Abstract
Introduction
KRAS mutations are poor prognostic markers for patients with non-small cell lung cancer (NSCLC). RALA and RALB GTPases lie downstream of RAS and are implicated in RAS mediated tumorigenesis. However, their biological or prognostic role in the context of KRAS mutation in NSCLC is unclear.
Methods
Using expression analysis of human tumors and a panel of cell lines coupled with functional in vivo and in vitro experiments, we evaluated the prognostic and functional importance of RAL in NSCLC and their relationship to KRAS expression and mutation.
Results
Immunohistochemical (N=189) and transcriptomic (N=337) analyses of NSCLC patients revealed high RALA and RALB expression was associated with poor survival. In a panel of 14 human NSCLC cell lines, RALA and RALB had higher expression in KRAS mutant cell lines while RALA but not RALB activity was higher in KRAS mutant cell lines. Depletion of RAL paralogs identified cell lines that are dependent on RAL expression for proliferation and anchorage independent growth. Overall, growth of NSCLC cell lines which carry a glycine to cystine (G12C) KRAS mutation were more sensitive to RAL depletion than those with wild-type KRAS. Using gene expression and outcome data from 337 human tumors, RAL-KRAS interaction analysis revealed that KRAS and RAL paralog expression jointly impact patient prognosis.
Conclusion
RAL GTPase expression carries important additional prognostic information to KRAS status in NSCLC patients. Simultaneously targeting RAL may provide a novel therapeutic approach in NSCLC patients harboring G12C KRAS mutations.
Keywords: RAL, RAS, non-small cell lung cancer, GTPase, prognosis
INTRODUCTION
In the United States, 228,190 new diagnoses and 159,480 lung cancer deaths are projected for 2013 1. Worldwide lung cancer is also the leading cause of cancer death 1. Historically therapy has been guided by tumor histology. Yet, despite recent advances, overall survival in the US remains 16% at five years for non-small-cell lung cancer (NSCLC), a group composed of adenocarcinoma, squamous cell carcinoma, and large-cell carcinoma subtypes. During the past decade, unique genetic changes have been observed in NSCLCs 2-5 and used for both prognostication and therapeutic decision making using targeted agents.
RAS mutations are found in 25-40% of NSCLCs 5-7 with KRAS mutations accounting for 90%. Approximately 97% of KRAS mutations in NSCLC involve codon 12 or 13 8 and are a negative prognostic marker for patients with NSCLC 9. Unfortunately, direct RAS targeted therapy is not clinically available 4, 5. An alternate strategy is targeting signal proteins downstream of RAS. RAS proteins signal primarily through three cascades; MAPK, PI3K/AKT and RAL GTPase 2, 5. Inhibitors of the MAPK and PI3K/AKT pathway are in various stages of clinical trials for treatment of NSCLC patients with KRAS mutations 5. Since no compelling clinical rationale exists for RAL targeting in cancer, no therapies have been developed.
RAL GTPases are critical drivers of human oncogenesis with vital roles in tumor growth and migration in pancreatic, prostate, colorectal and bladder cancers 10-13.The RAL GTPase family is comprised of RALA and RALB paralogs which share 85% amino acid sequence homology 14. Despite similar structures and downstream effectors they have differential effects on cancer cell phenotypes in different tumor models 14. Recently, a KRAS driven NSCLC mouse model showed RAL GTPase is required for tumorigenesis 15. However, the importance of RAL as a prognostic marker and its functional importance with respect to KRAS expression and mutation status in human NSCLC is unknown 16. Here we combine for the first time transcriptomic and immunohistochemical analyses on human samples with molecular manipulation and evaluation of RAS and RAL in human NSCLC cell lines to show that RALA and RALB are both important prognostic factors in NSCLC and drive tumor growth in vivo. Our work provides the rationale for thinking that simultaneous inhibition of MAPK, PI3K/AKT and RAL pathways would be effective treatment of patients with KRAS mutations 5 and impetus for drug development directed at the RAL pathway.
MATERIALS AND METHODS
Cell Lines and Biochemical Reagents
NSCLC cell lines H358, H2122, H460, A549, H157, Calu-6, SW1573, H2009, H2228, H1703, HCC4006, Calu-3, H322 and H292 were obtained from the American Type Culture Collection (ATCC, Manassas, VA) and cultured in RPMI-1640 (Invitrogen, Grand Island, NY) supplemented with 10% fetal bovine serum (FBS) as recommended by ATCC. Small interfering RNA (siRNA) targeting RALA (siRALA, 5′-GACAGGUUUCUGUAGAAGA-3′), RALB (siRALB, 5′-AAGCUGACAGUUAUAGAAA-3′) or both (siRALA+B, 5′-GACUAUGAACCUACCAAAG-3′) were obtained as previously described 17 . A second set of siRNA against RALA (siRALA II, 5′-CAGAGCUGAGCAGUGGAUU-3′) and RALB (siRALB II, 5′-GGUGAUCAUGGUUGGCAGC-3′) was also used. A non-specific siRNA (siCTL, 5′-CGTACGCGGAATACTTCGA-3′) was used as control for all the experiments17. All siRNAs were from Dharmacon (Lafayette, CO). Cells were transduced with siRNA (200nmol/L) using oligofectamine (Invitrogen, Grand Island, NY) according to the manufacturer's instructions. KRAS wild-type (WT) and KRAS G12C mutant constructs were from the Missouri S&T cDNA Center (Rolla, MO).
Western Blotting, RAL and KRAS Activation Assays
Cells were lysed using CelLytic™ Cell Lysis Reagent (Sigma-Aldrich) and Westerns performed as previously described 17. Equal amounts of protein were subjected to SDS-PAGE, transferred to PVDF membrane, and probed with antibodies against RALA (BD Transduction Laboratories, San Jose, CA), RALB (Millipore, Billerica, MA), α-Tubulin (Santa Cruz Biotechnology Inc, Santa Cruz, CA), ERK and phospho-ERK (pERK), AKT and phospho-AKT, and KRAS (all from Cell Signaling, Danvers, MA). RALA and RALB activity was measured using RAL Activation Assay Kit (Millipore, Billerica, MA). Briefly cell lysates were incubated with RALBP1 agarose slurry for 4 hrs at 4°C. After this the beads are washed and boiled in Laemmli Sample Buffer. The boiled samples are divided in half and run as two set of samples on SDS-PAGE followed by Western Blotting. One set of sample is probed for RALA and the second for RALB 17. HRP labeled mouse or rabbit secondary antibodies (Cell Signaling) were used to develop the blots by chemiluminescence using ECL (Pierce, Rockford, IL). KRAS activation assay was carried out on H2228 cells stably transfected with empty vector, KRAS WT and KRAS G12C constructs using RAS Activation ELISA Assay Kit (Millipore, Billerice, MA) following the manufacturer's protocol.
Cancer cell growth in vitro and in vivo
For assessment of monolayer proliferation, 104 cells were plated in 96 well plates in triplicate or greater 48 hrs after the cells were transduced with control siRNA (siCTL) or siRNA targeting RALA, RALB or both (RALA+B). Cell numbers were determined daily using the CYQUANT assay (Invitrogen, Grand Island, NY) as directed by the manufacturer. For anchorage independent growth assessment, lines were plated in triplicate in 0.4% agar (15,000 or 20,000 cells/well) 48 hrs after siRNA transduction. At selected time points based on the colony forming capacity of each cell line, colonies formed in soft agar were stained with Nitro-BT (Sigma-Aldrich) at 37°C overnight and counted using software ImageJ® 18. For xenograft experiments, 4-week-old female athymic NCr-nu/nu mice were obtained from the National Cancer Institute. (NCI-Frederick, Frederick, MD). At 6-8 weeks of age they were injected with H2122 cells (2x105 cells/site) transduced 48 hours earlier with RALA, RALB, RALA+RALB, or control siRNA in their left and right flanks, and monitored for subcutaneous tumor growth. Tumors were measured regularly as indicated in the results and tumor volume calculated as described 19.
RAL Immunohistochemical Analysis
Details on primary tumor samples and specific protocols for IHC sample preparation and RAL GTPase staining are in Supplementary Information. Membranous and cytoplasmic expression was scored separately and the association of RALA and RALB expression with patient outcomes (i.e., time to progression, overall survival, etc) was performed by the University of Colorado Cancer Center Biostatistics and Bioinformatics Shared Resource using SAS/BASE and SAS/STAT software, Version 9.2 of the SAS System for Windows (SAS Institute Inc., Cary, NC, USA). Cox proportional hazards model was fit and the proportional hazards assumption was checked and found to be met. The interaction between RALA and RALB protein expression was analyzed as well as the association to outcome of each protein alone, adjusting for patient characteristics, stage (1/2/3/4), age, histology (Adenocarcinoma, Squamous Cell Carcinoma, NSCLC, Not Otherwise Specified (NOS), Mixed (mixed adenocarcinoma and squamous cell carcinoma characteristics)), and sex (M/F). When analyzing the association between overall survival and the interaction between RALA and RALB, expression scores were dichotomized at their respective medians (i.e., low RALA+low RALB, low RALA+high RALB, high RALA+low RALB, high RALA+high RALB); when analyzing one protein alone, the expression score was categorized by its quartiles (lowest 25%, 25%-50%, 50%-75%, highest 25% of scores)
Microarray Analysis
Three publicly available NSCLC patient datasets (Supplementary Table 3) were used. To examine whether RALA or RALB gene expression could stratify patient survival, patients were divided into two groups according to gene expression or risk scores and were compared by Cox proportional hazards models and log-rank tests (see Supplementary Information). To analyze the interaction between KRAS and RAL genes, a patient is classified as either high- or low-expressing using a gene's median expression. Patient groups with different expression levels of KRAS and RAL genes were compared by Cox proportional hazards models and log-rank tests.
RESULTS
RAL expression stratifies prognosis in NSCLC patients
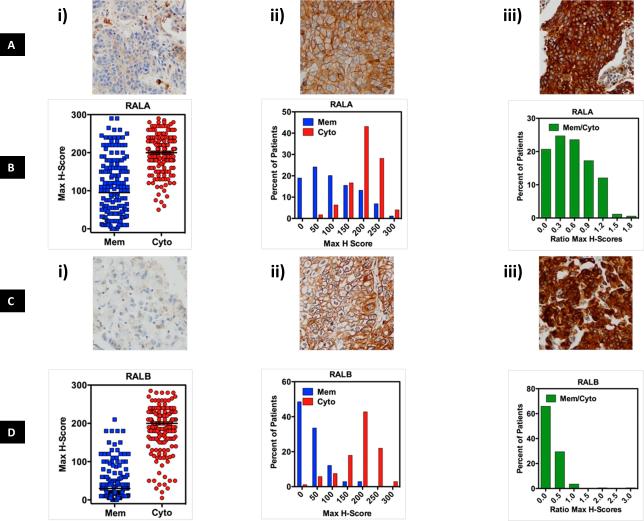
It is well established that in cancerous cells there is more RAL-GTP versus RAL-GDP compared to normal cells and its role in various tumor types is acknowledged10, 14, 17, 20. However RAL expression as a prognostic biomarker in cancers is unclear21. The role of RAL expression as a prognostic biomarker in NSCLC is yet to be determined. We addressed this gap in the literature by investigating whether RAL expression determined by either IHC or transcriptomic analysis was prognostic in patients with NSCLC. The clinical cohort used for the prognostic (IHC) study is described in Supp Table 1. Analysis of RALA and RALB membrane and cytoplasmic expression in the tumor samples revealed that NSCLC patients have variable degree of RAL GTPase membrane and cytoplasmic expression (Figure 1A, C). There is higher cytoplasmic expression of RALA and RALB compared to membrane expression (P<0.05, Figure 1B, D). Also RALA membrane expression is higher than RALB membrane expression (P<0.0001, Figure 1B, D). Since membrane localization of GTPases is canonically associated with higher activity 14, 20, 22, 23 these data suggest NSCLC specimens harbor more activated RALA than RALB. Interestingly, RAL cytoplasmic and membrane expression does not change as a function of tumor stage and histology (Supp Figure 1). With IHC techniques established, the correlation of RALA and RALB membrane and cytoplasmic expression with patient survival was evaluated. High RALA membrane expression (top 25% expression) trended towards poor overall survival when compared to low RALA membrane expression (low 25% expression) though the data is not statistically significant (Figure 2A, Supp Table 2) while high RALB membrane expression (top 25% expression) was associated with poor overall survival when compared to low RALB membrane expression (bottom 25% expression, P=0.04, Figure 2B, Supp Table 2). When we evaluated all four quartiles of RALA and B expression and effect on patient survival the data was not found to be statistically significant (Supp Figure 2A, B). Other mathematical constructs of RAL expression (with either each paralog separately or combined) such as, membrane (data not shown), membrane/cytoplasm (data not shown) or membrane + cytoplasm (Supp Figure 2C, D) etc. did not offer enhanced predictive ability over individual analysis.
Figure 1. RALA and B cytoplasmic and cell membrane immunohistochemical staining in the NSCLC patient tumor samples.
H-score was used to measure the staining for RAL protein expression (see Supplementary Materials and Methods). The average H-score of the triplicate cores per patient was highly correlated to the core with the highest score from the same patient, thus further analyses were performed using the maximum H-score. Panels (A) and (C) show typical examples of i) low or no cytoplasmic and membranous protein; ii) high membranous and low cytoplasmic; iii) high cytoplasmic and high membranous staining for RALA and RALB. (B) and (D) show three graphical depictions of RALA and B membrane and cytoplasmic staining in NSCLC specimens.
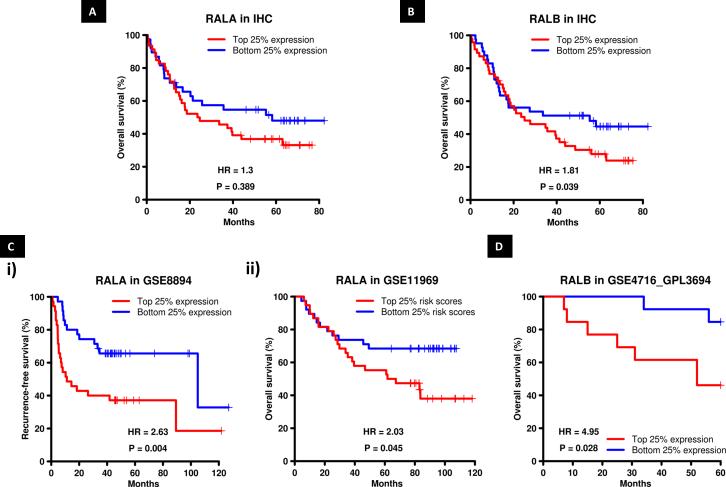
Figure 2. RALA and B protein and mRNA expression stratify NSCLC patient survival.
Overall NSCLC patient survival as a function of (A) RALA and (B) RALB membrane expression as evaluated by IHC. (C) Patient survival in NSCLC datasets GSE8894 and GSE11969 (Supp Table 3) as a function of RALA i) mRNA expression or ii) Risk Score (see Materials and Methods). (D) Overall patient survival in NSCLC dataset GSE4716_GPL3694 as a function of RALB expression.
Next, we examined the prognostic importance of RAL mRNA expression in predicting NSCLC patient survival. Three public gene expression datasets of lung cancer patients were analyzed (Supp Table 3). In 2 of 3 datasets, patients with high RALA expression had poorer recurrence-free survival and overall survival in comparison to low RALA expression (Figure 2Ci and ii). High RALB mRNA expression was also prognostic of poor overall survival in 1 of 3 datasets (Figure 2D). When we evaluated all four quartiles of RALA and B expression and impact on patient survival, the data was statistically significant in 2 of 3 datasets for RALA (Supp Figure 3A, B). RALB expression and impact on patient survival was not statistically significant when we investigate all four quartiles (Supp Figure 3C). However, RAL protein and mRNA expression was a strong prognostic marker of patient survival when we compared patients with very high RAL expression (top 25%) to patients with very low RAL expression (bottom 25%).
Expression and activation of RAL is related to KRAS mutation in human NSCLC cell lines
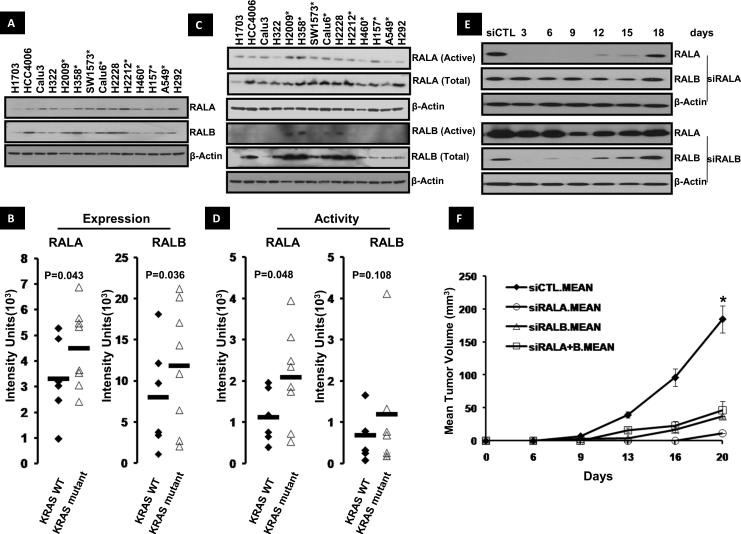
We next sought to determine the relationship of RAL expression and activity to the mutation status of its upstream regulator KRAS by evaluating RAL protein expression in 14 human NSCLC cell lines. RALA and RALB expression and activity were detected in all lines (Figure 3A, C). Eight cell lines (H358, H2122, A549, H2009, H460, SW1573, H157 and Calu6) have KRAS mutations while the remaining (H292, H322, H1703, H2228, H4006 and Calu3) are KRAS wild-type (WT), with H4006 harboring an epidermal growth factor receptor (EGFR) mutation. Quantification of expression by densitometry revealed average RALA and RALB expression was higher in KRAS mutant lines compared to WT lines (P=0.043 and 0.036 respectively, Figure 3B). Total RALA activation was also higher in KRAS mutant NSCLC cell lines compared to WT cells (P=0.048, Figure 3D). In contrast, minimal RALB activation was observed in these lines (Figure 3C) with no correlation between total RALB activity and KRAS mutation status (Figure 3D). There was no relationship between RAL paralog expression and their corresponding activation levels (P=0.08 for RALA and P=0.23 for RALB, Supp Figure 4). qRT-PCR analysis was also carried out for RALA and B mRNA expression in these cell lines (Supp Table 4). Cell lines with KRAS mutations trended towards high RALA (0.013 vs 0.011) and RALB (0.0024 vs 0.0015) relative mRNA expression normalized to β-actin compared to WT lines however the data was not statistically significant with P=0.11 and 0.28 respectively. We carried out RALA and RalB IHC on the above mentioned cell lines to study the relationship of Ral activation to RAL membrane and cytoplasmic expression and found no relationship.
Figure 3. RALA and RALB expression and activity in human NSCLC cell lines.
(A) RALA and RALB expression in 14 NSCLC cell lines by western blot. Cell lines with KRAS mutation indicated with asterisks. (B) Densitometric quantification of RALA and RALB expression shows their expression is higher in KRAS mutant NSCLC cell lines (P<0.05, student's t-test). (C) RALA and RALB activation levels in the 14 NSCLC cell lines. Cell lines with KRAS mutation indicated with asterisks. (D) Densitometric analysis revealed higher RALA activation in KRAS mutant NSCLC cell lines (P<0.05, student's t-test). No relationship to KRAS was found with RALB. (E) Knockdown of RAL GTPase in H2122 cells following transient transfection with siRNA against RALA (siRALA) and B (siRALB). Cells were lysed 3,6,9,12,15 and 18 days after transfection and RAL GTPase knockdown was determined by western blot. (F) Loss of RALA, B or A+B reduced subcutaneous tumor growth in mice. H2122 cells were transfected with siRNA against RALA (siRALA), B (siRALB) and A+B (siRALA+B), injected in mice and studied for in vivo tumor growth (see Materials and Methods). Luciferase siRNA transfected cells were used as control (siCTL). *P<0.05 by student's t-test.
RAL expression drives NSCLC growth in vitro and in vivo
To determine whether RAL has a functional role in human NSCLC we evaluated the dependency of cell line proliferation and anchorage independent growth on RAL expression. For these experiments we used KRAS WT NSCLC cell lines and cell lines with KRAS mutation in codon 12, the most common KRAS mutation site in NSCLC 3. RAL GTPase depletion was performed using specific RAL siRNAs targeting RALA, RALB or a motif common to both RALA and RALB (A+B) leading to simultaneous knockdown 17. RAL GTPase knockdown was evaluated 72 hrs after siRNA transduction by Western blotting for RALA and RALB and identified a 70-80% depletion of RALA, RALB or both proteins in all lines (Supp Figure 5). Depletion of RALA, RALB or both resulted in inhibition of anchorage dependent and independent growth of 4/6 KRAS WT and 5/6 KRAS mutant NSCLC cell line (P<0.05, Table 1). Loss of RALA vs. RALB vs. both proteins had different degrees of growth inhibition. Depletion of RALA had the greatest effect, with inhibition of monolayer growth in 2/6 KRAS WT NSCLC cell lines compared to 5/6 KRAS mutant cell lines (P=0.037) and inhibition of anchorage independent growth in 1/6 KRAS WT cell lines compared to 4/6 KRAS mutant cell lines (P=0.04).
TABLE 1. Monolayer and anchorage independent growth of NSCLC cell lines with knockdown of RAL GTPase.
Cells were transfected with siRNA against RAL GTPase and plated for proliferation and anchorage independent growth assay (see Materials and Methods). Monolayer growth was measured 4 days after transfection with results representing the mean of 2-3 studies with 4-5 replicates per cell line. Anchorage independent growth results are mean of three experiments.
| RALA Knockdown | RALB Knockdown | RALA+B Knockdown | ||||||
|---|---|---|---|---|---|---|---|---|
| Monolayer Growth | Anchorage Independent Growth | Monolayer Growth | Anchorage Independent Growth | Monolayer Growth | Anchorage Independent Growth | |||
| Cell Line | Histology | KRAS | ||||||
| H322 | Adenocarcinoma | WT | −52%* | −41.2%* | −33%* | −42.3%* | −46%* | −39%* |
| H1703 | Adenocarcinoma | WT | N.S. | N.S. | N.S. | N.S. | N.S. | −52.3%* |
| H2228 | Adenocarcinoma | WT | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. |
| H292 | Mucoepidermoid | WT | N.S. | No Colony | −23%* | No Colony | N.S. | No Colony |
| H4006 | Adenocarcinoma | WT | N.S. | No Colony | N.S. | No Colony | N.S. | No Colony |
| Calu3 | Adenocarcinoma | WT | −33%* | No Colony | −36%* | No Colony | −39%* | No Colony |
| H2122 | Adenocarcinoma | G12C | −91%* | −97%* | −61%* | −89.2%* | −42%* | −79.6%* |
| H358 | Adenocarcinoma | G12C | −24%* | −38%* | N.S. | −27.6%* | −86%* | −83.4%* |
| A549 | Adenocarcinoma | G12S | −26%* | −24.5%* | −29%* | N.S. | −27%* | −51.3%* |
| H2009 | Adenocarcinoma | G12A | −25%* | −35%* | −19%* | −28.7%* | 28%* | −15.8%* |
| H157 | Squamous Cell | G12R | −24%* | N.S. | −19%* | N.S. | N.S. | N.S. |
| SW1573 | Alveolar Cell | G12A | N.S. | No Colony | N.S. | No Colony | N.S. | No Colony |
P<0.05 (student's t-test)
(+) indicates increase compared to control
(−) indicates decrease compared to control
“N.S.” is no significant change
“No Colony” indicates cells did not exhibit anchorage independent growth.
The impact of RAL depletion on anchorage independent growth was greatest on the H358 (G12C KRAS) and H2122 (G12C KRAS) cell lines (Table 1). H2122 had near complete inhibition of colony formation with RALA knockdown while knock down of RALB or both proteins resulted in 90% and 79% inhibition. H358 had 38%, 27% and 83% decrease in anchorage independent growth with similar RAL depletions (Table 1). To confirm these observations were not due to nonspecific siRNA effects, the study was repeated in H2122 with a second set of RAL GTPase siRNAs (siRALA II and siRALB II) and similar results were observed (Supp Figure 6). Interestingly, investigation of the PI3K/AKT and MAPK pathways which also signal downstream of KRAS revealed that these alternate pathways had minimal to no activation in NSCLC cell lines which carry the KRAS G12C mutation (Supp Figure 7).
The H2122 cell line was then used to study RAL's effect on tumor growth in vivo. H2122 cells were transfected with control, RALA, RALB or RALA+RALB siRNAs and RAL expression evaluated at multiple time points following transfection (Figure 3E). RAL expression was found to be suppressed for up to 15 days (Figure 3E) supporting the notion that in vivo tumor growth would be suppressed during this time frame. With this data in hand, we repeated the depletion in H2122 cells and inoculated them into mice two days after transfection. As shown in Figure 3F, transient knockdown of RALA, RALB, or both proteins had significant impact on tumor growth with a 94%, 81% and 81% decrease respectively at 20 days (P<0.05). Together, these data suggest that RAL expression is especially important for tumor growth in NSCLC cell lines harboring G12C KRAS mutations.
G12C KRAS mutation regulates anchorage independent growth through RAL
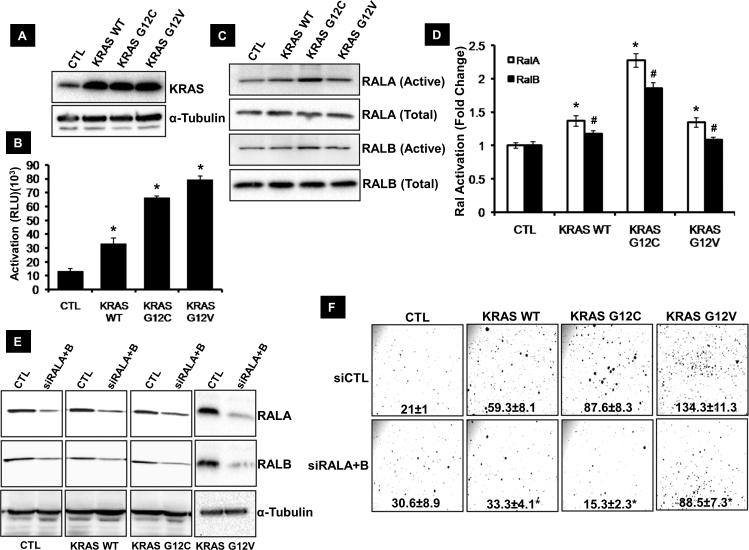
Functional assays revealed cell lines with KRAS G12C mutation were most dependent on RAL GTPase for tumor processes (proliferation, anchorage independent growth) (Table 1). Therefore, to determine whether KRAS G12C mutation results in tumor progression that is specifically dependent on the RAL pathway, H2228 cells were stably transfected with KRAS WT,KRAS G12C mutant (found in H2122) and KRAS G12V mutant (Figure 4A). The H2228 cell line was chosen because it is KRAS WT, has high RALA and B expression but its anchorage independent growth is independent of RAL expression (Table 1). H2228 cells overexpressing KRAS G12C and G12V mutants have high KRAS activation compared to cells transfected with KRAS WT (Figure 4B). An increase in RAL activation in H2228 cells overexpressing the KRAS G12C mutant was also seen when compared to H2228 cells overexpressing KRAS WT and KRAS G12V mutant (Figure 4C, D) suggesting increased signaling through RAL in cells having KRAS G12C mutation. Importantly, KRAS G12C and G12V overexpressing cells had a 48% and 127% increase in anchorage independent growth compared to KRAS WT (P<0.05) respectively. This increase in anchorage independent growth for cells overexpressing KRAS G12C was RAL dependent as shown by a 83% inhibition in anchorage independent growth with siRNA mediated depletion of RALA+RALB in H2228 cells expressing KRAS G12C compared to 44% and 34% inhibition in cells overexpressing WT KRAS and KRAS G12V mutant (Figure 4E, F). This suggests that NSCLC tumors with a KRAS G12C mutation become more reliant on the RAL pathway for tumor growth.
Figure 4. The role of RAL and KRAS in tumor growth.
(A) KRAS expression in H2228 cells stably transfected with empty vector (CTL), wild-type KRAS (KRAS WT) and KRAS G12C mutant (KRAS G12C) detected by western blot. (B) KRAS activation in the engineered H2228 cell lines as determined by RAS activation ELISA (see Materials and Methods). *P<0.05 by student's t-test. (C) RALA and B activation observed by western blot in H2228 cells transfected with empty vector (CTL), wild-type KRAS (KRAS WT) and KRAS G12C mutant (KRAS G12C) (see Materials and Methods). (D) Densitometric analysis of RALA and B activation observed in C. Asterisk and pound indicate P<0.05 for RALA and B by student's t-test) (E) RALA and B loss observed by western blot following transfection of engineered H2228 cell lines with siRNA targeting both RALs (siRALA+B). A luciferase siRNA transfected cells are used as control (siCTL). (F) Anchorage independent growth of engineered H2228 cell lines following loss of RAL GTPase. (*P<0.05 by student's t-test).
Contributions of KRAS and RAL in NSCLC tumor progression
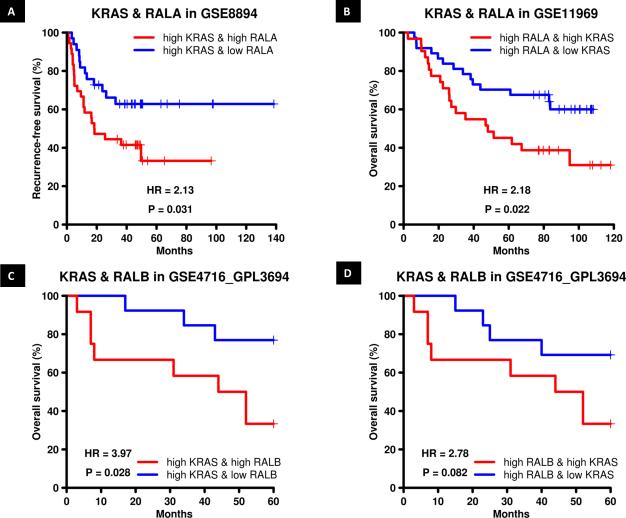
These data suggest RAL expression mediates KRAS driven NSCLC growth. This extends prior work showing RAL is required for KRAS induced tumor formation 20. To determine if this relationship is supported by clinical data, we examined the relationship of RAL paralog expression to that of KRAS mRNA expression in regard to survival of NSCLC patients. In dataset GSE8894 where we found high RALA mRNA predicted poor patient outcome (Figure 2C), stratification of patients by KRAS and RALA mRNA expression status found high KRAS and RALA mRNA expression was associated with poor recurrence free survival compared to patients with high KRAS and low RALA mRNA expression (P=0.031, Figure 5A). In addition, analysis of dataset GSE11969 where we had previously found high RALA risk score predicted poor patient outcome (Figure 2C) suggested that KRAS risk score impacts the prognostic stratification driven by the RALA risk score with high RALA and KRAS mRNA expression now associated with poor overall survival compared to patients with high RALA and low KRAS mRNA expression (P=0.022, Figure 5B). These analyses indicate that KRAS and RALA can contribute independently to patient prognosis despite their canonical hierarchical relationship. A similar analysis was carried out for KRAS and RALA in GSE4716_GPL3694 in which high RALB mRNA expression predicted poor overall patient survival (Figure 2D) but none of the interactions were statistically significant.
Figure 5. RAL and KRAS mRNA expression and risk score stratify overall and recurrence free survival in NSCLC patients.
(A) Recurrence free survival in NSCLC dataset GSE8894 as a function of RALA mRNA expression in patients with high KRAS mRNA expression. (B) Patient overall survival in NSCLC dataset GSE11969 as a function of KRAS risk score in patients with high RALA risk score. (C) Patient overall survival in NSCLC dataset GSE4716_GPL3694 as a function of RALB mRNA expression in patients with high KRAS mRNA expression. (D) Patient overall survival in NSCLC dataset GSE4716_GPL3694 as a function of KRAS mRNA expression in patients with high RALB mRNA expression.
We also evaluated the interaction of RALB and KRAS expression in the two datasets where high RALA mRNA (GSE8894) or risk score (GSE11969) is predictive of poor patient outcome but no interaction was observed. However in dataset GSE4716_GPL3694 patients with high KRAS and RALB mRNA expression had poor overall survival compared to patients with high KRAS and low RALB mRNA expression (P=0.028, Figure 5C). Patients with high KRAS and RALB mRNA expression trended (P=0.082) towards poor overall survival compared to patients with low KRAS and high RALB mRNA expression (Figure 5D). Since KRAS mutation status of patients in these datasets was unknown, we could not investigate the survival information carried by RAL GTPase in patients carrying KRAS mutations.
DISCUSSION
Recently RAL GTPase null and conditional knockout mice were crossed with mice developing KRAS driven NSCLC to show that RAL is important in KRAS driven pulmonary tumorigenesis 15. PI3K/AKT and MAPK pathways are also important for KRAS driven tumors 5. Here we move the field forward by using IHC, transcriptomic and human cell line based functional analysis to investigate the prognostic information carried by RAL GTPase and their role in NSCLC tumor progression as a function of KRAS. We began our investigations by evaluating the prognostic information carried by RAL protein and mRNA expression in human cancers. IHC analysis of RAL protein expression revealed that RALB cell membrane expression is a negative prognostic marker in NSCLC (Figure 2B). Since membrane localization suggests the GTPase is in an active state 14, 20, 22, 23 we suggest that active RALB is a prognostic marker for decreased survival in NSCLC. In addition, patients with high RALA membrane expression trended toward poorer overall survival but the data were not statistically significant (Figure 2A). Since RAL cytoplasmic expression (data not shown) and total RAL expression (Supp Figure 2C, D) is not predictive of survival we can advocate that membranous RAL is the functionally active form of RAL in NSCLC patient tumors. This protein analysis was followed up with an evaluation of RALA and RALB mRNA expression where higher levels were associated with shorter recurrence-free and overall survival (Figure 2C, D). Interestingly, we noted that in the patient datasets (IHC or microarray) either RALA or RALB primarily carried the prognostic information (Figure 2). We conclude that RAL carries important clinical information. However, we speculate that in each tumor only one of the two RAL paralogs is functionally dominant.
Investigating the functional role of RAL GTPase in NSCLC revealed loss of RALA, RALB or both resulted in variable changes in monolayer proliferation, anchorage independent growth and subcutaneous tumor formation in NSCLC cell lines (Table 1, Figure 3E, F). RALA expression appeared more important in driving growth in KRAS mutant NSCLC cell lines compared to KRAS WT cell lines suggesting that in this panel of NSCLC cell lines RALA is the functionally dominant isoform driving KRAS dependent tumor growth (Table 1, Figure 3A, B). Interestingly, cell lines with a G12C KRAS activating mutation had greater dependence on RAL for anchorage independent growth compared to lines with other codon 12 mutations or KRAS WT (Table 1, Figure 4). Furthermore, NSCLC cell lines (H2122 and H358) with the G12C mutation had minimal activation of PI3K/AKT and MAPK pathways downstream of RAS (Supp Figure 7). H2122 has no activation whereas H358 has minimal activation of these alternate pathways and high activation of RAL GTPase. The lack of alternate pathway activation in these cell lines suggested that they are dependent on RAL signaling for their tumorigenic phenotype while those cell lines with activation of PI3K/AKT and MAPK pathways downstream of KRAS are partially or not dependent on RAL GTPase due to the availability of these alternate pathways for tumor progression (Table 1). A549 was the only exception to this trend (Supp Figure 7). This line does not have activation of PI3K/AKT and MAPK pathways yet is only marginally dependent on RAL expression for tumorigenicity (Table 1). This may be explained by the finding that A549 is independent of RAS activity for tumor growth 24. We gained further support for the notion that KRAS G12C mutations drive tumor progression via RAL GTPase by stably overexpressing KRAS G12C mutant construct in H2228 NSCLC cell line which is KRAS WT and showing RAL loss inhibits anchorage independent growth (Figure 4). Our findings confirmed and validated initial observations made by Ihle et al.16.
To understand the functional correlation between RAL paralogs and KRAS in NSCLC we examined RAL-KRAS statistical interactions in microarray datasets. This analysis was limited by the lack of KRAS mutation status and thus we made the assumption that tumors with high KRAS expression had concomitantly elevated activation. Our analysis revealed that both KRAS and RAL expression are determinants of patient prognosis and suggest that RAL function in NSCLC is driven only in part by KRAS while KRAS also activated other prognostic pathways (Figure 5). This observation is consistent with the aforementioned finding in the NSCLC cell line panel where we observe activation of PI3K/AKT and ERK pathways in cells that were less dependent on RAL expression (Table 1, Supp Figure 7).
These studies have significant implications for human NSCLC such as the identification of their RAL dependence, the identification of RAL as a potential therapeutic target in this disease and the ability to stratify patients for future anti-RAS or anti-RAL therapy by virtue of RAS and RAL expression and mutation status. Since inhibitors of downstream RAS signaling pathways such as PI3K/AKT and MAPK pathways are in clinical investigation in KRAS mutant NSCLC 5, these studies serve as a strong impetus for the development of anti-RAL therapeutics to suppress KRAS driven signal propagation and improve poor clinical outcomes seen in NSCLC patients.
Supplementary Material
Acknowledgments
FUNDING
This work is supported in part by National Institutes of Health grant CA075115 and the University of Colorado Comprehensive Cancer Center NCI SPORE in Lung Cancer CA046934.
Footnotes
AUTHORS’ CONTRIBUTIONS
Overall conception of the project was by D. Theodorescu. Design of the experiments was by S. Guin, F. Hirsch, J.A. Kern, D. Theodorescu. The entire cell based and xenograft experiments were conducted by S. Guin with help from R. Mishra and C. Owens. qRT-PCR on the cell lines was carried out by V. Vasu. Patient dataset analysis was conducted by Y. Ru. IHC staining and scoring was overseen by M. Wynes. IHC data interpretation and statistical analysis was by X. Lu and A.E. Barόn.
SUPPLEMENTARY MATERIAL
Supplementary Methods, Supplementary Figure 1-7 and Supplementary Tables 1-4 are found in the Supplementary Material Section.
CONFLICT OF INTEREST: None.
REFERENCES
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Malumbres M, Barbacid M. RAS oncogenes: the first 30 years. Nat Rev Cancer. 2003;3:459–465. doi: 10.1038/nrc1097. [DOI] [PubMed] [Google Scholar]
- 3.Okudela K, Woo T, Kitamura H. KRAS gene mutations in lung cancer: particulars established and issues unresolved. Pathol Int. 2010;60:651–660. doi: 10.1111/j.1440-1827.2010.02580.x. [DOI] [PubMed] [Google Scholar]
- 4.Riely GJ, Marks J, Pao W. KRAS mutations in non-small cell lung cancer. Proc Am Thorac Soc. 2009;6:201–205. doi: 10.1513/pats.200809-107LC. [DOI] [PubMed] [Google Scholar]
- 5.Suda K, Tomizawa K, Mitsudomi T. Biological and clinical significance of KRAS mutations in lung cancer: an oncogenic driver that contrasts with EGFR mutation. Cancer Metastasis Rev. 2010;29:49–60. doi: 10.1007/s10555-010-9209-4. [DOI] [PubMed] [Google Scholar]
- 6.Santos E, Martin-Zanca D, Reddy EP, et al. Malignant activation of a K-ras oncogene in lung carcinoma but not in normal tissue of the same patient. Science. 1984;223:661–664. doi: 10.1126/science.6695174. [DOI] [PubMed] [Google Scholar]
- 7.Rodenhuis S, van de Wetering ML, Mooi WJ, et al. Mutational activation of the K-ras oncogene. A possible pathogenetic factor in adenocarcinoma of the lung. N Engl J Med. 1987;317:929–935. doi: 10.1056/NEJM198710083171504. [DOI] [PubMed] [Google Scholar]
- 8.Forbes S, Clements J, Dawson E, et al. Cosmic 2005. Br J Cancer. 2006;94:318–322. doi: 10.1038/sj.bjc.6602928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mascaux C, Iannino N, Martin B, et al. The role of RAS oncogene in survival of patients with lung cancer: a systematic review of the literature with meta-analysis. Br J Cancer. 2005;92:131–139. doi: 10.1038/sj.bjc.6602258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lim KH, O'Hayer K, Adam SJ, et al. Divergent roles for RalA and RalB in malignant growth of human pancreatic carcinoma cells. Curr Biol. 2006;16:2385–2394. doi: 10.1016/j.cub.2006.10.023. [DOI] [PubMed] [Google Scholar]
- 11.Yin J, Pollock C, Tracy K, et al. Activation of the RalGEF/Ral pathway promotes prostate cancer metastasis to bone. Mol Cell Biol. 2007;27:7538–7550. doi: 10.1128/MCB.00955-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin TD, Samuel JC, Routh ED, et al. Activation and involvement of Ral GTPases in colorectal cancer. Cancer Res. 2011;71:206–215. doi: 10.1158/0008-5472.CAN-10-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gildea JJ, Harding MA, Seraj MJ, et al. The role of Ral A in epidermal growth factor receptor-regulated cell motility. Cancer Res. 2002;62:982–985. [PubMed] [Google Scholar]
- 14.Bodemann BO, White MA. Ral GTPases and cancer: linchpin support of the tumorigenic platform. Nat Rev Cancer. 2008;8:133–140. doi: 10.1038/nrc2296. [DOI] [PubMed] [Google Scholar]
- 15.Peschard P, McCarthy A, Leblanc-Dominguez V, et al. Genetic Deletion of RALA and RALB Small GTPases Reveals Redundant Functions in Development and Tumorigenesis. Curr Biol. 2012;22:2063–2068. doi: 10.1016/j.cub.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 16.Ihle NT, Byers LA, Kim ES, et al. Effect of KRAS oncogene substitutions on protein behavior: implications for signaling and clinical outcome. J Natl Cancer Inst. 2012;104:228–239. doi: 10.1093/jnci/djr523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oxford G, Owens CR, Titus BJ, et al. RalA and RalB: antagonistic relatives in cancer cell migration. Cancer Res. 2005;65:7111–7120. doi: 10.1158/0008-5472.CAN-04-1957. [DOI] [PubMed] [Google Scholar]
- 18.Gagliardi PA, di Blasio L, Orso F, et al. 3-phosphoinositide-dependent kinase 1 controls breast tumor growth in a kinase-dependent but Akt-independent manner. Neoplasia. 2012;14:719–731. doi: 10.1593/neo.12856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang H, Owens C, Chandra N, et al. Phosphorylation of RalB is important for bladder cancer cell growth and metastasis. Cancer Res. 2010;70:8760–8769. doi: 10.1158/0008-5472.CAN-10-0952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feig LA. Ral-GTPases: approaching their 15 minutes of fame. Trends Cell Biol. 2003;13:419–425. doi: 10.1016/s0962-8924(03)00152-1. [DOI] [PubMed] [Google Scholar]
- 21.Smith SC, Baras AS, Owens CR, et al. Transcriptional signatures of Ral GTPase are associated with aggressive clinicopathologic characteristics in human cancer. Cancer Res. 2012;72:3480–3491. doi: 10.1158/0008-5472.CAN-11-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kishida S, Koyama S, Matsubara K, et al. Colocalization of Ras and Ral on the membrane is required for Ras-dependent Ral activation through Ral GDP dissociation stimulator. Oncogene. 1997;15:2899–2907. doi: 10.1038/sj.onc.1201473. [DOI] [PubMed] [Google Scholar]
- 23.Matsubara K, Kishida S, Matsuura Y, et al. Plasma membrane recruitment of RalGDS is critical for Ras-dependent Ral activation. Oncogene. 1999;18:1303–1312. doi: 10.1038/sj.onc.1202425. [DOI] [PubMed] [Google Scholar]
- 24.Singh A, Greninger P, Rhodes D, et al. A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer Cell. 2009;15:489–500. doi: 10.1016/j.ccr.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.