Abstract
On a large plasmid of Acinetobacter johnsonii strain XBB1 from hospital sewage, bla PER-1 and ISCR1 were found in a complex Tn402-like integron carrying an arr3-aacA4 cassette array. The integron was truncated by the same 439-bp miniature inverted-repeat transposable element (MITE) at both ends. bla PER-1 and its complex surroundings might have been mobilized by the MITEst into an orf of unknown function, evidenced by the presence of the characteristic 5-bp direct target repeats. The same 439-bp MITEs have also been found flanking class 1 integrons carrying metallo-β-lactamases genes bla IMP-1, bla IMP-5 and bla VIM-2 before but without ISCR1. Although the cassette arrays are different, integrons have always been truncated by the 439-bp MITEs at the exact same locations. The results suggested that MITEs might be able to mobilize class 1 integrons via transposition or homologous recombination and therefore represent a possible common mechanism for mobilizing antimicrobial resistance determinants.
Introduction
bla PER-1 encodes the extended-spectrum β-lactamase (ESBL) PER-1 conferring resistance to penicillins, cephalosporins and monobactams [1] and has been found in Aeromonas spp., Acinetobacter baumannii, Alcaligenes faecalis, Pseudomonas aeruginosa and the Enterobacteriaceae in Asia and Europe [2], [3]. The production of PER-1 by Gram-negative bacilli of clinical significance compromises the option for antimicrobial chemotherapy. It has been well established that the mobilization of antimicrobial resistance determinants such as ESBL-encoding genes is largely mediated by mobile genetic elements including insertion sequences, transposons, integrons and gene cassettes. Recently, miniature inverted-repeat transposable elements (MITEs), which are small non-autonomous mobile elements containing repeated sequences and are present in diverse bacteria [4], have also been found mediating the mobilization of antimicrobial resistance determinants [5]. bla PER-1 has been found in diverse genetic contexts and is generally located downstream of the insertion sequence ISPa12 [2]. However, bla PER-1 had not been found associated with MITEs before [2].
Acinetobacter johnsonii (Acinetobacter genospecies 7) is a bacterial species that has usually been found in the aquatic environment [6] but occasionally colonizes humans [7] or causes clinical infections [8]. A strain of A. johnsonii was recovered from hospital sewage in western China and was found carrying bla PER-1. Here the genetic context of bla PER-1 was examined in detail.
Materials and Methods
Strain and the Detection of bla PER
A. johnsonii XBB1 was obtained from sewage collected at the influx of the wastewater treatment plant in West China Hospital, Chengdu, China, on October 2010 and was identified as A. johnsonii by partially sequencing the 16S rRNA gene [9]. No specific permissions were required for the activities to obtain the sewage as the wastewater treatment plant is a closed unit in the hospital and the field studies did not involve endangered or protected species. A. johnsonii XBB1 was resistant to ceftriaxone and ceftazidime [9] and the presence of bla PER gene was detected using PCR with self-designed primers (PER-UF, 5′-CCTGACGATCTGGAACCTTT; PER-UR, 5′-TCATCGASGTCCAGTTTTGA). PCR was performed using ExTaq premix (Takara, Dalian, China) and the following conditions: 94°C for 5 min; 30 cycles of 94°C for 30 s, 55°C for 30 s, 72°C for 1 min; and a final elongation step at 72°C for 7 min.
Genome Sequencing and Assembly
A. johnsonii XBB1 was subjected to the whole genome sequencing. The genomic DNA of XBB1 was prepared using the QIAamp DNA Mini kit (Qiagen, Hilden, Germany) and was then sequenced using the Roche 454 GS FLX+ (Roche 454 Life Sciences, Branford, CT, USA) and the Illumina Paired-end (Illumina, San Diego, CA, USA) platforms at the Beijing Genomics Institute (Beijing, China). Reads containing at least 50-bp non-adapter non-primer sequence generated by the 454 platform were assembled using the Newbler program (version 2.6) using 40 bp as the minimum overlap length and 90% as the minimum overlap identity. The assembled sequences were further assembled with reads generated by the Illumina platform using the Sspace program (version 2.0). The sequence gap of the plasmid carrying bla PER-1 was filled in using PCR with primers designed based on sequences available and Sanger sequencing using an ABI 3730×l DNA Analyzer (Applied Biosystems, Foster City, CA, USA). Similarity searches of sequences obtained were carried out using BLAST programs (http://www.ncbi.nlm.nih.gov/BLAST/) and the function of proteins was predicted using the InterProScan (http://www.ebi.ac.uk/Tools/pfa/iprscan/).
Verification of the Genetic Context of bla PER-1
To confirm the genetic context of bla PER-1, two overlapping long-range PCR were employed with primer pair PER-UF/DWMITE-F1 and PER-UR/UPMITE-R1. Self-designed primers DWMITE-F1 (5′-TGGCTCAATGTCTGATTGCT) and UPMITE-R1 (5′-TTGTTTGGGATTTGGTCCTC) were located 141 and 166 bp away from each end of the 15.3 kb large region containing bla PER-1, respectively (Figure 1). The conditions of long-range PCR (Fermentas, Burlington, ON, Canada) were 94°C for 2 min; 10 cycles of 94°C for 10 s, 55°C for 30 s, 68°C for 12 min; then 25 cycles of 94°C for 15 s, 55°C for 30 s, 68°C for 6 to 12 min plus 10 s cycle elongation for each successive cycle; and a final elongation step at 68°C for 7 min. The amplicons were sequenced at both directions (see above).
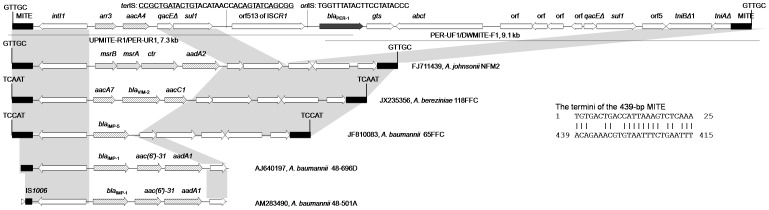
Figure 1. Structures flanked by the 439-bp MITE.
Two overlapping long-range PCR are shown by lines with primer names and amplicon sizes being indicated. The MITEs shown here have identical nucleotide sequences and are in the same direction. The 5-bp DR characteristics of the transposition of MITE-form composite transposon-like element are indicated. The GenBank accession numbers, the host species and strain number of each structure are listed. The identical regions are highlighted in grey. Only partial sequences are available for the integron carrying bla IMP-1 (GenBank accession numbers AM283490 and AJ640197). The oriIS and the putative terIS of ISCR1 are shown and the inverted repeats of terIS [23] are underlined. The 25-bp terminal sequences of this 439-bp MITE are shown with 18 bp matched.
Nucleotide sequences accession number
Sequence of the genetic context of bla PER-1 in A. johnsonii XBB1 has been deposited in GenBank as KF017283.
Results and Discussion
The genetic region containing bla PER-1 in XBB1 generated by assembling whole genome sequencing reads was verified by the overlapping long-range PCR and Sanger sequencing. In XBB1, bla PER-1 was carried by a 399 kb large plasmid, which had been completely sequenced, assembled and circularized. bla PER-1 was found downstream of the insertion sequence ISCR1 in a 5.7 kb region that included gts (encoding a glutathione S-transferase), abct (encoding an ABC-type transporter) and four genes of unknown function in a complex Tn402-like class 1 integron with the arr3 and aacA4 cassettes (Figure 1). The ISCR1-bla PER-1 structure was bracketed by two copies of 3′ conserved segment (3′CS), which comprised a truncated qacE gene encoding resistance to quaternary ammonium compounds and the sulphonamide resistance gene sul1. The 5′ conserved segment (5′CS) and the Tn402-like tni transposition module (tniBΔ1-tniA) were truncated by the same 439-bp MITE) (Figure 1). The two MITEs, which were in the same direction, might have formed a composite transposon-like mobile element mediating the transposition of the complex Tn402-like class 1 integron containing ISCR1 and bla PER-1 into a gene of unknown function as evidenced by the presence of characteristic 5 bp (GTTGC) direct target repeats (DR, Figure 1).
This MITE is AT-rich (AT content, 72.2%), has no orf and possesses terminal sequences without significant matches to inverted repeats of any known insertion sequences and transposons. The same (100% nucleotide identity) 439-bp MITEs have also been found flanking Tn402-like class 1 integrons carrying different cassette arrays but without ISCR1 in several Acinetobacter strains (Figure 1). In A. johnsonii strain NFM2 that was recovered from a prawn in Australia [10], the 439-bp MITEs flanked an unusual class 1 integron that carried methionine sulfoxide reductase genes and this MITE-flanked element was inserted into a location exactly the same as that seen in the present study. In A. baumannii clinical strain 65FFC, MITEs flanked a class 1 integron containing a single bla IMP-5 cassette and this MITE-flanked structure was transposed into the transposase-encoding tnpA gene of an ISAba14-like element, generating 5-bp (TCCAT) DR [5]. In Acinetobacter bereziniae strain 118FFC (GenBank accession number JX235356), a class 1 integron with the aacA7-bla VIM-2-aacC1 cassette array was flanked by MITEs, which were mobilized into the transposase-encoding tnpA gene of IS26 evidenced by the presence of 5-bp (TCAAT) DR (Figure 1). Partial MITEs have also been found to truncate the 5′CS of a class 1 integron carrying the bla IMP-1-aac(6′)-31-aadA1 cassette array in A. baumannii strain 48–501A (AM283490) and strain 48–696D (AJ640197). Only partial MITE sequence is provided for strain 48–696D, while MITE is truncated by IS1006 in strain 48–501A. No sequences downstream of 3′CS are available for both strains and therefore it remains unknown whether there is another copy of MITE at the other end of In86. bla IMP-1, bla IMP-5 and bla VIM-2 are genes encoding class B metallo-β-lactamases. Of note, the MITE-flanked structures containing Tn402-like integrons have been seen interrupting different genes with the presence of DR, suggesting that the MITEs indeed mediated the mobilization of integrons. Interestingly, MITE always truncated the 5′CS and tniA at the same locations in all known sequences regardless of the cassette array and host species or strains, suggesting that the truncation of Tn402-like integrons by MITEs at either ends might have only occurred once and the different cassette arrays might therefore have resulted from homologous recombination between integrons. The association of MITEs with a complex class 1 integron containing ISCR1 was not seen before and the ISCR1-bla PER-1 region seen in the present study might have also been introduced by homologous recombination with the 5′CS or 3′CS serving as one homologous region and the tni module serving as the other required for double crossover. There is also evidence that circular molecules created by recombination in the duplicated 3′CS flanking regions containing ISCR-antimicrobial resistance gene [11]. Although the MITEs-flanked structures seen in strain NFM2 from Australia and A. johnsonii XBB1 from China had different genetic components, they were present at the same location, suggesting that the acquisition of the ISCR1-bla PER-1 region might have occurred later than the transposition mediated by MITEs. As mobile genetic elements, MITEs might be able to mobilize bla PER-1 into different plasmids within the host strain and then could generate various genetic scaffolds to facilitate the horizontal transfer of the ESBL gene between clinical isolates and those of an environmental origin. The presence of bla PER-1 within the mobile genetic element formed by MITEs from an environment isolate in a hospital setting is therefore of significance.
To date, bla PER-1 has been found in genetic contexts either associated with the insertion sequence ISPa12 or with ISCR1 (Table 1). ISPa12 and its close relative ISPa13 formed a composite transposon termed Tn1213, which realized the mobilization of bla PER-1 and a truncated gts remnant as evidenced by the presence of characteristic 8-bp DR. Several variants of Tn1213 have also been identified (Table 1), including IS6100 or ISPpu17 truncating ISPa12 and ISPrst1 inserting into Tn1213 [2], [12], [13]. Tn1213 has been found in various species in Europe and Asia and therefore appears to be a common mechanism mediating the mobilization of bla PER-1. In particular, Tn1213 and its variants were the only type genetic context of bla PER-1 identified in P. aeruginosa so far. Another type of association of ISPa12 with bla PER-1 has been seen in A. baumannii strain C.A. and Salmonella enterica, in which the insertion of ISPa12 at 57 bp upstream of bla PER-1 was independently of the bla PER-1 acquisition as the presence of the characteristic 8-bp DR flanking ISPa12 suggested transposition of ISPa12 on its own [2]. In these cases, the mechanisms for the mobilization of bla PER-1 remain undetermined. The association of ISCR1 with bla PER-1 has been found in two A. baumannii isolates (Table 1) before but no further sequence data were available to identify the integrons in which ISCR1 and bla PER-1 might be embedded and to demonstrate whether MITEs were also involved like the case seen in the present study. Of note, there are two spacer sizes between ISCR1 and bla PER-1, suggesting that the acquisition of bla PER-1 by ISCR1 might have occurred more than once. As the spacer between ISCR1 and bla PER-1 was longer than that between ISPa12 and bla PER-1 and gts was truncated in Tn1213, it might be reasonable to propose that the acquisition of bla PER-1 by ISPa12 is a more recent event when compared to the acquisition by ISCR1. Both Tn1213 and the ISCR1-bla PER-1 context have been identified in A. baumannii, suggesting that A. baumannii might be the host species in which different platforms for mobilizing bla PER-1 had been formed.
Table 1. Genetic contexts of bla PER-1 a.
| Genetic contextb | Species and strain | Country | IS-bla PER-1spacer (bp) | Accessionno. | Reference |
| IS Pa12 -IS Pa13 composite transposon c | |||||
| ISPa12-bla PER-1-gtsΔ-ISPa13 | A. baumannii AMA-1 | France | 13 | [2], [14] | |
| A. baumannii 1656-2 | Korea | 13 | CP001921 | [15] | |
| A. baumannii strains 7 and 8 | Belgium | 13 | [16] | ||
| Aeromonas media A72 | Switzerland | 13 | [3] | ||
| Klebsiella pneumoniae CS1711 | Korea | 13 | [17] | ||
| P. aeruginosa RNL-1 | France | 13 | AY779042 | [2], [18] | |
| P. aeruginosa MUL | France | 13 | [2] | ||
| P. aeruginosa 1 | Turkey | 13 | [2] | ||
| P. aeruginosa PER12 | Belgium | 13 | [2], [19] | ||
| P. aeruginosa 2622 | Poland | 13 | [2] | ||
| P. aeruginosa PA2345 | France | 13 | AY866517 | [20] | |
| ISPa12-bla PER-1-gtsΔ-ISPrst1-gtsΔ-ISPa13 | Providencia stuartii BEN | France | 13 | [2] | |
| IS6100- ISPa12Δ-bla PER-1-gtsΔ-ISPa13 | P. aeruginosa P66 | Greece | 13 | FR847979 | [12] |
| ISPpu17- ISPa12Δ-bla PER-1-gtsΔ-ISPa13 | Alcaligenes faecalis FL-424/98 | Italy | 13 | AJ627643 | [13] |
| IS Pa12 - bla PER-1- gts - abct -? | A. baumannii C.A. | Turkey | 57 | [2], [14] | |
| Serovar Typhimurium TUR | Turkey | 57 | [2], [21] | ||
| Serovar Typhimurium 147 | Turkey | 57 | [2] | ||
| IS CR1 - bla PER-1 | |||||
| MITE-intI1-arr3-aacA4-3′CS-ISCR1-bla PER-1- gts- abct-4orfs-3′CS-orf5-tniBΔ-tniAΔ-MITE | A. johnsonii XBB1 | China | 80 | This study | |
| ?-ISCR1-bla PER-1-gts-abct-? | A. baumannii NF161710 | China | 72 | JQ780836 | |
| A. baumannii NF812784 | China | 80 | GU944725 | [22] |
The bla PER gene in Aeromonas punctata strain 169 (GenBank accession number GQ871757) has also been annotated as bla PER-1 but it has two nucleotide differences from bla PER-1 (Z21957), specifying one amino acid substitution compared to PER-1. Therefore, this bla PER gene was not included.
?, sequence remains unknown.
ISPa12 is called as IS1387a in Tn5393d and ISPa23 in P. aeruginosa PA2345. ISPpu17 is annotated as IS1066 in Tn5393d and ISPa13 is termed as ISPa24 in P. aeruginosa PA2345.
Conclusion
A unique MITE-flanked complex Tn402-like integron carrying ISCR1 and bla PER-1 was revealed in an A. johnsonii strain from hospital sewage. Such a complex integron flanked by a pair of 439-bp MITEs has never been reported before. The 439-bp MITEs might be able to mobilize class 1 integrons via direct transposition or homologous recombination. It appears that MITEs could serve as a common mechanism mediating the mobilization of antimicrobial resistance genes. This study has prompted an additional survey for the presence of MITEs in clinical isolates of Acinetobacter spp.
Acknowledgments
The author is grateful to Xiaohui Wang, Xingzhuo Zhang and Yanyu Gao for technical support and Dr. Sally Partridge, Westmead Hospital, The University of Sydney, Australia for helpful discussion about ISCR1.
Funding Statement
This work was supported by a grant from the National Natural Science Foundation of China (project no. 81222025). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. No additional external funding was received for this study.
References
- 1. Erac B, Hosgor-Limoncu M, Ermertcan S, Tasli H, Aydemir S (2013) Prevalence of bla PER-1 and integrons in ceftazidime-resistant Gram-negative bacteria at a university hospital in Turkey. Jpn J Infect Dis 66: 146–148. [DOI] [PubMed] [Google Scholar]
- 2. Poirel L, Cabanne L, Vahaboglu H, Nordmann P (2005) Genetic environment and expression of the extended-spectrum β-lactamase bla PER-1 gene in gram-negative bacteria. Antimicrob Agents Chemother 49: 1708–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Picao RC, Poirel L, Demarta A, Petrini O, Corvaglia AR, et al. (2008) Expanded-spectrum β-lactamase PER-1 in an environmental Aeromonas media isolate from Switzerland. Antimicrob Agents Chemother 52: 3461–3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Delihas N (2008) Small mobile sequences in bacteria display diverse structure/function motifs. Mol Microbiol 67: 475–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Domingues S, Nielsen KM, da Silva GJ (2011) The bla IMP-5-carrying integron in a clinical Acinetobacter baumannii strain is flanked by miniature inverted-repeat transposable elements (MITEs). J Antimicrob Chemother 66: 2667–2668. [DOI] [PubMed] [Google Scholar]
- 6. Guardabassi L, Dalsgaard A, Olsen JE (1999) Phenotypic characterization and antibiotic resistance of Acinetobacter spp. isolated from aquatic sources. J Appl Microbiol 87: 659–667. [DOI] [PubMed] [Google Scholar]
- 7. Seifert H, Dijkshoorn L, Gerner-Smidt P, Pelzer N, Tjernberg I, et al. (1997) Distribution of Acinetobacter species on human skin: comparison of phenotypic and genotypic identification methods. J Clin Microbiol 35: 2819–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Penzak SR, Gubbins PO, Stratton SL, Anaissie EJ (2000) Investigation of an outbreak of gram-negative bacteremia among hematology-oncology outpatients. Infect Control Hosp Epidemiol 21: 597–599. [DOI] [PubMed] [Google Scholar]
- 9. Zong Z, Zhang X (2013) bla NDM-1-carrying Acinetobacter johnsonii detected in hospital sewage. J Antimicrob Chemother 68: 1007–1010. [DOI] [PubMed] [Google Scholar]
- 10. Gillings MR, Labbate M, Sajjad A, Giguere NJ, Holley MP, et al. (2009) Mobilization of a Tn402-like class 1 integron with a novel cassette array via flanking miniature inverted-repeat transposable element-like structures. Appl Environ Microbiol 75: 6002–6004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Partridge SR, Hall RM (2003) In34, a complex In5 family class 1 integron containing orf513 and dfrA10 . Antimicrob Agents Chemother 47: 342–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ranellou K, Kadlec K, Poulou A, Voulgari E, Vrioni G, et al. (2012) Detection of Pseudomonas aeruginosa isolates of the international clonal complex 11 carrying the bla PER-1 extended-spectrum β-lactamase gene in Greece. J Antimicrob Chemother 67: 357–361. [DOI] [PubMed] [Google Scholar]
- 13. Mantengoli E, Rossolini GM (2005) Tn5393d, a complex Tn5393 derivative carrying the PER-1 extended-spectrum β-lactamase gene and other resistance determinants. Antimicrob Agents Chemother 49: 3289–3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Poirel L, Karim A, Mercat A, Le Thomas I, Vahaboglu H, et al. (1999) Extended-spectrum β-lactamase-producing strain of Acinetobacter baumannii isolated from a patient in France. J Antimicrob Chemother 43: 157–158. [PubMed] [Google Scholar]
- 15. Park JY, Kim S, Kim SM, Cha SH, Lim SK, et al. (2011) Complete genome sequence of multidrug-resistant Acinetobacter baumannii strain 1656–2, which forms sturdy biofilm. J Bacteriol 193: 6393–6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Naas T, Bogaerts P, Bauraing C, Degheldre Y, Glupczynski Y, et al. (2006) Emergence of PER and VEB extended-spectrum β-lactamases in Acinetobacter baumannii in Belgium. J Antimicrob Chemother 58: 178–182. [DOI] [PubMed] [Google Scholar]
- 17. Bae IK, Jang SJ, Kim J, Jeong SH, Cho B, et al. (2011) Interspecies dissemination of the bla gene encoding PER-1 extended-spectrum β-lactamase. Antimicrob Agents Chemother 55: 1305–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nordmann P, Ronco E, Naas T, Duport C, Michel-Briand Y, et al. (1993) Characterization of a novel extended-spectrum β-lactamase from Pseudomonas aeruginosa . Antimicrob Agents Chemother 37: 962–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Claeys G, Verschraegen G, de Baere T, Vaneechoutte M (2000) PER-1 β-lactamase-producing Pseudomonas aeruginosa in an intensive care unit. J Antimicrob Chemother 45: 924–925. [DOI] [PubMed] [Google Scholar]
- 20. Llanes C, Neuwirth C, El Garch F, Hocquet D, Plesiat P (2006) Genetic analysis of a multiresistant strain of Pseudomonas aeruginosa producing PER-1 β-lactamase. Clin Microbiol Infect 12: 270–278. [DOI] [PubMed] [Google Scholar]
- 21. Casin I, Hanau-Bercot B, Podglajen I, Vahaboglu H, Collatz E (2003) Salmonella enterica serovar Typhimurium bla PER-1-carrying plasmid pSTI1 encodes an extended-spectrum aminoglycoside 6′-N-acetyltransferase of type Ib. Antimicrob Agents Chemother 47: 697–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang F, Wu K, Sun J, Wang Q, Chen Q, et al. (2012) Novel ISCR1-linked resistance genes found in multidrug-resistant Gram-negative bacteria in southern China. Int J Antimicrob Agents 40: 404–408. [DOI] [PubMed] [Google Scholar]
- 23.Partridge S (2012) Identifying terIS of ISCR1 and new ISCR elements. 22nd European Congress of Clinical Microbiology and Infectious Diseases. London, UK. pp. Abstract P1242.