Abstract
The objective of this study was to determine the antimicrobial susceptibility patterns and clonal diversity of clinical Staphylococcus aureus isolates from Ghana. A total of 308 S. aureus isolates from six healthcare institutions located across Northern, Central and Southern Ghana were characterized by antibiotyping, spa typing and PCR detection of Panton Valentine leukocin (PVL) genes. Methicillin-resistant S. aureus (MRSA) were confirmed by PCR detection of mecA gene and further characterized by SCCmec and multi-locus sequence typing (MLST). The prevalence of antimicrobial resistance was below 5% for all agents tested except for penicillin (97%), tetracycline (42%) and erythromycin (6%). Ninety-one spa types were found, with t355 (ST152, 19%), t084 (ST15, 12%) and t314 (ST121, 6%) being the most frequent types. Based on established associations between spa and MLST types, isolates were assigned to 16 clonal complexes (CCs): CC152 (n = 78), CC15 (n = 57), CC121 (n = 39), CC8 (n = 36), CC5 (n = 33), CC1 (n = 29), CC45 (n = 9), CC88 (n = 8), CC30 (n = 4), CC9 (n = 3), CC25 (n = 2), CC97 (n = 2) CC20 (n = 2), CC707 (n = 2), CC7 (n = 3) and CC522 (n = 1). Most isolates (60%) were PVL-positive, especially those belonging to ST152, ST121, ST5, ST15, ST1, ST8, and ST88. Nine (3%) isolates were MRSA belonging to seven distinct clones: ST88-IV (n = 2), ST250-I (n = 2), ST8-IV (n = 1), ST72-V (n = 1), ST789-IV (n = 1), ST2021-V (n = 1), and ST239-III (n = 1). The study confirmed a high frequency of PVL-positive S. aureus in Africa, low prevalence of antimicrobial resistance and high diversity of MRSA lineages in Ghana compared to developed countries and other African countries. The detection of known pandemic MRSA clones in the absence of routine MRSA identification in most Ghanaian clinical microbiology laboratories calls for capacity building to strengthen surveillance and prevent spread of these clones.
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) is a major concern in clinical medicine due to the importance of β-lactams in the therapy of staphylococcal infections and the additional morbidity and mortality for MRSA patients compared to patients infected with methicillin-susceptible S. aureus (MSSA) [1]. Despite the importance of MRSA, MSSA are among the most common causative agents of bacteraemia and skin and soft tissue infections (SSTI) [2]. Epidemiological surveillance of MRSA and MSSA is of importance for the development and implementation of infection control programmes. Data on S. aureus epidemiology in African countries are limited and a common trait for MSSA strains from various African countries seems to be the carriage of the PVL genes: lukS/F-pv at much higher frequencies (>55%) than in the rest of the world (<10%) [2]–[6]. The high frequency of PVL among human MSSA strains is of special interest since the most successful community associated (CA) MRSA clones share this genetic marker, and could have MSSA ancestors associated with Africa as recently suggested [5], [6]. PVL is associated with SSTI and severe necrotising pneumonia and has been shown to be a characteristic feature of community acquired (CA) -MRSA clones disseminated in Europe and Middle East (ST-80), Australia and South America (ST30-IV), and United States (ST8-IV, also known as USA300) [2], [7], [8].
The objective of this study was to investigate the antimicrobial susceptibility and clonal diversity of clinical S. aureus isolates from Ghana. Antimicrobial resistance in S. aureus has previously been reported from Ghana with findings of a low MRSA prevalence in nasal swabs from patients and health care workers at the Korle-bu Hospital, Accra [9] however, treatment in Ghana is mainly empirical due to a relative lack of appropriate laboratory facilities [10] and therefore only few susceptibility data exists and so far no study has investigated the clonal structure of S. aureus in clinical samples. The study was part of a cooperation program on Antibiotic Drug use, Monitoring and Evaluation of Resistance (ADMER) in Ghana under the Danish Ministry of Foreign Affairs. This program was conceived to strengthen clinical microbiology and surveillance of antibiotic resistance, and ultimately to improve awareness of antimicrobial use in Ghana.
Materials and Methods
Ethics Statement
Ethical clearance was obtained from the University of Ghana Medical School Ethical and Protocol Review Board (reference no. MS-EI/M.9 - P.3.212010-11).
Bacterial Isolates
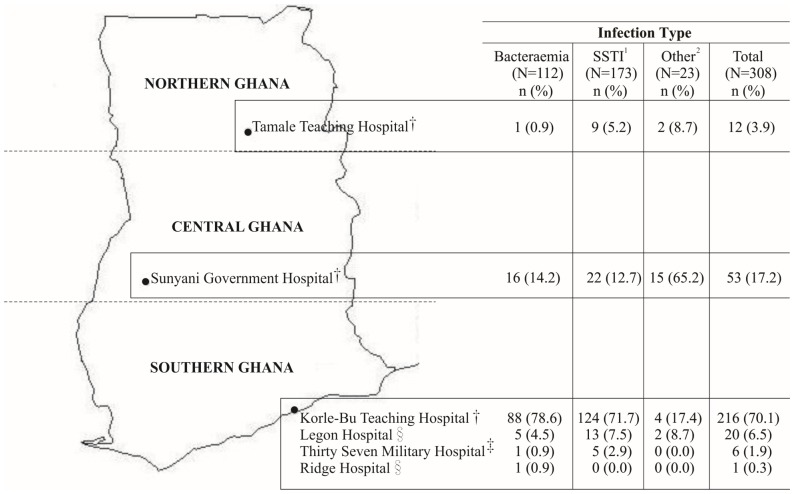
Staphylococcal isolates from clinical specimens were obtained in a prospective cross-sectional-like study between October 2010–June 2012 from six healthcare institution situated at Northern (Tamale Teaching Hospital), Central (Sunyani Government Hospital) and Southern Ghana (Korle bu Teaching Hospital, Thirty-seven Military Hospital, Ridge Hospital and Legon Hospital) (Figure 1). The majority of the isolates (70%) were obtained from Korle bu Teaching Hospital, which serves a population of over 3 million and acts as a major referral health facility for an estimated population of 24 million people across Ghana. Presumptive staphylococci identified by colony morphology at the hospital clinical microbiology laboratories were collected and sent to Noguchi Memorial Institute for Medical Research, where they were identified as S. aureus by Gram staining, catalase, tube coagulase and slidex staphplus test (bioMérieux, Marcy l’Etoile, France). Available patient demographic characteristics such as age and sex were retrieved from laboratory records.
Figure 1. Origins of the 308 clinical Staphylococcus aureus collected from six hospitals in Ghana, 2010–2012.
†Referral Hospitals. ‡Secondary Hospitals. §Primary Hospitals. 1SSTI: Skin and Soft Tissue Infections. 2Other: (Urinary Tract Infection (n = 9); Unknown Infections (n = 14)).
Antimicrobial Susceptibility Testing
Susceptibility testing was carried out by disc diffusion technique following the European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines (www.eucast.org) using 1U penicillin, 30 µg tetracycline, 30 µg cefoxitin, 2 µg clindamycin, 15 µg erythromycin, 10 µg norfloxacin, 10 µg gentamicin, 10 µg linezolid, 5 µg rifampicin, 1. 25 µg +23.75 µg trimethoprim-sulfamethoxazole, and 10 µg fusidic acid (Rosco NeoSenstabs, Taastrup, Denmark). Inducible clindamycin resistance was detected by placing clindamycin and erythromycin 12–20 mm apart (D-test). Brain Heart Infusion agar supplemented with teicoplanin (5 mg/L) (Becton Dickinson, Denmark) was used to screen MRSA isolates for glycopeptides resistance by a spot test; if 10 or more colonies were detected on these plates, E-tests (bioMérieux, Marcy I’Etoile, France) were used to determine the minimum inhibitory concentration of vancomycin and teicoplanin [11]. Multidrug resistance (MDR) was defined as resistance to at least three distinct antimicrobial classes or being MRSA [12].
Molecular Typing
Molecular characterization of the isolates was done at Statens Serum Institut (SSI), Denmark. A multiplex PCR was used for detection of spa, lukS/F-pv and mecA [13]. spa typing was performed as described by Harmsen et al. [14]. Using BioNumerics v.6.5 (Applied Maths, Sint-Martens-Latem, Belgium) with the Ridom spa server (http://spa.server.ridom.de) plug-in, spa sequences were automatically assigned to spa types and clonal complexes (CCs) based on spa repeats. Multi-Locus Sequence Typing (MLST) [15] was done on all MRSA and MSSA isolates whose CC could not be assigned by the Ridom spa server. Minimum Spanning Tree (MST) based on spa- types was made using BioNumerics V6.5 (Applied Maths, Sint-Martens-Latem Belgium). Staphylococcal cassette chromosome mec (SCCmec) typing was performed by multiplex PCR as described previously [16].
Statistical Analysis
Distributions of the various genotypes determined in the study (PVL-positivity, spa type, ST and CC) were associated to region, hospital, sex and infection type to determine if specific patterns existed. Only genotypes with more than 10 observations were included in statistical analysis. MRSA isolates were not evaluated due to the low prevalence of their genotypes. Associations were determined using the χ2 test, except for PVL-positivity, which was analysed by logistic regression. A significant association was deemed at p-values <0.05.
Results
Of the 903 presumed staphylococci collected from the six hospitals, 308 (34%) were identified as S. aureus and 595 (66%) as coagulase negative staphylococci. S. aureus isolates originated from SSTI (n = 173), bacteraemia (n = 112), and other (urinary tract infection, n = 9; unknown infections, n = 14) infections (n = 23). S. aureus was isolated from 143 females and 109 males. Sex origin of 56 isolates could not be traced from laboratory records. With regard to hospital origin, 12 isolates were from Tamale Teaching Hospital (TTH, Northern Ghana), 53 from Sunyani Government Hospital (SGH, Central Ghana) and 243 from the four hospitals in Southern Ghana. Details of hospital location (stratified into regions of study) and proportions of isolates from clinical infections are shown in Figure 1. None of the clinical laboratories used methods for MRSA detection and typing, and several pitfalls were recognized in routine microbiological procedures (e.g. poor identification to species/genus level, and low compliance with international standards for susceptibility testing).
The highest prevalence of resistance was for penicillin (97%), followed by tetracycline (42%) and erythromycin (6%). Lower percentages of resistance were observed for clindamycin (5%), norfloxacin (4%), trimethoprim-sulphamethoxazole (4%), gentamicin (3%), cefoxitin (3%) and fusidic acid (2%). Inducible clindamycin resistance was detected among seven (2%) isolates. Twenty-nine isolates (9%) were MDR, of which 9 (3%) were confirmed mecA positive MRSA. Details of MDR isolates have been shown in Table 1. MRSA isolates were susceptible to vancomycin and teicoplanin. Most of the MSSA isolates (88%, 264/299) were resistant to penicillin (n = 154), penicillin and tetracycline (n = 99) and penicillin and trimethoprim-sulphamethoxazole (n = 11). All isolates were susceptible to linezolid and rifampicin while three isolates (1%) were susceptible to all antimicrobial drugs tested.
Table 1. Origins and characteristics of 29 multi-drug resistant (MDR) Staphylococcus aureus isolated from healthcare institutions in Ghana, 2010–2012.
| ID | Hospitala | Infectionb | CC | ST | spa type | SCCmec | PVL | Antibiotypec | |
| MRSA | 5016 | KB | SSTI | CC1 | ST72 | t537 | V | − | Fox, Pen, Tet |
| 744 | KB | Blood | CC8 | ST2021 | t024 | V | − | Fox, Pen, Tet | |
| 2244 | KB | Blood | CC8 | ST239 | t037 | III | − | Fox, Pen,Tet, Fuc, Gen, Cli, Ery, | |
| 3464 | KB | Blood | CC8 | ST8 | t121 | IV | + | Fox, Pen, Nor, Cli, Ery | |
| 2207 | KB | SSTI | CC8 | ST250 | t928 | I | − | Fox, Pen, Tet, Gen, Nor, Cli, Ery | |
| 2224 | KB | SSTI | CC8 | ST250 | t928 | I | − | Fox, Pen, Tet, Gen, Nor, Cli, Ery | |
| 44 | SGH | Unknown | CC88 | ST88 | t186 | IV | − | Fox, Pen, Tet | |
| AU81 | SGH | SSTI | CC88 | ST88 | t186 | IV | − | Fox, Pen, Tet | |
| 11087 | KB | UTI | CC152 | ST789 | t547 | IV | + | Fox, Pen, Tet, Nor | |
| MSSA | 2639 | KB | Blood | CC1 | ST1 | t7835 | NA | + | Pen, Tet, Cli, Ery |
| AU93 | SGH | SSTI | CC1 | ST1 | t559 | NA | + | Pen, Tet, Fuc | |
| A6 | KB | Unknown | CC5 | ST5 | t311 | NA | + | Pen, Tet, Fuc | |
| 5095 | KB | SSTI | CC5 | ST5 | t071 | NA | + | Pen, Tet, Gen, TMS, Fuc, Nor, Cli, Ery | |
| T2845 | TTH | SSTI | CC8 | ST8 | t451 | NA | + | Pen, Tet, TMS | |
| 1455 | KB | SSTI | CC9 | ST9 | t2700 | NA | − | Pen, Gen, Cli, Ery | |
| 1050 | KB | Blood | CC15 | ST15 | t084 | NA | + | Pen, Tet, Gen | |
| 2320 | KB | Blood | CC45 | ST508 | t635 | NA | − | Pen, Cli, Ery | |
| 1548 | KB | SSTI | CC88 | ST88 | t10809 | NA | + | Pen, Tet, Nor, Cli, Ery | |
| 5270 | KB | SSTI | CC88 | ST88 | t10810 | NA | + | Pen, Tet, Nor, Cli, Ery | |
| NAB | KB | SSTI | CC121 | ST121 | t213 | NA | _ | Pen, Tet, Fuc | |
| 3209 | KB | Blood | CC121 | ST121 | t091 | NA | _ | Pen, Tet, Nor, Gen, Cli, Ery | |
| 2437 | KB | Blood | CC121 | ST121 | t091 | NA | _ | Pen, Tet, Nor | |
| 5293 | KB | Blood | CC121 | ST121 | t314 | NA | + | Pen, Tet, Nor | |
| 5775 | KB | Blood | CC121 | ST121 | t314 | NA | + | Pen, Tet, Nor | |
| 3984 | KB | SSTI | CC152 | ST152 | t1299 | NA | + | Pen, Tet, Cli, Ery | |
| 1544 | KB | SSTI | CC152 | ST152 | t355 | NA | + | Pen, Tet, Cli, Ery | |
| 4836 | KB | SSTI | CC152 | ST152 | t355 | NA | + | Pen, Tet, Cli, Ery | |
| 112242 | MH | SSTI | CC152 | ST152 | t355 | NA | + | Pen, Tet, Ery | |
| A71 | SGH | Blood | CC152 | ST152 | t355 | NA | + | Pen, Tet, Fuc |
ST: Sequence Type; CC: Clonal Complex; SCC: Staphylococcal Cassette Chromosome; PVL: Panton- Valentine leukocidin.
KB: Korle bu Teaching Hospital; SGH: Sunyani Government Hospital; TTH: Tamale Teaching Hospital; MH: Military Hospital.
SSTI: Skin and Soft Tissue Infection; UTI: Urinary Tract Infection; c TMS: trimethoprim-sulphamethoxazole.
Pen: penicillin; Fox: cefoxitin; Tet: tetracycline; Nor: norfloxacin; Gen: gentamicin; Fuc: Fucidic acid; Cli: clindamycin; Ery: erythromycin.
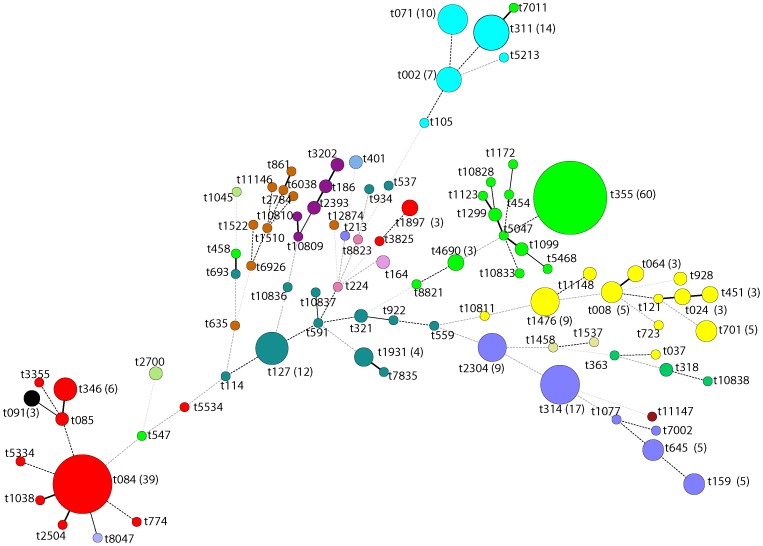
High genetic diversity was observed by spa typing, as indicated by the recovery of 91 spa types among all isolates tested. The most common spa types were t355 (19%), t084 (12%), t314 (6%) and t311 (5%). Fifty-six spa types were singletons and eight new spa types were detected: t10809 (ST88), t10810 (ST88), t10811 (ST8), t10828 (ST152), t10833 (ST152), t10836 (ST1), t10837 (ST1) and t10838 (ST30). A minimum spanning tree, including spa types and clonal complexes of the 308 isolates is shown in Figure 2.
Figure 2. Minimum spanning tree of 308 clinical Staphylococcus aureus isolates from healthcare institutions in Ghana.
Nodes indicate spa types and their size shows the relative number of isolates for each spa type. Numbers of frequent (three or more) spa types have been shown. Every colour represents a distinct clonal complex.
Based on spa typing, isolates (n = 308) were assigned to 16 MLST clonal complexes: CC152 (n = 78), CC15 (n = 57), CC121 (n = 39), CC8 (n = 36), CC5 (n = 33), CC1 (n = 29), CC45 (n = 9), CC88 (n = 8), CC30 (n = 4), CC9 (n = 3), CC7 (n = 3), CC25 (n = 2), CC97 (n = 2) CC20 (n = 2), CC707 (n = 2), and CC522 (n = 1). MRSA isolates belonged to ST88-IV (n = 2), ST8-IV (n = 1), ST789-IV (n = 1), ST72-V (n = 1) ST2021-V (n = 1), ST250-I (n = 2), and ST239-III (n = 1). Two of them were PVL-positive and belonged to t121 (ST8) and t547 (ST789) (Table 1). The most common MSSA lineages were ST152 (27%) and ST15 (18%).
Five spa types (t084, t127, t311, t314 and t355), six STs (ST1, ST5, ST8, ST15, ST121 and ST152) and six CCs (CC1, CC5, CC8, CC15, CC121 and CC152) were included in the statistical analysis. Surprisingly, spa type t311 occurred less frequently among females (0.3%) than among males (2%) and those of unknown sex (2.3%) (p = 0.0016). No other clinical or spatial associations were observed in the distribution of spa-types, STs and CCs with regard to infection type, sex, region and hospital of origin, Some spatial variations were observed in the distribution of spa types (e.g. t355 occurred in 34% and 13% isolates from Sunyani Government Hospital and Korle bu Teaching Hospital respectively) but such differences could not be proven to be significant. Isolates from Ridge Hospital (n = 1), Thirty-seven Military Hospital (n = 6) and Tamale Teaching Hospital (n = 12) were excluded from the statistical analysis due to low numbers.
PVL was detected in 60% (n = 184) of the isolates, mainly isolates from SSTIs (57%) and belonging to ST152 (38.5%), ST121 (21%), ST5 (13.5%), ST15 (11%), ST1 (7%), ST8 (3.8%) and ST88 (2.7%). Genotypes and clinical origins of PVL positive S. aureus are shown in Table 2. The patterns of PVL varied by region, with Central regions having a higher chance 2.2 (95% CI: 1.1–4.2) of seeing PVL-positive S. aureus (p = 0.02) than the Southern region.
Table 2. Clonal complex (CC), multi-locus sequence type (ST), spa type and clinical origin of 184 Staphylooccus aureus harbouring Panton-Valentine leukocidin (PVL) genes isolated in Ghana, 2010–2012. STs and CCs were inferred from spa types.
| CC | ST | spa type (N) | Clinical origin, N (%) | ||||
| Bacteraemia N = 65 | SSTI N = 104 | Other* N = 15 | TotalN = 184 | ||||
| CC1 | ST1 | t127 (3), t1931 (3), t693 (1), t559 (1), t10836 (1),t114 (1), t922 (1), t934 (1), t7835 (1) | 6 (9.2) | 7 (6.7) | 0 (0.0) | 13 (7.0) | |
| CC5 | ST5 | t071 (9) t311 (9), t002 (5), t105 (1) | 12 (18.5) | 11 (10.6) | 1 (6.7) | 24 (13.0) | |
| CC8 | ST8 | t1476 (3), t024 (1), t451 (1), t064 (1), °t121(1) | 2 (3.0) | 5 (4.8) | 0 (0.0) | 7 (3.8) | |
| CC15 | ST15 | t084 (19), t5534 (1), t774 (1) | 7 (10.8) | 13 (12.5) | 1 (6.7) | 21 (11.4) | |
| CC25 | ST25 | t401 (1) | 1 (1.5) | 0 (0.0) | 0 (0.0) | 1 (0.5) | |
| CC30 | ST30 | t10838 (1),t363 (1) | 0 (0.0) | 2 (1.9) | 0 (0.0) | 2 (1.1) | |
| CC88 | ST88 | t2393 (2) t3202 (1), t10809 (1), t10810 (1) | 1 (1.5) | 4 (3.8) | 0 (0.0) | 5 (2.7) | |
| CC121 | ST121 | t314 (15), t2304 (9), t159 (5), t645 (5), t1077(1), t7002(1) | 14 (21.5) | 21 (20.2) | 1 (6.7) | 36 (19.6) | |
| CC152 | ST152 | t355 (57), t4690 (3), t1096 (2), t1299 (2), Singletons (11)a | 22 (34.0) | 41 (39.4) | 12 (80.0) | 75 (40.8) | |
SSTI: Skin and Soft Tissue Infection;*Other: UTI: Urinary Tract Infection (n = 5: spa types t355 (3), t547(1) and t5534 (1); Unknown (n = 10; spa types: t311 (1), t645 (1), t4690 (1), t355 (7) aOther spa types associated with CC152: t454, t458, t5268, °t547, t1123, t1172, t5047, t7011, t8821, t10828, and t10833°MRSA.
Discussion
This study fills an important gap in the knowledge of the epidemiology of S. aureus in Ghana. As such, the study contributes to the current knowledge of the diversity and population structure of this important bacterial pathogen at the global level. Ghana and several other African countries have so far been black spots on the map due to lack of established national surveillance programmes and adequate clinical microbiology infrastructure [10], [17]. Our results show that the most common spa types among MSSA isolates are t355 (ST152) and t084 (ST15). The spa types were previously found to be predominant among S. aureus isolates from asymptomatic nasal carriers at Korle bu, the largest Teaching Hospital in Ghana [9], suggesting that they are well established in the human population of this country. In another African study, t084 (ST15) was also reported as one of the most frequent spa types among S. aureus isolated from seven tertiary hospitals located in five major African towns [3]. PVL-positive ST152 (t355) is also widely distributed in African countries [4], [5] and its frequent recovery from SSTI is consistent with studies in other countries [4], [18]. Other PVL-positive MSSA lineages found in this study such as ST121, ST30, ST15 and ST5 have also been reported elsewhere in Africa [19]. The observed high prevalence (60%) of PVL appears to be a distinguishing genetic trait of African MSSA [3], [4], [6] compared to USA, Asia and Europe, where this virulence factor is uncommon in MSSA [2], [19], [20]. This finding was correlated to the high frequency of PVL-positive ST152, which is a likely ancestor of the CA-MRSA ST152-V clone circulating in certain European regions, especially the Balkan area [21], [22].
The nine MRSA isolates belonged to seven unrelated spa types and STs harbouring four different SCCmec types (Table 1), indicating high clonal diversity. Some of the MRSA lineages identified in this study are widely distributed worldwide: ST239-III is a pandemic clone prevalent in Europe, Asia and South Africa [23]–[25] and ST789-IV is a single locus variant of the ST7 clone frequently reported in Asia [25]. ST88-IV, ST8-IV and ST72-V have been previously reported among inpatients and staff at Korle-bu Hospital in Ghana [9] and in communities and hospitals in other African countries [26], [27]. MRSA ST88 has been reported sporadically in some European countries like Portugal [28] and Sweden [29]. ST8-IV MRSA (spa type t121, PVL+) found in this study is related to the epidemic MRSA ST8-IV (USA300) clone in the USA [2]. Other African studies have reported this ST8-IV MRSA (spa type t121, PVL+) strain in communities and hospitals [26], [30]. ST250-I, also referred to as the “Archaic clone”, differs from ST8 by a point mutation in the yqiL gene and is related to ST247-I (Iberian clone), a major clone isolated in European hospitals [7], [31]. ST72 has been reported as a major MRSA clone from communities in Australia [32] and as MSSA in Nigeria and Gabon [6], [27]. The least known MRSA lineage found in this study was ST2021-V, which to the best of our knowledge has previously been reported in a single isolate from Nigeria (www.mlst.net; accessed on: 4th April 2013). Although ST5, ST30 and ST80 MRSA have been described in several African and other countries around the world [26], [27], none of these clones were detected among clinical MRSA isolates in Ghana. PVL-positive ST5 and ST30 were however detected among MSSA isolates (Table 2), indicating that these two S. aureus lineages are widespread in African countries, even though acquisition of methicillin resistance seems to be confined to some countries.
The prevalence of antimicrobial resistance in clinical S. aureus isolates from Ghana was generally low. Other African studies have reported similar levels of resistance to penicillin (86%–93%) and tetracycline (28%–48%) but higher levels of resistance to sulphonamides (22%–68%) compared to this study [3], [4], [9]. Comparatively, the prevalence of MRSA (3%) was lower than those reported in other African countries such as Nigeria (20%) [27], Algeria (45%) [33] and in a multicenter study (15%) involving five major African towns [26]. The low MRSA frequency reported in this study could be attributed to the low consumption of antimicrobial agents such as fluoroquinolones and third generation cephalosporins in Ghana, because they are expensive and are usually prescribed for acute infections [10]. Usage of the afore-mentioned antimicrobial agents has been shown to correlate with an increase in MRSA prevalence [34]–[36]. The observed MRSA prevalence among clinical isolates in Ghana is similar to those reported in European countries with low MRSA prevalence, such as the Scandinavian countries and The Netherlands [37].
Some apparent geographical variations in clonal distribution were observed, but the low number of isolates obtained from the Northern region made comparisons between hospitals or regions meaningless. The clinical information on the 308 S. aureus included in the study varied in quality due to incompleteness of the patient records collected from the various hospital clinical laboratories involved in the study. Thus, it was not possible to determine possible associations between antimicrobial therapy and resistance patterns.
We conclude that MRSA occurs at low prevalence among S. aureus investigated in this study. MRSA clones circulating in the country are genetically diverse and a number of them belong to known pandemic clones. The overall levels of antimicrobial resistance are generally low compared to other African countries and to most developed countries, most likely because of the low usage of antimicrobial agents in the country. On the other hand, the study also denotes absence of routine MRSA testing and poor performance standards in most clinical microbiology laboratories in Ghana, highlighting the need for infrastructures to support national antimicrobial policies and surveillance capacity.
Acknowledgments
The directors of hospitals are thanked for their support to the study. The authors are grateful to Dr. Jesper Larsen, Mr Michael Olu-Taiwoo, Mr Samuel Acquah, Christian Bonsu, Stephen Osei-Wusu, Sandra Sowah, Lone Ryste Kildevang Hansen and Julie Hindsberg Nielsen for their excellent assistance.
Funding Statement
This study was supported by Antibiotic Drug use, Monitoring and Evaluation of Resistance (ADMER) project funded by the Danish International Development Agency (DANIDA). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Cosgrove SE, Sakoulas G, Perencevich EN, Schwaber MJ, Karchmer AW, et al. (2003) Comparison of mortality associated with methicillin-resistant and methicillin-susceptible Staphylococcus aureus bacteremia: a meta-analysis. Clin Infect Dis 36: 53–59. [DOI] [PubMed] [Google Scholar]
- 2. David MZ, Boyle-Vavra S, Zychowski DL, Daum RS (2011) Methicillin-susceptible Staphylococcus aureus as a predominantly healthcare-associated pathogen: a possible reversal of roles? PloS One 6: e18217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Breurec S, Fall C, Pouillot R, Boisier P, Brisse S, et al. (2011) Epidemiology of methicillin-susceptible Staphylococcus aureus lineages in five major African towns: high prevalence of Panton-Valentine leukocidin genes. Clin Microbiol Infect 17: 633–639. [DOI] [PubMed] [Google Scholar]
- 4. Shittu AO, Okon K, Adesida S, Oyedara O, Witte W, et al. (2011) Antibiotic resistance and molecular epidemiology of Staphylococcus aureus in Nigeria. BMC Microbiol 11: 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ruimy R, Maiga A, Armand-Lefevre L, Maiga I, Diallo A, et al. (2008) The carriage population of Staphylococcus aureus from Mali is composed of a combination of pandemic clones and the divergent Panton-Valentine leukocidin-positive genotype ST152. J Bacteriol 190: 3962–3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schaumburg F, Köck R, Friedrich AW, Soulanoudjingar S, Ngoa UA, et al. (2011) Population structure of Staphylococcus aureus from remote African Babongo Pygmies. PLoS Neglect Trop Dis 5: e1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Deurenberg RH, Stobberingh EE (2009) The molecular evolution of hospital- and community-associated methicillin-resistant Staphylococcus aureus . Curr Mol Med 9: 100–115. [DOI] [PubMed] [Google Scholar]
- 8. Lina G, Piémont Y, Godail-Gamot F, Bes M, Peter MO, et al. (1999) Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis 29: 1128–1132. [DOI] [PubMed] [Google Scholar]
- 9.Egyir B, Guardabassi L, Nielsen SS, Larsen J, Addo KK, et al. (2013) Prevalence of nasal carriage and diversity of Staphylococcus aureus among inpatients and hospital staff at Korle Bu Teaching Hospital, Ghana. J Global Antimicrob Resist http://dx.doi.org/doi:10.1016/j.jgar.2013.05.006. [DOI] [PubMed]
- 10. Newman MJ, Frimpong E, Donkor ES, Opintan JA, Asamoah-Adu A (2011) Resistance to antimicrobial drugs in Ghana. Infect Drug Resist 4: 215–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fitzgibbon MM, Rossney AS, O’Connell B (2007) Investigation of reduced susceptibility to glycopeptides among methicillin-resistant Staphylococcus aureus isolates from patients in Ireland and evaluation of agar screening methods for detection of heterogeneously glycopeptide-intermediate S. aureus . J Clin Microbiol 45: 3263–3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Magiorakos A, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, et al. (2011) Multidrug resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance Clin Microbiol Infect 2012. 18: 268–281. [DOI] [PubMed] [Google Scholar]
- 13. Larsen AR, Stegger M, Sørum M (2008) spa typing directly from a mecA, spa and pvl multiplex PCR assay–a cost-effective improvement for methicillin-resistant Staphylococcus aureus surveillance. Clin Microbiol Infect 14: 611–614. [DOI] [PubMed] [Google Scholar]
- 14. Harmsen D, Claus H, Witte W, Claus H, Rothgänger J, et al. (2003) Typing of Methicillin-Resistant Staphylococcus aureus in a University Hospital Setting by Using Novel Software for spa Repeat Determination and Database Management. J Clin Microbiol 41: 5442–5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG (2000) Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus . J Clin Microbiol 38: 1008–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kondo Y, Ito T, Ma XX, Watanabe S, Kreiswirth BN, et al. (2007) Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob Agents Chemother 51: 264–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Falagas ME, Karageorgopoulos DE, Leptidis J, Korbila IP (2013) MRSA in Africa: Filling the Global Map of Antimicrobial Resistance. PLoS One 8: e68024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Monecke S, Slickers P, Ellington MJ, Kearns AM, Ehricht R (2007) High diversity of Panton-Valentine leukocidin-positive, methicillin-susceptible isolates of Staphylococcus aureus and implications for the evolution of community-associated methicillin-resistant S. aureus . Clin Microbiol Infect 13: 1157–1164. [DOI] [PubMed] [Google Scholar]
- 19. Rasigade J-P, Laurent F, Lina G, Meugnier H, Bes M, et al. (2010) Global distribution and evolution of Panton-Valentine leukocidin-positive methicillin-susceptible Staphylococcus aureus, 1981–2007. J Infect Dis 201 1589–1597. [DOI] [PubMed] [Google Scholar]
- 20. Mine Y, Nakasone I, Yamamoto Y, Utani A, Yamane N, et al. (2013) Dissemination of Panton-Valentine Leukocidin-positive methicillin-resistant Staphylococcus aureus in Okinawa, Japan. J Dermatol 40: 34–38. [DOI] [PubMed] [Google Scholar]
- 21. Francois P, Harbarth S, Huyghe A, Renzi G, Bento M, et al. (2008) Methicillin Resistant Staphylococcus aureus, Geneva, Switzerland 1993–2005. Emerg Infect Dis 14: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Monecke S, Berger-Bächi B, Coombs G, Holmes A, Kay I, et al. (2007) Comparative genomics and DNA array-based genotyping of pandemic Staphylococcus aureus strains encoding Panton-Valentine leukocidin. Clin Microbiol Infect 13: 236–249. [DOI] [PubMed] [Google Scholar]
- 23. Harris SR, Feil EJ, Holden MTG, Quail MA, Nickerson EK, et al. (2010) Evolution of MRSA During Hospital Transmission and Intercontinental Spread. Science 327: 469–474 doi:10.1126/science.1182395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Moodley A, Oosthuysen WF, Dusé AG, Marais E (2010) South Africa MRSA Surveillance Group (2010) Molecular characterization of clinical methicillin-resistant Staphylococcus aureus isolates in South Africa. J Clin Microbiol 48: 4608–4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Song Y, Du X, Li T, Zhu Y, Li M (2013) Phenotypic and molecular characterization of Staphylococcus aureus recovered from different clinical specimens of inpatients at a teaching hospital in Shanghai between 2005 and 2010. J Med Microbiol 62: 274–282. [DOI] [PubMed] [Google Scholar]
- 26. Breurec S, Zriouil SB, Fall C, Boisier P, Brisse S, et al. (2011) Epidemiology of methicillin-resistant Staphylococcus aureus lineages in five major African towns: emergence and spread of atypical clones. Clin Microbiol Infect 17: 160–165. [DOI] [PubMed] [Google Scholar]
- 27. Ghebremedhin B, Olugbosi MO, Raji AM, Layer F, Bakare RA, et al. (2009) Emergence of a Community-Associated Methicillin-Resistant Staphylococcus aureus Strain with a Unique Resistance Profile in Southwest Nigeria. J Clin Microbiol 47: 2975–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Aires-de-Sousa M, Conceição T, De Lencastre H (2006) Unusually high prevalence of nosocomial Panton-Valentine Leukocidin-positive Staphylococcus aureus isolates in Cape Verde Islands. J Clin Microbiol 44: 3790–3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fang H, Hedin G, Li G, Nord CE (2008) Genetic diversity of community-associated methicillin-resistant Staphylococcus aureus in southern Stockholm, 2000–2005: Clin Microbiol Infect. 14: 370–376. [DOI] [PubMed] [Google Scholar]
- 30. Ateba Ngoa U, Schaumburg F, Adegnika AA, Kösters K, Möller T, et al. (2012) Epidemiology and population structure of Staphylococcus aureus in various population groups from a rural and semi urban area in Gabon, Central Africa. Acta Trop 124: 42–47. [DOI] [PubMed] [Google Scholar]
- 31. Deurenberg RH, Vink C, Kalenic S, Friedrich AW, Bruggeman CA, et al. (2007) The molecular evolution of methicillin-resistant Staphylococcus aureus. Clin Microbiol Infect 13: 222–235. [DOI] [PubMed] [Google Scholar]
- 32. Coombs GW, Monecke S, Pearson JC, Tan H, Chew Y-K, et al. (2011) Evolution and diversity of community-associated methicillin-resistant Staphylococcus aureus in a geographical region. BMC Microbiol 11: 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bekkhoucha SN, Cady A, Gautier P, Itim F, Donnio P-Y (2009) A portrait of Staphylococcus aureus from the other side of the Mediterranean Sea: molecular characteristics of isolates from Western Algeria. Eur J Clin Microbiol Infect Dis 28: 553–555. [DOI] [PubMed] [Google Scholar]
- 34. Monnet DL, Mackenzie FM, López-lozano JM, Beyaert A, Camacho M, et al. (2004) Antimicrobial Drug Use and Methicillin-resistant Staphylococcus aureus, Aberdeen, 1996–2000 Emerg Infect Dis. 10: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Graffunder EM, Venezia RA (2002) Risk factors associated with nosocomial methicillin resistant Staphylococcus aureus (MRSA) infection including previous use of antimicrobials. J Antimicrob Chemother 49: 999–1005. [DOI] [PubMed] [Google Scholar]
- 36. Muller AA, Mauny F, Bertin M, Cornette C, Lopez-Lozano J-M, et al. (2003) Relationship between spread of methicillin-resistant Staphylococcus aureus and antimicrobial use in a French university hospital. Clin Infect Dis 36: 971–978. [DOI] [PubMed] [Google Scholar]
- 37. Johnson AP (2011) Methicillin-resistant Staphylococcus aureus: the European landscape. The J Antimicrob Chemother 66 Suppl 4iv43–iv48. [DOI] [PubMed] [Google Scholar]