Abstract
The role of signaling in regulating cholesterol homeostasis is gradually becoming more widely recognized. Here, we explored how kinases and phosphorylation sites regulate the activity of the enzyme involved in the final step of cholesterol synthesis, 3β-hydroxysterol Δ24-reductase (DHCR24). Many factors are known to regulate DHCR24 transcriptionally, but little is known about its posttranslational regulation. We developed a system to specifically test human ectopic DHCR24 activity in a model cell-line (Chinese hamster ovary-7) using siRNA targeted only to hamster DHCR24, thus ensuring that all activity could be attributed to the human enzyme. We determined the effect of known phosphorylation sites and found that mutating certain residues (T110, Y299, and Y507) inhibited DHCR24 activity. In addition, inhibitors of protein kinase C ablated DHCR24 activity, although not through a known phosphorylation site. Our data indicate a novel mechanism whereby DHCR24 activity is regulated by signaling.
Keywords: 3β-hydroxysterol Δ24-reductase, phosphorylation, bisindolylmaleimide I, protein kinase C, regulation, desmosterol, gas chromatography-mass spectrometry
Cholesterol is a vital raw material for the cell, which is toxic in excess and leads to a number of diseases. Thus, cellular cholesterol levels are tightly regulated through a balance of synthesis, uptake, and efflux. The final step in the Bloch cholesterol synthetic pathway is catalyzed by the enzyme 3β-hydroxysterol Δ24-reductase (DHCR24). DHCR24 is a 60.1 kDa endoplasmic reticulum membrane-bound oxidoreductase that converts desmosterol to cholesterol by saturating the C-24,25 double bond in the side-chain (1). DHCR24 activity is strictly dependent on nicotinamide adenine dinucleotide phosphate (NADPH) as the reducing agent (1), although no consensus sequence for NADPH binding is predicted. However, DHCR24 does contain a highly conserved flavin adenine dinucleotide (FAD) binding domain [amino acids 111-203] (2), characteristic of the well-defined family of FAD-dependent oxidoreductases (3); with reduction of desmosterol dependent on FAD, suggesting functionality of the conserved domain (1). Additionally, a substrate binding domain was predicted in DHCR24 (amino acids 236-491) by homology modeling on an FAD-dependent oxidoreductase, plant cytokinin dehydrogenase (4).
DHCR24 is involved in multiple cellular functions. Related to its role in cholesterol biosynthesis, DHCR24 modulates lipid raft formation, thus facilitating signal transduction and trafficking (5–7). It is involved in regulating oxidative stress (8, 9), and is neuroprotective (5, 10, 11), anti-apoptotic (8, 10), and anti-inflammatory (12, 13). DHCR24 is also implicated in many diseases, such as cardiovascular diseases (12, 14), cancers (e.g., prostate cancer) (15), and hepatitis C (16). Moreover, mutations in DHCR24 underlie the rare autosomal recessive disease, desmosterolosis, whereby patients have elevated desmosterol and lowered cholesterol, resulting in multiple congenital anomalies (17). Specifically, seven missense mutations have been described in desmosterolosis: R94H, R103C, E191K, N294T, K306N, Y471S, E480K (1, 18–21).
Like many cholesterol synthetic genes, DHCR24 is transcriptionally regulated by sterols via the sterol-regulatory element-binding protein-2 transcription factor (22), and we recently identified two sterol-regulatory elements and nuclear factor Y sites in the human DHCR24 promoter that mediate this regulation (23). Moreover, DHCR24 is regulated at the transcriptional level by sex steroids (24, 25), adrenocorticotropic hormone (26), thyroid hormone (27), and xenobiotics (28). Epigenetic factors such as methylation and acetylation also regulate DHCR24 expression (29). In contrast, relatively little is known about the posttranslational regulation of DHCR24 activity. We recently found that the oxysterol regulator, 24(S),25-epoxycholesterol, inhibits DHCR24 activity with potent effects on cellular cholesterol levels (30). Plant sterols (31) and progesterone and similar progestins (32, 33) similarly inhibit DHCR24 activity. However, the role of posttranslational modifications in DHCR24 regulation, such as phosphorylation, has yet to be investigated. Large-scale proteomic studies have identified phosphorylation sites on DHCR24; T110 (34) and Y321 (35), and there is evidence for additional phosphorylation sites identified by large-scale unpublished proteomics studies [PhosphoSitePlus (36)]. The function of these phosphorylation sites and the kinases/phosphatases involved are currently unknown due to a lack of mechanistic studies.
Cell signaling and phosphorylation is a major mode of regulating cellular processes, and is implicated in various facets of cholesterol homeostasis [reviewed in (37)]. Because DHCR24 catalyzes the ultimate step in cholesterol synthesis, we hypothesized that it is a likely target for feedback regulation via signaling, notably phosphorylation, as this would provide a rapid means of switching cholesterol synthesis on or off. In this study, we examined the effect of known phosphorylation sites on DHCR24 expression and activity. Our data demonstrate that particular putative phosphorylated residues (T110, Y299, and Y507) have significant effects on DHCR24 activity. In addition, inhibiting a major serine/threonine kinase, protein kinase C (PKC), ablated DHCR24 activity, although this effect was independent of T110, the only published serine/threonine phosphorylation site. Together, these results indicate a novel regulatory mechanism for DHCR24 by kinases and phosphorylation.
MATERIALS AND METHODS
Materials
Chinese hamster ovary (CHO)-7 cells (38) were a generous gift from Drs. Michael S. Brown and Joseph L. Goldstein (University of Texas Southwestern Medical Center, Dallas, TX). The Flp-In-CHO cell line (for stable-cell generation) was generated from CHO-7 cells by Gorjana Mitic. HeLaT cells were generously donated by Dr. Noel Whitaker (The University of New South Wales, Sydney, NSW, Australia). SensiMix SYBR No-Rox was from Bioline (London, UK). Fetal calf serum (FCS) was from Bovogen (East Keilor, VIC, Australia). Amersham Hyperfilm ECL was from GE Healthcare (Buckinghamshire, UK). Horseradish peroxidase-conjugated AffiniPure donkey anti-mouse IgG antibody was from Jackson ImmunoResearch Laboratories, Inc. (West Grove, PA). Anti-V5 antibody, Dulbecco's Modified Eagle's Medium-Ham's Nutrient Mixture F12 (1:1) (DMEM/F12), Lipofectamine RNAiMAX transfection reagent, newborn bovine serum (NBS), Roswell Park Memorial Institute 1640 medium (RPMI), and Superscript III reverse transcriptase were from Life Technologies (Carlsbad, CA). Akt inhibitor VIII was from Merck (Darmstadt, Germany). Immobilon Western chemiluminescent horseradish peroxidase substrate enhanced chemiluminescent detection system was from Millipore (Billerica, MA). Phusion high-fidelity buffer, Phusion Hot Start II high-fidelity polymerase, and purified BSA were from New England Biolabs (Ipswich, MA). [14C]acetate sodium salt (specific radioactivity: 55.3 mCi/mmol, 2.046 GBq/mmol) was from Perkin Elmer (Waltham, MA). α-Tubulin antibody, Bis-Tris, bisindolylmaleimide I (BIM), cycloheximide, compactin (mevastatin), desmosterol, H89, primers, mevalonate, MG132 (Z-Leu-Leu-Leu-al), MOPS, N-O-bis-(trimethylsilyl) trifluoroacetamide containing 1% trimethylchlorosilane (BSTFA), protease inhibitor cocktail, Ro-318220, siRNA, sodium bisulfite, zinc (II) nitrate, and Tri reagent were from Sigma-Aldrich (St. Louis, MO). [2H6]desmosterol was from Avanti Polar Lipids (Alabaster, AL). Phos-tag acrylamide pendant [10 mg supplied in 100 μl 3% (v/v) methanol] was from Wako Pure Chemical Industries (Osaka, Japan). Lipoprotein-deficient serum (LPDS) was prepared from heat-inactivated NBS as previously described (39).
Cell culture and treatments
CHO-7 cells were maintained in 5% (v/v) LPDS/DMEM/F12. CHO-7 stable cell-lines were generated and maintained in 5% (v/v) LPDS/DMEM/F12 supplemented with 150 μg/ml hygromycin B. HeLaT cells were maintained in 10% (v/v) FCS/RPMI. Where there were treatments, cells were treated in fresh media with various test agents [added in dimethyl sulfoxide or ethanol; desmosterol was complexed to methyl-β-cyclodextrin (CD) according to (40)], as indicated in the figure legends. Within an experiment, the final concentrations of solvent were kept constant between conditions and did not exceed 0.2% (v/v).
Generation of CHO overexpressing cell-lines
Primers were designed to perform megaprimed site-directed mutagenesis (41) on a plasmid encoding the human DHCR24 sequence (accession number NM_014762.3) containing a FRT recombination site (pcDNA5-FRT-DHCR24-V5) (30) to create plasmids encoding DHCR24 phosphorylation mutants T110A, Y299F, Y300F, Y321F, Y507F, and a T110 phosphomimetic (T110E). These plasmids were then used to generate stable cell-lines using the CHO-7 Flp-In system, as described in (30). DHCR24 protein expression was screened by Western blotting with the anti-V5 antibody, and stable cells expressing similar levels of DHCR24 were selected and characterized.
The empty vector stable cell-line (CHO-EV) was prepared as described (42), and the human wild-type DHCR24 overexpressing cell-line (CHO-DHCR24-WT) was prepared as described (30).
siRNA transfection
Cells were transfected with 25 nM control or hamster-specific DHCR24 siRNA (target sequence: GAGAGCCACGTGTGAAGCA; designed by Sigma-Aldrich) for 24 h using Lipofectamine RNAiMAX transfection reagent according to the manufacturer's instructions. The cells were washed with phosphate-buffered saline (PBS), and then refed fresh media with [14C]acetate labeling for argentation-thin-layer chromatography (Arg-TLC).
Quantitative real-time RT-PCR
Cells were seeded in triplicate wells per condition as described. Total RNA (1 μg) was reverse transcribed into cDNA using the SuperScript III First Strand cDNA synthesis kit. mRNA levels were determined by quantitative real-time reverse transcription PCR (qRT-PCR) using the Corbett Rotorgene 3000 (Corbett Life Sciences, Sydney, NSW, Australia) and analyzed using Rotor Gene version 6.0 (Build 27) (Qiagen, Doncaster, VIC, Australia). Primers were used to amplify the cDNA of hamster or human DHCR24 (43) and the housekeeping control, porphobilinogen deaminase (PBGD) (44, 45). Changes in DHCR24 gene expression levels were normalized to PBGD for each sample by the ΔΔCt method, and made relative to the CHO-7 cell-line, which was set to one.
Western blotting
After treatment, cells were harvested in 10% (w/v) SDS supplemented with 5% (v/v) protease inhibitor cocktail. Equal amounts of protein were mixed with loading buffer (final concentration: 50 mM Tris-HCl (pH 6.8), 2% (w/v) SDS, 5% (v/v) glycerol, 0.04% (w/v) bromophenol blue, and 1% (v/v) β-mercaptoethanol), boiled for 5 min, and subjected to SDS-PAGE. After electrophoresis, the proteins were transferred to a nitrocellulose membrane, blocked for 1 h, incubated with primary anti-V5 antibody (1:5,000) or anti-α-tubulin (1:200,000), and then further incubated with secondary antibody (1:20,000). The antibodies were visualized by the enhanced chemiluminescent detection system, and membranes were exposed to Hyperfilm. Proteins were identified by their predicted molecular mass (α-tubulin, 50 kDa; DHCR24, 60 kDa). Protein band intensities from Western blots were quantified by densitometry using ImageJ (version 1.47t).
Cholesterol synthesis assay
Cholesterol and desmosterol synthesis were measured as an indicator of DHCR24 activity using Arg-TLC as described previously (30). The relative intensities of bands were quantified using Sciencelab ImageGauge 4.0 software (Fujifilm).
Phos-tag SDS-PAGE
Phosphorylated proteins were visualized using the phos-tag SDS-PAGE method described in (46), with modifications. Cells were seeded in 60 mm or 100 mm dishes, and treated with various test agents, as indicated in the figure legends. After treatments, cells were washed twice with ice-cold PBS. The cells were scraped in PBS, then pelleted and lysed in 100 μl modified RIPA buffer [50 mM Tris-HCl (pH 7.3), 150 mM NaCl, 1% sodium deoxycholate, and 1% Triton X-100 (47)]. The lysates were passed through a 21 gauge needle 20 times, and centrifuged at 20,000 g at 4°C for 15 min. Equal amounts of protein were mixed with 0.25 volume 5× loading buffer and boiled for 5 min before subjecting to phos-tag SDS-PAGE (7.5% separation gel containing Zn2+-phos-tag complex and a 4% stacking gel), transferred to nitrocellulose membranes, and Western blotted as indicated in the figure legends. Purified BSA (0.5 μg) was prepared in the same volume of modified RIPA buffer containing loading buffer, and served as a molecular mass marker (66 kDa).
GC-MS
Cells were treated as indicated in the figure legends with or without 1 μg/ml [2H6]desmosterol/CD for 4 h. Cells were harvested and lipid extracts were prepared as described in (30).
Sterols were derivatized with BSTFA for 1 h at 60°C. Derivatized samples were analyzed using a Thermo Trace gas chromatograph coupled with a Thermo DSQII mass spectrometer and Thermo Triplus autosampler (Thermo Fisher Scientific, Waltham, MA). Samples (1 μl) were injected via a heated (290°C) splitless inlet into an Rxi-5Sil mass spectrometer with Integra-Guard, 30 m × 0.25 mm, df 0.25 μm film thickness, capillary GC column (Restek, Waltham, MA). The column oven was initially held at 80°C for 1 min, then heated to 260°C at 80°C min−1, then to 280°C at 10°C min−1, and then to 295°C at 2°C min−1. Finally, the oven was increased to 305°C at 10°C min−1 and held for 1 min. Helium was used as a carrier gas at a constant flow (1.3 ml min−1, with vacuum compensation on).
MS conditions were electron energy 70 eV, ion source temperature 200°C, and transfer line temperature 305°C. The emission current was set to 130 μA and the detector gain to 3.0 × 105. Samples were analyzed either in scan mode (35–520 Da, 2.5 scans s−1) to obtain mass spectra for peak identification, or in single ion monitoring (SIM) mode to measure DHCR24 activity. Method details and the m/z values of SIM ions monitored for 5α-cholestane, [2H6]cholesterol, cholesterol, [2H6]desmosterol, and desmosterol are listed in supplementary Table I. Thermo Xcalibur software (version 2.1.0.1140) was used to acquire and process the data. 5α-Cholestane (0.1 μg added during cell harvesting) was used as an internal standard. Peak identification was based on comparison of retention times with those of standards and comparison of mass spectra with the Wiley 9/NIST 2011 combined mass spectral library.
Statistical analysis
Statistical differences were determined by the Student's paired t-test (two-tailed), where P values of <0.01 (**) were considered statistically significant. Statistical analysis was performed in Fig. 5D using a two-way ANOVA with repeated measures (stability over time of WT vs. Y299F or Y300F).
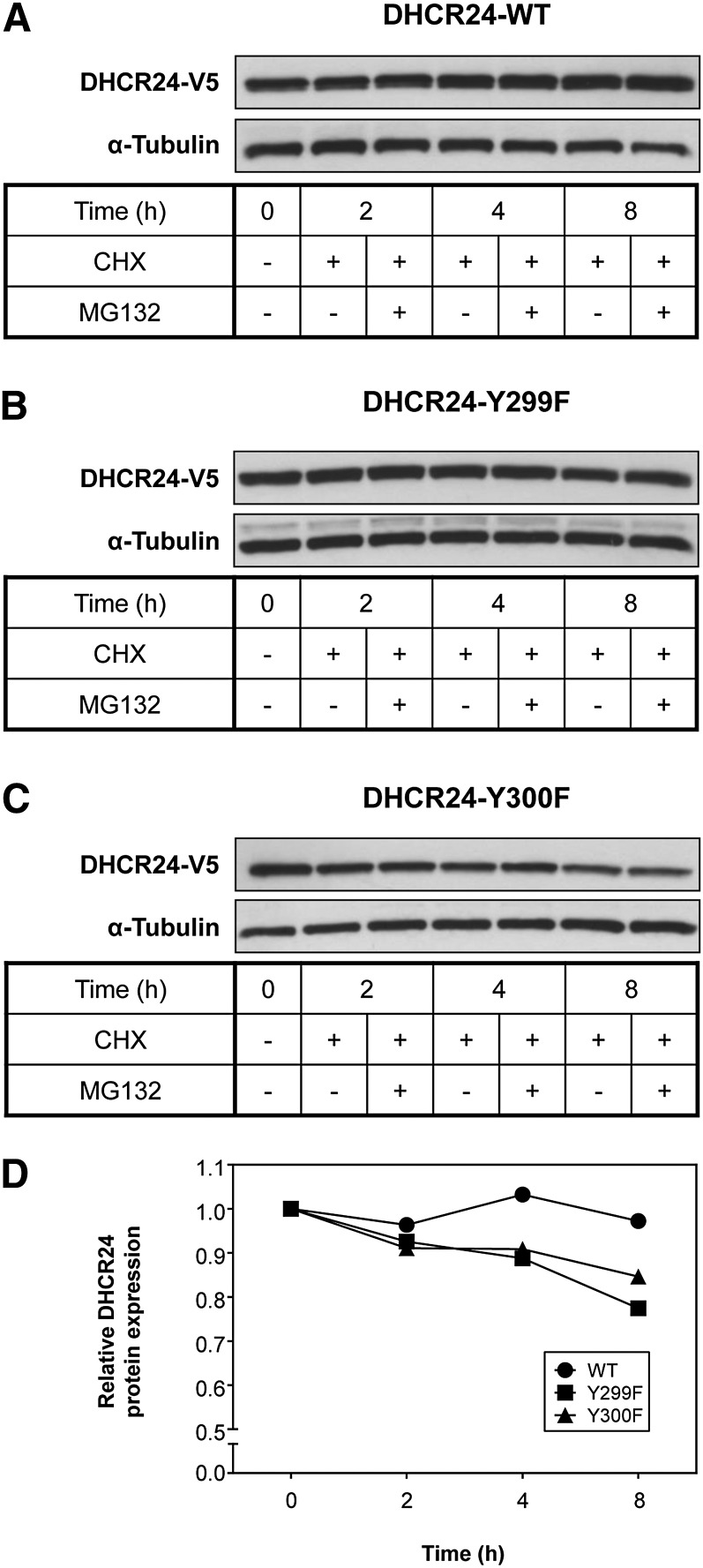
Fig. 5.
The effects of mutating DHCR24 at Y299 or Y300 on protein stability. A–C: CHO-7 cells stably overexpressing DHCR24 (WT, Y299F, and Y300F) were treated with cycloheximide (CHX) (10 μg/ml) in the presence (+) or absence (−) of MG132 (10 μM) for 0–8 h. Whole cell lysates were subjected to SDS-PAGE and Western blotting for DHCR24 (V5) and α-tubulin. Western blots are representative of four separate experiments. D: Line graph represents the mean of the −MG132 condition from (A) to (C), and indicates stability of DHCR24 over an 8 h period.
RESULTS
PKC inhibition decreases DHCR24 activity
To examine the effect of common protein kinases on DHCR24 activity, we used inhibitors of protein kinase A (PKA) [H89 (48)], protein kinase B (PKB) [also called Akt; AktiVIII (49)], and protein kinase C (PKC) [BIM (50) and Ro-318220 (51)]. CHO-7 cells stably transfected with epitope-tagged human DHCR24 (30) were radiolabeled with [14C]acetate, which feeds into the beginning of the cholesterol synthetic pathway. Cholesterol and desmosterol were resolved by Arg-TLC, and radioactive bands were visualized by phosphorimaging. The cholesterol to desmosterol ratio was used as an indicator of DHCR24 activity (30).
As shown previously (30), treatment with unlabeled desmosterol/CD decreased the cholesterol to desmosterol ratio, demonstrating competitive inhibition of DHCR24 enzyme. Inhibition of PKA (H89) or PKB (AktiVIII) did not have a selective effect on DHCR24 activity (Fig. 1A), although H89 affected cholesterol synthesis on a global level (supplementary Fig. I). In contrast, PKC inhibitors, BIM and Ro-318220, reduced cholesterol levels and accumulated desmosterol within 4 h, indicating decreased DHCR24 activity. BIM had the most robust effect, and abated DHCR24 activity in a dose-dependent manner (Fig. 1B). Conversely, PKC stimulation with phorbol myristate acetate increased cholesterol synthesis by ∼30% (±5%, P < 0.01, Student's t-test). Because inhibition did not affect DHCR24 protein levels (Fig. 1A), these results suggest that PKC may influence DHCR24 activity through modulating its phosphorylation state.
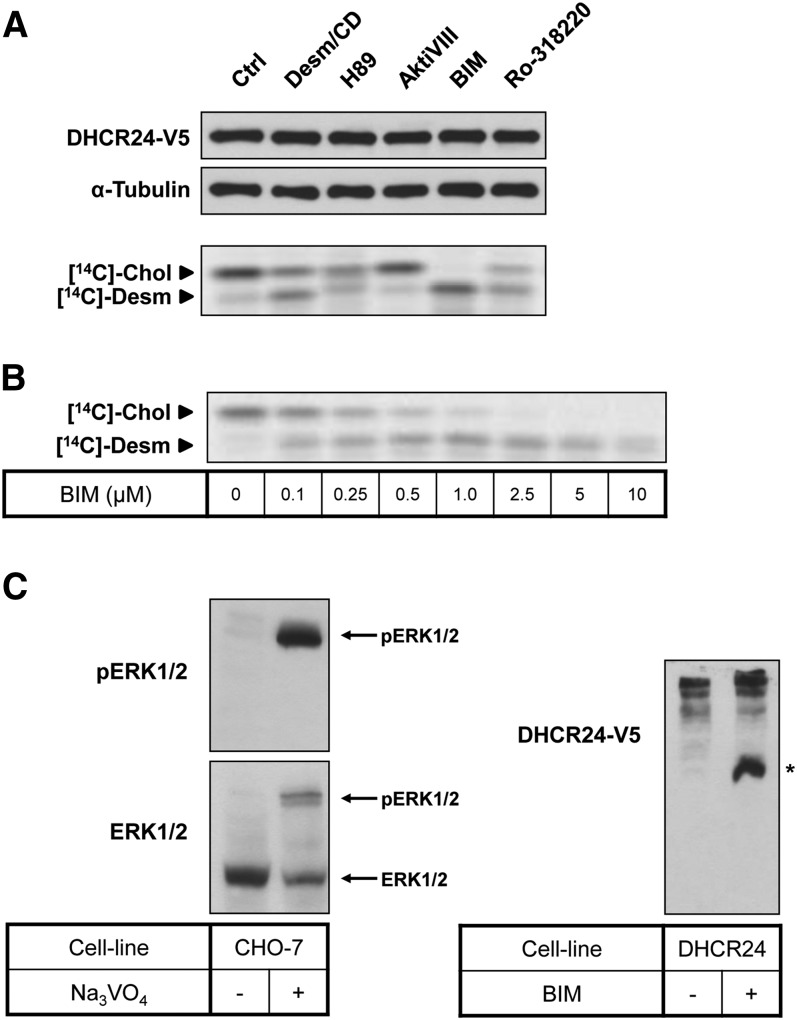
Fig. 1.
Effect of kinase inhibitors on DHCR24 activity. A: CHO-7 cells stably overexpressing DHCR24-V5 were seeded and treated for 4 h with desmosterol/CD (Desm/CD) (2 μg/ml), H89 (10 μM), Akt inhibitor VIII (AktiVIII) (5 μM), BIM (5 μM), or Ro-318220 (10 μM). B: CHO-7 cells stably overexpressing DHCR24-V5 were seeded, pretreated in NBS, and then treated for 4 h with BIM at the indicated concentrations. A, B: Cells were either harvested for Western blotting, or radiolabeled with [14C]acetate during the treatment for Arg-TLC. For Western blotting: whole cell lysates were subjected to SDS-PAGE and Western blotting for DHCR24 (V5) and α-tubulin. Western blots are representative of at least two separate experiments. For Arg-TLC: lipid extracts were separated by Arg-TLC, and bands corresponding to cholesterol (Chol) and desmosterol (Desm) were visualized by phosphorimaging. Arg-TLC is representative of at least two separate experiments. C: Left panel: CHO-7 cells were seeded and then treated with (+) or without (−) Na3VO4 for 4 h. Right panel: CHO-7 cells stably overexpressing DHCR24-WT were seeded and treated with or without BIM (5 μM) for 4 h. Whole cell lysates were subjected to 7.5% SDS-PAGE supplemented with Zn2+ phos-tag (50 μM), and Western blotted for pERK1/2 and total ERK1/2 (left panel) or DHCR24 (V5; right panel). * denotes ∼66 kDa where BSA migrates, and close to the calculated molecular mass of DHCR24, 60 kDa. These results were observed at least twice.
To confirm that DHCR24 was phosphorylated in our cell system, we employed the phos-tag SDS-PAGE method (46). This system utilizes gels supplemented with phos-tag acrylamide-bound metal complexes, which have affinity for phosphate groups on proteins. This, therefore, slows the migration of phosphorylated species during electrophoresis, producing a mobility shift. As a positive control, we treated cells with a phosphatase inhibitor (Na3VO4) and determined ERK1/2 phosphorylation. Na3VO4 produced two distinct bands when probed with total ERK1/2 antibody, with the bottom band being the nonphosphorylated ERK1/2, as confirmed with phosphorylated ERK1/2 (pERK1/2) antibody (Fig. 1C, left panel). In CHO-DHCR24-WT cells, nontreated cells resulted in multiple slower migrating DHCR24 bands (Fig. 1C, right panel). Interestingly, BIM treatment produced a faster migrating band, suggesting that DHCR24 is constitutively phosphorylated at more than one site.
PKC inhibition results in desmosterol accumulation
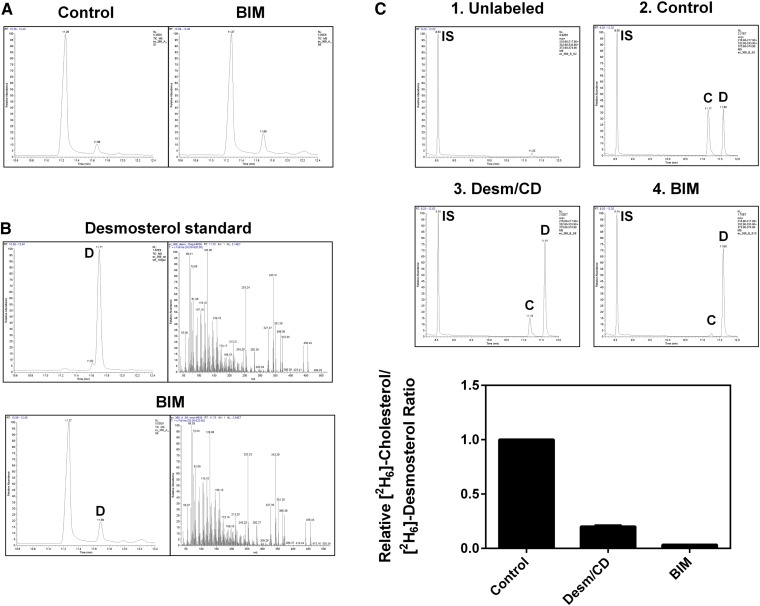
To confirm the identity of the desmosterol band accumulating with BIM treatment using Arg-TLC, samples were reanalyzed using GC-MS (Fig. 2). Two peaks were identified, where the major peak was cholesterol and a second minor peak, which increased with BIM treatment (Fig. 2A), was identified as desmosterol based on the mass spectra (Fig. 2B). To measure DHCR24 activity, samples were treated with BIM and [2H6]desmosterol. As observed with Arg-TLC (Fig. 1A), competition from unlabeled desmosterol inhibited the conversion of [2H6]desmosterol to [2H6]cholesterol, resulting in an increase in [2H6]desmosterol (Fig. 2C). BIM treatment had similar effects, strongly inhibiting DHCR24 activity, as evidenced by the [2H6]cholesterol to [2H6]desmosterol ratio (Fig. 2C).
Fig. 2.
BIM inhibits DHCR24 activity. CHO-7 cells stably overexpressing DHCR24-V5 were treated with desmosterol/CD (Desm/CD) (2 μg/ml) or BIM (5 μM) for 4 h. Lipid extracts and, for (B), a desmosterol standard, were analyzed by GC-MS. A, B: Show total ion chromatograms, and (B) also shows mass spectra obtained for the peak labeled ‘D’ and used to verify its identity as desmosterol. C: Cells were labeled with [2H6]desmosterol/CD (1 μg/ml) during the treatment, then SIM ion traces were used to quantify [2H6]cholesterol and [2H6]desmosterol relative to the internal standard (IS), 5α-cholestane. Data are mean + SEM (C) from at least three separate experiments.
DHCR24-T110 phosphorylation mutant decreases DHCR24 activity
From the panel of kinase inhibitors tested, only PKC inhibitors affected DHCR24 activity. Because PKC phosphorylates serine and threonine residues, it is possible that BIM inhibits DHCR24 activity through the only known serine/threonine phosphorylation site, T110 (34). Thus, we mutated this to alanine (T110A; mutant) or glutamic acid (T110E; phosphomimetic), and stably overexpressed these constructs in CHO-7 cells, as we did previously with the generation of the CHO-DHCR24-WT cells (30). DHCR24 mRNA and protein levels between the CHO-DHCR24 cell-lines were comparable (supplementary Fig. II).
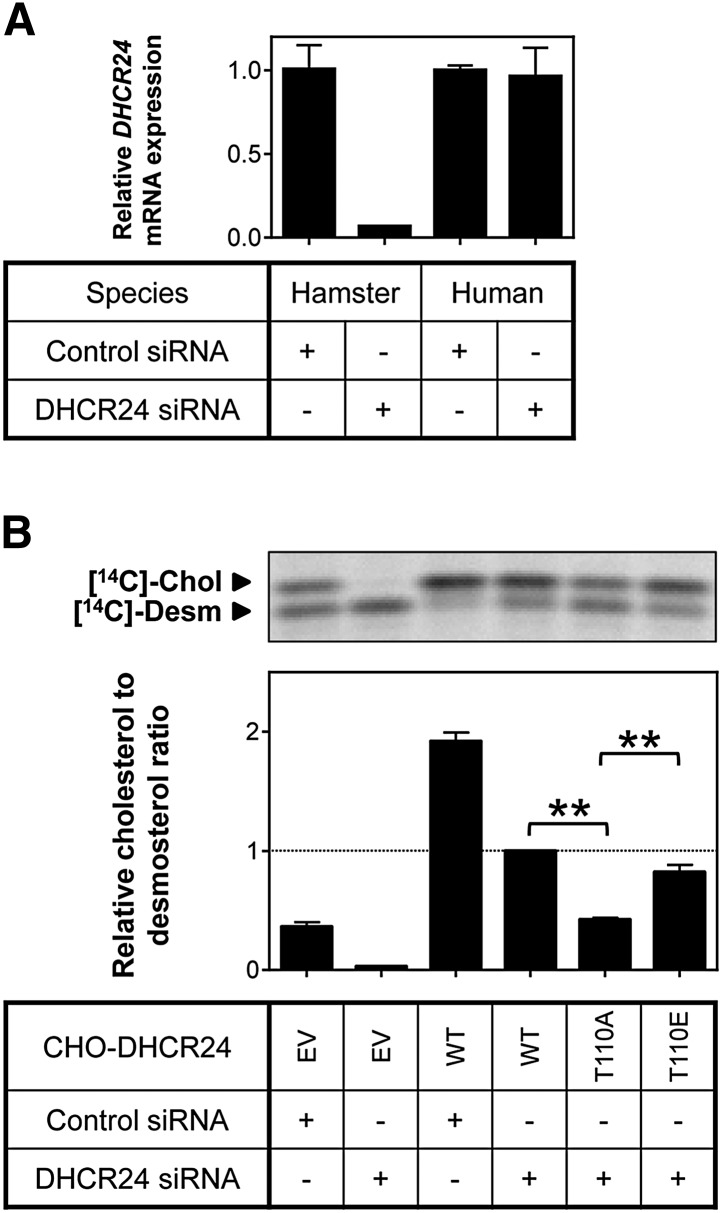
To measure the specific effects of the mutants on human DHCR24, endogenous hamster DHCR24 expression was reduced using RNA interference. Transfection of hamster-specific DHCR24 siRNA ablated endogenous hamster DHCR24 mRNA without affecting human DHCR24 expression (Fig. 3A). Thus, transfecting CHO-DHCR24 cells with hamster siRNA reduced background activity in our stable cells, with measured DHCR24 activity attributed to the stably expressed human DHCR24 only.
Fig. 3.
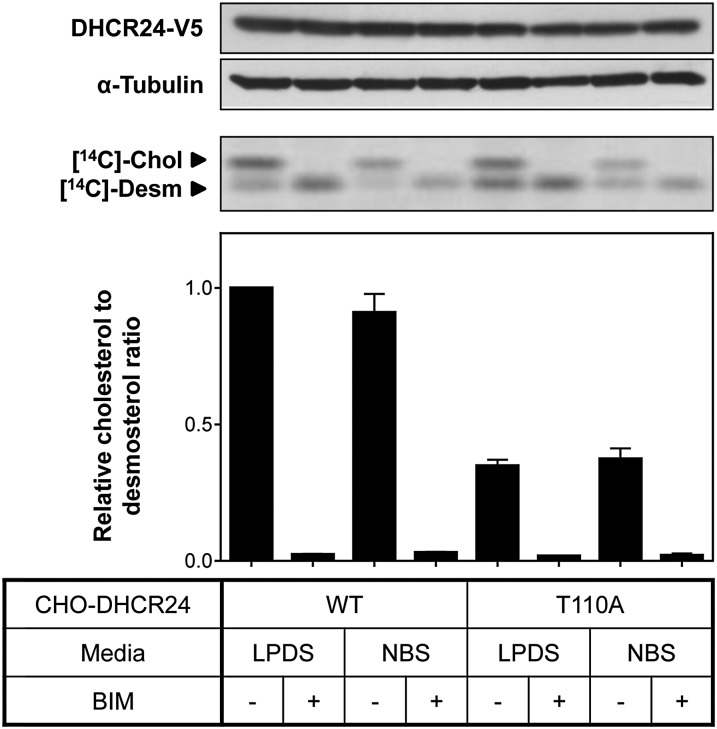
Effect of T110 mutants on DHCR24 activity. A: CHO-7 (hamster) and HeLaT (human) cells were transfected with control or hamster-specific DHCR24 siRNA (25 nM) for 24 h. The cells were washed with PBS and refed fresh media overnight. Total RNA was harvested and reverse transcribed to cDNA, and gene expression levels of DHCR24 were quantified using qRT-PCR, and normalized to the housekeeping gene, PBGD. Data are presented relative to the hamster control siRNA condition, which has been set to one, and are the mean + SD of triplicate cultures per condition. B: CHO-7 cells stably overexpressing EV or DHCR24 (WT, T110A, and T110E) were seeded and transfected with control or hamster-specific DHCR24 siRNA (25 nM) for 24 h. The cells were washed with PBS and refed fresh media overnight, and then radiolabeled with [14C]acetate in fresh media. Lipid extracts were separated by Arg-TLC, and bands corresponding to cholesterol (Chol) and desmosterol (Desm) were visualized by phosphorimaging. Arg-TLC is representative of four separate experiments, and the relative cholesterol to desmosterol ratio was quantified by densitometry, normalized to protein expression (supplementary Fig. IIB), and then normalized to the DHCR24 siRNA transfected CHO-DHCR24-WT condition, which has been set to one. **P < 0.01. Data are mean + SEM (n = 4).
Using this approach, we examined whether DHCR24-T110 mutants affect DHCR24 activity. As indicated in Fig. 3B, transfecting DHCR24 siRNA into empty vector (EV) cells ablated endogenous DHCR24 activity. Mutating T110 so that it cannot be phosphorylated (T110A), significantly decreased DHCR24 activity by ∼60% (Fig. 3B). Furthermore, DHCR24 activity of the phosphomimetic (T110E) mutant was increased relative to T110A, with activity not statistically different from WT cells. These results suggest that phosphorylation at T110 modulates DHCR24 activity.
PKC inhibition decreases DHCR24 activity independently of T110
Next, we determined whether BIM inhibition of DHCR24 activity occurs at T110. Cells were treated in low (LPDS) or high (NBS) cholesterol conditions to exclude any potential effects of cholesterol status. BIM had similar effects in DHCR24-T110A as WT cells: a decrease in DHCR24 activity, with no effect on DHCR24 protein levels (Fig. 4). This indicates that BIM targets a different residue in DHCR24, or that the effect is indirect.
Fig. 4.
BIM does not affect DHCR24-T110A activity. CHO-7 cells stably overexpressing DHCR24 (WT, T110A) were pretreated in either 5% (v/v) LPDS/DMEM/F12 or 5% (v/v) NBS/DMEM/F12 overnight, and treated in fresh media, with (+) or without (−) BIM (5 μM), for 4 h. Cells were either harvested for Western blotting, or radiolabeled with [14C]acetate during the treatment for Arg-TLC. Whole cell lysates were subjected to SDS-PAGE and Western blotting for DHCR24 (V5) and α-tubulin. Western blots are representative of two separate experiments. Lipid extracts were separated by Arg-TLC, and bands corresponding to cholesterol (Chol) and desmosterol (Desm) were visualized by phosphorimaging. Arg-TLC is representative of four separate experiments, and the relative cholesterol to desmosterol ratio was quantified by densitometry and normalized to the nontreated control CHO-DHCR24-WT condition (in LPDS), which has been set to one. Data are mean + SEM (n = 4).
Other DHCR24 phosphorylation sites
Another published phosphorylation site is Y321 (35) and PhosphoSitePlus (36) give evidence for three other phosphorylated tyrosine residues, Y299, Y300, and Y507. Thus, we similarly generated DHCR24 stable cell-lines containing these mutants, and characterized mRNA and protein expression. DHCR24 protein correlated with mRNA expression (supplementary Fig. III), with the exception of the DHCR24-Y299F and DHCR24-Y300F mutants. Compared with the DHCR24-WT, the DHCR24-Y299F stable cells contained lower mRNA expression while having comparable protein levels. Conversely, the DHCR24-Y300F stable cells had similar levels of mRNA to the DHCR24-WT stable cells, while containing less protein.
Examining the protein stability of DHCR24-Y299F and DHCR24-Y300F
Y299 and Y300 are neighboring residues, yet phosphorylation mutants at these sites have vastly different effects on DHCR24 mRNA and protein expression, suggesting that mutating DHCR24 at these residues may alter protein stability. In addition, there is evidence for DHCR24 ubiquitination (36), including at K301 (52), which is adjacent to Y299 and Y300. Therefore, we compared DHCR24 protein stability in DHCR24-WT, -Y299F, and -Y300F stable cells by inhibiting protein synthesis (using cycloheximide) and proteasomal degradation (using MG132). Both DHCR24-Y299F and DHCR24-Y300F were destabilized during the 8 h treatments, though only DHCR24-Y299F was statistically different compared with DHCR24-WT (P < 0.01); DHCR24-Y300F displayed a nonsignificant trend (P = 0.12) (Fig. 5). Moreover, proteasomal inhibition did not rescue this degradation. Importantly, DHCR24-WT was found to be stable throughout the treatments, suggesting that DHCR24 has a relatively long half-life under these culturing conditions.
Effect of tyrosine mutants on DHCR24 activity
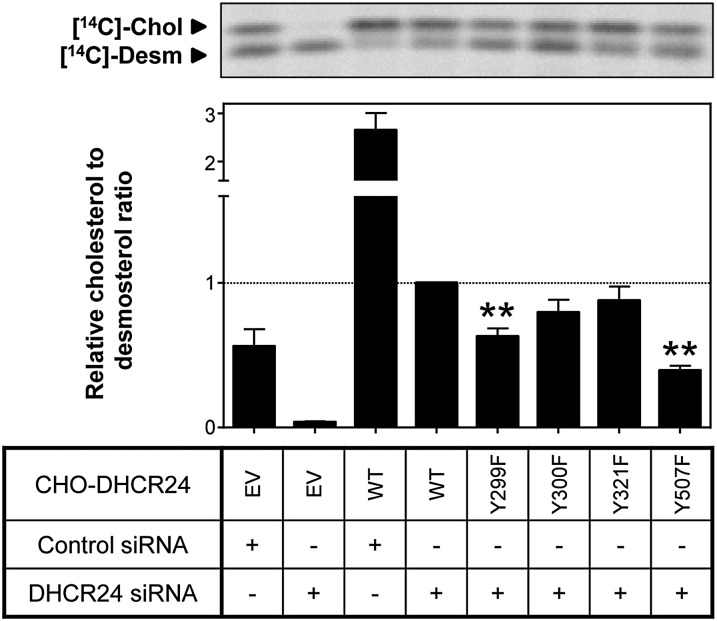
Next, we determined whether DHCR24 stable cell-lines containing mutant tyrosine residues affected DHCR24 activity. Mutating Y299 and Y507 significantly decreased DHCR24 activity by ∼40% and ∼60%, respectively, while mutating Y300 and the published site, Y321, had no effect (Fig. 6). Thus, Y299 and Y507, but not Y300 and Y321, may be phosphorylation sites that regulate DHCR24 activity.
Fig. 6.
Effect of mutating tyrosine residues on DHCR24 activity. CHO-7 cells stably overexpressing EV or DHCR24 (WT, Y299F, Y300F, Y321F, and Y507F) were seeded and transfected with control or hamster-specific DHCR24 siRNA (25 nM) for 24 h. The cells were washed with PBS and refed fresh media overnight, and then radiolabeled with [14C]acetate in fresh media. Lipid extracts were separated by Arg-TLC, and bands corresponding to cholesterol (Chol) and desmosterol (Desm) were visualized by phosphorimaging. Arg-TLC is representative of four separate experiments, and the relative cholesterol to desmosterol ratio was quantified by densitometry, normalized to protein expression (supplementary Fig. IIIB), and then normalized to the DHCR24 siRNA transfected CHO-DHCR24-WT condition, which has been set to one. **P < 0.01 compared with the DHCR24 siRNA transfected CHO-DHCR24-WT condition. Data are mean + SEM (n = 4).
DISCUSSION
We discovered a novel mode of regulation of a key protein in cholesterol synthesis, DHCR24. Recent studies on DHCR24 have identified a number of regulatory mechanisms, primarily at the transcriptional level and particularly via feedback regulation by sterol/steroid molecules, with only a few at the posttranslational level. Regulation by posttranslational modifications, such as phosphorylation, has yet to be fully investigated, despite evidence that DHCR24 is phosphorylated at multiple sites (36), which is in line with our phos-tag SDS-PAGE results (Fig. 1C).
One aim of this work was to identify protein kinase(s) involved in DHCR24 phosphorylation by looking at common protein kinases, using a panel of kinase inhibitors. While inhibiting serine/threonine kinases did not affect DHCR24 protein expression, inhibition of PKC (using BIM and Ro-318220) markedly decreased desmosterol to cholesterol conversion. A potential caveat when studying PKC with pharmacological inhibitors is the issue of specificity (53). For example, BIM and Ro-318220 have been shown to exhibit comparable inhibition of 90 kDa and 70 kDa ribosomal S6 kinase and PKC in vitro, but in cell culture, at least 10 times higher concentrations are required to inhibit the S6 kinases (54). BIM and Ro-318220 are therefore relatively specific inhibitors of the PKC family, with BIM reducing DHCR24 activity in a dose-dependent manner from concentrations as low as 0.1 μM. Thus, our results are consistent with the involvement of PKC in moderating DHCR24 activity. However, further experiments should confirm this, and determine which of the 10 distinct PKC isoforms is involved.
We then determined whether BIM inhibition of DHCR24 activity could be negated by mutating the only published serine/threonine phosphoresidue in DHCR24, T110. T110 is also predicted to be a low stringency PKC substrate by a matrix-based prediction program, Scansite (55) [see supplementary Table II for a list of sites examined here]. However, the BIM inhibitory effect was still observed in the DHCR24 T110 mutant cell-line (CHO-DHCR24-T110A), indicating that PKC does not phosphorylate DHCR24 at T110. This suggests that BIM may be affecting DHCR24 directly through an as yet unidentified phosphorylation site(s), or indirectly. Future work should involve testing other putative sites, as predicted by Scansite, for PKC phosphorylation and effect on DHCR24 activity.
Another aim of this study was to characterize the known phosphorylation sites in DHCR24 and examine their effects on DHCR24 expression and activity. We developed a novel method for measuring DHCR24 activity in cell culture, by stably expressing human DHCR24 in hamster (CHO-7) cells combined with metabolic labeling. To selectively examine the activity of stably expressed human DHCR24, hamster DHCR24 was knocked down by hamster-specific DHCR24 siRNA (Fig. 3A). Hence, this assay measured cholesterol conversion from desmosterol by ectopic human DHCR24 only.
We first examined T110, a published phosphoresidue [(34) and supplementary Table II], and created CHO cell-lines stably expressing a DHCR24 T110 mutant (T110A) or a phosphomimetic (T110E). DHCR24-T110A significantly decreased DHCR24 activity relative to DHCR24-WT (by ∼60%). Mutating T110 to a phosphomimetic (T110E) restored most of this activity (Fig. 3B), indicating that this is indeed a functional phosphorylation site. Although the physiological relevance of T110 phosphorylation needs to be elucidated, this study introduces a new mode of regulation for DHCR24 activity, and may explain settings where desmosterol accumulates, such as in cholesterol-laden macrophage foam cells (14) or in HCV infection (56).
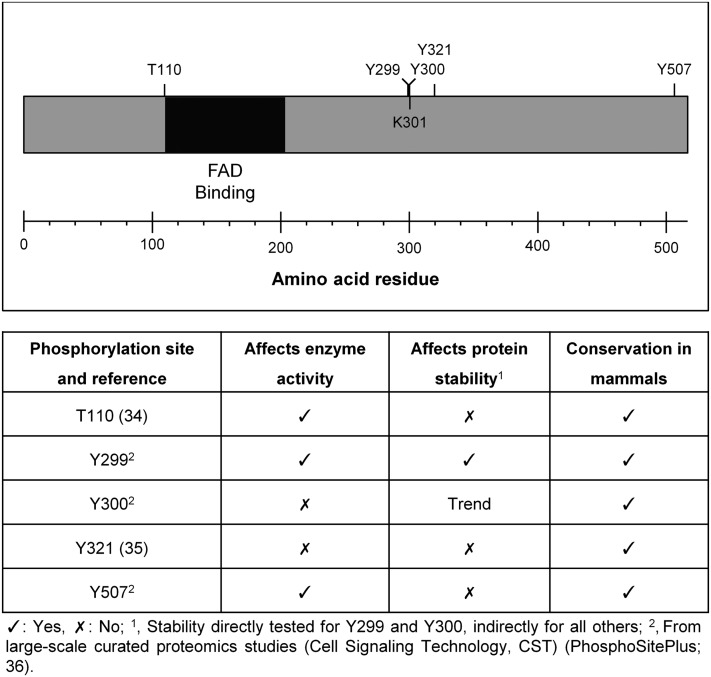
As T110 lies within the predicted FAD binding domain (amino acids 55-235) (4) and just outside of the highly conserved region for FAD binding (amino acids 111-203) [(2, 3) and Fig. 7], it is possible that phosphorylation of T110 affects DHCR24 enzyme activity by influencing cofactor interaction. Amino acids located near the highly conserved region of the FAD binding domain have been shown to be crucial for DHCR24 function; mutation of arginine 94 (R94H) resulted in an almost complete loss of activity, causing desmosterolosis (20). R94 has also been predicted to be involved in specific interactions between FAD phosphate groups and DHCR24 (4). T110 is preceded by three positively charged residues, which are all conserved in higher organisms, and may also be involved in electrostatic interactions with the negatively-charged phosphate groups in FAD. Further work testing FAD binding affinity in DHCR24-T110A is required to consolidate this hypothesis.
Fig. 7.
DHCR24 schematic with phosphoresidues marked. Human DHCR24 protein sequence with putative FAD binding domain (region matching the Pfam (2) entry for an FAD binding domain) and tested phosphoresidues, as well as a putative ubiquitination site, K301. T110, Y299, Y300, K301, Y321, and Y507 are conserved in mammalian DHCR24, including chimpanzee (ENSP00000360316), mouse (ENSMUSP00000038063), and rabbit (ENSOCUP00000004499). Sequences from Ensembl (64).
The other published phosphorylation site was a tyrosine residue (Y321), with three other tyrosine residues implicated. We created phosphorylation mutants by substituting the tyrosine with phenylalanine (Y to F), and stably expressed these in CHO cells. Similar to T110A, the Y299F and Y507F mutants significantly decreased DHCR24 activity, with a ∼60% reduction in activity observed for Y507F relative to DHCR24-WT (Fig. 6). Y507 is a highly conserved residue at the extreme C-terminal end of DHCR24 (Fig. 7), which lies within a small region predicted to interact with the cofactor FAD (4).
The kinases/phosphatases responsible for T110, Y299, and Y507 phosphorylation remain elusive and require further investigation. In this study, we have excluded the effects of PKA, PKB, and PKC (Figs. 1A, 4), as well as ERK1/2, PI3K, mTOR, JAK2, SHP1/2, and PTP1B (data not shown). We have also attempted to confirm these phosphorylation sites in our cell system by liquid chromatography-tandem mass spectrometry. While we had reasonable sequence coverage for DHCR24 (up to 32%), we were unable to detect the required peptides, as determining phosphorylation with this approach remains challenging in our hands (57). Nevertheless, the phosphorylation sites examined in this study were originally identified in large-scale phosphoproteomics studies, with T110 being a published phosphorylation site (34), as well as being identified in Cell Signaling Technology MS/MS studies. As discussed, T110 is very likely physiologically relevant, as the phosphorylation mutant decreased and the phosphomimetic restored DHCR24 activity. While Y507 is not a published phosphorylation site, PhosphoSitePlus indicated this site to be a phosphorylated residue in 12 data sets (36), increasing confidence that it is a bona fide phosphorylation site. Furthermore, our phos-tag result suggested that DHCR24 is phosphorylated at more than one site in our cell system (Fig. 1C).
Although this study focused on potential posttranslational modification by phosphorylation, there is a vast amount of evidence that DHCR24 is ubiquitinated. At present, there are 11 identified ubiquitination sites [9 published (52, 58–60) and 2 from PhosphoSitePlus (36)], suggesting that DHCR24 may be regulated by proteasomal degradation. One such site, K301 (52), is a highly conserved residue that neighbors putative phosphoresidues Y299 and Y300 (Fig. 7). Although the mutants are in close proximity, they have differing effects on DHCR24 mRNA and protein expression, with DHCR24-Y300F levels exhibiting a striking decrease from mRNA to protein (supplementary Fig. III). Therefore, we suspected that this mutant might be subject to proteasomal degradation. However, both DHCR24-Y299F and DHCR24-Y300F caused only minor destabilization that cannot explain the observed mRNA and protein levels, which are not due to differences in cell-lines, as we used a Flp-In system which introduces only one copy of a gene into each cell, and different clones gave the same effect (data not shown). Notably, DHCR24-WT was very stable under the conditions tested (Fig. 5).
There is emerging evidence that other enzymes in the cholesterol synthetic pathway besides HMG-CoA reductase (the classic rate-limiting step) are potential control points for rapid regulation of cholesterol synthesis (61), such as squalene monooxygenase (62). DHCR24 is a prime candidate (61) due to its position in the Bloch pathway as the final enzyme. Thus, regulating DHCR24 by phosphorylation would allow for a rapid means of attenuating cholesterol synthesis, with previous studies already demonstrating its ability for rapid inhibition by multiple feedback mechanisms (30, 32). Furthermore, modulating DHCR24 activity alters levels of desmosterol, a strong liver X receptor ligand (14, 63), further reducing cellular cholesterol status. Collectively, this study has demonstrated that signaling and phosphorylation have functional effects on DHCR24. Further work is required to investigate the precise kinase(s) responsible for the phosphorylation sites investigated in this study, as well as the phosphorylation site(s) required for PKC's action.
Supplementary Material
Acknowledgments
The authors thank members of the Brown Laboratory for critically reviewing this manuscript.
Footnotes
Abbreviations:
- Arg-TLC
- argentation-thin-layer chromatography
- BIM
- bisindolylmaleimide I
- CD
- methyl-β-cyclodextrin
- CHO
- Chinese hamster ovary
- DHCR24
- 3β-hydroxysterol Δ24-reductase
- EV
- empty vector
- FAD
- flavin adenine dinucleotide
- LPDS
- lipoprotein-deficient serum
- NBS
- newborn bovine serum
- PBGD
- porphobilinogen deaminase
- PKA
- protein kinase A
- PKB
- protein kinase B
- PKC
- protein kinase C
- SIM
- single ion monitoring
The Brown Laboratory is supported by grants from the National Health and Medical Research Council (1008081) and the National Heart Foundation of Australia (G11S5757).
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of three figures and two tables.
REFERENCES
- 1.Waterham H. R., Koster J., Romeijn G. J., Hennekam R. C., Vreken P., Andersson H. C., FitzPatrick D. R., Kelley R. I., Wanders R. J. 2001. Mutations in the 3beta-hydroxysterol Delta24-reductase gene cause desmosterolosis, an autosomal recessive disorder of cholesterol biosynthesis. Am. J. Hum. Genet. 69: 685–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Finn R. D., Mistry J., Tate J., Coggill P., Heger A., Pollington J. E., Gavin O. L., Gunasekaran P., Ceric G., Forslund K., et al. 2010. The Pfam protein families database. Nucleic Acids Res. 38: D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mushegian A. R., Koonin E. V. 1995. A putative FAD-binding domain in a distinct group of oxidases including a protein involved in plant development. Protein Sci. 4: 1243–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pedretti A., Bocci E., Maggi R., Vistoli G. 2008. Homology modelling of human DHCR24 (seladin-1) and analysis of its binding properties through molecular docking and dynamics simulations. Steroids. 73: 708–719 [DOI] [PubMed] [Google Scholar]
- 5.Crameri A., Biondi E., Kuehnle K., Lutjohann D., Thelen K. M., Perga S., Dotti C. G., Nitsch R. M., Ledesma M. D., Mohajeri M. H. 2006. The role of seladin-1/DHCR24 in cholesterol biosynthesis, APP processing and Abeta generation in vivo. EMBO J. 25: 432–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilk S. D., Cockrell D. C., Luterbach C., Hansen B., Knodler L. A., Ibarra J. A., Steele-Mortimer O., Heinzen R. A. 2013. Bacterial colonization of host cells in the absence of cholesterol. PLoS Pathog. 9: e1003107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jansen M., Pietiaïnen V. M., Pölönen H., Rasilainen L., Koivusalo M., Ruotsalainen U., Jokitalo E., Ikonen E. 2008. Cholesterol substitution increases the structural heterogeneity of caveolae. J. Biol. Chem. 283: 14610–14618 [DOI] [PubMed] [Google Scholar]
- 8.Kuehnle K., Crameri A., Kalin R. E., Luciani P., Benvenuti S., Peri A., Ratti F., Rodolfo M., Kulic L., Heppner F. L., et al. 2008. Prosurvival effect of DHCR24/Seladin-1 in acute and chronic responses to oxidative stress. Mol. Cell. Biol. 28: 539–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu C., Miloslavskaya I., Demontis S., Maestro R., Galaktionov K. 2004. Regulation of cellular response to oncogenic and oxidative stress by Seladin-1. Nature. 432: 640–645 [DOI] [PubMed] [Google Scholar]
- 10.Greeve I., Hermans-Borgmeyer I., Brellinger C., Kasper D., Gomez-Isla T., Behl C., Levkau B., Nitsch R. M. 2000. The human DIMINUTO/DWARF1 homolog seladin-1 confers resistance to Alzheimer's disease-associated neurodegeneration and oxidative stress. J. Neurosci. 20: 7345–7352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sarajärvi T., Haapasalo A., Viswanathan J., Mäkinen P., Laitinen M., Soininen H., Hiltunen M. 2009. Down-regulation of seladin-1 increases BACE1 levels and activity through enhanced GGA3 depletion during apoptosis. J. Biol. Chem. 284: 34433–34443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu B. J., Chen K., Shrestha S., Ong K. L., Barter P. J., Rye K. A. 2013. High-density lipoproteins inhibit vascular endothelial inflammation by increasing 3beta-hydroxysteroid-delta24 reductase expression and inducing heme oxygenase-1. Circ. Res. 112: 278–288 [DOI] [PubMed] [Google Scholar]
- 13.McGrath K. C., Li X. H., Puranik R., Liong E. C., Tan J. T., Dy V. M., DiBartolo B. A., Barter P. J., Rye K. A., Heather A. K. 2009. Role of 3beta-hydroxysteroid-delta 24 reductase in mediating antiinflammatory effects of high-density lipoproteins in endothelial cells. Arterioscler. Thromb. Vasc. Biol. 29: 877–882 [DOI] [PubMed] [Google Scholar]
- 14.Spann N. J., Garmire L. X., McDonald J. G., Myers D. S., Milne S. B., Shibata N., Reichart D., Fox J. N., Shaked I., Heudobler D., et al. 2012. Regulated accumulation of desmosterol integrates macrophage lipid metabolism and inflammatory responses. Cell. 151: 138–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Battista M. C., Guimond M. O., Roberge C., Doueik A. A., Fazli L., Gleave M., Sabbagh R., Gallo-Payet N. 2010. Inhibition of DHCR24/seladin-1 impairs cellular homeostasis in prostate cancer. Prostate. 70: 921–933 [DOI] [PubMed] [Google Scholar]
- 16.Takano T., Tsukiyama-Kohara K., Hayashi M., Hirata Y., Satoh M., Tokunaga Y., Tateno C., Hayashi Y., Hishima T., Funata N., et al. 2011. Augmentation of DHCR24 expression by hepatitis C virus infection facilitates viral replication in hepatocytes. J. Hepatol. 55: 512–521 [DOI] [PubMed] [Google Scholar]
- 17.Horvat S., McWhir J., Rozman D. 2011. Defects in cholesterol synthesis genes in mouse and in humans: lessons for drug development and safer treatments. Drug Metab. Rev. 43: 69–90 [DOI] [PubMed] [Google Scholar]
- 18.FitzPatrick D. R., Keeling J. W., Evans M. J., Kan A. E., Bell J. E., Porteous M. E., Mills K., Winter R. M., Clayton P. T. 1998. Clinical phenotype of desmosterolosis. Am. J. Med. Genet. 75: 145–152 [PubMed] [Google Scholar]
- 19.Andersson H. C., Kratz L., Kelley R. 2002. Desmosterolosis presenting with multiple congenital anomalies and profound developmental delay. Am. J. Med. Genet. 113: 315–319 [DOI] [PubMed] [Google Scholar]
- 20.Schaaf C. P., Koster J., Katsonis P., Kratz L., Shchelochkov O. A., Scaglia F., Kelley R. I., Lichtarge O., Waterham H. R., Shinawi M. 2011. Desmosterolosis-phenotypic and molecular characterization of a third case and review of the literature. Am. J. Med. Genet. A. 155A: 1597–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zolotushko J., Flusser H., Markus B., Shelef I., Langer Y., Heverin M., Bjorkhem I., Sivan S., Birk O. S. 2011. The desmosterolosis phenotype: spasticity, microcephaly and micrognathia with agenesis of corpus callosum and loss of white matter. Eur. J. Hum. Genet. 19: 942–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horton J. D., Shah N. A., Warrington J. A., Anderson N. N., Park S. W., Brown M. S., Goldstein J. L. 2003. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc. Natl. Acad. Sci. USA. 100: 12027–12032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zerenturk E. J., Sharpe L. J., Brown A. J. 2012. Sterols regulate 3beta-hydroxysterol Delta24-reductase (DHCR24) via dual sterol regulatory elements: cooperative induction of key enzymes in lipid synthesis by Sterol Regulatory Element Binding Proteins. Biochim. Biophys. Acta. 1821: 1350–1360 [DOI] [PubMed] [Google Scholar]
- 24.Bonaccorsi L., Luciani P., Nesi G., Mannucci E., Deledda C., Dichiara F., Paglierani M., Rosati F., Masieri L., Serni S., et al. 2008. Androgen receptor regulation of the seladin-1/DHCR24 gene: altered expression in prostate cancer. Lab. Invest. 88: 1049–1056 [DOI] [PubMed] [Google Scholar]
- 25.Luciani P., Deledda C., Rosati F., Benvenuti S., Cellai I., Dichiara F., Morello M., Vannelli G. B., Danza G., Serio M., et al. 2008. Seladin-1 is a fundamental mediator of the neuroprotective effects of estrogen in human neuroblast long-term cell cultures. Endocrinology. 149: 4256–4266 [DOI] [PubMed] [Google Scholar]
- 26.Battista M. C., Roberge C., Martinez A., Gallo-Payet N. 2009. 24-dehydrocholesterol reductase/seladin-1: a key protein differentially involved in adrenocorticotropin effects observed in human and rat adrenal cortex. Endocrinology. 150: 4180–4190 [DOI] [PubMed] [Google Scholar]
- 27.Ishida E., Hashimoto K., Okada S., Satoh T., Yamada M., Mori M. 2013. Thyroid hormone receptor and liver X receptor competitively up-regulate human selective Alzheimer's disease indicator-1 gene expression at the transcriptional levels. Biochem. Biophys. Res. Commun. 432: 513–518 [DOI] [PubMed] [Google Scholar]
- 28.Yoshinari K., Ohno H., Benoki S., Yamazoe Y. 2012. Constitutive androstane receptor transactivates the hepatic expression of mouse Dhcr24 and human DHCR24 encoding a cholesterogenic enzyme 24-dehydrocholesterol reductase. Toxicol. Lett. 208: 185–191 [DOI] [PubMed] [Google Scholar]
- 29.Drzewinska J., Walczak-Drzewiecka A., Ratajewski M. 2011. Identification and analysis of the promoter region of the human DHCR24 gene: involvement of DNA methylation and histone acetylation. Mol. Biol. Rep. 38: 1091–1101 [DOI] [PubMed] [Google Scholar]
- 30.Zerenturk E. J., Kristiana I., Gill S., Brown A. J. 2012. The endogenous regulator 24(S),25-epoxycholesterol inhibits cholesterol synthesis at DHCR24 (Seladin-1). Biochim. Biophys. Acta. 1821: 1269–1277 [DOI] [PubMed] [Google Scholar]
- 31.Fernández C., Suárez Y., Ferruelo A. J., Gómez-Coronado D., Lasunción M. A. 2002. Inhibition of cholesterol biosynthesis by Delta22-unsaturated phytosterols via competitive inhibition of sterol Delta24-reductase in mammalian cells. Biochem. J. 366: 109–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jansen M., Wang W., Greco D., Bellenchi G. C., di Porzio U., Brown A. J., Ikonen E. 2013. What dictates the accumulation of desmosterol in the developing brain? FASEB J. 27: 865–870 [DOI] [PubMed] [Google Scholar]
- 33.Lindenthal B., Holleran A. L., Aldaghlas T. A., Ruan B., Schroepfer G. J., Jr, Wilson W. K., Kelleher J. K. 2001. Progestins block cholesterol synthesis to produce meiosis-activating sterols. FASEB journal: official publication of the Federation of American Societies for Experimental Biology 15: 775–784 [DOI] [PubMed] [Google Scholar]
- 34.Molina H., Horn D. M., Tang N., Mathivanan S., Pandey A. 2007. Global proteomic profiling of phosphopeptides using electron transfer dissociation tandem mass spectrometry. Proc. Natl. Acad. Sci. USA. 104: 2199–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rigbolt K. T., Prokhorova T. A., Akimov V., Henningsen J., Johansen P. T., Kratchmarova I., Kassem M., Mann M., Olsen J. V., Blagoev B. 2011. System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation. Sci. Signal. 4: rs3. [DOI] [PubMed] [Google Scholar]
- 36.Hornbeck P. V., Kornhauser J. M., Tkachev S., Zhang B., Skrzypek E., Murray B., Latham V., Sullivan M. 2012. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 40: D261–D270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luu W., Sharpe L. J., Gelissen I. C., Brown A. J. 2013. The role of signalling in cellular cholesterol homeostasis. IUBMB Life. 65: 675–684 [DOI] [PubMed] [Google Scholar]
- 38.Metherall J. E., Goldstein J. L., Luskey K. L., Brown M. S. 1989. Loss of transcriptional repression of three sterol-regulated genes in mutant hamster cells. J. Biol. Chem. 264: 15634–15641 [PubMed] [Google Scholar]
- 39.Goldstein J. L., Basu S. K., Brown M. S. 1983. Receptor-mediated endocytosis of low-density lipoprotein in cultured cells. Methods Enzymol. 98: 241–260 [DOI] [PubMed] [Google Scholar]
- 40.Brown A. J., Sun L., Feramisco J. D., Brown M. S., Goldstein J. L. 2002. Cholesterol addition to ER membranes alters conformation of SCAP, the SREBP escort protein that regulates cholesterol metabolism. Mol. Cell. 10: 237–245 [DOI] [PubMed] [Google Scholar]
- 41.Tseng W. C., Lin J. W., Wei T. Y., Fang T. Y. 2008. A novel megaprimed and ligase-free, PCR-based, site-directed mutagenesis method. Anal. Biochem. 375: 376–378 [DOI] [PubMed] [Google Scholar]
- 42.Luu W., Sharpe L. J., Stevenson J., Brown A. J. 2012. Akt acutely activates the cholesterogenic transcription factor SREBP-2. Biochim. Biophys. Acta. 1823: 458–464 [DOI] [PubMed] [Google Scholar]
- 43.Sharpe L. J., Wong J., Garner B., Halliday G. M., Brown A. J. 2012. Is seladin-1 really a selective Alzheimer's disease indicator? J. Alzheimers Dis. 30: 35–39 [DOI] [PubMed] [Google Scholar]
- 44.Du X., Kristiana I., Wong J., Brown A. J. 2006. Involvement of Akt in ER-to-Golgi transport of SCAP/SREBP: a link between a key cell proliferative pathway and membrane synthesis. Mol. Biol. Cell. 17: 2735–2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kielar D., Dietmaier W., Langmann T., Aslanidis C., Probst M., Naruszewicz M., Schmitz G. 2001. Rapid quantification of human ABCA1 mRNA in various cell types and tissues by real-time reverse transcription-PCR. Clin. Chem. 47: 2089–2097 [PubMed] [Google Scholar]
- 46.Kinoshita E., Kinoshita-Kikuta E. 2011. Improved Phos-tag SDS-PAGE under neutral pH conditions for advanced protein phosphorylation profiling. Proteomics. 11: 319–323 [DOI] [PubMed] [Google Scholar]
- 47.Bogoyevitch M. A., Yeap Y. Y., Qu Z., Ngoei K. R., Yip Y. Y., Zhao T. T., Heng J. I., Ng D. C. 2012. WD40-repeat protein 62 is a JNK-phosphorylated spindle pole protein required for spindle maintenance and timely mitotic progression. J. Cell Sci. 125: 5096–5109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chijiwa T., Mishima A., Hagiwara M., Sano M., Hayashi K., Inoue T., Naito K., Toshioka T., Hidaka H. 1990. Inhibition of forskolin-induced neurite outgrowth and protein phosphorylation by a newly synthesized selective inhibitor of cyclic AMP-dependent protein kinase, N-[2-(p-bromocinnamylamino)ethyl]-5-isoquinolinesulfonamide (H-89), of PC12D pheochromocytoma cells. J. Biol. Chem. 265: 5267–5272 [PubMed] [Google Scholar]
- 49.Barnett S. F., Defeo-Jones D., Fu S., Hancock P. J., Haskell K. M., Jones R. E., Kahana J. A., Kral A. M., Leander K., Lee L. L., et al. 2005. Identification and characterization of pleckstrin-homology-domain-dependent and isoenzyme-specific Akt inhibitors. Biochem. J. 385: 399–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Toullec D., Pianetti P., Coste H., Bellevergue P., Grand-Perret T., Ajakane M., Baudet V., Boissin P., Boursier E., Loriolle F., et al. 1991. The bisindolylmaleimide GF 109203X is a potent and selective inhibitor of protein kinase C. J. Biol. Chem. 266: 15771–15781 [PubMed] [Google Scholar]
- 51.Davis P. D., Hill C. H., Keech E., Lawton G., Nixon J. S., Sedgwick A. D., Wadsworth J., Westmacott D., Wilkinson S. E. 1989. Potent selective inhibitors of protein kinase C. FEBS Lett. 259: 61–63 [DOI] [PubMed] [Google Scholar]
- 52.Wagner S. A., Beli P., Weinert B. T., Nielsen M. L., Cox J., Mann M., Choudhary C. 2011. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol. Cell. Proteomics. 10: 013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu-Zhang A. X., Newton A. C. 2013. Protein kinase C pharmacology: refining the toolbox. Biochem. J. 452: 195–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roberts N. A., Marber M. S., Avkiran M. 2004. Specificity of action of bisindolylmaleimide protein kinase C inhibitors: do they inhibit the 70 kDa ribosomal S6 kinase in cardiac myocytes? Biochem. Pharmacol. 68: 1923–1928 [DOI] [PubMed] [Google Scholar]
- 55.Obenauer J. C., Cantley L. C., Yaffe M. B. 2003. Scansite 2.0: proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 31: 3635–3641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rodgers M. A., Villareal V. A., Schaefer E. A., Peng L. F., Corey K. E., Chung R. T., Yang P. L. 2012. Lipid metabolite profiling identifies desmosterol metabolism as a new antiviral target for hepatitis C virus. J. Am. Chem. Soc. 134: 6896–6899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sharpe L. J., Luu W., Brown A. J. 2011. Akt phosphorylates Sec24: new clues into the regulation of ER-to-Golgi trafficking. Traffic. 12: 19–27 [DOI] [PubMed] [Google Scholar]
- 58.Danielsen J. M., Sylvestersen K. B., Bekker-Jensen S., Szklarczyk D., Poulsen J. W., Horn H., Jensen L. J., Mailand N., Nielsen M. L. 2011. Mass spectrometric analysis of lysine ubiquitylation reveals promiscuity at site level. Mol. Cell. Proteomics. 10: 003590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim W., Bennett E. J., Huttlin E. L., Guo A., Li J., Possemato A., Sowa M. E., Rad R., Rush J., Comb M. J., et al. 2011. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell. 44: 325–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wagner S. A., Beli P., Weinert B. T., Scholz C., Kelstrup C. D., Young C., Nielsen M. L., Olsen J. V., Brakebusch C., Choudhary C. 2012. Proteomic analyses reveal divergent ubiquitylation site patterns in murine tissues. Mol. Cell. Proteomics. 11: 1578–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sharpe L. J., Brown A. J. 2013. Controlling cholesterol synthesis beyond 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR). J. Biol. Chem. 288: 18707–18715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gill S., Stevenson J., Kristiana I., Brown A. J. 2011. Cholesterol-dependent degradation of squalene monooxygenase, a control point in cholesterol synthesis beyond HMG-CoA reductase. Cell Metab. 13: 260–273 [DOI] [PubMed] [Google Scholar]
- 63.Yang C., McDonald J. G., Patel A., Zhang Y., Umetani M., Xu F., Westover E. J., Covey D. F., Mangelsdorf D. J., Cohen J. C., et al. 2006. Sterol intermediates from cholesterol biosynthetic pathway as liver X receptor ligands. J. Biol. Chem. 281: 27816–27826 [DOI] [PubMed] [Google Scholar]
- 64.Flicek P., Ahmed I., Amode M. R., Barrell D., Beal K., Brent S., Carvalho-Silva D., Clapham P., Coates G., Fairley S., et al. 2013. Ensembl 2013. Nucleic Acids Res. 41: D48–D55 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.