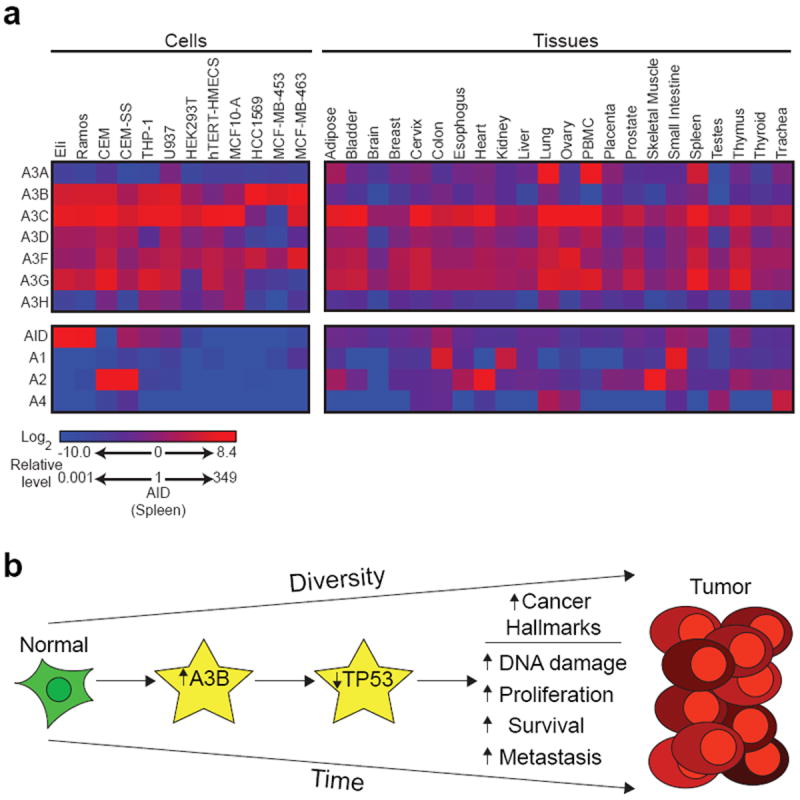
APOBEC expression and model of carcinogenesis. (a) APOBEC family mRNA expression in the indicated cell lines and normal human tissues. Data are relative to levels of AID in the spleen [reprinted with permission from (Burns et al. 2013)]. (b) Model depicting the effect of APOBEC3B overexpression over time.
In contrast, most APOBEC3s have much broader expression ranges that span most tissues in the human body (Fig. 3a) (Koning et al. 2009; Refsland et al. 2010; Burns et al. 2013). Broad expression profiles, potent DNA deaminase activity, and C-to-T transition biases in tumor genome sequences strongly suggested that one or more of the APOBEC3 proteins may be a source of mutation in different cancers (Harris et al. 2002; Nik-Zainal et al. 2012; Roberts et al. 2012; Burns et al. 2013). APOBEC3B became a leading candidate as it uniquely and constitutively localizes to the nucleus by inheriting a nuclear import mechanism from AID (Bogerd et al. 2006b; Bonvin et al. 2006; Muckenfuss et al. 2006; Stenglein and Harris 2006; Kinomoto et al. 2007; Stenglein et al. 2008; Hultquist et al. 2011; Pak et al. 2011; Lackey et al. 2012; Lackey et al. 2013). Recently, APOBEC3B was found overexpressed in some laboratory breast cancer cell lines, but not in available control cell lines (Fig.3a; e.g., compare HCC1569, MCF-MB-453, and MCF-MB-463 to telomerase immortalized human mammary epithelial cells, hTERT HMECS) (Burns et al. 2013). APOBEC3B up-regulation was shown to be responsible for elevated genomic uracil levels and increased mutation rates in breast cancer cell lines (Burns et al. 2013). APOBEC3B up-regulation was similarly robust in the majority of human breast tumors, in contrast to barely detectable levels in normal breast tissue (Burns et al. 2013). Remarkably, APOBEC3B overexpression correlated with a doubling in the tumor genomic mutation loads, and the majority of C-to-T transition mutations occurred within the preferred motif of recombinant APOBEC3B (Burns et al. 2013).
Overall, a model is emerging for how APOBEC3B provides genetic fuel for tumorigenesis, which, coupled with selection, may help explain many hallmarks of cancer such as increased DNA damage, elevated proliferation, decreased apoptosis, and massive heterogeneity (Fig. 3b). In particular, APOBEC3B up-regulation correlates with inactivation of the tumor suppressor gene TP53, which strongly suggests that it may be an early tumor-initiating event (Burns et al. 2013). Obviously, the potential benefits to encoding APOBEC3B must outweigh potential costs of carcinogenesis. An attractive explanation for this apparent conundrum may be that its innate immune function is important early in life and for the health of the species, for instance in germ cells or early development (Bogerd et al. 2006b; Wissing et al. 2011), whereas the toll of cancer is not imposed in most instances until after the reproductive years. In any event, much more work is now justified on APOBEC3B and its role in breast and, potentially, other human cancers.
5. Possible Avenues to APOBEC3-Based Therapeutics
5.1. Therapy by hypermutation
If left unimpeded by Vif, APOBEC3 proteins such as APOBEC3G can convert up to 10% of viral cDNA cytosines into uracils in a single-round of virus replication [e.g., (Harris et al. 2003; Yu et al. 2004b)]. The resulting massive levels of G-to-A mutations effectively ruin the genetic potential of the retrovirus in a process called lethal mutagenesis (Loeb et al. 1999; Haché et al. 2006). Moreover, the preferred context of APOBEC3G deamination events often results in the conversion of the tryptophan codon TGG into a premature stop codon TAG, which is more detrimental to the virus than a simple amino acid change. Overall, physiological levels of APOBEC3 proteins largely, if not fully, suppress the replication of Vif-deficient HIV.
This remarkable potency therefore raises the prospect of developing drugs to leverage the APOBEC3 restriction mechanism against HIV (Fig. 4a). Direct inhibition of Vif is certainly one strategy, but such an approach is destined to be susceptible to problems imposed by extensive natural HIV variation and the rapid evolution of drug resistance. A more appealing alternative may be to develop a drug toward one of the more genetically stable cellular proteins recruited to the APOBEC3 degradation complex. In particular, Vif requires at least four heterologous protein-protein interactions to successfully counteract the APOBEC3 proteins. The first strategy, of course, is targeting the direct interaction with the APOBEC3 proteins themselves, which may occur through conserved structural motifs (Albin et al. 2010; Kitamura et al. 2012a). Second is targeting the recently discovered interaction with CBFβ, which is essential for Vif stability and function (Jäger et al. 2012b; Zhang et al. 2012). Third and fourth are targeting the distinct Vif interaction motifs in ELOC (the SLQ motif) or CUL5, which are also essential for activity of the APOBEC3 Ub ligase complex (Marin et al. 2003; Yu et al. 2003). Finally, it may be possible to target an upstream component of the proteasomal pathway involved in APOBEC3 degradation [e.g., (Kim et al. 2013)].
Fig. 4.
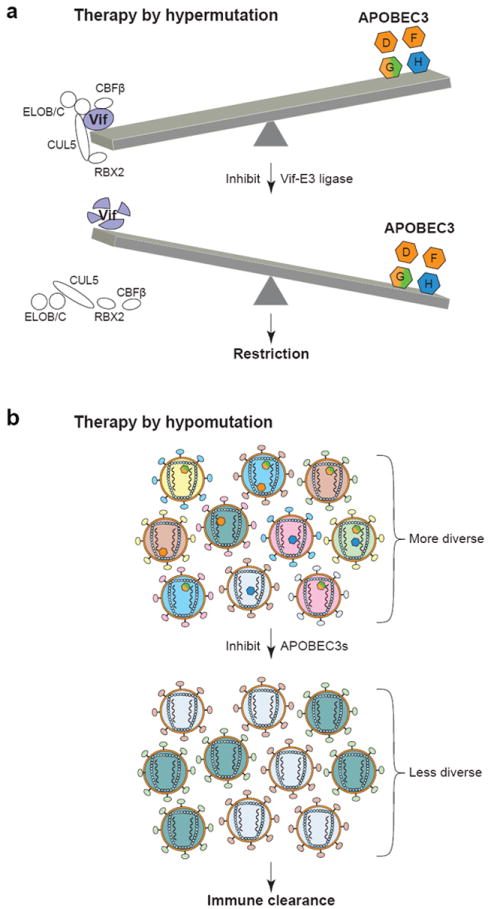
Potential therapies harnessing APOBEC3 innate immunity. (a) Inhibition of the Vif-E3 ligase complex could result in virus restriction. (b) Inhibiting the deaminase activity of APOBEC3 proteins could deprive HIV of a source of variation and allow immune clearance.
However, despite these clear opportunities for drug development, progress has been relatively underwhelming. Proof of concept experiments have been achieved through cell-based screens for preservation of APOBEC3G-GFP fluorescence in the presence of HIV Vif (Nathans et al. 2008). The lead molecule from these studies, RN18, however, has only been subject to modest additional development (Ali et al. 2012). An additional concern holding back development of RN18 is the major challenge of identifying the molecular target, which is essential for structural studies and rational improvements through medicinal chemistry.
Independent lead compounds have also been identified based on predicted fit into the Vif-binding pocket in ELOC, which would effectively outcompete the SLQ motif of Vif (Huang et al. 2013a; Huang et al. 2013b). However, the best of these indolizine type compounds has a modest IC50 value of 11 μM. This will likely need improvements in potency and solubility before efficacy can be achieved in live cell studies. In any event, much more work in this area is needed, including larger scale cellular screens, biochemical screens, and computational screens, followed by a comprehensive sets of secondary and tertiary screens to narrow in on the most effective drug candidates.
5.2. Therapy by hypomutation
Vif efficiently counteracts the HIV-relevant APOBEC3 repertoire (Conticello et al. 2003; Kao et al. 2003; Marin et al. 2003; Sheehy et al. 2003; Stopak et al. 2003; Hultquist et al. 2011). Nonetheless, evidence of APOBEC3 activity is found in the proviral sequences derived from infected patients [e.g., (Janini et al. 2001)]. This raises the possibilities that either Vif cannot completely neutralize all of the cytoplasmic APOBEC3 before encapsidation and/or HIV regulates its high mutation rate, in part, through controlled degradation of the APOBEC3 proteins with Vif acting as a molecular rheostat optimizing the levels of cytosine deamination necessary for immune evasion and potentially drug resistance (Haché et al. 2006; Harris 2008).
A counterintuitive but potentially more effective strategy to decrease the pathogenesis of HIV may be to inhibit the enzymatic activity of the APOBEC3s, thereby eliminating a potential source of variation for the virus (Fig. 4b) (Harris 2008; Li et al. 2012; Olson et al. 2013). Toward this end, high-throughput screens have resulted in proof-of-concept experiments that APOBEC3G activity can be blocked with small molecules (Li et al. 2012; Olson et al. 2013). However, much additional work is still necessary to improve the solubility, potency, and bioavailability of APOBEC3 inhibitors, and efficacy will need to be demonstrated in cell-based studies before critical animal experiments can be done. Nevertheless, a hypomutation strategy is attractive because decreasing the HIV mutation rate has the potential to limit the diversity of the viral population and render it susceptible to normal immune clearance mechanisms (Fig. 4b). Indeed, the adaptive T and B cell-mediated immune responses manage to keep the virus in-check initially in most patients, and it is tempting to speculate that even a slight tip in favor of host immunity may enable complete virus clearance (i.e., analogous to most other viral infections). Given the fact that existing anti-retroviral drugs are numerous and largely effective, a big challenge facing the field is the development of a curative therapy. It remains possible that promoting either virus hypermutation or hypomutation could be part of such a cure.
Acknowledgments
We apologize to colleagues whose work could not be cited due to space limitations. Support was provided by grants from the National Institutes of Health R01-AI064046 and P01-GM091743 to RSH and F31-DA033186 to EWR.
References
- Abudu A, Takaori-Kondo A, Izumi T, et al. Murine retrovirus escapes from murine APOBEC3 via two distinct novel mechanisms. Current biology : CB. 2006;16:1565–1570. doi: 10.1016/j.cub.2006.06.055. [DOI] [PubMed] [Google Scholar]
- Agranat L, Raitskin O, Sperling J, Sperling R. The editing enzyme ADAR1 and the mRNA surveillance protein hUpf1 interact in the cell nucleus. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:5028–5033. doi: 10.1073/pnas.0710576105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguiar RS, Lovsin N, Tanuri A, Peterlin BM. Vpr.A3A chimera inhibits HIV replication. J Biol Chem. 2008;283:2518–2525. doi: 10.1074/jbc.M706436200. [DOI] [PubMed] [Google Scholar]
- Albin JS, Harris RS. Interactions of host APOBEC3 restriction factors with HIV-1 in vivo: implications for therapeutics. Expert Rev Mol Med. 2010;12:e4. doi: 10.1017/S1462399409001343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albin JS, LaRue RS, Weaver JA, et al. A single amino acid in human APOBEC3F alters susceptibility to HIV-1 Vif. The Journal of biological chemistry. 2010;285:40785–40792. doi: 10.1074/jbc.M110.173161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alce TM, Popik W. APOBEC3G is incorporated into virus-like particles by a direct interaction with HIV-1 Gag nucleocapsid protein. The Journal of biological chemistry. 2004;279:34083–34086. doi: 10.1074/jbc.C400235200. [DOI] [PubMed] [Google Scholar]
- Ali A, Wang J, Nathans RS, Cao H, Sharova N, Stevenson M, Rana TM. Synthesis and structure-activity relationship studies of HIV-1 virion infectivity factor (Vif) inhibitors that block viral replication. ChemMedChem. 2012;7:1217–1229. doi: 10.1002/cmdc.201200079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anwar F, Davenport MP, Ebrahimi D. Footprint of APOBEC3 on the genome of retrotransposons. Journal of Virology. 2013 doi: 10.1128/JVI.00298-13. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betts SD, Hachigian TM, Pichersky E, Yocum CF. Reconstitution of the spinach oxygen-evolving complex with recombinant Arabidopsis manganese-stabilizing protein. Plant molecular biology. 1994;26:117–130. doi: 10.1007/BF00039525. [DOI] [PubMed] [Google Scholar]
- Bishop KN, Holmes RK, Malim MH. Antiviral potency of APOBEC proteins does not correlate with cytidine deamination. Journal of virology. 2006;80:8450–8458. doi: 10.1128/JVI.00839-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop KN, Holmes RK, Sheehy AM, Davidson NO, Cho SJ, Malim MH. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr Biol. 2004;14:1392–1396. doi: 10.1016/j.cub.2004.06.057. [DOI] [PubMed] [Google Scholar]
- Bishop KN, Verma M, Kim EY, Wolinsky SM, Malim MH. APOBEC3G inhibits elongation of HIV-1 reverse transcripts. PLoS pathogens. 2008;4:e1000231. doi: 10.1371/journal.ppat.1000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd HP, Cullen BR. Single-stranded RNA facilitates nucleocapsid: APOBEC3G complex formation. RNA. 2008;14:1228–1236. doi: 10.1261/rna.964708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd HP, Wiegand HL, Doehle BP, Lueders KK, Cullen BR. APOBEC3A and APOBEC3B are potent inhibitors of LTR-retrotransposon function in human cells. Nucleic Acids Res. 2006a;34:89–95. doi: 10.1093/nar/gkj416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd HP, Wiegand HL, Hulme AE, Garcia-Perez JL, O’Shea KS, Moran JV, Cullen BR. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc Natl Acad Sci U S A. 2006b;103:8780–8785. doi: 10.1073/pnas.0603313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohn M-F, Shandilya SMD, Albin JS, Kouno T, Anderson BD, McDougle RM, Carpenter MA, Rathore A, Davis AN, Zhang J, Lu Y, Somasundaran M, Matsuo H, Harris RS, Schiffer CA. Crystal structure of the HIV-1 Vif binding and catalytically active domain of APOBEC3F. Structure. 2013 doi: 10.1016/j.str.2013.04.010. in review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonvin M, Achermann F, Greeve I, et al. Interferon-inducible expression of APOBEC3 editing enzymes in human hepatocytes and inhibition of hepatitis B virus replication. Hepatology. 2006;43:1364–1374. doi: 10.1002/hep.21187. [DOI] [PubMed] [Google Scholar]
- Bransteitter R, Pham P, Scharff MD, Goodman MF. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:4102–4107. doi: 10.1073/pnas.0730835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brass AL, Dykxhoorn DM, Benita Y, et al. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- Britan-Rosich E, Nowarski R, Kotler M. Multifaceted counter-APOBEC3G mechanisms employed by HIV-1 Vif. Journal of molecular biology. 2011;410:1065–1076. doi: 10.1016/j.jmb.2011.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne EP, Allers C, Landau NR. Restriction of HIV-1 by APOBEC3G is cytidine deaminase-dependent. Virology. 2009;387:313–321. doi: 10.1016/j.virol.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett A, Spearman P. APOBEC3G multimers are recruited to the plasma membrane for packaging into human immunodeficiency virus type 1 virus-like particles in an RNA-dependent process requiring the NC basic linker. Journal of virology. 2007;81:5000–5013. doi: 10.1128/JVI.02237-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns MB, Lackey L, Carpenter MA, et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature. 2013;494:366–370. doi: 10.1038/nature11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caride E, Brindeiro RM, Kallas EG, de Sa CA, Eyer-Silva WA, Machado E, Tanuri A. Sexual transmission of HIV-1 isolate showing G-->A hypermutation. J Clin Virol. 2002;23:179–189. doi: 10.1016/s1386-6532(01)00218-9. [DOI] [PubMed] [Google Scholar]
- Carmi S, Church GM, Levanon EY. Large-scale DNA editing of retrotransposons accelerates mammalian genome evolution. Nature communications. 2011;2:519. doi: 10.1038/ncomms1525. [DOI] [PubMed] [Google Scholar]
- Carpenter MA, Rajagurubandara E, Wijesinghe P, Bhagwat AS. Determinants of sequence-specificity within human AID and APOBEC3G. DNA repair. 2010;9:579–587. doi: 10.1016/j.dnarep.2010.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cen S, Guo F, Niu M, Saadatmand J, Deflassieux J, Kleiman L. The interaction between HIV-1 Gag and APOBEC3G. The Journal of biological chemistry. 2004;279:33177–33184. doi: 10.1074/jbc.M402062200. [DOI] [PubMed] [Google Scholar]
- Chan L. Apolipoprotein B, the major protein component of triglyceride-rich and low density lipoproteins. The Journal of biological chemistry. 1992;267:25621–25624. [PubMed] [Google Scholar]
- Chen H, Lilley CE, Yu Q, et al. APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Curr Biol. 2006;16:480–485. doi: 10.1016/j.cub.2006.01.031. [DOI] [PubMed] [Google Scholar]
- Chen KM, Harjes E, Gross PJ, et al. Structure of the DNA deaminase domain of the HIV-1 restriction factor APOBEC3G. Nature. 2008;452:116–119. doi: 10.1038/nature06638. [DOI] [PubMed] [Google Scholar]
- Chen YJ, Li M, Carpenter MA, McDougle RM, Macdonald PJ, Harris RS, Mueller JD. APOBEC3 multimerization correlates with HIV-1 restriction activity in living cells. Journal of Molecular Biology. 2013 doi: 10.1016/j.jmb.2013.12.014. in preparation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu YL, Witkowska HE, Hall SC, et al. High-molecular-mass APOBEC3G complexes restrict Alu retrotransposition. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:15588–15593. doi: 10.1073/pnas.0604524103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conticello SG. The AID/APOBEC family of nucleic acid mutators. Genome biology. 2008;9:229. doi: 10.1186/gb-2008-9-6-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conticello SG, Harris RS, Neuberger MS. The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G. Curr Biol. 2003;13:2009–2013. doi: 10.1016/j.cub.2003.10.034. [DOI] [PubMed] [Google Scholar]
- Conticello SG, Langlois MA, Neuberger MS. Insights into DNA deaminases. Nature structural & molecular biology. 2007a;14:7–9. doi: 10.1038/nsmb0107-7. [DOI] [PubMed] [Google Scholar]
- Conticello SG, Langlois MA, Yang Z, Neuberger MS. DNA deamination in immunity: AID in the context of its APOBEC relatives. Advances in immunology. 2007b;94:37–73. doi: 10.1016/S0065-2776(06)94002-4. [DOI] [PubMed] [Google Scholar]
- Conticello SG, Thomas CJ, Petersen-Mahrt SK, Neuberger MS. Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol Biol Evol. 2005;22:367–377. doi: 10.1093/molbev/msi026. [DOI] [PubMed] [Google Scholar]
- Dang Y, Siew LM, Wang X, Han Y, Lampen R, Zheng YH. Human cytidine deaminase APOBEC3H restricts HIV-1 replication. J Biol Chem. 2008;283:11606–11614. doi: 10.1074/jbc.M707586200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang Y, Wang X, Esselman WJ, Zheng YH. Identification of APOBEC3DE as another antiretroviral factor from the human APOBEC family. J Virol. 2006;80:10522–10533. doi: 10.1128/JVI.01123-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delebecque F, Suspene R, Calattini S, et al. Restriction of foamy viruses by APOBEC cytidine deaminases. Journal of virology. 2006;80:605–614. doi: 10.1128/JVI.80.2.605-614.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Noia J, Neuberger MS. Altering the pathway of immunoglobulin hypermutation by inhibiting uracil-DNA glycosylase. Nature. 2002;419:43–48. doi: 10.1038/nature00981. [DOI] [PubMed] [Google Scholar]
- Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annual review of biochemistry. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- Doehle BP, Schäfer A, Cullen BR. Human APOBEC3B is a potent inhibitor of HIV-1 infectivity and is resistant to HIV-1 Vif. Virology. 2005a;339:281–288. doi: 10.1016/j.virol.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Doehle BP, Schafer A, Wiegand HL, Bogerd HP, Cullen BR. Differential sensitivity of murine leukemia virus to APOBEC3-mediated inhibition is governed by virion exclusion. Journal of virology. 2005b;79:8201–8207. doi: 10.1128/JVI.79.13.8201-8207.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutko JA, Schafer A, Kenny AE, Cullen BR, Curcio MJ. Inhibition of a yeast LTR retrotransposon by human APOBEC3 cytidine deaminases. Curr Biol. 2005;15:661–666. doi: 10.1016/j.cub.2005.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnault C, Heidmann O, Delebecque F, et al. APOBEC3G cytidine deaminase inhibits retrotransposition of endogenous retroviruses. Nature. 2005;433:430–433. doi: 10.1038/nature03238. [DOI] [PubMed] [Google Scholar]
- Esnault C, Millet J, Schwartz O, Heidmann T. Dual inhibitory effects of APOBEC family proteins on retrotransposition of mammalian endogenous retroviruses. Nucleic acids research. 2006;34:1522–1531. doi: 10.1093/nar/gkl054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher AG, Ensoli B, Ivanoff L, et al. The sor gene of HIV-1 is required for efficient virus transmission in vitro. Science. 1987;237:888–893. doi: 10.1126/science.3497453. [DOI] [PubMed] [Google Scholar]
- Fitzgibbon JE, Mazar S, Dubin DT. A new type of G-->A hypermutation affecting human immunodeficiency virus. AIDS Res Hum Retroviruses. 1993;9:833–838. doi: 10.1089/aid.1993.9.833. [DOI] [PubMed] [Google Scholar]
- Furukawa A, Nagata T, Matsugami A, et al. Structure, interaction and real-time monitoring of the enzymatic reaction of wild-type APOBEC3G. The EMBO journal. 2009;28:440–451. doi: 10.1038/emboj.2008.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabuzda DH, Lawrence K, Langhoff E, Terwilliger E, Dorfman T, Haseltine WA, Sodroski J. Role of vif in replication of human immunodeficiency virus type 1 in CD4+ T lymphocytes. J Virol. 1992;66:6489–6495. doi: 10.1128/jvi.66.11.6489-6495.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi SK, Siliciano JD, Bailey JR, Siliciano RF, Blankson JN. Role of APOBEC3G/F-mediated hypermutation in the control of human immunodeficiency virus type 1 in elite suppressors. J Virol. 2008;82:3125–3130. doi: 10.1128/JVI.01533-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber AP, Keller W. An adenosine deaminase that generates inosine at the wobble position of tRNAs. Science. 1999;286:1146–1149. doi: 10.1126/science.286.5442.1146. [DOI] [PubMed] [Google Scholar]
- Goila-Gaur R, Khan MA, Miyagi E, Kao S, Opi S, Takeuchi H, Strebel K. HIV-1 Vif promotes the formation of high molecular mass APOBEC3G complexes. Virology. 2008;372:136–146. doi: 10.1016/j.virol.2007.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goila-Gaur R, Khan MA, Miyagi E, Kao S, Strebel K. Targeting APOBEC3A to the viral nucleoprotein complex confers antiviral activity. Retrovirology. 2007;4:61. doi: 10.1186/1742-4690-4-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean H. DNA and RNA modification enzymes : structure, mechanism, function and evolution. Landes Bioscience; Austin, Tex: 2009. [Google Scholar]
- Haché G, Liddament MT, Harris RS. The retroviral hypermutation specificity of APOBEC3F and APOBEC3G is governed by the C-terminal DNA cytosine deaminase domain. The Journal of biological chemistry. 2005;280:10920–10924. doi: 10.1074/jbc.M500382200. [DOI] [PubMed] [Google Scholar]
- Haché G, Mansky LM, Harris RS. Human APOBEC3 proteins, retrovirus restriction, and HIV drug resistance. AIDS Rev. 2006;8:148–157. [PubMed] [Google Scholar]
- Harari A, Ooms M, Mulder LC, Simon V. Polymorphisms and splice variants influence the antiretroviral activity of human APOBEC3H. Journal of virology. 2009;83:295–303. doi: 10.1128/JVI.01665-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RS. Enhancing immunity to HIV through APOBEC. Nature biotechnology. 2008;26:1089–1090. doi: 10.1038/nbt1008-1089. [DOI] [PubMed] [Google Scholar]
- Harris RS, Bishop KN, Sheehy AM, et al. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113:803–809. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- Harris RS, Hultquist JF, Evans DT. The restriction factors of human immunodeficiency virus. The Journal of biological chemistry. 2012;287:40875–40883. doi: 10.1074/jbc.R112.416925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RS, Liddament MT. Retroviral restriction by APOBEC proteins. Nature reviews Immunology. 2004;4:868–877. doi: 10.1038/nri1489. [DOI] [PubMed] [Google Scholar]
- Harris RS, Petersen-Mahrt SK, Neuberger MS. RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol Cell. 2002;10:1247–1253. doi: 10.1016/s1097-2765(02)00742-6. [DOI] [PubMed] [Google Scholar]
- Holden LG, Prochnow C, Chang YP, et al. Crystal structure of the anti-viral APOBEC3G catalytic domain and functional implications. Nature. 2008;456:121–124. doi: 10.1038/nature07357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Zuo T, Jin H, Liu Z, Yang Z, Yu X, Zhang L. Design, synthesis and biological evaluation of indolizine derivatives as HIV-1 VIF-ElonginC interaction inhibitors. Molecular diversity. 2013a doi: 10.1007/s11030-013-9424-3. [DOI] [PubMed] [Google Scholar]
- Huang W, Zuo T, Luo X, et al. Indolizine derivatives as HIV-1 VIF-ElonginC interaction inhibitors. Chemical biology & drug design. 2013b doi: 10.1111/cbdd.12119. [DOI] [PubMed] [Google Scholar]
- Hultquist JF, Lengyel JA, Refsland EW, LaRue RS, Lackey L, Brown WL, Harris RS. Human and Rhesus APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H Demonstrate a Conserved Capacity to Restrict Vif-deficient HIV-1. J Virol. 2011 doi: 10.1128/JVI.05238-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda T, Abd El Galil KH, Tokunaga K, et al. Intrinsic restriction activity by apolipoprotein B mRNA editing enzyme APOBEC1 against the mobility of autonomous retrotransposons. Nucleic Acids Res. 2011;39:5538–5554. doi: 10.1093/nar/gkr124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda T, Ohsugi T, Kimura T, Matsushita S, Maeda Y, Harada S, Koito A. The antiretroviral potency of APOBEC1 deaminase from small animal species. Nucleic Acids Res. 2008;36:6859–6871. doi: 10.1093/nar/gkn802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ireton GC, Black ME, Stoddard BL. The 1.14 A crystal structure of yeast cytosine deaminase: evolution of nucleotide salvage enzymes and implications for genetic chemotherapy. Structure. 2003;11:961–972. doi: 10.1016/s0969-2126(03)00153-9. [DOI] [PubMed] [Google Scholar]
- Iwatani Y, Chan DS, Wang F, et al. Deaminase-independent inhibition of HIV-1 reverse transcription by APOBEC3G. Nucleic acids research. 2007;35:7096–7108. doi: 10.1093/nar/gkm750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwatani Y, Takeuchi H, Strebel K, Levin JG. Biochemical activities of highly purified, catalytically active human APOBEC3G: correlation with antiviral effect. Journal of virology. 2006;80:5992–6002. doi: 10.1128/JVI.02680-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäger S, Cimermancic P, Gulbahce N, et al. Global landscape of HIV-human protein complexes. Nature. 2012a;481:365–370. doi: 10.1038/nature10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäger S, Kim DY, Hultquist JF, et al. Vif hijacks CBF-beta to degrade APOBEC3G and promote HIV-1 infection. Nature. 2012b;481:371–375. doi: 10.1038/nature10693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janini M, Rogers M, Birx DR, McCutchan FE. Human immunodeficiency virus type 1 DNA sequences genetically damaged by hypermutation are often abundant in patient peripheral blood mononuclear cells and may be generated during near-simultaneous infection and activation of CD4(+) T cells. J Virol. 2001;75:7973–7986. doi: 10.1128/JVI.75.17.7973-7986.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmuz A, Chester A, Bayliss J, Gisbourne J, Dunham I, Scott J, Navaratnam N. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics. 2002;79:285–296. doi: 10.1006/geno.2002.6718. [DOI] [PubMed] [Google Scholar]
- Jern P, Coffin JM. Effects of retroviruses on host genome function. Annual review of genetics. 2008;42:709–732. doi: 10.1146/annurev.genet.42.110807.091501. [DOI] [PubMed] [Google Scholar]
- Johansson E, Mejlhede N, Neuhard J, Larsen S. Crystal structure of the tetrameric cytidine deaminase from Bacillus subtilis at 2.0 A resolution. Biochemistry. 2002;41:2563–2570. doi: 10.1021/bi011849a. [DOI] [PubMed] [Google Scholar]
- Johansson E, Neuhard J, Willemoes M, Larsen S. Structural, kinetic, and mutational studies of the zinc ion environment in tetrameric cytidine deaminase. Biochemistry. 2004;43:6020–6029. doi: 10.1021/bi035893x. [DOI] [PubMed] [Google Scholar]
- Jonsson SR, LaRue RS, Stenglein MD, Fahrenkrug SC, Andresdottir V, Harris RS. The restriction of zoonotic PERV transmission by human APOBEC3G. PloS one. 2007;2:e893. doi: 10.1371/journal.pone.0000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser SM, Emerman M. Uracil DNA glycosylase is dispensable for human immunodeficiency virus type 1 replication and does not contribute to the antiviral effects of the cytidine deaminase Apobec3G. Journal of virology. 2006;80:875–882. doi: 10.1128/JVI.80.2.875-882.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan NC, Franchini G, Wong-Staal F, DuBois GC, Robey WG, Lautenberger JA, Papas TS. Identification of HTLV-III/LAV sor gene product and detection of antibodies in human sera. Science. 1986;231:1553–1555. doi: 10.1126/science.3006245. [DOI] [PubMed] [Google Scholar]
- Kao S, Khan MA, Miyagi E, Plishka R, Buckler-White A, Strebel K. The human immunodeficiency virus type 1 Vif protein reduces intracellular expression and inhibits packaging of APOBEC3G (CEM15), a cellular inhibitor of virus infectivity. J Virol. 2003;77:11398–11407. doi: 10.1128/JVI.77.21.11398-11407.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao S, Miyagi E, Khan MA, Takeuchi H, Opi S, Goila-Gaur R, Strebel K. Production of infectious human immunodeficiency virus type 1 does not require depletion of APOBEC3G from virus-producing cells. Retrovirology. 2004;1:27. doi: 10.1186/1742-4690-1-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian HH., Jr Mobile elements: drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- Khan MA, Goila-Gaur R, Opi S, Miyagi E, Takeuchi H, Kao S, Strebel K. Analysis of the contribution of cellular and viral RNA to the packaging of APOBEC3G into HIV-1 virions. Retrovirology. 2007;4:48. doi: 10.1186/1742-4690-4-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MA, Kao S, Miyagi E, et al. Viral RNA is required for the association of APOBEC3G with human immunodeficiency virus type 1 nucleoprotein complexes. Journal of virology. 2005;79:5870–5874. doi: 10.1128/JVI.79.9.5870-5874.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer TL, Kwon P, Nettles RE, Han Y, Ray SC, Siliciano RF. G-->A hypermutation in protease and reverse transcriptase regions of human immunodeficiency virus type 1 residing in resting CD4+ T cells in vivo. J Virol. 2005;79:1975–1980. doi: 10.1128/JVI.79.3.1975-1980.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DY, Kwon E, Hartley PD, Crosby DC, Mann S, Krogan NJ, Gross JD. CBFbeta Stabilizes HIV Vif to Counteract APOBEC3 at the Expense of RUNX1 Target Gene Expression. Molecular cell. 2013 doi: 10.1016/j.molcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim U, Wang Y, Sanford T, Zeng Y, Nishikura K. Molecular cloning of cDNA for double-stranded RNA adenosine deaminase, a candidate enzyme for nuclear RNA editing. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:11457–11461. doi: 10.1073/pnas.91.24.11457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinomoto M, Kanno T, Shimura M, et al. All APOBEC3 family proteins differentially inhibit LINE-1 retrotransposition. Nucleic acids research. 2007;35:2955–2964. doi: 10.1093/nar/gkm181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura S, Ode H, Nakashima M, et al. The APOBEC3C crystal structure and the interface for HIV-1 Vif binding. Nature structural & molecular biology. 2012a;19:1005–1010. doi: 10.1038/nsmb.2378. [DOI] [PubMed] [Google Scholar]
- Kitamura S, Ode H, Nakashima M, et al. The APOBEC3C crystal structure and the interface for HIV-1 Vif binding. Nat Struct Mol Biol. 2012b;19:1005–1010. doi: 10.1038/nsmb.2378. [DOI] [PubMed] [Google Scholar]
- Klemm L, Duy C, Iacobucci I, et al. The B cell mutator AID promotes B lymphoid blast crisis and drug resistance in chronic myeloid leukemia. Cancer cell. 2009;16:232–245. doi: 10.1016/j.ccr.2009.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko TP, Lin JJ, Hu CY, Hsu YH, Wang AH, Liaw SH. Crystal structure of yeast cytosine deaminase. Insights into enzyme mechanism and evolution. The Journal of biological chemistry. 2003;278:19111–19117. doi: 10.1074/jbc.M300874200. [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Takaori-Kondo A, Shindo K, Abudu A, Fukunaga K, Uchiyama T. APOBEC3G targets specific virus species. Journal of virology. 2004;78:8238–8244. doi: 10.1128/JVI.78.15.8238-8244.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli RM, Abrams SR, Gajula KS, Maul RW, Gearhart PJ, Stivers JT. A portable hot spot recognition loop transfers sequence preferences from APOBEC family members to activation-induced cytidine deaminase. The Journal of biological chemistry. 2009;284:22898–22904. doi: 10.1074/jbc.M109.025536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli RM, Maul RW, Guminski AF, et al. Local sequence targeting in the AID/APOBEC family differentially impacts retroviral restriction and antibody diversification. The Journal of biological chemistry. 2010;285:40956–40964. doi: 10.1074/jbc.M110.177402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig R, Zhou Y, Elleder D, et al. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell. 2008;135:49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koning FA, Newman EN, Kim EY, Kunstman KJ, Wolinsky SM, Malim MH. Defining APOBEC3 expression patterns in human tissues and hematopoietic cell subsets. J Virol. 2009;83:9474–9485. doi: 10.1128/JVI.01089-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackey L, Demorest ZL, Land AM, Hultquist JF, Brown WL, Harris RS. APOBEC3B and AID have similar nuclear import mechanisms. Journal of molecular biology. 2012;419:301–314. doi: 10.1016/j.jmb.2012.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackey L, Law EK, Brown WL, Harris RS. Subcellular localization of the APOBEC3 proteins during mitosis and implications for genomic DNA deamination. Cell Cycle. 2013;12 doi: 10.4161/cc.23713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Land AM, Ball TB, Luo M, et al. Human immunodeficiency virus (HIV) type 1 proviral hypermutation correlates with CD4 count in HIV-infected women from Kenya. J Virol. 2008;82:8172–8182. doi: 10.1128/JVI.01115-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langlois MA, Beale RC, Conticello SG, Neuberger MS. Mutational comparison of the single-domained APOBEC3C and double-domained APOBEC3F/G anti-retroviral cytidine deaminases provides insight into their DNA target site specificities. Nucleic Acids Res. 2005;33:1913–1923. doi: 10.1093/nar/gki343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRue RS, Andrésdóttir V, Blanchard Y, et al. Guidelines for naming nonprimate APOBEC3 genes and proteins. J Virol. 2009;83:494–497. doi: 10.1128/JVI.01976-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRue RS, Jonsson SR, Silverstein KA, et al. The artiodactyl APOBEC3 innate immune repertoire shows evidence for a multi-functional domain organization that existed in the ancestor of placental mammals. BMC Mol Biol. 2008;9:104. doi: 10.1186/1471-2199-9-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecossier D, Bouchonnet F, Clavel F, Hance AJ. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science. 2003;300:1112. doi: 10.1126/science.1083338. [DOI] [PubMed] [Google Scholar]
- Lee TH, Coligan JE, Allan JS, McLane MF, Groopman JE, Essex M. A new HTLV-III/LAV protein encoded by a gene found in cytopathic retroviruses. Science. 1986;231:1546–1549. doi: 10.1126/science.3006243. [DOI] [PubMed] [Google Scholar]
- Lee YN, Bieniasz PD. Reconstitution of an infectious human endogenous retrovirus. PLoS pathogens. 2007;3:e10. doi: 10.1371/journal.ppat.0030010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YN, Malim MH, Bieniasz PD. Hypermutation of an ancient human retrovirus by APOBEC3G. Journal of virology. 2008;82:8762–8770. doi: 10.1128/JVI.00751-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levanon EY, Eisenberg E, Yelin R, et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nature biotechnology. 2004;22:1001–1005. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- Lever AML, Jeang K-T, Berkhout B. Recent advances in human retroviruses : principles of replication and pathogenesis : advances in retroviral research. World Scientific; Singapore ; Hackensack, NJ: 2010. [Google Scholar]
- Li JB, Levanon EY, Yoon JK, et al. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science. 2009;324:1210–1213. doi: 10.1126/science.1170995. [DOI] [PubMed] [Google Scholar]
- Li M, Shandilya SM, Carpenter MA, et al. First-in-class small molecule inhibitors of the single-strand DNA cytosine deaminase APOBEC3G. ACS chemical biology. 2012;7:506–517. doi: 10.1021/cb200440y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddament MT, Brown WL, Schumacher AJ, Harris RS. APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Curr Biol. 2004;14:1385–1391. doi: 10.1016/j.cub.2004.06.050. [DOI] [PubMed] [Google Scholar]
- Lindahl T. Suppression of spontaneous mutagenesis in human cells by DNA base excision-repair. Mutation research. 2000;462:129–135. doi: 10.1016/s1383-5742(00)00024-7. [DOI] [PubMed] [Google Scholar]
- Lochelt M, Romen F, Bastone P, et al. The antiretroviral activity of APOBEC3 is inhibited by the foamy virus accessory Bet protein. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:7982–7987. doi: 10.1073/pnas.0501445102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb LA, Essigmann JM, Kazazi F, Zhang J, Rose KD, Mullins JI. Lethal mutagenesis of HIV with mutagenic nucleoside analogs. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:1492–1497. doi: 10.1073/pnas.96.4.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longerich S, Basu U, Alt F, Storb U. AID in somatic hypermutation and class switch recombination. Current opinion in immunology. 2006;18:164–174. doi: 10.1016/j.coi.2006.01.008. [DOI] [PubMed] [Google Scholar]
- Losey HC, Ruthenburg AJ, Verdine GL. Crystal structure of Staphylococcus aureus tRNA adenosine deaminase TadA in complex with RNA. Nature structural & molecular biology. 2006;13:153–159. doi: 10.1038/nsmb1047. [DOI] [PubMed] [Google Scholar]
- Luo K, Liu B, Xiao Z, Yu Y, Yu X, Gorelick R, Yu XF. Amino-terminal region of the human immunodeficiency virus type 1 nucleocapsid is required for human APOBEC3G packaging. Journal of virology. 2004;78:11841–11852. doi: 10.1128/JVI.78.21.11841-11852.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksakova IA, Romanish MT, Gagnier L, Dunn CA, van de Lagemaat LN, Mager DL. Retroviral elements and their hosts: insertional mutagenesis in the mouse germ line. PLoS genetics. 2006;2:e2. doi: 10.1371/journal.pgen.0020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malim MH, Bieniasz PD. HIV Restriction Factors and Mechanisms of Evasion. Cold Spring Harbor perspectives in medicine. 2012;2:a006940. doi: 10.1101/cshperspect.a006940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malim MH, Emerman M. HIV-1 accessory proteins--ensuring viral survival in a hostile environment. Cell host & microbe. 2008;3:388–398. doi: 10.1016/j.chom.2008.04.008. [DOI] [PubMed] [Google Scholar]
- Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- Mariani R, Chen D, Schrofelbauer B, et al. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell. 2003;114:21–31. doi: 10.1016/s0092-8674(03)00515-4. [DOI] [PubMed] [Google Scholar]
- Marin M, Rose KM, Kozak SL, Kabat D. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat Med. 2003;9:1398–1403. doi: 10.1038/nm946. [DOI] [PubMed] [Google Scholar]
- Martin KL, Johnson M, D’Aquila RT. APOBEC3G complexes decrease human immunodeficiency virus type 1 production. Journal of virology. 2011;85:9314–9326. doi: 10.1128/JVI.00273-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbisa JL, Barr R, Thomas JA, et al. Human immunodeficiency virus type 1 cDNAs produced in the presence of APOBEC3G exhibit defects in plus-strand DNA transfer and integration. Journal of virology. 2007;81:7099–7110. doi: 10.1128/JVI.00272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehle A, Strack B, Ancuta P, Zhang C, McPike M, Gabuzda D. Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin-proteasome pathway. J Biol Chem. 2004;279:7792–7798. doi: 10.1074/jbc.M313093200. [DOI] [PubMed] [Google Scholar]
- Miyagi E, Opi S, Takeuchi H, Khan M, Goila-Gaur R, Kao S, Strebel K. Enzymatically active APOBEC3G is required for efficient inhibition of human immunodeficiency virus type 1. J Virol. 2007;81:13346–13353. doi: 10.1128/JVI.01361-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse DP, Aruscavage PJ, Bass BL. RNA hairpins in noncoding regions of human brain and Caenorhabditis elegans mRNA are edited by adenosine deaminases that act on RNA. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:7906–7911. doi: 10.1073/pnas.112704299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muckenfuss H, Hamdorf M, Held U, et al. APOBEC3 proteins inhibit human LINE-1 retrotransposition. The Journal of biological chemistry. 2006;281:22161–22172. doi: 10.1074/jbc.M601716200. [DOI] [PubMed] [Google Scholar]
- Münk C, Willemsen A, Bravo IG. An ancient history of gene duplications, fusions and losses in the evolution of APOBEC3 mutators in mammals. BMC evolutionary biology. 2012;12:71. doi: 10.1186/1471-2148-12-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- Muramatsu M, Sankaranand VS, Anant S, Sugai M, Kinoshita K, Davidson NO, Honjo T. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J Biol Chem. 1999;274:18470–18476. doi: 10.1074/jbc.274.26.18470. [DOI] [PubMed] [Google Scholar]
- Nathans R, Cao H, Sharova N, et al. Small-molecule inhibition of HIV-1 Vif. Nature biotechnology. 2008;26:1187–1192. doi: 10.1038/nbt.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman EN, Holmes RK, Craig HM, Klein KC, Lingappa JR, Malim MH, Sheehy AM. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Current biology : CB. 2005;15:166–170. doi: 10.1016/j.cub.2004.12.068. [DOI] [PubMed] [Google Scholar]
- Nik-Zainal S, Alexandrov LB, Wedge DC, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annual review of biochemistry. 2010;79:321–349. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowarski R, Britan-Rosich E, Shiloach T, Kotler M. Hypermutation by intersegmental transfer of APOBEC3G cytidine deaminase. Nature structural & molecular biology. 2008;15:1059–1066. doi: 10.1038/nsmb.1495. [DOI] [PubMed] [Google Scholar]
- OhAinle M, Kerns JA, Li MM, Malik HS, Emerman M. Antiretroelement activity of APOBEC3H was lost twice in recent human evolution. Cell Host Microbe. 2008;4:249–259. doi: 10.1016/j.chom.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki IM, Hiai H, Kakazu N, Yamada S, Muramatsu M, Kinoshita K, Honjo T. Constitutive expression of AID leads to tumorigenesis. The Journal of experimental medicine. 2003;197:1173–1181. doi: 10.1084/jem.20030275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson ME, Li M, Harris RS, Harki DA. Small-molecule APOBEC3G DNA cytosine deaminase inhibitors based on a 4-amino-1,2,4-triazole-3-thiol scaffold. ChemMedChem. 2013;8:112–117. doi: 10.1002/cmdc.201200411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opi S, Kao S, Goila-Gaur R, Khan MA, Miyagi E, Takeuchi H, Strebel K. Human immunodeficiency virus type 1 Vif inhibits packaging and antiviral activity of a degradation-resistant APOBEC3G variant. Journal of virology. 2007;81:8236–8246. doi: 10.1128/JVI.02694-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opi S, Takeuchi H, Kao S, et al. Monomeric APOBEC3G is catalytically active and has antiviral activity. Journal of virology. 2006;80:4673–4682. doi: 10.1128/JVI.80.10.4673-4682.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace C, Keller J, Nolan D, James I, Gaudieri S, Moore C, Mallal S. Population level analysis of human immunodeficiency virus type 1 hypermutation and its relationship with APOBEC3G and vif genetic variation. J Virol. 2006;80:9259–9269. doi: 10.1128/JVI.00888-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak V, Heidecker G, Pathak VK, Derse D. The role of amino-terminal sequences in cellular localization and antiviral activity of APOBEC3B. Journal of virology. 2011;85:8538–8547. doi: 10.1128/JVI.02645-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Duran P, de Yebenes VG, Ramiro AR. Oncogenic events triggered by AID, the adverse effect of antibody diversification. Carcinogenesis. 2007;28:2427–2433. doi: 10.1093/carcin/bgm201. [DOI] [PubMed] [Google Scholar]
- Petersen-Mahrt SK, Harris RS, Neuberger MS. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 2002;418:99–103. doi: 10.1038/nature00862. [DOI] [PubMed] [Google Scholar]
- Petit V, Guetard D, Renard M, Keriel A, Sitbon M, Wain-Hobson S, Vartanian JP. Murine APOBEC1 is a powerful mutator of retroviral and cellular RNA in vitro and in vivo. Journal of molecular biology. 2009;385:65–78. doi: 10.1016/j.jmb.2008.10.043. [DOI] [PubMed] [Google Scholar]
- Powell LM, Wallis SC, Pease RJ, Edwards YH, Knott TJ, Scott J. A novel form of tissue-specific RNA processing produces apolipoprotein-B48 in intestine. Cell. 1987;50:831–840. doi: 10.1016/0092-8674(87)90510-1. [DOI] [PubMed] [Google Scholar]
- Ramiro AR, Jankovic M, Eisenreich T, et al. AID is required for c-myc/IgH chromosome translocations in vivo. Cell. 2004;118:431–438. doi: 10.1016/j.cell.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Rathore A, Carpenter MA, Ikeda T, Li M, Law EK, Brown WL, Harris RS. A single amino acid governs the dinucleotide DNA cytosine deamination preference of the HIV restriction factor APOBEC3G. 2013 in preparation. [Google Scholar]
- Refsland EW, Hultquist JF, Harris RS. Endogenous Origins of HIV-1 G-to-A Hypermutation and Restriction in the Nonpermissive T Cell Line CEM2n. PLoS Pathog. 2012;8:e1002800. doi: 10.1371/journal.ppat.1002800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Refsland EW, Stenglein MD, Shindo K, Albin JS, Brown WL, Harris RS. Quantitative profiling of the full APOBEC3 mRNA repertoire in lymphocytes and tissues: implications for HIV-1 restriction. Nucleic Acids Res. 2010;38:4274–4284. doi: 10.1093/nar/gkq174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SA, Sterling J, Thompson C, et al. Clustered mutations in yeast and in human cancers can arise from damaged long single-strand DNA regions. Molecular cell. 2012;46:424–435. doi: 10.1016/j.molcel.2012.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogozin IB, Iyer LM, Liang L, et al. Evolution and diversification of lamprey antigen receptors: evidence for involvement of an AID-APOBEC family cytosine deaminase. Nat Immunol. 2007;8:647–656. doi: 10.1038/ni1463. [DOI] [PubMed] [Google Scholar]
- Rose KM, Marin M, Kozak SL, Kabat D. Regulated production and anti-HIV type 1 activities of cytidine deaminases APOBEC3B, 3F, and 3G. AIDS Res Hum Retroviruses. 2005;21:611–619. doi: 10.1089/aid.2005.21.611. [DOI] [PubMed] [Google Scholar]
- Russell RA, Wiegand HL, Moore MD, Schafer A, McClure MO, Cullen BR. Foamy virus Bet proteins function as novel inhibitors of the APOBEC3 family of innate antiretroviral defense factors. Journal of virology. 2005;79:8724–8731. doi: 10.1128/JVI.79.14.8724-8731.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santa-Marta M, da Silva FA, Fonseca AM, Goncalves J. HIV-1 Vif can directly inhibit apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like 3G-mediated cytidine deamination by using a single amino acid interaction and without protein degradation. The Journal of biological chemistry. 2005;280:8765–8775. doi: 10.1074/jbc.M409309200. [DOI] [PubMed] [Google Scholar]
- Sanville B, Dolan MA, Wollenberg K, et al. Adaptive evolution of Mus Apobec3 includes retroviral insertion and positive selection at two clusters of residues flanking the substrate groove. PLoS pathogens. 2010;6:e1000974. doi: 10.1371/journal.ppat.1000974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer SL, Emerman M, Malik HS. Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol. 2004;2:E275. doi: 10.1371/journal.pbio.0020275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer SL, Emerman M, Malik HS. Discordant evolution of the adjacent antiretroviral genes TRIM22 and TRIM5 in mammals. PLoS pathogens. 2007;3:e197. doi: 10.1371/journal.ppat.0030197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer A, Bogerd HP, Cullen BR. Specific packaging of APOBEC3G into HIV-1 virions is mediated by the nucleocapsid domain of the gag polyprotein precursor. Virology. 2004;328:163–168. doi: 10.1016/j.virol.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Schumacher AJ, Haché G, MacDuff DA, Brown WL, Harris RS. The DNA deaminase activity of human APOBEC3G is required for Ty1, MusD, and human immunodeficiency virus type 1 restriction. J Virol. 2008;82:2652–2660. doi: 10.1128/JVI.02391-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher AJ, Nissley DV, Harris RS. APOBEC3G hypermutates genomic DNA and inhibits Ty1 retrotransposition in yeast. Proc Natl Acad Sci U S A. 2005;102:9854–9859. doi: 10.1073/pnas.0501694102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severi F, Chicca A, Conticello SG. Analysis of reptilian APOBEC1 suggests that RNA editing may not be its ancestral function. Molecular biology and evolution. 2011;28:1125–1129. doi: 10.1093/molbev/msq338. [DOI] [PubMed] [Google Scholar]
- Shandilya SM, Nalam MN, Nalivaika EA, et al. Crystal structure of the APOBEC3G catalytic domain reveals potential oligomerization interfaces. Structure. 2010;18:28–38. doi: 10.1016/j.str.2009.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- Sheehy AM, Gaddis NC, Malim MH. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat Med. 2003;9:1404–1407. doi: 10.1038/nm945. [DOI] [PubMed] [Google Scholar]
- Shindo K, Li M, Gross PJ, et al. A Comparison of Two Single-Stranded DNA Binding Models by Mutational Analysis of APOBEC3G. Biology. 2012;1:260–276. doi: 10.3390/biology1020260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlyakhtenko LS, Lushnikov AY, Li M, Lackey L, Harris RS, Lyubchenko YL. Atomic force microscopy studies provide direct evidence for dimerization of the HIV restriction factor APOBEC3G. The Journal of biological chemistry. 2011;286:3387–3395. doi: 10.1074/jbc.M110.195685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlyakhtenko LS, Lushnikov AY, Miyagi A, Li M, Harris RS, Lyubchenko YL. Nanoscale structure and dynamics of ABOBEC3G complexes with single-stranded DNA. Biochemistry. 2012;51:6432–6440. doi: 10.1021/bi300733d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodroski J, Goh WC, Rosen C, Tartar A, Portetelle D, Burny A, Haseltine W. Replicative and cytopathic potential of HTLV-III/LAV with sor gene deletions. Science. 1986;231:1549–1553. doi: 10.1126/science.3006244. [DOI] [PubMed] [Google Scholar]
- Song C, Sutton L, Johnson ME, D’Aquila RT, Donahue JP. Signals in APOBEC3F N-terminal and C-terminal deaminase domains each contribute to encapsidation in HIV-1 virions and are both required for HIV-1 restriction. The Journal of biological chemistry. 2012;287:16965–16974. doi: 10.1074/jbc.M111.310839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soros VB, Yonemoto W, Greene WC. Newly synthesized APOBEC3G is incorporated into HIV virions, inhibited by HIV RNA, and subsequently activated by RNase H. PLoS pathogens. 2007;3:e15. doi: 10.1371/journal.ppat.0030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenglein MD, Burns MB, Li M, Lengyel J, Harris RS. APOBEC3 proteins mediate the clearance of foreign DNA from human cells. Nat Struct Mol Biol. 2010;17:222–229. doi: 10.1038/nsmb.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenglein MD, Harris RS. APOBEC3B and APOBEC3F inhibit L1 retrotransposition by a DNA deamination-independent mechanism. The Journal of biological chemistry. 2006;281:16837–16841. doi: 10.1074/jbc.M602367200. [DOI] [PubMed] [Google Scholar]
- Stenglein MD, Matsuo H, Harris RS. Two regions within the amino-terminal half of APOBEC3G cooperate to determine cytoplasmic localization. J Virol. 2008;82:9591–9599. doi: 10.1128/JVI.02471-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stopak K, de Noronha C, Yonemoto W, Greene WC. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol Cell. 2003;12:591–601. doi: 10.1016/s1097-2765(03)00353-8. [DOI] [PubMed] [Google Scholar]
- Strebel K, Daugherty D, Clouse K, Cohen D, Folks T, Martin MA. The HIV ‘A’ (sor) gene product is essential for virus infectivity. Nature. 1987;328:728–730. doi: 10.1038/328728a0. [DOI] [PubMed] [Google Scholar]
- Suspène R, Guetard D, Henry M, Sommer P, Wain-Hobson S, Vartanian JP. Extensive editing of both hepatitis B virus DNA strands by APOBEC3 cytidine deaminases in vitro and in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:8321–8326. doi: 10.1073/pnas.0408223102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svarovskaia ES, Xu H, Mbisa JL, et al. Human apolipoprotein B mRNA-editing enzyme-catalytic polypeptide-like 3G (APOBEC3G) is incorporated into HIV-1 virions through interactions with viral and nonviral RNAs. The Journal of biological chemistry. 2004;279:35822–35828. doi: 10.1074/jbc.M405761200. [DOI] [PubMed] [Google Scholar]
- Tareen SU, Sawyer SL, Malik HS, Emerman M. An expanded clade of rodent Trim5 genes. Virology. 2009;385:473–483. doi: 10.1016/j.virol.2008.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng B, Burant CF, Davidson NO. Molecular cloning of an apolipoprotein B messenger RNA editing protein. Science. 1993;260:1816–1819. doi: 10.1126/science.8511591. [DOI] [PubMed] [Google Scholar]
- Tian C, Wang T, Zhang W, Yu XF. Virion packaging determinants and reverse transcription of SRP RNA in HIV-1 particles. Nucleic acids research. 2007;35:7288–7302. doi: 10.1093/nar/gkm816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli P, Mangeat B, Jost S, Vianin S, Trono D. Inhibition of hepatitis B virus replication by APOBEC3G. Science. 2004;303:1829. doi: 10.1126/science.1092066. [DOI] [PubMed] [Google Scholar]
- Unniraman S, Zhou S, Schatz DG. Identification of an AID-independent pathway for chromosomal translocations between the Igh switch region and Myc. Nature immunology. 2004;5:1117–1123. doi: 10.1038/ni1127. [DOI] [PubMed] [Google Scholar]
- Vartanian JP, Meyerhans A, Sala M, Wain-Hobson S. G-->A hypermutation of the human immunodeficiency virus type 1 genome: evidence for dCTP pool imbalance during reverse transcription. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:3092–3096. doi: 10.1073/pnas.91.8.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Schwedler U, Song J, Aiken C, Trono D. Vif is crucial for human immunodeficiency virus type 1 proviral DNA synthesis in infected cells. Journal of virology. 1993;67:4945–4955. doi: 10.1128/jvi.67.8.4945-4955.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Rada C, Neuberger MS. Altering the spectrum of immunoglobulin V gene somatic hypermutation by modifying the active site of AID. The Journal of experimental medicine. 2010;207:141–153. doi: 10.1084/jem.20092238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Tian C, Zhang W, et al. 7SL RNA mediates virion packaging of the antiviral cytidine deaminase APOBEC3G. Journal of virology. 2007;81:13112–13124. doi: 10.1128/JVI.00892-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Abudu A, Son S, Dang Y, Venta PJ, Zheng YH. Analysis of human APOBEC3H haplotypes and anti-human immunodeficiency virus type 1 activity. J Virol. 2011;85:3142–3152. doi: 10.1128/JVI.02049-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedekind JE, Dance GS, Sowden MP, Smith HC. Messenger RNA editing in mammals: new members of the APOBEC family seeking roles in the family business. Trends in genetics : TIG. 2003;19:207–216. doi: 10.1016/S0168-9525(03)00054-4. [DOI] [PubMed] [Google Scholar]
- Wiegand HL, Doehle BP, Bogerd HP, Cullen BR. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 2004;23:2451–2458. doi: 10.1038/sj.emboj.7600246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissing S, Montano M, Garcia-Perez JL, Moran JV, Greene WC. Endogenous APOBEC3B restricts LINE-1 retrotransposition in transformed cells and human embryonic stem cells. J Biol Chem. 2011;286:36427–36437. doi: 10.1074/jbc.M111.251058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang S, Short SA, Wolfenden R, Carter CW., Jr The structure of the cytidine deaminase-product complex provides evidence for efficient proton transfer and ground-state destabilization. Biochemistry. 1997;36:4768–4774. doi: 10.1021/bi963091e. [DOI] [PubMed] [Google Scholar]
- Xie K, Sowden MP, Dance GS, Torelli AT, Smith HC, Wedekind JE. The structure of a yeast RNA-editing deaminase provides insight into the fold and function of activation-induced deaminase and APOBEC-1. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:8114–8119. doi: 10.1073/pnas.0400493101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka S, Balestra ME, Ferrell LD, et al. Apolipoprotein B mRNA-editing protein induces hepatocellular carcinoma and dysplasia in transgenic animals. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:8483–8487. doi: 10.1073/pnas.92.18.8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Chen K, Zhang C, Huang S, Zhang H. Virion-associated uracil DNA glycosylase-2 and apurinic/apyrimidinic endonuclease are involved in the degradation of APOBEC3G-edited nascent HIV-1 DNA. The Journal of biological chemistry. 2007;282:11667–11675. doi: 10.1074/jbc.M606864200. [DOI] [PubMed] [Google Scholar]
- Yeung ML, Houzet L, Yedavalli VS, Jeang KT. A genome-wide short hairpin RNA screening of jurkat T-cells for human proteins contributing to productive HIV-1 replication. J Biol Chem. 2009;284:19463–19473. doi: 10.1074/jbc.M109.010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Chen D, König R, Mariani R, Unutmaz D, Landau NR. APOBEC3B and APOBEC3C are potent inhibitors of simian immunodeficiency virus replication. J Biol Chem. 2004a;279:53379–53386. doi: 10.1074/jbc.M408802200. [DOI] [PubMed] [Google Scholar]
- Yu Q, Konig R, Pillai S, et al. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nature structural & molecular biology. 2004b;11:435–442. doi: 10.1038/nsmb758. [DOI] [PubMed] [Google Scholar]
- Yu X, Yu Y, Liu B, Luo K, Kong W, Mao P, Yu XF. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science. 2003;302:1056–1060. doi: 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- Zhang H, Yang B, Pomerantz RJ, Zhang C, Arunachalam SC, Gao L. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature. 2003;424:94–98. doi: 10.1038/nature01707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Webb DM. Rapid evolution of primate antiviral enzyme APOBEC3G. Human molecular genetics. 2004;13:1785–1791. doi: 10.1093/hmg/ddh183. [DOI] [PubMed] [Google Scholar]
- Zhang W, Du J, Evans SL, Yu Y, Yu XF. T-cell differentiation factor CBF-beta regulates HIV-1 Vif-mediated evasion of host restriction. Nature. 2012;481:376–379. doi: 10.1038/nature10718. [DOI] [PubMed] [Google Scholar]
- Zheng YH, Irwin D, Kurosu T, Tokunaga K, Sata T, Peterlin BM. Human APOBEC3F is another host factor that blocks human immunodeficiency virus type 1 replication. J Virol. 2004;78:6073–6076. doi: 10.1128/JVI.78.11.6073-6076.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Xu M, Huang Q, et al. Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe. 2008;4:495–504. doi: 10.1016/j.chom.2008.10.004. [DOI] [PubMed] [Google Scholar]
- Zrenner R, Stitt M, Sonnewald U, Boldt R. Pyrimidine and purine biosynthesis and degradation in plants. Annual review of plant biology. 2006;57:805–836. doi: 10.1146/annurev.arplant.57.032905.105421. [DOI] [PubMed] [Google Scholar]


