Abstract
The cystolic sulfotransferse 1C3 (SULT1C3) gene was identified by computational analysis of the human genome and suggested to contain duplications of its last two exons (7a/b and 8a/b). Although the SULT1C3 isoform containing the more downstream exons 7b and 8b (SULT1C3d) has been expressed in Escherichia coli, crystallized, and characterized for activity, there is currently no evidence that SULT1C3 is expressed in any human tissue. Using reverse-transcription polymerase chain reaction, we detected SULT1C3 mRNA in the colorectal adenocarcinoma cell line (LS180), colon, and small intestine, but the amplified fragment contained the more upstream exons 7a and 8a. 3′-Rapid amplification of cDNA ends (RACE) confirmed that the SULT1C3 transcript expressed in LS180 cells contained exons 7a/8a, whereas 5′-RACE identified a noncoding exon 1. Full-length SULT1C3 transcript containing exons 7a/8a was amplified from LS180 and intestinal RNA, and in vitro transcription-translation of the cloned cDNA indicated that translation primarily began at the first of three in-frame ATG codons. Since SULT1C3 containing exons 7a/8a (SULT1C3a) would differ by 30 amino acids from SULT1C3d containing exons 7b/8b, we considered the functional implications of expressing one or the other isoform by generating structural models based on the reported crystal structure for SULT1C3d. Comparison of the structures indicated that five of the residues forming the substrate-binding pocket differed between the two isoforms, resulting in a change in both electron density and charge distribution along the inner wall of the substrate-binding pocket. These data indicate that SULT1C3 is expressed in human intestine but suggest that the expressed isoform is likely to differ functionally from the isoform that has been previously characterized.
Introduction
The cytosolic sulfotransferases (SULTs) catalyze the sulfate conjugation of myriad xenobiotic and endogenous molecules. The SULTs include 13 human genes that are grouped into four gene families: SULT1, SULT2, SULT4, and SULT6. The SULT1 family is further divided into four subfamilies: SULT1A, 1B, 1C, and 1E. The SULT1A subfamily consists of “phenol sulfotransferases”; SULT1E1 is a high-affinity estrogen sulfotransferase, but the substrate specificities of the human SULT1B and SUL1C enzymes are less well defined. The first SULT1C to be cloned, rat ST1C1, was an efficient catalyst of N-hydroxy-2-acetylaminofluorene sulfation (Gong et al., 1991; Nagata et al., 1993), suggesting that SULT1C enzymes might play important roles in procarcinogen bioactivation.
The SULT1C subfamily includes three human genes, named SULT1C2, SULT1C3, and SULT1C4, according to the nomenclature recommended by Blanchard et al. (2004) and used by the National Center for Biotechnology Information (NCBI). These three genes and one pseudogene, SULT1C2P1, are located in a cluster on chromosome 2 (Freimuth et al., 2004; Runge-Morris and Kocarek, 2013). Of these genes, SULT1C2 (sometimes called SULT1C1 in the literature) and SULT1C4 (sometimes called SULT1C2 in the literature) have been cloned, expressed, and partially characterized (Her et al., 1997; Sakakibara et al., 1998; Stanley et al., 2005). By contrast, SULT1C3 is essentially an orphan enzyme.
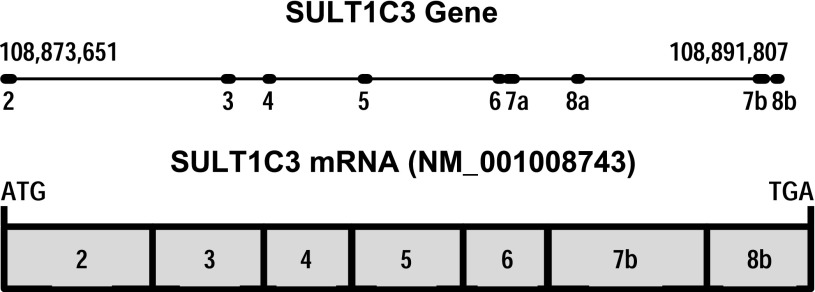
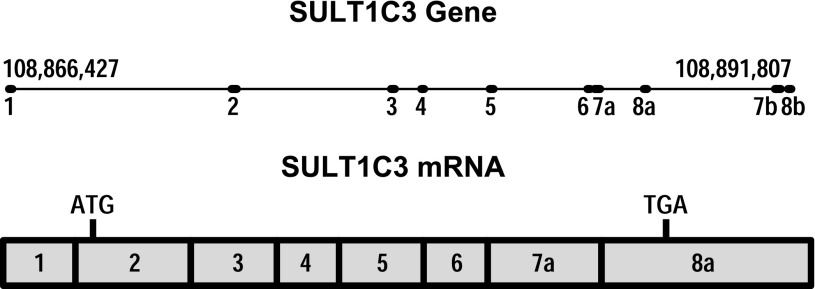
The existence of SULT1C3 was originally predicted by Freimuth et al. (2004) through a computational analysis of the human genome. These investigators noted the apparent duplication of two exons that they named exons 7 and 8 (the first named exon in this study was exon 2) in the order exon 7a, exon 8a, exon 7b, exon 8b (Fig. 1), suggesting the potential for alternative splicing in this region (Freimuth et al., 2004). Alternative splicing of SULT1C3 pre-mRNA theoretically could generate four transcripts containing exons 7a/8a, 7a/8b, 7b/8a (considered unlikely since exon 7b is downstream of 8a), or 7b/8b, which could then be translated into the corresponding proteins that were named SULT1C3 isoforms a, b, c, and d, respectively (Freimuth et al., 2004). Despite these predictions, Freimuth et al. (2004) did not find SULT1C3 mRNA sequence in the Expressed Sequence Tags (EST) database or detect SULT1C3 mRNA in samples from 20 human tissues by polymerase chain reaction (PCR), and they therefore suggested that SULT1C3 be considered a putative gene. Meinl et al. (2008) subsequently also used computational approaches to predict the existence of seven coding exons upstream of SULT1C2 (called SULT1C1 in that article), which could be assembled into the open reading frame corresponding to the isoform d described by Freimuth et al. (2004) (i.e., containing exons 7b and 8b). This is the mRNA sequence (beginning at the translation initiation codon and ending at the translation stop codon) that is listed as the reference sequence for human SULT1C3 (NM_001008743) in the NCBI database (Fig. 1). However, Meinl et al. (2008) were unable to amplify by PCR the cDNA corresponding to NM_001008743 from samples of liver or kidney RNA. These investigators therefore synthesized the cDNA sequence, expressed the recombinant protein in Escherichia coli, and raised an antibody against the protein. Using this antibody, they were unable to detect SULT1C3d in kidney, liver, platelets, testis, or prostate (Meinl et al., 2008). The expressed SULT1C3d protein did not catalyze the sulfonation of small phenols and had negligible activity toward paracetamol, ethanol, 5-hydroxymethylfurfural, 2-hydroxymethylpyrene, 2-(α-hydroxy)ethylpyrene, and corticosterone (Meinl et al., 2008). However, the enzyme did activate certain benzylic alcohols to mutagenic species in an Ames assay (Meinl et al., 2008). Independently, Allali-Hassani et al. (2007) also expressed recombinant SULT1C3d in E. coli, crystallized the recombinant protein (Protein Data Bank structure 2H8K), and tested the recombinant protein for its catalytic activities. They reported that SULT1C3d had the highest activities toward 2-ethylphenol and α-zearalenol. However, since SULT1C3 has not yet been reported to be expressed in any human cell or tissue, the physiologic relevance of these catalytic studies is unclear.
Fig. 1.
Schematic representations of the human SULT1C3 gene on chromosome 2 showing the exon-intron structure (nucleotide numbering is for NCBI reference sequence NC_000002.11) and the SULT1C3 mRNA (NCBI reference sequence NM_001008743), which contains exons 7b and 8b, begins with the translation initiation codon, and ends with the translation stop codon.
The purpose of this study was to investigate the expression of SULT1C3 in human tissues and cells and then to characterize the nature of the transcript that was found to be expressed in small and large intestine and in the LS180 colorectal adenocarcinoma cell line.
Materials and Methods
Reverse-Transcription PCR Analysis of SULT1C3 Expression in Human Tissues and LS180 Cells.
The First Choice Human Total RNA Survey Panel RNA was purchased from Life Technologies (Grand Island, NY). LS180 cells were purchased from the American Type Culture Collection (Manassas, VA) and cultured in Eagle’s minimum essential medium supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 1 mM sodium pyruvate, minimal essential medium nonessential amino acids, 100 U/ml penicillin, and 100 μg/ml streptomycin. Total RNA was isolated from LS180 cells using the RNeasy Mini kit (Qiagen, Valencia, CA). One microgram of total RNA was reverse-transcribed using random primers and the Omniscript Reverse Transcriptase kit (Qiagen). cDNA samples corresponding to 100 ng of input RNA were PCR-amplified using Taq DNA polymerase (Life Technologies) and the following SULT1C3 primers (purchased from Integrated DNA Technologies, Coralville, IA): forward primer, 5′-ACCATGTGAAAGGATGGTGGGCT-3′; reverse primer, 5′-AGTGGTCCATAATGCTGGTGGGC-3′. PCR cycling conditions were 3 minutes at 95°C, followed by 35 cycles of 30 seconds at 95°C, 30 seconds at 58°C, and 2 minutes at 72°C, followed by one cycle of 10 minutes at 72°C. Five-microliter aliquots of PCR reactions were resolved on a 0.8% agarose gel, and ethidium bromide-stained products were visualized and photographed under ultraviolet light.
Rapid Amplification of cDNA Ends.
The SMARTer Rapid Amplification of cDNA Ends (RACE) Kit was purchased from Clontech Laboratories, Inc. (Mountain View, CA). RACE-Ready cDNA was synthesized from LS180 cell RNA using the modified oligo(dT) primers supplied with the kit, and RACE PCR was performed using the Advantage 2 PCR kit (Clontech). The SULT1C3-specific reverse primer for 5′-RACE was 5′-CCTCGTAGAAGAGGTAGAGGATCCGGTGCA-3′, and the SULT1C3-specific forward primer for 3′-RACE was 5′-GGGCTGCAAAAGACATGCACCGGATCCTCTAC-3′. The PCR reactions were briefly run on a 0.8% agarose gel, but individual PCR fragments were not resolved. The fragments were then recovered from the gel and ligated into the pGEM-T Easy plasmid (Promega Corporation, Madison, WI). Individual clones were sequenced using the services of the Wayne State University Applied Genomics Technology Center. Computational analysis of the human chromosome 2 sequence in the vicinity of the 5′-RACE–determined transcription start site was performed using MatInspector (Genomatix Software, Inc., Ann Arbor, MI) (Cartharius et al., 2005).
In Vitro Transcription/Translation.
Full-length SULT1C3 cDNA was prepared by reverse-transcription (RT)-PCR using human small intestinal RNA or LS180 RNA as template, Herculase Enhanced DNA Polymerase (Agilent Technologies, Santa Clara, CA), and the following primers: Forward primer, 5′-GCGAAGCTTTGCTGTGCTCTTGCTTG-3′ (added HindIII site is underscored); reverse primer, 5′-GCGCTCGAGAACTAGCCACACTATAGGAG-3′ (added XhoI site is underscored). PCR cycling conditions were 1 minute at 95°C, followed by 30 cycles of 20 seconds at 95°C, 20 seconds at 58°C, and 1 minute at 68°C, followed by one cycle of 4 minutes at 68°C. After PCR, the amplified fragments were ligated into the HindIII and XhoI sites of pcDNA3.1 (Life Technologies) and sequenced completely. Since the sequences of the PCR fragments prepared from small intestinal RNA and LS180 RNA were identical, one of these clones was used for in vitro transcription/translation using the TNT Coupled Reticulocyte Lysate System (Promega) according to the manufacturer’s recommended protocol. Reactions (50 μl) contained 1 μg DNA template (either the SULT1C3 clone in pcDNA3.1 or Luciferase T7 Control DNA provided with the kit), 25 μl TNT rabbit reticulocyte lysate, 2 μl TNT reaction buffer, 1 μl T7 RNA polymerase, 1 μl of 1 mM amino acid mixture (minus methionine), 1 μl RNasin Ribonuclease Inhibitor (40 U/μl), and 2 μl PerkinElmer EasyTag [35S]L-methionine (1175 Ci/mmol). Reactions were performed at 30°C for 90 minutes. Proteins were separated by SDS-polyacrylamide (12.5%) gel electrophoresis and a constant current of 30 mA. After fixing in 50% methanol/10% acetic acid solution for 30 minutes, the gel was soaked in 1 M sodium salicylate for 30 minutes and then in drying solution (40% methanol/10% glycerol/7.5% acetic acid) for 5 minutes and dried overnight. The dried gel was exposed to Fuji Medical X-Ray film (FujiFilm USA, Stamford, CT) for 27 hours.
Structural Analysis.
The high amino acid sequence identify (89%) of SULT1C3a with SULT1C3d suggested that the crystal structure for SULT1C3d in complex with 3′-phosphoadenosine-5′-phosphate (PAP) reported by Allali-Hassani et al. (2H8K) (Allali-Hassani et al., 2007) would serve as the best template for constructing a structural model of SULT1C3a. Both the template structure and the amino acid sequence of interest were imported into Molecular Operating Environment (MOE), and the sequences were aligned (Chemical Computing Group Inc., 2012). The MOE Homology Model function was used to thread the amino acid sequence onto the template structure using the AMBER99 forcefield and Reaction Field treatment of electrostatics. Thirty independent intermediate models were constructed by the selection of loop candidates and side-chain rotamers. These models were briefly refined to minimize steric strain and then evaluated with the default MOE scoring function (GB/VI). The best scoring model was retained as the basic SULT1C3a model. 3′-Hosphoadenosine-5′-phosphosulfate (PAPS) was modeled into the active site of the newly constructed model by spatial alignment with the template structure. The AMBER12EHT force field was then allowed to guide the energy minimization of the cofactor. After cofactor “relaxation,” all atoms in the entire system were subjected to a final overall energy minimization procedure, completing the model construction process. For quality assessment, the model was uploaded to the NIH Structural Analysis and Verification Server (SAVS) and evaluated by each available program (http://nihserver.mbi.ucla.edu/SAVES/). The model received favorable scores, including an ERRAT score over 92%, furthering our confidence in the model’s quality. As a control, the same PAP to PAPS alteration and energy-minimization steps were performed on the template structure. The unaltered crystal structure, 2H8K, and the manipulated template structure were submitted to the National Institutes of Health H SAVS server for analysis. The programs indicated no substantial loss in model quality after force field–directed manipulation. The PyMOL Molecular Graphics System (MacPyMOL 2003; Schrödinger, LLC, Portland, OR) was used for alignment and comparison of the two models and served as the platform for the construction of structural figures.
Results
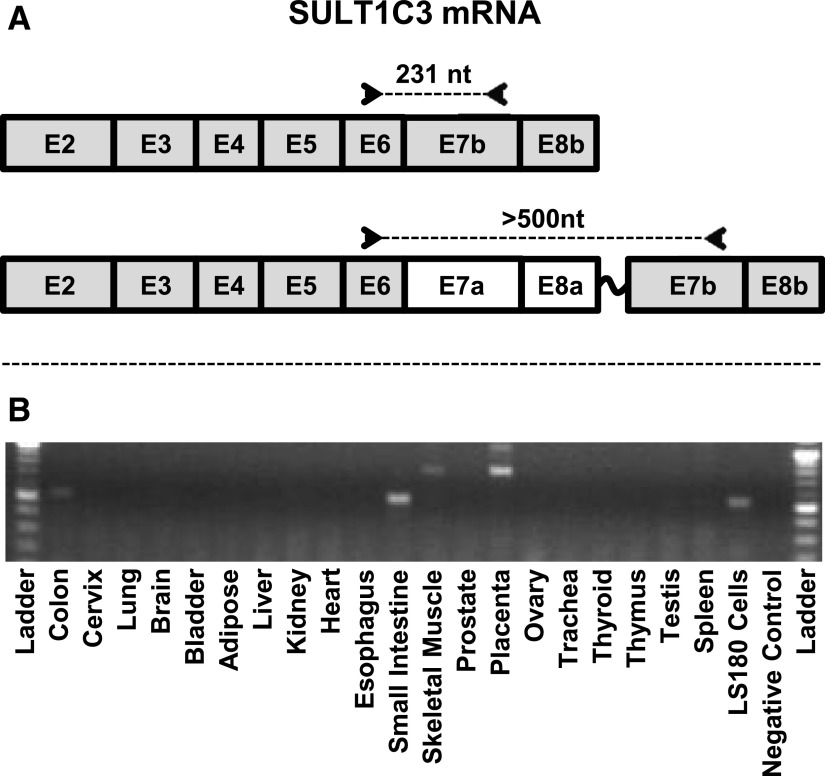
SULT1C3 mRNA has not previously been reported to be expressed in any human cell or tissue. During a characterization of SULT1C family member expression in the human colorectal cell line LS180, we found that SULT1C3 mRNA was readily detected using two different TaqMan gene expression assays that target different regions of the SULT1C3 mRNA sequence (Rondini et al., 2014). We then screened a panel of RNA samples from 20 human tissues using conventional RT-PCR with a forward primer that corresponded to nt 548–570 of NM_001008743, a region located within exon 6 of the sequence predicted by Freimuth et al. (2004) and a reverse primer that corresponded to nt 778–756, which is located within the predicted exon 7b (Fig. 2A). Using these primers, we amplified a fragment of comparable size from the colon and small intestine samples, as well as from LS180 cells (Fig. 2B), further suggesting that SULT1C3 is expressed in intestinal tissues and cells. However, if SULT1C3 was expressed as a transcript with the NM_001008743 sequence (i.e., containing exons 7b and 8b), an amplified fragment of 231 nt should have been produced by RT-PCR, and our amplified fragment was >500 nt long. An amplicon of this size could have been generated if the mRNA template contained exons 7a and 8a in addition to exon 7b. The amplicon that was generated from LS180 cells was ligated into the pGEM-T Easy plasmid, and sequencing confirmed the presence of the speculated upstream exons.
Fig. 2.
RT-PCR analysis of SULT1C3 mRNA content in human tissues and LS180 colon adenocarcinoma cells. PCR primers were designed to sequences in exons 6 and 7b (A). If the SULT1C3 transcript contained exon 7b, but not 7a, 8a, or other intervening sequence, the PCR reaction should have generated an amplicon of 231 nt. (B) A fragment of >500 nt was generated from the colon, small intestine, and LS180 RNA samples. Amplicons of ∼800 nt are visible in the skeletal muscle and placenta RNA samples.
In the RT-PCR analysis, we also noted that amplicons of ∼800 nt were generated from skeletal muscle and placenta RNA (Fig. 2B). These may represent additional alternative splicing variants in those tissues.
The preceding results indicated that SULT1C3 mRNA is expressed in human intestinal tissues and cells and that some fraction of the SULT1C3 transcript pool contains exons 7a and 8a, followed by at least a portion of exon 7b. However, these results do not indicate whether this is a common configuration. We therefore used 3′-RACE to determine the sequence of the SULT1C3 transcript in LS180 cells from part of exon 6 through the polyadenylation site. The SULT1C3-specific forward primer for 3′-RACE was located in exon 6, according to Freimuth et al. (2004) (Fig. 3A). When five SULT1C3 3′-RACE clones were sequenced, all contained exons 7a and 8a, which extended 160 to 207 nt beyond the translation stop codon, followed by a poly(A) tract, and none of the clones contained any portion of exons 7b or 8b. The five 3′-RACE clone sequences aligned against human chromosome 2 (NC_000002.11) are shown from the TGA stop codon in exon 8a to the polyadenylation site (Fig. 3B).
Fig. 3.
3′-RACE analysis of SULT1C3 mRNA in LS180 cells. 3′-RACE was performed using a SULT1C3 gene-specific primer located in exon 6 (A). The PCR products were briefly run, visualized (A), and recovered from an agarose gel. The products were then ligated into the pGEM-T Easy plasmid, and 5 clones were sequenced (B). The sequence of the SULT1C3 gene beginning at the predicted translation stop codon in exon 8a (TGA, nt 108,887,136 of NC_000002.11) and ending at nt 108,887,343 is shown aligned with the corresponding regions of the 3′-RACE clones.
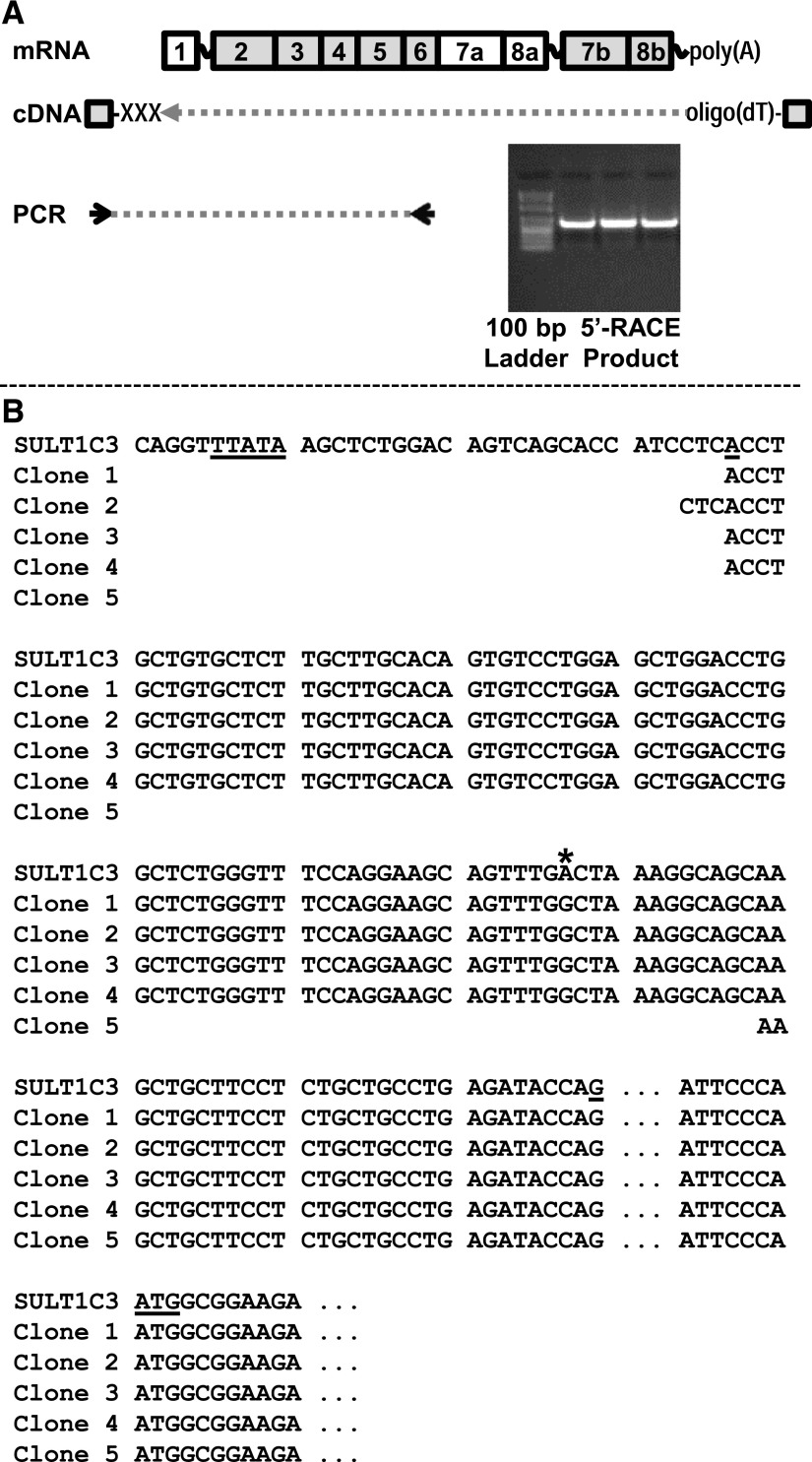
We also performed 5′-RACE to determine the transcription start site and upstream exon structure of SULT1C3. The SULT1C3-specific reverse primer was located in exon 6 (Fig. 4A). When five SULT1C3 5′-RACE clones were sequenced, the 5′ ends of four of the sequences began 120 to 123 nt upstream of the NM_001008743 translation initiation codon. Alignment of the 5′-RACE clone sequences against human chromosome 2 indicated that the SULT1C3 gene contains a noncoding exon 1 of ∼113 nt, followed by an intron of approximately 7.1 kb, followed by exon 2, which begins 7 nt upstream of the translation initiation codon (Fig. 4B). Computational analysis indicated the presence of a TATA box approximately 25 nt upstream of the 5′-RACE–identified transcription start site (MatBase matrix O$MTATA.01 at nt −24 to −40; matrix O$ATATA.01 at nt −35 to −19).
Fig. 4.
5′-RACE analysis of SULT1C3 mRNA in LS180 cells. 5′-RACE was performed using a SULT1C3 gene-specific primer located in exon 6 (A). The PCR products were briefly run, visualized (A) and recovered from an agarose gel. The products were then ligated into the pGEM-T Easy plasmid, and 5 clones were sequenced (B). The sequence of the SULT1C3 gene beginning just upstream of a computationally predicted TATA box (TTATA) and ending just downstream of the predicted translation start codon in exon 2 (ATG) is shown aligned with the corresponding regions of the 5′-RACE clones. The approximate transcription start site is at nt 108,866,427 of NC_000002.11 (A), the end of exon 1 is at nt 108,866,539 (G), and the translation start site is at nt 108,873,651. The position of a nucleotide difference that was consistently detected in the 5′-RACE clones relative to the corresponding gene sequence is marked with an asterisk.
To provide additional evidence that SULT1C3 is expressed in human intestine and LS180 cells with the 5′- and 3′ sequences identified by RACE, PCR primers were designed to amplify the SULT1C3 mRNA sequence from 4 nt downstream of the transcription start site, as indicated by 5′-RACE, to 127 nt downstream of the translation stop codon in exon 8a. The RT-PCR products that were generated from human small intestinal and LS180 RNA were ligated into the pcDNA3.1 expression plasmid and sequenced. The sequences of the PCR products from both sources were identical and contained a coding region that included exons 7a and 8a together with the expected 5′- and 3′-untranslated region sequences (data not shown).
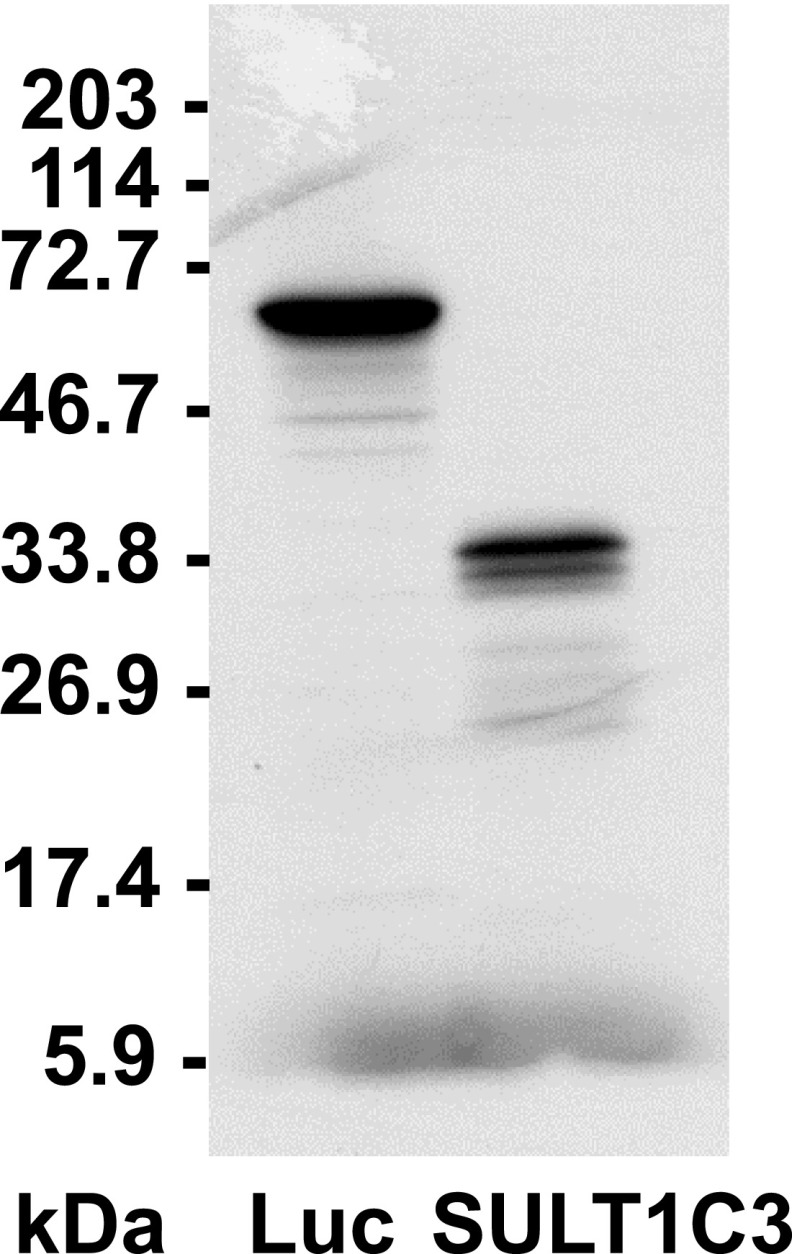
The SULT1C3 mRNA sequence was also noted to contain three in-frame translation start codons within exon 2 (Freimuth et al., 2004). As stated already, the first of these ATG codons is located 7 nt downstream of the beginning of exon 2, whereas the other two ATG codons begin 28 and 58 nt downstream from the first ATG. Computational analysis of the SULT1C3 mRNA sequence with the online tool NetStart 1.0 (Pedersen and Nielsen, 1997) suggested that the first and second ATG codons were likely to be functional translation start sites. The preceding described SULT1C3 clone containing exons 7a/8a in pcDNA3.1 was evaluated by in vitro transcription/translation and found to generate one major protein product. However, a smaller amount of a second, distinct product was also formed, suggesting that translation primarily begins at a single site (presumably the first ATG codon) but that some translation can also initiate at a downstream ATG codon (Fig. 5).
Fig. 5.
In vitro transcription/translation of a SULT1C3 cDNA clone containing exons 7a and 8a. A plasmid (pcDNA3.1) containing SULT1C3 cDNA that had been amplified from LS180 cells, from 4 nt downstream of the transcription start site indicated by 5′-RACE to 127 nt downstream of the translation stop codon in exon 8a, was analyzed by in vitro transcription/translation in a rabbit reticulocyte system. After in vitro transcription/translation, proteins were separated by SDS-PAGE and 35S-labeled SULT1C3 was visualized by fluorography. Lane one shows in vitro transcription/translation of a positive control luciferase DNA (Luc, ∼61 kDa). Locations of molecular weight size markers are indicated to the left of the image.
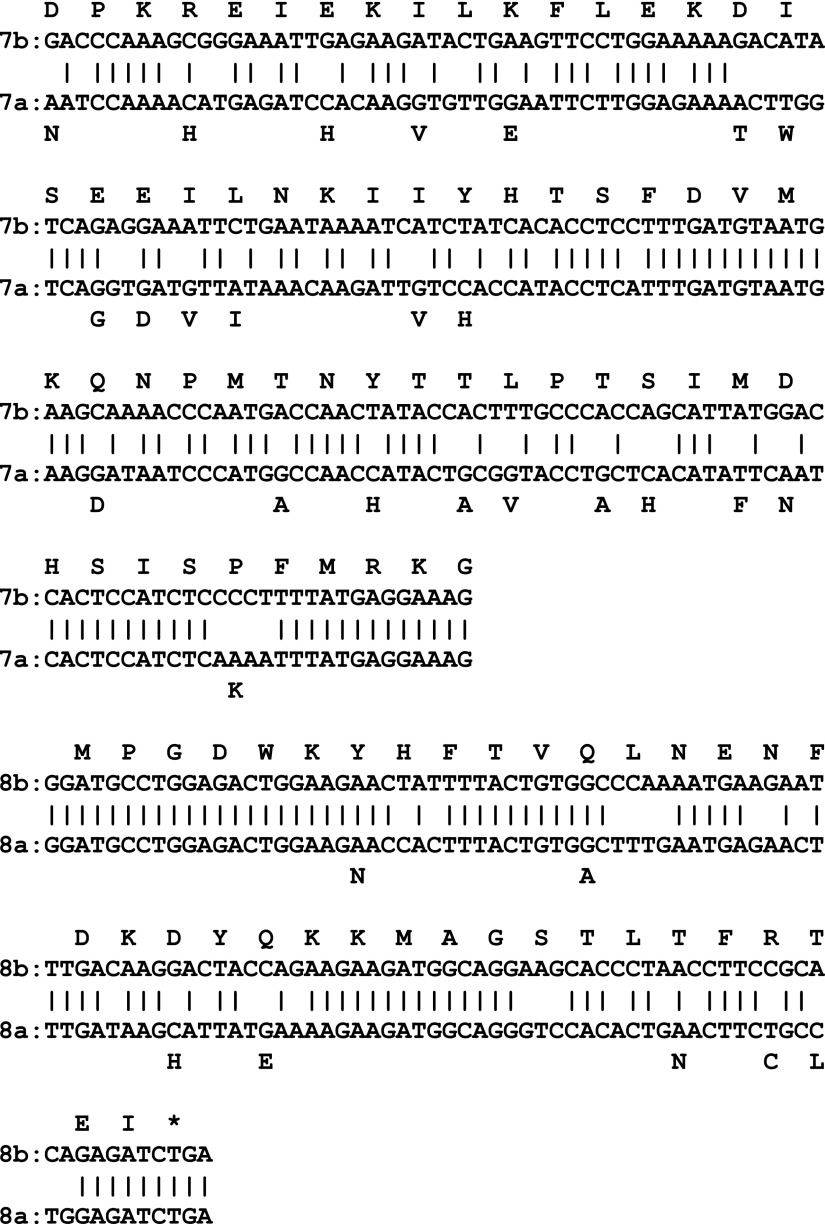
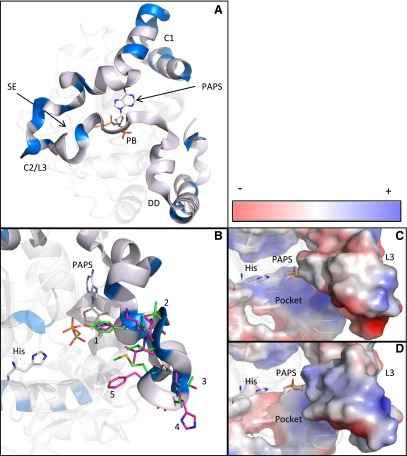
As noted, the SULT1C3 reference sequence (NM_001008743) contains exons 7b and 8b, and this sequence has been synthesized, expressed, crystallized, and used in some catalytic studies (Allali-Hassani et al., 2007; Meinl et al., 2008). However, we have shown here that the SULT1C3 transcript that is expressed in human intestinal cells contains exons 7a and 8a. When exons 7a/8a are aligned against exons 7b/8b, the nucleotide sequences differ at 86 of 291 positions (29.6%), and the deduced amino acid sequences differ at 30 of 97 positions (30.9%) (Fig. 6). To address the impact of exon 7/8 composition on the SULT1C3 three-dimensional structure, we constructed a SULT1C3 model containing exons 7a and 8a (SULT1C3a) using the crystal structure that has been reported for SULT1C3 containing exons 7b and 8b (SULT1C3d) as template. Overall, the position of the backbone of the protein was not drastically affected after energy minimization (alignment not shown). However, from a structure/function perspective, the location of altered residues is intriguing. Exons 7 and 8 span a region of the SULTs that is important for enzymatic activity, including the external wall of the PAPS binding domain, Loop 3, and the dimerization domain (Fig. 7A) (Allali-Hassani et al., 2007). The highly conserved PAPS binding motif RKGXXGDW (AA 264–271) and essential dimerization residues KXXXTVXXXE (AA 272–282) are identically conserved between both SULT1C3d and a variants, just as in SULTs across numerous species (Petrotchenko et al., 2001; Allali-Hassani et al., 2007). The altered residues are largely clustered in two regions, one region with no reported functional contribution (AA 206–232) and the other with a partially characterized functional role, Loop 3 (AA 241–263) (Fig. 7A). Sound functional inferences for residue alterations in the first region cannot be formulated because of a lack of evidence in the literature for the domain’s contribution to SULT function. In the second cluster of variable residues along Loop 3, five residues that form the substrate-binding pocket differ, including positions 247 (Y→H), 249 (T→A), 250 (L→V), 253 (S→H), and 255 (M→F) (Fig. 7B). These alterations, only one of which is conservative, collectively cause both a change in electron density and charge distribution of the inner wall of the substrate binding pocket, as shown in Fig. 7, C and D, effectively changing the shape and overall layout of the pocket.
Fig. 6.
Alignment of the nucleotide and translated amino acid sequences of SULT1C3 exons 7a and 8a versus 7b and 8b. The complete single-letter amino acid sequence is shown for exons 7b and 8b, with each letter placed above the first nucleotide the corresponding codon. Only the amino acids that are different for exons 7a and 8a are shown.
Fig. 7.
Translucent ribbon model of PAPS-bound SULT1C3 with amino acids encoded by exons 7b and 8b represented as an opaque ribbon. Altered amino acids are highlighted in blue, found largely separated into two clusters, Cluster 1 (C1) and Cluster 2/Loop 3 (C2/L3) near the substrate entry site (SE), whereas amino acids in the vicinity of the PAPS binding (PB) and dimerization domains (DD) are unaltered between the two SULT1C3 variants (A). Looking into the substrate entry site, with the active site histidine (His) on the left and PAPS in a distal position, residues in Loop 3, which form a large portion of the active site, are represented as green sticks for SULT1C3 exons 7b and 8b and magenta sticks for exons 7a and 8a. Residue numbers and altered residues are [1] 247 (Y→H), [2] 249 (T→A), [3] 250 (L→V), [4] 253 (S→H), and [5] 255 (M→F) (B). As viewed from a similar vantage point, these five residue alterations change the shape and charge distribution of the substrate-binding pocket, shown by comparing the Loop 3 regions in (C) (exons 7b and 8b) and (D) (exons 7a and 8a), both represented as a charge smoothened surface overlay with the same relative charge scale.
Discussion
Although a reference sequence for SULT1C3 mRNA exists in the NCBI nucleotide database, there are no published reports that SULT1C3 is actually expressed in any human tissue. There is currently one EST sequence (GenBank entry BY798486) that significantly aligns to SULT1C3 mRNA; this EST is a 5′ sequence that was derived from a human eye library. Our data demonstrate that SULT1C3 mRNA is expressed in human small and large intestine and in LS180 human colorectal cells. However, the SULT1C3 transcript that is expressed in intestinal tissues and cells is not the transcript variant that has been previously studied (i.e., containing exons 7b/8b and encoding isoform d); rather, it is the transcript that contains exons 7a/8a, representing isoform a.
The evidence for this conclusion comes primarily from conventional RT-PCR and 3′-RACE analysis. However, the results obtained from these two analyses differed somewhat, indicating that SULT1C3 is actually subject to different modes of pre-mRNA processing. The RT-PCR analysis indicated that SULT1C3 pre-mRNA was processed to an mRNA that contained exon 7a followed by an exon 8a sequence that extended to a splice site located 21 nt downstream of the stop codon, which was then followed directly by exon 7b. By contrast, the 3′-RACE analysis indicated that SULT1C3 mRNA included exon 7a, followed by an exon 8a sequence containing a 3′-untranslated region of 160–207 nt, which was followed by a poly(A) tail without inclusion of any exon 7b or 8b sequence. It is possible that the sequencing of a larger number of 3′-RACE clones could have led to the identification of clones containing features corresponding to the RT-PCR result. It is also possible that the use of an exon 7b–specific reverse primer for our RT-PCR analysis allowed us to detect a relatively rare transcript variant. In any case, our analyses indicated that the SULT1C3 mRNA that is expressed in intestinal cells consistently contained exons 7a and 8a. Since exon 8a contains a translation stop codon, any mRNA sequence downstream of this stop codon, whether derived from immediately downstream exon 8a sequence or from more distant downstream sequence introduced by splicing (i.e., from exon 7b), would simply represent a 3′-untranslated region. Therefore, the SULT1C3 protein that is expressed in intestinal cells should always be isoform a.
Our data also indicate that SULT1C3 contains a noncoding exon 1 that is located approximately 7.1 kb upstream of the first coding exon. Our updated representation of the SULT1C3 and transcript that is expressed in intestinal cells is shown in Fig. 8.
Fig. 8.
Revised representation of the SULT1C3 gene and the predominant mRNA sequence expressed in intestinal cells.
Although our data demonstrate that SULT1C3a is the isoform that is expressed in human intestine, the existence of duplicated exons implies the possibility that SULT1C3d or even SULT1C3b, containing exons 7a and 8b, might be expressed, possibly in a tissue not ordinarily surveyed in panels or during a particular stage of human development. SULT1C enzymes are likely to play roles during development since the existing data suggest that both SULT1C2 and SULT1C4 are more highly expressed in fetal than in adult tissues (Her et al., 1997; Sakakibara et al., 1998; Stanley et al., 2005).
Whereas SULT1C3 appears to be the only human SULT gene with duplicated downstream exons, differential splicing does factor into the expression of other SULTs. For example, differential splicing of SULT1C2 has been reported to yield two isoforms, SULT1C2a and SULT1C2b (Freimuth et al., 2000) (SULT1C2 was called SULT1C1 in that article). SULT1C2a is the prevalent isoform, whereas expression of SULT1C2b would require the activation of cryptic splice sites in intron 3 and exon 4 (Freimuth et al., 2000). However, the functional significance of SULT1C2b is currently not clear since this isoform has not been detected as an immunoreactive protein (Freimuth et al., 2000).
The three human SULT1A family members, SULT1A1, SULT1A2, and SULT1A3, all display heterogeneity in their 5′-untranslated regions because of the use of multiple promoters and differential splicing upstream of the translation start codons (Hempel et al., 2005). Although this differential promoter use has obvious implications for the regulation of SULT1A expression, it does not lead to any amino acid changes in the proteins. SULT2B1 provides another example of a gene that is subject to differential promoter use and 5′-splicing, but in this case different protein isoforms (SULT2B1a and SULT2B1b) are produced since the alternative transcription/splicing introduces different amino terminal sequences into the SULT2B1 protein (Her et al., 1998).
Structural analyses allow us to consider the functional implications of SULT1C3 exon composition. The backbones of the SULT1C3a and SULT1C3d models were nearly superimposable after energy-minimization procedures, suggesting that the overall architecture of the SULT1C3 isoforms is unaltered by the 30-residue difference. However, it is important to note that energy minimization does not fully reflect the dynamic potential of the two protein models, possibly masking valuable structural knowledge. Molecular dynamic simulation could provide further insight into structural alterations between the variants that we cannot infer from our results. Regardless, comparison between the models allowed for the identification of the topological location of each altered residue. Highly conserved regions throughout most faunal SULTs, such as the dimerization and PAPS-binding domains, are conserved in both isoforms, suggesting that each isoform is armed with the essential components to act as a sulfation catalyst. As suggested by this observation, SULT1C3d has been shown to be an active enzyme (Allali-Hassani et al., 2007; Meinl et al., 2008). The altered residues in Loop 3 are the most interesting changes owing to reports of Loop 3 contribution to SULT function. The loop forms a large portion of the substrate-binding pocket, as shown in numerous SULT crystal structures. Alterations in the residues surrounding the SULT substrate-binding site, including Loops 1 and 3, likely provide each enzyme with unique substrate specificity (Barnett et al., 2004; Berger et al., 2011). Any alteration to these residues could result in a change in this substrate specificity. Cook et al. (2013) recently reported that Loop 3 undergoes large dynamic shifts upon PAPS binding, restricting larger compounds from entering the active site while allowing smaller chemicals to dock. Our models suggest that the residue differences between SULT1C3 isoforms a and d are likely to cause relatively complex deviations in substrate specificity because of changes in both size or shape and electrostatic distribution along one side of the substrate-binding pocket. Further enzymatic investigation is needed to define the effects of the residue alterations on substrate specificity between the transcript variants.
In this study, we demonstrated that SULT1C3, specifically SULT1C3a, is expressed in intestinal tissues, strongly suggesting that this enzyme plays a role in human intestinal physiology. Also, our modeling data indicate that although SULT1C3a is probably a functional sulfotransferase, its substrate specificity is likely to differ from that of SULT1C3d. These findings provide the foundation for further studies on the regulation and catalytic properties of SULT1C3a that will enable its formal adoption as a functional member of the SULT1 family.
Abbreviations
- EST
Expressed Sequence Tags
- LS180
colorectal adenocarcinoma cell line
- MOE
Molecular Operating Environment
- NCBI
National Center for Biotechnology Information
- PAP
3′-phosphoadenosine-5′-phosphate
- PAPS
3′-phosphoadenosine-5′-phosphosulfate
- RACE
rapid amplification of cDNA ends
- RT-PCR
reverse-transcription polymerase chain reaction
- SULT
cytosolic sulfotransferase
Authorship Contributions
Participated in research design: Tibbs, Falany, Runge-Morris, Kocarek.
Conducted experiments: Duniec-Dmuchowski, Rondini, Tibbs, Kocarek.
Contributed new reagents or analytic tools: Falany.
Performed data analysis: Duniec-Dmuchowski, Tibbs, Falany, Kocarek.
Wrote or contributed to the writing of the manuscript: Duniec-Dmuchowski, Rondini, Tibbs, Falany, Runge-Morris, Kocarek.
Footnotes
This research was supported by the National Institutes of Health National Heart, Lung, and Blood Institute [Grant R01 HL050710] (to T.A.K.); the National Institutes of Health National Institute of Environmental Health Sciences [Grant R01 ES005823] (to M.R.-M.); and the National Institutes of Health National Institute of General Medical Sciences [Grant R01 GM038953] (to C.N.F.).
References
- Allali-Hassani A, Pan PW, Dombrovski L, Najmanovich R, Tempel W, Dong A, Loppnau P, Martin F, Thornton J, Edwards AM, et al. (2007) Structural and chemical profiling of the human cytosolic sulfotransferases. PLoS Biol 5:e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett AC, Tsvetanov S, Gamage N, Martin JL, Duggleby RG, McManus ME. (2004) Active site mutations and substrate inhibition in human sulfotransferase 1A1 and 1A3. J Biol Chem 279:18799–18805 [DOI] [PubMed] [Google Scholar]
- Berger I, Guttman C, Amar D, Zarivach R, Aharoni A. (2011) The molecular basis for the broad substrate specificity of human sulfotransferase 1A1. PLoS ONE 6:e26794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard RL, Freimuth RR, Buck J, Weinshilboum RM, Coughtrie MW. (2004) A proposed nomenclature system for the cytosolic sulfotransferase (SULT) superfamily. Pharmacogenetics 14:199–211 [DOI] [PubMed] [Google Scholar]
- Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. (2005) MatInspector and beyond: Promoter analysis based on transcription factor binding sites. Bioinformatics 21:2933–2942 [DOI] [PubMed] [Google Scholar]
- Chemical Computing Group Inc (2012) Molecular Operating Environment (MOE) Chemical Computing Group, Montreal, QC, Canada. [Google Scholar]
- Cook I, Wang T, Almo SC, Kim J, Falany CN, Leyh TS. (2013) The gate that governs sulfotransferase selectivity. Biochemistry 52:415–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freimuth RR, Raftogianis RB, Wood TC, Moon E, Kim UJ, Xu J, Siciliano MJ, Weinshilboum RM. (2000) Human sulfotransferases SULT1C1 and SULT1C2: cDNA characterization, gene cloning, and chromosomal localization. Genomics 65:157–165 [DOI] [PubMed] [Google Scholar]
- Freimuth RR, Wiepert M, Chute CG, Wieben ED, Weinshilboum RM. (2004) Human cytosolic sulfotransferase database mining: identification of seven novel genes and pseudogenes. Pharmacogenomics J 4:54–65 [DOI] [PubMed] [Google Scholar]
- Gong DW, Ozawa S, Yamazoe Y, Kato R. (1991) Purification of hepatic N-hydroxyarylamine sulfotransferases and their regulation by growth hormone and thyroid hormone in rats. J Biochem 110:226–231 [DOI] [PubMed] [Google Scholar]
- Hempel N, Negishi M and McManus ME (2005) Human SULT1A genes: cloning and activity assays of the SULT1A promoters. Methods Enzymol 400:147–165. [DOI] [PubMed]
- Her C, Kaur GP, Athwal RS, Weinshilboum RM. (1997) Human sulfotransferase SULT1C1: cDNA cloning, tissue-specific expression, and chromosomal localization. Genomics 41:467–470 [DOI] [PubMed] [Google Scholar]
- Her C, Wood TC, Eichler EE, Mohrenweiser HW, Ramagli LS, Siciliano MJ, Weinshilboum RM. (1998) Human hydroxysteroid sulfotransferase SULT2B1: two enzymes encoded by a single chromosome 19 gene. Genomics 53:284–295 [DOI] [PubMed] [Google Scholar]
- Meinl W, Donath C, Schneider H, Sommer Y, Glatt H. (2008) SULT1C3, an orphan sequence of the human genome, encodes an enzyme activating various promutagens. Food Chem Toxicol 46:1249–1256 [DOI] [PubMed] [Google Scholar]
- Nagata K, Ozawa S, Miyata M, Shimada M, Gong DW, Yamazoe Y, Kato R. (1993) Isolation and expression of a cDNA encoding a male-specific rat sulfotransferase that catalyzes activation of N-hydroxy-2-acetylaminofluorene. J Biol Chem 268:24720–24725 [PubMed] [Google Scholar]
- Pedersen AG and Nielsen H (1997) Neural network prediction of translation initiation sites in eukaryotes: perspectives for EST and genome analysis. Proc Int Conf Intell Syst Mol Biol 5:226–233. [PubMed]
- Petrotchenko EV, Pedersen LC, Borchers CH, Tomer KB, Negishi M. (2001) The dimerization motif of cytosolic sulfotransferases. FEBS Lett 490:39–43 [DOI] [PubMed] [Google Scholar]
- Rondini EA, Fang H, Runge-Morris M, Kocarek TA. (2014) Regulation of human cytosolic sulfotransferases (SULTs) 1C2 and 1C3 by nuclear signaling pathways in LS180 colorectal adenocarcinoma cells. Drug Metab Dispos 42:361–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runge-Morris M, Kocarek TA. (2013) Expression of the sulfotransferase 1C family: implications for xenobiotic toxicity. Drug Metab Rev 45:450–459 [DOI] [PubMed] [Google Scholar]
- Sakakibara Y, Yanagisawa K, Katafuchi J, Ringer DP, Takami Y, Nakayama T, Suiko M, Liu MC. (1998) Molecular cloning, expression, and characterization of novel human SULT1C sulfotransferases that catalyze the sulfonation of N-hydroxy-2-acetylaminofluorene. J Biol Chem 273:33929–33935 [DOI] [PubMed] [Google Scholar]
- Stanley EL, Hume R, Coughtrie MW. (2005) Expression profiling of human fetal cytosolic sulfotransferases involved in steroid and thyroid hormone metabolism and in detoxification. Mol Cell Endocrinol 240:32–42 [DOI] [PubMed] [Google Scholar]