Abstract
Transcriptome alterations in liver and adipose tissue of cows with subclinical endometritis (SCE) at 29 d postpartum were evaluated. Bioinformatics analysis was performed using the Dynamic Impact Approach by means of KEGG and DAVID databases. Milk production, blood metabolites (non-esterified fatty acids, magnesium), and disease biomarkers (albumin, aspartate aminotransferase) did not differ greatly between healthy and SCE cows. In liver tissue of cows with SCE, alterations in gene expression revealed an activation of complement and coagulation cascade, steroid hormone biosynthesis, apoptosis, inflammation, oxidative stress, MAPK signaling, and the formation of fibrinogen complex. Bioinformatics analysis also revealed an inhibition of vitamin B3 and B6 metabolism with SCE. In adipose, the most activated pathways by SCE were nicotinate and nicotinamide metabolism, long-chain fatty acid transport, oxidative phosphorylation, inflammation, T cell and B cell receptor signaling, and mTOR signaling. Results indicate that SCE in dairy cattle during early lactation induces molecular alterations in liver and adipose tissue indicative of immune activation and cellular stress.
Keywords: uterine infection, liver, adipose, cow genomics
Introduction
Endometritis is an inflammation of the uterine internal lining that develops in many cows soon after parturition (“calving”).1 Clinical endometritis is characterized by the presence of a purulent uterine discharge (>50% pus) in the first 21 days postpartum or mucupurulent discharge (50% pus, 50% mucus) after 26 days postpartum.2 It affects approximately 20% of lactating dairy cows, with a prevalence varying from 5% to >30% in certain herds.3,4 Subclinical endometritis (SCE) is typically defined as an elevated percentage of polymorphonuclear (PMN) cells in the uterus.5,6 The SCE is characterized by the presence of >18% PMN in uterine cytology samples collected at 21 to 33 days postpartum, or >10% PMN in samples collected at 34 to 47 days postpartum.2 The incidence of SCE is the most prevalent of all uterine pathologies, affecting approximately 30% of lactating dairy cows, with a prevalence varying from 11% to >70% in certain herds.7–9 During the endometritis process, dairy cows experience a decrease in blood PMN function when compared with healthy cows.5 Furthermore, cows afflicted with endometritis have reduced fertility10,11 and may have an impairment of both the innate and adaptive immune system12,13 rendering them more susceptible to other diseases.
Elevated non-esterified fatty acids (NEFA), beta-hydroxybutyrate (BHBA) and lower glucose and calcium plasma concentrations are some of the biomarkers often associated with periparturient immune function suppression and uterine health disorders.5,14,15 However, in a recent study with grazing cows, no change was observed in plasma concentrations of NEFA, BHBA, and glucose in cows with signs of SCE, whereas, a lower albumin plasma concentration indicated impaired liver function.16 Consistent with this, other studies have reported an association between higher plasma concentrations of positive acute-phase proteins and clinical endometritis,17,18 suggesting a pro-inflammatory response involving the liver in affected cows.
In addition to the liver, recent research on the immune role of adipose depots confirmed that the tissue (and resident immune cells) is capable of synthesizing and releasing adipokines, chemokines, cytokines and inflammatory mediators upon exposure to an inflammatory condition.30,66 This peculiarity of the adipose tissue might be relevant during early lactation when there is extensive mobilization of all fat depots.19 Therefore, knowledge of both hepatic and adipocyte responses to a pro-inflammatory condition, such as SCE, might be important in understanding the cow’s systemic adaptations to the disease. It is noteworthy that a recent study demonstrated that SCE leads to alterations in gene expression profiles of both endometrium and embryo, hence, underscoring the potential for localized uterine inflammation to affect other tissues or cells.20
Previous work focusing on specific target genes associated with inflammation in dairy cows has demonstrated the relevance of the molecular approach in helping uncover the cellular processes induced by endometritis.21 However, large-scale gene expression profiling (ie, transcriptomics) coupled with bioinformatics are better suited for integrating host-tissue responses to changes in disease state.22 Therefore, in addition to blood metabolites and enzymes, we sought to study transcriptome-wide changes in liver and subcutaneous adipose tissue using a bovine microarray and bioinformatics analysis.
Methods
Animal experimental design
All animal procedures were approved by the Ruakura Animal Ethics Committee (Hamilton, New Zealand). Cows grazed pasture, managed in an intensive rotational manner.23 Uterine sampling was undertaken twice a week between 22 and 25 days postpartum for evaluation of polymorphonuclear cells (PMN) and uterine bacteriology. Cows with calving difficulties, retained placenta, or those treated with antibiotics during the first 30 days postpartum were excluded from the study.
Selection of cows
The criterion for cow selection was based on the percentage of PMN in the uterine swab. Cows with no PMN or macrophages identified were classified as no uterine infection (NUI), whereas cows with >18% PMN with an absence of macrophages were classified as SCE. Cows were paired (a NUI and a SCE cow), ensuring a balance (as best as possible) for calving date, breed, and age.6
Body condition score (BCS) and milk yield
Body condition was measured on the New Zealand 1 to 10 scale, where 1 was emaciated and 10 obese.24 Individual body condition and body weight were measured weekly pre- and post-calving. Individual milk yields (kg/d) were recorded using the DeLaval Harmony and ALPRO® milking system with milking point controller (MPC™) at each bail recording for each cow.
Blood metabolites
Blood was collected by jugular venipuncture into multiple evacuated blood tubes (EDTA, heparin, clot-activator; Becton and Dickinson, New York, USA) on the day of slaughter and immediately placed in iced water. After collection, samples were centrifuged within 30 min (12 min at 1,500 × g) and the aspirated plasma and serum fractions were stored at −20 °C until further analysis. Plasma samples were analyzed for concentrations of NEFA (WAKO, Osaka, Japan), Mg (xlidyl blue reaction), albumin (bromocresol green reaction at pH 4.1), and total protein (Biuret reaction method), with globulin calculated by subtracting albumin from total protein. Glutamate dehydrogenase (GDH, IU/L; catalyzing activity of NADH-dependent conversion of α-ketoglutarate to glutamate), and aspartate aminotransferase (AST, IU/L; catalyzing activity of transamination of L-aspartate to oxalo-acetate) were also analyzed. All assays were colorimetric, and performed at 37 °C using a Roche Modular P800 analyzer (Roche Diagnostics, Indianapolis, IN) by Gribbles Veterinary Pathology Ltd (Hamilton, New Zealand). The inter-assay and intra-assay CV for all assays were ≤11% and ≤2%, respectively.
Tissues collection and RNA extraction
Animals (6 in each NUI and SCE) were slaughtered at a commercial abattoir (AgResearch Abattoir, Hamilton, New Zealand) at 29 days postpartum (range of 27–31 days). Samples of liver and subcutaneous adipose tissue from the shoulder (~1.0 g) were stored in 2 mL cryo-vials and placed in liquid nitrogen. All tissue samples were subsequently stored at −80 °C within 30 to 50 min after exsanguination. RNA was extracted from frozen tissue using established protocols in our laboratory.25 Briefly, tissue was weighed (~0.3–0.5 g) and placed in a 15 mL centrifuge tube (Corning Inc.®, Cat. No. 430052, Corning, NY, USA) containing 1 μL linear acrylamide (Ambion® Cat. No. 9520, Austin, TX, USA) as a co-precipitant, and 5 mL of ice-cold Trizol reagent (Invitrogen Corp., Carlsbad, CA, USA). Samples were homogenized using a hand-held polytron homogenizer. This extraction procedure also utilizes acid-phenol chloroform (Ambion® Cat. No. 9720, TX, USA), which removes residual DNA. Any residual genomic DNA was removed from RNA with DNase using RNeasy Mini Kit columns (Qiagen, Hilden, Germany). The RNA concentration was measured using a Nano-Drop ND-1000 spectrophotometer (Nano-Drop Technologies, Wilmington, DE, USA). The purity of RNA (A260/A280) for all samples was above 1.81. The quality of RNA was evaluated using the Agilent Bioanalyzer system (Agilent 2100 Bioanalyzer, Agilent Technologies, Santa Clara, CA, USA). The average RNA integrity number for samples was 8.0 ± 0.4.
Microarrays
Transcript profiling was conducted using a bovine oligonucleotide microarray containing ~13,000 unique elements described elsewhere.25 The methods used for hybridization and scanning were consistent with those of Loor et al.25 Annotation was based on similarity searches using BLASTN and TBLASTX against human and mouse UniGene databases and the human genome. Briefly, cDNA was labeled with Cy3 and Cy5 fluorescent dyes (Amersham, Piscataway, NJ). A dye-swap loop microarray design was used; briefly, the Cy3 labeled cDNA (T1) from SCE was co-hybridized with Cy5 labeled cDNA (C1) from NUI liver samples to generate fluorescence ratios as a measurement of relative expression across the treatments. Moreover, in the next microarray slide the Cy3-labeled cDNA (C1) of the same NUI sample was co-hybridized with another Cy5-labeled (T2) SCE sample to form a loop (Suppl 1. Fig. 1).
Figure 1.
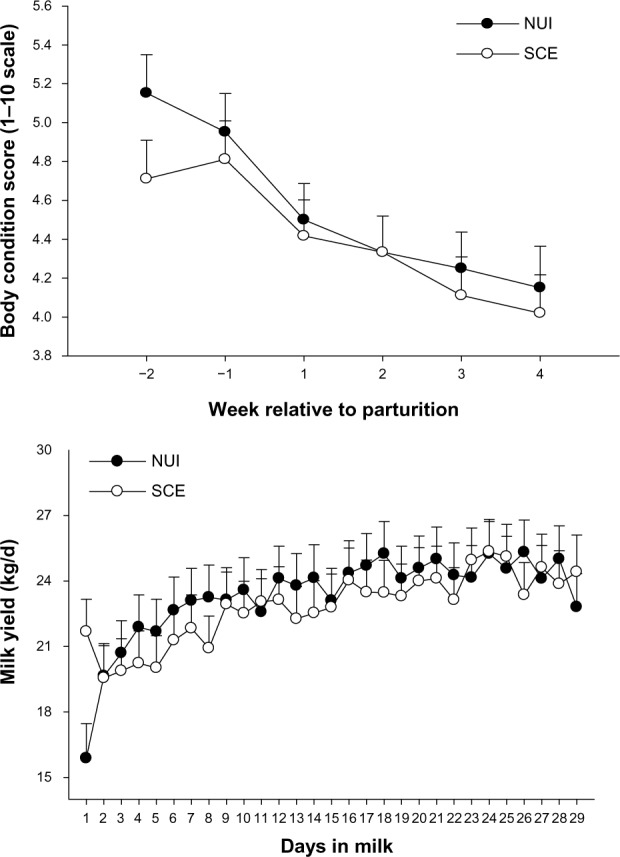
Body condition score and daily milk yield in healthy cows (no uterine infection, NUI) and cows with subclinical endometritis (SCE). Body condition score is reported on a 1–10 scale, where 1 is emaciated and 10 obese.24,83
Notes: The bar associated with each mean denotes the standard error of the mean.
Quantitative PCR
Quantitative real-time PCR (qPCR) was performed to verify microarray results and analyze other genes of interest. Protocols for primer design, primer testing, selection of internal control genes (ICG) for normalization, and details of the qPCR analysis were as previously described.27 Detailed information of the primers is provided in Suppl 1. Table 1 and 2. Briefly, for liver tissue the genes analyzed by qPCR included transcription regulators (PPARA, RXRA, NFIL3), genes associated with fatty acid oxidation (ACOX1, CPT1A), ketogenesis (HMGCS2), growth hormones/IGF-1-axis (total GHR, IGF1), cytokines/inflammatory mediators (IL10, IL1B, TNF, STAT3, SOCS2, STAT5B), acute-phase proteins (ORM1, HP, SAA3, PON1), oxidative stress (SOD1, GPX1), hepatokines (ANGPTL4, FGF21) and insulin signaling (AKT1). These targets are central components of the pathways under study.28 The final data were normalized using the geometric mean (V2/3 = 0.20; geNorm) of ubiquitously-expressed transcript (UXT), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and ribosomal protein S9 (RPS9), detailed information of qPCR efficiency and% relative RNA abundance can be found in Suppl 1. Table 3.
Table 1.
Plasma metabolite concentration on the day of slaughter in healthy cows (no uterine infection, NUI) or cows with subclinical endometritis (SCE).
| GROUP
|
||||
|---|---|---|---|---|
| ITEM | SCE | NUI | SEM | P = |
|
| ||||
| NEFA, mmol/L | 1.50 | 1.69 | 0.18 | 0.46 |
|
| ||||
| Protein | 82.1 | 77.1 | 2.8 | 0.23 |
|
| ||||
| Albumin, g/L | 36.8 | 37.5 | 25.8 | 0.75 |
|
| ||||
| Globulin, g/L1 | 45.3 | 39.6 | 2.2 | 0.11 |
|
| ||||
| Albumin/Globulin ratio | 0.83 | 0.95 | 0.06 | 0.23 |
|
| ||||
| Mg, mmol/L | 0.66 | 0.70 | 10.6 | 0.70 |
|
| ||||
| GDH, IU/L2 | 24.3 | 28.3 | 5.5 | 0.62 |
|
| ||||
| AST, IU/L3 | 78.5 | 8 7. 5 | 4.6 | 0.20 |
Notes:
Globulin is calculated when albumin is subtracted from total protein.
GDH = glutamate dehydrogenase.
AST = aspartate aminotransferase.
Table 2.
List of all genes that were differentially expressed in liver tissue due to subclinical endometritis.1
| GENE ID | SYMBOL | EXPRESSION RATIO SCE/NUI | P = | GENE ID | SYMBOL | EXPRESSION RATIO SCE/NUI | P = |
|---|---|---|---|---|---|---|---|
| 281358 | NPR3 | 0.43 | 0.001 | – | KIAA1639 | 1.54 | 0.05 |
| 782932 | C2orf53 | 0.46 | 0.04 | 282578 | ATP5A1 | 1.54 | 0.03 |
| 541142 | LANCL2 | 0.48 | 0.01 | 526937 | DCXR | 1.54 | 0.001 |
| 338074 | AOX1 | 0.52 | 0.03 | 504664 | NRBP2 | 1.54 | 0.001 |
| 286794 | MMP7 | 0.52 | 0.04 | 534579 | TFDP1 | 1.55 | 0.01 |
| 281938 | MYOD1 | 0.52 | 0.001 | 534799 | TOP1 | 1.55 | 0.001 |
| 538785 | STAMBPL1 | 0.53 | 0.03 | 514465 | SMAP1L | 1.56 | 0.001 |
| 522346 | LOC522346 | 0.55 | 0.03 | 767942 | GRN | 1.56 | 0.001 |
| 616425 | SUPT4H1 | 0.55 | 0.01 | 280815 | NPC2 | 1.56 | 0.01 |
| 524334 | L3MBTL3 | 0.55 | 0.001 | 538975 | CNN3 | 1.57 | 0.05 |
| 282092 | TIMP1 | 0.55 | 0.04 | 511852 | ASB11 | 1.57 | 0.03 |
| 530184 | SSBP3 | 0.56 | 0.04 | 525619 | LOC525619 | 1.57 | 0.02 |
| 615833 | IFITM3 | 0.56 | 0.04 | 535362 | GRM7 | 1.58 | 0.02 |
| 286822 | CKM | 0.58 | 0.02 | 514514 | CELSR3 | 1.58 | 0.01 |
| – | CN437645 | 0.59 | 0.01 | 444859 | ST3GAL3 | 1.58 | 0.03 |
| 768316 | NRBF2 | 0.60 | 0.03 | 535258 | HSPC148 | 1.58 | 0.001 |
| 338074 | LOC618565 | 0.61 | 0.05 | 533483 | CMAS | 1.59 | 0.03 |
| 282684 | CSNK1A1 | 0.61 | 0.05 | 282385 | TMSB10 | 1.60 | 0.05 |
| 780809 | VTI1B | 0.61 | 0.01 | 522795 | KLF10 | 1.61 | 0.04 |
| 282100 | TRPC1 | 0.61 | 0.001 | 530352 | SLC39A1 | 1.63 | 0.01 |
| 535232 | LOC535232 | 0.62 | 0.01 | 783871 | PI3 | 1.64 | 0.01 |
| 616676 | GALM | 0.62 | 0.03 | 444874 | UBC | 1.64 | 0.02 |
| 524743 | LOC524743 | 0.62 | 0.01 | 614936 | ILF3 | 1.65 | 0.03 |
| – | BF043596 | 0.62 | 0.001 | 536863 | MPDZ | 1.65 | 0.04 |
| 574056 | TNFRSF8 | 0.63 | 0.02 | 534366 | C6orf62 | 1.65 | 0.03 |
| 100139141 | SHROOM3 | 0.63 | 0.01 | 281040 | CAV1 | 1.66 | 0.001 |
| – | CN438887 | 0.63 | 0.01 | 286883 | MAP2K6 | 1.66 | 0.02 |
| – | CN435533 | 0.64 | 0.01 | 100125835 | PCYOX1 | 1.67 | 0.01 |
| 507060 | WBP2 | 0.64 | 0.01 | 280997 | AMD1 | 1.68 | 0.04 |
| 507503 | THAP4 | 0.64 | 0.03 | 282711 | EPAS1 | 1.70 | 0.01 |
| 408019 | CCR4 | 0.64 | 0.03 | 282146 | ATP2B4 | 1.72 | 0.01 |
| 512648 | ELL3 | 0.65 | 0.01 | 614280 | C7orf23 | 1.73 | 0.001 |
| 514788 | ACOT7 | 0.65 | 0.05 | 530076 | GC | 1.76 | 0.03 |
| 508167 | LOC508167 | 0.66 | 0.04 | 327715 | GABARAP | 1.76 | 0.001 |
| 319095 | ADCYAP1R1 | 0.66 | 0.01 | 510102 | RARRES1 | 1.78 | 0.001 |
| – | LOC727737 | 1.50 | 0.03 | 767925 | RPL13A | 1.80 | 0.02 |
| 505031 | C6orf49 | 1.51 | 0.001 | 100298683 | FAM43B | 1.85 | 0.02 |
| 533894 | LRP1 | 1.51 | 0.001 | 415113 | HSPA5 | 1.91 | 0.03 |
| 511425 | LOC511425 | 1.51 | 0.01 | 618041 | APOC4 | 1.95 | 0.001 |
| 504506 | RNPS1 | 1.51 | 0.001 | 280988 | AHSG | 2.01 | 0.04 |
| 506149 | C22orf32 | 1.51 | 0.03 | 534961 | C1QA | 2.07 | 0.001 |
| 508722 | RBM39 | 1.52 | 0.04 | 768005 | Slc9a3r2 | 2.11 | 0.001 |
| 512841 | Crk | 1.52 | 0.01 | 540176 | ATP5G3 | 2.15 | 0.001 |
| – | BF045974 | 1.53 | 0.001 | 281760 | FABP5 | 2.31 | 0.02 |
| – | SLC2A3P1 | 1.53 | 0.01 | 510522 | FGB | 2.42 | 0.01 |
| 613930 | MGC128212 | 1.54 | 0.03 | 513055 | RGS2 | 2.52 | 0.001 |
| 540007 | FAAH | 1.54 | 0.001 | 539334 | IL1RAP | 2.54 | 0.001 |
| 507107 | SLC3A2 | 1.54 | 0.001 | 338048 | CYP11A1 | 2.65 | 0.04 |
Notes:
SCE = subclinical uterine infection (endometritis). NUI = no uterine infection.
Table 3.
List of all genes that were differentially expressed in subcutaneous adipose tissue due to subclinical endometritis.1
| GENE ID | SYMBOL | EXPRESSION RATIO SCE/NUI | P = | GENE ID | SYMBOL | EXPRESSION RATIO SCE/NUI | P = |
|---|---|---|---|---|---|---|---|
| 326334 | CA11 | 0.06 | 0.001 | 100140537 | POLR3E | 0.65 | 0.01 |
| 506415 | RSAD2 | 0.30 | 0.01 | 281187 | GDF8 | 0.65 | 0.03 |
| 527934 | CORO7 | 0.32 | 0.001 | 338079 | NDUFS7 | 0.65 | 0.01 |
| 281127 | DSC1 | 0.34 | 0.04 | 100138153 | CNNM3 | 0.65 | 0.02 |
| – | 9430053O09Rik | 0.41 | 0.02 | 281781 | B4GALT1 | 0.65 | 0.02 |
| 515954 | HYI | 0.41 | 0.01 | 514263 | TMEM39B | 0.65 | 0.01 |
| 513765 | TRABD | 0.43 | 0.03 | 519163 | KLF16 | 0.67 | 0.05 |
| 615833 | IFITM3 | 0.43 | 0.01 | – | SCRAMBLE2 | 1.50 | 0.04 |
| 507525 | VTN | 0.44 | 0.02 | 527491 | SLC38A1 | 1.50 | 0.001 |
| 509430 | SRP72 | 0.45 | 0.01 | 530401 | LOC530401 | 1.51 | 0.01 |
| 786644 | LOC786644 | 0.46 | 0.001 | 518675 | BZRAP1 | 1.51 | 0.03 |
| 508527 | FAM44A | 0.46 | 0.03 | 767924 | MST150 | 1.51 | 0.001 |
| 280830 | IVL | 0.47 | 0.01 | 507707 | TOMM70A | 1.51 | 0.05 |
| 531369 | TTF2 | 0.47 | 0.02 | – | LOC729867 | 1.52 | 0.04 |
| 516232 | ZNF653 | 0.48 | 0.02 | – | CN441658 | 1.52 | 0.05 |
| 527928 | UGT2A1 | 0.48 | 0.001 | 281564 | UGDH | 1.53 | 0.01 |
| 510377 | SP140 | 0.49 | 0.01 | 503554 | COX6B2 | 1.53 | 0.01 |
| 529049 | MRC2 | 0.49 | 0.05 | 516762 | LOC516762 | 1.53 | 0.04 |
| – | LOC512590 | 0.50 | 0.001 | 505738 | MCOLN1 | 1.53 | 0.01 |
| 516866 | ALPK3 | 0.51 | 0.01 | 514957 | KIAA0020 | 1.55 | 0.001 |
| 768005 | Slc9a3r2 | 0.52 | 0.03 | 509050 | C15orf40 | 1.55 | 0.01 |
| – | BM363375 | 0.52 | 0.001 | 508935 | SYMPK | 1.55 | 0.04 |
| 527903 | LOC653968 | 0.52 | 0.03 | 282524 | SLC25A16 | 1.55 | 0.03 |
| 614868 | KHK | 0.53 | 0.01 | 613479 | MGC133516 | 1.56 | 0.02 |
| – | C14orf73 | 0.53 | 0.02 | 281944 | NR2E3 | 1.57 | 0.01 |
| 533984 | SUB1 | 0.53 | 0.03 | 538444 | SET | 1.59 | 0.03 |
| – | OAS1 | 0.53 | 0.01 | 540605 | CREM | 1.59 | 0.04 |
| 512972 | HMHA1 | 0.54 | 0.001 | 525040 | LOC525040 | 1.60 | 0.01 |
| 541100 | PLEKHH3 | 0.55 | 0.001 | 281751 | EIF4E | 1.61 | 0.05 |
| 504889 | GMPPA | 0.56 | 0.04 | 338082 | ATP6V1B2 | 1.61 | 0.03 |
| 286850 | GNG12 | 0.56 | 0.02 | 536417 | SEMA3D | 1.62 | 0.04 |
| – | BF043373 | 0.56 | 0.04 | 613644 | CD99L2 | 1.62 | 0.001 |
| 516456 | MVP | 0.56 | 0.04 | 614521 | Rab35 | 1.62 | 0.03 |
| 615501 | LOC615501 | 0.56 | 0.04 | 614583 | LOC614583 | 1.62 | 0.04 |
| 514859 | AKT1S1 | 0.57 | 0.03 | – | CN438914 | 1.63 | 0.001 |
| 532997 | AOF2 | 0.57 | 0.04 | 521854 | NID2 | 1.64 | 0.02 |
| 615146 | SOCS6 | 0.57 | 0.03 | – | TC359671 | 1.65 | 0.001 |
| 529131 | LOC529131 | 0.58 | 0.02 | 532209 | LOC532209 | 1.65 | 0.02 |
| 521868 | PTPRH | 0.58 | 0.05 | 505727 | NOV | 1.66 | 0.04 |
| 514291 | EMILIN2 | 0.59 | 0.02 | 524854 | THUMPD1 | 1.66 | 0.02 |
| 534280 | RARA | 0.59 | 0.03 | 531535 | MGC139383 | 1.67 | 0.001 |
| 516318 | PER1 | 0.59 | 0.04 | – | CK394167 | 1.74 | 0.02 |
| 536731 | LOC536731 | 0.59 | 0.001 | 507102 | SREBF2 | 1.75 | 0.02 |
| 505328 | OSBPL2 | 0.60 | 0.001 | 450214 | CENPC1 | 1.76 | 0.01 |
| 783452 | NRG2 | 0.60 | 0.05 | 534923 | AKT2 | 1.76 | 0.04 |
| 513577 | LOC513577 | 0.60 | 0.01 | 768209 | 5730406M06Rik | 1.77 | 0.02 |
| 512069 | MGC137476 | 0.60 | 0.02 | 529149 | MGC52110 | 1.79 | 0.04 |
| 511254 | NRD1 | 0.60 | 0.03 | 540525 | GTF2E1 | 1.79 | 0.001 |
| 522346 | LOC522346 | 0.60 | 0.02 | 504483 | BLMH | 1.79 | 0.01 |
| 327685 | ANXA6 | 0.61 | 0.02 | 280981 | ADFP | 1.80 | 0.05 |
| 507065 | RBM19 | 0.61 | 0.03 | 282188 | COL1A2 | 1.80 | 0.01 |
| 513300 | SENP8 | 0.61 | 0.03 | 282484 | SLC34A2 | 1.80 | 0.02 |
| 518833 | CYFIP2 | 0.61 | 0.02 | 782059 | SCYL2 | 1.82 | 0.01 |
| 786191 | HIC1 | 0.61 | 0.001 | 780788 | ITGB1BP3 | 1.85 | 0.02 |
| 282431 | PROKR2 | 0.62 | 0.03 | 617358 | ASB1 | 1.85 | 0.03 |
| – | CN438353 | 0.62 | 0.001 | 540172 | MGC10433 | 1.91 | 0.04 |
| 539675 | ZNF415 | 0.62 | 0.03 | 505889 | PPYR1 | 1.94 | 0.04 |
| – | FLJ45422 | 0.62 | 0.04 | 281598 | ACVR2A | 1.97 | 0.04 |
| 532645 | MAN1A2 | 0.62 | 0.02 | – | NG010008B10D06 | 1.98 | 0.02 |
| 280946 | TST | 0.62 | 0.01 | 282127 | ZFP36 | 2.01 | 0.04 |
| 511691 | LOC511691 | 0.63 | 0.02 | 539795 | ABHD3 | 2.04 | 0.001 |
| 512308 | MMRN2 | 0.63 | 0.01 | 525795 | AGRN | 2.04 | 0.02 |
| 617894 | LOC617894 | 0.63 | 0.02 | 281490 | JARID1C | 2.06 | 0.02 |
| – | CR452857 | 0.63 | 0.01 | – | TC345541 | 2.11 | 0.01 |
| 505884 | KLF6 | 0.63 | 0.01 | 327691 | NDUFS6 | 3.32 | 0.001 |
| 534394 | NIP30 | 0.63 | 0.03 | 614759 | LSM3 | 3.48 | 0.001 |
| 616482 | RANBP3 | 0.63 | 0.03 | 337916 | IRF2 | 4.13 | 0.001 |
| 535203 | NTHL1 | 0.63 | 0.01 | 617720 | NUDT12 | 4.41 | 0.001 |
| – | BF440371 | 0.63 | 0.02 | – | CR551628 | 7.15 | 0.001 |
| 407996 | KRIT1 | 0.64 | 0.02 | ||||
| 532721 | USP42 | 0.64 | 0.02 | ||||
| 518458 | SEMA6D | 0.64 | 0.02 | ||||
| 539250 | KCNJ1 | 0.64 | 0.04 | ||||
| 536818 | LOC536818 | 0.64 | 0.03 | ||||
| 534063 | EIF2B3 | 0.65 | 0.001 |
Notes:
SCE = subclinical uterine infection (endometritis). NUI = no uterine infection.
For subcutaneous adipose tissue the genes analyzed by qPCR included transcription regulators (RXRA, PPARG, SREBF1, TP53, NFIL3), lipogenic enzymes (LP1N1, SCD, FASN, PLIN2), lipolysis (ABHD5, LIPE, PNPLA2), insulin signaling pathway (GHR, SLC2A4, IRS1), adipokines (ADIPOQ, ANGPTL4, CCL5, BCL2, CCL2), cytokines/inflammatory mediators (IL6, HP, SAA3, NFKB1, STAT3) and oxidative stress (SOD2). These targets are central components of the pathways under study and some have been reported to respond to inflammatory stimuli.29,30 The final data were normalized using the geometric mean (V2/3 = 0.20; geNorm) of FUN14 domain containing 2 (FUNDC2), secretion regulating guanine nucleotide exchange factor (SERGEF), solute carrier family 35 (adenosine 3′-phospho 5′-phosphosulfate transporter), member B2 (SLC35B2), and prokineticin receptor 1 (PROKR1). The detailed information for qPCR efficiency and% relative RNA abundance of the measured genes is provided in Suppl 1. Table 4.
Table 4.
Expression of genes associated with inflammation, oxidative stress and metabolism in liver tissue from healthy cows (no uterine infection, NUI) or cows with subclinical endometritis (SCE). Data were generated via quantitative RT-PCR.
| GROUP
|
||||
|---|---|---|---|---|
| GENE | SCE | NUI | SEM | P = |
|
| ||||
| Transcription regulators | ||||
|
| ||||
| PPARA | −0.19 | −0.09 | 0.24 | 0.78 |
|
| ||||
| RXRA | 0.38(↑) | 0.11 | 0.10 | 0.06 |
|
| ||||
| NFIL3 | 0.60 | 0.54 | 0.30 | 0.87 |
|
| ||||
| Fatty acid oxidation | ||||
|
| ||||
| ACOX1 | 0.23 | 0.32 | 0.14 | 0.64 |
|
| ||||
| CPT1A | 0.25 | 0.20 | 0.17 | 0.82 |
|
| ||||
| Ketogenesis | ||||
|
| ||||
| HMGCS2 | 0.23 | 0.35 | 0.24 | 0.72 |
|
| ||||
| GH/IGF-1 axis | ||||
|
| ||||
| GHR | 0.53(↑) | 0.19 | 0.16 | 0.05 |
|
| ||||
| IGF1 | 0.25 | 0.24 | 0.27 | 0.96 |
|
| ||||
| Cytokines/inflammatory mediators | ||||
|
| ||||
| IL10 | −0.29 | 0.33 | 0.51 | 0.38 |
|
| ||||
| IL1B | 0.22 | 0.32 | 0.35 | 0.84 |
|
| ||||
| TNF | 0.28 | 0.57 | 0.37 | 0.57 |
|
| ||||
| STAT3 | 0.50 | 0.21 | 0.18 | 0.25 |
|
| ||||
| SOCS2 | 0.23 | −0.40 | 0.40 | 0.27 |
|
| ||||
| STAT5B | 0.08 | 0.13 | 0.08 | 0.71 |
|
| ||||
| Acute-phase proteins | ||||
|
| ||||
| ORM1 | 0.13 | 0.10 | 0.49 | 0.96 |
|
| ||||
| HP | 0.41(↑) | −1.03 | 0.69 | 0.04 |
|
| ||||
| SAA3 | −0.36 | −0.75 | 0.68 | 0.69 |
|
| ||||
| PON1 | 0.32 | 0.10 | 0.15 | 0.30 |
|
| ||||
| Oxidative stress | ||||
|
| ||||
| SOD2 | 0.42(↑) | 0.05 | 0.13 | 0.04 |
|
| ||||
| GPX1 | 0.44(↑) | −0.05 | 0.20 | 0.08 |
|
| ||||
| Hepatokines | ||||
|
| ||||
| ANGPTL4 | 0.11 | −0.05 | 0.35 | 0.74 |
|
| ||||
| FGF21 | −0.65 | −0.51 | 0.92 | 0.91 |
|
| ||||
| Insulin signaling | ||||
|
| ||||
| AKT1 | −0.63 | −0.69 | 0.11 | 0.72 |
Statistical analysis
Data from a total of 12 microarrays were normalized for dye and microarray effects (ie, Lowess normalization and microarray centering) and used for statistical analysis.25 Data were analyzed using the Proc MIXED procedure of SAS (SAS, SAS Inst. Inc., Cary, NC). Fixed effects were treatment (SCE = infection and NUI = control), and random effects included cow and microarray. Quantitative PCR data normalized relative to the internal control genes were first log2-transformed prior to statistical analysis. Differentially expressed genes were determined as reported recently31 using a cutoff of P < 0.05 and a fold-change greater or lower than 1.5 in the comparison of SCE versus NUI. Validation for this approach, which does not take into account multiple testing corrections, was reported previously.26,102
Bioinformatics
The entire microarray data set with associated statistical P-values was imported into the Database for Annotation, Visualization and Integrated Discovery (DAVID)32 analyses tools (http://david.abcc.ncifcrf.gov) and Kyoto Encyclopedia of Genes and Genomes (KEGG) software to obtain significant gene ontology (GO) terms in different categories (molecular functions, biological pathways and cellular components). The whole annotated microarray was used as the reference dataset (ie, background) for pathway analysis. In other words, only pathways represented by the genes on the microarray platform were used for the bioinformatics analysis.
Functional enrichment analysis
The functional enrichment analysis was performed using the Dynamic Impact Approach (DIA).33 The DIA is a novel functional analysis tool that allows visualizing the impact and the direction of the impact of DEG on several annotation databases including GO and KEGG pathways. The DIA has been described in detail previously;33 briefly, the alterations in expression of proteins in a metabolic or signaling pathway determine the flux (or overall direction, ie, activation/up-regulation, inhibition/down-regulation, or no change) of the respective pathway, whereas, the extent of the impact on a pathway is directly depicted by the number of proteins affected by a particular treatment. The cumulative flux of pathways is determined by total number of up-regulated and down-regulated proteins involved in a pathway. If the ratio of up-regulated/down-regulated = 1, the flux can be taken as unchanged overall, regardless of the fact that the treatment had a large impact on the pathway.34 The impact and the flux for KEGG pathways were calculated for only those terms which were represented by at least 30% in the microarray compared with the whole annotated bovine genome.
Results
Body condition score (BCS), milk production and blood metabolites
There was a postpartum (P < 0.01) decrease in BCS, but no effect of health status was observed (P = 0.45) (Fig. 1). There were no health status × week relative to parturition interactions (P = 0.76) for BCS. Irrespective of health status (P = 0.78), milk yield increased between day 1 and 29 postpartum (P = 0.07). However, there was a trend (P = 0.07) for an interaction of health status × week for daily milk yield due to healthy cows producing more milk through the first 3 weeks postpartum (Fig. 1). There was no effect (P = 0.11 to 0.70) of health status on the concentration of blood biomarkers measured (Table 1).
Differential expression of genes using microarrays
The analysis of microarray data revealed a total of 97 differentially expressed genes (DEG; P < 0.05, fold-change cut off ≥1.5 or ≤1/1.5 SCE vs. NUI) in liver (35 down-regulated, 62 up-regulated) (Table 2). The primary up-regulated DEG included genes involved in steroid biosynthesis (CYP11A1), acute-phase and pro-inflammatory proteins synthesis (IL1RAP), GTPase activity and leukemogenesis (RGS2), beta component of fibrinogen (FGB), lipid uptake, transport and metabolism (FABP5, APOC4), oxidative phosphorylation (ATP5A1, ATP5G3) and cell growth (GRN) and complement system (C1QA). Whereas, the primary down-regulated DEG included genes involved in peptide hormone binding (NPR3), immune system and inflammation (IFITM3, CCR4, AOX1), breakdown of extracellular matrix (MMP7), ATP binding (CKM), and fatty acid catabolism (ACOT7) (Table 2).
Microarray analysis of adipose uncovered 144 DEG (P < 0.05, fold-change cut off ≥1.5 or ≤1/1.5 SCE vs. NUI) (82 down-regulated, 62 up-regulated) (Table 3). The primary up-regulated DEG included genes involved in endogenous cholesterol synthesis (SREBF2), NAD metabolism (NUDT12, UGDH), cytokine-mediated signaling (IRF2), oxidative phosphorylation (NDUFS6), and insulin signaling (AKT2). Whereas, the primary down-regulated DEG included genes involved in golgi complex morphology and function (CORO7), cytokine-mediated signaling (IFITM3), calcium ion binding (ANXA6), oxidative phosphorylation (NDUFS7), and apoptosis (HIC1) (Table 3).
Differential expression of genes by qPCR
Expression analysis by qPCR in liver tissue revealed a significant up-regulation of total GHR (somatotropic axis), HP (acute-phase protein), SOD2 and GPX1 (oxidative stress), and RXRA (fatty acid oxidation) in cows with SCE (Table 4). In subcutaneous adipose tissue, infection was associated with up-regulation of TP53 (transcription factor), IL6 and CCL2 (adipokines), and tended (P = 0.16) to increase STAT3 (inflammation). The expression of the lipid droplet-associated gene PLIN2, SLC2A4 and IRS1 (insulin signaling), and SOD2 (oxidative stress) in adipose was down-regulated in SCE (Table 5).
Table 5.
Expression of genes associated with inflammation, oxidative stress and metabolism in subcutaneous adipose from healthy cows (no uterine infection, NUI) or cows with subclinical endometritis (SCE). Data were generated via quantitative RT-PCR.
| GROUP
|
||||
|---|---|---|---|---|
| GENE | SCE | NUI | SEM | P = |
|
| ||||
| Transcription regulators | ||||
|
| ||||
| RXRA | −1.30 | −0.71 | 0.37 | 0.28 |
|
| ||||
| PPARG | 1.49 | 1.63 | 0.18 | 0.60 |
|
| ||||
| SREBF1 | 0.80 | 1.15 | 0.31 | 0.44 |
|
| ||||
| TP53 | 1.23(↑) | −0.20 | 0.44 | 0.04 |
|
| ||||
| NFIL3 | 0.68 | 0.44 | 0.12 | 0.20 |
|
| ||||
| Lipogenic enzymes | ||||
|
| ||||
| LPIN1 | −0.15 | 0.16 | 0.32 | 0.50 |
|
| ||||
| SCD | −1.21 | −0.12 | 0.80 | 0.36 |
|
| ||||
| FASN | 0.26 | 0.31 | 0.33 | 0.91 |
|
| ||||
| PLIN2 | −1.86(↓) | 0.20 | 0.57 | 0.02 |
|
| ||||
| Lipolytic-related (lipolysis) | ||||
|
| ||||
| ABHD5 | 0.09 | 0.08 | 0.26 | 0.98 |
|
| ||||
| LIPE | 0.14 | −0.10 | 0.19 | 0.38 |
|
| ||||
| PNPLA2 | 0.15 | 0.45 | 0.19 | 0.26 |
|
| ||||
| Insulin signaling | ||||
|
| ||||
| GHR | −0.09 | −0.08 | 0.30 | 0.98 |
|
| ||||
| SLC2A4 | 0.82(↓) | 1.44 | 0.21 | 0.03 |
|
| ||||
| IRS1 | −1.69(↓) | −1.02 | 0.13 | <0.01 |
|
| ||||
| Adipokines and hepatokines | ||||
|
| ||||
| ADIPOQ | 0.73 | 0.77 | 0.68 | 0.97 |
|
| ||||
| ANGPTL4 | −0.07 | −0.06 | 0.27 | 0.98 |
|
| ||||
| CCL5 | −1.23 | −1.26 | 0.52 | 0.98 |
|
| ||||
| BCL2 | −0.70 | −0.48 | 0.31 | 0.62 |
|
| ||||
| CCL2 | −1.34(↑) | −3.62 | 0.46 | <0.01 |
|
| ||||
| Oxidative stress | ||||
|
| ||||
| SOD2 | −2.44(↓) | 0.99 | 1.22 | 0.06 |
|
| ||||
| Cytokines/inflammatory mediators | ||||
|
| ||||
| IL6 | 0.36(↑) | −2.27 | 0.33 | <0.01 |
|
| ||||
| HP | −1.74 | −1.20 | 0.51 | 0.46 |
|
| ||||
| SAA3 | −2.64 | −0.68 | 1.26 | 0.29 |
|
| ||||
| NFKB1 | 1.26 | 0.98 | 0.28 | 0.98 |
|
| ||||
| STAT3 | 1.68 | 0.96 | 0.35 | 0.16 |
Functional analysis of KEGG pathways using DIA
Liver
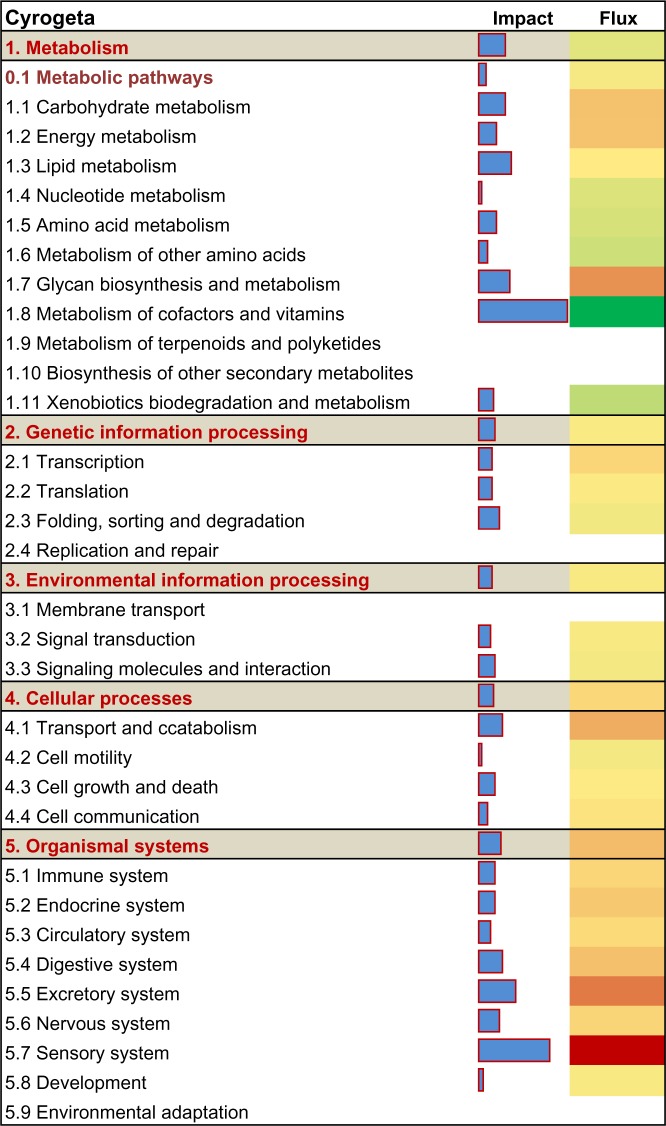
A functional analysis of DEG (P < 0.05, fold-change cut off ≥1.5 or ≤1/1.5 SCE vs. NUI) was undertaken with DIA using the KEGG pathway database. In liver, the metabolic pathways and organismal systems categories were the most impacted due to SCE; whereas, environmental information processing, genetic information processing and cellular processes were the least impacted categories (Fig. 2). Among the top metabolism subcategories affected by SCE, there was an overall inhibition of vitamin B6 metabolism, nicotinate and nicotinamide metabolism (metabolism of cofactors and vitamins), and arginine and proline metabolism (amino acid metabolism) (Fig. 3). Biosynthesis of unsaturated fatty acids (lipid metabolism), tyrosine metabolism, tryptophan metabolism, and drug metabolism— cytochrome P450 also were among the top 50 inhibited metabolism subcategories (Suppl 2: KEGG Liver sheet). The analysis further uncovered that steroid hormone biosynthesis (lipid metabolism), pentose inter-conversion (carbohydrate metabolism), glycosaminoglycan biosynthesis– keratan sulfate (glycan biosynthesis and metabolism), and oxidative phosphorylation (energy metabolism) were the most activated subcategories (Fig. 3; Suppl 2: KEGG Liver sheet).
Figure 2.
Impact and flux of main KEGG pathways categories and sub-categories affected by subclinical endometritis constructed from liver differentially expressed genes (DEG) as calculated by the Dynamic Impact Approach. Reported are the total impact (Blue horizontal bars; larger the bars higher the impact) and the direction of the impact (or flux; green bars expanding left denote inhibition and red bars expanding right denote activation) of DEG on the categories and subcategories.
Figure 3.
Impact and flux of the top 20 KEGG pathways affected by subclinical endometritis in liver tissue uncovered by the Dynamic Impact Approach (DIA). Reported are the total impact (blue horizontal bars; larger the bars higher the impact) and the direction of the impact (or flux; green shade denotes inhibition and red shade denotes activation).
Among the top subcategories within hepatic organismal system that were highly-impacted and activated with SCE were complement and coagulation cascades and chemokine signaling pathway (immune system), aldosterone-regulated sodium reabsorption (excretory system), and PPAR signaling pathway (endocrine system) (Fig. 3). Another activated subcategory with a role in the immune response was cytokine-cytokine receptor interaction within the environmental information processing category (Suppl 2: KEGG Liver sheet). Within cellular processes, the most induced pathways included regulation of autophagy, lysosome and endocytosis (transport and catabolism), and apoptosis (cellular growth and death) (Suppl 2: KEGG Liver sheet). Within the genetic information processing category, transcription, translational and protein export were the most activated and the SNARE interactions in vesicular transport was the most inhibited pathway (Fig. 3). Subclinical endometritis also was associated with induction of calcium signaling pathway, GnRH signaling pathway, and hedgehog signaling pathway among the top 50 subcategories (Suppl 2: KEGG Liver sheet).
Adipose
Functional analysis of the adipose microarray data revealed that metabolism, genetic information processing, and organismal system categories were the most impacted; whereas, cellular processes and environmental information processing were the least impacted categories (Fig. 4). Within the metabolism category, the most activated terms in the top 20 subcategories affected (Fig. 5) include nicotinate and nicotinamide metabolism (metabolism of cofactors and vitamins), pentose and glucuronate interconversions, starch and sucrose metabolism, ascorbate and aldarate metabolism (carbohydrate metabolism), amino sugar and nucleotide sugar metabolism, and oxidative phosphorylation (energy metabolism) (Fig. 5). The DIA also uncovered an inhibition of fructose and mannose metabolism among the top 20 affected subcategories (Fig. 5).
Figure 4.
Impact and flux of main KEGG pathways categories and sub-categories affected by subclinical endometritis constructed from adipose differentially expressed genes (DEG) as calculated by the Dynamic Impact Approach. Reported are the total impact (Blue horizontal bars; larger the bars higher the impact) and the direction of the impact (or flux; green bars expanding left denote inhibition and red bars expanding right denote activation) of DEG on the categories and subcategories.
Figure 5.
Impact and flux of top 20 KEGG pathways affected by subclinical endometritis in subcutaneous adipose tissue uncovered by the Dynamic Impact Approach (DIA). Reported are the total impact (blue horizontal bars; larger the bars higher the impact) and the direction of the impact (or flux; green shade denotes inhibition and red shade denotes activation).
In addition, galactose metabolism (carbohydrate metabolism), cysteine and methionine metabolism, (amino acid metabolism), glycosaminoglycan biosynthesis—keratan sulfate, N-glycan biosynthesis, and glycosphingolipid biosynthesis— lacto and neolacto series (Glycan biosynthesis and metabolism) were among the top 50 metabolism subcategories affected (Suppl 2: KEGG Adipose sheet). Within the top 20 genetic information processing subcategories affected, the RNA degradation and basal transcription factor were the most impacted and activated, while RNA polymerase, protein export and sulfur-relay system were the most impacted and inhibited pathways (Fig. 5).
Among the top 20 affected subcategories, the peroxisome (cellular process) was the second most affected and, along with spliceosome, was overall activated (Fig. 5). Within adipose organismal system and among the top 50 affected subcategories the most impacted and activated pathways include T cell and B cell receptors signaling and Fc gamma R-mediated phagocytosis (immune system) (Suppl 2: KEGG Adipose sheet); whereas, aldosterone-regulated sodium reabsorption (excretory system) and circadian rhythm (environmental adaptation) were the most impacted and inhibited pathways among the top 20 affected subcategories (Fig. 5). Among the top 20 affected subcategories, extracellular matrix (ECM)-receptor interaction (signaling and molecular interaction) was overall activated (Fig. 5). Among the top 50 affected subcategories, SCE was associated with an overall activation of mTOR signaling and VEGF signaling (Suppl 2: KEGG Adipose sheet).
Gene ontology (GO) functional categories affected by subclinical endometritis as uncovered by the DIA
We used the DIA to uncover the impact and direction (flux) of the pathways within the terms obtained from DAVID (Suppl 2: DAVID Liver Up and Down sheets) as it relates to GO Biological Process (GOTERM_BP_FAT), Molecular Function (GOTERM_MF_FAT), Cellular Components (GOTERM_CC_FAT), KEGG_PATHWAY, and SP_PIR_KEYWORDS. A cutoff of the mean plus 1 standard deviation was used to obtain the top impacted pathways in each term. Detailed results from DIA are reported in supplementary file 2 (Liver DIA sheet).
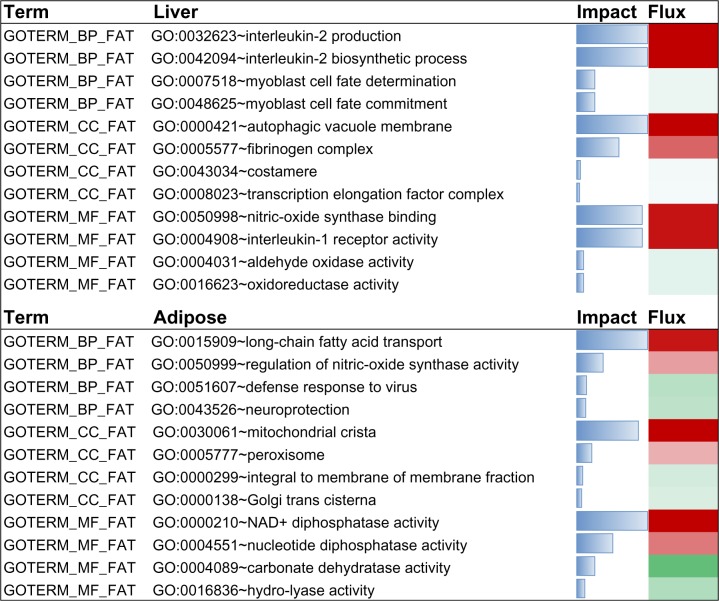
The functional enrichment analysis of GO biological process (GOTERM_BP_FAT) with DIA revealed that the top 10 most impacted and activated pathways in this category were biosynthesis and production of interleukin-2, response to progesterone stimulus, gas homeostasis, nitric oxide homeostasis, negative regulation of tyrosine phosphorylation of STAT protein, negative regulation of peptidyl-tyrosine phosphorylation, positive regulation of sequestering of triglyceride, post-embryonic body morphogenesis, and vitamin D metabolic process (Fig. 6; Suppl 2: Liver DIA sheet). Other pathways among the top 20 with important roles in metabolism included positive regulation of lipid storage, negative regulation of nitric oxide biosynthetic process, negative regulation of JAK-STAT cascade, and positive regulation of lipid storage (Fig. 6; Suppl 2: Liver DIA sheet).
Figure 6.
Most up- and down-regulated pathways due to subclinical endometritis in the Biological process (BP), Cellular components (CC) and Molecular processes (MP) GO categories uncovered by DIA using differentially expressed genes in liver and adipose tissue. Reported are the impact (blue horizontal bars; the larger the bar the higher the impact) and the direction of the impact (or flux; green shade denotes inhibition and red shade denotes activation).
The functional enrichment analysis of GO cellular component (GOTERM_CC_FAT) with DIA revealed that the top 10 most impacted and activated pathways by SCE in this category were autophagic vacuole membrane, fibrinogen complex, smooth endoplasmic reticulum, perikaryon, integral to membrane of membrane fraction, very-low-density lipoprotein particle, triglyceride-rich lipoprotein particle, proton-transporting ATP synthase complex, coupling factor F(o), high-density lipoprotein particle, and autophagic vacuole (Fig. 6; Suppl 2: Liver DIA sheet).
The functional enrichment analysis of GO molecular functions (GOTERM_MF_FAT) with DIA revealed that the top 10 most impacted and activated pathways by SCE in this category were nitric-oxide synthase binding, interleukin-1 receptor activity, GABA receptor binding, cholesterol monooxygenase (side-chain-cleaving) activity, prenylcysteine oxidase activity, interleukin-1 binding, L-xylulose reductase (NADP+) activity, formate transmembrane transporter activity, N-acylneuraminate cytidylyltransferase activity, and adenosylmetathionine decarboxylase activity (Fig. 6; Suppl 2: liver DIA sheet).
Adipose
Among the relevant GO biological process (GOT-ERM_BP_FAT) that were among the top 20 most impacted and activated pathways in this category were positive regulation of activin receptor signaling pathway, muscarinic acetylcholine receptor signaling pathway, long-chain fatty acid transport, and negative regulation of myoblast differentiation (Fig. 6; Suppl 2: Adipose DIA sheet). Although with a much lower impact, defense response to virus, neuroprotection, and calcium-dependent cell-cell adhesion were among the top 50 pathways and were inhibited (Suppl 2: Adipose DIA sheet). Among the top 10 most impacted and activated terms within cellular components (GOT-ERM_CC_FAT) were mitochondrial crista, transcription factor TFIIE complex, collagen type I, peroxisome, and lipid particle (Fig. 6; Suppl 2: Adipose DIA Sheet).
Within the molecular function (GOTERM_MF-FAT) category, among the top ten impacted pathways were NAD+diphosphatase activity and nucleotide diphosphatase activity which were activated (Fig. 6). In contrast, carbonate dehydratase activity was among the top 10 inhibited pathways, and retinoic acid receptor activity among the top 25 inhibited pathways.
Discussion
Despite the marked differences in uterine swab PMN percentage, neither the BCS nor the blood concentrations of most biomarkers differed between healthy cows and those with SCE. However, those responses contrast with the lower milk production (Fig. 1) during the first three weeks of lactation, numerically greater plasma globulin concentration, and changes in gene expression in the subcutaneous adipose and liver tissue of cows with SCE. The bioinformatics analyses of affected genes revealed alterations in several pathways encompassing nutrient metabolism, cell signaling, inflammation, and oxidative stress, among others. Furthermore, the up-regulation of total GHR without a change in IGF1 expression in liver suggested a further or more persistent uncoupling of the somatotropic axis in the SCE cows. Molecular data from this study are among the first to demonstrate a response in peripheral tissues to the onset of subclinical inflammation of the uterine tissue.
Body condition score (BCS), blood metabolites, and subclinical endometritis
In the current study, despite the small number of animals, the lack of a health status effect on BCS pre or postpartum suggests that loss of BCS due to onset of lactation, per se, is not a major risk factor for developing SCE. The lack of differences in concentration of NEFA in the present study agrees with previous similar studies.16,45,84,85 Overall, the observations from the present study seem to confirm the suggestion of Burke et al. that energy status per se is not a risk factor for SCE.16
Serum globulin concentrations can provide an indication of an animal’s humoral immune status or response;35 a high concentration of globulin and globulin: albumin ratio is suggestive of lymphocyte proliferation and greater levels of circulating antibodies.35 The numerically greater concentration of globulin (Table 1) is in accordance with previous data from cows with endometritis,16,36 and is indicative of an activation of adaptive immune responses.
The lack of clear differences in concentrations of Mg, AST, and GDH between groups is opposite to previous reports of lower Mg concentrations in cows with liver dysfunction;37 whereas, Burke et al.16 reported greater concentrations of AST and GDH and lower Mg concentration in cows with SCE. Therefore, in the current study the onset of SCE did not seem to cause a severe impairment in liver function and likely explains the similar BCS after calving and the small difference in milk production between groups.
Molecular links between adipose and liver tissue during subclinical endometritis
Subclinical endometritis induced a state of local inflammation and oxidative stress in adipose tissue as indicated by the greater expression of CCL2 and IL6 and the lower expression of metabolic-related genes such as PLIN2, SLC2A4, and IRS1 (Table 5). At the molecular level such changes would have resulted in sustained adipose tissue lipolysis leading to activation of long-chain fatty acid transport (Fig. 6), either for efflux into the blood or as a means to recycle fatty acids within the adipose tissue.38 Despite the lack of statistical difference in the expression of lipogenic genes (SCD, FASN) and transcription regulators (SREBF1), the lack of difference in blood NEFA could be taken as indication of greater recycling of long-chain fatty acids within adipose tissue. Alternatively, the lack of increase in blood NEFA of cows with SCE resulted from metabolism in liver and/or mammary gland to prevent excessive buildup in the circulation.
The greater hepatic expression of RXRA and the overall activation and high impact of the PPAR signaling pathway (Fig. 3) in SCE cows provides some evidence of greater NEFA utilization in liver despite the fact that expression of genes associated with fatty acid oxidation (ACOX1, CPT1A) and ketogenesis (HMGCS2) did not differ (Table 4). Together with most of the blood data, eg the lack of change in AST and GDH, which are markers of liver function,18 it appears that SCE did not cause long-term negative effects on metabolic activity of liver.
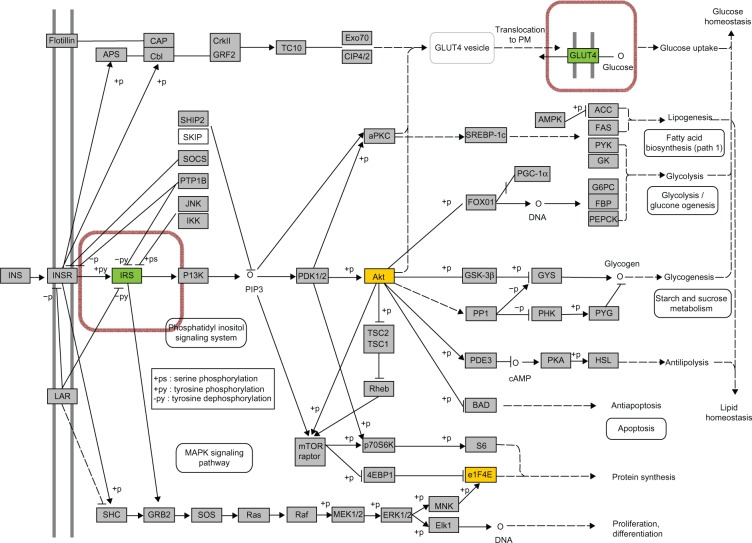
The tumor suppressor TP53 is a transcription factor that preserves genomic stability and prevents oncogenesis. Various genotoxic stresses such as DNA damage, oxidative stress, hypoxia, and heat shock can activate TP53, causing changes in the expression of its target genes.39 In non-ruminants, in addition to maintaining genomic integrity, TP53 can induce proinflammatory cytokines in adipose and cause insulin resistance.40,41 In dairy cows, negative energy balance and inflammatory conditions were reported to induce TP53 expression and its signaling in various tissues, eg liver, adipose, and spleen.30,86–88 In addition to eliciting effects on other genes, TP53 can affect glucose metabolism in insulin-sensitive tissues by repressing the expression of SLC2A4,42 which is the insulin-sensitive glucose transporter. The up-regulation of TP53 expression (Table 5) in adipose tissue and the lower milk production in cows with SCE suggests that the availability of glucose for the immune system might have increased at least in part by limiting the intracellular glucose transport via direct repression of glucose uptake (Table 5, Fig. 7).42,43 The expression of SLC2A4 in dairy cattle adipose tissue can be up-regulated by feeding diets with greater non-structural carbohydrate, hence, allowing for greater glucose uptake for lipogenesis.44
Figure 7.
Insulin signaling KEGG pathway in adipose tissue. Green shade denotes down-regulation while red/yellow tones denote up-regulation of genes in SCE cows. The genes enclosed in a colored box were not present on the microarray platform but were analyzed by qPCR.
A recent study also did not find differences in glucose concentration between healthy and endometritic cows.16 In contrast, Senosy et al. reported that low blood glucose is a risk factor in cows diagnosed with endometritis.45 The difference in the response of glucose to endometritis among different studies could be due to the time of sampling, eg, Senosy et al.45 observed differences in glucose concentration at 28 days postpartum in cows diagnosed with endometritis at 42 days postpartum but in the present study the SCE was evaluated at 22 or 25 days postpartum.
Subclinical endometritis and cholesterol metabolism
Cholesterol and its derivatives are essential for vital functions in the body; the intracellular quantity of cholesterol and its distribution to subcellular compartments (eg, ER, Golgi) are two key regulatory points that help maintain an optimal concentration of cholesterol at the cellular level.46 As in other mammals, the cow achieves cholesterol homeostasis through the endogenous synthesis of cholesterol and regulating its turn over via lipoprotein metabolism; hepatic gene expression is an important part of the control mechanisms involved in this process.89,90
Transcriptomic data from liver in the present study indicate that SCE induced intracellular cholesterol homeostasis and transport, at least in part, by up-regulating the expression of CAV1 and NPC2 (Table 2); both genes are involved in intracellular cholesterol flux and have important roles in the regulation of intracellular cholesterol homeostasis.46–48 Sub-clinical endometritis increased hepatic RXRA (Table 4), which in addition to enhancing long-chain fatty acid oxidation leads to an increase in CAV1 expression (cholesterol homeostasis).49 Similar to a previous study,50 the overall activation of apoptosis (Fig. 3) in liver also could have partly mediated the observed change in cholesterol homeostasis and the up-regulation of CAV1. Although we did not measure it in the study, such response might have increased hepatic lipid accumulation.
Cholesterol is the precursor for steroid hormone synthesis, and the process is initiated by cleavage of cholesterol to produce pregnenolone. The observed induction in cholesterol monooxygenase activity (the key enzyme) and MAPK activity (Suppl 2: Liver DIA sheet) in cows with SCE are noteworthy. The MAPK proteins are known to be involved in the regulation of steroid production by different mechanisms; among those, the induction of expression of CYP11A1 is very prominent.51 Subclinical endometritis induced hepatic CYP11A1 (Table 2), which catalyzes the conversion of cholesterol to pregnenolone, the first and rate-limiting step in the synthesis of the steroid hormones. Therefore, MAPK appears to play a role in regulating cholesterol homeostasis in liver during endometritis.52 The expression data support observations from a recent study, which reported lower cholesterol at wk 4 postpartum in cows diagnosed with endometritis at wk 5.45 Taking into account that samples in our study were harvested on day 29 postpartum, we speculate that the decrease in cholesterol at wk 4 in the study of Senosy et al.45 could have been associated with greater cleavage of cholesterol to steroids.
Molecular signatures of inflammation
The activation of local and systemic host defense mechanisms requires interactions between numerous types of immune cells and inflammatory mediators, such as nitric oxide (NO), prostaglandins, and cytokines.53 Signal transduction occurs via several intracellular pathways, including the Janus kinase (JAK)–STAT pathway, the phosphoinositide 3-kinase (PI3K)–AKT pathway and the MAPK pathway.54 Moreover, the complement and coagulation cascades are the first line of defense that helps the host against injurious stimuli and inflammation.91,92
During pathophysiological situations, the activation of these cascades (both complement and coagulation cascades) occurs simultaneously and is intended to act locally; however, systemic activation of these pathways has been reported in situations when the relevant control mechanism at the site of infection fail to respond.55 Therefore it is speculated that the increase in the hepatic expression of genes associated with complement and coagulation cascades in SCE cows (Fig. 3; Suppl 2: Liver DIA) could have been associated with a failure of control mechanisms within the endometrium. This idea seems to be supported by previous data demonstrating a decrease in PMN function5 and potential modifications of both the innate and adaptive immune systems in cows with endometritis.12,13
Cytokines play an important role in a wide range of reproductive-related processes via regulation of a complex metabolic network.56 The activation of pathways leading to biosynthesis of IL-2 and activity of IL-1 in cows with SCE indicates an activation of both innate and adaptive immune systems.54,57 The decrease in hepatic expression of chemokine (C-C motif) receptor 4 (CCR4) (Table 2) with SCE could have had a major contribution to the activation of the innate immune system.58 For instance, in CCR4−/− mice receiving LPS,58 there was a marked induction of the JNK and p38 MAPK pathways, which we also observed in liver (Suppl 2: Liver DIA).
Nitric oxide is an inflammatory mediator that among other effects mediates cytoimmunity and inflammation toxicity.59 An increase of NO has been reported with endometritis in both cows59 and mares.60 Although we did not measure the concentration of NO in tissue or blood, the activation of pathways associated with hepatic NO homeostasis and negative regulation of nitric oxide biosynthetic process (Suppl 2: Liver DIA) indicates changes in NO concentrations in SCE cows. As such, a local increase in NO within liver tissue could, at least in part, be associated with the changes in the expression of inflammatory genes.
Haptoglobin is an acute-phase protein primarily synthesized in the liver that accomplishes its function by acting as an antioxidant, anti-inflammatory agent, bacteriostat, and by regulating the maturation and activity of immune cells.61 The increase in HP expression in liver (Table 4) from cows with SCE agrees with previous work reporting greater serum concentration of HP in Holstein cows with acute puerperal metritis.62 A greater concentration of HP also was observed in cows with retained fetal membranes, which is one of the key risk factors associated with uterine infection.63 From a mechanistic standpoint, the numerical up-regulation of SOCS2 (1.5-fold) and STAT3 (~2-fold) (Table 4) not only suggests an augmentation of cytokine signaling64 with SCE but also a potential impairment of growth hormone signaling.65
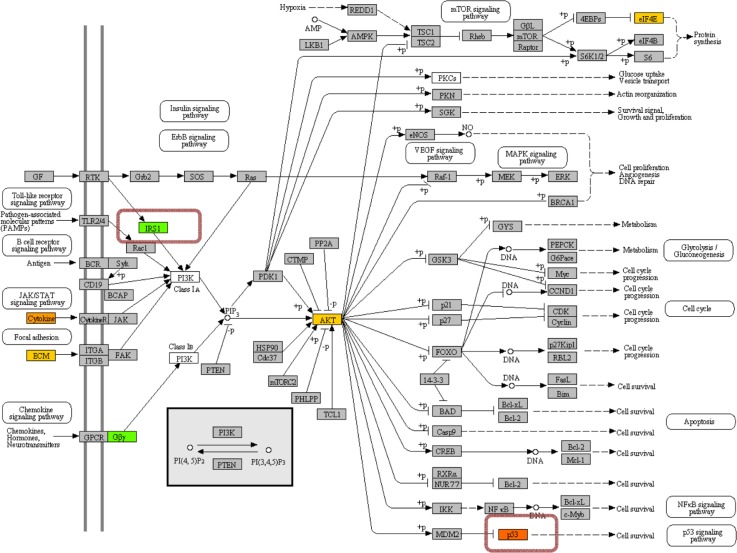
The immune responsive role of adipose depots has been studied previously,30,66 and data from the present study support this role, eg there was activation of B cell receptor signaling, cytokine-cytokine receptor interaction, and T cell receptor signaling pathways in subcutaneous adipose from cows with SCE. A novel response in this study was the induction of the mTOR signaling pathway in cows with SCE (Suppl 2: Adipose DIA); mTOR is a large molecular-weight protein that mediates intracellular signaling related to cell growth, proliferation, and differentiation.67,68 In non-ruminants the increase in mTOR signaling pathway in adipocytes is mediated by PI3K-ATK signaling, which is activated by cytokines and ECM (Fig. 8).69,70
Figure 8.
PI3K-AKT signaling KEGG pathway in adipose tissue. Green shade denotes down-regulation while red/yellow tones denote up-regulation of genes in SCE cows. The genes enclosed in a colored box were not present on the microarray platform but were analyzed by qPCR.
In addition to other regulatory mechanisms, PI3K can also be activated by insulin through IRS1/2(Insulin receptor substrate-1/2); PI3K then leads to activation of mTOR via Akt.71 In adipose tissue, AKT2 is the predominant isoform of Akt;72 the down-regulation of IRS1 and SLC2A4 (insulin-induced glucose transporter) indicates that the up-regulation of AKT2 (Table 3) might have been mediated by PI3 K via the activation of cytokines and ECM rather than insulin signaling (Fig. 8). Increased signaling through AKT2 then could have regulated mTOR signaling by phosphorylating the AKT1S1 (PPAS40) (Table 3). Moreover, the tendency for greater expression of STAT3 in adipose tissue (Table 5) suggests that mTOR might have elicited its effect by phosphorylation at Ser727.93,94 Following phosphorylation, STAT protein translocates to the nucleus to regulate transcription or activate the transcription of target genes associated with cell-cycle progression and apoptosis. As a result, STAT can promote cellular transformation as well as abnormal cell proliferation.95,96 In addition to important roles in cytokine signaling pathways, STAT3 has been reported to contribute to the phenotypic variation in embryonic survival in cattle.97,98
Molecular adaptations in Vitamin B3 and B6 metabolism
In the present study, an activation of genes coding for enzymes associated with oxidative phosphorylation in both liver (eg, ATP5A1) and adipose (eg, ATP6V1B2, COX6B2, NDUFS6) (Tables 2 and 3) was observed. As precursors of the coenzymes nicotinamide-adenine dinucleotide (NAD+) and nicotinamide-adenine dinucleotide phosphate (NADP+), both nicotinic acid (anionic form: nicotinate) and nicotinamide (amide derivative of nicotinic acid) are essential for organisms.73 Nicotinic acid and nicotinamide are often grouped together under the generic term niacin, and are also known as vitamin B3.74
The reduction in flux of nicotinate and nicotinamide metabolism in liver did not seem to alter mitochondrial respiration per se because expression of genes associated with fatty acid oxidation (CPT1A, ACOX1) did not change. It could be possible that this reduction in flux in SCE cows reflected a state of oxidative stress (up-regulated SOD2 and GPX1; Table 4), at least in part, because of a decrease in niacin availability.75 In contrast, the induction of this pathway in adipose tissue suggests the tissue might have an inherently greater ability to counteract oxidative stress compared with liver, potentially as a function of niacin accumulation. Additionally, cows in the present study were fed predominately forage diets, with potentially greater availability of niacin.76 The amino acid tryptophan is the sole substrate for de novo NAD+ and NADP+ synthesis in the absence of nicotinamide or nicotinic acid. All species are able to synthesize niacin from tryptophan and quinolinate.73 The decrease in tryptophan metabolism (Suppl 2: Liver KEGG sheet) in cows with SCE is additional evidence of a state of oxidative stress in the liver.
Because of its integral involvement in the synthesis of nucleic acid, and consequently in mRNA and protein synthesis, vitamin B6 influences acquired and humoral immunity, and the production of cytokines and inflammatory mediators.77 The vitamin B3 metabolite N-methylnicotinamide may exert anti-inflammatory and anti-oxidative stress effects.78 The observed inhibition of vitamin B3 and B6 metabolism was associated with activation of cytokine and proinflammatory cytokine production (Suppl 2: Liver DIA sheet) and oxidative stress (SOD2 and GPX1 up-regulation; Table 4). In contrast, the activation of vitamin B3 (nicotinamide) metabolism (Fig. 5) in adipose tissue may have served a protective role against oxidative stress (SOD2 up-regulation) (Table 5) to protect cells against reactive oxygen species.79
Intracellular energy status and oxidative stress
Mitochondria are responsible for ~90% of oxygen consumption and ATP production;80 however, they are also major sources of intracellular reactive oxygen metabolites (ROM).81 In dairy cows, excessive production of ROM may lead to oxidative stress when the body antioxidant defenses are insufficient, which is a major risk factor associated with endometritis.82
The protein ATP6V1B2, which transports H+ across the membrane of intracellular organelles, also is a component of the B2 subunit of the H+ transporting AT-Pase (VATPase).99,100 Previous studies reported that an increase in V-ATPase led to the accumulation of ROM.100,101 Therefore, it could be possible that in the present study the evident changes enhanced oxidative phosphorylation and ROM production, which coupled with the greater hepatic expression of SOD2 and GPX1 (Table 4), suggests that SCE can enhance the oxidative stress status of peripheral tissues.
Conclusions
The local uterine inflammation appears to induce a marked inflammatory response at the level of liver, whereas in adipose, there was a more pronounced response in oxidative stress and apoptosis coupled with impaired insulin signaling. The lack of change in BCS, plasma NEFA concentration, and gene expression data associated with lipolysis and lipogenesis provides additional evidence that energy status of the cow is not a primary risk factor for SCE. Despite the observed changes at the molecular level, there were no evident systemic long-term negative effects on liver function.
Supplementary Data
Supplementary file 1. Has information about primers, qPCR efficiency, relative % mRNA abundance and microarray designed used.
Supplementary file 2. Had complete out put obtained from the KEGG and DIA analysis run with Liver and Adipose microarray data.
Acknowledgments
The help of Dr. M. Jawad Khan during microarray and bioinformatics analyses is greatly appreciated.
Footnotes
ACADEMIC EDITOR: JT Efird, Associate Editor
FUNDING: This research was supported by New Zealand dairy farmers through DairyNZ Inc. (AN808, AN1202), and the New Zealand Ministry of Business, Innovation and Employment (UOAX0814).
COMPETING INTERESTS: Author(s) disclose no potential conflicts of interest.
Author Contributions
Conceived and designed the experiments: SM, JRR, SMc, CB, MM, JJL. Conceived and performed the analyses: HA, SLR. Wrote the manuscript: HA, JJL, FCC, JRR. Contributed reagents: REE, HAL. Agree with manuscript results and conclusions: HA, FCC, JRR, JJL. Jointly developed the structure and arguments for the paper: HA, FCC, JJL, JRR. Made critical revisions and approved final version: HA, FCC, JJL, JRR, SM, REE, CB, CW. All authors reviewed and approved of the final manuscript.
DISCLOSURES AND ETHICS
As a requirement of publication the authors have provided signed confirmation of their compliance with ethical and legal obligations including but not limited to compliance with ICMJE authorship and competing interests guidelines, that the article is neither under consideration for publication nor published elsewhere, of their compliance with legal and ethical guidelines concerning human and animal research participants (if applicable), and that permission has been obtained for reproduction of any copyrighted material. This article was subject to blind, independent, expert peer review. The reviewers reported no competing interests.
REFERENCES
- 1.Sheldon IM, Cronin J, Goetze L, Donofrio G, Schuberth HJ. Defining postpartum uterine disease and the mechanisms of infection and immunity in the female reproductive tract in cattle. Biol Reprod. 2009 Dec;81(6):1025–32. doi: 10.1095/biolreprod.109.077370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sheldon IM, Lewis GS, LeBlanc S, Gilbert RO. Defining postpartum uterine disease in cattle. Theriogenology. 2006 May;65(8):1516–30. doi: 10.1016/j.theriogenology.2005.08.021. [DOI] [PubMed] [Google Scholar]
- 3.LeBlanc SJ, Duffield TF, Leslie KE, et al. Defining and diagnosing postpartum clinical endometritis and its impact on reproductive performance in dairy cows. J Dairy Sci. 2002 Sep;85(9):2223–36. doi: 10.3168/jds.S0022-0302(02)74302-6. [DOI] [PubMed] [Google Scholar]
- 4.Galvao KN, Greco LF, Vilela JM, Sa Filho MF, Santos JE. Effect of intrauterine infusion of ceftiofur on uterine health and fertility in dairy cows. J Dairy Sci. 2009 Apr;92(4):1532–42. doi: 10.3168/jds.2008-1615. [DOI] [PubMed] [Google Scholar]
- 5.Hammon DS, Evjen IM, Dhiman TR, Goff JP, Walters JL. Neutrophil function and energy status in Holstein cows with uterine health disorders. Vet Immunol Immunopathol. 2006 Sep 15;113(1–2):21–9. doi: 10.1016/j.vetimm.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 6.Kasimanickam R, Duffield TF, Foster RA, et al. Endometrial cytology and ultrasonography for the detection of subclinical endometritis in postpartum dairy cows. Theriogenology. 2004 Jul;62(1–2):9–23. doi: 10.1016/j.theriogenology.2003.03.001. [DOI] [PubMed] [Google Scholar]
- 7.Gilbert RO, Shin ST, Guard CL, Erb HN, Frajblat M. Prevalence of endometritis and its effects on reproductive performance of dairy cows. Theriogenology. 2005 Dec;64(9):1879–88. doi: 10.1016/j.theriogenology.2005.04.022. [DOI] [PubMed] [Google Scholar]
- 8.Galvao KN, Frajblat M, Brittin SB, Butler WR, Guard CL, Gilbert RO. Effect of prostaglandin F2alpha on subclinical endometritis and fertility in dairy cows. J Dairy Sci. 2009 Oct;92(10):4906–13. doi: 10.3168/jds.2008-1984. [DOI] [PubMed] [Google Scholar]
- 9.Cheong SH, Nydam DV, Galvao KN, Crosier BM, Gilbert RO. Cow-level and herd-level risk factors for subclinical endometritis in lactating Holstein cows. J Dairy Sci. 2011 Feb;94(2):762–70. doi: 10.3168/jds.2010-3439. [DOI] [PubMed] [Google Scholar]
- 10.McDougall S, Macaulay R, Compton C. Association between endometritis diagnosis using a novel intravaginal device and reproductive performance in dairy cattle. Anim Reprod Sci. 2007 May;99(1–2):9–23. doi: 10.1016/j.anireprosci.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 11.Pleticha S, Drillich M, Heuwieser W. Evaluation of the Metricheck device and the gloved hand for the diagnosis of clinical endometritis in dairy cows. J Dairy Sci. 2009 Nov;92(11):5429–35. doi: 10.3168/jds.2009-2117. [DOI] [PubMed] [Google Scholar]
- 12.Bondurant RH. Inflammation in the bovine female reproductive tract. J Anim Sci. 1999;77(Suppl 2):101–10. doi: 10.2527/1999.77suppl_2101x. [DOI] [PubMed] [Google Scholar]
- 13.Levkut M, Pisti J, Revajova V, Choma J, Levkutova M, David V. Comparison of immune parameters in cows with normal and prolonged involution time of uterus. Vet Med-Czech. 2002 Oct-Nov;47(10–1):277–82. [Google Scholar]
- 14.Rukkwamsuk T, Kruip TAM, Wensing T. Relationship between overfeeding and overconditioning in the dry period and the problems of high producing dairy cows during the postparturient period. Vet Quart. 1999 Jun;21(3):71–7. doi: 10.1080/01652176.1999.9694997. [DOI] [PubMed] [Google Scholar]
- 15.Galvao KN, Flaminio MJBF, Brittin SB, et al. Association between uterine disease and indicators of neutrophil and systemic energy status in lactating Holstein cows. Journal of Dairy Science. 2010 Jul;93(7):2926–37. doi: 10.3168/jds.2009-2551. [DOI] [PubMed] [Google Scholar]
- 16.Burke CR, Meier S, McDougall S, Compton C, Mitchell M, Roche JR. Relationships between endometritis and metabolic state during the transition period in pasture-grazed dairy cows. Journal of Dairy Science. 2010 Nov;93(11):5363–73. doi: 10.3168/jds.2010-3356. [DOI] [PubMed] [Google Scholar]
- 17.Sheldon IM, Rycroft AN, Zhou C. Association between postpartum pyrexia and uterine bacterial infection in dairy cattle. Vet Rec. 2004 Mar 6;154(10):289–93. doi: 10.1136/vr.154.10.289. [DOI] [PubMed] [Google Scholar]
- 18.Bertoni G, Trevisi E, Han X, Bionaz M. Effects of inflammatory conditions on liver activity in puerperium period and consequences for performance in dairy cows. J Dairy Sci. 2008 Sep;91(9):3300–10. doi: 10.3168/jds.2008-0995. [DOI] [PubMed] [Google Scholar]
- 19.Butlerhogg BW, Wood JD, Bines JA. Fat Partitioning in British Friesian Cows—the Influence of Physiological-State on Dissected Body-Composition. J Agr Sci. 1985;104(Jun):519–28. [Google Scholar]
- 20.Hoelker M, Salilew-Wondim D, Drillich M, et al. Transcriptional response of the bovine endometrium and embryo to endometrial polymorphonuclear neutrophil infiltration as an indicator of subclinical inflammation of the uterine environment. Reprod Fertil Dev. 2012;24(6):778–93. doi: 10.1071/RD11171. [DOI] [PubMed] [Google Scholar]
- 21.Gabler C, Fischer C, Drillich M, Einspanier R, Heuwieser W. Time-dependent mRNA expression of selected pro-inflammatory factors in the endometrium of primiparous cows postpartum. Reprod Biol Endocrinol. 2010;8:152. doi: 10.1186/1477-7827-8-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loor JJ, Moyes KM, Bionaz M. Functional adaptations of the transcriptome to mastitis-causing pathogens: the mammary gland and beyond. J Mammary Gland Biol Neoplasia. 2011 Dec;16(4):305–22. doi: 10.1007/s10911-011-9232-2. [DOI] [PubMed] [Google Scholar]
- 23.Roche JR, Berry DP. Periparturient climatic, animal, and management factors influencing the incidence of milk Fever in grazing systems. J Dairy Sci. 2006 Jul;89(7):2775–83. doi: 10.3168/jds.S0022-0302(06)72354-2. [DOI] [PubMed] [Google Scholar]
- 24.Roche JR, Dillon PG, Stockdale CR, Baumgard LH, VanBaale MJ. Relationships among international body condition scoring systems. J Dairy Sci. 2004 Sep;87(9):3076–9. doi: 10.3168/jds.S0022-0302(04)73441-4. [DOI] [PubMed] [Google Scholar]
- 25.Loor JJ, Everts RE, Bionaz M, et al. Nutrition-induced ketosis alters metabolic and signaling gene networks in liver of periparturient dairy cows. Physiol Genomics. 2007 Dec 19;32(1):105–16. doi: 10.1152/physiolgenomics.00188.2007. [DOI] [PubMed] [Google Scholar]
- 26.Guo L, Lobenhofer EK, Wang C, et al. Rat toxicogenomic study reveals analytical consistency across microarray platforms. Nat Biotechnol. 2006 Sep;24(9):1162–69. doi: 10.1038/nbt1238. [DOI] [PubMed] [Google Scholar]
- 27.Akbar H, Bionaz M, Carlson DB, et al. Feed restriction, but not L-carnitine infusion, alters the liver transcriptome by inhibiting sterol synthesis and mitochondrial oxidative phosphorylation and increasing gluconeogenesis in mid-lactation dairy cows. Journal of Dairy Science. 2013 Apr;96(4):2201–13. doi: 10.3168/jds.2012-6036. [DOI] [PubMed] [Google Scholar]
- 28.Loor JJ. Genomics of metabolic adaptations in the peripartal cow. Animal. 2010 Jul;4(7):1110–39. doi: 10.1017/S1751731110000960. [DOI] [PubMed] [Google Scholar]
- 29.Ji P, Osorio JS, Drackley JK, Loor JJ. Overfeeding a moderate energy diet prepartum does not impair bovine subcutaneous adipose tissue insulin signal transduction and induces marked changes in peripartal gene network expression. J Dairy Sci. 2012 Aug;95(8):4333–51. doi: 10.3168/jds.2011-5079. [DOI] [PubMed] [Google Scholar]
- 30.Mukesh M, Bionaz M, Graugnard DE, Drackley JK, Loor JJ. Adipose tissue depots of Holstein cows are immune responsive: Inflammatory gene expression in vitro. Domest Anim Endocrin. 2010 Apr;38(3):168–78. doi: 10.1016/j.domaniend.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 31.Piantoni P, Daniels KM, Everts RE, et al. Level of nutrient intake affects mammary gland gene expression profiles in preweaned Holstein heifers. Journal of Dairy Science. 2012 May;95(5):2550–61. doi: 10.3168/jds.2011-4539. [DOI] [PubMed] [Google Scholar]
- 32.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 33.Bionaz M, Periasamy K, Rodriguez-Zas SL, Hurley WL, Loor JJ. A novel dynamic impact approach (DIA) for functional analysis of time-course omics studies: validation using the bovine mammary transcriptome. PLoS One. 2012;7(3):e32455. doi: 10.1371/journal.pone.0032455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morandini P. Rethinking metabolic control. Plant Science. 2009;176:441–51. doi: 10.1016/j.plantsci.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 35.Chorfi Y, Lanevschi-Pietersma A, Girard V, Tremblay A. Evaluation of variation in serum globulin concentrations in dairy cattle. Vet Clin Path. 2004;33(3):122–7. doi: 10.1111/j.1939-165x.2004.tb00360.x. [DOI] [PubMed] [Google Scholar]
- 36.Green MP, Ledgard AM, Berg MC, Peterson AJ, Back PJ. Prevalence and identification of systemic markers of sub-clinical endometritis in postpartum dairy cows. Proc N Z Soc Anim Prod. 2009;69:37–42. [Google Scholar]
- 37.Bertoni G, Trevisi E, Han X, Bionaz M. Effects of Inflammatory Conditions on Liver Activity in Puerperium Period and Consequences for Performance in Dairy Cows. Journal of Dairy Science. 9// 2008;91(9):3300–10. doi: 10.3168/jds.2008-0995. [DOI] [PubMed] [Google Scholar]
- 38.Nye CK, Hanson RW, Kalhan SC. Glyceroneogenesis is the dominant pathway for triglyceride glycerol synthesis in vivo in the rat. J Biol Chem. 2008 Oct 10;283(41):27565–74. doi: 10.1074/jbc.M804393200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oren M. Regulation of the p53 tumor suppressor protein. Journal of Biological Chemistry. 1999 Dec 17;274(51):36031–4. doi: 10.1074/jbc.274.51.36031. [DOI] [PubMed] [Google Scholar]
- 40.Minamino T, Orimo M, Shimizu I, et al. A crucial role for adipose tissue p53 in the regulation of insulin resistance. Nat Med. 2009 Sep;15(9):1082–U1140. doi: 10.1038/nm.2014. [DOI] [PubMed] [Google Scholar]
- 41.Yahagi N, Shimano H, Matsuzaka T, et al. p53 Activation in adipocytes of obese mice. J Biol Chem. 2003 Jul 11;278(28):25395–400. doi: 10.1074/jbc.M302364200. [DOI] [PubMed] [Google Scholar]
- 42.Schwartzenberg-Bar-Yoseph F, Armoni M, Karnieli E. The tumor suppressor p53 down-regulates glucose transporters GLUT1 and GLUT4 gene expression. Cancer Res. 2004 Apr 1;64(7):2627–33. doi: 10.1158/0008-5472.can-03-0846. [DOI] [PubMed] [Google Scholar]
- 43.Maddocks ODK, Vousden KH. Metabolic regulation by p53 (vol 89, pg 237, 2011) J Mol Med. 2011 May;89(5):531–1. doi: 10.1007/s00109-011-0735-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ji P, Osorio JS, Drackley JK, Loor JJ. Overfeeding a moderate energy diet prepartum does not impair bovine subcutaneous adipose tissue insulin signal transduction and induces marked changes in peripartal gene network expression. Journal of Dairy Science. 2012 Aug;95(8):4333–51. doi: 10.3168/jds.2011-5079. [DOI] [PubMed] [Google Scholar]
- 45.Senosy WS, Izaike Y, Osawa T. Influences of Metabolic Traits on Subclinical Endometritis at Different Intervals Postpartum in High Milking Cows. Reprod Domest Anim. 2012 Aug;47(4):666–74. doi: 10.1111/j.1439-0531.2011.01941.x. [DOI] [PubMed] [Google Scholar]
- 46.Storch J, Xu Z. Niemann-Pick C2 (NPC2) intracellular cholesterol trafficking. Biochim Biophys Acta. 2009 Jul;1791(7):671–8. doi: 10.1016/j.bbalip.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smart EJ, Ying YS, Donzell WC, Anderson RGW. A role for caveolin in transport of cholesterol from endoplasmic reticulum to plasma membrane. Journal of Biological Chemistry. 1996 Nov 15;271(46):29427–35. doi: 10.1074/jbc.271.46.29427. [DOI] [PubMed] [Google Scholar]
- 48.Frank PG, Cheung MWC, Pavlides S, Llaverias G, Park DS, Lisanti MP. Caveolin-1 and regulation of cellular cholesterol homeostasis. Am J Physiol-Heart C. 2006 Aug;291(2):H677–H686. doi: 10.1152/ajpheart.01092.2005. [DOI] [PubMed] [Google Scholar]
- 49.Llaverias G, Vazquez-Carrera M, Sanchez RM, et al. Rosiglitazone upregulates caveolin-1 expression in THP-1 cells through a PPAR-dependent mechanism. J Lipid Res. 2004 Nov;45(11):2015–24. doi: 10.1194/jlr.M400049-JLR200. [DOI] [PubMed] [Google Scholar]
- 50.Gargalovic P, Dory L. Cellular apoptosis is associated with increased caveolin-1 expression in macrophages. J Lipid Res. 2003 Sep;44(9):1622–32. doi: 10.1194/jlr.M300140-JLR200. [DOI] [PubMed] [Google Scholar]
- 51.Singh RP, Dhawan P, Golden C, Kapoor GS, Mehta KD. One-way cross-talk between p38(MAPK) and p42/44(MAPK). Inhibition of p38(MAPK) induces low density lipoprotein receptor expression through activation of the p42/44(MAPK) cascade. The Journal of biological chemistry. 1999 Jul 9;274(28):19593–600. doi: 10.1074/jbc.274.28.19593. [DOI] [PubMed] [Google Scholar]
- 52.Go GW, Mani A. Low-density lipoprotein receptor (LDLR) family orchestrates cholesterol homeostasis. Yale J Biol Med. 2012 Mar;85(1):19–28. [PMC free article] [PubMed] [Google Scholar]
- 53.Sordillo LM, Contreras GA, Aitken SL. Metabolic factors affecting the inflammatory response of periparturient dairy cows. Animal Health Research Reviews. 2009;10(1):53–63. doi: 10.1017/S1466252309990016. [DOI] [PubMed] [Google Scholar]
- 54.Boyman O, Sprent J. The role of interleukin-2 during homeostasis and activation of the immune system. Nat Rev Immunol. 2012 Mar;12(3):180–90. doi: 10.1038/nri3156. [DOI] [PubMed] [Google Scholar]
- 55.Markiewski MM, Nilsson B, Ekdahl KN, Mollnes TE, Lambris JD. Complement and coagulation: strangers or partners in crime? Trends Immunol. 2007 Apr;28(4):184–92. doi: 10.1016/j.it.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 56.Orsi NM, Tribe RM. Cytokine networks and the regulation of uterine function in pregnancy and parturition. J Neuroendocrinol. 2008 Apr;20(4):462–9. doi: 10.1111/j.1365-2826.2008.01668.x. [DOI] [PubMed] [Google Scholar]
- 57.Sims JE, Smith DE. The IL-1 family: regulators of immunity. Nat Rev Immunol. 2010 Feb;10(2):89–102. doi: 10.1038/nri2691. [DOI] [PubMed] [Google Scholar]
- 58.Ness TL, Ewing JL, Hogaboam CM, Kunkel SL. CCR4 is a key modulator of innate immune responses. J Immunol. 2006 Dec 1;177(11):7531–9. doi: 10.4049/jimmunol.177.11.7531. [DOI] [PubMed] [Google Scholar]
- 59.Li DJ, Liu YF, Li YF, Lv Y, Pei XY, Guo DZ. Significance of nitric oxide concentration in plasma and uterine secretes with puerperal endometritis in dairy cows. Vet Res Commun. 2010 Apr;34(4):315–21. doi: 10.1007/s11259-010-9355-8. [DOI] [PubMed] [Google Scholar]
- 60.Christoffersen M, Woodward E, Bojesen AM, et al. Inflammatory responses to induced infectious endometritis in mares resistant or susceptible to persistent endometritis. Bmc Vet Res. 2012 Mar 29;:8. doi: 10.1186/1746-6148-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sabedra DA. Serum Haptoglobin as an Indicator for Calving Difficulties and Postpartal Diseases in Transition Dairy Cows. Undergraduate Thesis. 2012 [Google Scholar]
- 62.Drillich M, Voigt D, Forderung D, Heuwieser W. Treatment of acute puerperal metritis with flunixin meglumine in addition to antibiotic treatment. J Dairy Sci. 2007 Aug;90(8):3758–63. doi: 10.3168/jds.2007-0052. [DOI] [PubMed] [Google Scholar]
- 63.Mordak R. Postpartum Serum Concentration of Haptoglobin in Cows with Fetal Membranes Retention. Cattle Pract. 2009 Mar;17:100–2. [Google Scholar]
- 64.Nicholson SE, Hilton DJ. The SOCS proteins: a new family of negative regulators of signal transduction. J Leukoc Biol. 1998 Jun;63(6):665–8. doi: 10.1002/jlb.63.6.665. [DOI] [PubMed] [Google Scholar]
- 65.Udy GB, Towers RP, Snell RG, et al. Requirement of STAT5b for sexual dimorphism of body growth rates and liver gene expression. Proc Natl Acad Sci USA. 1997 Jul 8;94(14):7239–44. doi: 10.1073/pnas.94.14.7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ingvartsen KL, Boisclair YR. Leptin and the regulation of food intake, energy homeostasis and immunity with special focus on periparturient ruminants. Domest Anim Endocrin. 2001 Nov;21(4):215–50. doi: 10.1016/s0739-7240(02)00119-4. [DOI] [PubMed] [Google Scholar]
- 67.Kim DH, Sarbassov DD, Ali SM, et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002 Jul 26;110(2):163–75. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 68.Rosner M, Hanneder M, Siegel N, Valli A, Fuchs C, Hengstschlager M. The mTOR pathway and its role in human genetic diseases. Mutat Res. 2008 Sep-Oct;659(3):284–92. doi: 10.1016/j.mrrev.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 69.Bhatt AP, Damania B. AKTivation of PI3K/AKT/mTOR signaling pathway by KSHV. Front Immunol. 2012;3:401. doi: 10.3389/fimmu.2012.00401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Friedl P, Wolf K. Tumour-cell invasion and migration: diversity and escape mechanisms. Nat Rev Cancer. 2003 May;3(5):362–74. doi: 10.1038/nrc1075. [DOI] [PubMed] [Google Scholar]
- 71.Pirola L, Bonnafous S, Johnston AM, Chaussade C, Portis F, Van Obberghen E. Phosphoinositide 3-kinase-mediated reduction of insulin receptor substrate-1/2 protein expression via different mechanisms contributes to the insulin-induced desensitization of its signalling pathways in L6 muscle cells. Journal of Biological Chemistry. 2003 May 2;278(18):15641–51. doi: 10.1074/jbc.M208984200. [DOI] [PubMed] [Google Scholar]
- 72.Choi SM, Tucker DF, Gross DN, et al. Insulin Regulates Adipocyte Lipolysis via an Akt-Independent Signaling Pathway. Molecular and Cellular Biology. 2010 Nov;30(21):5009–20. doi: 10.1128/MCB.00797-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Niehoff ID, Huther L, Lebzien P. Niacin for dairy cattle: a review. Br J Nutr. 2009 Jan;101(1):5–19. doi: 10.1017/S0007114508043377. [DOI] [PubMed] [Google Scholar]
- 74.Moffett JR, Namboodiri MA. Tryptophan and the immune response. Immunol Cell Biol. 2003 Aug;81(4):247–65. doi: 10.1046/j.1440-1711.2003.t01-1-01177.x. [DOI] [PubMed] [Google Scholar]
- 75.Kamanna VS, Vo A, Kashyap ML. Nicotinic acid: recent developments. Current Opinion in Cardiology. 2008 Jul;23(4):393–8. doi: 10.1097/HCO.0b013e3283021c82. [DOI] [PubMed] [Google Scholar]
- 76.Schwab EC, Schwab CG, Shaver RD, Girard CL, Putnam DE, Whitehouse NL. Dietary forage and nonfiber carbohydrate contents influence B-vitamin intake, duodenal flow, and apparent ruminal synthesis in lactating dairy cows. J Dairy Sci. 2006 Jan;89(1):174–87. doi: 10.3168/jds.S0022-0302(06)72082-3. [DOI] [PubMed] [Google Scholar]
- 77.Friso S, Jacques PF, Wilson PWF, Rosenberg IH, Selhub J. Low circulating vitamin B-6 is associated with elevation of the inflammation marker C-reactive protein independently of plasma homocysteine levels. Circulation. 2001 Jun 12;103(23):2788–91. doi: 10.1161/01.cir.103.23.2788. [DOI] [PubMed] [Google Scholar]
- 78.Domagala TB, Szeffler A, Dobrucki LW, et al. Nitric oxide production and endothelium-dependent vasorelaxation ameliorated by N1-methylnicotinamide in human blood vessels. Hypertension. 2012 Apr;59(4):825–32. doi: 10.1161/HYPERTENSIONAHA.111.183210. [DOI] [PubMed] [Google Scholar]
- 79.Sauve AA. NAD+ and vitamin B3: from metabolism to therapies. J Pharmacol Exp Ther. 2008 Mar;324(3):883–93. doi: 10.1124/jpet.107.120758. [DOI] [PubMed] [Google Scholar]
- 80.Kelly AK, Waters SM, McGee M, Fonseca RG, Carberry C, Kenny DA. mRNA expression of genes regulating oxidative phosphorylation in the muscle of beef cattle divergently ranked on residual feed intake. Physiological Genomics. 2011 Jan;43(1):12–23. doi: 10.1152/physiolgenomics.00213.2009. [DOI] [PubMed] [Google Scholar]
- 81.Wei YH, Lee HC. Oxidative stress, mitochondrial DNA mutation, and impairment of antioxidant enzymes in aging. Exp Biol Med. 2002 Oct;227(9):671–82. doi: 10.1177/153537020222700901. [DOI] [PubMed] [Google Scholar]
- 82.Heidarpour MMM, Fallah-Rad AH, Dehghan Shahreza F, Mohammadi M. Oxidative stress and trace elements before and after treatment in dairy cows with clinical and subclinical endometritis. Revue Méd Vét. 2012;163(12):628–33. [Google Scholar]
- 83.Roche JR, Friggens NC, Kay JK, Fisher MW, Stafford KJ, Berry DP. Invited review: Body condition score and its association with dairy cow productivity, health, and welfare. J Dairy Sci. 2009 Dec;92(12):5769–801. doi: 10.3168/jds.2009-2431. [DOI] [PubMed] [Google Scholar]
- 84.Dubuc J, Duffield TF, Leslie KE, Walton JS, LeBlanc SJ. Risk factors for postpartum uterine diseases in dairy cows. J Dairy Sci. 2010 Dec;93(12):5764–71. doi: 10.3168/jds.2010-3429. [DOI] [PubMed] [Google Scholar]
- 85.Valergakis GE, Oikonomou G, Arsenos G, Banos G. Phenotypic association between energy balance indicators and reproductive performance in primiparous Holstein cows. Veterinary Record. 2011 Feb 19;168(7):189. doi: 10.1136/vr.c6259. [DOI] [PubMed] [Google Scholar]
- 86.McCabe M, Waters S, Morris D, Kenny D, Lynn D, Creevey C. RNA-seq analysis of differential gene expression in liver from lactating dairy cows divergent in negative energy balance. BMC Genomics. 2012;13:193. doi: 10.1186/1471-2164-13-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Morris DG, Waters SM, McCarthy SD, et al. Pleiotropic effects of negative energy balance in the postpartum dairy cow on splenic gene expression: repercussions for innate and adaptive immunity. Physiol Genomics. 2009 Sep 9;39(1):28–37. doi: 10.1152/physiolgenomics.90394.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Loor JJ, Dann HM, Guretzky NAJ, et al. Plane of nutrition prepartum alters hepatic gene expression and function in dairy cows as assessed by longitudinal transcript and metabolic profiling. Physiological Genomics. 2006 Oct 3;27(1):29–41. doi: 10.1152/physiolgenomics.00036.2006. [DOI] [PubMed] [Google Scholar]
- 89.Cummins CL, Volle DH, Zhang Y, et al. Liver X receptors regulate adrenal cholesterol balance. J Clin Invest. 2006 Jul;116(7):1902–12. doi: 10.1172/JCI28400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Viturro E, Koenning M, Kroemer A, et al. Cholesterol synthesis in the lactating cow: Induced expression of candidate genes. J Steroid Biochem Mol Biol. 2009 May;115(1–2):62–7. doi: 10.1016/j.jsbmb.2009.02.011. [DOI] [PubMed] [Google Scholar]
- 91.Choi G, Schultz MJ, Levi M, van der Poll T. The relationship between inflammation and the coagulation system. Swiss Medical Weekly. 2006 Mar 4;136(9–10):139–44. doi: 10.4414/smw.2006.11059. [DOI] [PubMed] [Google Scholar]
- 92.Amara U, Rittirsch D, Flierl M, et al. Interaction Between the Coagulation and Complement System. Current Topics in Complement Ii. 2008;632:71–9. doi: 10.1007/978-0-387-78952-1_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang Y, Zhang JW, Lv GY, Xie SL, Wang GY. Effects of STAT3 gene silencing and rapamycin on apoptosis in hepatocarcinoma cells. Int J Med Sci. 2012;9(3):216–24. doi: 10.7150/ijms.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yokogami K, Wakisaka S, Avruch J, Reeves SA. Serine phosphorylation and maximal activation of STAT3 during CNTF signaling is mediated by the rapamycin target mTOR. Curr Biol. 2000 Janauary;10(1):47–50. doi: 10.1016/s0960-9822(99)00268-7. [DOI] [PubMed] [Google Scholar]
- 95.Hsieh FC, Cheng G, Lin J. Evaluation of potential Stat3-regulated genes in human breast cancer. Biochem Bioph Res Co. 2005 Sep 23;335(2):292–9. doi: 10.1016/j.bbrc.2005.07.075. [DOI] [PubMed] [Google Scholar]
- 96.Seita J, Asakawa M, Ooehara J, et al. Interleukin-27 directly induces differentiation in hematopoietic stem cells. Blood. 2008 Feb 15;111(4):1903–12. doi: 10.1182/blood-2007-06-093328. [DOI] [PubMed] [Google Scholar]
- 97.Khatib H, Huang W, Mikheil D, Schutzkus V, Monson RL. Effects of signal transducer and activator of transcription (STAT) genes STAT1 and STAT3 genotypic combinations on fertilization and embryonic survival rates in Holstein cattle. J Dairy Sci. 2009 Dec;92(12):6186–91. doi: 10.3168/jds.2009-2439. [DOI] [PubMed] [Google Scholar]
- 98.Kisseleva T, Bhattacharya S, Braunstein J, Schindler CW. Signaling through the JAK/STAT pathway, recent advances and future challenges. Gene. 2002 Feb 20;285(1–2):1–24. doi: 10.1016/s0378-1119(02)00398-0. [DOI] [PubMed] [Google Scholar]
- 99.Nishi T, Forgac M. The vacuolar (H+)-atpases – Nature’s most versatile proton pumps. Nature Reviews Molecular Cell Biology. 2002 Feb;3(2):94–103. doi: 10.1038/nrm729. [DOI] [PubMed] [Google Scholar]
- 100.Dong Z, Wang Y, Chang M, Li GY, Hu LS. Valproic acid alters differential protein expression in SH-SY5Y cells. Neural Regeneration Research. 2011 Sep 25;6(27):2134–9. [Google Scholar]
- 101.Migliore L, Coppede F. Environmental-induced oxidative stress in neurodegenerative disorders and aging. Mutation Research-Genetic Toxicology and Environmental Mutagenesis. 2009 Mar 31;674(1–2):73–84. doi: 10.1016/j.mrgentox.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 102.Shi L, Jones WD, Jensen RV, et al. The balance of reproducibility, sensitivity, and specificity of lists of differentially expressed genes in microarray studies. Bmc Bioinformatics. 2008;9(Suppl 9):S10. doi: 10.1186/1471-2105-9-S9-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary file 1. Has information about primers, qPCR efficiency, relative % mRNA abundance and microarray designed used.
Supplementary file 2. Had complete out put obtained from the KEGG and DIA analysis run with Liver and Adipose microarray data.