Abstract
We aimed to characterize microbiota of the gastric mucosa as it progress to intestinal type of cancer. Study included five patients each of non-atrophic gastritis (NAG), intestinal metaplasia (IM) and intestinal-type gastric cancer (GC). Gastric tissue was obtained and DNA extracted for microbiota analyses using the microarray G3 PhyloChip. Bacterial diversity ranged from 8 to 57, and steadily decreased from NAG to IM to GC (p = 0.004). A significant microbiota difference was observed between NAG and GC based on Unifrac-presence/absence and weighted-Unifrac-abundance metrics of 283 taxa (p < 0.05). HC-AN analyses based on presence/absence of 238 taxa revealed that GC and NAG grouped apart, whereas IM overlapped with both. An ordinated analyses based on weighted-Unifrac distance given abundance of 44 taxa showing significance across categories revealed significant microbiota separation between NAG and GC. This study is the first to show a gradual shift in gastric microbiota profile from NAG to IM to GC.
H. pylori infection is recognized as the main risk factor for distal gastric cancer (GC), although just a fraction of those infected (<3%) ever develop GC, suggesting that other factors also play a role1. Pathological studies of GC show that this is an inflammation-driven disease, and all factors influencing the mucosal immune response may become involved in this multifactorial model2. Thus, host genetics, environment, and H. pylori genetics have a role to play. Polymorphisms in inflammation-related genes like IL-1β 511T, Interferon-γR1 -56C/T, or in TLRs have been reported to be associated with GC3; reduced consumption of food rich in anti-oxidants (vegetables, fruits), smoking, alcohol or obesity also increase the risk4. Most of the above risk factors may have a role on the mucosal inflammatory and immune response, thus modulating the driving force for tissue damage and development of gastric cancer.
Distal GC may be of two types, the intestinal and the diffuse, each following different development routes. For the intestinal type, the development model predicted by Correa states that an initial gastric inflammation may go uncontrolled and lead to mucosal atrophy and hypochlorydria, which in turn increases the risk for intestinal metaplasia, dysplasia and finally intestinal type GC5. Although little is known about the development of diffuse GC, it is accepted that H. pylori and inflammation may also play a role6.
The stomach microbiota may also modulate the intensity and type of inflammatory and immune responses in the gastric mucosa. Studies on the microbiota of the stomach are scarce, and one study found that the human stomach is colonized by a complex microbiota including mainly Proteobacteria, Firmicutes, Actinobacteria and Fusobacterium phyla, and showed clear differences with microbiota described in the mouth and esophagus7. The study also showed that patients positive for H. pylori culture showed significant increased colonization of Proteobacteria and decrease of Actinobacteria. Initial attempts to compare microbiota in GC vs dyspeptic patients reported no significant differences in bacterial communities, although the authors observed that Streptococcus, Lactobacillus, Veillonella and Prevotella dominated in GC patients8.
We know that H. pylori infection usually does not alter the acid barrier of the gastric mucosa, unless an unregulated inflammatory response in the corpus leads to atrophy and hypochlorydria9. Alteration of acidity may result in a more permissive milieu for colonization with other bacteria8. We hypothesize that this shift in microbiota adds to the inflammatory response already in place and increases the risk for more atrophy and intestinal metaplasia, thus increasing the risk to develop GC. In this work we aimed to study changes in stomach microbiota in gastric tissue of patients with progressive histologic stages leading to gastric cancer, from non-atrophic gastritis (NAG), to intestinal metaplasia (IM) and to GC.
Results
Gastric microbiota diversity is low, with 9 families representing >50% of all OTUs
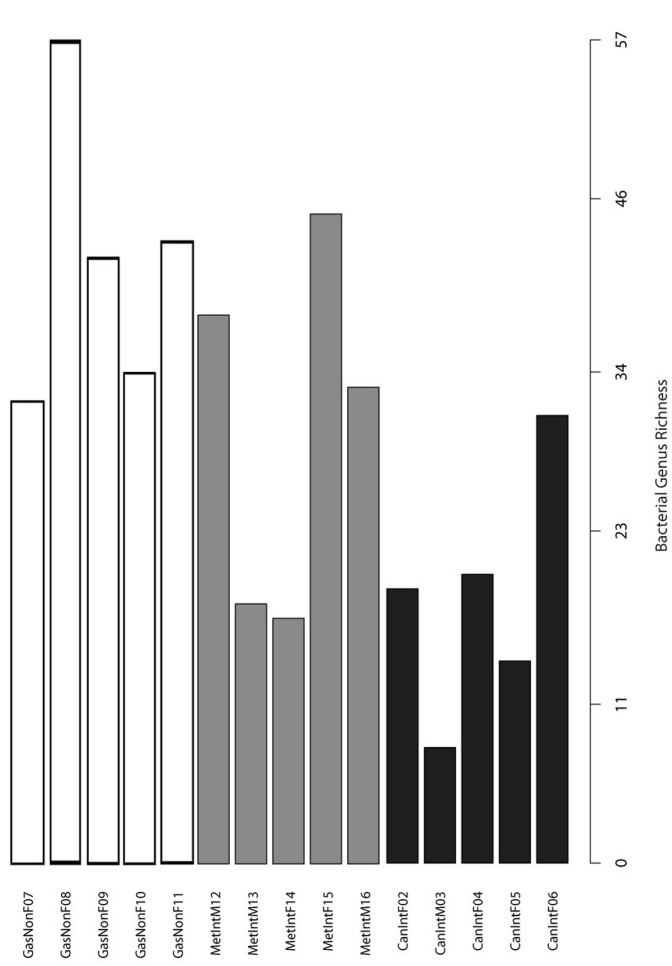
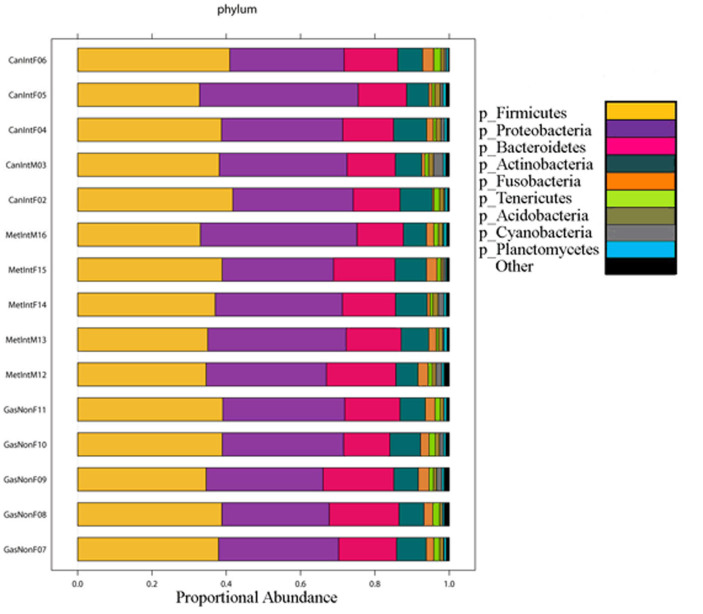
Bacterial genus diversity was low in all samples and ranged from 8 in GC patient M03, to 57 in NAG patient F08 (Figure 1). Diversity was significantly different between patients with NAG and patients with GC (p = 0.004, two-tailed heteroscadastic t-test), but not between NAG and IM; still, we observed a trend of diversity that diminished from NAG to IM to GC (Figure 1). The proportion of OTUs for each phylum is depicted in Figure 2, which shows that two phyla, Firmicutes and Proteobacteria represented almost 70% of phyla in all samples. We also found that the top 9 families represented on average as much as 55.6% of each sample's OTUs, with Lachnospiraceae and Streptococcaceae representing over 20% of families in patients from all three disease groups.
Figure 1. Microbial diversity.
Bacterial taxon diversity at the genus level in the gastric microbiota of patients with non-atrophic gastritis (GasNon), intestinal metaplasia (MetInt) and gastric cancer of the intestinal type (CanInt). Bacterial diversity was significantly higher in non-atrophic gastritis than in gastric cancer patients (p = 0.004, heteroscadastic t-test).
Figure 2. Abundance of OTUs.
Proportion of OTUs at the phylum level in the gastric microbiota of patients with non-atrophic gastritis (GasNon), intestinal metaplasia (MetInt) and gastric cancer of the intestinal type (CanInt). Firmicutes and Proteobacteria represented almost 70% of phyla in each sample.
A whole microbiota profile showed separation between GC and NAG, but not between IM and GC or IM and NAG
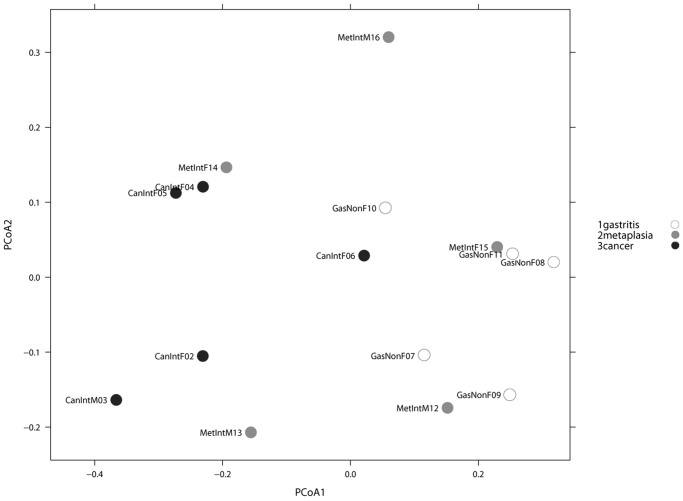
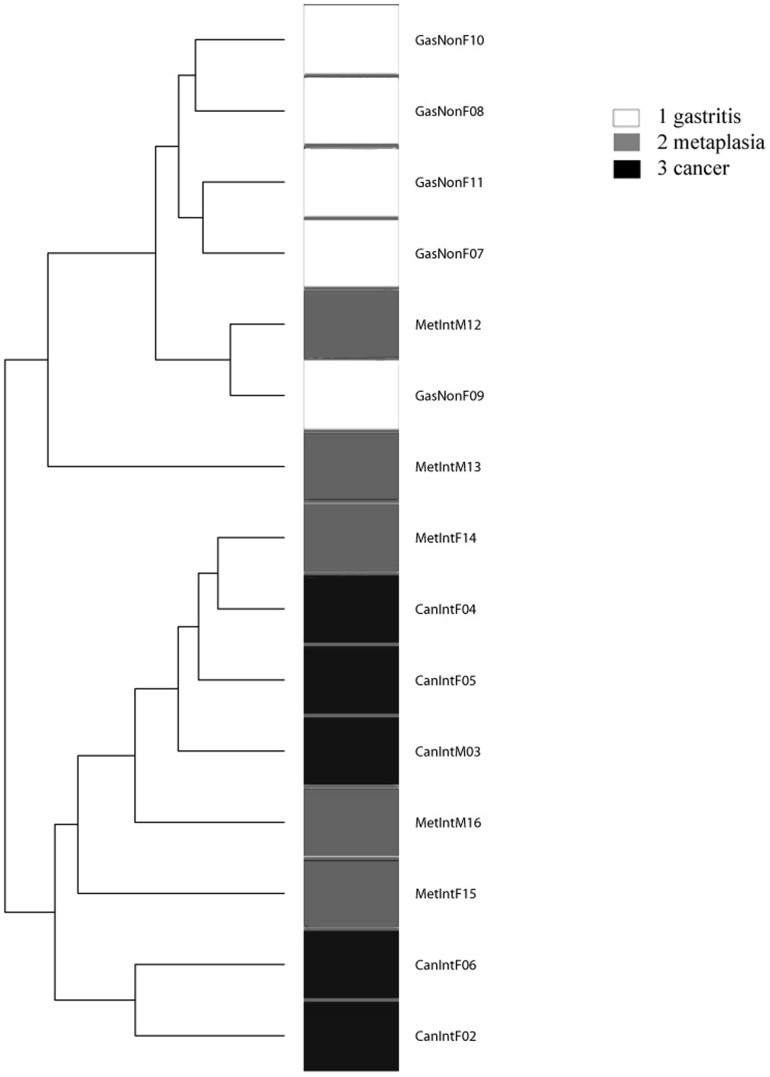
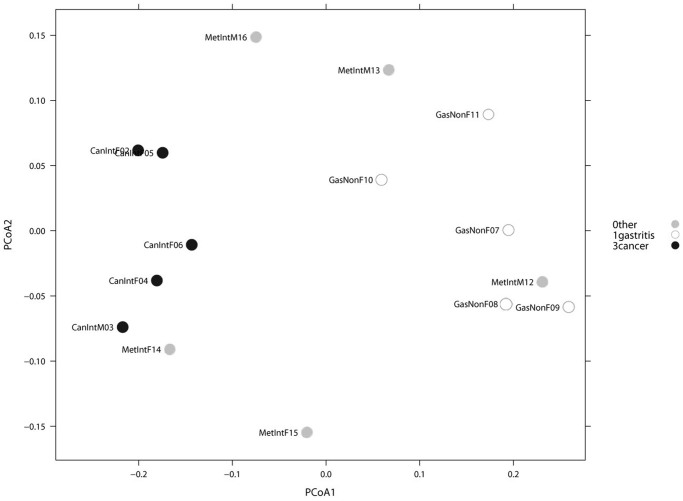
We first analyzed the possible effect of sex and age on microbiota composition given presence/absence of 283 taxa present in at least one sample. A Principal Coordinate Analysis (PCoA) including all 15 patients, and based on unweighted Unifrac distance showed no significant microbiota differences based on either age or sex (p-value > 0.50, Adonis test). A similar PCoA analyses comparing NAG vs IM vs GC, given presence/absence of 283 taxa present in at least one sample was done (Figure 3). The Adonis test yielded a p-value of 0.026, indicating a significant microbiota difference between at least one of the disease groups from the others. In addition, a binary metric analysis revealed separation of GC microbiota from NAG, whereas microbiota of IM was not separated from either GC or NAG groups. To further confirm these findings, a PCoA comparing NAG vs GC was done, and the Adonis test yielded a p-value of 0.005, indicating a significant microbiota difference between these two groups. In addition, comparison between IM and NAG (p = 0.42) and between IM and GC (p = 0.129) showed no difference between groups. Hierarchical clustering analysis (HC-AN) based on presence/absence of 283 taxa revealed two clusters, one containing only GC and IM and the other NAG and IM, with one GC possible outlier (case CanIntF06).
Figure 3. PCoA analyses Unweighted Unifrac.
PCoA analyses of the gastric microbiota in non-atrophic gastritis vs intestinal metaplasia vs gastric cancer, based on Unweighted Unifrac distance between samples given presence/absence of 283 taxa present in at least one sample. Axis 1 explained 30% of variation and axis 2, 13%. Adonis test yielded a p-value of 0.026, indicating significant microbiota difference between at least one of the sample categories from the others.
We next did PCoA analyses based on wUnifrac distance between samples given abundance of 283 taxa present in at least one sample. Whereas abundance metric analysis showed no significant separation of microbiota according to disease group, the presence/absence metric analyses showed a significant difference between NAG and GC (p-value = 0.03, Adonis test). The same analyses revealed no significant differences between NAG and IM and between GC and IM; although together GC and IM showed significant microbiota difference when compared with the NAG group (p = 0.048, Adonis test).
Analysis based on taxa with significant abundance differences across groups confirmed microbiota separation between GC and NAG, and overlapping of IM with both
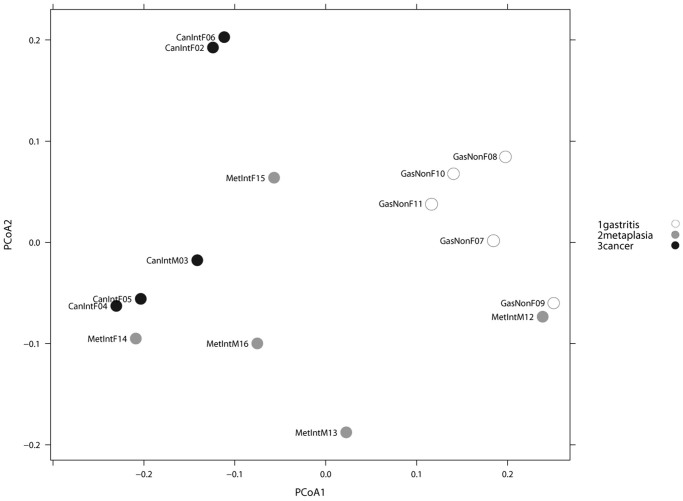
We performed a parametric Welch test to identify OTUs that were significantly increased or decreased across at least one disease group, and found 27 taxa with significant abundance difference across groups. A PCoA weighted Unifrac distance between groups based on the 27 taxa, showed no significant separation of microbiota between all three groups (NAG vs IM vs GC). However, when considering only NAG and GC patients, an entire separation of the microbiota along PCoA1 axis was observed (Figure 4). HC-AN analysis based on the same 27 taxa showed that NAG and GC clustered apart, whereas IM cases overlapped with the two NAG and GC groups (Figure 5).
Figure 4. PCoA analyses Weighted Unifrac.
PCoA analyses of the gastric microbiota in non-atrophic gastritis vs intestinal metaplasia vs gastric cancer, based on Weighted Unifrac distance between samples given abundance of 27 taxa with significant abundance difference across at least one of the categories. Axis 1, explained 55% of variation and axis 2, 22%. An entire separation of the microbiota of non-atrophic gastritis and gastric cancer groups along the PCoA1 axis was observed.
Figure 5. Hierarchical Clustering of the gastric microbiota.
Analysis based on Weighted Unifrac distance between samples given abundance of 27 taxa with significant abundance differences across at least one of the categories. A distinct clustering was observed between non-atrophic gastritis and gastric cancer, whereas intestinal metaplasia overlapped with these two groups.
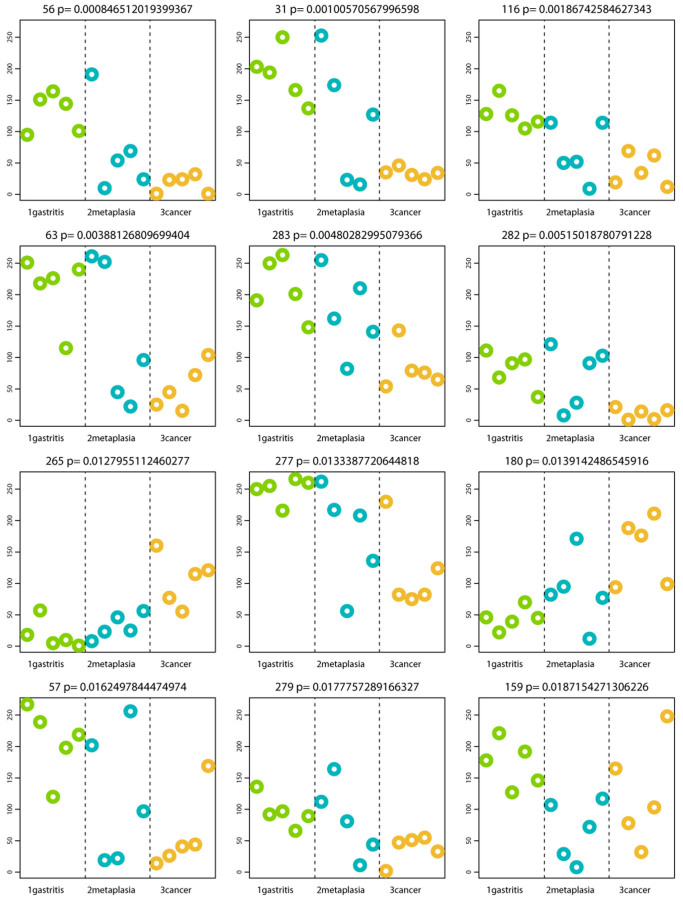
From the 27 taxa with significant abundance differences across groups, we selected the 12 with the more significant p-values (Table 1) and plotted their abundance in each disease group (Figure 6). The results showed that 5 of these top 12 displayed a trend to decrease from NAG to IM to GC (2 TM7, 2 Porphyromonas and 1 Neisseria), whereas 2 significantly increased from NAG to IM to GC (Lactobacillus coleohominis and Lachnospiraceae).
Table 1. Description of the top 12 OTUs with the lowest p-value (parametric Welch test) that were significantly increased or decreased across at least one disease group; comparison of gastric cancer with intestinal metaplasia and with non-atrophic gastritis.
| Taxa ID | Phylum | Class | Order | Family | Genus | Species | Trend |
|---|---|---|---|---|---|---|---|
| 56 | TM7 | TM7-3 | CW040 | F16 | unclassified | unclassified | Decrease |
| 31 | Bacteroidetes | Bacteroidia | Bacteroidales | Porphyromonadaceae | Porphyromonas | unclassified | Decrease |
| 116 | TM7 | TM7-3 | EW055 | unclassified | unclassified | unclassified | Decrease |
| 63 | Proteobacteria | Betaproteo-bacteria | Neisseriales | Neisseriaceae | Neisseria | unclassified | Decrease |
| 283 | Bacteroidetes | Bacteroidia | Bacteroidales | Porphyromonadaceae | Porphyromonas | unclassified | Decrease |
| 282 | Bacteroidetes | Bacteroidia | Bacteroidales | Prevotellaceae | Prevotella | pallens | Decrease |
| 265 | Firmicutes | Bacilli | Lactobacillales | Lactobacillaceae | Lactobacillus | coleohominis | Increase |
| 277 | Proteobacteria | Gammaproteo-bacteria | Pasteurellales | Pasteurellaceae | Haemophilus | unclassified | Decrease |
| 180 | Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | unclassified | unclassified | Increase |
| 57 | Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | Streptococcus | sinensis | Decrease |
| 279 | Proteobacteria | Betaproteo-bacteria | Neisseriales | Neisseriaceae | Bergeriella | denitrificans | Decrease |
| 159 | Tenericutes | Erysipelotrichi | Erysipelotrichales | Erysipelotrichaceae | Bulleidia | unclassified | Decrease |
Figure 6. Profiles of the top 12 OTUs generating the lowest p-values distinguishing disease groups.
P-values shown at top of each OTU plot are unadjusted for multiple testing. The y-axis represents the HybScore (integers of fluorescence intensity). Samples are grouped and colored by category along the x-axis in the following order: GasNonF08, GasNonF09, GasNonF10, GasNonF11; MetIntM12, MetIntM13, MetIntF14, MetIntF15, MetIntM16; CanIntF02, CanIntM03, CanIntF04, CanIntF05, CanIntF06.
Analysis based on taxa with significant abundance differences between NAG and GC confirmed significant microbiota separation of these groups
To further document the observed differences between NAG and GC, a parametric Welch test was done to look for OTUs that significantly increased or decreased between these two groups. The test identified 44 OTUs with a p-value < 0.05 and with these a PCoA weighted Unifrac distance analyses showed a significant separation of the microbiota of NAG vs GC (Figure 7). A HC-AN with the above 44 OTUs also showed separate clusters of NAG and GC (results not shown). A profile of the top 12 with the lowest p-values showed that 9 of the 12 displayed a significant increase in NAG patients, whereas Lactobacillus coleohominis and Lachnospiraceae were significantly more abundant in GC patients (Table 2).
Figure 7. Non-atrophic gastritis vs gastric cancer.
PCoA analyses of the gastric microbiota in non-atrophic gastritis vs gastric cancer, based on Weighted Unifrac distance between samples, given abundance of 44 taxa with significant abundance difference across at least one of the categories. Axis 1 explained 62% of variation and axis 2, 14%. A significant separation of microbiota between non-atrophic gastritis and gastric cancer was observed.
Table 2. Description of the top 12 OTUs with the lowest p-value (parametric Welch test) that were significantly increased or decreased across at least one disease group; comparison of gastric cancer with non-atrophic gastritis.
| Taxa No | Phylum | Class | Order | Family | Genus | Species | Trend |
|---|---|---|---|---|---|---|---|
| 56 | TM7 | TM7-3 | CW040 | F16 | unclassified | unclassified | Decrease |
| 116 | TM7 | TM7-3 | EW055 | unclassified | unclassified | unclassified | Decrease |
| 31 | Bacteroidetes | Bacteroidia | Bacteroidales | Porphyromonadaceae | Porphyromonas | unclassified | Decrease |
| 63 | Proteobacteria | Betaproteo-bacteria | Neisseriales | Neisseriaceae | Neisseria | unclassified | Decrease |
| 283 | Bacteroidetes | Bacteroidia | Bacteroidales | Porphyromonadaceae | Porphyromonas | unclassified | Decrease |
| 57 | Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | Streptococcus | sinensis | Decrease |
| 282 | Bacteroidetes | Bacteroidia | Bacteroidales | Prevotellaceae | Prevotella | pallens | Decrease |
| 279 | Proteobacteria | Betaproteo-bacteria | Neisseriales | Neisseriaceae | Bergeriella | denitrificans | Decrease |
| 265 | Firmicutes | Bacilli | Lactobacillales | Lactobacillaceae | Lactobacillus | coleohominis | Increase |
| 180 | Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | unclassified | unclassified | Increase |
| 277 | Proteobacteria | Gammaproteo-bacteria | Pasteurellales | Pasteurellaceae | Haemophilus | unclassified | Decrease |
| 222 | Proteobacteria | Alphaproteo-bacteria | Rhodospirillales | Rhodospirillaceae | unclassified | unclassified | Increase |
Discussion
In this study we tested the hypothesis that the microbiota of the gastric mucosa changes from patients with NAG to patients with IM and to patients with GC. To test this we used the generation G3 PhyloChip™, with an increased sensitivity for low abundance bacteria10. This is particularly relevant in the study of stomach samples where the microenvironment is restrictive for bacterial colonization. Indeed, we found low microbiota diversity, with a total of 12 phyla and 283 OTUs detected, whereas genus diversity varied from 8 up to 57 in all patients. Our results are not comparable with previous studies because of differences in techniques and populations; still, agreement exist in the low number of phyla detected and in the predominance of Firmicutes, Proteobacteria, Bacteroidetes and Actinobacteria in stomach microbiota7,11,12. However, with the use of generation 3 PhyloChip we were able to detect more OTUs and higher diversity at the genus level. Thus, it has been reported that in the gastric mucosa the most abundant families are Streptococcaceae and Prevotellaceae8,11, which contrast with our results where Lachnospiraceae was the most abundant, representing almost 20% of the microbiota in all patients. The Lachnospiraceae family is formed by strict anaerobes, of which a new genera has been recently described in the mouth13. In the intestine, members of this family have been reported to exert a protective role against C. difficile colonization after microbiota has been modified with antibiotics14. In addition, it has been suggested that members of Lachnospiraceae family may be associated with a reduced intestinal carcinogenesis15.
We also found in several of the patients the species Haemophilus parainfluenzae and Veillonella ratti, which have been reported previously only in the urogenital tract16,17, and members of the families Desulfobacteraceae and Acidobacteriaceae (data not shown), which have been described as free living bacteria18,19. The presence of these low abundance bacteria in several cases suggests they are not a contaminant, and their identification might be due to the improved sensitivity of G3 PhyloChip10. Results from this and other studies suggest that the gastric mucosa may be colonized with bacteria previously undescribed in the stomach or even in humans7.
The low diversity observed in this study contrasts with a recent report using the G2 PhyloChip that detected substantially higher diversity in adults with gastritis20, as many as 44 phyla, many of them in very low frequencies, although there was agreement in the dominant phyla. Differences with our study might be explained in part because of differences in population, since they sampled Amerindians from the Venezuelan Amazon with a particular ancestry and poor socioeconomic and hygiene conditions.
Concerning changes in the microbiota, we found evidence of a gradual change in patients from NAG to IM to GC, bacterial diversity showed a trend to diminish from NAG to IM to GC, and was significantly higher in NAG than in GC patients. These results would suggest that changes in the gastric mucosa as patients move to preneoplasia and to cancer render the environment more restrictive to bacterial growth, contrary to what others and we have hypothesized21. Mucins are produced by normal epithelial cells along the gastrointestinal tract and serve as a protective barrier and as specialized niche for colonization of microbiota22. A healthy gastric mucosa produces MUC5AC and MUC6 mucins23, which the gastric microbiota colonizes; but when normal mucosa is replaced by atrophy or pre-neoplastic lesions the mucins change to metaplastic expression of intestinal MUC224, and we suggest this makes the mucosa less suitable for colonization by the normal microbiota.
When whole microbiota profile was studied, ordination analyses based on both, presence/absence or abundance, revealed a significant separation of GC from NAG, whereas IM did not separate from either NAG or GC. A cluster analyses also showed two separate groups for NAG and GC with the IM cases intermingled with GC and NAG groups. These findings would suggest a change from NAG to GC, with IM as an intermediate group. This observation was possible because our study included IM patients, in contrast to a previous report where GC was compared only with NAG patients8. Recent studies in a transgenic hypergastrinemic mouse model found that infection with H. pylori in the presence of intestinal flora caused the development of gastrointestinal intraepithelial neoplasia25; even colonization with a very restricted intestinal flora was found sufficient to induce neoplasia26. These observations suggest that presence of other bacteria in addition to H. pylori are needed for the development of gastric cancer, although they do not really help elucidate whether an increased or a decreased bacterial diversity is responsible for this adjuvant effect.
Of particular interest is to highlight that one of the taxa that was significantly more abundant in GC than in NAG was the genus Pseudomonas (data not shown), which is relevant in light of a recent report in patients with stomach adenocarcinoma that found specific integration of Pseudomonas-like DNA in somatic cells, in the 5′-UTR and 3′-UTR of four proto-oncogenes that are up-regulated in GC27. In fact, this is the first study to document bacterial-human somatic cell lateral gene transfer in cancer patients, and our results on predominance of Pseudomonas in GC would be in agreement with this important report.
A further selection of the OTUs with the lowest p-values showed 6 taxa with a clear decreasing trend from NAG to IM to GC (2 TM7, 2 Porphyromonas sp, Neisseria sp and Streptococcus sinensis), and 2 with an increasing trend (Lactobacillus coleohominis and Lachnospiraceae). TM7 is a recently described phylum present in the mouth and intestine of humans; and a small fraction of its genes have sequence similarity to genes found in members of the classes Bacilli, Clostridia and Fusobacteria28. TM7 encodes genes for Type IV pili that is recognized as a virulence factor in some pathogenic bacteria, and may also have a role in inflammatory diseases such as Crohn's disease29 and colitis30. Streptococcus sinensis has been reported in the oral cavity31 and associated with infective endocarditis32. On the other hand, L. coleohominis is a relatively novel species recovered from the vagina33 and from urine of healthy woman34, but no reports exist on any disease association. Lachnospiracea family has been reported to significantly diminish in patients with inflammatory disease35. Thus, many of the bacterial groups we found altered in the gastric mucosa of patients with GC have been reported associated to inflammatory processes, and may conceivable play a role in the regulation of the inflammatory response observed in the gastric wall associated with the development of GC.
A previous study reported no difference in stomach microbiota between GC and dyspeptic patients, although in some cancer patients species of Streptococcus, Lactobacillus and Veillonella were more prevalent8. This study was done based on cloning, sequencing and restriction fragment length polymorphisms of 16S rRNA, which has important limitations in sensitivity and coverage compared with more recently developed techniques, including the G3 PhyloChip. Still, in agreement with our results, they also reported dominance of Lactobacillus in GC. To our knowledge, there are no additional reports describing microbiota in patients with GC, and studies are necessary to learn more on the role of stomach microbiota in health and disease. Studies of microbiota in other human tissues revealed important diversity between individuals and significant differences among different human populations, even in studies using the same sample handling and technological platform36,37. We need to address the study of microbiota in the stomach using a similar approach, with studies involving larger groups and different human populations. We acknowledge that an important draw back of our study is the reduced number of patients, which allowed us finding differences between disease groups, but fall short to study the effect of other variables within groups; e.g., we had a small number of males in the NAG and GC groups. These issues should be addressed by studying a larger number of cases.
The current study is the first to show differences in the stomach microbiota of patients with gastric cancer, and suggest a gradual shift in microbiota profile from gastritis to pre-neoplastic lesion to cancer. The study also suggests that a decrease in Porphyromonas, Neisseria, TM7 group, and S. sinensis, as well as an increase in L. coleohominis and Lachnospiraceae might favor development of gastric cancer. The role of these taxa and its mechanisms of pathogenesis in gastric cancer remain to be studied.
Methods
Patients
We included patients consulting the Oncology and General hospitals in the Medical Center SXXI, Instituto Mexicano del Seguro Social (IMSS), in Mexico City. Five patients each with NAG, IM, and GC of the intestinal type were selected for the study. For both, the NAG and IM patients we selected tissue samples from the antrum, whereas for the GC group we worked with tissue from the lesion, which in 4 cases extended in both antrum and corpus, and one involved fundus and corpus. The characteristics of these patients are described in Table 3; the groups of patients studied did not differ significantly in body mass index or school years, although there were limited number of males, and patients with NAG were younger than those with IM and GC. We excluded patients with immunodeficiencies, diabetes or other chronic diseases, and patients who received antibiotics or proton pump inhibitors during the last three months, or patients who previously received therapy for H. pylori eradication. H. pylori diagnosis was done using an enzyme-linked immunosorbent assay previously validated in our population38. All patients were Mexican-mestizo with a medium to low socioeconomic level. Patients were informed about the study and those willing to participate signed a consent letter. The IRB committee from IMSS approved the study.
Table 3. General characteristics of the patients included in the study.
| Group, ID | Age | Sex | H. pylori, serology | Body mass Index | School years |
|---|---|---|---|---|---|
| Non-atrophic gastritis | |||||
| F07 | 76 | Female | + | 22.8 | 12 |
| F08 | 37 | Female | + | 27.5 | 8 |
| F09 | 43 | Female | + | 21.3 | 16 |
| F10 | 32 | Female | + | 21.6 | 8 |
| F11 | 32 | Female | − | 24.8 | 3 |
| Intestinal metaplasia | |||||
| M12 | 63 | Male | + | 23.6 | 6 |
| M13 | 71 | Male | + | 20.7 | 2 |
| F14 | 71 | Female | + | 24.6 | 12 |
| F15 | 60 | Female | + | 27.1 | 6 |
| M16 | 70 | Male | + | 31.9 | 12 |
| Intestinal Gastric cancer | |||||
| F02 | 79 | Female | + | 24.25 | 4 |
| M03 | 59 | Male | − | 26.66 | 8 |
| F04 | 82 | Female | + | 26.56 | 2 |
| F05 | 52 | Female | + | 27.39 | 6 |
| F06 | 81 | Female | + | 19.77 | 6 |
Gastric samples
Patients with NAG and with IM were subjected to endoscopy and biopsies from antrum and corpus were taken, whereas patients with GC were subjected to surgery to remove the tumor. Samples of either NAG, IM or GC were processed as follows: one biopsy or a fraction of the tumor lesion was immediately placed in formalin for histological studies, and another tissue fraction placed in liquid nitrogen for transportation to our lab, where both biopsies and tissues were stored at −70°C until tested. Fixed tissue was stained with Hematoxylin and Eosin and modified Giemsa, and with periodic acid.Schiff/Alcian blue staining when metaplasia was suspected. Preparations were examined by a single experienced pathologist and final diagnosis assigned using previously reported criteria39,40,41.
DNA extraction and PCR amplification for 16S rRNA microarray
The tissue samples from the 15 patients were thawed and DNA extracted using the QIAamp DNA easy mini kit (Qiagen, Hilden, Germany). Isolated lyophilized DNA was then hydrated and quantified using the PicroGreen kit (Life technologies, Carlsbad, CA). Samples were subjected to amplification using degenerate 16S rRNA gene primers 27F.1 (5′-AGRGTTTGATCMTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′) for bacteria and 4Fa and 1492R for archaea as described in Hazen et al.10. Amplified PCR products were concentrated, purified and quantified, and 500 ng of amplified product were fragmented to a range of 50–200 bp and then biotin labeled (Affymetrix WT Double Stranded DNA Terminal Labeling) for hybridization.
Microarray hybridization
The Phylochip Generation 3 (G3) microarray was used for hybridization, and samples were run at Second Genome, INC. (San Francisco, CA). This includes 1.1 million DNA probes able to categorize all known bacteria and archaeal operational taxonomic units (OTUs) into over 50,000 taxa using 59,959 clusters of 17 nucleotides as probes representing 147 phyla, 1123 classes, 1219 orders and 1464 families, for a total of 27938 OTUs. Staining and scanning were performed according to the manufacturer instructions (Affymetrix) and following previously described procedures10. PhyloChip has built-in controls for all steps of the microarray hybridization process and normalization of the probe intensity values. Fluorescence intensity is coded as integers ranging from 0 to 65,536. Hybridization score (HybScore) values are in the range of 0 to 16000 and changes by 1000 in the OUT's HybScore indicate a two-fold increase in the fluorescence intensity.
Data reduction and statistical analysis
Taxa are filtered to those present in at least one of the samples, or to taxa significantly increased in their abundance in one category compared to the alternate categories. For the abundance filter, the parametric Welch test was employed to calculate p-values. Additionally, q-values were calculated using the Benjamini-Hochberg procedure to correct p-values, controlling for false discovery rates10.
After the taxa were identified for inclusion in the analysis, the values used for each taxa-sample intersection were defined in two distinct ways. In the first case, the abundance metrics were used directly (AT). In the second case, binary metrics were created (absence, 0; presence, 1) (BT). A non-paired, heteroscadastic student's t-test assuming unequal variances was used to test for differences in microbial diversity and hierarchical summarization between two sample categories.
To evaluate sample-to-sample distance all profiles were inter-compared in a pair-wise fashion to determine a dissimilarity score and stored it in a distance dissimilarity matrix. The distance functions were chosen to allow similar biological samples to produce only small dissimilarity scores. The Unifrac distance metric utilizes the phylogenetic distance between OTUs to determine the dissimilarity between communities42. Unifrac was used for presence/absence, whereas Weighted Unifrac (WUnifrac) was used for abundance data.
Two-dimensional ordinations and hierarchical clustering maps of the samples in the form of dendrograms were created to graphically summarize the inter-sample relationships. To create dendrograms, the samples from the distance matrix were clustered hierarchically using the average-neighbor (HC-AN) method. Principal Coordinate Analyses (PCoA) is a method of two-dimensional ordination plotting used to visualize complex relationships between samples. PCoA uses the dissimilarity values to position the points relative to each other. The Adonis test was used to evaluate statistically significant differences. In this randomization/Monte Carlo permutation test, the samples are randomly reassigned to the various sample categories, and the between-category differences are compared to the true between-category differences. Adonis utilizes the sample-to-sample distance matrix directly. Data analyses were done using the PhyCA-Stats™ analyses software (Second Genome, San Francisco, CA).
Author Contributions
J.T. design the study; J.T. and F.A.J. analyzed data and wrote the main manuscript; F.V.J. processed clinical samples for microbiota analyses; R.M.G. was responsible for patient's selection and clinical diagnosis; A.M. was responsible for selection of tissue samples and pathology studies. All authors reviewed the manuscript.
Acknowledgments
This work was supported by CONACYT (2007-C01-69450) and the Coordinación de Investigación en Salud, IMSS (Fis/G12/1102), Mexico. J.T. is a recipient of a exclusivity scholarship from Fundación IMSS, Mexico.
References
- Bjorkholm B., Falk P., Engstrand L. & Nyren O. Helicobacter pylori: resurrection of the cancer link. J Intern Med 253, 102–119, 1119 (2003). [DOI] [PubMed] [Google Scholar]
- Allison C. C. & Ferrero R. L. Role of virulence factors and host cell signaling in the recognition of Helicobacter pylori and the generation of immune responses. Future Microbiol 5, 1233–1255, 10.2217/fmb.10.84 (2010). [DOI] [PubMed] [Google Scholar]
- Gianfagna F., De Feo E., van Duijn C. M., Ricciardi G. & Boccia S. A systematic review of meta-analyses on gene polymorphisms and gastric cancer risk. Curr Genomics 9, 361–374, 10.2174/138920208785699544 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsugane S. & Sasazuki S. Diet and the risk of gastric cancer: review of epidemiological evidence. Gastric Cancer 10, 75–83, 10.1007/s10120-007-0420-0 (2007). [DOI] [PubMed] [Google Scholar]
- Correa P. & Houghton J. Carcinogenesis of Helicobacter pylori. Gastroenterology 133, 659–672, 10.1053/j.gastro.2007.06.026 (2007). [DOI] [PubMed] [Google Scholar]
- Uemura N. et al. Helicobacter pylori infection and the development of gastric cancer. N Engl J Med 345, 784–789, 10.1056/NEJMoa001999 (2001). [DOI] [PubMed] [Google Scholar]
- Bik E. M. et al. Molecular analysis of the bacterial microbiota in the human stomach. Proc Natl Acad Sci USA 103, 732–737, 10.1073/pnas.0506655103 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dicksved J. et al. Molecular characterization of the stomach microbiota in patients with gastric cancer and in controls. J Med Microbiol 58, 509–516, 10.1099/jmm.0.007302-0 (2009). [DOI] [PubMed] [Google Scholar]
- Talley N. J., Ormand J. E., Frie C. A. & Zinsmeister A. R. Stability of pH gradients in vivo across the stomach in Helicobacter pylori gastritis, dyspepsia, and health. Am J Gastroenterol 87, 590–594 (1992). [PubMed] [Google Scholar]
- Hazen T. C. et al. Deep-sea oil plume enriches indigenous oil-degrading bacteria. Science 330, 204; 10.1126/science.1195979 (2010). [DOI] [PubMed] [Google Scholar]
- Li X. X. et al. Bacterial microbiota profiling in gastritis without Helicobacter pylori infection or non-steroidal anti-inflammatory drug use. PloS One 4, e7985, 10.1371/journal.pone.0007985 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson A. F. et al. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PloS One 3, e2836, 10.1371/journal.pone.0002836 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sizova M. V. et al. Stomatobaculum longum gen. nov., sp. nov., an obligately anaerobic bacterium from the human oral cavity. Int J Syst Evol Microbiol 63, 1450–1456, 10.1099/ijs.0.042812-0 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves A. E., Koenigsknecht M. J., Bergin I. L. & Young V. B. Suppression of Clostridium difficile in the gastrointestinal tracts of germfree mice inoculated with a murine isolate from the family Lachnospiraceae. Infect Immun 80, 3786–3794, 10.1128/IAI.00647-12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai V., Colbert L. H., Perkins S. N., Schatzkin A. & Hursting S. D. Intestinal microbiota: a potential diet-responsive prevention target in ApcMin mice. Mol Carcinog 46, 42–48, 10.1002/mc.20233 (2007). [DOI] [PubMed] [Google Scholar]
- Drouet E. B., Denoyel G. A., Boude M. M., Boussant G. & de Montclos H. P. Distribution of Haemophilus influenzae and Haemophilus parainfluenzae biotypes isolated from the human genitourinary tract. Eur J Clin Microbiol Infect Dis 8, 951–955 (1989). [DOI] [PubMed] [Google Scholar]
- Marchandin H. et al. Molecular identification of the first human isolate belonging to the Veillonella ratti-Veillonella criceti group based on 16S rDNA and dnaK gene sequencing. Res Microbiol 156, 603–607, 10.1016/j.resmic.2005.01.013 (2005). [DOI] [PubMed] [Google Scholar]
- Kjeldsen K. U., Jakobsen T. F., Glastrup J. & Ingvorsen K. Desulfosalsimonas propionicica gen. nov., sp. nov., a halophilic, sulfate-reducing member of the family Desulfobacteraceae isolated from a salt-lake sediment. Int J Syst Evol Microbiol 60, 1060–1065, 10.1099/ijs.0.014746-0 (2010). [DOI] [PubMed] [Google Scholar]
- Yang S., Wen X., Jin H. & Wu Q. Pyrosequencing investigation into the bacterial community in permafrost soils along the China-Russia Crude Oil Pipeline (CRCOP). PloS One 7, e52730, 10.1371/journal.pone.0052730 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado-Contreras A. et al. Structure of the human gastric bacterial community in relation to Helicobacter pylori status. ISME J 5, 574–579, 10.1038/ismej.2010.149 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engstrand L. & Lindberg M. Helicobacter pylori and the gastric microbiota. Best Pract Res Clin Gastroenterol 27, 39–45, 10.1016/j.bpg.2013.03.016 (2013). [DOI] [PubMed] [Google Scholar]
- Lievin-Le Moal V. & Servin A. L. The front line of enteric host defense against unwelcome intrusion of harmful microorganisms: mucins, antimicrobial peptides, and microbiota. Clin Microbiol Rev 19, 315–337, 10.1128/CMR.19.2.315-337.2006 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillipson M. et al. The gastric mucus layers: constituents and regulation of accumulation. Am J Physiol Gastrointest Liver Physiol 295, G806–812, 10.1152/ajpgi.90252.2008 (2008). [DOI] [PubMed] [Google Scholar]
- Hollingsworth M. A. & Swanson B. J. Mucins in cancer: protection and control of the cell surface. Nat Rev Cancer 4, 45–60, 10.1038/nrc1251 (2004). [DOI] [PubMed] [Google Scholar]
- Lofgren J. L. et al. Lack of commensal flora in Helicobacter pylori-infected INS-GAS mice reduces gastritis and delays intraepithelial neoplasia. Gastroenterology 140, 210–220, 10.1053/j.gastro.2010.09.048 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lertpiriyapong K. et al. Gastric colonisation with a restricted commensal microbiota replicates the promotion of neoplastic lesions by diverse intestinal microbiota in the Helicobacter pylori INS-GAS mouse model of gastric carcinogenesis. Gut 63, 54–63, 10.1136/gutjnl-2013-305178 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley D. R. et al. Bacteria-Human Somatic Cell Lateral Gene Transfer is Enriched in Cancer Samples. PLoS Comput Biol 9, e1003107, 10.1371/journal.pcbi.1003107 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcy Y. et al. Dissecting biological “dark matter” with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth. Proc Natl Acad Sci U S A 104, 11889–11894, 10.1073/pnas.0704662104 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehbacher T. et al. Intestinal TM7 bacterial phylogenies in active inflammatory bowel disease. J Med Microbiol 57, 1569–1576, 10.1099/jmm.0.47719-0 (2008). [DOI] [PubMed] [Google Scholar]
- Elinav E. et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell 145, 745–757, 10.1016/j.cell.2011.04.022 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P. C. et al. The oral cavity as a natural reservoir for Streptococcus sinensis. Clin Microbiol Infect 14, 1075–1079 (2008). [DOI] [PubMed] [Google Scholar]
- Faibis F. et al. Streptococcus sinensis: an emerging agent of infective endocarditis. J Med Microbiol 57, 528–531, 10.1099/jmm.0.47528-0 (2008). [DOI] [PubMed] [Google Scholar]
- Nikolaitchouk N. et al. Lactobacillus coleohominis sp. nov., isolated from human sources. Int J Syst Evol Microbiol 51, 2081–2085 (2001). [DOI] [PubMed] [Google Scholar]
- Siddiqui H., Nederbragt A. J., Lagesen K., Jeansson S. L. & Jakobsen K. S. Assessing diversity of the female urine microbiota by high throughput sequencing of 16S rDNA amplicons. BMC Microbiol 11, 244, 10.1186/1471-2180-11-244 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry D. & Reinisch W. Intestinal microbiota: A source of novel biomarkers in inflammatory bowel diseases? Best Pract Res. Clin Gastroenterol 27, 47–58, 10.1016/j.bpg.2013.03.005 (2013). [DOI] [PubMed] [Google Scholar]
- Eckburg P. B. et al. Diversity of the human intestinal microbial flora. Science 308, 1635–1638, 10.1126/science.1110591 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumugam M. et al. Enterotypes of the human gut microbiome. Nature 473, 174–180, 10.1038/nature09944 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres J. et al. A community-based seroepidemiologic study of Helicobacter pylori infection in Mexico. J Infect Dis 178, 1089–1094 (1998). [DOI] [PubMed] [Google Scholar]
- Dixon M. F., Genta R. M., Yardley J. H. & Correa P. Classification and grading of gastritis. The updated Sydney System. International Workshop on the Histopathology of Gastritis, Houston 1994. The American journal of surgical pathology 20, 1161–1181 (1996). [DOI] [PubMed] [Google Scholar]
- Lauren P. The Two Histological Main Types of Gastric Carcinoma: Diffuse and So-Called Intestinal-Type Carcinoma. An Attempt at a Histo-Clinical Classification. Acta Pathol Microbiol Scand 64, 31–49 (1965). [DOI] [PubMed] [Google Scholar]
- Ming S. C. Gastric carcinoma. A pathobiological classification. Cancer 39, 2475–2485 (1977). [DOI] [PubMed] [Google Scholar]
- Lozupone C., Hamady M. & Knight R. UniFrac--an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics 7, 371, 10.1186/1471-2105-7-371 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]