Abstract
NMDA receptors (NMDAR), ligand-gated ion channels, play important roles in various neurological disorders, including epilepsy. Here we show the functional analysis of a de novo missense mutation (L812M) in a gene encoding NMDAR subunit GluN2A (GRIN2A). The mutation, identified in a patient with early-onset epileptic encephalopathy and profound developmental delay, is located in the linker region between the ligand-binding and transmembrane domains. Electrophysiological recordings revealed that the mutation enhances agonist potency, decreases sensitivity to negative modulators including magnesium, protons and zinc, prolongs the synaptic response time course, and increases single channel open probability. The functional changes of this amino acid apply to all other NMDAR subunits, suggesting an important role of this residue on the function of NMDARs. Taken together, these data suggest that the L812M mutation causes over-activation of NMDARs and drives neuronal hyperexcitability. We hypothesize that this mechanism underlies the patient’s epileptic phenotype as well as cerebral atrophy.
Keywords: Glutamate receptors, NMDA receptors, GluN2A, NR2A, refractory epilepsy, intractable seizures, epileptic encephalopathy, neurodegeneration, heterozygous, deactivation time course, single channel recording, outside out
INTRODUCTION
N-methyl-D-aspartate receptors (NMDARs) are ligand-gated ion channels that mediate excitatory neurotransmission in the brain. NMDARs are activated following the binding of glutamate and glycine to the GluN1 and GluN2 subunits, respectively. Simultaneous binding of both co-agonists opens a cation-selective pore that results in both depolarization and an increase in the intracellular Ca2+ concentration. The GluN1 subunit expressed universally throughout the brain, whereas four different GluN2 subtypes (A–D) have varying temporal and spatial expression in the central nervous system1,2. GluN2A and GluN2C subunits express mainly after birth, whereas the expression of GluN2B and GluN2D subunits is at high level at embryonic stages and is modestly diminished after birth in most (but not all) brain regions. NMDARs play important roles in neuronal development, synaptic plasticity, learning and memory, as well as in a wide range of pathological conditions, including epilepsy3,4. GRIN2A encodes GluN2A and has been genetically associated with autism, schizophrenia, Huntington and Parkinson diseases5, 6, 7, 8, 9, 10, 11. Recently, pathogenic mutations in genes encoding NMDARs subunits (GRIN1, GRIN2A, and GRIN2B) have been identified in human neurodevelopmental phenotypes, i.e. epilepsy, developmental delay, or intellectual disability12, 13, 14, 15, 16.
Here, we present functional analyses for a missense mutation in GRIN2A (GluN2A-L812M) identified in a child with intractable seizures and early-onset epileptic encephalopathy with profound developmental delay. In vitro studies reveal that the L812M mutated receptor has uniquely altered properties that increases NMDAR activity by every measure evaluated, leading to profound hyper-activation of NMDARs, which is almost certainly pathogenic. This residue is closely associated with gating in a conserved region in M4, which is adjacent to the conserved residues SYTANLAAF in the GluN1 M3 transmembrane helix as well as the GluN1 pre-M1 helix, both of which are proposed to participate in gating17. Given the conservation of this residue in all NMDAR subunits, we investigated whether the functional changes of the leucine to methionine mutation transfer to other NMDAR subunits. Our results show that this position is a key structural element of gating that is conserved across the NMDA receptor family.
RESULTS
Identification of GRIN2A L812M mutation
A six-year-old boy was admitted to the NIH Undiagnosed Diseases Program with a history of intractable infantile-onset epilepsy and profound global developmental delay with no attainment of any milestones, not even head control. At presentation he was having daily, generalized seizures that had been refractory to multiple anticonvulsants including lacosamide, rufinamide, and valproic acid. Brain MRI at 6 years of age showed diffuse cerebral parenchymal volume loss and a thin corpus callosum proportionate to the loss of cortical grey matter. There were no dysmorphic features and ophthalmological anomalies. Overall, his clinical history and features were consistent with an early-onset epileptic encephalopathy.
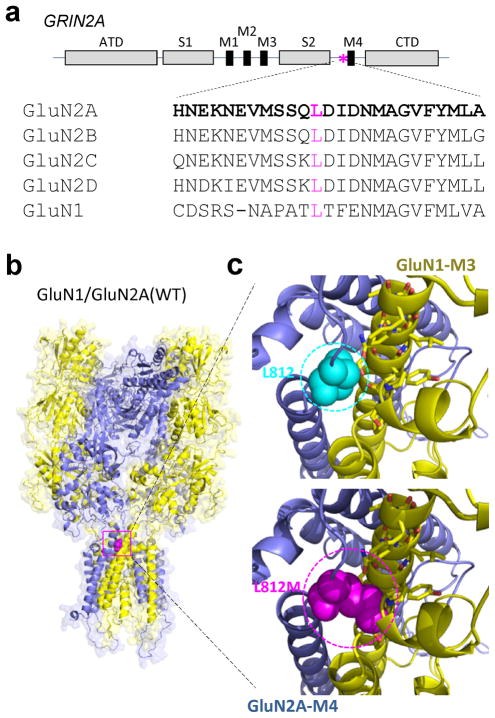
Since many early-onset epileptic encephalopathies are due to de novo mutations in ion channels or receptors18, 19, we screened for de novo potentially dominant variants and paired potentially recessive variants with whole exome sequencing. The only remarkable deleterious variant identified was a heterogeneous, de novo GRIN2A mutation: NCBI nucleotide accession number NM_001134407.1: c.2434C>A (p.Leu812Met, hereafter referred to as L812M), absent in his unaffected sibling as well as his unaffected parents. The Leu812 residue is highly conserved across vertebrate species and GluN1 and all GluN2 NMDAR subunits (Fig. 1a), indicating a possible critical role in NMDAR function. GluN2A Leu812 resides in the linker region between the lower portion of the agonist-binding domain (S2) and the transmembrane domain (M4) (Fig. 1a, b). Using the AMPA receptor structure as a guide17, this residue is predicted to reside close enough (within 5 Angstroms) to interact with two potential gating regions in the GluN1 subunit, the M3 transmembrane helix and the pre-M1 helix (Fig. 1c). Multiple lines of evidence suggest that a conserved motif (SYTANLAAF) in the M3 transmembrane helix controls gating17, 20, 21, 22, 23, 24, 25. Moreover, in AMPA receptors the pre-M1 helix lies in van der Waals contact with the gate and has been suggested to act as a cuff around gating elements that may stabilize the closed state17. Interestingly, this site is also a locus for allosteric regulation in NMDA, AMPA, and kainate receptors17, 26, 27, 28. Our working hypothesis is that the mutation GluN2A-L812M influences a conserved gating control element to both reduce the activation energy to reach the open state and stabilize the open state. A series of electrophysiological experiments were performed to evaluate this hypothesis, and explore the possibility that this mutation could account for this patient’s phenotype.
Figure 1. Identification of a GRIN2A missense mutation in a boy with intractable seizures and epileptic encephalopathy.
(a) A schematic linear representation of GluN2A with portions of the polypeptide chain that fold into semiautonomous domains labeled is shown. The position of the patient’s mutation is indicated by an asterisk. Leu812 is conserved through all GluN1 and GluN2 subunits. ATD (amino terminal domain), S1 and S2 (agonist binding domains), M1–4 (transmembrane domains 1–4), CTD (carboxy terminal domain). (b) A homology model of the GluN1/GluN2A receptor70 built from the GluA2 crystallographic data is shown as ribbon superimposed on a space-filled representation of the receptor. The position of the alteration p. L812M is indicated by magenta color in the linker region between ligand-binding and transmembrane domains. (c) An expanded view of the location of residue Leu812 (cyan color; upper panel) and the possible interaction between L812M (magenta color; lower panel) and transmembrane domain M3 of GluN1 subunit and a GluN1 pre-M1 helix are predicted from the homomeric GluA2 structure.
GluN2A L812M enhances agonist potency
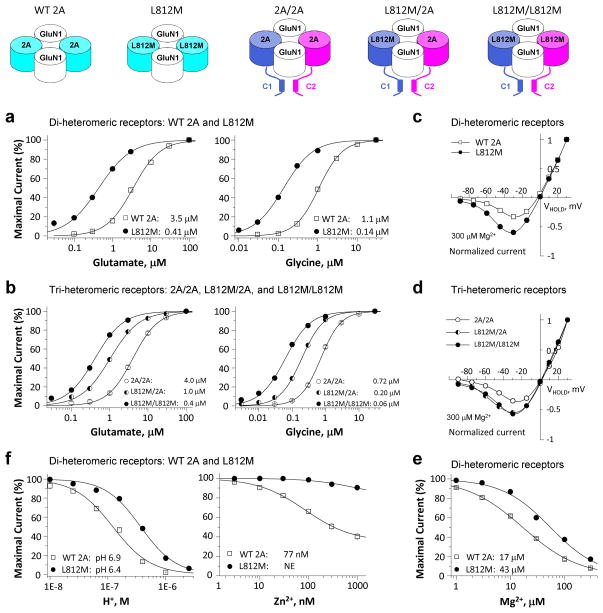
To investigate whether the mutation GluN2A-L812M influences NMDAR function, site-directed mutagenesis was used to introduce L812M into cDNA encoding the human GluN2A gene product (hereafter hGluN2A). We subsequently expressed wild-type (WT) and mutant hGluN2A with human GluN1 (hGluN1) in Xenopus oocytes and evaluated the concentration-effect curves for glutamate and glycine using a two-electrode voltage-clamp (TEVC). The mutation L812M increased the glutamate potency, as measured by reduction in the half-maximally effective concentration of agonist (EC50) from 3.5 μM to 0.41 μM (Fig. 2a, left; Table 1). Glycine potency was similarly increased, with the EC50 decreasing from 1.1 μM in WT to 0.14 μM in mutant receptors (Fig. 2a, right; Table 1). These data showed that the L812M mutation enhanced the potency of glutamate and glycine by 8-fold, which will allow the mutant receptor to be activated by much lower concentration of the agonists.
Figure 2. The L812M mutation changes the pharmacology of NMDARs.
(a) and (b) Composite concentration-response curves for agonists were determined by TEVC recordings from Xenopus oocytes expressing di-heteromeric GluN1/GluN2A (WT 2A) or GluN1/GluN2A-L812M (L812M) receptors and tri-heteromeric receptors with identity of each GluN2A in the tetrameric complex controlled (e.g. GluN1/GluN2A/GluN2A (2A/2A), GluN1/GluN2A-L812M/GluN2A (L812M/2A), and GluN1/GluN2A-L812M/GluN2A-L812M (L812M/L812M)) (see Methods and Supplemental Figure S2). a (left) and b (left) show the composite glutamate (in the presence of 100 μM glycine) concentration-response curves for di-heteromeric receptors and tri-heteromeric receptors. a (right) and b (right) show the composite glycine (in the presence of 100 μM glutamate) concentration-response curves for di-heteromeric receptors and tri-heteromeric receptors. (c) Mg2+ current voltage (I–V) curves reveal a decreased Mg2+-inhibition for L812M di-heteromeric receptors. (d) Mg2+ current-voltage (I–V) curves reveal a dominant decreased Mg2+-inhibition in the receptors containing a single copy of L812M. (e) Mg2+ concentration-response curves for di-heteromeric receptors are shown. (f) Composite concentration-response curves for proton (left) and zinc (right) inhibition show a decreased inhibition for L812M in di-heteromeric receptors.
Table 1.
Summary of pharmacological data for L812M
| Diheteromeric Receptors | Triheteromeric Receptors | ||||
|---|---|---|---|---|---|
|
| |||||
| WT 2A | 2A-L812M | 2A/2A | L812M/2A | L812M/L812M | |
| Glu EC50, μM (n) | 3.5 ± 0.21 (15) | 0.41 ± 0.04 (24)* | 4.0 ± 0.20 (10) | 1.0 ± 0.07 (10)# | 0.39 ± 0.04 (11)# |
| Gly, EC50, μM (n) | 1.1 ± 0.05 (24) | 0.14 ± 0.01 (39)* | 0.72 ± 0.04 (10) | 0.20 ± 0.01 (8)# | 0.06 ± 0.007 (8)# |
| Mg2+ IV& | 33 ± 1.3% (7) | 60 ± 6.8% (8)* | 35 ± 6.8% (8) | 55 ± 5.0% (13)# | 57 ± 2.8% (8)# |
| Mg2+, IC50, μM (n)ξ | 17 ± 1.2 (19) | 43 ± 2.9 (21)* | 15 ± 2.6 (7) | 38 ± 2.8 (8)# | 41 ± 2.8 (8)# |
| Proton, IC50, pH (n) | 6.9 (6–10) | 6.4 (6–11) | 6.9 (8–18) | 6.5 (6–9) | 6.4 (6–7) |
| Zinc, IC50, nM (n) | 77 ± 7.8 (7) | NE | 91 ± 3.9 (9) | NE | NE |
| %inhibition by Zinc$ | 49 ± 3.2% (7) | 6.1 ± 1.7% (6)* | 34 ± 1.8% (14) | 7.8 ± 0.9% (10)# | 4.8 ± 1.1% (6)# |
The data were expressed as Mean ± SEM (n).
Percentage current remaining in the presence of 0.3 mM MgCl2 at −30 mV
at −60 mV
at 300 nM of zinc
NE: no effect
p < 0.05 compared to WT 2A, unpaired t-test
p < 0.05 compared to 2A/2A; one way ANOVA, Tukey post hoc
Since the L812M mutation of this patient is heterozygous and the tetrameric NMDAR complex contains two GluN2 subunits, we anticipate that some receptors in the patient will contain only a single copy of the mutation. We therefore employed a strategy to control GluN2 trafficking and thus the subunit composition of cell surface NMDARs. We used GluN2A with heterodimeric coiled-coiled domains appended to their C-termini along with a short dilysine ER retention/retrieval signal comprised of KKTN (see Methods). We made a similar pair of constructs harboring the GluN2A-L812M mutation, which allowed us to express surface receptors with 0, 1, or 2 copies of the mutant GluN2A. We subsequently explored the pharmacology and biophysical properties of these tri-heteromeric receptors. The receptors that contained a single copy of the mutant subunit GluN2A-L812M (referred to as L812M/2A) showed an intermediate increase in glutamate potency (4-fold shift in EC50, to 1.0 μM from 4.0 μM for 2A/2A; Fig. 2b, left; Table 1) and glycine potency (3.6-fold decrease in EC50 to 0.20 μM from 0.72 μM for 2A/2A; Fig. 2b, right; Table 1). Thus, NMDAR activation was enhanced at lower extracellular concentrations of agonist even for receptors with a single copy of the mutation.
GluN2A L812M attenuates voltage-dependent Mg2+ block
Voltage-dependent channel block of the NMDAR by extracellular Mg2+ is an important feature of NMDARs, rendering current flow voltage-dependent29, 30. Two experiments were performed to test if the GluN2A-L812M alters Mg2+ inhibition. Evaluation of the current-voltage relationship (Fig. 2c; Table 1) shows that nearly twice as much current (60% vs. 33% of WT) will flow in the mutant receptors at negative holding potentials (−30 mV). The concentration-response curves for Mg2+ block (Fig. 2e; Table 1) show that this mutation increased the Mg2+ IC50 value to 43 μM from 17 μM for WT receptors at the holding potential of −60 mV; similar decreases in potency were observed at other holding potentials (WT vs. L812M IC50 was 455 vs. 820 μM at −20 mV, 74 vs. 174 μM at −40 mV, and 4.7 vs. 11 μM at −80 mV). In contrast to the results for agonist potency, a single copy of L812M (L812M/2A) produced a dominant reduction in the Mg2+ inhibition. The current-voltage relationship (Fig. 2d; Table 1) shows that more current flows in the single copy of mutant receptors at the negative holding potentials (e.g. 55% vs. 35% of 2A/2A at −30 mV). A single copy of GluN2A-L812M (Table 1) increased Mg2+ IC50 to 38 μM from 15 μM for 2A/2A at the holding potential of −60 mV.
Because Mg2+ block occurs deep within the ion channel pore and involves the reentrant pore loops that also control Ca2+ permeability3, 31, 32, 33, we tested whether the L812M mutation alters the calcium permeability. Measurement of the reversal potentials for wild type hGluN2A, L812M and wild type hGluN2D receptors (as a positive control) in bi-ionic conditions (Ca2+ or Cs+) allows the determination of relative Ca2+ permeability. The data in Supplementary Fig. 1 show that L812M has no detectable effect on the relative Ca2+ permeability.
GluN2A L812M diminishes negative allosteric regulation
Endogenous proton and zinc can inhibit NMDAR function in the central nervous system3, and these endogenous ions likely serve as important regulatory mechanisms that limit NMDAR activation. We therefore tested whether the L812M mutation can influence regulation by extracellular zinc or protons. GluN2A-L812M reduced the proton sensitivity, with mutant receptors showing an IC50 corresponding to pH 6.4, compared with pH 6.9 for WT (Fig. 2f, left; Table 1). The mutation also virtually eliminated high-affinity zinc inhibition for GluN2A-L812M, compared to WT receptors that were inhibited with an IC50 of 77 nM (Fig. 2f, right; Table 1). Interestingly, a single copy of L812M (L812M/2A) produced a dominant reduction in proton sensitivity with IC50 of pH 6.5, compared with the pH 6.9 of 2A/2A (Table 1). The single copy of the mutation also produced a dominant decrease in the sensitivity of the receptor to 300 nM extracellular zinc (2A/2A: 34%; L812M/2A: 7.8%; L812M/L812M: 4.8%; Table 1).
GluN2A L812M prolongs deactivation time course
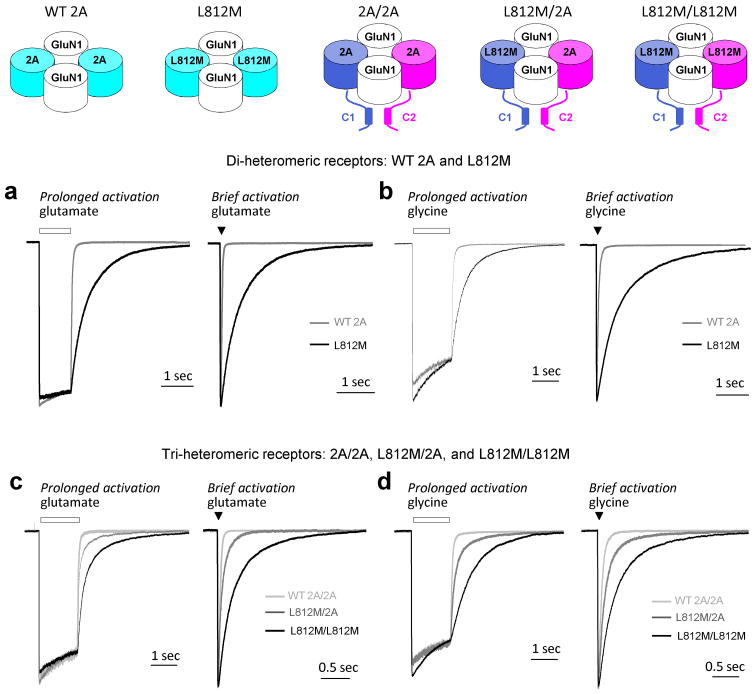
We next assessed the effect of L812M mutation on the response time course of NMDARs. The deactivation rate following removal from glutamate of NMDARs is thought to control the time course of the NMDAR component of the excitatory postsynaptic current (EPSC)34. To evaluate whether the mutation altered the deactivation time course, the current response following glutamate removal was measured using a rapid solution exchange system in whole-cell patch-clamp recordings of WT GluN2A or mutant L812M co-expressed with GluN1 in HEK 293 cells. The deactivation time course of WT 2A following rapid removal of glutamate was fitted by two exponential components with a weighted tau (τW) of 47 ms (n = 11; Fig. 3a; Table 2). The mutant L812M receptors significantly prolonged the glutamate deactivation time course, which was fitted by two exponential components with a τW of 443 ms (n=10; Fig. 3a, left; Table 2). We also briefly moved the cell into the agonist solution for 3–5 milliseconds (brief application) to mimic synaptic events in the brain. As with the long pulse of agonist, the mutant receptors deactivated with a slower time course compared to WT NMDARs (τW 425 ms vs. 44 ms of WT; Fig. 3a, right). The deactivation time course following glycine removal was measured, and could be fitted by two exponential components with a τW of 63 ms (n = 5; Fig. 3b; Table 2). The mutant GluN2A-L812M receptors showed a slower glycine deactivation time course, which was fitted by two exponential components with a τW of 691 ms (n=5; Fig. 3b; Table 2).
Figure 3. The L812M mutation prolongs deactivation time course.
(a) and (b) The L812M mutation prolongs deactivation time course of di-heteromeric GluN2A receptors. (a) The normalized representative current response of di-heteromeric NMDARs (WT 2A and L812M) to 1 mM glutamate (Left: long application (1 sec) is shown; Right: brief application (5 msec); 50 μM glycine in all solutions) (VHOLD −60 mV). L812M (black) prolongs deactivation time course compared with the WT 2A receptors (gray). (b) The normalized representative current response of di-heteromeric NMDARs to 1 mM glycine is shown (Left: long application; Right: brief application; 30 μM glutamate in all solutions) (VHOLD −60 mV). (c, d) The L812M mutation prolongs deactivation time course of tri-heteromeric GluN2A receptors. (c) A representative current response is shown for tri-heteromeric NMDARs with 0, 1, or 2 copies of the L812M mutation in each complex (2A/2A, L812M/2A, and L812M/L812M) to 1 mM glutamate (Left: long application; Right: brief application; 50 μM glycine in all solutions). (d) The normalized representative current response is shown for tri-heteromeric NMDA receptors to 1 mM glycine (Left: long application; Right: brief application; 30 μM glutamate in all solutions) (VHOLD −60 mV).
Table 2.
Summary of deactivation time course
| Diheteromeric Receptors | Triheteromeric Receptors | |||||
|---|---|---|---|---|---|---|
|
| ||||||
| WT 2A | 2A-L812M | 2A/2A | L812M/2A | L812M/L812M | ||
| Glutamate Deactivation | Amplitude (peak, pA/pF) | 121 ± 19 | 123 ± 19 | 98 ± 26 | 90 ± 22 | 72 ± 17 |
| Amplitude (SS, pA/pF) | 96 ± 17 | 99 ± 13 | 67 ± 16 | 50 ± 9.7 | 67 ± 16 | |
| ISS/IPEAK | 0.83 ± 0.05 | 0.86 ± 0.05 | 0.71 ± 0.04 | 0.67 ± 0.06 | 0.92 ± 0.02 | |
| Rise time (ms) | 6.3 ± 0.5 | 7.1 ± 0.4 | 6.9 ± 0.6 | 6.8 ± 0.3 | 6.6 ± 1.1 | |
| τFAST (ms) | 37 ± 4.1 | 264 ± 28 | 30 ± 2.2 | 56 ± 3.9 | 201 ± 18 | |
| τSLOW (ms) | 488 ± 177 | 1362 ± 152 | 179 ± 27 | 354 ± 27 | 1201 ± 143 | |
| %τFAST | 87% | 82% | 87% | 74% | 74% | |
| τW(ms) | 47 ± 4.6 | 443 ± 47 * | 40 ± 2.9 | 112 ± 8.9 # | 438 ± 32 # | |
| Charge transfer, pA×ms/pF | 4,565 | 44,008 | 2,687 | 5,589 | 29,378 | |
| n | 11 | 10 | 10 | 12 | 6 | |
|
| ||||||
| Glycine Deactivation | τFAST (ms) | 44 ± 6.5 | 431 ± 45 | 56 ± 4.2 | 138 ± 7.9 | 413 ± 109 |
| τSLOW (ms) | 449 ± 78 | 1237 ± 130 | 428 ± 91 | 698 ± 96 | 1467 ± 103 | |
| %τFAST | 95% | 68% | 90% | 68% | 67% | |
| τW(ms) | 63 ± 9.6 | 691 ± 66 * | 83 ± 10 | 315 ± 41 # | 784 ± 112 # | |
| Charge transfer, pA×ms/pF | 3,906 | 60,775 | 1,851 | 6,938 | 17,725 | |
| n | 5 | 5 | 6 | 7 | 5 | |
The data were expressed as Mean ± SEM.
p < 0.05 compared to WT 2A, unpaired t-test.
p < 0.05 compared to 2A/2A; one way ANOVA, Tukey post hoc.
The data of glutamate and glycine deactivation time course tau were from prolonged application (1–1.5 sec). There were no significant effects of the mutation on the response rise time (9.5 ms for WT 2A; 9.3 ms for 2A-L812M; 8.0 ms for 2A/2A; 10 ms for L812M/2A; 11 ms L812M/L812M) and amplitude (62 pA for WT 2A; 88 pA for 2A-L812M; 22 pA for 2A/2A; 22 pA for L812M/2A; 23 pA for L812M/L812M) when glycine was applied with background glutamate.
We additionally assessed the time course of deactivation following removal of glutamate or glycine for receptors containing one copy of GluN2A-L812M. As expected, the glutamate and glycine deactivation time constants of receptors with two copies of mutant GluN2A (L812M/L812M) and receptors with no mutation (2A/2A) were comparable to those determined for GluN2A-L812M and WT GluN2A, respectively (Fig. 3c; Table 2). However, the receptors containing a single copy of mutant GluN2A (L812M/2A) showed an intermediate dual exponential glutamate deactivation time course with a τW of 112 ms (n = 12; Fig. 3c, left; Table 2). The NMDAR current response upon briefly moving the cell into the glutamate solution to mimic the synaptic activation showed a similar tendency of the GluN2A-L812M mutation to slow the deactivation time course (Fig. 3c, right). Furthermore, the receptors containing a single copy of mutant GluN2A (L812M/2A) showed an intermediate slowing of the glycine deactivation time course that could also be described by two exponential components with τW of 315 ms (n = 7; Fig. 3d, left; Table 2), compared with τW 83 ms for 2A/2A and 784 ms for L812M/L812M. The data from experiments moving the cell briefly into the glycine solution show a similar intermediate effect of the GluN2A-L812M mutation on slowing the glycine deactivation time course (Fig. 3d, right). These data show that even a single copy of the mutant subunit prolong the deactivation time course for both agonists, and thus the time course of the NMDAR component of the EPSC.
GluN2A L812M alters single channel properties
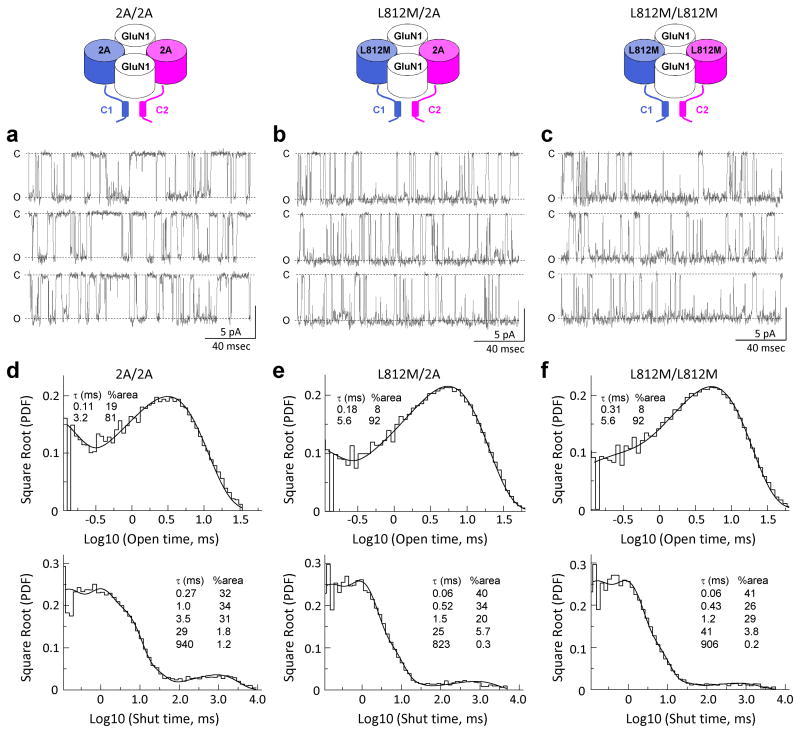
We expressed in HEK cells GluN1/GluN2A receptors that contained 0, 1, or 2 copies of the L812M mutation, and excised outside-out patches from these cells. Steady-state single channel unitary currents were recorded from 2A/2A, L812M/2A, and L812M/L812M co-expressed with GluN1 in response to maximally effective concentrations of glutamate and glycine (1000 μM and 30 μM). Analysis of open time and overall open probability from patches that contained a single active receptor suggested that one copy of the mutant GluN2A subunit within only a single receptor has a dominant effect on open time, shut time, and open probability.
Figure 4a shows 2A/2A single channel currents that have a complex bursting behavior with one predominant conductance state (84 pS), and an open probability of 0.21. Open duration and shut duration histograms were fitted with two and five exponential components, respectively. A parallel set of recordings for GluN1 co-expressed with L812M/2A (Fig. 4b) and L812M/L812M (Fig. 4c) showed identical single channel conductance (L812M/2A 85 pS; L812M/L812M 85 pS), but significantly more channel openings with prolonged open periods and shortened shut periods, resulting in an increased open probability. Figure 4d-f and Table 3 summarize the pooled data describing single channel properties from patches that contained one active channel with GluN2 subunit composition of 2A/2A, L812M/2A and L812M/L812M. MTSEA modification of a mutation in GluN1 SYTANLAAF gating region, can lock receptors open21, 23, rendering the degree of potentiation reciprocally related to the open probability prior to MTSEA treatment. Use of this approach to estimate open probability yielded the same result as data from one active channel in excised outside-out patches (Supplementary Fig. 2), confirming a dominant effect on channel gating by a single copy of the mutant receptor. These data indicate that the mutant channels open more often resulting in a profound overactivation of the NMDARs and enhanced neuronal excitability. This situation almost certainly renders neurons vulnerable to excitotoxic neuronal death35, 36.
Figure 4. The L812M mutation alters the single channel properties.
Steady-state recordings from an outside-out patch containing a single active NMDAR with two wild type leucine residues at GluN2A position 812 (2A/2A; a), a single copy of mutation L812M (L812M/2A; b), and two copies of L812M (L812M/L812M; c). Unitary currents were activated in these excised patches by 1 mM glutamate and 50 μM glycine, data are shown on two different time scales. (d, e, f) The pooled open duration and shut duration histogram are shown for 4 single channel recordings. Open duration histograms were made from 42302 openings for 2A/2A, 80303 openings for L812M/2A, 74138 openings for L812M/L812M. Closed duration histograms were made from 42299 events for 2A/2A, 80301 events for L812M/2A, 74134 events for L812M/L812M.
Table 3.
Summary of single channel properties
| 2A/2A | L812M/2A | L812M/L812M | |
|---|---|---|---|
| Open probability | 0.23 ± 0.04 | 0.58 ± 0.03 # | 0.64 ± 0.03 # |
| Mean open time, ms | 2.6 ± 0.17 | 5.1 ± 0.33 # | 5.2 ± 0.34 # |
| τ1, ms (%) | 0.11 ± 0.02 (19%) | 0.18 ± 0.07# (8%#) | 0.31 ± 0.11# (8%#) |
| τ2, ms (%) | 3.2 ± 0.16 (81%) | 5.6 ± 0.34# (92%#) | 5.6 ± 0.34# (92%#) |
| Mean shut time, ms | 13 ± 4.2 | 3.6 ± 1.2 # | 2.0 ± 0.04 # |
| τ1, ms (%) | 0.27 ± 0.01 (32%) | 0.06 & (40%) | 0.06 & (41%) |
| τ2, ms (%) | 1.0 ± 0.11 (34%) | 0.52 ± 0.07# (34%) | 0.43 ± 0.08# (26%) |
| τ3, ms (%) | 3.5 ± 0.38 (31%) | 1.5 ± 0.16# (20%) | 1.2 ± 0.27# (29%) |
| τ4, ms (%)ξ | 29 ± 7.9 (1.8%) | 25 ± 21 (5.7%) | 41 ± 31 (3.8%) |
| τ5, ms (%)ξ | 940 ± 193 (1.2%) | 823 ± 210 (0.3%) | 906 ± 66 (0.2%) |
| Major amplitude (pA) | 6.7 ± 0.1 | 6.9 ± 0.1 | 6.9 ± 0.1 |
| γ, pS | 84 ± 1.6 | 85 ± 1.7 | 85 ± 1.2 |
| n | 4 | 4 | 4 |
The data were expressed as Mean ± SEM.
p < 0.05 compared to 2A/2A; one way ANOVA, Tukey post hoc
The briefest shut time component was not well described due to the imposed resolution
Statistical tests were not run on the two slowest shut time components given the limited number of events underlying these components
Molecular basis for enhanced NMDA receptor function
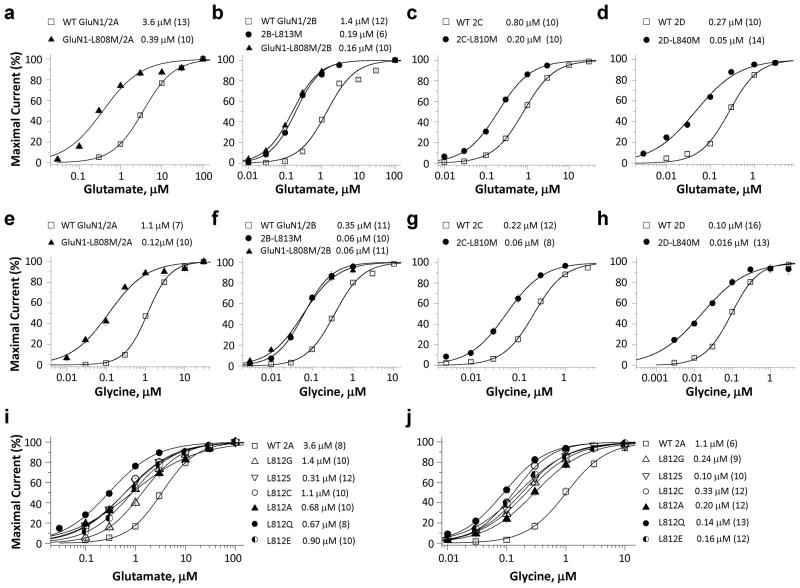
The residue corresponding to GluN2A-Leu812 is conserved for the GluN1 and GluN2 NMDAR subunits. We hypothesized that this region may play an important role in channel gating37, 38, 39. To test this idea, we first determined whether changes in agonist potency were associated with the L812M mutation transferred into other GluN subunits at the same position. We substituted Met for Leu in other GluN subunits at the corresponding position, leading to the generation of GluN1-L808M, GluN2B-L813M, GluN2C-L810M, GluN2D-L840M. The potency (EC50 values) of glutamate and glycine were evaluated for recombinant NMDARs expressed in Xenopus oocytes. The GluN1-L808M mutation coexpressed with GluN2A increased glutamate potency by 9.2-fold (EC50: 0.39 μM vs. 3.6 μM for WT 2A) and glycine potency by 9.1-fold (EC50: 0.12 μM vs. 1.1 μM for WT) (Fig. 5a, e; Table 4). GluN1-L808M when coexpressed with GluN2B similarly increased glutamate potency by 6.8-fold (Fig. 5b; 0.19 μM vs. 1.3 μM for WT 2B). Likewise, all GluN2 subunits harboring the L-M mutation showed strongly enhanced potency of both glutamate and glycine (Fig. 5b–d, f–h; Table 4). These data suggested that the function of the L812 residue is conserved across all GluN1 and GluN2 NMDAR subunits.
Figure 5. The L812M mutation changes agonist potency of other NMDAR subunits.
Composite concentration-response curves were determined by TEVC recordings from Xenopus oocytes expressing different GluN subunits-containing receptors (a and e, GluN1/GluN2A and GluN1-L808M/GluN2A; b and f, GluN1/GluN2B, GluN1/GluN2B-L813M, and GluN1-L808M/GluN2B; c and g, GluN1/GluN2C and GluN1/GluN2C-L810M; d and h, GluN1/GluN2D and GluN1/GluN2D-L840M). (a, b, c, d) show the composite glutamate concentration-response curves determined in the presence of 100 μM glycine. (e, f, g, h) show the composite glycine concentration-response curves determined in the presence of 100 μM glutamate. (i, j) show the composite glutamate (i, in the presence of 100 μM glycine) and glycine (j, in the presence of 100 μM glutamate) concentration-response curves of GluN2A-L812G, -L812S, -L812C, -L812A, -L812Q, -L812E constructs.
Table 4.
Summary of agonist EC50 values for Leu-Met mutations at the position corresponding to GluN2A Leu812 in all other GluN1 and GluN2 subunits.
| Glutamate EC50, μM | Glycine EC50, μM | |
|---|---|---|
| WT 2A | 3.6 ± 0.26 (13) | 1.1 ± 0.04 (7) |
| L808M/2A | 0.39 ± 0.04 (10)* | 0.12 ± 0.01 (10)* |
| WT 2B | 1.3 ± 0.10 (12) | 0.35 ± 0.01 (11) |
| 2B-L813M | 0.19 ± 0.01 (6)# | 0.07 ± 0.007 (10)# |
| L808M/2B | 0.16 ± 0.01 (10)# | 0.06 ± 0.005 (11)# |
| WT 2C | 0.82 ± 0.06 (10) | 0.23 ± 0.2 (12) |
| 2C-L808M | 0.20 ± 0.01 (10)* | 0.06 ± 0.01 (8)* |
| WT 2D | 0.27 ± 0.01 (10) | 0.10 ± 0.03 (16) |
| 2D-L840M | 0.05 ± 0.006 (14)* | 0.016 ± 0.002 (13)* |
The data were expressed as Mean ± SEM (n).
p < 0.05 compared to corresponding WT; unpaired t-test
p<0.05 compared to WT 2B; one way ANOVA, Tukey post hoc
We also introduced a series of substitutions (L-G, L-A, L-C, L-S, L-Q, L-E) at this position in GluN2A and assessed the agonist potency. We found increased co-agonist potency in all six mutant receptors (Fig. 5i, j; Table 5), indicating that the leucine residue at position 812 is critical for NMDAR function. The fact that all perturbations of the side chain tested at this position enhanced NMDAR function may point towards a selective pressure to maintain Leu at this position to limit NMDAR overactivation.
Table 5.
Summary of agonist EC50 values for multiple substitutions at GluN2A Leu812
| Glutamate EC50, μM | Glycine EC50, μM | |
|---|---|---|
| WT 2A | 3.6 ± 0.4 (8) | 1.1 ± 0.2 (6) |
| 2A-L812G | 1.4 ± 0.03 (10) | 0.24 ± 0.03 (9) |
| 2A-L812S | 0.31 ± 0.04 (12) | 0.10 ± 0.01 (10) |
| 2A-L812C | 1.1 ± 0.25 (10) | 0.33 ± 0.04 (12) |
| 2A-L812A | 0.68 ± 0.10 (10) | 0.20 ± 0.02 (12) |
| 2A-L812Q | 0.67 ± 0.09 (8) | 0.14 ± 0.01 (13) |
| 2A-L812E | 0.90 ± 0.07 (10) | 0.16 ± 0.01 (12) |
The data were expressed as Mean ± SEM (n).
DISCUSSION
Idiopathic epilepsies are considered to be associated with genetic alterations or genetic susceptibility40. Several types of epilepsy are due to mutations in single genes, especially those encoding ion channels40, 41. Numerous genes have been associated with early-infantile epileptic encephalopathy42, 43. Recently, mutations of GRIN2A have been identified in several cases of early-onset epileptic encephalopathy12, 14, 15, 16, 44. In some of these cases (as in the case we report), the children did not attain basic milestones. Based on the profound increase in charge transfer predicted for EPSCs from our data as well as the increase in activation by low levels of non-synaptic agonist, we hypothesize that these mutations cause NMDAR hyperactivity and consequently intractable seizures, abnormal development, and possible excitotoxic neuronal loss.
For this patient, it seems likely that the early-onset epileptic encephalopathy could be due to the reduced block by extracellular Mg2+, over a 10-fold predicted increase in charge transfer occurring during synaptic transmission, as well as the strong increase in open probability. The greatly increased current flow through GluN2A-containing NMDARs into principle neurons should provide the excessive excitatory drive, facilitating or triggering the development of clinically relevant epileptiform activity, and perhaps seizures. Compounding this excessive excitatory drive will be activation of NMDARs by low levels of glutamate and/or glycine that would normally be insufficient to induce NMDAR-mediated current flow. Our results with NMDARs that contain a single copy of the mutant receptor show that overactivation does not require two copies of the mutant subunit. This enhanced excitatory drive from all receptors with at least one copy of the mutation could lead to excitotoxic neuronal death, a process long known to occur following overactivation of NMDARs35, 36. This neuronal loss may result in cognitive impairment, motor dysfunction, and other neurological problems.
Our functional data show that the GluN2A-L812M linker region mutation influences several key features of NMDAR function37, 38, 39. We interpret all of these effects to be consistent with enhanced gating that could reflect a decrease in the activation energy for pore opening and an increase in stability of the open pore. The pre-M4 linker resides on the immediate extracellular end of M4, placing it between the agonist binding domain and the transmembrane pore-forming elements. It is therefore a key region for translating the conformational changes induced by agonist binding to opening of the ion channel pore. Previous work with pre-M4 mutations, cross-linking, and covalent modification all have suggested a role for the M4 region in gating37, 38, 39, 45 and in allosteric modulation by the physiologically-relevant concentrations of protons46. This position has also shown anomalous behavior in previous studies of covalent modification of Cys residues37, 39.
Assuming the NMDARs share similar pore structure as AMPA receptors, examination of the position of Leu812 in GluN2 suggests that it resides within 5 Angstroms of the SYNTANLAAF gating motif in GluN1 M3. The M3 helices are thought to form the helical bundle crossing that occludes ion flow through the pore, and the transmembrane domain M3 is hypothesized to couple agonist binding and channel opening as a transduction element17, 21. M3 residues are also hypothesized to affect cleft closure of the ligand binding domain, and modification of the M3 conformation by MTSEA (for example the lurcher site47) influences LBD cleft closure and induces higher agonist potency, slower deactivation time course, and decreased sensitivity to competitive antagonists and the allosteric modulators (e.g. proton inhibition)20, 21, 23, 48, 49.
Lastly, Leu812 is within 5 Angstroms of the GluN1 pre-M1 helix; both the GluN1 and GluN2 pre-M1 helices lie parallel to the extracellular membrane and are in van der Waals contact with the M3 gate. The pre-M1 region is also a locus for allosteric regulation by exogenous molecules17, 28, 50. Thus, Leu812 lies at an ideal position to interact with two key elements (the conserved SYTANLAAF gating motif and the pre-M1 helix) hypothesized from structural and functional work to be involved in channel gating.
In summary, we present in-depth functional analyses for a unique case of a mutation identified from a patient with intractable seizures, early-onset epileptic encephalopathy associated with progressive cerebral parenchymal volume loss. We report a novel de novo missense mutation in the GRIN2A gene, which dramatically increases the receptor response to agonists, decreases the actions of endogenous negative modulators, increases channel open probability and prolongs deactivation time course and by analogy synaptic duration. The result of this gain-of-function mutation was an increase of over 10-fold in the current mediated by NMDARs, which we predict will contribute to the refractory seizures, cognitive impairment, and cortical cell loss in the grey matter. Importantly, this mutation has also allowed the identification of a conserved gating element (e.g. Leu812 in GluN2A) that plays a role in the activation of all NMDARs. The proximity of this mutation to a site for allosteric regulation suggests a possible avenue for therapeutic exploration51. Moreover, the results suggest that future evaluation of children with refractory seizures and early-onset epileptic encephalopathies might include analysis of the glutamate receptors subunit genes.
METHODS
Whole exome sequencing
DNA was obtained and sequenced based upon clinical protocol 76-HG-0238, “Diagnosis and Treatment of Patients with Inborn Errors of Metabolism and other Genetic Disorders”, approved by the NHGRI Institutional Review Board. The parents gave written, informed consent for medical and genetic studies designed to reach diagnoses in the affected children.
Exome sequencing was performed using genomic DNA extracted from peripheral blood. In-solution exome capture was performed according to the manufacturer’s protocol using SureSelect Human All Exon Kits (Agilent Technologies, Santa Clara, CA) or the TruSeq Exome Enrichment Kit (Illumina, San Diego, CA). Alignment to the human genome reference sequence (UCSC assembly hg18, NCBI build 36) was carried out using the Efficient Local Alignment of Nucleotide Data algorithm (Eland, Illumina, Inc., San Diego, CA). Eland was used in such a way that PE reads were aligned independently, and those that aligned uniquely were grouped into genomic sequence intervals of about 100 kb. Reads that failed to align were binned with their PE mates without Eland using the PE information. Reads that mapped equally well in more than one location were discarded. Crossmatch, a Smith-Waterman based local alignment algorithm, was used to align binned reads to their respective 100 kb genomic sequence, using the parameters –minscore 21 and –masklevel 0 (http://www.phrap.org)52. Genotypes were called using a Bayesian genotype-caller, Most Probable Genotype (MPG)53.
To facilitate identification of de novo variants we restricted analysis of exome sequence data to regions of the genome that were routinely well covered and consistently genotyped in 147 exomes from the UDP cohort and 561 exomes from the Clinseq cohort (unaffected individuals). De novo mutations in these regions were identified using a Boolean exclusion filter that selected for variants not present in any other UDP exome and that were at positions for which more than 50% of the ClinSeq exomes had a successful genotype call. The de novo variants identified were then further filtered to exclude variants present in sequence data derived from unaffected individuals (i.e., the ClinSeq cohort or the 1K genome database of April 2011), in known pseudogenes, or in known artifacts in exome sequencing54. Among the variants identified by this process was the c.2434C>A (p.L812M) mutation in exon13 of GRIN2A.
Mutagenesis
Site-directed mutagenesis was performed using the QuikChange protocol with Pfu DNA polymerase (Stratagene, La Jolla, CA) to replicate the parental strand with desired mismatch incorporated into the primer23. Methylated parental DNA template was digested with DpnI. The nicked double-stranded mutant DNA was transformed into TOP10 Competent Cells (Life Tech, Grand Island, NY). The mutations were verified by sequencing through the region of the mutations. The cDNA for wild type human NMDA subunit GluN1-1a (hereafter GluN1), GluN2A, GluN2B, and GluN2D were in pCI-neo55 (GenBank accession codes: NP_015566, NP_000824, NP_000825, and NP_000827.1). The cDNA for wild type rat GluN2C (GenBank accession codes: M91563) was provided by Dr. P. Seeburg (University of Heidelberg). The constructs for tri-heteromeric receptors (GluN1/GluN2A/GluN2A, GluN1/GluN2A-L812M/GluN2A, and GluN1/GluN2A-L812M/GluN2A-L812M) were prepared by fusing engineered peptide tags, termed C1 and C2, to the C-terminus of the GluN2A subunit. The two different C1 and C2 tags were composed of a short polypeptide linker containing four repeats of amino acids EAAAK followed by the leucine zipper motifs from GABAB1 or GABAB2, respectively56, and immediately followed by C-terminal di-lysine KKTN endoplasmic reticulum retention/retrieval motifs57, 58, 59. C1 and C2 were fused to the C-terminus of the wild type GluN2A subunit and the mutant GluN2A-L812M to form the following subunits: GluN2A-C1, GluN2A-C2, GluN2A-C1-L812M, GluN2A-C2-L812M. These subunits were then co-expressed with GluN1 to form the following receptors: GluN1/GluN2A-C1/GluN2A-C2 (abbreviated 2A/2A), GluN1/GluN2A-C1-L812M/GluN2A-C2 (abbreviated L812M/2A), and GluN1/GluN2A-C1-L812M/GluN2A-C2-L812M (abbreviated L812M/L812M). The C1 and C2 tags can form a parallel heterodimeric coiled-coil structure, and the KKTN motifs at the distal C-termini will localize the subunits in the endoplasmic reticulum unless masked by coiled-coil formation between C1 and C256, 60, 61. Consequently, the C1 and C2 tags should allow selective surface expression of NMDA receptors composed of two GluN1 subunits, one C1-tagged GluN2A subunit, and one C2-tagged GluN2A subunit as evidenced by the control experiments evaluating the escape of non-triheteromeric receptors from the ER retention shown in Supplementary Fig. 3. This evaluation estimated that <4% of the current responses in the tri-heteromeric experiments were mediated by non-triheteromeric receptors.
Two-electrode voltage clamp recordings
cRNA for WT and mutants was synthesized from linearized template of cDNA according to manufacturer’s specification (Ambion, Austin, TX) and injection of cRNA, as well as two-electrode voltage-clamp recordings from Xenopus laevis oocytes, were performed62. Oocytes were obtained from Ecocyte (Austin, Texas) and were injected with 5–10 ng cRNAs synthesized in vitro from linearized template cDNA. Following injection, the oocytes were stored at 15–19°C in Barth’s solution containing (in mM) 88 NaCl, 2.4 NaHCO3, 1 KCl, 0.33 Ca(NO3)2, 0.41 CaCl2, 0.82 MgSO4, 5 Tris/HCl (pH 7.4 with NaOH). The ratio of injected di-heteromeric GluN1 to GluN2 cRNA was 1:2. The ratio of injected cRNA for experiments involving tri-heteromeric receptors was 1:6:6 for GluN1: GluN2A-C1: GluN2A-C2, and the cRNA was diluted with RNase-free water to a concentration that would result in ~10 ng total injected cRNA. Voltage clamp recordings were performed 2–4 days post-injection at room temperature (23°C). The recording solution contained (in mM) 90 NaCl, 1 KCl, 10 HEPES, 0.5 BaCl2, 0.01 EDTA (pH 7.4 with NaOH). Solution exchange was computer controlled through an 8-modular valve positioner (Digital MVP Valve, Hamilton, CT). Voltage and current electrodes were filled with 0.3 and 3.0 M KCl, respectively, and current responses were recorded at a holding potential of −40 mV unless otherwise stated. Data acquisition and voltage control were accomplished with a two-electrode voltage-clamp amplifier (OC725, Warner Instrument, Hamden, CT). Zinc experiments were performed in the tricine solution at (10 mM) pH 7.3. ZnCl2 solutions (10 mM) were made fresh every day and added directly to the recording solution to obtain the desired nominal Zn2+ concentration62. 100 μM glutamate and 100 μM glycine were used in all oocyte experiments unless otherwise stated. EC50 values and IC50 values were obtained by fitting the concentration-response curve with Equation 1 and Equation 2, respectively:
| [Equation 1] |
where EC50 is the concentration of agonist that causes a half maximal response, and nH is a slope factor of the response curve.
| [Equation 2] |
where minimum is the residual percent response in saturating concentration (constrained to > 0) of Mg2+, protons or zinc, IC50 is the concentration of antagonist that causes half maximal inhibition, and nH is the slope factor.
Preparation and transfection of HEK293 cells
HEK 293 cells (CRL 1573, ATCC, Manassas, VA) or HEK 293 Tet-On Advanced cells (Clontech Laboratories, Mountain View, CA) were plated onto glass coverslips coated in 100 μg/ml poly-D-lysine and incubated at 37°C (5% CO2 in DMEM/GlutaMax) supplemented with 10% dialyzed fetal bovine serum, and 10 μg/ml streptomycin. G418 sulfate (150 μg/ml; InvivoGen, San Diego, CA) was added into the medium for HEK 293 Tet-On Advanced cells. HEK 293 cells were co-transfected using the calcium phosphate method63 with plasmid cDNAs encoding green fluorescent protein (GFP), GluN1, and GluN2A or GluN2A-L812M (ratio was 5:1:1) for di-heteromeric receptor experiments. HEK 293 Tet-On Advanced cells were used for tri-heteromeric receptor experiments and plasmid cDNAs encoding GFP, GluN1, 2A/2A, or L812M/2A, or L812M/L812M were transfected at a ratio of 1:1:1:1. In di-heteromeric receptor experiments all receptor subunit cDNA were in the pCI-neo, whereas GluN1 expression was under the control of the Tet-On inducible promoter in the pTRE-tight vector (Clontech Laboratories, Mountain View, CA) in tri-heteromeric receptor experiments. The mixed plasmid DNA (0.5 ug) was mixed with 6.25 μl of 1 M CaCl2, 18.75 μl dH2O, 25 μl of 2× BES solution (50 mM BES, N,N-Bis(2-hydroxyethyl)-2-aminoethanesulfonic acid, N,N-Bis(2-hydroxyethyl)taurine; 280 mM NaCl; 1.5 mM Na2HPO4, pH adjusted to 6.95 with HCl) was added, and the mixture was incubated for 5 min at room temperature. Calcium phosphate-DNA solution (50 μl per well) was added dropwise to the plate of cells, and the mixture was swirled gently and incubated for 4 h at 37°C. After transfection, the cells were incubated overnight post-transfection in standard media supplemented with NMDAR antagonists (200 μM D,L-2-amino-5-phosphonovalerate, 100 μM 7-chlorokynurenic acid, and 1000 μM MgCl2) for both HEK 293 cells and HEK293 Tet-On Advanced cells. In addition, doxycycline (1–3 μg/ml) was added to the HEK293 Tet-On Advanced cells to induce expression of GluN1. The following day, approximately 24 hours after transfection, the cells were used for patch-clamp recording (whole-cell voltage-clamp recordings and outside-out single channel recordings).
Whole cell and single channel voltage-clamp recordings
Whole-cell voltage-clamp recordings were performed on transfected HEK 293 cells at -60 mV (23°C) using an Axopatch 200B amplifier (Molecular Devices, Union City, CA) with micropipettes that contained (in mM) 110 D-gluconate, 110 CsOH, 30 CsCl, 5 HEPES, 4 NaCl, 0.5 CaCl2, 2 MgCl2, 5 BAPTA, 2 NaATP, and 0.3 NaGTP (pH 7.35). Recording electrodes (3–5 MΩ) were made from thin wall glass micropipettes (TW150F-4, World Precision Instruments, Sarasota, FL) pulled using a vertical puller (Narishige P-10, Tokyo, Japan). The external solution contained (in mM) 150 NaCl, 10 HEPES, 30 D-mannitol, 3 KCl, 1.0 CaCl2, 0.01 EDTA at 23°C and pH 7.4. This level of EDTA will chelate with exceptionally high affinity nM levels of contaminant transition metals (zinc, iron, manganese, etc.) but not significantly alter the extracellular Ca2+ concentration. Rapid solution exchange with open tip exchange times (<1 ms) for the solutions around the cell was achieved with a two-barreled theta-glass pipette controlled by a piezoelectric translator (Burleigh Instruments, Newton, NJ) and the data were analyzed by ChanneLab (Synaptosoft, Decatur, GA)64. Deactivation time course following removal of glutamate was fitted with Equation 3:
| [Equation 3] |
Single channel recordings of outside-out patches excised from transfected HEK 293 cells were recorded at −80 mV (digitized at 40 kHz, filtered at 8 kHz) using an Axopatch 200B amplifier at room temperature (23°C). Recording electrodes were made from thick-wall glass pipettes (G150F-4, Warner Instruments Inc., Hamden, CT) pulled using a vertical puller (Narishige P-10), coated with Sylgard (Dow Corning, Midland, MI), and then fire-polished to 7–9 MΩ. The internal solution was the same as that used for the whole-cell voltage-clamp recording. The channel was activated by 1 mM glutamate and 50 μM glycine in an external solution composed of (in mM) 150 NaCl, 10 HEPES, 3 KCl, 0.5 CaCl2, and 0.01 EDTA at pH 7.4. For analysis, the recordings were pre-filtered at 8 kHz (−3 dB), digitized at 40 kHz, and idealized using the Viterbi algorithm in QUB (http://www.qub.buffalo.edu)65, 66. Maximum likelihood fitting of open and shut duration histograms was accomplished using ChanneLab (Synaptosoft, Decatur, GA). The single channel open probability POPEN was calculated as the ratio of total open duration divided by the sum of total open and closed duration.
Evaluation of relative Ca2+ permeability
The relative Ca2+ permeability (PCa/PCs) was assessed by measuring reversal potential (Vrev) of WT and mutant NMDARs in bi-ionic conditions (Ca2+ or Cs+). The extracellular Ca2+ solution (1.8 mM) contained 140 mM NMDG, 10 mM HEPES and 1.8 mM Ca2+, adjusted to pH 7.2 by addition of HCl. The extracellular Cs+ solution (143 mM) consisted of 140 mM CsCl and 10 mM HEPES, adjusted to pH 7.2 by addition of CsOH. The intracellular recording solution contained (in mM) 125 CsCl, 10 HEPES, 10 EGTA (pH 7.2 by addition of CsOH). The solution was bath-applied by gravity-fed perfusion system and agonists (100 μM glutamate and 100 μM glycine) were applied by fast-perfusion system same with the experiments of deactivation time course. The whole-cell voltage clamp recordings were performed on HEK 293 cells co-transfected with GluN1, GFP, and either WT GluN2A, GluN2A(L812M), or WT GluN2D as a positive control. Current-voltage curves were generated by applying a ramp protocol over −90 mV to 30 mV (800 msec) after stepping to −90 mV from a holding potential of −60 mV. To minimize voltage changes, a 3 M KCl agarose (0.5%) bridge was used. Liquid junction potential between the extracellular and internal solution were measured (−12.6 mV in 1.8 mM Ca2+ extracellular solution) and used to correct the holding potential. The current-voltage relationship was fitted by the following Equation 4:
| [Equation 4] |
The reversal potential was determined as the voltage at which the fitted curve crossed the abscissa. Relative Ca2+ permeability was calculated with a modified Lewis equation (Equation 5)67,68, 69:
| [Equation 5] |
All reagents were purchased from Sigma. All experiments were performed on two or more separate cell preparations (injected oocytes from different frogs or separate cultures of HEK cells). Data are expressed as mean ± SEM and analyzed statistically using unpaired t test and one-way ANOVA with Tukey post hoc test. Significance for all tests was set at p < 0.05. Error bars in all figures are SEM.
Supplementary Material
Acknowledgments
We are grateful to Anel Tankovic, Phuong Le, and Kevin Ogden for technical assistance. We are also grateful to Eric D. Marsh and Dimitre R. Simeonov for assistance with the clinical and molecular evaluation of the patient.
This work was supported by the NINDS (NS036654, S.F.T.), by the NIH Undiagnosed Disease Program (HSN268201300162P, H.Y.), and by the Intramural Research Program of the National Human Genome Research Institute. TMP was also supported by the Cedars-Sinai Diana and Steve Marienhoff Fashion Industries Guild Endowed Fellowship in Pediatric Neuromuscular Diseases.
Footnotes
Author contributions HY: designed and performed experiments, analyzed data and wrote the paper. KBH: performed experiments and wrote the paper. JZ: performed experiments. TMP: clinical evaluation, patient phenotyping and wrote the paper. TM: supervised genomic data mining. KFF: discovered mutation. CMH: validated mutation. GG: clinical evaluation and patient phenotyping. DRA: clinical evaluation and patient phenotyping. CFB: project supervision and wrote the paper. WAG: managed UDP and wrote the paper. SFT: designed experiments, analyzed data and wrote the paper. All authors discussed the results and implications and commented on the manuscript.
Potential Conflicts of Interest The authors have no competing interests.
References
- 1.Akazawa C, Shigemoto R, Bessho Y, Nakanishi S, Mizuno N. Differential expression of five N-methyl-D-aspartate receptor subunit mRNAs in the cerebellum of developing and adult rats. J Comp Neurol. 1994;347:150–160. doi: 10.1002/cne.903470112. [DOI] [PubMed] [Google Scholar]
- 2.Monyer H, Burnashev N, Laurie DJ, Sakmann B, Seeburg PH. Developmental and regional expression in the rat brain and functional properties of four NMDA receptors. Neuron. 1994;12:529–540. doi: 10.1016/0896-6273(94)90210-0. [DOI] [PubMed] [Google Scholar]
- 3.Traynelis SF, et al. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol Rev. 2010;62:405–496. doi: 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ghasemi M, Schachter SC. The NMDA receptor complex as a therapeutic target in epilepsy: a review. Epilepsy Behav. 2011;22:617–640. doi: 10.1016/j.yebeh.2011.07.024. [DOI] [PubMed] [Google Scholar]
- 5.Coyle JT, Tsai G, Goff D. Converging evidence of NMDA receptor hypofunction in the pathophysiology of schizophrenia. Ann N Y Acad Sci. 2003;1003:318–327. doi: 10.1196/annals.1300.020. [DOI] [PubMed] [Google Scholar]
- 6.Hallett PJ, Standaert DG. Rationale for and use of NMDA receptor antagonists in Parkinson’s disease. Pharmacol Ther. 2004;102:155–174. doi: 10.1016/j.pharmthera.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 7.Arning L, Kraus PH, Valentin S, Saft C, Andrich J, Epplen JT. NR2A and NR2B receptor gene variations modify age at onset in Huntington disease. Neurogenetics. 2005;6:25–28. doi: 10.1007/s10048-004-0198-8. [DOI] [PubMed] [Google Scholar]
- 8.Fan MM, Raymond LA. N-methyl-D-aspartate (NMDA) receptor function and excitotoxicity in Huntington’s disease. Prog Neurobiol. 2007;81:272–293. doi: 10.1016/j.pneurobio.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 9.Saft C, et al. NMDA receptor gene variations as modifiers in Huntington disease: a replication study. PLoS Curr. 2011;3:RRN1247. doi: 10.1371/currents.RRN1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamza TH, et al. Genome-wide gene-environment study identifies glutamate receptor gene GRIN2A as a Parkinson’s disease modifier gene via interaction with coffee. PLoS Genet. 2011;7:e1002237. doi: 10.1371/journal.pgen.1002237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tarabeux J, et al. Rare mutations in N-methyl-D-aspartate glutamate receptors in autism spectrum disorders and schizophrenia. Transl Psychiatry. 2011;1:e55. doi: 10.1038/tp.2011.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Endele S, et al. Mutations in GRIN2A and GRIN2B encoding regulatory subunits of NMDA receptors cause variable neurodevelopmental phenotypes. Nat Genet. 2010;42:1021–1026. doi: 10.1038/ng.677. [DOI] [PubMed] [Google Scholar]
- 13.Hamdan FF, et al. Excess of de novo deleterious mutations in genes associated with glutamatergic systems in nonsyndromic intellectual disability. Am J Hum Genet. 2011;88:306–316. doi: 10.1016/j.ajhg.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Ligt J, et al. Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med. 2012;367:1921–1929. doi: 10.1056/NEJMoa1206524. [DOI] [PubMed] [Google Scholar]
- 15.Lemke JR, et al. Mutations in GRIN2A cause idiopathic focal epilepsy with rolandic spikes. Nat Genet. 2013;45:1067–1072. doi: 10.1038/ng.2728. [DOI] [PubMed] [Google Scholar]
- 16.Lesca G, et al. GRIN2A mutations in acquired epileptic aphasia and related childhood focal epilepsies and encephalopathies with speech and language dysfunction. Nat Genet. 2013;45:1061–1066. doi: 10.1038/ng.2726. [DOI] [PubMed] [Google Scholar]
- 17.Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009;462:745–756. doi: 10.1038/nature08624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heron SE, et al. De novo SCN1A mutations in Dravet syndrome and related epileptic encephalopathies are largely of paternal origin. J Med Genet. 2010;47:137–141. doi: 10.1136/jmg.2008.065912. [DOI] [PubMed] [Google Scholar]
- 19.Mastrangelo M, Leuzzi V. Genes of early-onset epileptic encephalopathies: from genotype to phenotype. Pediatr Neurol. 2012;46:24–31. doi: 10.1016/j.pediatrneurol.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 20.Kohda K, Wang Y, Yuzaki M. Mutation of a glutamate receptor motif reveals its role in gating and delta2 receptor channel properties. Nat Neurosci. 2000;3:315–322. doi: 10.1038/73877. [DOI] [PubMed] [Google Scholar]
- 21.Jones KS, VanDongen HM, VanDongen AM. The NMDA receptor M3 segment is a conserved transduction element coupling ligand binding to channel opening. J Neurosci. 2002;22:2044–2053. doi: 10.1523/JNEUROSCI.22-06-02044.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sobolevsky AI, Rooney L, Wollmuth LP. Staggering of subunits in NMDAR channels. Biophys J. 2002;83:3304–3314. doi: 10.1016/S0006-3495(02)75331-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuan H, Erreger K, Dravid SM, Traynelis SF. Conserved structural and functional control of N-methyl-D-aspartate receptor gating by transmembrane domain M3. J Biol Chem. 2005;280:29708–29716. doi: 10.1074/jbc.M414215200. [DOI] [PubMed] [Google Scholar]
- 24.Sobolevsky AI, Prodromou ML, Yelshansky MV, Wollmuth LP. Subunit-specific contribution of pore-forming domains to NMDA receptor channel structure and gating. J Gen Physiol. 2007;129:509–525. doi: 10.1085/jgp.200609718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kazi R, Gan Q, Talukder I, Markowitz M, Salussolia CL, Wollmuth LP. Asynchronous movements prior to pore opening in NMDA receptors. J Neurosci. 2013;33:12052–12066. doi: 10.1523/JNEUROSCI.5780-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Regalado MP, Villarroel A, Lerma J. Intersubunit cooperativity in the NMDA receptor. Neuron. 2001;32:1085–1096. doi: 10.1016/s0896-6273(01)00539-6. [DOI] [PubMed] [Google Scholar]
- 27.Lazzaro JT, et al. Functional characterization of CP-465,022, a selective, noncompetitive AMPA receptor antagonist. Neuropharmacology. 2002;42:143–153. doi: 10.1016/s0028-3908(01)00170-8. [DOI] [PubMed] [Google Scholar]
- 28.Ogden KK, Traynelis SF. Contribution of the M1 transmembrane helix and pre-M1 region to positive allosteric modulation and gating of N-methyl-D-aspartate receptors. Mol Pharmacol. 2013;83:1045–1056. doi: 10.1124/mol.113.085209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nowak L, Bregestovski P, Ascher P, Herbet A, Prochiantz A. Magnesium gates glutamate-activated channels in mouse central neurones. Nature. 1984;307:462–465. doi: 10.1038/307462a0. [DOI] [PubMed] [Google Scholar]
- 30.Mayer ML, Westbrook GL, Guthrie PB. Voltage-dependent block by Mg2+ of NMDA responses in spinal cord neurones. Nature. 1984;309:261–263. doi: 10.1038/309261a0. [DOI] [PubMed] [Google Scholar]
- 31.Sakurada K, Masu M, Nakanishi S. Alteration of Ca2+ permeability and sensitivity to Mg2+ and channel blockers by a single amino acid substitution in the N-methyl-D-aspartate receptor. J Biol Chem. 1993;268:410–415. [PubMed] [Google Scholar]
- 32.Wollmuth LP, Kuner T, Sakmann B. Adjacent asparagines in the NR2-subunit of the NMDA receptor channel control the voltage-dependent block by extracellular Mg2+ J Physiol. 1998;506 (Pt 1):13–32. doi: 10.1111/j.1469-7793.1998.013bx.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dingledine R, Borges K, Bowie D, Traynelis SF. The glutamate receptor ion channels. Pharmacol Rev. 1999;51:7–61. [PubMed] [Google Scholar]
- 34.Lester RA, Clements JD, Westbrook GL, Jahr CE. Channel kinetics determine the time course of NMDA receptor-mediated synaptic currents. Nature. 1990;346:565–567. doi: 10.1038/346565a0. [DOI] [PubMed] [Google Scholar]
- 35.Choi DW. Glutamate receptors and the induction of excitotoxic neuronal death. Prog Brain Res. 1994;100:47–51. doi: 10.1016/s0079-6123(08)60767-0. [DOI] [PubMed] [Google Scholar]
- 36.Rothman SM, Olney JW. Excitotoxicity and the NMDA receptor--still lethal after eight years. Trends Neurosci. 1995;18:57–58. doi: 10.1016/0166-2236(95)93869-y. [DOI] [PubMed] [Google Scholar]
- 37.Talukder I, Borker P, Wollmuth LP. Specific sites within the ligand-binding domain and ion channel linkers modulate NMDA receptor gating. J Neurosci. 2010;30:11792–11804. doi: 10.1523/JNEUROSCI.5382-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Talukder I, Wollmuth LP. Local constraints in either the GluN1 or GluN2 subunit equally impair NMDA receptor pore opening. J Gen Physiol. 2011;138:179–194. doi: 10.1085/jgp.201110623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salussolia CL, Prodromou ML, Borker P, Wollmuth LP. Arrangement of subunits in functional NMDA receptors. J Neurosci. 2011;31:11295–11304. doi: 10.1523/JNEUROSCI.5612-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weber YG, Lerche H. Genetic mechanisms in idiopathic epilepsies. Dev Med Child Neurol. 2008;50:648–654. doi: 10.1111/j.1469-8749.2008.03058.x. [DOI] [PubMed] [Google Scholar]
- 41.Nicita F, et al. The genetics of monogenic idiopathic epilepsies and epileptic encephalopathies. Seizure. 2012;21:3–11. doi: 10.1016/j.seizure.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 42.Noh GJ, Jane Tavyev Asher Y, Graham JM., Jr Clinical review of genetic epileptic encephalopathies. Eur J Med Genet. 2012;55:281–298. doi: 10.1016/j.ejmg.2011.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tavyev Asher YJ, Scaglia F. Molecular bases and clinical spectrum of early infantile epileptic encephalopathies. Eur J Med Genet. 2012;55:299–306. doi: 10.1016/j.ejmg.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 44.Carvill GL, et al. GRIN2A mutations cause epilepsy-aphasia spectrum disorders. Nat Genet. 2013;45:1073–1076. doi: 10.1038/ng.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beck C, Wollmuth LP, Seeburg PH, Sakmann B, Kuner T. NMDAR channel segments forming the extracellular vestibule inferred from the accessibility of substituted cysteines. Neuron. 1999;22:559–570. doi: 10.1016/s0896-6273(00)80710-2. [DOI] [PubMed] [Google Scholar]
- 46.Low CM, et al. Molecular determinants of proton-sensitive N-methyl-D-aspartate receptor gating. Mol Pharmacol. 2003;63:1212–1222. doi: 10.1124/mol.63.6.1212. [DOI] [PubMed] [Google Scholar]
- 47.Taverna F, Xiong ZG, Brandes L, Roder JC, Salter MW, MacDonald JF. The Lurcher mutation of an alpha-amino-3-hydroxy-5-methyl- 4-isoxazolepropionic acid receptor subunit enhances potency of glutamate and converts an antagonist to an agonist. J Biol Chem. 2000;275:8475–8479. doi: 10.1074/jbc.275.12.8475. [DOI] [PubMed] [Google Scholar]
- 48.Sobolevsky AI, Beck C, Wollmuth LP. Molecular rearrangements of the extracellular vestibule in NMDAR channels during gating. Neuron. 2002;33:75–85. doi: 10.1016/s0896-6273(01)00560-8. [DOI] [PubMed] [Google Scholar]
- 49.Blanke ML, VanDongen AM. The NR1 M3 domain mediates allosteric coupling in the N-methyl-D-aspartate receptor. Mol Pharmacol. 2008;74:454–465. doi: 10.1124/mol.107.044115. [DOI] [PubMed] [Google Scholar]
- 50.Villarroel A, Regalado MP, Lerma J. Glycine-independent NMDA receptor desensitization: localization of structural determinants. Neuron. 1998;20:329–339. doi: 10.1016/s0896-6273(00)80460-2. [DOI] [PubMed] [Google Scholar]
- 51.Ogden KK, Traynelis SF. New advances in NMDA receptor pharmacology. Trends Pharmacol Sci. 2011;32:726–733. doi: 10.1016/j.tips.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wei X, et al. Exome sequencing identifies GRIN2A as frequently mutated in melanoma. Nat Genet. 2011;43:442–446. doi: 10.1038/ng.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Teer JK, et al. Systematic comparison of three genomic enrichment methods for massively parallel DNA sequencing. Genome Res. 2010;20:1420–1431. doi: 10.1101/gr.106716.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fuentes Fajardo KV, et al. Detecting false-positive signals in exome sequencing. Hum Mutat. 2012;33:609–613. doi: 10.1002/humu.22033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hedegaard M, Hansen KB, Andersen KT, Brauner-Osborne H, Traynelis SF. Molecular pharmacology of human NMDA receptors. Neurochem Int. 2012;61:601–609. doi: 10.1016/j.neuint.2011.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kniazeff J, Bessis AS, Maurel D, Ansanay H, Prezeau L, Pin JP. Closed state of both binding domains of homodimeric mGlu receptors is required for full activity. Nat Struct Mol Biol. 2004;11:706–713. doi: 10.1038/nsmb794. [DOI] [PubMed] [Google Scholar]
- 57.Jackson MR, Nilsson T, Peterson PA. Retrieval of transmembrane proteins to the endoplasmic reticulum. J Cell Biol. 1993;121:317–333. doi: 10.1083/jcb.121.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jackson MR, Nilsson T, Peterson PA. Identification of a consensus motif for retention of transmembrane proteins in the endoplasmic reticulum. EMBO J. 1990;9:3153–3162. doi: 10.1002/j.1460-2075.1990.tb07513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zerangue N, et al. Analysis of endoplasmic reticulum trafficking signals by combinatorial screening in mammalian cells. Proc Natl Acad Sci U S A. 2001;98:2431–2436. doi: 10.1073/pnas.051630198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Margeta-Mitrovic M, Jan YN, Jan LY. A trafficking checkpoint controls GABA(B) receptor heterodimerization. Neuron. 2000;27:97–106. doi: 10.1016/s0896-6273(00)00012-x. [DOI] [PubMed] [Google Scholar]
- 61.Pagano A, et al. C-terminal interaction is essential for surface trafficking but not for heteromeric assembly of GABA(b) receptors. J Neurosci. 2001;21:1189–1202. doi: 10.1523/JNEUROSCI.21-04-01189.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Traynelis SF, Burgess MF, Zheng F, Lyuboslavsky P, Powers JL. Control of voltage-independent zinc inhibition of NMDA receptors by the NR1 subunit. J Neurosci. 1998;18:6163–6175. doi: 10.1523/JNEUROSCI.18-16-06163.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hansen KB, et al. Structural determinants of agonist efficacy at the glutamate binding site of N-methyl-D-aspartate receptors. Mol Pharmacol. 2013;84:114–127. doi: 10.1124/mol.113.085803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yuan H, Hansen KB, Vance KM, Ogden KK, Traynelis SF. Control of NMDA receptor function by the NR2 subunit amino-terminal domain. J Neurosci. 2009;29:12045–12058. doi: 10.1523/JNEUROSCI.1365-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Qin F, Auerbach A, Sachs F. Estimating single-channel kinetic parameters from idealized patch-clamp data containing missed events. Biophys J. 1996;70:264–280. doi: 10.1016/S0006-3495(96)79568-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Erreger K, Dravid SM, Banke TG, Wyllie DJ, Traynelis SF. Subunit-specific gating controls rat NR1/NR2A and NR1/NR2B NMDA channel kinetics and synaptic signalling profiles. J Physiol. 2005;563:345–358. doi: 10.1113/jphysiol.2004.080028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Siegler Retchless B, Gao W, Johnson JW. A single GluN2 subunit residue controls NMDA receptor channel properties via intersubunit interaction. Nat Neurosci. 2012;15:406–413. S401–402. doi: 10.1038/nn.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lewis CA. Ion-concentration dependence of the reversal potential and the single channel conductance of ion channels at the frog neuromuscular junction. J Physiol. 1979;286:417–445. doi: 10.1113/jphysiol.1979.sp012629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wollmuth LP, Kuner T, Sakmann B. Intracellular Mg2+ interacts with structural determinants of the narrow constriction contributed by the NR1-subunit in the NMDA receptor channel. J Physiol. 1998;506 (Pt 1):33–52. doi: 10.1111/j.1469-7793.1998.00033.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Acker TM, et al. Mechanism for noncompetitive inhibition by novel GluN2C/D N-methyl-D-aspartate receptor subunit-selective modulators. Mol Pharmacol. 2011;80:782–795. doi: 10.1124/mol.111.073239. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.