Abstract
In Arabidopsis thaliana, the R2R3 MYB-like transcription factor MYB30 is a positive regulator of the pathogen-induced hypersensitive response and of brassinosteroid and abscisic acid signaling. Here, we show that MYB30 expressed under the control of the strong phloem-specific SUC2 promoter accelerates flowering both in long and short days. Early flowering is mediated by elevated expression of FLOWERING LOCUS T (FT), which can be observed in the absence and presence of CONSTANS (CO), the main activator of FT. CO-independent activation by high MYB30 expression results in FT levels that remain below those observed in the wild-type plants, which show an additive CO-dependent activation. In contrast, TWIN SISTER OF FT (TSF) is repressed in plants expressing high levels of MYB30 in the phloem. In transient assays, MYB30 and CO additively increase the activity of a reporter construct driven by a 1 kb FT promoter. Acceleration of flowering by MYB30 does not require the presence of salicylic acid and is independent of FLC. Taken together, increased levels of MYB30, which was reported to be induced in response to the perception of pathogens, can accelerate flowering and MYB30 may thus be a candidate to mediate cross-talk between gene networks involved in biotic stress perception and flowering time.
Introduction
Optimal timing of the transition from vegetative to reproductive development is a critical step for the successful reproduction of flowering plants. Independent endogenous and environmental cues combine to determine flowering time through converging pathways. In Arabidopsis, many regulatory inputs are channeled into the transcriptional regulation of FLOWERING LOCUS T (FT) [1], [2]. FT encodes a major component of florigen, which transmits a flowering stimulus from the leaves to the apical meristem [3], [4]. Arabidopsis plants carrying mutations in the FT locus flower late in long days (LD), but almost as wild-type plants if grown in short days (SD) [5]. Under inductive conditions, FT is transcribed in the companion cells of the distal leaf veins, and FT protein is translocated with the assistance of FT-INTERACTING PROTEIN 1 (FTIP1) to sieve elements [6], [7], [8]. After the long distance movement, FT protein is unloaded to the shoot apical meristem cells, where it is proposed to migrate to the nuclei to form a complex with the bZIP transcription factor FLOWERING LOCUS D (FD) and 14-3-3 proteins [9], [10], [11]. The FT/FD complex reprograms the transcriptional networks in the meristem to induce the floral transition. SUPPRESSOR OF OVEREXPRESSOR OF CONSTANS 1 (SOC1) and APETALA1 (AP1) are induced in the meristem through FT [3], [10], [11], [12], [13].
Several key activators and repressors of FT have been identified. In the photoperiod pathway, CONSTANS (CO) activates FT by binding to one of several TGTG(N2-3)AT(G) motifs located in the proximal promoter [7], [14]. CO protein is stabilized at the end of LDs but does not accumulate under SDs [15]. CO activation of FT depends on the presence of accessory binding partners belonging to two related histone-fold protein families, NF-YB and NF-YC [16], [17]. The MADS-domain transcription factor FLOWERING LOCUS C (FLC), whose levels decrease gradually during vernalization, represses FT by binding to a CArG-box in the first intron in leaves [18], [19], [20]. In ambient low temperature, another MADS-domain factor, SHORT VEGETATIVE PHAGE (SVP), represses FT by binding to a CArG-box upstream of the FT proximal promoter [21]. FLC and SVP can form a complex and it seems that they can collectively but also individually repress FT [20]. In high ambient temperatures, the bHLH domain transcription factor PHYTOCHROME-INTERACTING FACTOR 4 (PIF4) activates flowering dependent on sequences found in the 5′UTR of FT [22]. TEMPRANILLO1 (TEM1), an AP2-like protein counteracts the CO-mediated activation of FT and directly binds to the FT 5′ UTR in vivo [23]. Interestingly, GIGANTEA (GI) as an activator of CO seems to activate FT directly in mesophyll cells by binding to FT repressors and associating with the FT proximal promoter [24], and also other upstream regulators of CO such as FKF1 and CDFs have been reported to directly regulate FT based on ChIP results [25]. Similar to FT, its closest relative TWIN SISTER OF FT (TSF) plays a role as flowering integrator [26], [27], [28]. In seedlings, TSF is mainly expressed in the vascular tissue of leaf petioles and the hypocotyls [28]. Although tsf single mutants do not show obvious later flowering in LD or SD, a loss of tsf enhances the ft flowering phenotype in LDs and SDs and TSF transcription is also responsive to CO [26], [27], [28].
Plant hormones also provide an input to flowering time regulation. Under non-inductive SDs, Gibberellin (GA) plays an important role as demonstrated by the observation that the ga1 mutant, which is strongly impaired in GA biosynthesis, never flowers [29]. In LDs, overexpression of GA2OX7, which encodes a protein that decreases the amount of active GA in vascular leaf tissues delays flowering through decreased FT and TSF expression [30]. Brassinosteroids (BR) and salicylic acid (SA) are reported to promote flowering at least partially by repressing FLC [31]. Plants constitutively expressing the bacterial salicylic acid hydrolase encoding NahG gene and the SA-biosynthetic mutant salicylic acid induction deficient 2 (sid2) have been shown to flower late in LDs and SDs [32]. SIZ1, a PIAS-type SUMO E3 ligase related to yeast Siz (SAP and Miz) proteins [33], facilitates SUMO modification of FLD, which represses FLC through the autonomous pathway. Plants mutated in siz1 flower early and express reduced levels of FLC indicating that sumoylation is important for FLD’s repressive function [34]. In addition, the early flowering in siz1 mutant plants is dependent on the presence of SA [34].
Plants are likely also to coordinate the transition to reproductive development with adverse environmental factors such as biotic and abiotic stresses although they may have to differentiate their response to the severity of the stress [35]. However, relatively little is known about the cross-talk and potential overlap between the gene networks regulating flowering time, pathogen defense and response to biotic stress. In a systematic screen for transcription factors that accelerate flowering if highly expressed in the phloem, we identified the transcription factor MYB30 as novel factor with a potential to regulate flowering from the phloem. MYB30 was first characterized for its role in cell death during the hypersensitive response (HR) to pathogens [36]. This R2R3 MYB-like factor is a positive regulator of HR and induces cell death through SA accumulation and the regulation of SA-responsive genes [37], [38], [39]. MYB30 expression is transiently upregulated during bacterial pathogen recognition [37]. In the BR signaling pathway, MYB30 interacts with BRI1-EMS-SUPPRESSOR 1 (BES1) and, as a cofactor, positively regulates BR-responsive target genes [40]. During germination, MYB30 is a target of SIZ1-mediated sumoylation, which appears to stabilize the protein possibly by protecting it from ubiquitin-mediated degradation [41], [42]. Seeds carrying myb30 loss-of-function alleles are hypersensitive to ABA indicating that MYB30 acts on the crossroad of yet another hormonal pathway [41]. Taken together, MYB30 is an interesting candidate, which has the potential to mediate cross-talk between the flowering and stress pathways. In the following, we show how MYB30 could integrate into the flowering time regulatory network.
Materials and Methods
Plant Growth
For expression studies, seeds were sterilized in 75% ethanol and 100% ethanol for 5 min each and sowed on GM medium supplemented by 1% sucrose. After stratification at 4°C for 3 days, plants were grown in climate chambers at 22°C in LDs (16 hours light/8 hours dark). Time of transfer to climate chambers represents day 0. During the collection of samples, ZEITGEBER TIME (ZT) 16 was in the light and ZT24 was in the dark before the switch of light condition. For flowering time measurements, seeds were sowed on soil and stratified at 4°C for 3 days. Soil trays were transferred to LDs (16 hours light/8 hours dark) or SDs (8 hours light/16 hours dark) Percival cabinets or the green house as indicated.
Mutant and Transgenic Plants
The SUC2prom::MYB30 fusion was constructed by recombining the MYB30 open reading frame into a modified pGREEN vector in which the SUC2 promoter was inserted upstream of the GATEWAY recombination site. The original MYB30-pENTRY clone containing the full open reading frame was generated by the REGIA consortium [43]. The MYB30prom::GUS plants were generated by amplifying 3.8 kb of the upstream MYB30 regions with GATEWAY compatible primers and recombining the product into GW-GUS-pGREEN vector previously described [7]. The constructs were introduced into Agrobacterium tumefaciens GV3101 that carried the pSOUP helper plasmid. Transgenic plants were generated using the floral dip method [44]. Only lines that segregated a single locus T-DNA in the T2 generation were further analyzed. For SUC2prom::MYB30, two of three lines were early flowering of which one was selected for crosses and further analysis. Plants of ft-10, tsf-1, co-sail, NahG and flc-3 are all Colombia-0 ecotype and have been described (http://www.arabidopsis.org/). The myb30-1 (SALK_027644C) and myb30-2 (GK_022F04) mutants were ordered from NASC. Primers for plasmid construction and genotyping are listed in Table S1.
Bombardment
30 µg gold particles were prepared for every 10 bombardments. First, 70% ethanol was used to wash the gold particles followed by three times sterile water. Finally the gold particles were suspended in 500 µl of 50% sterile glycerol.
In each bombardment, 15 µg DNA in total was used including 5 µg 35Sprom::RedLUC-pJAN, 5 µg 1.0 kb FTprom::GreenLUC-pGREEN and 5 µg of either 35Sprom::CO, 35Sprom::MYB30 or 35Sprom::GW cassette vector. The DNA was mixed with 50 µl prepared gold beads, 50 µl 2.5 M CaCl2 and 20 µl 0.1 M spermidine. After two washes with ethanol, the DNA-gold mix was resuspended in 50 µl 100% ethanol which was used for two independent bombardments. 5–10 mm Arabidopsis leaves were transformed by the Biolistic™ Particle Delivery System (BIO-RAD, PDS-1000/HE). After 12–24 hours incubation of the samples in constant light conditions, 1 mM luciferin was sprayed on the leaves and after one minute the emitted light was immediately measured with the help of a cooled CCD-camera adapted with optical filters to detect RedLUC and GreenLUC independently. The ratios of GreenLUC/RedLUC signals were calculated with the help of the excel macro Chroma-LUC™ Calculator version 1.0 (Promega).
Reverse Transcribed Quantitative-PCR (RT-qPCR) and Semi-quantitative PCR
Total RNA was isolated using the RNeasy Mini kit according to instructions (Qiagen, Cat. no. 74104). 5 µg RNA was used for reverse transcription. After treatment with DNA-free™ DNAse I according to manufacturer’s instructions (Ambion, Cat. no. AM1906), cDNA was generated at 42°C for 2 hours using Superscript II reverse transcriptase (Life Technologies, Cat. no. 18080-044). The reaction was terminated by incubation at 75°C for 10 min.
Quantitative PCR (qPCR) was performed using a real-time PCR cycler (Roche LC480). 10 µl PCR reactions included cDNA corresponding to 70 ng RNA template, 10 pmole forward primer, 10 pmole reverse primer and 5 µl 2×SYBR Green qPCR buffer (Biorad, Car. no.18080-044). PCR products varied in length of 200∼300 bp, and the program was composed of step 1∶95°C, 3 min; step 2: [95°C 10 sec; 58°C 10 sec; 72°C, 20 sec] 40 cycles; and step 3: [95°C 3 min; 50°C 1 min; rise 0.5°C every 10 sec until up to 95°C]. Absence of primer dimers or unspecific PCR products was confirmed by melting curve analysis. PCR products were quantified against an internal standard generated by diluting a cDNA sample (5-fold dilution series from 1 to 0.008) prepared from SUC2::MYB30 expressing Col plants grown in LD. Standard deviations were calculated from three technical replicates. PP2A was used for sample normalization.
Semi-quantitative PCR was performed using a thermal PCR cycler (Eppendorf). 10 µl PCR reactions included cDNA corresponding to 70 ng RNA template, 10 pmole forward primer andreverse primer, 1×reaction buffer and 0.5 U LA Taq DNA polymerase (Takara). PCR products MYB30 and PP2A were between 200–300 bp, and the program was composed of step 1∶95°C, 3 min; step 2: [95°C 10 sec; 58°C 10 sec; 72°C, 20 sec] 25 cycles.
GUS Histochemical Staining
Young seedlings were incubated in 90% Acetone on ice for 30 min, rinsed with 50 mM sodium phosphate buffer and incubated for 24 hours at 37°C in GUS staining solution (0.5 mg/ml X-Gluc, 50 mM sodium phosphate buffer, 0.5 mM potassium ferrocyanide, 0.5 mM potassium ferricyanide, 0.1% Triton X-100). After incubation, samples were washed with 50 mM sodium phosphate buffer for 30 min and 70% ethanol several times until leaves turned white. The GUS staining was visualized and photographed under a stereomicroscope (Leica).
Results
Strong Expression of MYB30 in Phloem Companion Cells Promotes Flowering under LDs and SDs in Arabidopsis
The key flowering time components of the photoperiod pathway, CO and FT, are expressed in phloem companion cells of the leaf [6], [7], [45]. In a flowering time screen under LD conditions we used the SUCROSE-PROTON SYMPORTER 2 (SUC2) promoter, which drives expression in all phloem companion cells, to identify novel factors from a library of 800 transcription factors (TF) that have a potential to influence flowering from the phloem [46]; [43], [47], [48]. Two independent SUC2prom::MYB30 transformed lines (#1 and #2) flowered earlier than wild type (WT) in LD growth conditions (Figure S1). We selected line #2 for further analysis and confirmed the earlier flowering phenotype, which was even more pronounced under SDs compared to Col plants grown in the same condition (Figure 1a). To establish that MYB30 (AT3G28910) was expressed in phloem companion cells in wild-type plants, we generated MYB30prom::GUS reporter lines in the Col background. Strong GUS signal was observed in vascular tissues of leaves, hypocotyl and roots, but expression was not restricted to the phloem (Figure S2). In our growth conditions, two independent T-DNA insertion lines of myb30 (Figure 1b), which did not express full length MYB30 transcript (Figure 1c) showed similar flowering time to WT under both LD and SD conditions showing that MYB30 is not required to promote flowering in WT plants under these conditions (Figure 1d).
Figure 1. Flowering time of SUC2prom::MYB30 and myb30 mutant plants in LD and SD conditions.
(a)WT and SUC2prom::MYB30 expressing Col plants were grown in LDs (16 h light) and SDs (8 h light) in a temperature controlled climate chamber. The number of rosette and cauline leaves was counted to determine flowering time. Pictures were taken after the WT plants had started to bolt in each condition. Statistical significance was determined using the Student’s t-test (p<0.01). Significant differences are indicated by different letters above the bars, SD and LD treatments are tested as separate experiments. (b) Position of T-DNA insertion lines of myb30. A 400 bp PCR fragment used to detect expression is indicated. (c) Semi-quantitative RT-PCR confirms myb30 mutants as loss-of function alleles. (d) Flowering time of Col, myb30_1, myb30_2 and SUC2prom::MYB30 plants were measured in LD (left) and SD (right) conditions in a temperature controlled climate chamber. Statistical significance was determined using the Student’s t-test by comparing each genotype to the respective Col control (p<0.01). Significant differences are indicated by different letters above the bars. The number of plants for each genotype (n) is indicated below the graph.
Overexpression of MYB30 in the Phloem Increases FT and Reduces TSF Levels
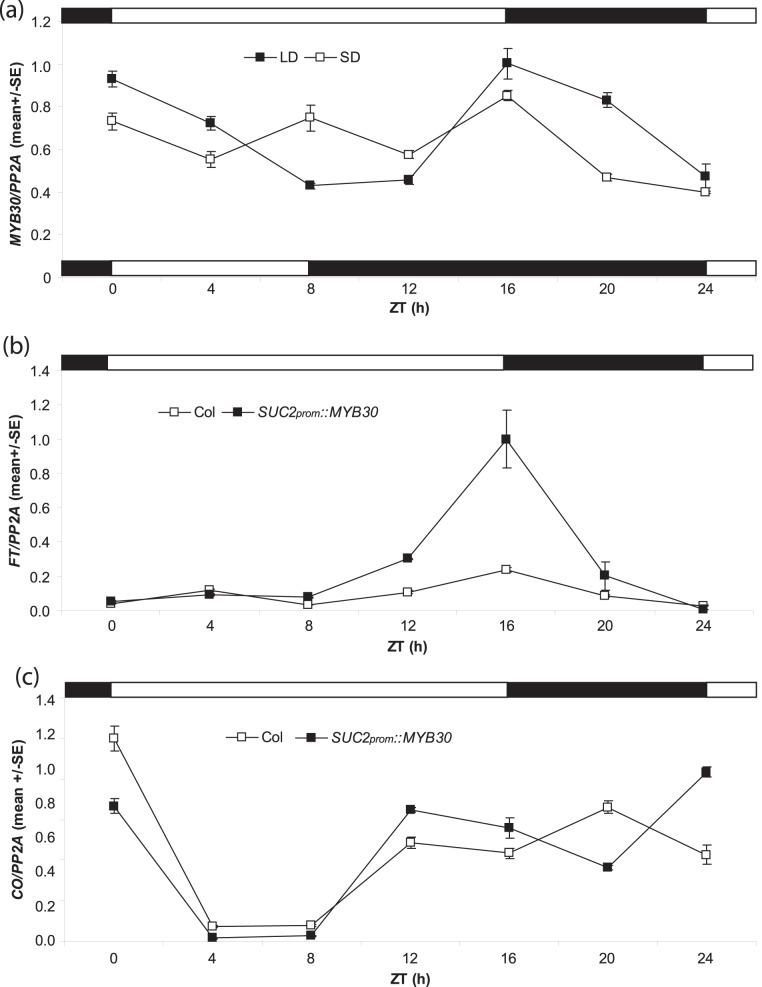
Day-time specific expression is a key feature of the photoperiodic pathway genes CO and FT. To test whether MYB30 transcripts accumulate rhythmically, we measured steady-state mRNA levels during 24 h cycles in LDs and SDs. WT seedlings showed a diurnal rhythm of MYB30 transcripts with a peak at ZT16 and a relatively similar pattern under LDs and SDs (Figure 2a). Since transcript levels were high throughout the day despite the observed diurnal pattern, MYB30 is likely to be present throughout the day unless the protein is subjected to post-transcriptional regulation. In LDs, MYB30’s highest expression overlaps with the strongest accumulation of CO protein.
Figure 2. Diurnal expression of MYB30, FT and CO.
(a) Diurnal expression pattern of MYB30 was measured in Col grown in LDs and SDs by RT-qPCR. Total RNA was prepared from 10-day old seedlings sampled every 4 hours (ZT0-ZT24). (b) FT expression was measured comparing WT to SUC2prom::MYB30 expressing Col plants. 10-day-plants in LDs were collected every 4 hours from ZT0-ZT24. (c) CO expression was measured comparing WT to SUC2prom::MYB30 expressing Col plants. 10-day-plants in LDs were collected every 4 hours from ZT0-ZT24. Error bars represent the standard error of three technical replicates relative to the expression of PHOSPHATASE 2A (PP2A). The experiment was repeated three times with similar results.
Next, we tested whether expression of flowering time marker genes such as FT, TSF and CO changed in SUC2prom::MYB30 plants. We measured steady-state mRNA levels during 24 h cycles in LDs. FT transcripts at ZT16 were ∼5 times enriched comparing SUC2prom::MYB30 to wild-type Col plants (Figure 2b and Figure S3a). An increase of FT expression can likely contribute to the earlier flowering time of SUC2prom::MYB30 under LDs. In contrast, CO mRNA did not change significantly, especially during daytime (Figure 2c and Figure S3b). This suggested that SUC2prom::MYB30 activated flowering through transcriptional regulation of FT that was not conveyed by changes in CO mRNA levels.
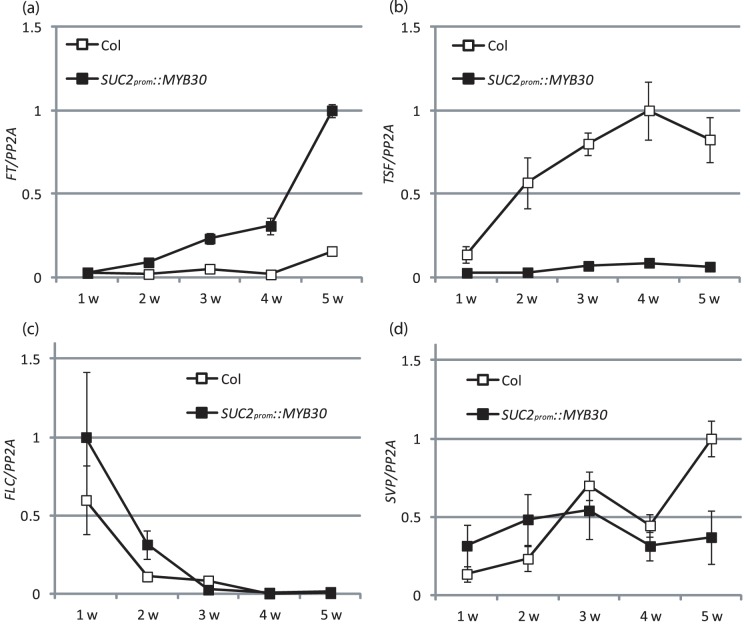
We also measured the expression level of different flowering-time genes during the course of Arabidopsis development. With increasing age from one to five weeks, FT mRNA transcripts accumulated to significantly higher levels in SUC2prom::MYB30 plants (Figure 3a), whereas SVP and FLC mRNA did not obviously differentiate the genotypes during the first 4 weeks. Therefore, increase of FT mRNA by strong MYB30 expression in the phloem does not correlate with reduced FLC and SVP levels. At 5 weeks SVP is differentially expressed in SUC2prom::MYB30 and Col plants, which could be due to earlier transition into flowering that is observed in the transgenic plants. Surprisingly, TSF mRNA showed a pattern opposite to FT in that it was down-regulated in SUC2prom::MYB30 plants during development (Figure 3b). Thus, FT and TSF, which are both part of ‘florigen’ perform agonistically in SUC2prom::MYB30 expressing plants grown in LDs.
Figure 3. Expression of flowering time genes in SUC2 prom ::MYB30 expressing plants during development.
Material was collected once per week at ZT16 from WT and SUC2prom::MYB30 expressing Col plants grown in LDs in climate chamber. Samples of 1 week and 2 week old plants were collected from all aerial parts, those of 3–5 weeks from leaves. Expression levels were measured by RT-qPCR for FT (a), TSF (b), FLC (c), and SVP (d). Values are shown as Mean ± SD after normalization of expression with values obtained for PP2A for technical triplicates. A biological replicate of the experiment gave similar results.
Ectopic Expression of MYB30 Accelerates Flowering and Impacts FT and TSF Expression in Absence and Presence of CO
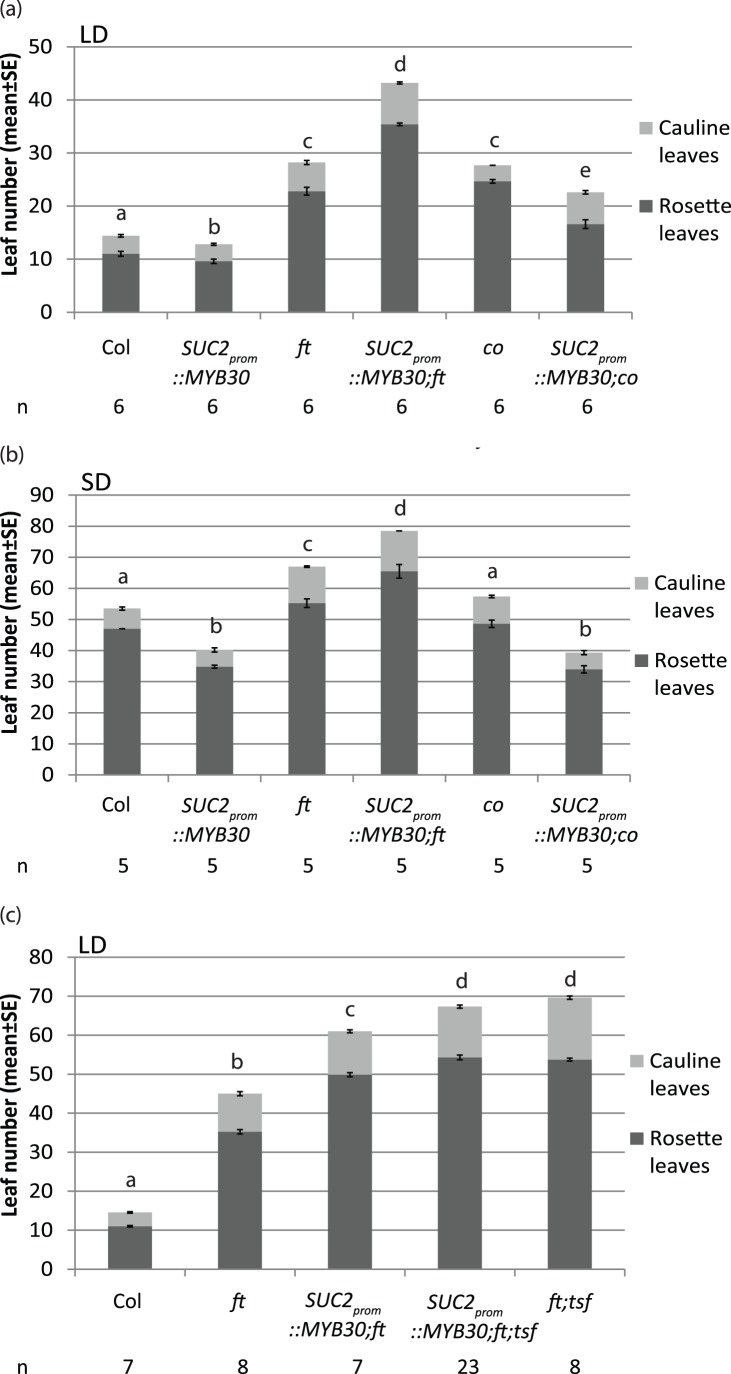
In the photoperiod pathway, CO is the major activator of FT and TSF [26]. To answer whether ectopic expression of MYB30 accelerates flowering via the photoperiod pathway, we crossed SUC2prom::MYB30 plants to ft and co mutants. SUC2prom::MYB30;ft double mutants flowered even later than ft single mutants under LDs, whereas SUC2prom::MYB30;co flowered obviously earlier than co mutants (Figure 4a). This corroborated the observation that MYB30 acts upstream of FT and suggested that other genes are affected that caused an enhanced late flowering phenotype in the ft-10 mutants. Since ectopic expression of MYB30 can accelerate flowering in the co mutant, it can be concluded that MYB30 does not require CO to impact flowering although plants with defective co flowered much later than wild-type plants. Accelerated flowering in SUC2prom::MYB30 was also observed under SDs (Figure 4b), which indicated that MYB30 can promote flowering independent of the photoperiod.
Figure 4. SUC2prom::MYB30 expression accelerates flowering dependent on FT and independent on CO.
Flowering time measurement of Col, SUC2prom::MYB30, ft, SUC2prom::MYB30;ft, co and SUC2prom::MYB30;co under LDs (a) and SDs (b) in the greenhouse. Plants SUC::MYB30;ft and SUC2prom::MYB30;ft;tsf were grown in LDs greenhouse (c). Statistical significance was determined using one way Analysis of Variance (ANOVA) followed by multiple comparison of Holm-Sidak method (p<0.01). Significant differences are indicated by different letters above the bars. The number of plants for each genotype (n) is indicated below the graph.
SUC2prom::MYB30;ft flowered later than ft plants in both LD and SD growth conditions (Figure 4a, b). Considering that TSF was down-regulated in SUC2prom::MYB30, TSF was a candidate gene to explain an enhanced late flowering phenotype in SUC2prom::MYB30;ft plants. To further test this, we generated SUC2prom::MYB30;ft;tsf triple mutants, which showed a flowering time that was not significantly different to that of ft;tsf double mutants (Figure 4c and Figure S4). This confirms that altered TSF transcription likely explains the flowering time differences between ft, SUC2prom::MYB30;ft and SUC2prom::MYB30;ft;tsf plants. In sum, FT and TSF are both downstream factors of MYB30, but while FT is activated, TSF is repressed.
FT transcript levels were ∼8 times higher in SUC2prom::MYB30;co plants compared to co single mutants under LDs, which is similar to the ∼5-fold increase observed in SUC2prom::MYB30 plants compared to WT (Figure 5a). Therefore, the effect of SUC2prom::MYB30 on FT expression is additive to that of CO, which is obviously much stronger (∼40-fold at ZT16). This suggests that under LDs, although CO is the main activator of FT, MYB30 can activate FT through a parallel pathway (Figure 5a).
Figure 5. Expression of FT and TSF in WT, ft and co plants.
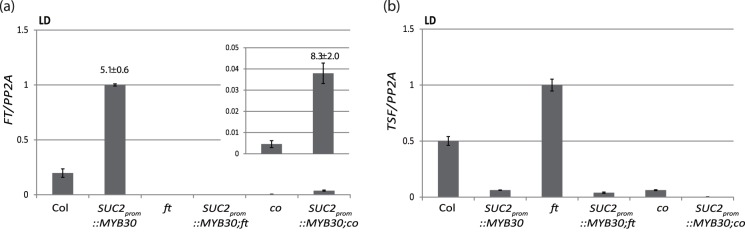
Material from 13-day-seedlings grown on soil in LDs in the greenhouse was collected at ZT16. FT (a) and TSF mRNA (b) levels were determined by RT-qPCR in different genotypes as indicated. Insert in (a) shows values for co and SUC2prom::MYB30;co at a lower scale. Fold-change FT expression in the SUC2prom::MYB30 lines compared to the respective control is indicated above the bar. Values are shown as Mean ± SD after normalization of expression with values obtained for PP2A. Experiments were repeated twice with similar results.
MYB30 Induces 1.0 kb FT Promoter Activity in Transient Bombardment Assay of Leaves
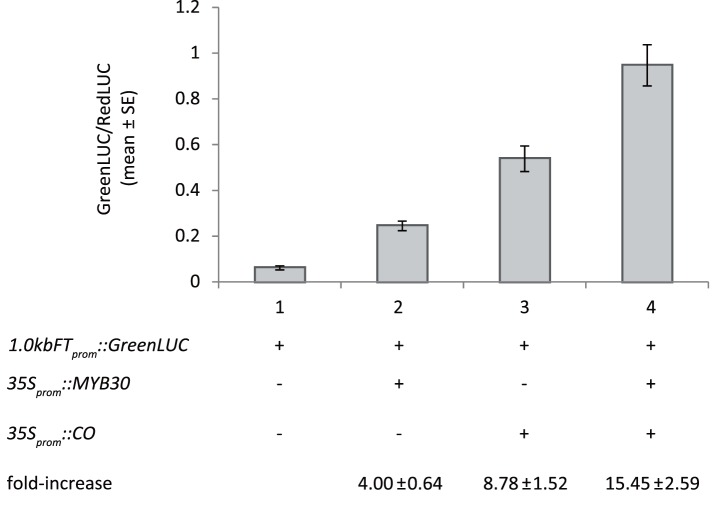
To test FT promoter activity in association with MYB30 and CO, we used transient bombardment of Arabidopsis leaves. In this assay, a 1 kb FT promoter was previously shown to be inducible by CO [7]. Co-bombardment of either CO or MYB30 alone with the 1 kb FT reporter showed that MYB30 alone could activate the expression of the reporter although the effect of CO was stronger (Figure 6). Combining MYB30 and CO in the bombardment additively increased the fold-expression of the promoter over the control (Figure 6). Thus, the bombardment assays confirmed that MYB30 and CO can affect FT expression in parallel pathways.
Figure 6. MYB30 increases 1.0 kb FT promoter activity.
Leaves from Col plants grown in SDs were bombarded with particles carrying 1.0kbFTprom::GreenLUC combined with 35Sprom::MYB30,35Sprom::CO or both as indicated. 35Sprom::RedLUC was included to measure transformation efficiencies. Values are shown as relative GreenLUC compared to RedLUC signals (top panel) after a 16 h–24 h incubation in constant light conditions. Fold-induction over the baseline level obtained for 1.0kbFTprom::GreenLUC alone is indicated in the table below the graph. Values are averages of measurements of five independent leaves from two technical replicates of one bombardment experiment. The experiment was repeated three times with similar results.
MYB30 Promotes Flowering Independently of SA Levels and FLC
MYB30 has previously been shown to be a positive regulator of pathogen defense, which is in part explained through increased accumulation of SA upon increased MYB30 expression [38]. In addition, MYB30 may directly regulate genes in the SA signaling pathway. SA was shown to be required for accelerated flowering observed in siz1 mutants [34] and the absence of SA in NahG plants was shown to delay flowering compared to Col-0 plants in LD and SD [32]. To test whether MYB30 promotes flowering through an SA pathway and dependent on FLC, we introduced the SUC2prom::MYB30 construct into NahG and flc3 mutant backgrounds by crossing. In our LD growth conditions, flowering was not changed significantly in the SA-deficient 35Sprom::NahG plants as compared to WT. In addition, SUC2prom::MYB30;35Sprom::NahG plants flowered as early as SUC2prom::MYB30 plants, which suggested that MYB30 accelerated flowering independently of its reported effect on increasing SA levels (Figure 7a). Expression analysis of FT, TSF and the pathogenesis response marker gene PR1 further supported the flowering-time data and confirmed a role of MYB30 in inducing PR1 expression (Figure S5). FT was super-induced at ZT16 in SUC2prom::MYB30;35Sprom::NahG plants confirming that SA was not required for the effect of MYB30 on FT expression (Figure S5). As before, strong MYB30 expression in the phloem repressed the expression of TSF, a response which was also detected in the NahG background (Figure S5).
Figure 7. SUC2prom::MYB30 promotes flowering independently of SA and FLC.
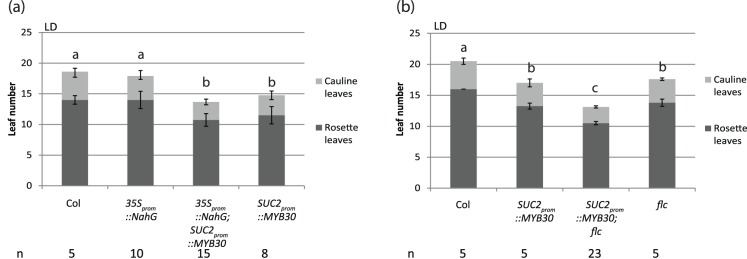
(a) Flowering time measurement of Col, NahG, SUC2prom::MYB30 and NahG;SUC2prom::MYB30 under LD conditions. (b) Flowering time measurement of Col, SUC2prom::MYB30, flc and SUC2prom::MYB30;flc under LD conditions. Statistical significance was determent using one way Analysis of Variance (ANOVA) followed by multiple comparison with the Holm-Sidak procedure (p<0.01). Significant differences are indicated by different letters above the bars. The number of plants for each genotype (n) is indicated below the graph.
As indicated by the unaltered expression of FLC in SUC2prom::MYB30 expressing plants (Figure 3), early flowering mediated by MYB30 was not dependent on FLC. Compared to the flowering observed in both parents, SUC2prom::MYB30;flc3 double mutants showed an additive early flowering phenotype (Figure 7b).
In conclusion, increased expression of MYB30 in the phloem can modulate flowering by increasing the expression of FT through a pathway that acts either independent of or additively to CO. Increased FT expression by MYB30 is independent of flc and does not require the presence of SA.
Discussion
Working Model for the Effect of MYB30 on Flowering Time
MYB30 can enter the flowering-time regulatory network by influencing the transcription rate of FT. As suggested by transient expression assays, the activation of FT by MYB30 requires sequences contained in the proximal 1 kb of the FT promoter. It is unclear, whether FT is a direct target of MYB30 or whether other factors mediate between the transcription factor and FT. In transgenic plants, MYB30 activates FT in absence of CO approximately 5-fold and a co-activation leads to a 5-fold higher induction of FT over CO-induced levels. This additive induction indicates that CO and MYB30 represent parallel inputs for FT induction. Although MYB30 overexpression reportedly leads to increased SA levels, which have been shown to accelerate flowering, we showed that MYB30-mediated induction of FT in the phloem does not depend on the presence of SA (Figure 7; [32], [39]). Furthermore, neither SVP nor FLC transcript levels are significantly altered in SUC2prom::MYB30 plants at different developmental stages and we have shown that the induction of FT by MYB30 is independent of FLC (Figures 3 and 7).
Opposed Effects of MYB30 on the Expression of FT and TSF
Under LD conditions, FT and TSF are affected by MYB30 overexpression but their transcripts are regulated in opposite patterns in SUC2prom::MYB30 plants (Figure 5). The accelerated flowering observed in wild-type Col and co mutant plants expressing SUC2prom::MYB30 regulate is explained through an activation of FT, which is epistatic to the repression of TSF. In contrast, the delay of flowering visible in SUC2prom::MYB30;ft mutant plants is explained by the repression of TSF (Figure 4 and 8).
FT and TSF are paralogous genes in Arabidopsis and both encode for proteins that contribute to the florigen function [3], [26], [27], [28]. In LDs, genetic defects of FT are epistatic to those of TSF and this is correlated with the significantly higher expression levels of the former in LDs. Both genes show substantial conservation in their proximal promoter sequences, but TSF lacks the distal enhancer that is important for high activation of FT by CO [7]. Nevertheless, FT and TSF are usually co-regulated and share CO as upstream activator as well as SVP and FLC as transcriptional repressors [26], [28]. Based on promoter::GUS reporter plants, both genes are predominantly expressed in the phloem but FT expression is usually restricted to the distal leaf veins, whereas TSF mainly expresses in the veins of hypocotyls and leaf petioles [28].
MYB30 is the first upstream factor that regulates FT and TSF transcript levels in an obvious opposite way, where FT is up-regulated and TSF is down-regulated (Figure 3). FT and TSF have redundant and independent roles in the floral transition. For example, TSF but not FT is required for the acceleration of flowering in response to external cytokinin application and only TSF expression is increased in these conditions [49].
High FT levels may also cause TSF repression. Such a negative feed-back has been observed in a previous study where TSF was down-regulated in plants that contained an activation-tagged allele of FT. This is also supported by our observation that TSF levels were increased in the ft mutant compared to WT (Figure 5b). In contrast, FT expression was not obviously changed when TSF was activated [26], [27]. Neither ft nor tsf loss-of function mutants show altered expression of their respective paralog but this could be due to the small overlap of their expression domains [28].
MYB30 as a New Component in Flowering Time Control?
Our data show that high levels of the transcription factor MYB30 in the phloem companion cells affect flowering time (Figure 1). Based on GUS signals controlled by the upstream intergenic region of MYB30, the gene is present in the phloem in wild-type plants (Figure S3). In addition, the diurnal expression pattern of MYB30 mRNA levels showed a peak at ZT16 in LDs, which overlaps with the moment of the strongest FT induction by CO in LDs (Figure 2; [15], [50]). Since MYB30 expression is induced by biotic stress [38], it is possible that accelerated flowering observed under stress condition may be dependent on MYB30. Stress accelerated flowering has been reported in Arabidopsis as response to nutrient depletion [51], pathogen perception [52], temperature [53], UVC-stress and external SA application [32]. In Pharbitis nil and Lemna paucicostata, induction of flowering in response to nutrient depletion was abolished by the addition of aminooxyacetic acid, an effect that was reversed by adding SA externally [54], [55], [56]. However, we could not confirm the delayed flowering that was reported earlier for 35Sprom::NahG plants (Figure 7; [32]). In addition, we did not observe a delay in flowering in two independent myb30 loss-of-function mutants grown under normal LD and SD greenhouse conditions (Figure 1). It is probable that multiple factors act together to cause a distinct stress-induced acceleration of flowering and as long as these factors are not entirely known, different experimental observations are expected considering variant culture conditions between laboratories.
Alternatively, a more constitutive role of MYB30-like genes could be masked by redundancy within the large MYB transcription factor family. MYB30 is part of a clade of 10 proteins, with MYB96 and MYB94 as closest relatives [57]. Similarly, regulation of FT by CCAAT-box binding NF-Y complexes, which are encoded by multi-gene families, was first shown by ectopic expression and required the generation of multiple stacked loss-of-function lines to demonstrate their role as genuine components of the photoperiod pathway [16], [17], [47].
In conclusion, the multi-pathway regulator MYB30 seems well positioned to connect gene networks regulating flowering, hormone signaling and stress perception, because it regulates genes involved in these various pathways and perceives signal inputs from the stress-related pathways. Further work will first have to focus on determining the conditions under which cross-talk between the signals is strongest, which will greatly facilitate uncovering whether MYB30 truly plays a biologically relevant role in flowering.
Supporting Information
Two independent transgenic lines of SUC2prom::MYB30 flower early in LDs. Two transgenic lines of SUC2prom::MYB30 and Col-0 were grown in LDs, and their rosette and cauline leaves were counted. Statistical significance was determined using the Student’s t-test (p<0.01). Significant differences are indicated by different letters above the bars. The number of plants for each genotype (n) is indicated below the graph.
(EPS)
GUS staining of MYB30prom::GUS plant. 10-day-seedlings transformed with MYB30prom::GUS in LDs were stained in X-GLUC solution to detect GUS activity.
(EPS)
Diurnal expression of FT and CO –2nd biological replicate. (a) FT expression was measured comparing WT to SUC2prom::MYB30 expressing Col plants. 12-day-plants in LDs were collected every 4 hours from ZT0-ZT24. (b) CO expression was measured comparing WT to SUC2prom::MYB30 expressing Col plants. 12-day-plants in LDs were collected every 4 hours from ZT0-ZT24. Error bars represent the standard error of three technical replicates relative to the expression PP2A (AT1G13320).
(EPS)
Phenotype of SUC2prom::MYB30 ; ft;tsf plants grown in LDs.
(EPS)
SUC2prom::MYB30 promotes flowering independently of SA. FT (a), TSF (b) and PR1 (c) mRNA levels were measured in Col, NahG, SUC2prom::MYB30 and NahG;SUC2prom::MYB30 plants. Samples were collected at ZT 16 from 10-day old LD grown seedlings.
(EPS)
List of oligonucleotides used in this study.
(DOCX)
Funding Statement
The authors thank the Chinese Academy of Sciences (CAS) and the Max Planck Society (MPS) for funding. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Pin PA, Nilsson O (2012) The multifaceted roles of FLOWERING LOCUS T in plant development. Plant Cell Environ 35: 1742–1755. [DOI] [PubMed] [Google Scholar]
- 2. Andres F, Coupland G (2012) The genetic basis of flowering responses to seasonal cues. Nat Rev Genet 13: 627–639. [DOI] [PubMed] [Google Scholar]
- 3. Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, et al. (2007) FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033. [DOI] [PubMed] [Google Scholar]
- 4. Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K (2007) Hd3a protein is a mobile flowering signal in rice. Science 316: 1033–1036. [DOI] [PubMed] [Google Scholar]
- 5. Gomez-Mena C, Pineiro M, Franco-Zorrilla JM, Salinas J, Coupland G, et al. (2001) early bolting in short days: An Arabidopsis mutation that causes early flowering and partially suppresses the floral phenotype of leafy. Plant Cell 13: 1011–1024. [PMC free article] [PubMed] [Google Scholar]
- 6. Takada S, Goto K (2003) TERMINAL FLOWER2, an Arabidopsis homolog of HETEROCHROMATIN PROTEIN1, counteracts the activation of FLOWERING LOCUS T by CONSTANS in the vascular tissues of leaves to regulate flowering time. Plant Cell 15: 2856–2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Adrian J, Farrona S, Reimer JJ, Albani MC, Coupland G, et al. (2010) cis-Regulatory elements and chromatin state coordinately control temporal and spatial expression of FLOWERING LOCUS T in Arabidopsis. Plant Cell 22: 1425–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu L, Liu C, Hou XL, Xi WY, Shen LS, et al.. (2012) FTIP1 Is an Essential Regulator Required for Florigen Transport. Plos Biol 10. [DOI] [PMC free article] [PubMed]
- 9. Taoka K, Ohki I, Tsuji H, Furuita K, Hayashi K, et al. (2011) 14–3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 476: 332–U397. [DOI] [PubMed] [Google Scholar]
- 10. Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, et al. (2005) FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309: 1052–1056. [DOI] [PubMed] [Google Scholar]
- 11. Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, et al. (2005) Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309: 1056–1059. [DOI] [PubMed] [Google Scholar]
- 12. Yoo SK, Chung KS, Kim J, Lee JH, Hong SM, et al. (2005) CONSTANS activates SUPPRESSOR OFOVEREXPRESSION OFCONSTANS 1 through FLOWERING LOCUS T to promote flowering in Arabidopsis. Plant Phys 139: 770–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Torti S, Fornara F, Vincent C, Andres F, Nordstrom K, et al. (2012) Analysis of the Arabidopsis shoot meristem transcriptome during floral transition identifies distinct regulatory patterns and a leucine-rich repeat protein that promotes flowering. Plant Cell 24: 444–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tiwari SB, Shen Y, Chang HC, Hou YL, Harris A, et al. (2010) The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis-element. New Phytol 187: 57–66. [DOI] [PubMed] [Google Scholar]
- 15. Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, et al. (2004) Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303: 1003–1006. [DOI] [PubMed] [Google Scholar]
- 16. Kumimoto RW, Zhang Y, Siefers N, Holt BF 3rd (2010) NF-YC3, NF-YC4 and NF-YC9 are required for CONSTANS-mediated, photoperiod-dependent flowering in Arabidopsis thaliana. Plant J 63: 379–391. [DOI] [PubMed] [Google Scholar]
- 17. Kumimoto RW, Adam L, Hymus GJ, Repetti PP, Reuber TL, et al. (2008) The Nuclear Factor Y subunits NF-YB2 and NF-YB3 play additive roles in the promotion of flowering by inductive long-day photoperiods in Arabidopsis. Planta 228: 709–723. [DOI] [PubMed] [Google Scholar]
- 18. Sheldon CC, Rouse DT, Finnegan EJ, Peacock WJ, Dennis ES (2000) The molecular basis of vernalization: The central role of FLOWERING LOCUS C (FLC). Proc Natl Acad Sci U S A 97: 3753–3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Searle I, He YH, Turck F, Vincent C, Fornara F, et al. (2006) The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Gen Devel 20: 898–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li D, Liu C, Shen L, Wu Y, Chen H, et al. (2008) A repressor complex governs the integration of flowering signals in Arabidopsis. Dev Cell 15: 110–120. [DOI] [PubMed] [Google Scholar]
- 21. Lee JH, Yoo SJ, Park SH, Hwang I, Lee JS, et al. (2007) Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes Dev 21: 397–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kumar SV, Lucyshyn D, Jaeger KE, Alos E, Alvey E, et al. (2012) Transcription factor PIF4 controls the thermosensory activation of flowering. Nature 484: 242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Castillejo C, Pelaz S (2008) The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Curr Biol 18: 1338–1343. [DOI] [PubMed] [Google Scholar]
- 24. Sawa M, Kay SA (2011) GIGANTEA directly activates Flowering Locus T in Arabidopsis thaliana. Proc Natl Acad Sci U S A 108: 11698–11703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Song YH, Smith RW, To BJ, Millar AJ, Imaizumi T (2012) FKF1 Conveys Timing Information for CONSTANS Stabilization in Photoperiodic Flowering. Science 336: 1045–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jang S, Torti S, Coupland G (2009) Genetic and spatial interactions between FT, TSF and SVP during the early stages of floral induction in Arabidopsis. Plant J 60: 614–625. [DOI] [PubMed] [Google Scholar]
- 27. Michaels SD, Himelblau E, Kim SY, Schomburg FM, Amasino RM (2005) Integration of flowering signals in winter-annual Arabidopsis. Plant Physiol 137: 149–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yamaguchi A, Kobayashi Y, Goto K, Abe M, Araki T (2005) TWIN SISTER OF FT (TSF) acts as a floral pathway integrator redundantly with FT. Plant and Cell Physiology 46: 1175–1189. [DOI] [PubMed] [Google Scholar]
- 29. Wilson RN, Heckman JW, Somerville CR (1992) Gibberellin Is Required for Flowering in Arabidopsis-Thaliana under Short Days. Plant Physiol 100: 403–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Porri A, Torti S, Romera-Branchat M, Coupland G (2012) Spatially distinct regulatory roles for gibberellins in the promotion of flowering of Arabidopsis under long photoperiods. Development 139: 2198–2209. [DOI] [PubMed] [Google Scholar]
- 31. Davis SJ (2009) Integrating hormones into the floral-transition pathway of Arabidopsis thaliana. Plant Cell Environ 32: 1201–1210. [DOI] [PubMed] [Google Scholar]
- 32. Martinez C, Pons E, Prats G, Leon J (2004) Salicylic acid regulates flowering time and links defence responses and reproductive development. Plant J 37: 209–217. [DOI] [PubMed] [Google Scholar]
- 33. Miura K, Rus A, Sharkhuu A, Yokoi S, Karthikeyan AS, et al. (2005) The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc Natl Acad Sci U S A 102: 7760–7765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jin JB, Jin YH, Lee J, Miura K, Yoo CY, et al. (2008) The SUMO E3 ligase, AtSIZ1, regulates flowering by controlling a salicylic acid-mediated floral promotion pathway and through affects on FLC chromatin structure. Plant J 53: 530–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takeno K (2012) Stress-Induced Flowering. Abiotic Stress Responses in Plants: Metabolism, Productivity and Sustainability: 331–345.
- 36. Daniel X, Lacomme C, Morel JB, Roby D (1999) A novel myb oncogene homologue in Arabidopsis thaliana related to hypersensitive cell death. Plant J 20: 57–66. [DOI] [PubMed] [Google Scholar]
- 37. Vailleau F, Daniel X, Tronchet M, Montillet JL, Triantaphylides C, et al. (2002) A R2R3-MYB gene, AtMYB30, acts as a positive regulator of the hypersensitive cell death program in plants in response to pathogen attack. Proc Natl Acad Sci U S A 99: 10179–10184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Raffaele S, Vailleau F, Leger A, Joubes J, Miersch O, et al. (2008) A MYB transcription factor regulates very-long-chain fatty acid biosynthesis for activation of the hypersensitive cell death response in Arabidopsis. Plant Cell 20: 752–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Raffaele S, Rivas S, Roby D (2006) An essential role for salicylic acid in AtMYB30-mediated control of the hypersensitive cell death program in Arabidopsis. FEBS letters 580: 3498–3504. [DOI] [PubMed] [Google Scholar]
- 40. Li L, Yu XF, Thompson A, Guo M, Yoshida S, et al. (2009) Arabidopsis MYB30 is a direct target of BES1 and cooperates with BES1 to regulate brassinosteroid-induced gene expression. Plant J 58: 275–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zheng Y, Schumaker KS, Guo Y (2012) Sumoylation of transcription factor MYB30 by the small ubiquitin-like modifier E3 ligase SIZ1 mediates abscisic acid response in Arabidopsis thaliana. Proc Natl Acad Sci U S A 109: 12822–12827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Marino D, Froidure S, Canonne J, Ben Khaled S, Khafif M, et al. (2013) Arabidopsis ubiquitin ligase MIEL1 mediates degradation of the transcription factor MYB30 weakening plant defence. Nat Commun 4: 1476. [DOI] [PubMed] [Google Scholar]
- 43. Castrillo G, Turck F, Leveugle M, Lecharny A, Carbonero P, et al. (2011) Speeding cis-trans regulation discovery by phylogenomic analyses coupled with screenings of an arrayed library of Arabidopsis transcription factors. PLoS One 6: e21524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743. [DOI] [PubMed] [Google Scholar]
- 45. An H, Roussot C, Suarez-Lopez P, Corbesier L, Vincent C, et al. (2004) CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131: 3615–3626. [DOI] [PubMed] [Google Scholar]
- 46. Imlau A, Truernit E, Sauer N (1999) Cell-to-cell and long-distance trafficking of the green fluorescent protein in the phloem and symplastic unloading of the protein into sink tissues. Plant Cell 11: 309–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wenkel S, Turck F, Singer K, Gissot L, Le Gourrierec J, et al. (2006) CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell 18: 2971–2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fornara F, Panigrahi KC, Gissot L, Sauerbrunn N, Ruhl M, et al. (2009) Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev Cell 17: 75–86. [DOI] [PubMed] [Google Scholar]
- 49. D’Aloia M, Bonhomme D, Bouche F, Tamseddak K, Ormenese S, et al. (2011) Cytokinin promotes flowering of Arabidopsis via transcriptional activation of the FT paralogue TSF. Plant J 65: 972–979. [DOI] [PubMed] [Google Scholar]
- 50. Imaizumi T, Tran HG, Swartz TE, Briggs WR, Kay SA (2003) FKF1 is essential for photoperiodic-specific light signalling in Arabidopsis. Nature 426: 302–306. [DOI] [PubMed] [Google Scholar]
- 51. Kolar J, Senkova J (2008) Reduction of mineral nutrient availability accelerates flowering of Arabidopsis thaliana. J Plant Physiol 165: 1601–1609. [DOI] [PubMed] [Google Scholar]
- 52. Korves TM, Bergelson J (2003) A developmental response to pathogen infection in Arabidopsis. Plant Physiol 133: 339–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Balasubramanian S, Sureshkumar S, Lempe J, Weigel D (2006) Potent induction of Arabidopsis thaliana flowering by elevated growth temperature. Plos Genet 2: 980–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hatayama T, Takeno K (2003) The metabolic pathway of salicylic acid rather than of chlorogenic acid is involved in the stress-induced flowering of Pharbitis nil. J Plant Physiol 160: 461–467. [DOI] [PubMed] [Google Scholar]
- 55. Wada KC, Yamada M, Shiraya T, Takeno K (2010) Salicylic acid and the flowering gene FLOWERING LOCUS T homolog are involved in poor-nutrition stress-induced flowering of Pharbitis nil. J Plant Physiol 167: 447–452. [DOI] [PubMed] [Google Scholar]
- 56. Shimakawa A, Shiraya T, Ishizuka Y, Wada KC, Mitsui T, et al. (2012) Salicylic acid is involved in the regulation of starvation stress-induced flowering in Lemna paucicostata. J Plant Physiol 169: 987–991. [DOI] [PubMed] [Google Scholar]
- 57. Matus JT, Aquea F, Arce-Johnson P (2008) Analysis of the grape MYB R2R3 subfamily reveals expanded wine quality-related clades and conserved gene structure organization across Vitis and Arabidopsis genomes. BMC Plant Biol 8: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Two independent transgenic lines of SUC2prom::MYB30 flower early in LDs. Two transgenic lines of SUC2prom::MYB30 and Col-0 were grown in LDs, and their rosette and cauline leaves were counted. Statistical significance was determined using the Student’s t-test (p<0.01). Significant differences are indicated by different letters above the bars. The number of plants for each genotype (n) is indicated below the graph.
(EPS)
GUS staining of MYB30prom::GUS plant. 10-day-seedlings transformed with MYB30prom::GUS in LDs were stained in X-GLUC solution to detect GUS activity.
(EPS)
Diurnal expression of FT and CO –2nd biological replicate. (a) FT expression was measured comparing WT to SUC2prom::MYB30 expressing Col plants. 12-day-plants in LDs were collected every 4 hours from ZT0-ZT24. (b) CO expression was measured comparing WT to SUC2prom::MYB30 expressing Col plants. 12-day-plants in LDs were collected every 4 hours from ZT0-ZT24. Error bars represent the standard error of three technical replicates relative to the expression PP2A (AT1G13320).
(EPS)
Phenotype of SUC2prom::MYB30 ; ft;tsf plants grown in LDs.
(EPS)
SUC2prom::MYB30 promotes flowering independently of SA. FT (a), TSF (b) and PR1 (c) mRNA levels were measured in Col, NahG, SUC2prom::MYB30 and NahG;SUC2prom::MYB30 plants. Samples were collected at ZT 16 from 10-day old LD grown seedlings.
(EPS)
List of oligonucleotides used in this study.
(DOCX)