Abstract
Objectives
Triazole resistance in Aspergillus fumigatus has been increasing. We explored the A. fumigatus azole resistance profiles in bronchoalveolar lavage (BAL) fluid samples from Danish patients examined for aspergillosis.
Methods
A total of 94 BAL samples from 87 patients were evaluated by galactomannan (GM) test and A. fumigatus CYP51A profiling by PCR.
Results
Aspergillus spp. were isolated from 27/48 (56.3%) cultured samples, including 23 A. fumigatus with one resistant strain (4.3%). Samples were classified into GM-positive (≥3.0), GM-intermediate (0.5 to <3.0) and GM-negative (<0.5) groups, where the CYP51A PCR was positive in 81.8% (36/44), 56.3% (18/32) and 38.9% (7/18) of samples, respectively. Nine CYP51A PCR-positive samples (9/61, 14.8%) were found to have mutations resulting in amino acid substitutions. M220V was detected from a sample culture positive for susceptible A. fumigatus and P216L was found in a culture-negative BAL sample. Conversely, no mutation was found in one sample culture positive for azole-resistant A. fumigatus. The tandem repeat/L98H mutation was not detected.
Conclusions
Our study shows that azole resistance in A. fumigatus can be cryptic and may go undiagnosed. The combination of improved culture/susceptibility tests and the direct molecular detection of resistance markers will facilitate prompt institution of appropriate antifungal therapy.
Keywords: A. fumigatus, azole resistant, GM, CYP51A PCR
Introduction
Aspergillus species cause a wide range of diseases including invasive aspergillosis (IA), chronic pulmonary aspergillosis and allergic syndromes. Triazole antifungal drugs represent first-line therapy for all forms of aspergillosis, comprising the only orally active group of antifungal drugs within the limited therapeutic options.1 Aspergillus fumigatus, the species accounting for ∼80% of invasive infections, is normally susceptible to triazole antifungal drugs;2 however, clinical failures have been increasingly reported to be associated with triazole-resistant A. fumigatus.3,4 The frequency of azole resistance in clinical A. fumigatus isolates by patient was 0% in 2002 and 2003, significantly increased to 17% in 2007, 14% in 2008 and 20% in 2009 at a referral centre for chronic aspergillosis in the UK.3,5 Azole resistance in Aspergillus can be detected by susceptibility tests. However, susceptibility testing is not performed for Aspergillus at many laboratories for clinical microbiology. Furthermore, aspergilli are notorious for their low culturability from the infection sites and culture-positive rates in confirmed IA typically range between 25% and ∼50%.6–11 Hence resistance rates found in clinical isolates may only represent the tip of the global Aspergillus azole resistance iceberg.
Target-site modification is the principal mechanism underlying azole resistance in A. fumigatus. The CYP51A gene encodes the target lanosterol 14α-demethylase, which catalyses a central step in the biosynthetic pathway of ergosterol, an essential cell membrane component of filamentous fungi. Mutations in CYP51A result in structural alterations to the enzyme, which are presumed to reduce or block the binding of drugs.12,13 Mutational hotspots confirmed to cause resistance have been characterized at amino acid positions G54, L98, G138, P216, M220 and G448, of which the L98 alterations require a tandem repeat (TR) in the promoter region of CYP51A to cause resistance.14,15 A wide range of CYP51A mutations have been described in resistant isolates although other mechanisms may also confer resistance. Yet the above-mentioned hotspots are dominant at many centres and this, in combination with the low culturability, suggests that highly sensitive molecular diagnostic tools may be a preferred approach to better understand the nature and the true frequency of azole resistance. In a recent study we determined resistance rates using classical culture and susceptibility testing versus direct PCR combined with detection of hotspot alteration from respiratory samples of patients with pulmonary Aspergillus infections in Innsbruck, Austria and Manchester, UK. Unexpectedly, we found that azole resistance hotspot mutations were detected at an astonishingly high rate of 55.1% in culture-negative PCR-positive samples.16 In a significant proportion of these samples a 34 TR or the L98H alteration alone or in combination was found, which was particularly surprising as the TR/L98H resistance mechanism is not the dominant one in the UK.3,5 In the light of these findings we further explored the A. fumigatus azole resistance profiles in bronchoalveolar lavage (BAL) fluid samples from patients with various underlying diseases in Denmark who were being examined for aspergillosis. Denmark was chosen as the TR/L98H resistance genotype has been found by culture in environmental and clinical samples.17,18
Methods
Sample collection, patient information and microbiological tests
A total of 94 BAL samples (from 87 patients) received at the mycology reference laboratory in Copenhagen (Statens Serum Institut [SSI]) for galactomannan (GM) testing were included in the study (Table S1, available as Supplementary data at JAC Online). All specimens were divided upon arrival in a laminar airflow cabinet and an aliquot for PCR testing frozen immediately. The ratio of 1 : 5 for samples with a GM index below and above 0.5 was used for sample selection. Clinical and mycological data were retrieved from the laboratory data management systems. Underlying diseases were categorized into the following groups: (i) cystic fibrosis (CF; n = 23); (ii) haematological disorders (HAEM; n = 22); (iii) pulmonary disorders other than CF (PULM; n = 38); and (iv) critically ill patients without CF or HAEM (ICU; n = 11) (Table 1). GM was tested using a commercial kit (Platelia Aspergillus, Bio-Rad, France) according to the recommendations of the manufacturer. For BAL samples, an index of <0.5 was considered negative, an index of 0.5 to <3.0 was considered intermediate and an index of ≥3.0 was considered positive for GM.19 Intermediate BAL GM results were classified as true positives when supported by additional microbiological data (e.g. positive culture or positive serum GM) in patients at risk for Aspergillus infection (Table S1, available as Supplementary data at JAC Online).
Table 1.
BAL and serum GM antigen test results by BAL GM index and patient category
| BAL GM result | No. of BAL samples (GM-positive serum samples/no. of serum samples) |
||||
|---|---|---|---|---|---|
| patient category |
Total | ||||
| CF | HAEM | PULM | ICU | ||
| Negative (GM index <0.5) | 3 (0/0) | 3 (0/3) | 10 (0/3) | 2 (0/0) | 18 (0/6) |
| Intermediate (GM index 0.5 to <3.0) | 13 (0/1) | 7 (1/7) | 12 (0/6) | 0 (0/0) | 32 (1/14) |
| subgroup classified as false positives | 1 (0/0) | 0 | 3 (0/2) | 0 | 4 (0/2) |
| subgroup classified as true positives | 12 (0/1) | 7 (1/7) | 9 (0/4) | 0 | 28 (1/12) |
| Positive (GM index ≥3) | 7 (0/2) | 12 (6/11) | 16 (4/8) | 9 (3/4) | 44 (13/25) |
| Total | 23 (0/3) | 22 (7/21) | 38 (4/17) | 11 (3/4) | 94 (14/45) |
Most of the BAL samples had been submitted to culture and microscopy either locally or at the SSI (Table S1, available as Supplementary data at JAC Online). Mycological investigations at the SSI included:
(i) Culture: BAL samples were centrifuged and the sediment resuspended in 500 μL of BAL and split between microscopy (using Blankophor) and culture [using Sabouraud agar (SSI Diagnostika, Hillerød, Denmark) and incubation at 37°C for a period of ≥3 days]. Plates were checked for growth every 24 h. Mould colonies were identified to species level according to macroscopic and microscopic criteria and thermotolerance at 48°C.20
(ii) Antifungal susceptibility testing: the microbroth dilution method for itraconazole, voriconazole and posaconazole was used according to EUCAST E.DEF 9.1 guidelines.21 Drug stocks and working solutions were prepared as described previously.18 MICs of azoles were determined visually as a no-growth endpoint at 48 h of incubation.
(iii) Serum/plasma GM testing: cut-off of ≥0.5.22
DNA extraction and CYP51A profiling
The volume of BAL samples varies in our collection. When available, 1 mL of BAL fluid was pelleted at 16 000 g for 10 min at 4°C. Otherwise, the entire volume of BAL fluid was pelleted under the same condition. DNA extraction was performed on pellets using MasterPureTM Yeast DNA Purification kit (Epicentre Biotechnologies, Madison, WI, USA) with modifications of adding an extra mechanical lysis after the cell lysis solution treatment. Briefly, the lysing mixture was processed on a FastPrepTM instrument (MP Biomedicals, Solon, OH, USA) in lysing matrix tubes (MP Biomedicals), after which proteinase K (100 mg/L) was added and cell lysates were incubated at 56°C for 45 min followed by a 10 min centrifugation at 16 000 g. The supernatant was then processed until the final elution according to the manual of the extraction kit. Final elution volume was 50 μL. Nuclease-free water was extracted in parallel as a negative extraction control.
Both pan-Aspergillus real-time PCR and nested PCR amplification for the A. fumigatus CYP51A gene were performed blinded on all samples. The pan-Aspergillus real-time PCR targets the 18S rDNA, using molecular beacon technology to detect Aspergillus spp.23 This earlier validated PCR assay was able to detect total nucleic acids from as few as four spores of A. fumigatus conidia with 96.7% amplification efficiency. It demonstrated superior sensitivity to culture for detection and burden quantification on BAL fluid samples and lung tissues from an experimental IA rat model.23 In the current study, a negative extraction control and human genomic DNA (100 ng/assay) were tested as negative PCR controls. Nested PCR was employed to amplify CYP51A due to the modest amount of Aspergillus DNA in most samples. The initial three PCR fragments cover the entire CYP51A coding region as well as the majority of the promoter region. Each fragment was subjected to two nested PCRs for smaller products as previously described.16 PCR amplicons were evaluated by real-time PCR assays with allele-specific molecular beacons directed at key single-nucleotide polymorphisms (SNPs) linked with azole resistance (G54, TR/L98, G138, M220 and G448).24 All results were confirmed by DNA sequencing.
Statistical analysis
One-way analysis of variance was used to compare three groups of continuous data. Dunn's test was used for multiple comparisons between each two groups. A P value <0.05 was considered statistically significant.
Results
GM testing and culture
Based on GM index there were 18 (19%) negative, 32 (34%) intermediate and 44 (46.8%) positive samples (Table 1). Intermediate samples were further classified as true positive [n = 28, median GM index 1.5 (range 0.7–2.9)] or false positive [n = 4, 1.3 (0.6–1.8)] according to a review of additional clinical and microbiological data (Table S1, available as Supplementary data at JAC Online). Thus, overall, 72 samples were considered indicative for Aspergillus disease/colonization and 22 were considered to derive from patients without Aspergillus infection (Table 1).
Of 48 cultured samples, Aspergillus spp. were isolated in 27/48 (56.3%) (Table 2), with the following species distribution: A. fumigatus (n = 23), A. flavus (n = 3) and A. terreus (n = 1). All A. fumigatus isolates exhibited an azole wild-type susceptibility pattern except for one, which was multiazole resistant (itraconazole >4 mg/L; posaconazole >1 mg/L; voriconazole >2 mg/L). The resistance rate of A. fumigatus was 1/23 (4.3%). One GM-negative sample was culture positive for Penicillium sp.
Table 2.
BAL GM index and culture result
| GM index | Culture |
Culture NDb | Total | |||||
|---|---|---|---|---|---|---|---|---|
| A. fumigatus | A. flavus | A. terreus | Penicillium sp. | negative | total (Aspergillus %a) | |||
| <0.5 | 0 | 0 | 0 | 1 | 6 | 7 (0) | 11 | 18 |
| 0.5 to <3.0 | 7 | 1 | 0 | 0 | 8 | 16 (50) | 16 | 32 |
| ≥3 | 16 | 2 | 1 | 0 | 6 | 25 (76) | 19 | 44 |
| Total | 23 | 3 | 1 | 1 | 20 | 48 (60) | 46 | 94 |
aPercentage of positive Aspergillus culture.
bCulture not done at the SSI (see Table S1, available as Supplementary data at JAC Online).
Pan-Aspergillus and A. fumigatus PCR results and CYP51A mutations
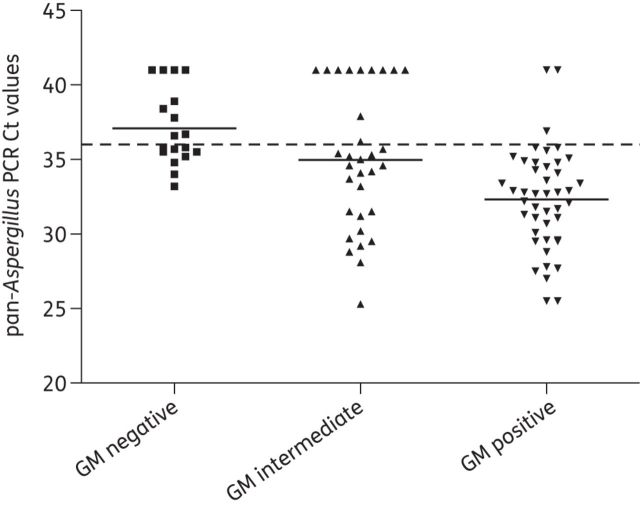
Using a Ct (threshold cycle) cut-off of 36, Aspergillus DNA was detected by pan-Aspergillus real-time PCR in 41/44 (93.2%) GM-positive, 21/32 (65.6%) GM-intermediate and 9/18 (50.0%) GM-negative BAL samples, with Ct values averaging 32.3 (IQR 29.7–34.7), 35.0 (31.3–41) and 37.1 (35.4–39.4), respectively (Figure 1 and Table 3). The mean Ct value of GM-positive samples was significantly lower than that of GM-negative (P < 0.05) and GM-intermediate (P < 0.05) samples, but there was no statistical difference in Ct values between GM-intermediate and GM-negative samples. The detection rate of pan-Aspergillus PCR was 92.0% (23/25), 81.3% (13/16) and 57.1% (4/7) in cultured GM-positive, GM-intermediate and GM-negative samples, respectively, compared with 76.0% (19/25), 50.0% (8/16) and 0.0% (0/7) by culture (Table 3). If PCR and culture results were evaluated using the classification of BAL samples into those indicative or not for Aspergillus disease/colonization, 84.7% (61/72) were PCR positive and 67.5% (27/40) culture positive, respectively, for samples associated with Aspergillus disease/colonization, whereas this was the case for 43.5% (10/23) and 0% (0/8) of the BAL samples from patients with no indication of Aspergillus disease or colonization.
Figure 1.
Distribution of pan-Aspergillus PCR Ct values by categories of GM index. The broken line represents a Ct cut-off of 36 and horizontal bars represent mean Ct values. The mean Ct value of GM-positive samples was significantly lower than that of GM-negative (P < 0.05) and GM-intermediate samples (P < 0.05).
Table 3.
Pan-Aspergillus PCR, A. fumigatus CYP51A PCR and culture results classified by BAL GM index
| PCR assays | GM index |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| positive (≥3.0) |
intermediate (0.5 to <3.0) |
negative (<0.5) |
|||||||||||||
|
Aspergillus culture |
cultured samples | culture NDa | total (%) |
Aspergillus culture |
cultured samples | culture NDa | total (%) |
Aspergillus culture |
cultured samples | culture NDa | total (%) | ||||
| neg. | pos. | neg. | pos. | neg. | pos. | ||||||||||
| Pan-Aspergillus PCR | |||||||||||||||
| negative | 2 | 0 | 2 | 1 | 3 (6.8) | 2 | 1 | 3 | 8 | 11 (34.4) | 3 | 0 | 3 | 6 | 9 (50.0) |
| positive | 4 | 19 | 23 | 18 | 41 (93.2) | 6 | 7 | 13 | 8 | 21 (65.6) | 4 | 0 | 4 | 5 | 9 (50.0) |
| total | 6 | 19 | 25 | 19 | 44 | 8 | 8 | 16 | 16 | 32 | 7 | 0 | 7 | 11 | 18 |
| CYP51A nested PCR | |||||||||||||||
| negative | 5 | 0 | 5 | 3 | 8 (18.2) | 3 | 1 | 4 | 10 | 14 (43.8) | 3 | 0 | 3 | 8 | 11 (61.1) |
| positive | 4 | 16 | 20 | 16 | 36 (81.8) | 6 | 6 | 12 | 6 | 18 (56.4) | 4 | 0 | 4 | 3 | 7 (38.9) |
| total | 9 | 16 | 25 | 19 | 44 | 9 | 7 | 16 | 16 | 32 | 7 | 0 | 7 | 11 | 18 |
aCulture not done at the SSI (see Table S1, available as Supplementary data at JAC Online).
When A. fumigatus CYP51A was profiled in specimens using nested PCR the detection rate was 81.8% (36/44), 56.3% (18/32) and 38.9% (7/18) in GM-positive, GM-intermediate and GM-negative samples, respectively, somewhat lower than that of pan-Aspergillus PCR (Table 3). Ten samples were positive in pan-Aspergillus PCR but failed to amplify in CYP51A PCR, of which two samples were positive by both culture and real-time PCR (data not shown) for A. flavus. One sample had a Ct of 30.2 in pan-Aspergillus PCR but was negative in species-specific real-time PCR for A. fumigatus, A. flavus and A. terreus (data not shown), indicative of other non-tested Aspergillus species. The remaining seven samples had an average Ct value of 34.2 (range 33.2–34.8). All but one of the A. fumigatus culture-positive BAL samples were CYP51A PCR positive and so were one A. flavus culture-positive BAL sample and one A. terreus culture-positive BAL sample. Two other A. flavus culture-positive BAL samples were negative in A. fumigatus CYP51A PCR. Positive CYP51A PCR was also obtained in 7/18 (38.9%) of GM-negative samples.
The majority of CYP51A PCR-positive samples (45/61, 73.8%) had synonymous mutations only (relative to sequenced wild-type strain Af293), whereas 9 (14.8%) were detected with single or multiple mutations resulting in amino acid substitutions (Table 4). No TR/L98H mutation was detected; neither was any mutation occurring at codon G54, G138 or G448. Alterations confirmed to be associated with azole resistance M220V and P216L were found in two individual patients. Interestingly, the M220V sample was culture positive for a susceptible A. fumigatus strain and the P216L sample was culture negative. Other mutations causing amino acid substitutions in the Cyp51A protein were novel, except M172V and F46Y, which were previously reported in both susceptible and resistant isolates.5,21
Table 4.
Mutations in the CYP51A gene of A. fumigatus detected directly from BAL fluid samples
| No. of samples | Amino acid substitutions caused by non-synonymous mutations | Polymorphisms caused by synonymous mutations | Culture/azole susceptibility |
|---|---|---|---|
| Samples carrying single or multiple non-synonymous mutations | |||
| 1 | N425S | A. fumigatus/susceptible | |
| 1 | M172Va, F46Ya | A. fumigatus/susceptible | |
| 1 | L206P, L210P | G89G | negative/NA |
| 1 | K67E | G89G | Penicillium species/POS = 1, VRC = 4, ITC > 4 |
| 1 | Y107C | D70D, G89G | negative/NA |
| 1 | M220Vb | G89G | A. fumigatus/susceptible |
| 1 | N33D, K80E | G89G | ND/NA |
| 1 | F41S, E66G | D70D, G89G | ND/NA |
| 1 | P216Lc | G89G | negative/NA |
| Samples carrying synonymous mutations only | |||
| 1 | G89G, F495F | ND/NA | |
| 4 | D70D, G89G | 3 ND; 1 negative/NA | |
| 1 | V44V, G89G | negative/NA | |
| 1 | G89G, F165F | A. fumigatus/susceptible | |
| 38 | G89G | one resistant A. fumigatus isolate; POS = 1, VRC = 2, ITC > 4 | |
NA, not available; ND, culture not done; ITC, itraconazole; POS, posaconazole; VRC, voriconazole.
bMutation confirmed to be associated with azole resistance in A. fumigatus;40 shown in bold due to its confirmed association with azole resistance in A. fumigatus.
cMutation confirmed to be associated with azole resistance in A. fumigatus;15 shown in bold due to its confirmed association with azole resistance in A. fumigatus.
Discussion
The prevalence of azole resistance in clinical A. fumigatus is largely undefined because susceptibility testing is not routinely performed in most clinical laboratories. Some variability was seen from 13 studies comprising almost 5000 isolates, with an average of 2% (0%–6%).14 In the present study 4.3% (1/23) of A. fumigatus isolates displayed an azole resistance phenotype, consistent with findings in Danish CF patients, where 4.5% of A. fumigatus isolates in respiratory samples had reduced susceptibility to azoles,18 and also consistent with findings from Denmark as part of the global SCARE study.25 However, a significant number of cases involving azole-resistant isolates may go unnoticed. It is well known that culture-positive rates in Aspergillus-infected patients are low (around 50%) even in confirmed cases of chronic pulmonary aspergillosis.26 In our study 76% of GM-positive and 50% of GM-intermediate BAL samples submitted for culture isolation were culture positive for Aspergillus, although the majority of these by review of the clinical and additional microbiological data were consistent with Aspergillus infection/colonization. To discover the cryptic resistance hidden in the non-culturable aspergilli we employed molecular diagnostic tools to detect azole resistance markers directly from the BAL samples.
A number of studies have shown that PCR-based detection of Aspergillus could be a valuable adjunctive tool for diagnosis of Aspergillus infections,27–30 although issues such as the risk of environmental/procedural contamination and standardization from sample preparation to data analysis need to be carefully addressed before its implementation in clinical practice.31,32 In our study the pan-Aspergillus PCR had a higher detection rate than culture, confirming the sensitivity of this diagnostic approach. Moreover, a fair correlation between GM index and Ct values was observed, which supports the incentive of using PCR as an adjunctive tool for diagnosis of Aspergillus infections. Of note, there were 10 samples positive in pan-Aspergillus PCR that failed to amplify in the A. fumigatus-specific CYP51A detection. Apart from two samples that were undoubtedly positive for A. flavus and one sample that was possibly positive for an Aspergillus species other than A. fumigatus, A. flavus or A. terreus, the remaining seven samples had relatively high Ct values, suggesting a low amount of Aspergillus DNA in these samples, which may account for the negative CYP51A PCR results. The lower detection rate of CYP51A PCR is not surprising given the species-specific nature of this PCR and, importantly, that the CYP51A gene is present only in a single copy in the genome33 while as many as 38–91 copies per genome of 18S rDNA34 are targeted by pan-Aspergillus PCR. The presence of A. fumigatus was confirmed in the majority of GM-positive and GM-intermediate BAL samples, including all but one of the A. fumigatus culture-positive specimens.
However, the finding of A. fumigatus DNA in 39% of the GM-negative specimens is less straightforward to interpret in the clinical context. Certainly, this number would be significantly reduced to 16.7% (3/18) if a lower Ct cut-off of 35.5 was adopted, with some sacrifice on the detection rate in GM-positive (dropping from 82% to 75%) and GM-intermediate (from 56% to 53%) samples. A recent study investigating the probability of aspergillosis after a BAL GM test found that a GM index cut-off of <0.5 corresponded to a high specificity, virtually always ruling out the disease.19 In light of this finding we cannot exclude the possibility that at least some of the positive PCR results among the GM-negative samples may reflect contamination or transient colonization rather than infection. In fact the high sensitivity of PCR may tend to overestimate disease burdens in non-sterile specimens such as BAL samples and underestimate them in sterile specimens like blood, where the presence of pathogen-specific DNA has been poorly defined. More controlled clinical studies are needed to define PCR threshold detection levels for various specimen types. Furthermore, the detection of specific expressed genes associated with hyphal growth may aid in distinguishing between contaminating ungerminated spores and rapidly growing infecting organisms causing infection.
Perhaps the most powerful application of molecular diagnostic technology is the rapid elucidation of genetic variants associated with resistance.35 Analysis of the CYP51A amplicons identified two mutations confirmed to be associated with azole resistance—M220V and P216L—in one culture-positive sample from a CF patient and one culture-negative sample from an elderly patient receiving meropenem for Streptococcus pneumoniae pneumonia, respectively. Interestingly, the sample harbouring the M220V alteration was culture positive for an azole-susceptible A. fumigatus isolate. It is plausible that a mixed population of both susceptible and resistant A. fumigatus existed in the same BAL sample, consistent with previous reports of patients harbouring both susceptible and resistant isolates.5,18 Together with the fact that the P216L alteration was found from a culture-negative BAL sample, both cases highlight the potential of detecting cryptic resistant A. fumigatus with the aid of molecular diagnostic tools. Because patients infected with resistant strains often respond poorly to therapy, the presence of a mixed (susceptible/resistant) population or an unknown resistant strain is expected to result in a diminished response to azole therapy, despite adequate therapy and favourable immune status of the patient. Molecular detection of azole resistance markers may therefore provide important information for clinical care.
A remarkably high rate of hotspot mutations causing azole resistance, primarily TR/L98H, was seen in the previous study with direct detection of resistance markers using our nested PCR and respiratory samples from patients with chronic aspergillosis in Manchester.16 This was not the case in the present study; no TR/L98H mutation was observed in the BAL collection from Danish patients, although this is the predominant mutation recovered from 8% of environmental A. fumigatus isolates in Denmark and has been found in Danish CF patients.17 The difference is possibly due to different sample types and different compositions of studied patient populations. Whereas sputum samples in the former study in the UK were collected from patients diagnosed with chronic or allergic aspergillosis, all of whom had positive serological tests for Aspergillus,16 BAL samples in the present study were from a wider spectrum of underlying diseases and inclusion was based on the GM index, with GM-negative BAL samples comprising 19% of the total specimens. The human lung may be a filter of airborne aspergilli, whereby what is found in the respiratory samples reflects recent exposure (i.e. light and transient colonization) as well as potentially acute or chronic infections. This may particularly be the case for sputum samples that include upper airway material and saliva and in patients who cannot clear this fungus from their respiratory tract. In the UK study a majority of the samples (24/29) tested for resistance markers had azole exposure, which might contribute to the high detection rate of azole resistance. However, 23 patients in the present study had underlying CF, a condition associated with allergic bronchopulmonary aspergillosis and frequent azole treatment in Denmark. For 13 patients (9 of whom were not CF patients), previous azole treatment was documented because samples from these patients had been received for therapeutic drug monitoring, suggesting ∼50% of the study population had been exposed to azoles.
Another key consideration here is whether the fungal flora in the upper airway is possibly more diverse, with wild-type and resistant strains often in coexistence. Such diversity and coexistence becomes less common when the flora migrates to the lower airway because wild-type strains are likely to outcompete the mutants in the more stringent environment of the lower respiratory tract, assuming the acquisition of a mutation driven by azole therapy carries a fitness cost in A. fumigatus. Reduced virulence of azole-resistant A. fumigatus isolates has been observed;36 however, it does not seem to apply to the TR/L98H mutation and in the scenario of ongoing or previous azole exposure the wild-type isolate may be less competitive.37 Nevertheless, further studies including different geographical areas and patient categories are warranted to better understand the impact of using different sample types to uncover the true frequency of azole resistance. However, at least currently it seems that the surprisingly high frequency of azole resistance mutations in non-culturable Aspergillus in the Manchester cohort, which potentially questions whether voriconazole is an appropriate first-line agent, is not a ubiquitous phenomenon.
Several novel mutations were also found in our study. Except for N425S, which was identified from a BAL sample culture positive for susceptible A. fumigatus, others (L206P + L210P; K67E; Y107C; N33D + K80E; F41S + E66G) were detected from A. fumigatus culture-negative BAL samples or where culture was not requested at our laboratory. Substitutions M172V and F46Y have previously been detected in both susceptible and resistant A. fumigatus isolates,14 but a later study demonstrated that isolates carrying serial polymorphisms (F46Y-M172V ± N248T ± D255E ± E427K) had higher voriconazole MICs, although ≤1 mg/L using EUCAST methodology.38 The clinical significance of and relationship between these polymorphisms and azole exposure needs to be explored. A silent mutation, G89G, was seen in 85% of CYP51A PCR-positive samples. Our finding echoes a recent study, which found that four of five clinical A. fumigatus isolates with at least one mutation in the CYP51A gene had G89G.39 Yet we cannot draw any conclusions on the clinical or phylogenetic significance of this mutation due to the limited data.
In conclusion, our study confirms that azole resistance in A. fumigatus can be cryptic and may go undiagnosed even when culture/susceptibility testing are performed. The presence of a mixed (susceptible/resistant) population may increase this risk. Under such circumstances a couple of precautions should be employed for standard culture and susceptibility testing in order to improve our capability to detect azole resistance in clinical A. fumigatus: (i) sampling several colonies for susceptibility tests to increase the chance of capturing the resistant strain in the mixed population and (ii) including an azole-containing screening agar to the culture isolation in order to detect resistant isolates even when present in a low proportion. These modifications might enhance the likelihood of detecting azole-resistant isolates in positive cultures independently of the nature of the underlying molecular mechanism. However, in order to detect resistant cases where fungal isolates are not available, rapid molecular detection of resistance markers directly from respiratory samples offers a useful tool. The combination of improved culture/susceptibility tests and the direct molecular detection of resistance markers will facilitate prompt institution of appropriate antifungal therapy.
Funding
This study was supported by internal funding.
Transparency declarations
D. S. P. has received past support from the US National Institute of Allergy and Infectious Diseases and has received past support from Pfizer, Merck and Astellas, and participates in expert panels for these companies. M. C. A. has been paid for talks on behalf of Astellas, Merck, Pfizer, Schering-Plough and Gilead, has acted as a consultant for Merck, Astellas, Spepharm, Pcovery and Pfizer, has received research grants from Astellas, Pfizer, Gilead and Merck, and has received travel grants from Astellas, Merck and Pfizer. Y. Z. and C.R. S.: none to declare.
Supplementary data
Table S1 is available as Supplementary data at JAC Online (http://jac.oxfordjournals.org/).
References
- 1.Walsh TJ, Anaissie EJ, Denning DW, et al. Treatment of aspergillosis: clinical practice guidelines of the Infectious Diseases Society of America. Clin Infect Dis. 2008;46:327–60. doi: 10.1086/525258. [DOI] [PubMed] [Google Scholar]
- 2.Messer SA, Jones RN, Fritsche TR. International surveillance of Candida spp. and Aspergillus spp. report from the SENTRY Antimicrobial Surveillance Program (2003) J Clin Microbiol. 2006;44:1782–7. doi: 10.1128/JCM.44.5.1782-1787.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bueid A, Howard SJ, Moore CB, et al. Azole antifungal resistance in Aspergillus fumigatus: 2008 and 2009. J Antimicrob Chemother. 2010;65:2116–8. doi: 10.1093/jac/dkq279. [DOI] [PubMed] [Google Scholar]
- 4.van der Linden JW, Snelders E, Kampinga GA, et al. Clinical implications of azole resistance in Aspergillus fumigatus, The Netherlands, 2007–2009. Emerg Infect Dis. 2011;17:1846–54. doi: 10.3201/eid1710.110226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Howard SJ, Cerar D, Anderson MJ, et al. Frequency and evolution of azole resistance in Aspergillus fumigatus associated with treatment failure. Emerg Infect Dis. 2009;15:1068–76. doi: 10.3201/eid1507.090043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horvath JA, Dummer S. The use of respiratory-tract cultures in the diagnosis of invasive pulmonary aspergillosis. Am J Med. 1996;100:171–8. doi: 10.1016/s0002-9343(97)89455-7. [DOI] [PubMed] [Google Scholar]
- 7.Einsele H, Quabeck K, Muller KD, et al. Prediction of invasive pulmonary aspergillosis from colonisation of lower respiratory tract before marrow transplantation. Lancet. 1998;352:1443. doi: 10.1016/s0140-6736(05)61265-2. [DOI] [PubMed] [Google Scholar]
- 8.Reichenberger F, Habicht J, Matt P, et al. Diagnostic yield of bronchoscopy in histologically proven invasive pulmonary aspergillosis. Bone Marrow Transplant. 1999;24:1195–9. doi: 10.1038/sj.bmt.1702045. [DOI] [PubMed] [Google Scholar]
- 9.Maschmeyer G, Beinert T, Buchheidt D, et al. Diagnosis and antimicrobial therapy of lung infiltrates in febrile neutropenic patients: guidelines of the infectious diseases working party of the German Society of Haematology and Oncology. Eur J Cancer. 2009;45:2462–72. doi: 10.1016/j.ejca.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 10.Nguyen MH, Leather H, Clancy CJ, et al. Galactomannan testing in bronchoalveolar lavage fluid facilitates the diagnosis of invasive pulmonary aspergillosis in patients with hematologic malignancies and stem cell transplant recipients. Biol Blood Marrow Transplant. 2011;17:1043–50. doi: 10.1016/j.bbmt.2010.11.013. [DOI] [PubMed] [Google Scholar]
- 11.Arendrup MC, Bille J, Dannaoui E, et al. ECIL-3 classical diagnostic procedures for the diagnosis of invasive fungal diseases in patients with leukaemia. Bone Marrow Transplant. 2012;47:1030–45. doi: 10.1038/bmt.2011.246. [DOI] [PubMed] [Google Scholar]
- 12.Snelders E, Karawajczyk A, Schaftenaar G, et al. Azole resistance profile of amino acid changes in Aspergillus fumigatus CYP51A based on protein homology modeling. Antimicrob Agents Chemother. 2010;54:2425–30. doi: 10.1128/AAC.01599-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fraczek MG, Bromley M, Bowyer P. An improved model of the Aspergillus fumigatus CYP51A protein. Antimicrob Agents Chemother. 2011;55:2483–6. doi: 10.1128/AAC.01651-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Howard SJ, Arendrup MC. Acquired antifungal drug resistance in Aspergillus fumigatus: epidemiology and detection. Med Mycol. 2011;49(Suppl 1):S90–5. doi: 10.3109/13693786.2010.508469. [DOI] [PubMed] [Google Scholar]
- 15.Camps SM, van der Linden JW, Li Y, et al. Rapid induction of multiple resistance mechanisms in Aspergillus fumigatus during azole therapy: a case study and review of the literature. Antimicrob Agents Chemother. 2012;56:10–6. doi: 10.1128/AAC.05088-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Denning DW, Park S, Lass-Florl C, et al. High-frequency triazole resistance found in nonculturable Aspergillus fumigatus from lungs of patients with chronic fungal disease. Clin Infect Dis. 2011;52:1123–9. doi: 10.1093/cid/cir179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mortensen KL, Mellado E, Lass-Florl C, et al. Environmental study of azole-resistant Aspergillus fumigatus and other aspergilli in Austria, Denmark, and Spain. Antimicrob Agents Chemother. 2010;54:4545–9. doi: 10.1128/AAC.00692-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mortensen KL, Jensen RH, Johansen HK, et al. Aspergillus species and other molds in respiratory samples from patients with cystic fibrosis: a laboratory-based study with focus on Aspergillus fumigatus azole resistance. J Clin Microbiol. 2011;49:2243–51. doi: 10.1128/JCM.00213-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.D’Haese J, Theunissen K, Vermeulen E, et al. Detection of galactomannan in bronchoalveolar lavage fluid samples of patients at risk for invasive pulmonary aspergillosis: analytical and clinical validity. J Clin Microbiol. 2012;50:1258–63. doi: 10.1128/JCM.06423-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Hoog GS, Guarro J, Gene J., et al. Atlas of Clinical Fungi. Washington, DC: ASM Press; 2000. [Google Scholar]
- 21.Rodriguez-Tudela JL, Alcazar-Fuoli L, Mellado E, et al. Epidemiological cutoffs and cross-resistance to azole drugs in Aspergillus fumigatus. Antimicrob Agents Chemother. 2008;52:2468–72. doi: 10.1128/AAC.00156-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marr KA, Balajee SA, McLaughlin L, et al. Detection of galactomannan antigenemia by enzyme immunoassay for the diagnosis of invasive aspergillosis: variables that affect performance. J Infect Dis. 2004;190:641–9. doi: 10.1086/422009. [DOI] [PubMed] [Google Scholar]
- 23.Zhao Y, Park S, Warn P, et al. Detection of Aspergillus fumigatus in a rat model of invasive pulmonary aspergillosis by real-time nucleic acid sequence-based amplification. J Clin Microbiol. 2010;48:1378–83. doi: 10.1128/JCM.02214-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia-Effron G, Dilger A, Alcazar-Fuoli L, et al. Rapid detection of triazole antifungal resistance in Aspergillus fumigatus. J Clin Microbiol. 2008;46:1200–6. doi: 10.1128/JCM.02330-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van der Linden J, Arendrup M, Verweij P, et al. Prospective international surveillance of azole resistance (AR) in Aspergillus fumigatus (Af) (SCARE-Network). Abstracts of the Fifty-first Interscience Conference on Antimicrobial Agents and Chemotherapy; Chicago, IL. Washington, DC, USA: American Society for Microbiology; 2011. Abstract M-490. [Google Scholar]
- 26.Denning DW, Riniotis K, Dobrashian R, et al. Chronic cavitary and fibrosing pulmonary and pleural aspergillosis: case series, proposed nomenclature change, and review. Clin Infect Dis. 2003;37(Suppl 3):S265–80. doi: 10.1086/376526. [DOI] [PubMed] [Google Scholar]
- 27.Luong ML, Clancy CJ, Vadnerkar A, et al. Comparison of an Aspergillus real-time polymerase chain reaction assay with galactomannan testing of bronchoalvelolar lavage fluid for the diagnosis of invasive pulmonary aspergillosis in lung transplant recipients. Clin Infect Dis. 2011;52:1218–6. doi: 10.1093/cid/cir185. [DOI] [PubMed] [Google Scholar]
- 28.Walsh TJ, Wissel MC, Grantham KJ, et al. Molecular detection and species-specific identification of medically important Aspergillus species by real-time PCR in experimental invasive pulmonary aspergillosis. J Clin Microbiol. 2011;49:4150–7. doi: 10.1128/JCM.00570-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Torelli R, Sanguinetti M, Moody A, et al. Diagnosis of invasive aspergillosis by a commercial real-time PCR assay for Aspergillus DNA in bronchoalveolar lavage fluid samples from high-risk patients compared to a galactomannan enzyme immunoassay. J Clin Microbiol. 2011;49:4273–8. doi: 10.1128/JCM.05026-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aquino VR, Nagel F, Andreolla HF, et al. The performance of real-time PCR, galactomannan, and fungal culture in the diagnosis of invasive aspergillosis in ventilated patients with chronic obstructive pulmonary disease (COPD) Mycopathologia. 2012;174:163–9. doi: 10.1007/s11046-012-9531-1. [DOI] [PubMed] [Google Scholar]
- 31.White PL, Bretagne S, Klingspor L, et al. Aspergillus PCR: one step closer to standardization. J Clin Microbiol. 2010;48:1231–40. doi: 10.1128/JCM.01767-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harrison E, Stalhberger T, Whelan R, et al. Aspergillus DNA contamination in blood collection tubes. Diagn Microbiol Infect Dis. 2010;67:392–4. doi: 10.1016/j.diagmicrobio.2010.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mellado E, Diaz-Guerra TM, Cuenca-Estrella M, et al. Identification of two different 14-α sterol demethylase-related genes (cyp51A and cyp51B) in Aspergillus fumigatus and other Aspergillus species. J Clin Microbiol. 2001;39:2431–8. doi: 10.1128/JCM.39.7.2431-2438.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herrera ML, Vallor AC, Gelfond JA, et al. Strain-dependent variation in 18S ribosomal DNA copy numbers in Aspergillus fumigatus. J Clin Microbiol. 2009;47:1325–32. doi: 10.1128/JCM.02073-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perlin DS. Antifungal drug resistance: do molecular methods provide a way forward? Curr Opin Infect Dis. 2009;22:568–73. doi: 10.1097/QCO.0b013e3283321ce5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arendrup MC, Mavridou E, Mortensen KL, et al. Development of azole resistance in Aspergillus fumigatus during azole therapy associated with change in virulence. PLoS One. 2010;5:e10080. doi: 10.1371/journal.pone.0010080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mavridou E, Bruggemann RJ, Melchers WJ, et al. Efficacy of posaconazole against three clinical Aspergillus fumigatus isolates with mutations in the cyp51A gene. Antimicrob Agents Chemother. 2010;54:860–5. doi: 10.1128/AAC.00931-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alanio A, Cabaret O, Sitterle E, et al. Azole preexposure affects the Aspergillus fumigatus population in patients. Antimicrob Agents Chemother. 2012;56:4948–50. doi: 10.1128/AAC.05990-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Escribano P, Recio S, Pelaez T, et al. Aspergillus fumigatus strains with mutations in the cyp51A gene do not always show phenotypic resistance to itraconazole, voriconazole, or posaconazole. Antimicrob Agents Chemother. 2011;55:2460–2. doi: 10.1128/AAC.01358-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mellado E, Garcia-Effron G, Alcazar-Fuoli L, et al. Substitutions at methionine 220 in the 14α-sterol demethylase (Cyp51A) of Aspergillus fumigatus are responsible for resistance in vitro to azole antifungal drugs. Antimicrob Agents Chemother. 2004;48:2747–50. doi: 10.1128/AAC.48.7.2747-2750.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.