Abstract
Cytosolic sulfotransferases (SULTs) catalyze the sulfate conjugation of a myriad of endogenous and xenobiotic substrates. Among the 13 human SULTs, little is known regarding regulation of the SULT1C subfamily. We evaluated the effects of a panel of transcription factor activators on levels of SULT1C mRNA (1C2 and 1C3) and protein (1C2) in LS180 colorectal adenocarcinoma cells. Treatment with 3-[3-[N-(2-chloro-3-trifluoromethylbenzyl)-(2,2-diphenylethyl)amino]propyloxy]phenylacetic acid hydrochloride [GW3965, liver X receptor (LXR) activator], 3-(2,6-dichlorophenyl)-4-(3′-carboxy-2-chlorostilben-4-yl)oxymethyl-5-isopropylisoxazole [GW4064, farnesoid X receptor (FXR)], or rifampicin [pregnane X receptor (PXR)] moderately (≤2-fold) increased both SULT1C2 and SULT1C3 mRNA levels. 1α,25-Dihydroxyvitamin D3 [1,25(OH)2D3, vitamin D receptor (VDR)] selectively upregulated SULT1C2, whereas ciprofibrate [peroxisome proliferator-activated receptor α (PPARα)], rosiglitazone (PPARγ), and 2,3,7,8-tetrachlorodibenzo-p-dioxin [aryl hydrocarbon receptor (AhR)] selectively increased SULT1C3 mRNA levels. SULT1C2 protein content was strongly increased by 1,25(OH)2D3 treatment and moderately increased by GW3965, GW4064, and rifampicin. To evaluate SULT1C2 transcriptional regulation, treatment effects were determined on reporter activity from transfected constructs containing ∼10 kb of the SULT1C2 gene. Treatment with GW3965, GW4064, or 1,25(OH)2D3 increased reporter activity ∼2-, 5-, and 5.5-fold, respectively, from a construct containing mostly intron 1 of the SULT1C2 gene. Expression of AhR, LXRα, LXRβ, PPARα, PPARγ, PXR, and VDR was confirmed in LS180 cells using quantitative reverse-transcription polymerase chain reaction; however, FXR expression was negligible, suggesting that GW4064 increased SULT1C expression through an FXR-independent mechanism. Collectively, our findings are the first to characterize the regulation of human SULT1C2 and SULT1C3 expression by several transcription factor activators. Further, we determined that responsive regions for LXR and VDR are likely contained within intron 1 of the SULT1C2 gene.
Introduction
Sulfotransferase enzymes catalyze the transfer of a sulfonate group from 3′-phosphoadenosine 5′-phosphosulfate to an accepting molecule (Strott, 2002). Two major classes of sulfotransferases have been identified. The membrane-bound sulfotransferases function on larger biomolecules, including proteins and carbohydrates, and play important roles in the maintenance of tissue structure and in cell signaling (Strott, 2002). In contrast, enzymes of the cytosolic sulfotransferase (SULT) superfamily catalyze the phase II metabolism of xenobiotics as well as various endogenous molecules, including cholesterol, sterol derivatives, neurotransmitters, and hormones (Strott, 2002). SULTs are widely expressed in both hepatic and extrahepatic tissues (Dooley et al., 2000). Generally, SULTs facilitate inactivation and elimination of compounds via the urine or bile and thus are important in detoxification processes (Gamage et al., 2006). Sulfonation reactions also regulate the synthesis and biologic activity of a number of different hormones (Strott, 2002). However, as an undesirable effect, SULTs can catalyze the activation of promutagens into more mutagenic derivatives (Glatt, 2000).
A total of 13 SULT genes have been identified in humans that are grouped into four families (SULT1, 2, 4, and 6) based on amino acid sequence similarity (Blanchard et al., 2004). Most of the structural variation among SULTs occurs in the substrate-binding region, which is unique for each individual enzyme (Wang and James, 2006). Although there is some overlap in substrate specificity within family members, individual SULTs exhibit a characteristic expression pattern and can be distinguished somewhat based on their relative substrate affinities, thermal stability, and sensitivity to inhibitors (Nowell and Falany, 2006; Runge-Morris and Kocarek, 2009). In general, the SULT1 family members metabolize a range of phenolic substrates and include the high-affinity catecholamine-sulfating (SULT1A2) and estrogen-sulfating (SULT1E1) enzymes, whereas the SULT2 family exhibits selectivity toward molecules with a steroid or sterol nucleus, such as dehydroepiandrosterone (SULT2A1), bile acids (SULT2A1), pregnenolone (SULT2B1a), and cholesterol (SULT2B1b) (Gamage et al., 2006; Lindsay et al., 2008). Substrate specificities for other human SULTs, including SULT1A2, SULT1B1, SULT1C2/C3/C4, SULT4A1, and SULT6B1, have not been well characterized (Lindsay et al., 2008; Runge-Morris and Kocarek, 2009; Runge-Morris et al., 2013).
Members of the human SULT1C subfamily are located in a cluster on chromosome 2q12 and have been among the less-studied SULTs in terms of regulation and function. SULT1C2 (previously designated SULT1C1) was the first human SULT1C member to be cloned (Her et al., 1997). SULT1C2 mRNA was detected in adult stomach, kidney, and thyroid and in fetal liver and kidney (Her et al., 1997). SULT1C4 (previously designated SULT1C2) was later identified and found to be expressed at the mRNA level in adult spinal cord, kidney, and ovary and in fetal lung, heart, and kidney (Sakakibara et al., 1998). At the protein level, SULT1C4 was only detected in fetal tissues (Stanley et al., 2005). SULT1C2 and SULT1C4 both exhibited sulfonation activity toward p-nitrophenol and the promutagen N-hydroxy-2-acetylaminofluorene, suggesting a role in carcinogen activation (Sakakibara et al., 1998). Freimuth et al. (2004) later predicted the existence of a third enzyme, SULT1C3, with three plausible splice variants (a, b, d) based on the arrangements of duplicated exons 7 and 8. Despite demonstrations that recombinant SULT1C3d exhibited sulfotransferase activity (Allali-Hassani et al., 2007; Meinl et al., 2008), neither SULT1C3 mRNA (Freimuth et al., 2004) nor protein (Meinl et al., 2008) was detectable in human tissues.
Regulation of the human SULT1C subfamily has not been previously studied. Several members of the nuclear receptor superfamily, including peroxisome proliferator-activated receptors (PPARs), constitutive androstane receptor (CAR), pregnane X receptor (PXR), vitamin D receptor (VDR), liver X receptor (LXR), and farnesoid X receptor (FXR), have been identified as transcriptional regulators of other SULT family members (Echchgadda et al., 2004, 2007; Fang et al., 2005, 2007; Jiang et al., 2005; Miyata et al., 2006; Kodama et al., 2011). Using TaqMan primer/probes, we detected mRNA expression of SULT1C2 and SULT1C3, but not SULT1C4, in LS180 colorectal adenocarcinoma cells. Therefore, the purpose of this study was to evaluate the regulation of SULT1C2 and SULT1C3 using a panel of transcription factor activators. Additional studies were performed on SULT1C2 to assess treatment effects on protein levels as well as activation of cloned regions of the gene using reporter assays.
Materials and Methods
Chemicals and Reagents.
Ciprofibrate (Cipro) was a gift from Sterling Winthrop Pharmaceuticals Research Division (Rensselaer, NY). 3-[3-[N-(2-Chloro-3-trifluoromethylbenzyl)-(2,2-diphenylethyl)amino]propyloxy]phenylacetic acid hydrochloride (GW3965) and 3-(2,6-dichlorophenyl)-4-(3′-carboxy-2-chlorostilben-4-yl)oxymethyl-5-isopropylisoxazole (GW4064) were purchased from Tocris Biosciences (Minneapolis, MN), and 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) from Midwest Research Institute (Kansas City, MO). All other chemicals were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise stated. Cell culture medium, supplements, and Lipofectamine 2000 reagent were purchased from Invitrogen (Carlsbad, CA); TaqMan primer/probes from Applied Biosystems (Foster City, CA); SULT1C2 antibody (clone 5A) from Origene (Rockville, MD); β-actin antibody from Sigma-Aldrich; and horseradish peroxidase–conjugated secondary antibodies from Santa Cruz Biotechnology (Santa Cruz, CA). Additional materials were obtained from the sources indicated later.
Cell Culture and Treatments.
LS180 cells were purchased from the American Type Culture Collection (Manassas, VA) and maintained in minimal essential medium (MEM) supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 1 mM sodium pyruvate, MEM nonessential amino acids, 100 U/ml penicillin, and 100 μg/ml streptomycin. Cells were subcultured using 5 mM EDTA and seeded onto 12-well plates. Sixty hours following plating, transcription factor activators were added to the culture medium as concentrated (1000×) stock solutions dissolved in either dimethylsulfoxide (DMSO) [rifampicin (PXR), rosiglitazone (PPARγ), GW3965 (LXR), GW4064 (FXR), chenodeoxycholic acid (CDCA; FXR), Cipro (PPARα), 6-(4-chlorophenyl)imidazo[2,1-b][1,3]thiazole-5-carbaldehyde-O-(3,4-dichlorobenzyl)oxime (CITCO; CAR), 9-cis-retinoic acid (9cRA, RXR), all-trans-retinoic acid (ATRA; retinoic acid receptor), 3,3′,5′-triiodo-l-thyronine (T3, thyroid hormone receptor), and TCDD (aryl hydrocarbon receptor, AhR)] or ethanol (1α,25-dihydroxyvitamin D3 [1,25(OH)2D3; VDR]) at the concentrations indicated in the individual figure legends.
Quantitative Reverse-Transcription Polymerase Chain Reaction.
TaqMan primer/probes to detect SULT1C2 (Hs00602560_m1), SULT1C3 (Hs01371045_m1, Hs01371044_m1), AhR (Hs00169233_m1), CAR (Hs00901571_m1), FXR (Hs01026590_m1), LXRα (Hs00172885_m1), LXRβ (Hs00173195_m1), VDR (Hs01045847_m1), PXR (Hs01114267_m1), PPARα (Hs00947539_m1), PPARγ (Hs01115513_m1), and 18S ribosomal RNA were purchased from Applied Biosystems, and human reference cDNA was purchased from Clontech Laboratories (Mountain View, CA). The TaqMan primer/probe sets that were used to detect SULT1C3 mRNA targeted exons 3 and 4 (Hs01371045_m1) or exons 2 and 3 (Hs01371044_m1). Beginning 60 hours after plating, cells (∼50%–60% confluency) were either left untreated or treated with vehicle or transcription factor activators at the concentrations indicated in the individual figure legends. Culture medium containing treatments was renewed once after 24 hours. Forty-eight hours after the initial treatment, total RNA was extracted using RNeasy columns (Qiagen, Carlsbad, CA), and cDNA was synthesized from 1.5 µg of total RNA using the High Capacity Reverse Transcription kit (Applied Biosystems). Quantitative determination of mRNA levels was performed with the StepOne Plus Real Time Polymerase Chain Reaction (PCR) System (Applied Biosystems) using Gene Expression Master Mix (Applied Biosystems), 50 ng RNA equivalents of reverse transcription reactions, and TaqMan primer/probes. The cycling conditions were as follows: PCR initial activation step at 95°C for 15 minutes and a total of 40 cycles for melting (95°C, 15 seconds) and annealing/extension (60°C, 1 minute). Assays were performed in duplicate and were repeated in at least three independent cell culture experiments for SULT1C and two experiments for transcription factors. Relative fold changes in SULT1C2 and SULT1C3 mRNA levels were quantified using the comparative Ct (ΔΔCt) method (User bulletin no.2; Applied Biosystems) and are expressed as fold change relative to vehicle-treated control. For transcription factor assays, the cycle threshold (Ct) was compared with that of a human reference cDNA standard.
Immunodetection of SULT1C2 by Western Blotting.
LS180 cells were plated onto 12-well plates and treated with select nuclear receptor activators as described earlier. Forty-eight hours after the initial treatment, cells were scraped and pelleted at 1000 × g for 5 minutes in ice-cold phosphate-buffered saline. Whole-cell lysates were prepared by sonicating cell pellets in ice-cold radioimmunoprecipitation assay buffer (50 mM Tris, 150 mM NaCl, 0.2% SDS, 0.25% sodium deoxycholate, 1% Triton X-100, 1 mM EDTA) containing protease inhibitors (Thermo Fisher, Rockford, IL). Lysates were centrifuged at 13,000 × g for 15 minutes at 4°C, and the protein concentrations of the supernatants were measured using the bicinchoninic acid protein assay (Sigma-Aldrich). Proteins (15 µg) were resolved on 12.5% SDS-polyacrylamide gels and then transferred onto polyvinylidene difluoride membranes (BioRad, Hercules, CA). Following transfer, membranes were incubated in blocking buffer [2.5% nonfat dry milk dissolved in 20 mM Tris-buffered saline (pH 7.4) containing 0.05% Tween-20] for 1 hour at room temperature and then probed with anti-SULT1C2 antibody (1:2000) diluted in blocking buffer overnight at 4°C. After washing in 20 mM Tris buffer (pH 7.4) containing 0.05% Tween-20, blots were incubated with secondary antibody (1:25,000) linked to horseradish peroxidase for 1 hour at room temperature, and immunoreactive bands were detected using enhanced chemiluminescence. Images were captured on radiographic film (GE Healthcare Biosciences, Pittsburgh, PA). Following detection, membranes were stripped in glycine stripping buffer (pH 2.2) and reprobed for β-actin using the procedures described earlier. Films were subsequently scanned and band densities were quantified using ImageJ software (Rasband, 2012).
Preparation of SULT1C2 Reporter Plasmids and Transient Transfection of LS180 Cells.
Two constructs of the SULT1C2 gene spanning ∼10 kb upstream of the translation start site were prepared by PCR using human genomic DNA as a template, HotStar HiFidelity Polymerase (Qiagen, Valencia, CA), and primers corresponding to nucleotides (nt) 108,900,218 to 108,900,239 (5′-GGGGGTACCAGAGATGAGTATGGCTAACACA-3′) and 108,905,209 to 108,905,186 (5′-GGGGCTAGCTCATATGAACTAAGGGCACACTCT-3′) of National Center for Biotechnology Information chromosome 2 reference sequence NC_000002.11 for construct SULT1C2#1 and nt 108,905,126 to 108,905,145 (5′-GGGGGTACCCAGGCAGCTGAGGGCCAGGA-3′) and 108,910,123 to 108,910,104 (5′-GGGGCTAGCAGTGTCTCAGGGTTGGGGTC-3′) for construct SULT1C2#2. The underscored nucleotides of the primers indicate restriction sites for ligation into the pGL4.24[luc2P/minP] firefly luciferase reporter plasmid (Promega Corporation, Madison, WI), which contains a minimal promoter. Construct SULT1C2#1 is 4992 base pairs in size, spans nt −9906 to −4915 relative to the translation start site, and contains mostly 5′-flanking region of the SULT1C2 gene (Freimuth et al., 2000). SULT1C2#2 (4998 base pairs) spans nt −4998 to −1 relative to the translation start site and contains all of intron 1 and most of the noncoding exon 1 of the SULT1C2 gene (Freimuth et al., 2000). The sequences of all cloned fragments were verified by the Applied Genomics Technology Center at Wayne State University.
To determine the abilities of transcription factor activators to regulate SULT1C2 transcription, LS180 cells were transiently transfected with each of the reporter constructs. Following plating (∼60%–70% confluency), culture medium was replaced with 0.2 ml of Opti-MEM containing a premixed complex of 4 µl of Lipofectamine 2000, 1.6 µg of SULT1C2 reporter plasmid, and 1 ng of a Renilla luciferase reporter plasmid driven by a cytomegalovirus promoter (Promega Corporation) to allow for normalization. Plates were incubated in transfection medium for 5 hours, after which the medium was replaced with MEM. The following day, fresh medium, either alone or containing a transcription factor activator, was added to each well, as described in the individual figure legends. Medium was replaced once after 24 hours. Cells were harvested for measurement of luciferase activities 48 hours following initial treatment using the Dual Luciferase Reporter Assay System and a GloMax luminometer (Promega), each according to the manufacturer’s instructions. Data are expressed as normalized fold changes relative to vehicle-treated controls. All treatments were performed in triplicate and repeated at least three independent times.
Statistical Analysis.
Statistical analyses were performed using GraphPad Prism software (version 4; GraphPad, La Jolla, CA). Data for Western blotting were analyzed using one-way analysis of variance, and when statistical differences were detected with the F statistic (P < 0.05), individual comparisons were made using the Student-Newman-Keuls test. Data for quantitative reverse-transcription polymerase chain reaction (qRT-PCR) and transient transfections were analyzed using the Kruskal-Wallis test followed by Dunn’s multiple comparison test. All results were considered significant at P < 0.05 and are presented as the mean ± S.E.M.
Results
Regulation of SULT1C2 and SULT1C3 Expression by Transcription Factor Activators.
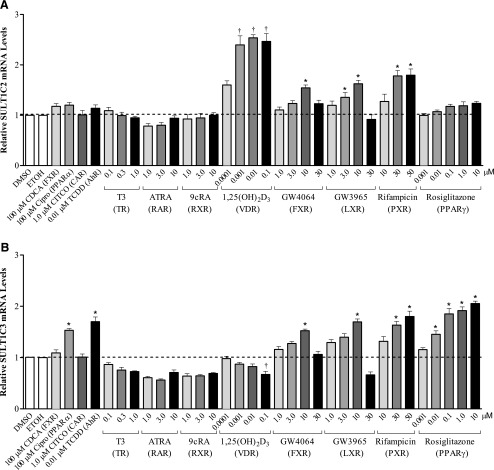
To determine the transcriptional regulation of SULT1C2 and SULT1C3, LS180 colorectal adenocarcinoma cells were initially treated with a panel of 12 prototypical transcription factor activators at varying concentrations for 48 hours, and the relative mRNA levels of SULT1C2 and SULT1C3 were assessed by qRT-PCR. Results are presented in Fig. 1. Treatment with 10 μM GW4064 (FXR), 10 μM GW3965 (LXR), or 30–50 μM rifampicin (PXR) moderately (≤2-fold) increased the mRNA levels of both SULT1C2 and SULT1C3 compared with DMSO-treated controls (P < 0.05). Treatment with 1,25(OH)2D3 (VDR) strongly and selectively increased SULT1C2 mRNA levels over a wide concentration range (0.001–0.1 μM), whereas Cipro (100 μM, PPARα), TCDD (0.01 μM, AhR), and rosiglitazone (0.01–10 μM, PPARγ) were selective inducers of SULT1C3 mRNA (Fig. 1; P < 0.05). No significant changes in mRNA levels were observed for any of the other nuclear receptor activators (CDCA, CITCO, T3, ATRA, and 9cRA) at the concentrations tested. Because initial attempts to identify SULT1C3 expression in human tissue samples were unsuccessful (Freimuth et al., 2004) and the potential existence of splice variants has been suggested (Freimuth et al., 2004), we further evaluated SULT1C3 mRNA levels using an additional primer/probe set spanning exons 2 and 3 (Supplemental Fig. 1). We found comparable induction of SULT1C3 mRNA by GW4064 (10 μM), GW3965 (10 μM), rifampicin (30–50 μM), Cipro (100 μM), TCDD (0.01 μM), and rosiglitazone (0.01–10 μM) with both primer/probe sets tested, further strengthening our findings on SULT1C3 transcriptional regulation.
Fig. 1.
Effects of transcription factor activators on SULT1C2 (A) and SULT1C3 (B) mRNA levels in LS180 cells. LS180 cells were subcultured onto 12-well plates, and 60 hours later were treated with nuclear receptor activators at the indicated concentrations. DMSO (0.1%) and ethanol (0.1%) were used as vehicle controls. Medium containing the drug treatments was replaced once after 24 hours. Forty-eight hours following initial treatment, cells were harvested and relative mRNA levels of SULT1C2 and SULT1C3 were quantified by qRT-PCR. The SULT1C3 TaqMan assay targeted exons 3 and 4. All assays were performed in duplicate and repeated at least three independent times (n = 6–26/treatment). Bars represent the mean ± S.E.M. *Significantly different from DMSO-treated control (P < 0.05); †significantly different from ethanol-treated control (P < 0.05). ETOH, ethanol.
SULT1C2 Protein Is Induced by Select Nuclear Receptor Activators in LS180 Cells.
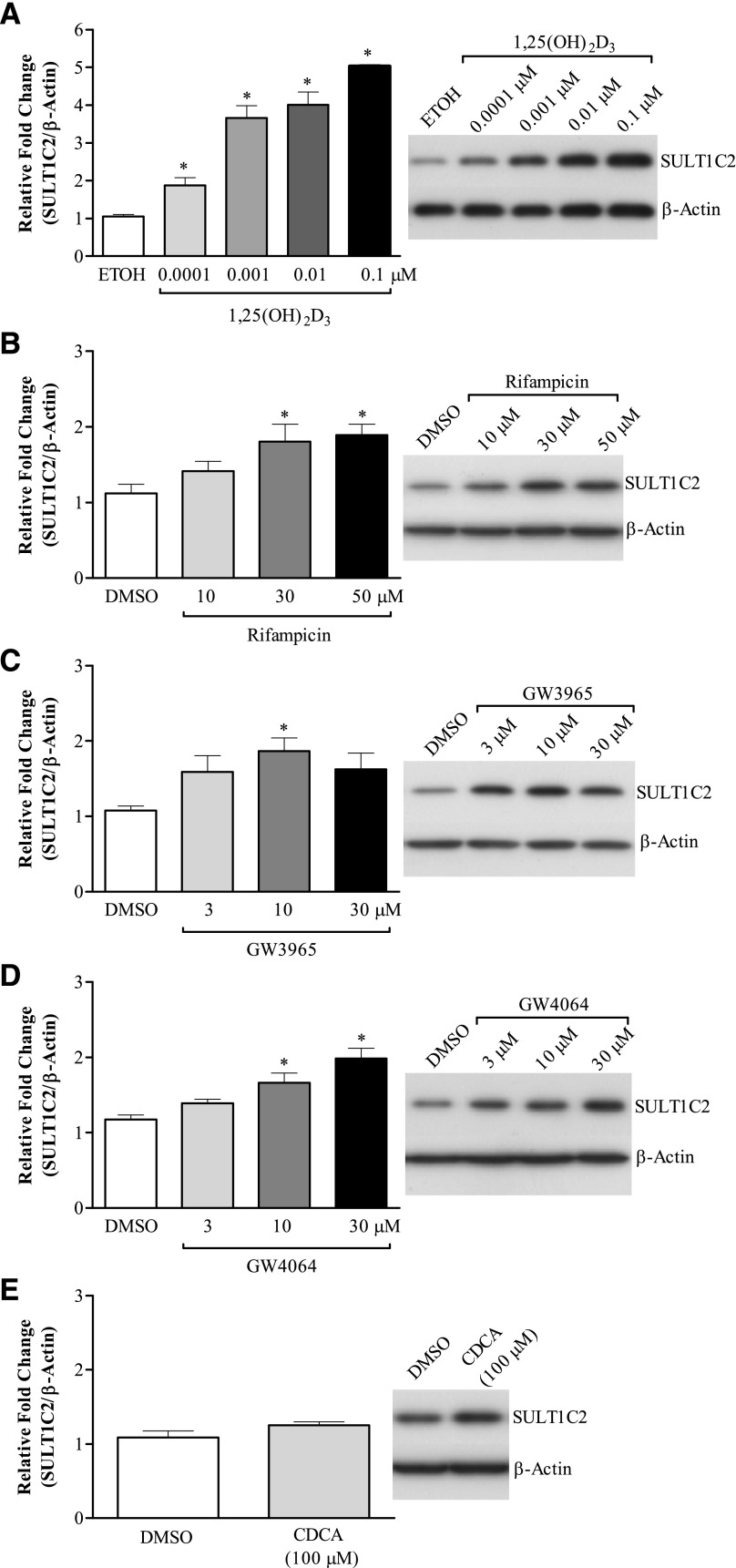
Changes in SULT1C2 expression were further evaluated at the protein level by Western blotting, and results are presented in Fig. 2. In general, 1,25(OH)2D3 (0.001–0.1 μM), GW4064 (10–30 μM), GW3965 (10 μM), and rifampicin (30–50 μM) all induced SULT1C2 protein levels, consistent with the changes observed for mRNA. However, the magnitude of change induced by 1,25(OH)2D3 was larger than that observed for mRNA, with a ∼5-fold increase at the highest concentration tested (0.1 μM; Fig. 2A). Despite the increase observed with GW4064, CDCA, a physiologic ligand for FXR, did not significantly alter SULT1C2 protein levels compared with vehicle-treated control (Fig. 2E).
Fig. 2.
Effects of select nuclear receptor activators on SULT1C2 protein levels in LS180 cells. LS180 cells were subcultured onto 12-well plates, and 60 hours later were treated with medium containing 1,25(OH)2D3 (VDR) (A), rifampicin (PXR) (B), GW3965 (LXR) (C), GW4064 (FXR) (D), or CDCA (FXR) (E) at the indicated concentrations. DMSO (0.1%) and ethanol (0.1%) were used as vehicle controls. Medium containing the drug treatments was replaced once after 24 hours. Forty-eight hours following the initial treatment, whole-cell lysates were prepared and SULT1C2 protein levels were determined by Western blotting. β-Actin was used as a loading control. Shown are representative images from two to three independent experiments. The densities of bands were quantified using ImageJ software and values normalized to vehicle-treated controls. Bars represent the mean ± S.E.M. (n = 4–6/treatment). *Significantly different from vehicle-treated controls (P < 0.05). ETOH, ethanol.
Activation of SULT1C2 Reporter Constructs by Nuclear Receptor Activators.
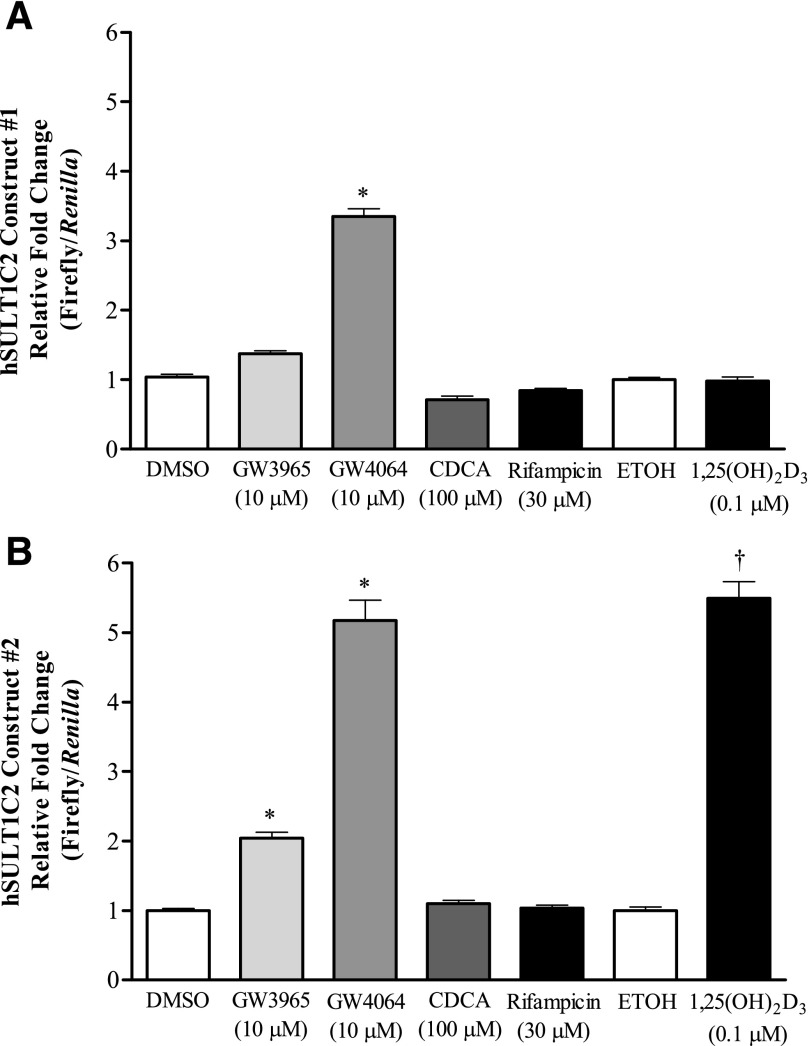
Additional experiments were conducted to evaluate the transcriptional regulation of SULT1C2 by testing whether FXR, LXR, VDR, and PXR agonists could alter the expression of a luciferase reporter driven by gene sequences located within ∼10 kb upstream of the SULT1C2 translation start site. Construct SULT1C2#1 (4992 nt) contains mostly 5′-flanking region of the SULT1C2 gene, whereas SULT1C2#2 (4998 nt) begins at nucleotide −1 relative to the translation start site and contains all of intron 1 and most of the noncoding exon 1 of the SULT1C2 gene. LS180 cells were transiently transfected with each reporter plasmid, and luciferase activities were measured 48 hours after treatment with activators. As shown in Fig. 3, we found that most of the nuclear receptor responsive elements are contained within construct #2 (Fig. 3B). Consistent with results for mRNA and protein expression, treatment of cells with either 1,25(OH)2D3 (0.1 μM), GW4064 (10 μM), or GW3965 (10 μM) significantly increased luciferase reporter activity from construct SULT1C2#2 compared with the vehicle-treated control (P < 0.05), whereas only GW4064 (10 μM) increased the activity of SULT1C2#1 (Fig. 3A). However, despite the changes observed at the mRNA and protein level by rifampicin, no significant effect on reporter activity for either construct was observed. CDCA also failed to induce luciferase expression compared with vehicle-treated control.
Fig. 3.
Modulation of SULT1C2 reporter activity by select nuclear receptor activators. Two regions of the SULT1C2 gene spanning ∼10 kb upstream of the translation start site [construct SULT1C2#1 (nt −9906 to −4915 relative to the translation start site) (A) and SULT1C2#2 (nt −4998 to −1) (B)] were PCR-amplified and ligated into the pGL4.24 luciferase reporter plasmid. LS180 cells were subcultured onto 12-well plates, and 60 hours after plating were transfected with the reporter plasmids as described in Materials and Methods. The following day, medium containing nuclear receptor activators at the indicated concentrations was added to the wells. Medium containing treatments was replaced once after 24 hours. Ninety-six hours after initial plating, cells were harvested for the measurement of luciferase activity. Bars represent the mean ± S.E.M. of normalized luciferase measurements (firefly/Renilla) combined from at least two independent experiments (n = 3 wells/treatment/experiment). *Significantly different from DMSO-treated control (P < 0.05); †significantly different from ethanol-treated control (P < 0.05). ETOH, ethanol; hSULT1C2, human SULT1C2.
Expression of Nuclear Receptors and AhR in LS180 Cells.
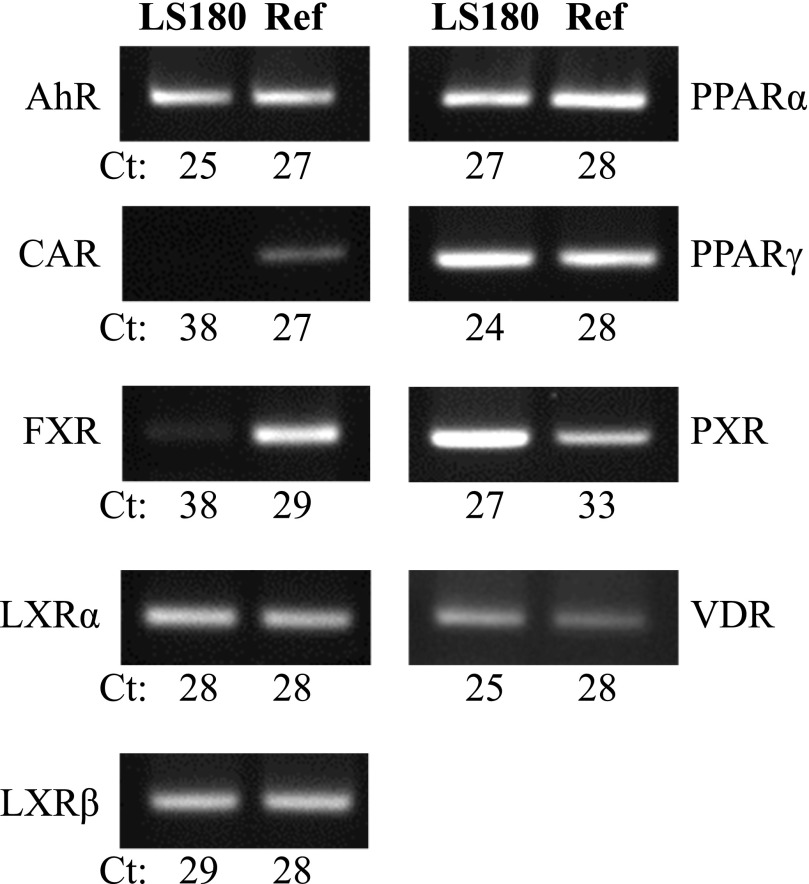
LS180 cells have been previously suggested to be a good model for studying PXR- and VDR-responsive gene expression (Gupta et al., 2008; Meyer et al., 2012). However, given a report that GW4064 is also an agonist for estrogen-related receptor (ERR) α, β, and γ (Dwivedi et al., 2011) and additional evidence that FXR mRNA and protein are expressed at low levels in several colon cancer cell lines (Torres et al., 2013), we evaluated the basal expression of FXR and other transcription factors in LS180 cells compared with a commercial human reference cDNA standard synthesized from RNA collected from a mixture of normal human adult tissues. We found that AhR, LXRα, LXRβ, PPARα, PPARγ, PXR, and VDR were all basally expressed in LS180 cells at levels comparable to or higher than that detected in the human reference cDNA standard (Fig. 4). Consistent with a previous report (Gupta et al., 2008), CAR was negligibly expressed in LS180 cells. Also, FXR expression was barely detectable (Ct = 38) compared with the reference standard (Fig. 4).
Fig. 4.
Expression of select nuclear receptors and AhR in LS180 cells compared with a human reference cDNA standard. LS180 cells were subcultured onto 12-well plates, and 108 hours later, RNA was isolated and cDNA synthesized as described in Materials and Methods. Basal expression of select nuclear receptors and AhR was assessed in LS180 cells compared with a human reference cDNA standard (Ref) using TaqMan qRT-PCR assays. Endpoint PCR products (20 μl) were resolved on 2% agarose gels, and ethidium bromide–stained bands were visualized using a UV transilluminator. Shown are representative images. Values represent the mean Ct from two independent experiments (n = 2–6/group).
Discussion
Enzymes of the SULT superfamily are involved in the metabolism of xenobiotics and endogenous compounds as well as in modulating the biologic activities of hormones (Strott, 2002). Given their diverse roles, evaluating the expression and regulation of SULTs has implications for understanding individual responses to pharmacological and toxicological agents as well as in disease etiology (Wang et al., 2002; Suzuki et al., 2003; Cotterchio et al., 2008; Hirata et al., 2008). The regulation of and endogenous substrates for the SULT1C subfamily have not been well characterized. Nuclear receptors have been demonstrated to regulate several human, murine, and rat SULTs (reviewed in Runge-Morris et al., 2013). Therefore the purpose of the current study was to determine if similar molecular signals also regulate expression of the SULT1C enzymes.
Transcriptional responses of SULT1C2 and SULT1C3 were evaluated by treating LS180 cells with prototypical transcription factor activators. Our results indicate both common and selective regulation of SULT1C expression. For example, activators of PXR (rifampicin), LXR (GW3965), and FXR (GW4064) moderately induced the mRNA levels of both SULT1C enzymes. However, whereas SULT1C2 expression was strongly upregulated by 1,25(OH)2D3, SULT1C3 mRNA levels were unaffected. Conversely, activators of AhR (TCDD), PPARα (Cipro), and PPARγ (rosiglitazone) were selective in inducing the expression of SULT1C3. These selective effects imply both contextual and nonoverlapping regulation of these enzymes. Other prototypical activators including CDCA (FXR), 9cRA (RXR), ATRA (retinoic acid receptor), and CITCO (CAR) did not influence SULT1C expression. Additionally, although SULT1C2 is expressed in thyroid tissue (Her et al., 1997) and exhibits activity toward iodothyronines in vitro (Li et al., 2000), neither SULT1C2 nor SULT1C3 was regulated by T3 in LS180 cells.
Treatment effects on SULT1C2 were further investigated using Western blotting and reporter assays. SULT1C2 protein levels were increased by rifampicin, GW4064, and GW3965 and strongly upregulated by 1,25(OH)2D3, whereas no significant effect on SULT1C2 protein content was observed with CDCA. Effective concentrations of GW4064, GW3965, and 1,25(OH)2D3 also increased reporter activity from a construct containing intron 1 and most of the noncoding exon 1 of the SULT1C2 gene. Computational analysis of the SULT1C2#2 sequence for potential transcription factor binding sites using MatInspector (Cartharius et al., 2005) identified four matches (matrix similarity greater than optimized matrix threshold) to LXR•RXR binding sites (i.e., MatBase matrices V$LXRE.01 and V$LXRE.02) and ten matches to VDR•RXR binding sites (i.e., matrices V$VDR_RXR.03, V$VDR_RXR.04, V$VDR_RXR.05, and V$VDR_RXR.06), supporting the likelihood that detailed analysis of this SULT1C2 gene region will lead to the identification of functional LXR and VDR response elements.
GW4064 is widely used as a selective agonist for evaluating FXR-mediated responses, although additional reports imply agonist-specific responses in addition to those mediated by FXR (Downes et al., 2003; Pircher et al., 2003; Dwivedi et al., 2011). The effect of GW4064 observed in this study was generally modest at the mRNA level for SULT1C2 and SULT1C3. We did observe a concentration-dependent induction of SULT1C2 protein by Western blotting and strong activation of both SULT1C2 reporter constructs. However, treatment with CDCA, an endogenous ligand for FXR, did not elicit similar responses. This, together with the low basal expression of FXR in LS180 cells, implies that the effects of GW4064 on SULT1C expression are likely not FXR-mediated. The target for GW4064-mediated SULT1C regulation in this study was not conclusively determined. Based on findings by Dwivedi et al. (2011), attempts were made to reduce basal luciferase activity of the SULT1C2 constructs using 3-[4-(2,4-bis-trifluoromethylbenzyloxy)-3-methoxyphenyl]-2-cyano-N-(5-trifluoromethyl-1,3,4-thiadiazol-2-yl)acrylamide (XCT790), an inverse ERRα agonist, or increase expression with 2-[[4-(1-methylethyl)phenyl]methylene]hydrazide 4-hydroxybenzoic acid (GSK4716), an activator of both ERRβ and γ (data not shown) (Zuercher et al., 2005). We found that 5 μM XCT790, a concentration that produced maximal inhibition in the study by Dwivedi et al. (2011), did not inhibit, but rather slightly increased, luciferase activity on construct #2. Additionally, GSK4716 did not significantly augment reporter activity at a concentration up to 5 μM. Therefore, although we cannot rule out a potential involvement of FXR and/or ERR in SULT1C2 regulation, the effects of GW4064 observed in this study do not appear to be directly mediated through either receptor.
Findings that SULT1C3 is expressed and regulated in LS180 cells are novel given the paucity of information available on this subfamily member. SULT1C3 was originally identified through computational analyses with three plausible splice variants (a, b, d) predicted based on the arrangements of duplicated exons 7 and 8 (Freimuth et al., 2004). Subsequent studies using a recombinant protein for SULT1C3d (containing exons 7b and 8b) demonstrated that this variant exhibited sulfotransferase activity in vitro and activated promutagens in an Ames assay (Meinl et al., 2008), although neither mRNA nor protein was detectable in human tissues (Freimuth et al., 2004; Meinl et al., 2008). In a concurrent study (Duniec-Dmuchowski et al., 2014), we report that the predominant transcript expressed in LS180 and intestinal cells is SULT1C3a (containing exons 7a and 8a). It should be noted that the primers used in the current study target more upstream (exons 2 and 3 and 3 and 4) regions of SULT1C3 mRNA and are therefore likely to detect all splice variants, if present. Although we found SULT1C3a to be expressed and transcriptionally regulated in this cell line, it is possible that other SULT1C3 splice variants are expressed in different tissues or at different times during development.
The intestine is exposed to numerous compounds derived from the diet and environment, as well as to endogenous and bacterial metabolites. In humans, SULT enzymes are highly expressed in intestinal tissue, with SULT1B1, 1A3, and 1A1 the major members detected (Riches et al., 2009). In the current study, we identified that SULT1C2 and SULT1C3 are additionally expressed in intestinal cells and are regulated by select ligand-dependent transcription factors. Additional detailed promoter analysis, mutational studies, and binding assays will more conclusively determine the specific regulatory elements for both enzymes. Our findings will guide future research that is designed to understand the regulation and physiologic role of the SULT1C class of enzymes.
Supplementary Material
Abbreviations
- AhR
aryl hydrocarbon receptor
- ATRA
all-trans-retinoic acid
- CAR
constitutive androstane receptor (NR1H4)
- CDCA
chenodeoxycholic acid
- Cipro
ciprofibrate
- CITCO
6-(4-chlorophenyl)imidazo[2,1-b][1,3]thiazole-5-carbaldehyde-O-(3,4-dichlorobenzyl)oxime
- 9cRA
9-cis-retinoic acid
- Ct
cycle threshold
- DMSO
dimethylsulfoxide
- ERR
estrogen-related receptor (NR3B)
- FXR
farnesoid X receptor (NR1H4)
- GSK4716
2-[[4-(1-methylethyl)phenyl]methylene]hydrazide 4-hydroxybenzoic acid
- GW3965
3-[3-[N-(2-chloro-3-trifluoromethylbenzyl)-(2,2-diphenylethyl)amino]propyloxy]phenylacetic acid hydrochloride
- GW4064
3-(2,6-dichlorophenyl)-4-(3′-carboxy-2-chlorostilben-4-yl)oxymethyl-5-isopropylisoxazole
- LXRα
liver X receptor α (NR1H3)
- LXRβ
liver X receptor β (NR1H2)
- MEM
minimal essential medium
- nt
nucleotide(s)
- 1,25(OH)2D3
1α,25-dihydroxyvitamin D3
- PCR
polymerase chain reaction
- PPARα
peroxisome proliferator-activated receptor α (NR1C1)
- PPARγ
peroxisome proliferator-activated receptor γ (NR1C3)
- PXR
pregnane X receptor (NR1I2)
- qRT-PCR
quantitative reverse-transcription polymerase chain reaction
- RXR
retinoid X receptor (NR2B)
- SULT
cytosolic sulfotransferase
- TCDD
2,3,7,8-tetrachlorodibenzo-p-dioxin
- T3
3,3′,5′-triiodo-l-thyronine
- VDR
vitamin D receptor (NR1I1)
- XCT790
3-[4-(2,4-bis-trifluoromethylbenzyloxy)-3-methoxyphenyl]-2-cyano-N-(5-trifluoromethyl-1,3,4-thiadiazol-2-yl)acrylamide
Authorship Contributions
Participated in research design: Rondini, Runge-Morris, Kocarek.
Conducted experiments: Rondini.
Contributed new reagents or analytic tools: Fang.
Performed data analysis: Rondini.
Wrote or contributed to the writing of the manuscript: Rondini, Runge-Morris, Kocarek.
Footnotes
This research was supported by the National Institutes of Health National Heart, Lung, and Blood Institute [Grant R01 HL050710] (to T.A.K.) and the National Institutes of Health National Institute of Environmental Health Sciences [Grant R01 ES005823] (to M.R.-M.).
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
References
- Allali-Hassani A, Pan PW, Dombrovski L, Najmanovich R, Tempel W, Dong AP, Loppnau P, Martin F, Thornton J, Edwards AM, et al. (2007) Structural and chemical profiling of the human cytosolic sulfotransferases. PLoS Biol 5:e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard RL, Freimuth RR, Buck J, Weinshilboum RM, Coughtrie MWH. (2004) A proposed nomenclature system for the cytosolic sulfotransferase (SULT) superfamily. Pharmacogenetics 14:199–211 [DOI] [PubMed] [Google Scholar]
- Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. (2005) MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics 21:2933–2942 [DOI] [PubMed] [Google Scholar]
- Cotterchio M, Boucher BA, Manno M, Gallinger S, Okey AB, Harper PA. (2008) Red meat intake, doneness, polymorphisms in genes that encode carcinogen-metabolizing enzymes, and colorectal cancer risk. Cancer Epidemiol Biomarkers Prev 17:3098–3107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley TP, Haldeman-Cahill R, Joiner J, Wilborn TW. (2000) Expression profiling of human sulfotransferase and sulfatase gene superfamilies in epithelial tissues and cultured cells. Biochem Biophys Res Commun 277:236–245 [DOI] [PubMed] [Google Scholar]
- Downes M, Verdecia MA, Roecker AJ, Hughes R, Hogenesch JB, Kast-Woelbern HR, Bowman ME, Ferrer JL, Anisfeld AM, Edwards PA, et al. (2003) A chemical, genetic, and structural analysis of the nuclear bile acid receptor FXR. Mol Cell 11:1079–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duniec-Dmuchowski Z, Rondini EA, Tibbs ZE, Falany CN, Runge-Morris M, Kocarek TA. (2014) Expression of the orphan cytosolic sulfotransferase SULT1C3 in human intestine: characterization of the transcript variant and implications for function. Drug Metab Dispos 42:352–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi SKD, Singh N, Kumari R, Mishra JS, Tripathi S, Banerjee P, Shah P, Kukshal V, Tyagi AM, Gaikwad AN, et al. (2011) Bile acid receptor agonist GW4064 regulates PPARγ coactivator-1α expression through estrogen receptor-related receptor α. Mol Endocrinol 25:922–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echchgadda I, Song CS, Oh T, Ahmed M, De La Cruz IJ, Chatterjee B. (2007) The xenobiotic-sensing nuclear receptors pregnane X receptor, constitutive androstane receptor, and orphan nuclear receptor hepatocyte nuclear factor 4alpha in the regulation of human steroid-/bile acid-sulfotransferase. Mol Endocrinol 21:2099–2111 [DOI] [PubMed] [Google Scholar]
- Echchgadda I, Song CS, Roy AK, Chatterjee B. (2004) Dehydroepiandrosterone sulfotransferase is a target for transcriptional induction by the vitamin D receptor. Mol Pharmacol 65:720–729 [DOI] [PubMed] [Google Scholar]
- Fang HL, Strom SC, Cai H, Falany CN, Kocarek TA, Runge-Morris M. (2005) Regulation of human hepatic hydroxysteroid sulfotransferase gene expression by the peroxisome proliferator-activated receptor alpha transcription factor. Mol Pharmacol 67:1257–1267 [DOI] [PubMed] [Google Scholar]
- Fang HL, Strom SC, Ellis E, Duanmu Z, Fu J, Duniec-Dmuchowski Z, Falany CN, Falany JL, Kocarek TA, Runge-Morris M. (2007) Positive and negative regulation of human hepatic hydroxysteroid sulfotransferase (SULT2A1) gene transcription by rifampicin: roles of hepatocyte nuclear factor 4alpha and pregnane X receptor. J Pharmacol Exp Ther 323:586–598 [DOI] [PubMed] [Google Scholar]
- Freimuth RR, Raftogianis RB, Wood TC, Moon E, Kim UJ, Xu JP, Siciliano MJ, Weinshilboum RM. (2000) Human sulfotransferases SULT1C1 and SULT1C2: cDNA characterization, gene cloning, and chromosomal localization. Genomics 65:157–165 [DOI] [PubMed] [Google Scholar]
- Freimuth RR, Wiepert M, Chute CG, Wieben ED, Weinshilboum RM. (2004) Human cytosolic sulfotransferase database mining: identification of seven novel genes and pseudogenes. Pharmacogenomics J 4:54–65 [DOI] [PubMed] [Google Scholar]
- Gamage N, Barnett A, Hempel N, Duggleby RG, Windmill KF, Martin JL, McManus ME. (2006) Human sulfotransferases and their role in chemical metabolism. Toxicol Sci 90:5–22 [DOI] [PubMed] [Google Scholar]
- Glatt H. (2000) Sulfotransferases in the bioactivation of xenobiotics. Chem Biol Interact 129:141–170 [DOI] [PubMed] [Google Scholar]
- Gupta A, Mugundu GM, Desai PB, Thummel KE, Unadkat JD. (2008) Intestinal human colon adenocarcinoma cell line LS180 is an excellent model to study pregnane X receptor, but not constitutive androstane receptor, mediated CYP3A4 and multidrug resistance transporter 1 induction: studies with anti-human immunodeficiency virus protease inhibitors. Drug Metab Dispos 36:1172–1180 [DOI] [PubMed] [Google Scholar]
- Her CT, Kaur GP, Athwal RS, Weinshilboum RM. (1997) Human sulfotransferase SULT1C1: cDNA cloning, tissue-specific expression, and chromosomal localization. Genomics 41:467–470 [DOI] [PubMed] [Google Scholar]
- Hirata H, Hinoda Y, Okayama N, Suehiro Y, Kawamoto K, Kikuno N, Rabban JT, Chen LM, Dahiya R. (2008) CYP1A1, SULT1A1, and SULT1E1 polymorphisms are risk factors for endometrial cancer susceptibility. Cancer 112:1964–1973 [DOI] [PubMed] [Google Scholar]
- Jiang YJ, Kim P, Elias PM, Feingold KR. (2005) LXR and PPAR activators stimulate cholesterol sulfotransferase type 2 isoform 1b in human keratinocytes. J Lipid Res 46:2657–2666 [DOI] [PubMed] [Google Scholar]
- Kodama S, Hosseinpour F, Goldstein JA, Negishi M. (2011) Liganded pregnane X receptor represses the human sulfotransferase SULT1E1 promoter through disrupting its chromatin structure. Nucleic Acids Res 39:8392–8403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Clemens DL, Anderson RJ. (2000) Sulfation of iodothyronines by human sulfotransferase 1C1 (SULT1C1)*. Biochem Pharmacol 60:1713–1716 [DOI] [PubMed] [Google Scholar]
- Lindsay J, Wang LL, Li Y, Zhou SF. (2008) Structure, function and polymorphism of human cytosolic sulfotransferases. Curr Drug Metab 9:99–105 [DOI] [PubMed] [Google Scholar]
- Meinl W, Donath C, Schneider H, Sommer Y, Glatt H. (2008) SULT1C3, an orphan sequence of the human genome, encodes an enzyme activating various promutagens. Food Chem Toxicol 46:1249–1256 [DOI] [PubMed] [Google Scholar]
- Meyer MB, Goetsch PD, Pike JW. (2012) VDR/RXR and TCF4/β-catenin cistromes in colonic cells of colorectal tumor origin: impact on c-FOS and c-MYC gene expression. Mol Endocrinol 26:37–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata M, Matsuda Y, Tsuchiya H, Kitada H, Akase T, Shimada M, Nagata K, Gonzalez FJ, Yamazoe Y. (2006) Chenodeoxycholic acid-mediated activation of the farnesoid X receptor negatively regulates hydroxysteroid sulfotransferase. Drug Metab Pharmacokinet 21:315–323 [DOI] [PubMed] [Google Scholar]
- Nowell S, Falany CN. (2006) Pharmacogenetics of human cytosolic sulfotransferases. Oncogene 25:1673–1678 [DOI] [PubMed] [Google Scholar]
- Pircher PC, Kitto JL, Petrowski ML, Tangirala RK, Bischoff ED, Schulman IG, Westin SK. (2003) Farnesoid X receptor regulates bile acid-amino acid conjugation. J Biol Chem 278:27703–27711 [DOI] [PubMed] [Google Scholar]
- Rasband WS. (2012) ImageJ, U. S. National Institutes of Health, Bethesda, Maryland [Google Scholar]
- Riches Z, Stanley EL, Bloomer JC, Coughtrie MWH. (2009) Quantitative evaluation of the expression and activity of five major sulfotransferases (SULTs) in human tissues: the SULT “pie”. Drug Metab Dispos 37:2255–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runge-Morris M, Kocarek TA. (2009) Regulation of sulfotransferase and UDP-glucuronosyltransferase gene expression by the PPARs. PPAR Res 2009:728941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runge-Morris M, Kocarek TA, Falany CN. (2013) Regulation of the cytosolic sulfotransferases by nuclear receptors. Drug Metab Rev 45:15–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara Y, Yanagisawa K, Katafuchi J, Ringer DP, Takami Y, Nakayama T, Suiko M, Liu MC. (1998) Molecular cloning, expression, and characterization of novel human SULT1C sulfotransferases that catalyze the sulfonation of N-hydroxy-2-acetylaminofluorene. J Biol Chem 273:33929–33935 [DOI] [PubMed] [Google Scholar]
- Stanley EL, Hume R, Coughtrie MWH. (2005) Expression profiling of human fetal cytosolic sulfotransferases involved in steroid and thyroid hormone metabolism and in detoxification. Mol Cell Endocrinol 240:32–42 [DOI] [PubMed] [Google Scholar]
- Strott CA. (2002) Sulfonation and molecular action. Endocr Rev 23:703–732 [DOI] [PubMed] [Google Scholar]
- Suzuki T, Nakata T, Miki Y, Kaneko C, Moriya T, Ishida T, Akinaga S, Hirakawa H, Kimura M, Sasano H. (2003) Estrogen sulfotransferase and steroid sulfatase in human breast carcinoma. Cancer Res 63:2762–2770 [PubMed] [Google Scholar]
- Torres J, Bao X, Iuga AC, Chen A, Harpaz N, Ullman T, Cohen BL, Pineton de Chambrun G, Asciutti S, Odin JA, et al. (2013) Farnesoid X receptor expression is decreased in colonic mucosa of patients with primary sclerosing cholangitis and colitis-associated neoplasia. Inflamm Bowel Dis 19:275–282 [DOI] [PubMed] [Google Scholar]
- Wang LQ, James MO. (2006) Inhibition of sulfotransferases by xenobiotics. Curr Drug Metab 7:83–104 [DOI] [PubMed] [Google Scholar]
- Wang YF, Spitz MR, Tsou AMH, Zhang KR, Makan N, Wu XF. (2002) Sulfotransferase (SULT) 1A1 polymorphism as a predisposition factor for lung cancer: a case-control analysis. Lung Cancer 35:137–142 [DOI] [PubMed] [Google Scholar]
- Zuercher WJ, Gaillard S, Orband-Miller LA, Chao EY, Shearer BG, Jones DG, Miller AB, Collins JL, McDonnell DP, Willson TM. (2005) Identification and structure-activity relationship of phenolic acyl hydrazones as selective agonists for the estrogen-related orphan nuclear receptors ERRbeta and ERRgamma. J Med Chem 48:3107–3109 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.