Abstract
MALDI imaging mass spectrometry (IMS) was used to characterize lipid species within sections of human eyes. Common phospholipids that are abundant in most tissues were not highly localized and observed throughout the accessory tissue, optic nerve, and retina. Triacylglycerols were highly localized in accessory tissue, whereas sulfatide and plasmalogen glycerophosphoethanolamine (PE) lipids with a monounsaturated fatty acid were found enriched in the optic nerve. Additionally, several lipids were associated solely with the inner retina, photoreceptors, or retinal pigment epithelium (RPE); a plasmalogen PE lipid containing DHA (22:6), PE(P-18:0/22:6), was present exclusively in the inner retina, and DHA-containing glycerophosphatidylcholine (PC) and PE lipids were found solely in photoreceptors. PC lipids containing very long chain (VLC)-PUFAs were detected in photoreceptors despite their low abundance in the retina. Ceramide lipids and the bis-retinoid, N-retinylidene-N-retinylethanolamine, was tentatively identified and found only in the RPE. This MALDI IMS study readily revealed the location of many lipids that have been associated with degenerative retinal diseases. Complex lipid localization within retinal tissue provides a global view of lipid organization and initial evidence for specific functions in localized regions, offering opportunities to assess their significance in retinal diseases, such as macular degeneration, where lipids have been implicated in the disease process.
Keywords: lipid localization, lipid characterization, docosahexaenoic acid, very long chain polyunsaturated fatty acids, ceramide
The retina is a light-sensitive multi-layered tissue that lines the back of the eye and is responsible for converting light into a neural signal. The innermost layer of the retina, which is closest to the lens, contains ganglion cells that are the output neurons which comprise the optic nerve and conduct signals to the visual centers of the brain (1). The central region is made up of several layers that include the horizontal, bipolar, and amacrine cells, which collectively process visual information from the photoreceptors, transmitting these signals to the ganglion cells. The outermost retinal neurons, the photoreceptors, are responsible for phototransduction (2). There are two types of photoreceptors, which include rods that are present in the peripheral retina and cones that are present in the central (macular) region of the retina. Rods function in dim light and manage peripheral and night vision, whereas cones are responsible for central high acuity bright light vision. The outermost layer of the retina, the retinal pigment epithelium (RPE), serves many functions, including the isomerization of all-trans retinol into 11-cis retinal in the visual cycle, phagocytosis and degradation of shed photoreceptor tips, and the phagocytic clearance of cellular debris resulting from apoptotic and necrotic processes (3).
The lipids present in the retina are highly unique and play a critical role in retina function and disease. The retina is a tissue that is highly enriched in PUFAs with DHA (22:6) (22 carbons with 6 double bonds) accounting for approximately 50% of the fatty acids in the photoreceptors (4). This large amount of DHA results in a highly fluid membrane that permits efficient conformational changes to occur in rhodopsin and its associated G-protein during phototransduction (5). Moreover, DHA is converted into neuroprotectin D1, a potent mediator that evokes counteracting cell-protective anti-inflammatory pro-survival repair signaling, including the induction of anti-apoptotic proteins and inhibition of pro-apoptotic proteins, activating signaling pathway(s) that modulate(s) pro-apoptotic signals, promoting cell survival (6–8). Another unique feature of retinal lipids is the abundance of phospholipids containing two PUFAs esterified to the glycerol backbone, where a very long chain (24–36 carbons) (VLC)-PUFA) is located at the sn-1 position and DHA is esterified at the sn-2 position (9). The functional importance of VLC-PUFAs in the retina is not well understood but VLC-PUFAs are reduced in retinas undergoing age-related macular degeneration (AMD) (10), and a key role for VLC-PUFAs has been suggested in both phototransduction regulation and photoreceptor survival (11, 12).
Retinal phospholipids also play an important role in the visual cycle and the conversion of all-trans retinal to 11-cis retinal. In some cases, the N-retinylidene-glycerophosphatidylethanolamine (PE) formed during the visual cycle reacts with a second molecule of all-trans-retinal instead of hydrolyzing to PE and all-trans-retinal, and leads to the formation of the cytotoxic compound N-retinylidene-N-retinylethanolamine (A2E) (13). This side reaction product can progressively accumulate with age in the RPE and may contribute to retinal degenerative diseases such as AMD (14, 15). Additionally, the production of ceramides, which are bioactive sphingolipids that are critical mediators for initiating oxidative stress-induced apoptotic cell death of both photoreceptors and RPE cells, may lead to several retinal diseases including retinitis pigmentosa and AMD (16–19).
Retinal phospholipids have been well-characterized by gas chromatography-mass spectrometry (20–22) and electrospray mass spectrometry (23–26) of extracted tissue, but the regional distribution of phospholipid molecular species in the various regions within the human retina has remained uncertain. MALDI imaging mass spectrometry (IMS) has been developed as a powerful method for investigating the distribution of phospholipids within tissues (27). Phospholipid molecular ions are produced directly from a tissue section coated with MALDI matrix, and sequential mass spectra are acquired by rastering the laser beam across the tissue surface; images can be obtained by plotting the intensity of the analytes of interest as a function of the x-y coordinates. Previously, MALDI IMS has been applied to the porcine lens (28), to salamander (29), rat, and mouse retinas (30–32), and to mouse optic nerve tissue (33), with the major focus generally on the localization of glycerophosphatidylcholine (PC) lipid molecular ions. In the current study, MALDI IMS is used to report the localization of PC, PE, glycerophosphatidylinositol (PI), and glycerophosphatidylserine (PS) lipid molecular species, as well as sphingolipids present in the human retina.
METHODS
Reagents
Polyvinyl alcohol 6-98, 2,5-dihydroxybenzoic acid, and 2,6-dihydroxyacetophenone (DHAP) were purchased from Sigma-Aldrich Chemical Co. (St. Louis, MO). OCT compound was obtained from Ted Pella, Inc. (Redding, CA). Polypropylene glycol, average molecular weight 2,000 g/mol, was purchased from Fisher Scientific (Fair Lawn, NJ).
Human eyes
Human eyes (whole globes) were obtained from the National Disease Research Interchange (Philadelphia, PA) and the Lions Eye Institute (Tampa, FL). Retinal tissue used in this study was from the right and left eyes of nine individuals, including a 45-year-old Caucasian female, a 47-year-old Caucasian male, a 55-year-old Hispanic male, a 56-year-old Caucasian female, a 60-year-old Caucasian male, a 73-year-old Caucasian female, a 79-year-old Black female, an 81-year-old Caucasian male, and an 82-year-old Caucasian female (Table 1). At procurement, globes were removed, wrapped in moist gauze, placed in “eye bank” plastic containers on wet ice in Styrofoam coolers, and sent to Louisiana State University for arrival at the laboratory by 10:00 AM the following day. Upon arrival, globes were immediately dissected and prepared (unfixed) for cryosectioning, or tissue samples collected for mass spectrometry. Frozen sections were placed on glass coverslips and immediately stored at −80°C. Frozen sections were then moved to plastic containers on dry ice and shipped to the University of Denver for delivery the following morning. These coverslips were kept frozen until processing for MALDI imaging began. Tissue samples for mass spectrometry were placed in 1.5 ml microtubes in methanol containing butylated hydroxytoluene (0.1%), and stored at −80°C until extraction.
TABLE 1.
Human donor information
| Age (years) | Sex | Race | Cause of Death | Time from Death to Procurement (h: min) | Time from Death to Arrival at LSU (h: min) | Source |
| 45 | F | C | Sepsis | 5: 29 | 23: 00 | Lions |
| 47 | M | C | Cardiac arrest | 7: 00 | 21: 08 | NDRI |
| 55 | M | H | Cardiac arrest | 6: 00 | 24: 35 | NDRI |
| 56 | F | C | Cardiac arrest | 6: 20 | 28: 04 | NDRI |
| 60 | M | C | Liver failure | 5: 20 | 23: 40 | Lions |
| 73 | F | C | Lung cancer | 4: 00 | 29: 50 | NDRI |
| 79 | F | B | Cardiac arrest | 8: 00 | 24: 25 | NDRI |
| 81 | M | C | Lung cancer | 3: 25 | 29: 00 | Lions |
| 82 | F | C | Cardiac arrest | 4: 32 | 24: 42 | Lions |
At procurement, globes were removed, wrapped in moist gauze, placed in eye bank plastic containers on wet ice in Styrofoam coolers at NDRI or the Lions Eye Institute, and sent to Louisiana State University for arrival at the laboratory by 10:00 AM the following day. Upon arrival, globes were immediately dissected and prepared (unfixed) for cryosectioning, or tissue samples collected for mass spectrometry. Frozen sections were placed on glass coverslips and immediately stored at −80°C. Frozen sections were then moved to plastic containers on dry ice and shipped to the University of Denver for delivery the following morning. These coverslips were kept frozen until processing for MALDI imaging began. Tissue samples for mass spectrometry were placed in 1.5 ml microtubes in methanol containing butylated hydroxytoluene (0.1%), and stored at −80°C. LSU, Louisiana State University; F, female; M, male; C, Caucasian; H, Hispanic; B, Black; Lions, Lions Eye Institute, Tampa, FL; NDRI, National Disease Research Institute, National Resource Center, Philadelphia, PA.
Preparation of human retinas
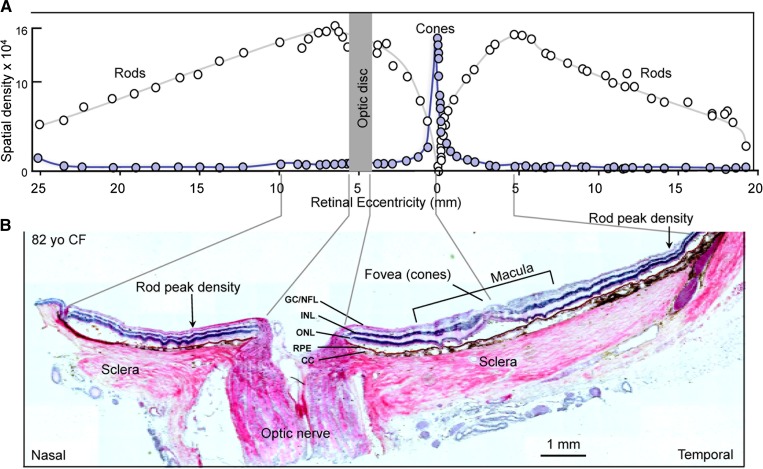
The anterior segment was removed, radial cuts were made to flatten each eye, and a horizontal strip containing the optic nerve and macular region was obtained and embedded in modified OCT. The preparation of modified OCT has been described in detail previously (34). The embedded tissue was frozen, attached to a sectioning stub, and cryosectioned to obtain 20 μm-thick vertical sections showing all layers of the retina, choroid, optic nerve, and sclera (Fig. 1B). Sections were attached to glass coverslips and stored at −80°C until needed for MALDI imaging. Alternate sections from each block were placed on conventional microscope slides and stained with hematoxylin and eosin (H&E) for orientation purposes. Rod and cone density across the human retina varies with photoreceptor type and location; cones are concentrated within the central macular region while rods occupy the peripheral retina (Fig. 1A) (35). Regions of interest within the eye, such as retina, optic nerve, sclera, accessory tissue, and RPE/choroid complex, as well as specific retinal layers, were labeled. For example, Fig. 1B shows an H&E-stained section that represents a typical tissue sample in this study. In total, more than 30 individual sections of eye tissue from the nine human subjects listed above were analyzed in both positive and negative ion mode by MALDI IMS for this study.
Fig. 1.
Organization of the human retina. A: Diagram illustrating the distribution of rod and cone photoreceptors. The origin of the optic nerve in the retina, the optic disk, lacks photoreceptors, thus creating a blind spot within the visual field. Cone photoreceptors become concentrated around the optic axis in a central region, the macula, which measures 4.5–6.0 mm in diameter. The center-most area of the macula, the fovea, contains the highest concentration of cones, which produce high acuity bright light color vision. As cones diminish peripherally, rod photoreceptors accumulate and extend to the retina margin. The peripheral regions of the retina function in dim light and produce monochromatic vision. Features along the retinal surface are measured in millimeters (retinal eccentricity) from the fovea. The vertical axis denotes photoreceptor density. [Redrawn and modified with permission from (35)]. B: H&E-stained retinal section (82-year-old Caucasian female). The orientation of the section is nasal to temporal to reveal both the optic nerve and the cone-rich macular region, surrounded by the rod-rich periphery. Guide lines relate relevant features on the histological section to corresponding areas on the diagram above. The fovea is indicated at the center of the macula and several retinal layers are indicated [ganglion cell/nerve fiber layer (GC/NFL), INL, ONL, choriocapillaris (CC), sclera, and optic nerve].
Matrix application
The glass coverslips containing the tissue sections were attached to stainless steel MALDI plate inserts (OptiToF, Applied Biosystems) using copper tape, and the matrix was deposited by sublimation (36). The sublimation of 2,5-dihydroxybenzoic acid and DHAP matrix onto tissues has been described in detail previously (34).
MALDI IMS
A quadrupole-TOF tandem mass spectrometer with an orthogonal MALDI source (QSTAR XL; Applied Biosystems/MDS Sciex, Thornhill, Ontario, Canada) was used to acquire mass spectral data from which images were constructed. MALDI mass spectra were obtained using a solid state laser (355 nm) at an energy of 7.4 μJ and a pulse rate of 200 Hz for positive ion MALDI imaging, and an energy of 7.6 μJ and a pulse rate of 500 Hz for negative ion imaging. The energy and pulse rate of the laser were selected to maximize the lipid signal from the tissue slice. Both the positive and negative ion MALDI imaging experiments had an accumulation time of 243 ms per image spot. The MALDI plate was moved at a rate of 12.75 mm/min and after each horizontal line was completed the plate was moved vertically 50 μm. The lateral resolution of this MALDI IMS technique has been estimated to be approximately 50 μm. The mass spectrometric data were processed using a specialized script for Analyst software (Applied Biosystems/MDS Sciex) at a mass resolution of 0.1 amu, and images were visualized using TissueView software (Applied Biosystems/MDS Sciex). Collision-induced dissociation (CID) of selected ions in both positive and negative ion modes was carried out after the MALDI imaging experiments on the same tissue slice using relative collision energy of 40 V with argon as collision gas. The shorthand notation used for lipid species in this paper follows those outlined by Liebisch et al. (37). The lipids observed in positive ion mode [PC, SM, and triacylglycerol (TAG)] were identified to the lipid species level by CID and are indicated by the number of acyl carbons and the number of double bonds, for example PC(34:1). The lipids observed in negative ion mode (PS, PI, and PE) were identified to the fatty acyl level by CID and are indicated by the fatty acyl groups esterified to the sn-1 and sn-2 position, such as PE(18:0/22:6).
RESULTS
Rod and cone density across the human retina varies with photoreceptor type and location; cones are concentrated within the central macular region while rods occupy the peripheral retina (Fig. 1A). Regions of interest within the eye, such as connective tissue, sclera, optic nerve, RPE, choroid, and retina, as well as the retinal layers, are labeled in an H&E-stained section which represents a typical tissue sample (Fig. 1B). In this study the localization of lipid molecular species was determined using MALDI IMS. The positive ion mass spectra in the mass range m/z 650–900 (Fig. 2B–D) and the negative ion mass spectra in the mass range m/z 650–1,000 (Fig. 3B–D) in different regions of eye tissue (accessory tissue, optic nerve, retina) demonstrated that the lipid molecular species varied extensively within discrete regions of eye tissue.
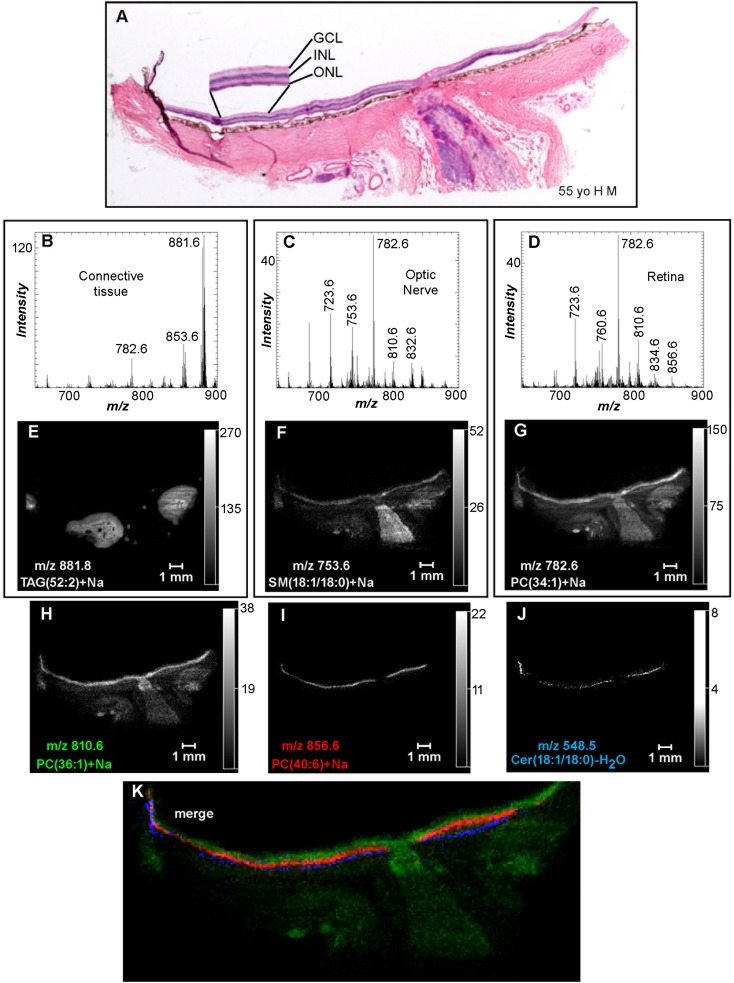
Fig. 2.
Positive ion MALDI IMS of PC and SM lipids in human ocular tissue. A: H&E stain of ocular tissue section immediately adjacent to the section used for MALDI imaging. Total positive ion MALDI mass spectra of the accessory tissue (B), optic nerve (C), and retina (D) were obtained directly from the ocular section. Extracted positive ion MALDI images of the [M+Na]+ of TAG(52:2) (m/z 881.8) (E), SM(d18:1/18:0) (m/z 753.6) (F), PC(34:1) (m/z 782.6) (G), PC(36:1) (m/z 810.6) (H), PC(40:6) (m/z 856.6) (I), and the [M+H-H2O]+ of Cer(d18:1/18:0) (m/z 548.5) (J). K: Merged positive ion MALDI image of Cer(d18:1/18:0) (blue), PC(40:6) (red), and PC(36:1) (green). GCL, ganglion cell layer.
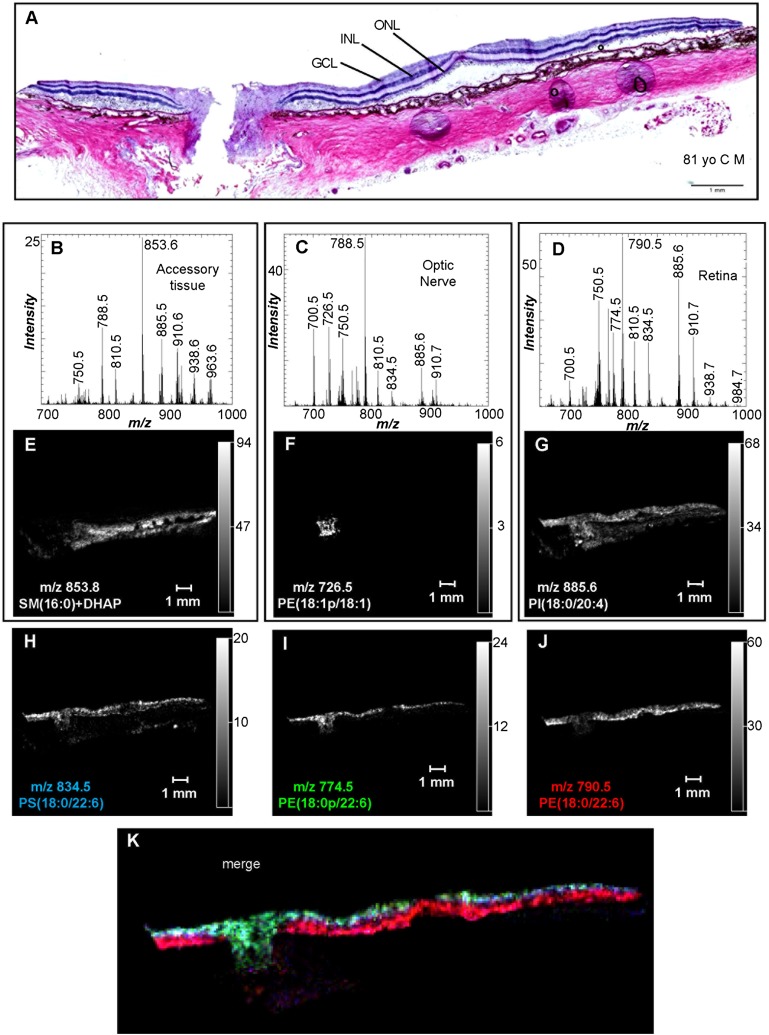
Fig. 3.
Negative ion MALDI IMS of PI, PS, and PE lipids in human ocular tissue. A: H&E stain of ocular tissue section immediately adjacent to the section used for MALDI imaging. Total negative ion MALDI mass spectra of the accessory tissue (B), optic nerve (C), and retina (D) were obtained directly from the ocular section. Extracted negative ion MALDI images of the [M-H]− of SM(d16:0/18:0)+DHAP (m/z 853.8) (E), PE(P-18:1/18:1) (m/z 726.5) (F), PI(18:0/20:4) (m/z 885.6) (G), PS(18:0/22:6) (m/z 834.5) (H), PE(P-18:0/22:6) (m/z 774.5) (I), and PE(18:0/22:6) (m/z 790.5) (J). K: Merged positive ion MALDI image of PE(P-18:0/22:6) (green), PE(18:0/22:6) (red), and PS(18:0/22:6) (blue). GCL, ganglion cell layer.
Positive ion data obtained for the MALDI IMS experiments revealed the distribution of PC, SM, and TAG lipids in the ocular tissue. The positive ion MALDI mass spectrum from accessory tissue (e.g., adipose and connective tissue) indicated unique ions in these regions of the ocular section (Fig. 2B) with the most intense ion at m/z 881.8. The positive ion MALDI image of m/z 881.8 suggested that this ion was present within the tissue behind the sclera (outside the eye ball) and surrounding the optic nerve (Fig. 2E). The black areas in the image within this region of tissue are cross-sections of blood vessels. The CID of the tissue-derived MALDI ion at m/z 881.8 (supplementary Fig. IA, Table 2) revealed product ions at m/z 625.5, 603.5, 599.5, and 577.5, a behavior that was consistent with this ion being the [M+Na]+ of a TAG, specifically a mixture of TAGs that had 52 acyl group carbon atoms with two double bonds residing collectively in the three fatty acyl groups and denoted as TAG(52:2) (38). Additionally, other sodiated TAG lipids were found with very similar regional distributions to m/z 881.8, including m/z 851.8 [TAG(50:3)+Na], 853.8 [TAG(50:2)+Na], 855.8 [TAG(50:1)+Na], 877.8 [TAG(52:4)+Na], 879.8 [TAG(52:3)+Na], and 883.8 [TAG(52:1)+Na] (data not shown). The positive ion MALDI mass spectrum of this accessory tissue (Fig. 2B) revealed the presence of other lipids in this region, such as m/z 782.6, that were not unique to this location. The CID of m/z 782.6 resulted in product ions at m/z 146.9, 184.1, and 599.6 (neutral loss of phosphocholine) (supplementary Fig. IB, Table 2) that are characteristic of a sodiated PC lipid and are consistent with the identification of m/z 782.6 as the [M+Na]+ of PC(34:1) (39). The positive ion MALDI image of m/z 782.6 (Fig. 2G) indicated the presence of this ion in the optic nerve and accessory tissues, but with the highest signal intensity occurring in the inner retina and the lumen of blood vessels behind the eye.
TABLE 2.
Summary of the positive ion CID obtained directly from the ocular sections post-MALDI IMS
| Observed (m/z) | Product Ions | Identification |
| 548.5 | 264.4 | Cer(d18:1/18:0)-H2O |
| 723.6 | 146.9, 540.5, 184.1 | SM(d18:1/16:1)+Na |
| 753.6 | 146.9, 570.6, 184.1 | SM(d18:1/18:0)+Na |
| 756.6 | 146.9, 184.1, 621.5 | PC(32:0)+Na |
| 760.6 | 184.1 | PC(34:1) |
| 782.6 | 146.9, 184.1, 599.5 | PC(34:1)+Na |
| 788.6 | 184.1 | PC(36:1) |
| 810.6 | 146.9, 627.5, 184.1 | PC(36:1)+Na |
| 828.6 | 146.9, 645.5, 184.1 | PC(38:6)+Na |
| 832.6 | 146.9, 649.5, 184.1 | PC(38:4)+Na |
| 834.6 | 184.1 | PC(40:6) |
| 856.6 | 673.6, 146.9, 184.1 | PC(40:6)+Na |
| 881.8 | 599.5, 577.5, 625.5, 603.5 | TAG(52:2)+Na |
| 900.7 | 147.0, 717.6, 184.1 | PC(44:12)+Na |
| 1,040.7 | 147.0, 184.1, 857.6 | PC(54:12)+Na |
| 1,042.7 | 147.0, 859.6, 184.1 | PC(54:11)+Na |
| 1,044.7 | 147.0, 184.1, 861.6 | PC(54:10)+Na |
| 1,070.7 | 147.0, 184.1, 887.6 | PC(56:11)+Na |
The presence of a wide variety of lipid molecular species in the optic nerve was revealed in the positive ion MALDI mass spectrum of this region of the ocular tissue section (Fig. 2C). The most abundant ion in the positive ion MALDI mass spectrum of the optic nerve was at m/z 782.6 [PC(34:1)+Na] and the MALDI image suggested that the lipid generating this ion was uniformly distributed in the optic nerve with a slightly higher signal intensity at the optic disc (Fig. 2G). The presence of additional lipids in the optic nerve at m/z 810.6 [PC(36:1)+Na] and 832.6 [PC(38:4)+Na], which were identified as sodiated PC lipids by CID (Table 2), is indicated in Fig. 2C. The positive ion MALDI images of these sodiated PC ions also localized these lipids to the optic nerve, as well as sclera, and retina (Fig. 2H, supplementary Fig. IIA). Additionally, the positive ion MALDI image of m/z 753.6 indicated that this particular ion was uniformly distributed throughout the optic nerve as well as in the retina and sclera, but with lower intensity, and that this particular lipid molecular species was not present at the optic disc (Fig. 2F). CID of m/z 753.6 yielded product ions at m/z 570.6 (neutral loss of phosphocholine), m/z 146.9 (sodiated cyclic 1,2-phosphodiester), and m/z 184.1 (phosphocholine) (Table 2), and was consistent with the identification of m/z 753.6 as SM(d18:1/18:0)+Na. Another sodiated SM at m/z 723.6, which was identified as SM(d18:1/16:1)+Na by CID (Table 2), occurred with a similar distribution described above in the optic nerve, sclera, and retina (supplementary Fig. IIB). Unlike the sodiated TAG lipids that were uniquely observed in the tissue behind the sclera, none of the positive ion MALDI images described above indicated that any of the lipid molecular species observed in the optic nerve were exclusive to this region of ocular tissue.
The positive ion MALDI mass spectrum of the retina indicated the presence of common, as well as some novel, phospholipid molecular species (Fig. 2D). The MALDI image of the most prevalent positive ion found in the retina, m/z 782.6 [identified as PC(34:1)+Na above], indicated the presence of this ion with varying intensity throughout the entire retina, as well as the sclera, optic nerve, and accessory tissues (Fig. 2G). The most intense signal observed for m/z 782.6 in the retina occurred in the rod photoreceptor-dominant periphery, but this ion was also present in the cone photoreceptor-dominant macular region (Fig. 2G). Discrete retinal layers were not observed in the positive ion MALDI image of m/z 782.6, but the distribution of this ion was apparent within all the retinal layers. Many other ions present in the MALDI mass spectrum of the retina (Fig. 2D) were sodiated PC or SM lipids that contained saturated or monounsaturated fatty acids (Table 2), such as m/z 723.6 [SM(18:1/16:1)+Na], m/z 756.6 [PC(32:0)+Na], and m/z 760.6 [PC(34:1)], and had a similar distribution to m/z 782.6 [PC(34:1)+Na] in the retina (supplementary Fig. IIB–D). Additionally, none of these PC or SM lipids containing only saturated or monounsaturated fatty acids esterified to the glycerol backbone were uniquely present in the retina.
Some of the lipid molecular ions present in the retina were highly localized and are described below. Within the inner retina, the neuronal portion proximal to the photoreceptor layer, a positive ion MALDI image of m/z 810.6 [identified as PC(36:1)+Na above] indicated that the signal for this lipid molecular species was strong, with similar intensity in both the peripheral and macular regions, at the optic disc, and within the astrocyte-rich optic nerve head and lamina cribrosa (Fig. 2H). Additionally, a similar localization in the inner retina was also observed in the MALDI images of m/z 832.6 [PC(38:4)+Na] and m/z 788.6 [PC(36:1)] (supplementary Fig. IIA, E), which were identified by CID (Table 2).
Within the photoreceptor layer, the positive ion MALDI image of m/z 856.6 indicated that this lipid was uniquely localized and was present with similar intensity in both the peripheral and macular regions (Fig. 2I). Also, this image indicates that this ion was not present at the optic disc. The CID spectrum of m/z 856.6 obtained directly from the tissue contained product ions at m/z 146.9, 184.1, and 673.6 (Table 2), indicating that this ion was derived from PC(40:6)+Na. Additionally, a similar localization in the retina was also observed in the MALDI images of m/z 828.6 [PC(38:6)+Na] and m/z 834.6 [PC(40:6)] (supplementary Fig. IIF, G), which were identified by CID (Table 2).
The RPE, a monolayer of cells associated with the tips of the photoreceptors at the back of the retina, contained a lipid at m/z 548.5 that demonstrated unique localization in the eye tissue (Fig. 2J). This lipid molecular ion displayed a higher intensity in the peripheral region compared with the macular region of the retina and was also notably absent at the optic disc. CID of m/z 548.5 yielded a product ion at m/z 264.4 (supplementary Fig. IC, Table 2), which was characteristic of ceramide lipids and occurred from amide bond cleavage with neutral loss of the fatty acid from [M+H-H2O]+ (40). From this information, the ion at m/z 548.5 was identified as the [M+H-H2O]+ of ceramide (Cer) (d18:1/18:0), which is consistent with a previous study which determined that ceramide lipids analyzed by MALDI yielded no molecular ions, but only the [M+H-H2O]+ ion (41). The merged images of the lipid molecular ions at m/z 810.6 [PC(36:1)+Na], m/z 856.6 [PC(40:6)+Na], and m/z 548.5 [Cer(d18:1/18:0)-H2O] nicely demonstrated the stratification of these particular lipids in the retina (Fig. 2K). Specifically, PC(36:1)+Na was observed within the retinal ganglion cell layer, the axons of which comprise the optic nerve, PC(40:6)+Na had panretinal localization but was only found within the photoreceptor layer, and Cer(d18:1/18:0)-H2O was specifically located in the RPE/choriocapillaris complex.
Negative ion data obtained from the MALDI IMS experiments revealed the distribution of many lipid classes, including PI, PS, and PE, in the ocular tissue. The negative ion MALDI mass spectrum of accessory tissue did not reveal many lipids that were unique to this region of the tissue section (Fig. 3B). The negative ion MALDI image of the most abundant [M-H]− in accessory tissue, m/z 853.8, revealed that this particular ion was present only in the sclera (Fig. 3E). CID of m/z 853.8 resulted in a product ion at m/z 151.1 and a product ion resulting from the neutral loss of choline (59 amu) (Table 3). Due to these product ions observed during CID, it was determined that this ion, m/z 853.8, was a DHAP matrix adduct of SM(d18:1/16:0). Additionally, a PC(16:0/18:1) standard was spotted on a MALDI plate followed by DHAP matrix sublimation and the only [M-H]− observed in the MALDI mass spectrum corresponded to PC(16:0/18:1)+DHAP (supplementary Fig. IIIA), and the CID spectrum of this DHAP adduct contained a product ion at m/z 151.1 and a product ion corresponding to neutral loss of choline (supplementary Fig. IIIB). Furthermore, matrix adducts of PC lipids are quite common and have been reported previously in a negative ion MALDI MS analysis of human LDL (42).
TABLE 3.
Summary of the negative ion CID obtained directly from the ocular sections post-MALDI IMS
| Observed (m/z) | Product Ions | Identification |
| 700.5 | 281.1, 418.4, 436.4 | PE(P-16:0/18:1) |
| 726.5 | 281.1, 444.4, 462.4 | PE(P-18:1/18:1) |
| 728.5 | 281.1, 446.4, 464.4 | PE(P-18:0/18:1) |
| 750.5 | 259.1, 303.1, 446.4, 464.4 | PE(P-18:0/20:4) |
| 762.5 | 255.1, 283.1, 327.1, 452.5 | PE(16:0/22:6) |
| 766.5 | 259.1, 283.1, 303.1, 462.4, 480.4 | PE(18:0/20:4) |
| 774.5 | 283.1, 327.1, 464.5 | PE(P-18:0/22:6) |
| 788.5 | 153.1, 281.1, 283.1, 419.3, 437.3, 701.5 | PS(18:0/18:1) |
| 790.5 | 283.1, 327.1, 480.5 | PE(18:0/22:6) |
| 810.5 | 153.1, 283.1, 303.1, 419.3, 437.3, 723.5 | PS(18:0/20:4) |
| 834.5 | 283.1, 327.1, 419.4, 437.4, 747.5 | PS(18:0/22:6) |
| 853.8 | 151.1, 794.7 | SM(d18:1/16:0)+DHAP |
| 885.6 | 223.1, 241.1, 283.1, 303.1, 419.3, 581.5 | PI(18:0/20:4) |
| 888.7 | 97.1, 153.1, 241.1 | ST(d18:1/24:1) |
| 904.7 | 97.1, 153.1, 241.1 | ST(d18:1/h24:1) |
Additional lipid molecular species were observed in the negative ion MALDI spectrum of accessory tissue, including m/z 788.5, 810.5, and 885.6 (Fig. 3B). CID of m/z 788.5 and 810.5 yielded product ions indicative of PS lipids (Table 3), identifying these lipid molecular species as PS(18:0/18:1) and PS(18:0/20:4), respectively (43). The product ions observed in the CID of m/z 885.6 directly off the ocular tissue, identified this lipid as PI(18:0/20:4) (supplementary Fig. ID; Table 3) (44). The negative ion MALDI images of PI(18:0/20:4), PS(18:0/18:1), and PS(18:0/20:4) revealed that these lipid molecular species were not unique to the accessory tissues, but were widespread across the entire tissue section (Fig. 3G; supplementary Fig. IVA, B).
The negative ion MALDI mass spectrum of the optic nerve in the mass range m/z 650–1,000 indicated that a wide variety of lipid molecular species were present (Fig. 3C). The negative ion MALDI image of one of the most abundant ions observed in the optic nerve, m/z 726.5, indicated that the lipid generating this particular ion was located only in the optic nerve and was not present in the retina or the sclera (Fig. 3F). CID of this lipid ion yielded product ions at m/z 281.1 (oleic acid), m/z 444.4 ([M-H-R2COOH]−), and m/z 462.4 ([M-H-R2C=O]−) (supplementary Fig. IE; Table 3), and was identified as PE(P-18:1/18:1) (45). Additionally, the negative ion MALDI images of other monounsaturated PE plasmalogen lipids, m/z 700.5 [PE(P-16:0/18:1)] and m/z 728.5 [PE(P-18:0/18:1)], and two sulfatide lipids, m/z 904.7 [ST(d18:1/h24:1)] and m/z 888.7 [ST(d18:1/24:1)], occurred with a similar distribution described above for m/z 726.5, and were only present in the optic nerve (supplementary Fig. IVC–F). Furthermore, these lipids were identified by CID directly from the optic nerve tissue (Table 3).
The negative ion MALDI mass spectrum of the retina indicated a wide variety of phospholipid molecular species (Fig. 3D). The MALDI image of one of the most abundant negative ions found in the retina, m/z 885.6, which was identified above as PI(18:0/20:4), indicates that this particular ion is found in the optic nerve, retina, and sclera (Fig. 3G). Furthermore, the negative ion MALDI image of m/z 834.5 indicated that this molecular species was present in the retina, except at the fovea, and at the optic disc (Fig. 3H). CID of this lipid ion at m/z 834.5 resulted in product ions at m/z 153.1, 283.1, 327.1, 419.4, 437.4, and 747.5 (supplementary Fig. IF; Table 3) and was identified as PS(18:0/22:6). Another lipid at m/z 750.5 had a similar distribution mentioned above and was present in both the optic nerve and the retina (supplementary Fig. IVG), and this ion was identified by negative ion CID as PE(P-18:0/20:4) (Table 3).
The distributions of some of the lipid molecular species observed in the negative ion MALDI images of the retina were highly localized. The negative ion MALDI image of m/z 774.5 indicated that this ion was observed at the optic disc and within the inner retina, except for a notable absence of signal intensity at the fovea (Fig. 3I). CID of this ion at m/z 774.5 resulted in product ions at m/z 283.1 (stearic acid), m/z 327.1 (DHA), and m/z 464.5 ([M-H-R2C=O]−) (Table 3), and was identified as PE(P-18:0/22:6) (45). Very similar localization was also observed in the negative ion MALDI image of m/z 766.5 (supplementary Fig. IVH), which was identified as PE(18:0/20:4) by CID (Table 3). The negative ion MALDI image of m/z 790.5 indicated that this lipid molecular species was found only in the photoreceptor layer, including the fovea, but not at the optic disc or the optic nerve (Fig. 3J). CID of this ion at m/z 790.5 resulted in product ions at m/z 283.1 (stearic acid), m/z 327.1 (DHA), and m/z 480.5 ([M-H-R2C=O]−) (Table 3), and was identified as PE(18:0/22:6). This lipid is thought to be present uniquely in the photoreceptor cells due to its specific localization in the outermost retinal layers, but not in the optic nerve, optic disc, or innermost retinal layers where photoreceptors are absent. Furthermore another lipid at m/z 762.5 had a similar distribution to PE(18:0/22:6) and was only present as a narrow line of signal intensity across the length of the retina, but not within the optic nerve or optic disc (supplementary Fig. IVI). This ion at m/z 762.5 was identified by negative ion CID as PE(16:0/22:6) (Table 3).
The merged image of the lipid molecular ions at m/z 834.5 [PS(18:0/22:6)], m/z 774.5 [PE(P-18:0/22:6)], and m/z 790.5 [PE(18:0/22:6)] clearly revealed the stratification of these particular lipids in the retina (Fig. 3K). Specifically, PS(18:0/22:6) was located throughout both the inner and outer retinal layers and was not specifically present in any one layer, PE(P-18:0/22:6) was specifically located in the retinal ganglion cells and the optic nerve head, and PE(18:0/22:6) was specifically located in the outer retinal layers, which include photoreceptors and the RPE cells (Fig. 3K).
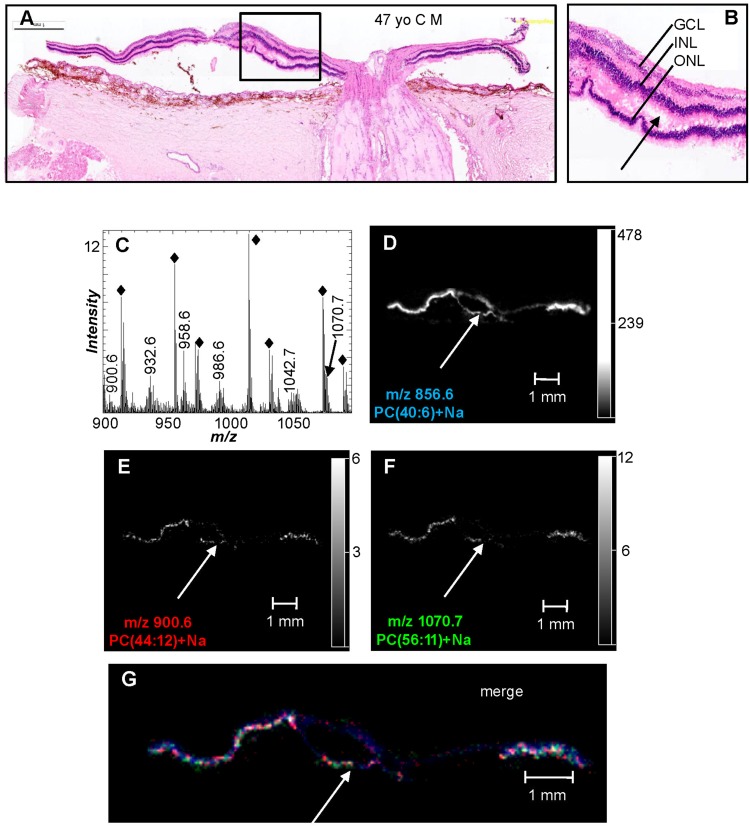
The retina is known to contain many dipolyunsaturated lipids, with the majority present as PC molecular species that contain VLC-PUFAs from the n-3 and n-6 series having 24–36 carbon atoms and four to six double bonds in the mass range of m/z 900–1,100 (11). The most abundant ions in the positive ion MALDI mass spectrum in the mass range of m/z 900–1,100 occurred at m/z 909.3, 951.3, 1009.3, and 1067.3 (Fig. 4C) and were determined to be nonlipid species due to the low mass defect and the CID data that was not indicative of lipids (data not shown). It was thought that these ions originated from the embedding compound and the tissue preparation process as the MALDI ion images indicate that these ions occur both on and off the tissue section (supplementary Fig. VA–D). The positive ion MALDI mass spectrum of the retina in this higher mass range (m/z 900–1,100) revealed the presence of several ions with an even mass-to-charge ratio and a high mass defect of 0.6 or 0.7 amu (Fig. 4C), indicative of lipid molecular species. The ions at m/z 900.6 and 1,070.7 were collisionally activated after the MALDI IMS experiment and displayed product ions at m/z 147.0 and 184.1 and a product ion corresponding to neutral loss of 183 (phosphocholine) (Table 2). This CID data indicates that the ions at m/z 900.6 and 1,070.7 were sodiated PC lipids and were therefore identified as a dipolyunsaturated PC, PC(44:12)+Na, and a VLC-PUFA PC lipid, PC(56:11)+Na, respectively. The positive ion MALDI images of PC(44:12)+Na and PC(56:11)+Na indicate that these PC lipids were uniquely localized to the retina (Fig. 4E, F). When the positive ion MALDI images of PC(40:6)+Na (blue), PC(44:12)+Na (red), and PC(56:11)+Na (green) were merged, the colocalization of these lipids in the photoreceptor layer was apparent (Fig. 4G). Additionally, the presence of DHA- and VLC-PUFA-containing PCs in the photoreceptor layer was further confirmed in this particular section of tissue where the photoreceptor layer was detached from the inner retina (Fig. 4A, B, and arrows ). This fortuitous tissue sectioning artifact allowed the photoreceptor layer to be distinguished and imaged/analyzed separately from the inner retina and aided in determining that DHA-containing and VLC-PUFA-containing PCs are indeed present in the photoreceptor layer. Additionally, similar distributions were also observed in the MALDI images of the VLC-PUFA PC lipids at m/z 1,040.7, 1,042.7, and 1,044.7 (supplementary Fig. VE–G), which were identified as PC(54:12)+Na, PC(54:11)+Na, and PC(54:10)+Na (Table 2).
Fig. 4.
Positive ion MALDI IMS of VLC-PUFA-containing PC lipids in human ocular tissue. A: H&E stain of ocular tissue section immediately adjacent to the section used for MALDI imaging. B: Inset enlarged from (A) showing a region where the photoreceptor layer is detached from the inner retina (arrow). C: Total positive ion MALDI mass spectra of the retina directly from the ocular section. The solid black diamonds indicate background ions from the embedding agent. Extracted positive ion MALDI images of the [M+Na]+ of PC(40:6) (m/z 856.6) (D), PC(44:12) (m/z 900.6) (E), and PC(56:11) (m/z 1,070.7) (F). G: Merged positive ion MALDI image of PC(56:11) (green), PC(44:12) (red), and PC(40:6) (blue). Arrows denote the layer of photoreceptors that have been detached from the inner retina. GCL, ganglion cell layer;
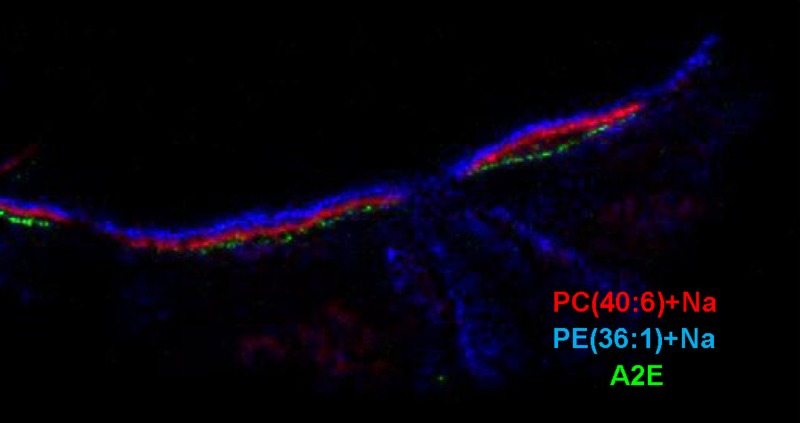
The reaction of all-trans retinal with PE to form a Schiff base is the initial step in formation of the toxic bis-retinoid, A2E (14), which has been found to accumulate in the RPE. Because this positive ion is of relatively low molecular weight, it seemed possible that it could be observed by MALDI IMS at m/z 592.5. A signal for this ion was localized to the RPE and clearly separated from the photoreceptor layer as indicated by the ion abundance at m/z 856.6 [PC(40:6)+Na] and the neuronal layer indicated by m/z 810.6 [PC(36:1)+Na] (Fig. 5). However, the signal intensity of this ion was too low to obtain CID data and the identification of this ion as A2E is tentative. The molecular ion of A2E was also observed in the RPE in a MALDI IMS study in mice that directly correlated lipofuscin with A2E (46).
Fig. 5.
Merged positive ion MALDI image of A2E (m/z 592.5) (RPE, green), PC(40:6)+Na (m/z 856.6) (photoreceptors, red), and PC(36:1)+Na (m/z 810.6) (inner retina, blue), illustrating stratification of lipid species into discrete identifiable retinal layers.
DISCUSSION
The lipid biochemistry present in the human retina is remarkable from many aspects, not the least of which is the unique accumulation of DHA (22:6) esterified to various phospholipid classes and molecular species within the retina. The high concentration of DHA esters within the photoreceptor cells of the retina has been known for a considerable length of time (4, 21, 22, 47, 48), however the exact molecular species and diverse phospholipid classes of esterified DHA has not been previously described. MALDI IMS readily revealed the location of DHA-containing phospholipids and indicated that most were only observed in the photoreceptor layer of the retina (Figs. 2I, 3J). However, MALDI IMS revealed that there were some DHA-containing phospholipids that were either in both the ganglion cell layer and the RPE/choriocapillaris complex, such as PS(18:0/22:6) shown in Fig. 3H, or highly localized to the ganglion cell layer and the optic disc, like PE(P-18:0/22:6) shown in Fig. 3I. In addition to the glycerophospholipids that contain this omega-3 fatty acid, there were also VLC-PUFAs produced by the elongation of DHA to fatty acyl chains containing 26–40 carbon atoms by the enzyme elongase of very long chain fatty acid 4 (ELOVL4) in the retina (49). While the structure of these VLC-PUFA-containing phospholipids has been previously established (22), determination of their exact location required careful dissection to uniquely isolate individual cellular structures within the retina. Our MALDI IMS studies did demonstrate the presence of VLC-PUFA-containing PC lipids exclusively in the photoreceptor layer, but the signal intensity of these ions was not robust due to the relatively low abundance of these lipids in general in the retina (10).
Several unique features of the retina could be directly observed by the abundance of specific lipids. In tissues localized behind the eye, a high abundance of neutral lipids such as TAG molecular species could be observed, but there was little if any PUFA content in this region rich in adipose tissue (Fig. 2E). In the structures containing neuronal cells, for example, the ganglion cell layer and optic nerve head, a relatively high concentration of phospholipids containing fatty acyl chains derived from saturated and monounsaturated precursors was observed (Fig. 2G, H). These lipid molecular species, including the phosphatidylcholine lipid, PC(34:1), are typically observed as the most abundant molecular species in most other neuronal tissues such as the brain (50, 51). Additionally, SM (Fig. 2F) lipids were also readily apparent in the retinal layers containing the neuronal cells and the optic nerve. The phospholipids that were unique to the photoreceptor layer of the retina contained DHA in several classes of glycerophospholipids including PC and PE. The highly unsaturated species of PI and PS, containing esterified DHA and arachidonate, were not found to be highly localized and were observed in all of the retinal layers (Fig. 3G, H). In the RPE/choriocapillaris complex, lipid mediators such as the bioactive sphingolipid ceramide could be detected. Ceramide has been shown to play a role in cell cycle arrest (52) and, conversely, is critical as a key mediator of oxidative stress-induced apoptosis in retinal photoreceptor cells (17, 18). Of interest is increasing evidence that suggests that the phosphorylation of ceramide to ceramide-1-phosphate regulates cell proliferation and apoptosis (53), suggesting a dual role for ceramide in regulating cellular homeostasis.
MALDI IMS studies of nonhuman retinas have also revealed stratification of specific phospholipids. In salamander retinas, PC(32:1), PC(32:0), and PC(34:1) are present in the outer and inner plexiform layers, and PC(36:4), PC(38:6), and PC(40:6) are found within the photoreceptor inner and outer segments, and the RPE (29). Due to the large size (10 μm or larger in diameter) of salamander photoreceptors this study was able to show that PC(36:4), PC(38:6), and PC(40:6) were differentially present within a single rod outer segment cross-section. Unfortunately, single 1.5–2.0 μm diameter rods found in human retinas cannot be detected by the method used in the current study, which has a resolution of 50 μm. A MALDI-based study of mouse retinas also demonstrated that PC(16:0/18:1) was localized to the inner nuclear layer (INL) and outer plexiform layer, PC(16:0/16:0) to the outer nuclear layer (ONL) and photoreceptor inner segment, and PC(16:0/22:6) and PC(18:0/22:6) to the photoreceptor outer segment and RPE (30), demonstrating 22:6 enrichment within photoreceptors as previously reported (4).
The accumulation of lipofuscin within the RPE occurs throughout life, is considered an indication of aging, and may correlate with the onset and/or progression of AMD (54, 55). While the complete makeup of lipofuscin is not known, it is highly enriched in bis-retinoids where the retinoid visual cycle is most active (13). The most completely characterized of the intermediate retinaldehyde adducts is A2E. In animal models with increased photoreceptor retinaldehyde, lipofuscin and A2E are also increased, suggesting that the A2E precursors, phosphatidylethanolamine and retinaldehydes, accumulate in the RPE from phagosomes and undergo incomplete degradation with gradual formation and accumulation of A2E. Evidence associates A2E with retinal degeneration (56). Recent studies utilizing MALDI imaging to compare lipofuscin and A2E localization in human and mouse RPE found good correlation in murine RPE (45), but only minimal correlation within human RPE (57). This would suggest that A2E metabolism may vary between human and mouse, and that variation in photoreceptor distribution may play a role in localization.
An additional MALDI IMS study utilized mice and rats to characterize lipids of the optic nerve (33). Mass spectra of optic nerve sections were generated from m/z 700–970 in both positive and negative ion mode, and peaks identified, revealing a wide variety of PC, PS, SM, and Cer lipids. Specifically, the sulfatide (d18:1/18:0) was localized to a region corresponding to the meninges around the nerve bundle and the sulfatide (d18:1/24:0) within the optic nerve, PC(18:1/18:0) was found around the optic nerve as it exited the retina, and PC(16:0/16:0) near the optic nerve head-vitreous interface. PC(18:0/20:4) was identified in the meninges, the connective tissue surrounding the optic nerve, and within the wall of a blood vessel. Several PS and PI lipids were also imaged, indicating unique labeling patterns for each lipid, demonstrating the specificity of MALDI IMS in revealing retinal lipid distribution.
Generally, lipid characterization by MALDI IMS in ocular tissue has been limited to mass spectra that extend to roughly m/z 900, eliminating the possibility of detecting and identifying the VLC-PUFA-containing PC lipids, which occur at very low concentrations in photoreceptors. Electrospray mass spectrometry analysis of retinas [salamander (29), rat (11), and human (10, 24)] has revealed the presence of VLC-PUFA-containing PC lipids, but localization of these lipids by MALDI imaging has not been presented. It is now understood that 22:6 is required for a healthy retina (6–8). It is important in homeostatic regulation of photoreceptor cell integrity and serves as the precursor for the potent mediator neuroprotectin D1. It plays a role in managing neuroinflammation, and diminished 22:6 has been linked to macular degeneration. It can also be the precursor molecule for the generation of VLC-PUFAs which are tightly associated with opsin in photoreceptor outer segment disk membranes. Moreover, a mutation in the ELOVL4 gene produces a dysfunctional enzyme that is incapable of elongating PUFAs (12, 58). Thus, it is extremely important to characterize the long chain and very long chain PUFAs of the retina and to determine their localization in young and aging human retinas, and in retinas exhibiting degeneration. MALDI imaging provides the means with which this can be accomplished.
While unique lipid biochemistry can be determined with this technique, application of this to study disease processes is limited to the availability of human eyes. The information presented in this manuscript, obtained from nine human subjects, revealed virtually identical distributions of major phospholipids in the inner retina, the photoreceptor cell layer, and the RPE/choriocapillaris complex, but this number of human subjects would be insufficient to ascertain subtle changes in lipid biochemistry attributable to disease processes. Nonetheless, MALDI IMS holds considerable promise if an appropriate number of age-matched control (normal) and ocular-diseased eyes could be obtained and imaged in the same study. This approach of determining the molecular species of phospholipids present in retinal tissue provides a global view of complex lipid content and evidence for unique biochemical processes taking place in localized regions.
In conclusion, the unique remodeling of omega-3 PUFAs within human retinal tissues was readily observed by MALDI IMS. The advantage of MALDI IMS is that the location of many different lipids, including glycerophospholipids, sphingolipids, and TAGs, can be readily assessed in tissue slices and relative abundance of these lipids can be determined based upon the intensities of their observed signals as long as they are within the same phospholipid family (59). Unfortunately, the 45–82 year time-span represented by the small number of specimens analyzed in this study was too small to determine age-specific differences. However, the results of this study revealed unique biochemical events related to cells within specific tissue regions of the retina and perhaps hold the promise that such an analytical approach could be used to assess the localization of lipids in age-related studies or in different diseases such as macular degeneration, retinitis pigmentosa, and diabetic retinopathy, where the engagement of lipids in the disease process has been implicated.
Supplementary Material
Acknowledgments
The authors would like to acknowledge the histological expertise of Cornelius E. Regan, Jr., for his preparation of retinal sections for orientation and MALDI IMS analysis.
Footnotes
Abbreviations:
- A2E
- N-retinylidene-N-retinylethanolamine
- AMD
- age-related macular degeneration
- Cer
- ceramide
- CID
- collision-induced dissociation
- DHAP
- 2,6-dihydroxyacetophenone
- ELOVL4
- elongase of very long chain fatty acid 4
- H&E
- hematoxylin and eosin
- IMS
- imaging mass spectrometry
- INL
- inner nuclear layer
- ONL
- outer nuclear layer
- PC
- glycerophosphatidylcholine
- PE
- glycerophosphatidylethanolamine
- PI
- glycerophosphatidylinositol
- PS
- glycerophosphatidylserine
- RPE
- retinal pigment epithelium, TAG, triacylglycerol, VLC-PUFA, very long chain PUFA
This work was supported in part by National Institutes of Health Grants AT002782 and HL-034303 (R.C.M. and K.Z.B.) and EY005121 and GM103340 (N.G.B.). It was also supported in part by the Louisiana Lions Eye Foundation, New Orleans, LA and Research to Prevent Blindness, New York, NY.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of five figures.
REFERENCES
- 1.Yu D-Y., Cringle S. J., Balaratnasingam C., Morgan W. H., Yu P. K., Su E-N. 2013. Retinal ganglion cells: energetics, compartmentation, axonal transport, cytoskeletons and vulnerability. Prog. Retin. Eye Res. 36: 217–246 [DOI] [PubMed] [Google Scholar]
- 2.Kawamura S., Tachibanaki S. 2008. Rod and cone photoreceptors: molecular basis of the difference in their physiology. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 150: 369–377 [DOI] [PubMed] [Google Scholar]
- 3.Simó R., Villarroel M., Corraliza L., Hernández C., Garcia-Ramírez M. 2010. The retinal pigment epithelium: something more than a constituent of the blood-retinal barrier–implications for the pathogenesis of diabetic retinopathy. J. Biomed. Biotechnol. 2010: 190724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fliesler S. J., Anderson R. E. 1983. Chemistry and metabolism of lipids in the vertebrate retina. Prog. Lipid Res. 22: 79–131 [DOI] [PubMed] [Google Scholar]
- 5.Jastrzebska B., Debinski A., Filipek S., Palczewski K. 2011. Role of membrane integrity on G protein-coupled receptors: rhodopsin stability and function. Prog. Lipid Res. 50: 267–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bazan N. G., Molina M. F., Gordon W. C. 2011. Docosahexaenoic acid signalolipidomics in nutrition: significance in aging, neuroinflammation, macular degeneration, Alzheimer’s, and other neurodegenerative diseases. Annu. Rev. Nutr. 31: 321–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bazan N. G., Calandria J. M., Serhan C. N. 2010. Rescue and repair during photoreceptor cell renewal mediated by docosahexaenoic acid-derived neuroprotectin D1. J. Lipid Res. 51: 2018–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bazan N. G. 2007. Homeostatic regulation of photoreceptor cell integrity: significance of the potent mediator neuroprotectin D1 biosynthesized from docosahexaenoic acid: the Proctor Lecture. Invest. Ophthalmol. Vis. Sci. 48: 4866–4881 [DOI] [PubMed] [Google Scholar]
- 9.Agbaga M. P., Mandal M. N., Anderson R. E. 2010. Retinal very long-chain PUFAs: new insights from studies on ELOVL4 protein. J. Lipid Res. 51: 1624–1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu A., Chang J., Lin Y., Shen Z., Bernstein P. S. 2010. Long-chain and very long-chain polyunsaturated fatty acids in ocular aging and age-related macular degeneration. J. Lipid Res. 51: 3217–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McMahon A., Jackson S. N., Woods A. S., Kedzierski W. 2007. A Stargardt disease-3 mutation in the mouse Elovl4 gene causes retinal deficiency of C32-C36 acyl phosphatidylcholines. FEBS Lett. 581: 5459–5463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Agbaga M. P., Brush R. S., Mandal M. N., Henry K., Elliott M. H., Anderson R. E. 2008. Role of Stargardt-3 macular dystrophy protein (ELOVL4) in the biosynthesis of very long chain fatty acids. Proc. Natl. Acad. Sci. USA. 105: 12843–12848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sparrow J. R., Wu Y., Kim C. Y., Zhou J. 2010. Phospholipid meets all-trans-retinal: the making of RPE bisretinoids. J. Lipid Res. 51: 247–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sparrow J. R., Fishkin N., Zhou J., Cai B., Jang Y. P., Krane S., Itagaki Y., Nakanishi K. 2003. A2E, a byproduct of the visual cycle. Vision Res. 43: 2983–2990 [DOI] [PubMed] [Google Scholar]
- 15.Molday R. S., Zhang K. 2010. Defective lipid transport and biosynthesis in recessive and dominant Stargardt macular degeneration. Prog. Lipid Res. 49: 476–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen H., Tran J. T., Brush R. S., Saadi A., Rahman A. K., Yu M., Yasumura D., Matthes M. T., Ahern K., Yang H., et al. 2012. Ceramide signaling in retinal degeneration. Adv. Exp. Med. Biol. 723: 553–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanvicens N., Cotter T. G. 2006. Ceramide is the key mediator of oxidative stress-induced apoptosis in retinal photoreceptor cells. J. Neurochem. 98: 1432–1444 [DOI] [PubMed] [Google Scholar]
- 18.German O. L., Miranda G. E., Abrahan C. E., Rotstein N. P. 2006. Ceramide is a mediator of apoptosis in retina photoreceptors. Invest. Ophthalmol. Vis. Sci. 47: 1658–1668 [DOI] [PubMed] [Google Scholar]
- 19.Strettoi E., Gargini C., Novelli E., Sala G., Piano I., Gasco P., Ghidoni R. 2010. Inhibition of ceramide biosynthesis preserves photoreceptor structure and function in a mouse model of retinitis pigmentosa. Proc. Natl. Acad. Sci. USA. 107: 18706–18711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bretillon L., Thuret G., Grégoire S., Acar N., Joffre C., Bron A. M., Gain P., Creuzot-Garcher C. P. 2008. Lipid and fatty acid profile of the retina, retinal pigment epithelium/choroid, and the lacrimal gland, and associations with adipose tissue fatty acids in human subjects. Exp. Eye Res. 87: 521–528 [DOI] [PubMed] [Google Scholar]
- 21.Aveldaño M. I., Bazán N. G. 1983. Molecular species of phosphatidylcholine, -ethanolamine, -serine, and -inositol in microsomal and photoreceptor membranes of bovine retina. J. Lipid Res. 24: 620–627 [PubMed] [Google Scholar]
- 22.Aveldaño M. I., Sprecher H. 1987. Very long chain (C24 to C36) polyenoic fatty acids of the n-3 and n-6 series in dipolyunsaturated phosphatidylcholines from bovine retina. J. Biol. Chem. 262: 1180–1186 [PubMed] [Google Scholar]
- 23.Acar N., Berdeaux O., Grégoire S., Cabaret S., Martine L., Gain P., Thuret G., Creuzot-Garcher C. P., Bron A. M., Bretillon L. 2012. Lipid composition of the human eye: are red blood cells a good mirror of retinal and optic nerve fatty acids? PLoS ONE. 7: e35102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berdeaux O., Juaneda P., Martine L., Cabaret S., Bretillon L., Acar N. 2010. Identification and quantification of phosphatidylcholines containing very-long-chain polyunsaturated fatty acid in bovine and human retina using liquid chromatography/tandem mass spectrometry. J. Chromatogr. A. 1217: 7738–7748 [DOI] [PubMed] [Google Scholar]
- 25.Han X., Yang K., Cheng H., Fikes K. N., Gross R. W. 2005. Shotgun lipidomics of phosphoethanolamine-containing lipids in biological samples after one-step in situ derivatization. J. Lipid Res. 46: 1548–1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ford D. A., Monda J. K., Brush R. S., Anderson R. E., Richards M. J., Fliesler S. J. 2008. Lipidomic analysis of the retina in a rat model of Smith-Lemli-Opitz syndrome: alterations in docosahexaenoic acid content of phospholipid molecular species. J. Neurochem. 105: 1032–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berry K. A., Hankin J. A., Barkley R. M., Spraggins J. M., Caprioli R. M., Murphy R. C. 2011. MALDI imaging of lipid biochemistry in tissues by mass spectrometry. Chem. Rev. 111: 6491–6512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vidová V., Pól J., Volny M., Novák P., Havlícek V., Wiedmer S. K., Holopainen J. M. 2010. Visualizing spatial lipid distribution in porcine lens by MALDI imaging high-resolution mass spectrometry. J. Lipid Res. 51: 2295–2302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roy M. C., Nakanishi H., Takahashi K., Nakanishi S., Kajihara S., Hayasaka T., Setou M., Ogawa K., Taguchi R., Naito T. 2011. Salamander retina phospholipids and their localization by MALDI imaging mass spectrometry at cellular size resolution. J. Lipid Res. 52: 463–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hayasaka T., Goto-Inoue N., Sugiura Y., Zaima N., Nakanishi H., Ohishi K., Nakanishi S., Naito T., Taguchi R., Setou M. 2008. Matrix-assisted laser desorption/ionization quadrupole ion trap time-of-flight (MALDI-QIT-TOF)-based imaging mass spectrometry reveals a layered distribution of phospholipid molecular species in the mouse retina. Rapid Commun. Mass Spectrom. 22: 3415–3426 [DOI] [PubMed] [Google Scholar]
- 31.Hayasaka T., Goto-Inoue N., Zaima N., Shrivas K., Kashiwagi Y., Yamamoto M., Nakamoto M., Setou M. 2010. Imaging mass spectrometry with silver nanoparticles reveals the distribution of fatty acids in mouse retinal sections. J. Am. Soc. Mass Spectrom. 21: 1446–1454 [DOI] [PubMed] [Google Scholar]
- 32.Hayasaka T., Goto-Inoue N., Ushijima M., Yao I., Yuba-Kubo A., Wakui M., Kajihara S., Matsuura M., Setou M. 2011. Development of imaging mass spectrometry (IMS) dataset extractor software, IMS convolution. Anal. Bioanal. Chem. 401: 183–193 [DOI] [PubMed] [Google Scholar]
- 33.Anderson D. M., Mills D., Spraggins J., Lambert W. S., Calkins D. J., Schey K. L. 2013. High-resolution matrix-assisted laser desorption ionization-imaging mass spectrometry of lipids in rodent optic nerve tissue. Mol. Vis. 19: 581–592 [PMC free article] [PubMed] [Google Scholar]
- 34.Berry K. A. Z., Li B., Reynolds S. D., Barkley R. M., Gijón M. A., Hankin J. A., Henson P. M., Murphy R. C. 2011. MALDI imaging MS of phospholipids in the mouse lung. J. Lipid Res. 52: 1551–1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodieck R. W. 1998. Sinauer Associates: Sunderland, MA. [Google Scholar]
- 36.Hankin J. A., Barkley R. M., Murphy R. C. 2007. Sublimation as a method of matrix application for mass spectrometric imaging. J. Am. Soc. Mass Spectrom. 18: 1646–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liebisch G., Vizcaíno J. A., Köfeler H., Trötzmüller M., Griffiths W. J., Schmitz G., Spener F., Wakelam M. J. O. 2013. Shorthand notation for lipid structures derived from mass spectrometry. J. Lipid Res. 54: 1523–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herrera L. C., Potvin M. A., Melanson J. E. 2010. Quantitative analysis of positional isomers of triacylglycerols via electrospray ionization tandem mass spectrometry of sodiated adducts. Rapid Commun. Mass Spectrom. 24: 2745–2752 [DOI] [PubMed] [Google Scholar]
- 39.Stübiger G., Pittenauer E., Allmaier G. 2008. MALDI seamless postsource decay fragment ion analysis of sodiated and lithiated phospholipids. Anal. Chem. 80: 1664–1678 [DOI] [PubMed] [Google Scholar]
- 40.Chen Y., Allegood J., Liu Y., Wang E., Cachón-Gonzalez B., Cox T. M., Merrill A. H., Jr, Sullards M. C. 2008. Imaging MALDI mass spectrometry using an oscillating capillary nebulizer matrix coating system and its application to analysis of lipids in brain from a mouse model of Tay-Sachs/Sandhoff disease. Anal. Chem. 80: 2780–2788 [DOI] [PubMed] [Google Scholar]
- 41.Hankin J. A., Farias S. E., Barkley R. M., Heidenreich K., Frey L. C., Hamazaki K., Kim H. Y., Murphy R. C. 2011. MALDI mass spectrometric imaging of lipids in rat brain injury models. J. Am. Soc. Mass Spectrom. 22: 1014–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Petkovic M., Schiller J., Müller M., Benard S., Reichl S., Arnold K., Arnhold J. 2001. Detection of individual phospholipids in lipid mixtures by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry: phosphatidylcholine prevents the detection of further species. Anal. Biochem. 289: 202–216 [DOI] [PubMed] [Google Scholar]
- 43.Hsu F. F., Turk J. 2005. Studies on phosphatidylserine by tandem quadrupole and multiple stage quadrupole ion-trap mass spectrometry with electrospray ionization: structural characterization and the fragmentation processes. J. Am. Soc. Mass Spectrom. 16: 1510–1522 [Erratum. 2006. J. Am. Soc. Mass Spectrom. 17: 640.] [DOI] [PubMed] [Google Scholar]
- 44.Hsu F. F., Turk J. 2000. Characterization of phosphatidylinositol, phosphatidylinositol-4-phosphate, and phosphatidylinositol-4,5-bisphosphate by electrospray ionization tandem mass spectrometry: a mechanistic study. J. Am. Soc. Mass Spectrom. 11: 986–999 [DOI] [PubMed] [Google Scholar]
- 45.Pulfer M., Murphy R. C. 2003. Electrospray mass spectrometry of phospholipids. Mass Spectrom. Rev. 22: 332–364 [DOI] [PubMed] [Google Scholar]
- 46.Grey A. C., Crouch R. K., Koutalos Y., Schey K. L., Ablonczy Z. 2011. Spatial localization of A2E in the retinal pigment epithelium. Invest. Ophthalmol. Vis. Sci. 52: 3926–3933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anderson R. E. 1970. Lipids of ocular tissues. IV. A comparison of the phospholipids from the retina of six mammalian species. Exp. Eye Res. 10: 339–344 [DOI] [PubMed] [Google Scholar]
- 48.Akino T., Tsuda M. 1979. Characteristics of phospholipids in microvillar membranes of octopus photoreceptor cells. Biochim. Biophys. Acta. 556: 61–71 [DOI] [PubMed] [Google Scholar]
- 49.Harkewicz R., Du H., Tong Z., Alkuraya H., Bedell M., Sun W., Wang X., Hsu Y. H., Esteve-Rudd J., Hughes G., et al. 2012. Essential role of ELOVL4 protein in very long chain fatty acid synthesis and retinal function. J. Biol. Chem. 287: 11469–11480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murphy R. C., Hankin J. A., Barkley R. M. 2009. Imaging of lipid species by MALDI mass spectrometry. J. Lipid Res. 50(Suppl): S317–S322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jackson S. N., Wang H. Y. J., Woods A. S. 2005. In situ structural characterization of phosphatidylcholines in brain tissue using MALDI-MS/MS. J. Am. Soc. Mass Spectrom. 16: 2052–2056 [DOI] [PubMed] [Google Scholar]
- 52.Jayadev S., Liu B., Bielawska A. E., Lee J. Y., Nazaire F., Pushkareva M. Yu., Obeid L. M., Hannun Y. A. 1995. Role for ceramide in cell cycle arrest. J. Biol. Chem. 270: 2047–2052 [DOI] [PubMed] [Google Scholar]
- 53.Arana L., Gangoiti P., Ouro A., Trueba M., Gómez-Muñoz A. 2010. Ceramide and ceramide 1-phosphate in health and disease. Lipids Health Dis. 9: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Winkler B. S., Boulton M. E., Gottsch J. D., Sternberg P. 1999. Oxidative damage and age-related macular degeneration. Mol. Vis. 5: 32. [PMC free article] [PubMed] [Google Scholar]
- 55.Sparrow J. R., Boulton M. 2005. RPE lipofuscin and its role in retinal pathobiology. Exp. Eye Res. 80: 595–606 [DOI] [PubMed] [Google Scholar]
- 56.Sparrow J. R., Gregory-Roberts E., Yamamoto K., Blonska A., Ghosh S. K., Ueda K., Zhou J. 2012. The bisretinoids of retinal pigment epithelium. Prog. Retin. Eye Res. 31: 121–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ablonczy Z., Higbee D., Grey A. C., Koutalos Y., Schey K. L., Crouch R. K. 2013. Similar molecules spatially correlate with lipofuscin and N-retinylidene-N-retinylethanolamine in the mouse but not in the human retinal pigment epithelium. Arch. Biochem. Biophys. 539: 196–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grayson C., Molday R. S. 2005. Dominant negative mechanism underlies autosomal dominant Stargardt-like macular dystrophy linked to mutations in ELOVL4. J. Biol. Chem. 280: 32521–32530 [DOI] [PubMed] [Google Scholar]
- 59.Hankin J. A., Murphy R. C. 2010. Relationship between MALDI IMS intensity and measured quantity of selected phospholipids in rat brain sections. Anal. Chem. 82: 8476–8484 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.