Abstract
DNA repair genes that have been inactivated by promoter methylation offer potential therapeutic targets either by targeting the specific repair deficiency, or by synthetic lethal approaches. This study evaluated promoter methylation status for eight selected DNA repair genes (ATM, BRCA1, ERCC1, MGMT, MLH1, NEIL1, RAD23B and XPC) in 56 non-small cell lung cancer (NSCLC) tumours and 11 lung cell lines using the methylation-sensitive high resolution melting (MS-HRM) methodology. Frequent methylation in NEIL1 (42%) and infrequent methylation in ERCC1 (2%) and RAD23B (2%) are reported for the first time in NSCLC. MGMT methylation was detected in 13% of the NSCLCs. Contrary to previous studies, methylation was not detected in ATM, BRCA1, MLH1 and XPC. Data from The Cancer Genome Atlas (TCGA) was consistent with these findings. The study emphasises the importance of using appropriate methodology for accurate assessment of promoter methylation.
There is accumulating evidence that tumour response to DNA-damaging agents is associated with the expression levels of DNA repair genes. BRCA1 mRNA expression has shown to be associated with cisplatin resistance1,2,3 and docetaxel sensitivity4,5 in breast, ovarian, lung and gastric cancers. Tailored chemotherapy based on BRCA1 mRNA levels has also shown to improve patient survival in lung2,6, bladder7 and ovarian cancers8. Similarly, high ERCC1 and MGMT mRNA expression has shown to confer resistance to platinum compounds9 and sensitivity to alkylating agents10,11. Expression profiling of selected DNA repair genes thus has potential for the better stratification of cancer patients who are likely to respond to DNA-damaging agents12.
Formalin-fixed paraffin-embedded (FFPE) tissues are commonly the only available clinical material for diagnostic analysis and for cancer biomarker studies. When FFPE tissue is used, expression profiling of DNA repair genes is often limited by low quantity and degraded RNA due to the detrimental effects of formalin fixation on nucleic acids. Moreover, normal cell contamination that is almost inevitably occurring during RNA extraction, even after macrodissection of tumour-enriched regions, confounds the interpretation of gene expression data.
Promoter methylation is an epigenetic mechanism leading to transcriptional silencing of gene expression and thus has been used for indirect assessment of gene expression. The use of relatively stable DNA for testing of molecular biomarkers provides an important advantage over RNA-based testing with respect to extraction, handling and storage conditions13. In human cancers, a number of DNA repair genes have been shown to undergo transcriptional silencing by DNA methylation14.
Previous studies have reported the occurrence of promoter methylation in several DNA repair genes in non-small cell lung cancer (NSCLC). However, the reported frequency of promoter methylation in NSCLC varies markedly between studies; ATM (0–47%), BRCA1 (4–30%), MGMT (8–50%) and MLH1 (0–68%) (Table 1). This high variation reflects the different methodologies used and emphasises that the DNA methylation status of clinically relevant genes needs to be validated using reliable and reproducible methodology before testing for their methylation has any clinical validity.
Table 1. Methylation of DNA repair genes in NSCLC reported in the literature.
| Gene | Sample | Frequency (%) | Method | Reference |
|---|---|---|---|---|
| ATM | 105 tumours | 47 | MSP | 50 |
| 37 tumours | 0 | MSP | 17 | |
| 180 tumours | 19 | MSP | 51 | |
| BRCA1 | 28 tumours | 18 | 3-D microarray with linker-PCR | 52 |
| 98 tumours | 30 | MSP | 39 | |
| 158 tumours | 4 | MSP | 38 | |
| MGMT | 220 tumours | 50 | MSP | 53 |
| 49 tumours | 8 | MethyLight | 54 | |
| 122 tumours | 30 | MSP | 55 | |
| 72 tumours | 17 | MSP | 56 | |
| 105 tumours | 10 | MSP | 50 | |
| MLH1 | 31 tumours | 0 | MSP | 57 |
| 105 tumours | 59 | MSP | 50 | |
| 239 tumours | 35 | MSP | 58 | |
| 77 tumours | 56 | COBRA | 59 | |
| 116 tumours | 68 | COBRA | 60 | |
| 49 tumours | 2 | MethyLight | 54 | |
| XPC | 158 tumours | 33 | MSP | 23 |
| ERCC1 | Not reported | - | - | - |
| NEIL1 | Not reported | - | - | - |
| RAD23B | Not reported | - | - | - |
It is important to note that most of the previous studies used methylation-specific PCR (MSP), a method that has multiple limitations15,16. Detection of methylation status by MSP is based on end-point analysis of PCR amplification by gel electrophoresis, which only provides qualitative results. As a consequence, the level of methylation and the pattern of methylation (homogeneous or heterogeneous) cannot be assessed. Furthermore, false positive results can be generated depending on primer design and the stringency of the assay conditions17.
It is thus important to critically re-evaluate previous methylation reports using methodologies with a low risk of false positives. In this study, we used methylation sensitive–high resolution melting (MS-HRM) methodology for detection of promoter methylation. MS-HRM is a sensitive and closed-tube methodology developed for detection of methylation in a locus-specific manner, utilizing the different melting temperatures (Tm) of methylated and unmethylated DNA after bisulfite modification18. Due to the high Tm difference between methylated and unmethylated DNA, methylated samples are readily identifiable by analysing melting profiles. MS-HRM allows semi-quantitative assessment of methylation levels when all the examined CpG sites are methylated, but like other methods cannot quantify when the methylation is heterogeneous16.
The aim of this study was to assess the promoter region methylation status of DNA repair genes of potential clinical importance. One set of genes had been previously investigated in NSCLC (ATM, BRCA1, MLH1, MGMT and XPC). We also assessed two DNA repair genes that had been reported as methylated in other tumour types (NEIL1 and RAD23B), and a gene (ERCC1) whose gene expression levels had been reported as having predictive significance in NSCLC19.
Results
Re-assessing the methylation status of DNA repair genes reported to be methylated in NSCLC
DNA methylation of the promoter region CpG islands in the ATM, BRCA1, MGMT, MLH1 and XPC genes has been previously reported in lung cancer. We assessed the methylation status of these CpG islands in 11 lung cancer cell lines and 56 NSCLC tumours using methylation sensitive–high resolution melting (MS- HRM). The MS-HRM assay conditions for ATM, BRCA1, MGMT and MLH1 were robust and published in our previous studies18,20,21,22. We designed our ERCC1 and XPC MS-HRM assay to screen the region where methylation was reported in a previous study23. A series of dilutions of methylated DNA in unmethylated DNA (100%, 50%, 25%, 10%, 5%, 1%, 0%) were used in each MS-HRM run as controls.
We did not find promoter methylation in four of these five DNA repair genes (ATM, BRCA1, MLH1 and XPC) in any of the 11 lung cancer cell lines (Table 2) or in the 56 NSCLC tumours (Figure 1). Although XPC methylation was previously reported in the H1355 and Calu-1 cell lines, we did not find any XPC methylation in these cell lines (Figure 1). All methylated controls tested in each MS-HRM run were readily interpreted as methylated as their melting profiles differed significantly from that of unmethylated controls.
Table 2. Methylation status of two control genes and eight DNA repair genes in 11 lung cancer cell lines.
| Control genes | DNA repair genes | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cell line | APC | CDKN2A | ATM | BRCA1 | ERCC1 | MGMT | MLH1 | NEIL1 | RAD23B | XPC |
| H1395 | UM | UM | UM | UM | UM | UM | UM | Low | UM | UM |
| H1650 | 100% | NA | UM | UM | UM | UM | UM | Moderate | UM | UM |
| H460 | 50% | NA | UM | UM | UM | UM | UM | 100% | UM | UM |
| H1755 | UM | NA | UM | UM | UM | Low | UM | Low | UM | UM |
| H1666 | UM | NA | UM | UM | UM | Moderate | UM | Low | UM | UM |
| H69 | UM | UM | UM | UM | UM | Low | UM | Low | UM | UM |
| H2087 | 50% | UM | UM | UM | Low | UM | UM | Moderate | UM | UM |
| H2228 | 5% | NA | UM | UM | UM | UM | UM | Low | UM | UM |
| H1975 | UM | UM | UM | UM | UM | UM | UM | Moderate | UM | UM |
| H1355 | 100% | 100% | UM | UM | UM | UM | UM | ~100% | UM | UM |
| Calu-1 | UM | 100% | UM | UM | UM | UM | UM | Low | UM | UM |
NA; no amplification, UM; unmethylation.
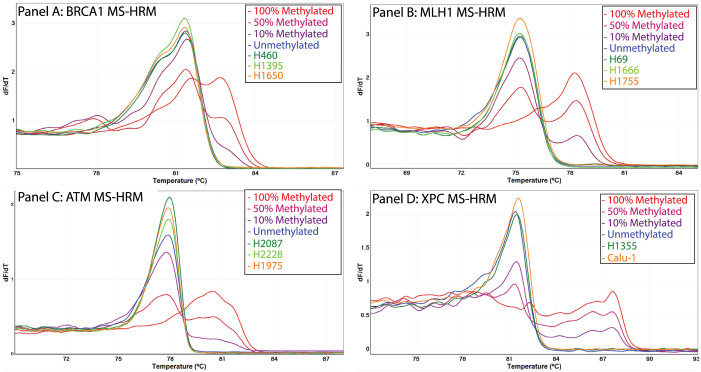
Figure 1. Absence of ATM, BRCA1, MLH1 and XPC methylation in lung cancer cell lines and tumours.
DNA methylation in the promoter regions of the ATM, BRCA1, MLH1 and XPC genes was assessed in 11 lung cancer cell lines and 56 NSCLC tumours using MS-HRM. After bisulfite modification, methylated DNA that retains cytosines has a higher melting temperature compared with unmethylated DNA that contains thymine (uracil before PCR). All samples having different melting patterns compared with unmethylated DNA control (in blue) are considered as methylated. Promoter methylation in ATM, BRCA1, MLH1 and XPC was not detected in any of the lung cancer cell lines and the NSCLC tumours. The negative first derivative plot of three representative lung cancer cell line samples are shown for BRCA1 (Panel A), MLH1 (Panel B), ATM (Panel C) and XPC (Panel D). Absence of XPC methylation is seen for the two lung cancer cell lines (H1355 and Calu-1) that were previously reported to be methylated.
MGMT methylation was found in three lung cancer cell lines (H69, H1666, and H1755) (Table 2) and seven of the 56 NSCLC tumours (13%) (Figure 2. Panel A). The melting patterns of the methylated lung cancer cell lines and NSCLC tumours were indicative of heterogeneous methylation i.e. the methylation status of individual CpGs varied across the amplicon. The level of heterogeneous methylation cannot be readily estimated by visual examination of the melting curves as the methylation dilution controls are only useful for estimation of methylated alleles of homogeneously methylated samples. However, the melting profiles of the lung cancer cell lines were indicative of low (H69, H1975) and moderate (H1666) numbers of methylated cytosines in the interrogated region. Similarly, only low or moderate levels of heterogeneous MGMT methylation were detected in seven NSCLC tumours.
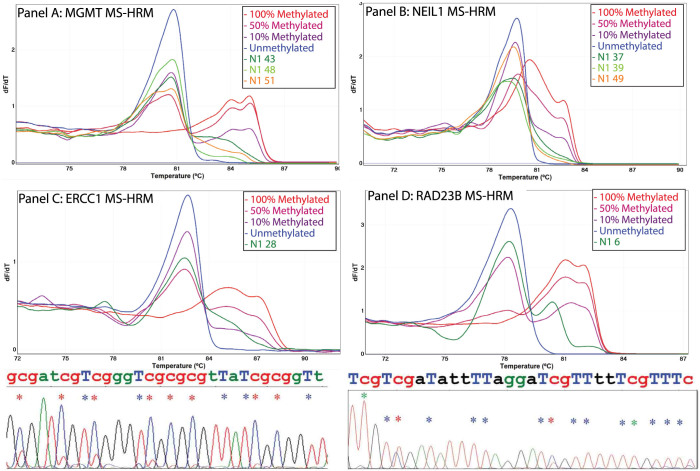
Figure 2. Detection of methylation in MGMT, NEIL1, ERCC1 and RAD23B in NSCLC tumours.
DNA methylation in the promoter regions of the MGMT, NEIL1, ERCC1 and RAD23B genes was assessed in 56 NSCLC tumours using MS-HRM. Methylation was detected in MGMT (25%), NEIL1 (42%), ERCC1 (2%) and RAD23B (2%). The negative first derivative plots of representative methylated NSCLC tumours for MGMT (Panel A) and NEIL1 (Panel B), ERCC1 (Panel C) and RAD23B (Panel D) are shown. Bisulfite Sanger sequencing traces confirming the ERCC1 and RAD23B methylation status are shown below the negative first derivative plots. Red, green and blue asterisks indicate the positions of methylated cytosines, unmethylated cytosines and bisulfite conversion controls respectively.
Assessing the methylation status of DNA repair genes not previously tested in NSCLC
We also assessed the promoter methylation of NEIL1, ERCC1 and RAD23B as their methylation has been reported in other cancers, but has not previously been examined in NSCLC24,25,26.
Frequent NEIL1 methylation was found in lung cancer cell lines and in NSCLC tumours. In the lung cancer cell lines, high levels of NEIL1 methylation were detected in H460 (100%) and H1355 (nearly 100%). The other cell lines showed heterogeneous methylation either at moderate levels (H1650, H2087 and H1975) or at low levels (H1395, H1755, H1666, H69, H2228, and Calu-1). Bisulfite Sanger sequencing of selected lung cancer cell lines confirmed the presence of NEIL1 methylation. NEIL1 methylation was also detected in 23 of the 56 NSCLC tumours (42%). All methylated NSCLC tumours showed heterogeneous methylation pattern either at moderate levels (13 samples) or at low levels (10 samples) (Figure 2. Panel B).
Methylation in ERCC1 and RAD23B was rarely detected. ERCC1 methylation was found in H2087 and one of the NSCLC tumours, the former showing a low-level heterogeneous methylation pattern and the latter showing a moderate-level heterogeneous methylation pattern. RAD23B methylation was detected in one NSCLC tumour and was not found in any of the lung cancer cell lines. Bisulfite Sanger sequencing of the tumour samples confirmed the methylation status (Figure 2. Panels C and D).
Methylation status of two control genes using MS-HRM
Given the low frequency of methylation of our target DNA repair gene set, we also assessed the promoter methylation status of the APC and CDKN2A genes in the cell lines and tumours to confirm that neither the bisulfite modification protocol nor the MS-HRM analysis precluded the detection of promoter methylation. The APC and CDKN2A genes were chosen as a relatively high methylation frequency for each has been previously found by multiple laboratories27,28,29. A high proportion of samples were methylated for either one or both of the genes, indicating that methylation, where present, could be readily identified by our MS-HRM assays.
APC methylation was found in five lung cancer cell lines (H460, H1355, H1650, H2087, and H2228) (Figure 3. Panel A). The melting patterns of all methylated lung cancer cell lines were suggestive of homogeneous methylation. By comparing with methylation dilution controls, the level of APC methylation was estimated to be 100% in H1650 and H1355, and 50% in H460 and H2987. Interestingly, APC methylation was estimated to be about 5–10% in H2228, suggestive of differential APC methylation status within the H2228 cells.
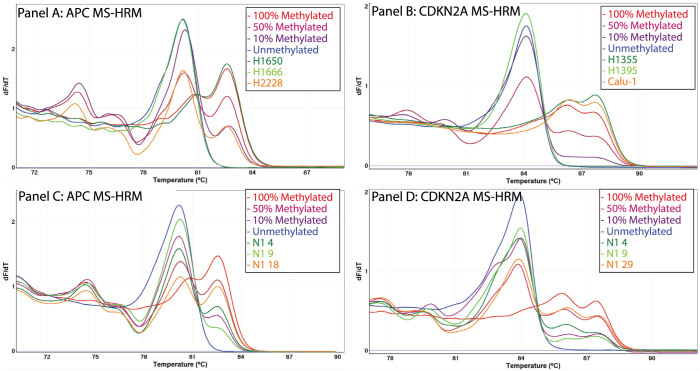
Figure 3. Assessment of methylation status for APC and CDKN2A in lung cancer cell lines and tumours using MS-HRM.
DNA methylation in the promoter regions of the APC and CDKN2A genes was assessed in 11 lung cancer cell lines and 56 NSCLC tumours using MS-HRM. APC was found in 5 lung cancer cell lines and 14 NSCLC tumours. CDKN2A methylation was detected in 2 lung cancer cell lines and 14 NSCLC tumours. The negative first derivative plots of three representative methylated lung cancer cell lines for APC (Panel A) and CDKN2A (Panel B) and three representative methylated NSCLC tumours for APC (Panel C) and CDKN2A (Panel D) are shown.
Homogenous CDKN2A methylation was found in two lung cancer cell lines (H1355 and Calu-1) and was estimated to be 100% (Figure 3. Panel B). Five of the lung cancer cell lines (H460, H1650, H1666, H1755, and H2228), that were previously found to have a homozygous CDKN2A deletion (CONAN database), were not amplified by the CDKN2A MS-HRM assay, confirming the absence of CDKN2A template. Four cell lines (H1395, H69, H2987 and H1975) were negative for CDKN2A methylation.
We also tested the 56 NSCLC tumours for methylation in the APC and CDKN2A promoter regions. APC and CDKN2A methylation was detected in 14 NSCLC tumours each (25%) (Figure 3. Panels C and D). A total of 23 NSCLC tumours (41%) were methylated for at least one of the genes and methylation in both genes was detected in 5 NSCLC tumours.
External validation using The Cancer Genome Atlas (TCGA) database
As we observed a considerable discrepancy in the methylation frequency of tested DNA repair genes between our MS-HRM results and the literature, we sought to validate our results using a second dataset generated by another methodology with a low false positive rate. We analysed the methylation data from the TCGA NSCLC database that provides genome-wide methylation status assessed by the HumanMethylation 450 k beadchip (Illumina). Methylation data were available from 568 NSCLC tumours, comprising of 341 adenocarcinomas and 227 squamous cell carcinomas.
We searched for HM450k probes overlapping the CpG sites within our eight MS- HRM amplicons. No overlapping CpG sites were detected for ATM, ERCC1 and MGMT. Eight probes were identified for the remaining genes; two probes for BRCA1, one for MLH1, one for NEIL1, three for RAD23B and one for XPC (Figure 4).
Figure 4. The TCGA methylation data of five DNA repair genes.
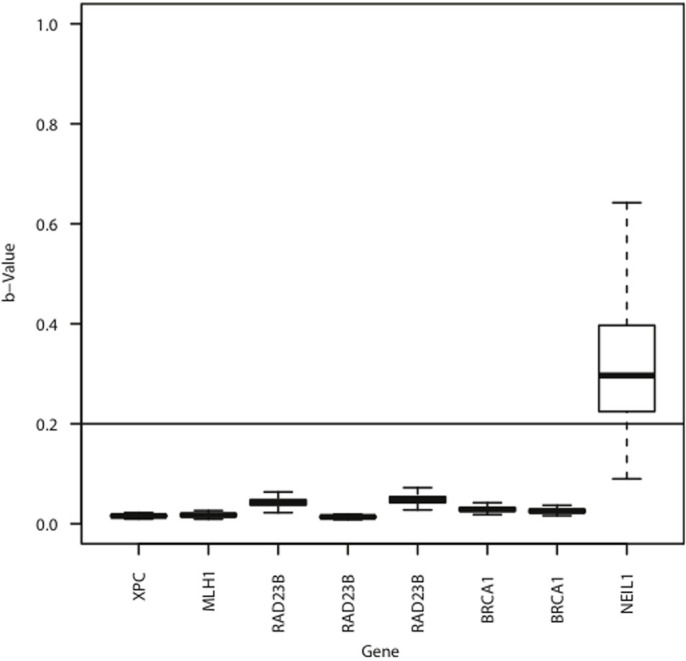
TCGA methylation data from 568 non-small cell lung cancers for the eight overlapping CpG sites with our MS-HRM amplicons is presented as boxplots. Two overlapping CpG sites for BRCA1, one for MLH1, one for NEIL1, three for RAD23B and one for XPC were analysed. A β-value of greater than 0.2 was used to define the presence of DNA methylation as shown by the horizontal line. Consistent with our MS-HRM results, absent or very rare methylation was found for BRCA1, MLH1, RAD23B and XPC, and highly frequent methylation was found for NEIL1.
Methylation was completely absent in the three CpG sites of RAD23B and the CpG site of XPC in the 568 NSCLC samples. Very low frequency methylation was seen for MLH1 (3/567, 0.5%). Three primary squamous cell carcinomas of lung showed methylation at both CpG sites of BRCA1 (3/568, 0.5%) although one of the sites in the third sample was just below the 20% cut-off. These results were consistent with our MS-HRM results finding either absent or very rare methylation in the BRCA1, MLH1, RAD23B and XPC promoters.
The CpG site in the NEIL1 promoter was frequently methylated (478/568, 84%), although the level of methylation varied among the methylated samples. This TCGA methylation data again confirmed our MS-HRM results demonstrating high frequency NEIL1 methylation in NSCLC.
Discussion
Epigenetic alterations in cancer are a potential source of therapeutic targets for personalised cancer treatment. MGMT methylation in glioma is the best known example where it predicts a durable response to treatment with alkylating agents30. Methylation of other DNA repair genes also has been considered for the selection of optimal chemotherapeutic agents for the treatment of cancer, although these have not been clinically implemented up to now31. Before clinical implementation, individual methylation markers need to be rigorously validated, ideally by using different methodologies13. In this study, we sought to validate a range of previously reported and to examine novel methylated DNA repair markers that could potentially be therapeutically exploited.
In the literature, highly variable estimates of the frequency of methylation for the ATM (0–47%), BRCA1 (4–30%), MLH1 (0–68%), MGMT (8–50%), and XPC (33%) genes have been reported in NSCLC tumours (Table 1). As most of the previous studies used the MSP method to determine the methylation status of candidate genes, the previous findings needed to be validated using other methodologies that are less prone to give false results. When we assessed DNA methylation using MS-HRM, we did not find methylation in any of these four DNA repair genes (ATM, BRCA1, MLH1, and XPC) in our 11 lung cancer cell lines and 56 NSCLC samples. This was consistent with the TCGA data which showed either absent or very low frequency of methylation for these promoters.
There are several possible explanations for the discrepant results, including differences in ethnicity or clinicopathological features of samples tested in each study. Several studies have reported varying methylation frequencies in cancer between different ethnic groups, including in the promoters of the IGFBP332, TMS133 and GSTP1 genes34. However, the real reasons for the discrepant results are likely to be technical such as scoring of low-level methylation, false positives due to the use of inadequately designed primers or amplification of methylated pseudogene sequences.
Low levels of methylation, especially present at ≤1%, can cause discrepant results due to the different analytic sensitivity of detection methods. The lower limit of MSP can be close to 0.1%35, allowing samples with low level methylation to be interpreted as methylated.
A high frequency of false positive ATM methylation calls deriving from the use of inadequate MSP conditions has been previously demonstrated in NSCLC17. ATM methylation was not detected in NSCLC when strict guidelines for performance of MSP are used17. We did not find ATM methylation in our 11 lung cancer cell lines and 56 lung tumours, confirming the absence of ATM methylation.
The BRCA1 pseudogene (BRCA1P1), a duplicated region of BRCA1 exons 1A, 1B, and 2, has a strong sequence homology to the BRCA1 gene. As methylation of BRCA1P1 has been previously reported in cancers36,37, there is a risk of false positives due to amplification of the methylated BRCA1P1 sequence. Two previous studies have assessed the BRCA1 methylation status in NSCLC tumours using MSP as a detection method38,39. The MSP primers designed by Lee et al. have 19 (19/21 in the forward) and 17 (17/20 bases in the reverse) matched bases, including the nine consecutive bases from the 3′ end of both primers, to the BRCA1 pseudogene39. The frequency of BRCA1 methylation (30%) reported by Lee et al. was 7-fold higher than that of being reported by Marsit et al. (4%) where more stringent MSP primers were used to avoid the amplification of the methylated BRCA1P1 sequence.
XPC methylation was initially reported in NSCLC tumours and lung cancer cell lines23. Wu et al. reported that XPC methylation was detectable in 34% of NSCLC tumours by HpaII-based PCR and in four lung cancer cell lines harboring TP53 mutations. Surprisingly, XPC methylation was not detected in this cohort of 56 NSCLC tumours and in two of the lung cancer cell lines (H1355 and Calu-1) previously reported as methylated23. The absence of XPC methylation was confirmed in the TCGA data. None of the 568 NSCLC tumours of the TCGA study had XPC methylation at the overlapping CpG site with our MS-HRM amplicon. As our XPC MS-HRM assay was designed to amplify those methylated CpG sites in the Wu et al. study, the discordant results was thus not likely to be caused by the examination of different CpG sites. To assess the XPC methylation status in NSCLC tumours, Wu et al. used the HpaII restriction endonuclease for selective cleavage of unmethylated DNA before PCR amplification23. However, there is a risk of incomplete enzymatic digestion of unmethylated DNA by HpaII, potentially resulting in false positives40,41.
This study is the first report showing methylation of ERCC1, RAD23B and NEIL1 in NSCLC. The excision repair cross-complementing group 1 (ERCC1) is a rate-limiting protein involved in the recognition and excision of DNA adducts. Recently, Chen et al. reported that ERCC1 methylation was significantly associated with chemosensitivity to cisplatin in glioma cell lines and glioma tumours25. In NSCLC, low levels of ERCC1 expression were correlated with favorable clinical outcomes of prolonged survival and sensitivity to platinum-based chemotherapies19. Therefore, ERCC1 methylation may serve as a predictive biomarker for identification of NSCLC patients who are highly sensitive to platinum-based chemotherapies. These patients potentially would have a more durable response than patients with high levels of ERCC1 expression that were not methylated.
The RAD23B protein forms a DNA damage recognition complex with the XPC and centrin 2 proteins. The RAD23B/XPC/centrin 2 complex recognises and interacts with the damaged bases or the sugar-phosphate backbone of DNA in the NER pathway42. High RAD23B expression has been suggested as a promising biomarker associated with response to histone deacetylase (HDAC) inhibitors in cutaneous T-cell lymphoma patients43. As anti-tumour activities of HDAC inhibitors have been demonstrated in NSCLC44, it can be speculated that silencing of RAD23B expression through promoter methylation makes tumour cells more resistant to HDAC inhibitors. On the other hand, RAD23B methylation may make tumour cells more sensitive to DNA-damaging agents.
Nei-like 1 (NEIL1), an ortholog of E.coli Nei (endonuclease VIII), is a bifunctional DNA glycosylase that repairs oxidative DNA damage of 8-hydroxyguanine and thymine glycol45 and protects cells from radiation-mediated cell death46. Recently, epigenetic silencing of NEIL1 through promoter methylation was reported in head and neck squamous cell carcinomas24. In this study, we found that the NEIL1 promoter is methylated in NSCLC at a high frequency (42%). This may identify patients that are sensitive to radiotherapy which deserves further study.
In conclusion, this study showed that methylation frequency of ATM, BRCA1, MLH1 and XPC in NSCLC is likely to be overestimated in the literature, emphasising the importance of rigorous validation of previous data on DNA methylation. In particular, the use of adequate methodology is critical to avoid false positive results. DNA methylation in the ERCC1, RAD23B and especially the NEIL1 DNA repair genes may serve as useful biomarkers for the determination of molecularly tailored therapies in a subset of NSCLC patients.
Methods
Samples
Fifty-six N1 stage NSCLC tumours were collected at the Austin Hospital, Melbourne, Australia with the approval of the Austin Human Research Ethics Committee (project title and approval number “Biomarkers in the Australian Non Small Cell Lung Cancer Population” – H2006-02394). The methylation study was approved by the Ethics of Human Research Committee at the Peter MacCallum Cancer Centre, Melbourne, Australia (project title and approval number “Molecular Pathology of Cancer: Methylation, Mutation & Expression” - 02/26).
DNA extraction from lung cancer cell lines
Lung cancer cell lines were cultured in RPMI 1640 medium with 25 mM HEPES supplemented with 10% fetal bovine serum, and 0.1 units/mL of penicillin and 0.1 μg/mL of streptomycin. Cells were maintained at 37°C in a humidified chamber containing 5% CO2. Cultured cells were harvested and washed twice with Dulbecco's phosphate-buffered saline buffer, followed by centrifugation at the speed of 10,000 rpm for 10 minutes. Cell pellets were then suspended in 200 μL of Dulbecco's phosphate-buffered saline buffer. After addition of 20 μL of proteinase K (20 mg/mL) and 200 μL of buffer AL, the suspended cells were incubating at 56°C for overnight. Genomic DNA was extracted from cultured lung cancer cell lines using the QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany).
DNA extraction from NSCLC tumours
The tumour purity of individual NSCLC cases was assessed by pathologists at Peter MacCallum Cancer Centre and was estimated to be 40–95% with a median of 67%. Two to five 5 μm formalin-fixed tissue sections were washed with 1 mL of xylene to remove paraffin and were incubated at 40°C for 10 minutes. Supernatant was removed after a centrifugation at 13,000 rpm for 10 minutes and tissue pellets were sequentially washed with 100% and 70% ethanol. The tissue pallets were then resuspended with 100 μL of ATL buffer of the DNeasy Tissue kit (Qiagen) and incubated at 97°C for 15 mins, followed by proteinase K digestion for 3 days at 56°C. Genomic DNA was then extracted using the DNeasy Tissue kit according to the manufacturer's instructions.
Bisulfite modification
One microgram of genomic DNA was bisulfite modified using the MethylEasy Xceed kit (Human Genetic Signatures, North Ryde, Australia) according to the manufacturer's instructions. Bisulfite modified DNA was eluted twice with 50 μL of elution buffer in an estimated concentration of 10 ng/μL.
Preparation of methylation standards
Commercially available methylated DNA (Millipore, Billerica, MA) was used as a fully methylated control. To prepare unmethylated DNA, peripheral blood DNA obtained from a healthy individual underwent two rounds of whole genome amplification (WGA) using the Illustra GenomiPhi V2 DNA Amplification Kit (GE Healthcare, Giles, UK) according to the manufacturer's instructions. The first round of WGA was performed with 1 ng of peripheral blood DNA. One microliter of a 10-fold dilution of the first round WGA product was used for the second round of WGA. After bisulfite modification, a qPCR assay that amplified a region lacking CpG dinucleotides within the COL2A1 gene was performed to quantify the amount of template47. A series of methylation dilution standards of 100%, 50%, 20%, 10%, 5% and 1% were prepared by mixing of the fully methylated DNA with the WGA DNA.
Methylation sensitive-high resolution melting assays
MS-HRM assays were designed either to target the promoter region or to overlap the regions used in previously reported assays. To better work with fragmented FFPE DNA, MS-HRM assays were designed to amplify a short amplicon size of less than 150 bp. Primer sequences, amplicon sizes, MgCl2 concentration, annealing temperatures, and the number of CpG dinucleotides analysed are summarised in Table 3. PCR amplification and MS-HRM were performed on the RotorGene Q (Qiagen). The PCR mixture was prepared in a final volume of 20 μL and contained: 10 ng of bisulfite modified template, 1× PCR buffer, 2.5–4 mM MgCl2, 200–400 nM each primer, 200 μM dNTPs, 5 μM SYTO9 and 0.5 U HotStar Taq (Qiagen). PCR cycling and melting conditions were as follows; one cycle of 95°C for 15 minutes; 50–55 cycles of 95°C for 10 seconds, annealing temperature of each assay for 20 seconds, 72°C for 25 seconds; one cycle of 97°C for one minute and a melt from 70°C to 95°C rising 0.2°C per second. The melting profiles of amplicons were analysed using RotorGene Q Software (v1.7). All samples were tested in duplicate.
Table 3. Primer sequences and amplicon information for each MS-HRM assay.
| Name | Primer Sequence | Genomic region∧ | Number of CpG# | Annealing Tm (°C) | Amplicon (bp) |
|---|---|---|---|---|---|
| APC_F | 5′-cggggttttgtgttttattg-3′ | chr5:112,073,406-112,073,476 | 4 | 58.5 | 71 |
| APC_R | 5′-tccaacgaattacacaactac-3′ | ||||
| ATM_F | !5′-gtttgcgttawgtttattaatggtt-3′ | chr11:108,092,818-108,094,287 | 12 | 55 | 147 |
| ATM_R | 5′-acttccgtcctcaaacttaaaa-3′ | ||||
| BRCA1_F | *5′-ttgttgtttagcggtagttttttggtt-3′ | chr17:41,277,396-41,277,474 | 4 | 63 | 79 |
| BRCA1_R | *5′-caatcgcaattttaatttatctataattccc-3′ | ||||
| CDKN2A_F | 5′-cggaggaagaaagaggaggggt-3′ | chr9:21,974,843-21,974,935 | 7 | 70 | 93 |
| CDKN2A_R | 5′-cgctacctactctccccctct-3′ | ||||
| ERCC1_F | *,†5′-gagtcgtttttttttattIgggtttttttg -3′ | chr19:45,932,040-45,932,132 | 12 | 65 | 93 |
| ERCC1_R | *5′-cgcccgcctctaaacttaacc -3′ | ||||
| MGMT_F | 5′-gcgtttcggatatgttgggatagt -3′ | chr10:131,265,469-131,265,578 | 12 | 61 | 110 |
| MGMT_R | 5′-aacgacccaaacactcaccaaa -3′ | ||||
| MLH1_F | 5′-agtttttaaaaacgaattaataggaagag-3′ | chr3:37,034,741-37,034,821 | 5 | 59 | 81 |
| MLH1_R | 5′-actacccgctacctaaaaaaatatac-3′ | ||||
| NEIL1_F | *5′-aatttttttttcgttttagggtttttgtattgg-3′ | chr15:75,639,328-75,639,472 | 9 | 59 | 145 |
| NEIL1_R | *5′-aaaacgaaaaaataaacccacttaaaataattc-3′ | ||||
| RAD23B_F | 5′-gtcgttaggaggaagttttaggagt-3′ | chr9:110,045,332-110,045,451 | 11 | 57 | 124 |
| RAD23B_R | †5′-gaaaactccgccccaaIattaaaaac-3′ | ||||
| XPC_F | 5′-ttttaacgaaggggcgtggttaag-3′ | chr3:14,220,061-14,220,178 | 13 | 62 | 118 |
| XPC_R | 5′-cgaaccatattacttatctaaacaaattcca-3′ |
∧UCSC Genome Browser on Human Feb. 2009 (GRCh37/hg19) Assembly was used.
*M13 sequences were attached to these primers.
#CpG dinucleotides flanked by the MS-HRM primers were counted.
†An inosine base (I) was used at the cytosine site of a CpG dinucleotide in the ERCC1 and RAD23B reverse primers to reduce the bias toward methylated templates.
!The ATM forward primer contains W (A/T) at the position of the SNP rs4987880 (A/T) to avoid any amplification bias.
Bisulfite Sanger sequencing
To confirm the MS-HRM results for ERCC1, NEIL1 and RAD23B methylation, MS- HRM products were sequenced using the Big Dye Terminator v3.1 chemistry (Applied Biosystems). Sequencing products were purified with the Agencourt CleanSEQ reagent (Beckman Coulter) and analysed by capillary electrophoresis on an ABI3730 sequencer (Applied Biosystems). The sequencing data was analysed using Sequencher 4.6 (Gene Codes Corporation).
Analysis of DNA methylation from publically available data
Marmal-aid (http://www.marmal-aid.org) is a data repository of publically available genome-scale DNA methylation analysis using the Illumina HumanMethylation 450 K (HM450K) beadchip. At the time of download, 8,654 data sets were available from marmal-aid and also contained data from The Cancer Genome Atlas (TCGA) study48. At the time of download, a total of 568 non-small cell lung cancers (341 adenocarcinomas and 227 squamous carcinomas) were available from marmal-aid from TCGA. We identified 11 HM450K probes interrogating CpG sites within our MS-HRM amplicons. Raw beta values were extracted using marmal-aid in R. Data were extracted according to their annotations in marmal-aid. Samples annotated with “lung” and “cancer” and our probes of interest were used for downstream analysis and visualization in R (http://www.cran.org). Using guidelines previously described49, a β-value of greater than 0.2 was used to define the presence of DNA methylation.
Author Contributions
Suitable samples were identified by P.M., T.J., B.S. and C.M. The experimental work was carried out by H.D. and the TCGA data was analysed by N.W., H.D. and N.W. prepared figures. H.D. and A.D. co-wrote the manuscript. All authors approved the final manuscript.
Acknowledgments
We would like to thank Thomas Mikeska for critical discussion. This project was supported by funding from Cancer Australia to A.D., the Cancer Council of Victoria to A.D., and the National Health and Medical Research Council of Australia to P.M. and A.D. H.D. is a recipient of Postdoctoral Fellowship from the Cancer Council of Victoria.
References
- Taron M. et al. BRCA1 mRNA expression levels as an indicator of chemoresistance in lung cancer. Hum Mol Genet 13, 2443–9 (2004). [DOI] [PubMed] [Google Scholar]
- Rosell R. et al. Customized treatment in non-small-cell lung cancer based on EGFR mutations and BRCA1 mRNA expression. PLoS One 4, e5133 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadaki C. et al. ERCC1 and BRAC1 mRNA expression levels in the primary tumor could predict the effectiveness of the second-line cisplatin-based chemotherapy in pretreated patients with metastatic non-small cell lung cancer. J Thorac Oncol 7, 663–71 (2012). [DOI] [PubMed] [Google Scholar]
- Papadaki C. et al. Correlation of BRCA1, TXR1 and TSP1 mRNA expression with treatment outcome to docetaxel-based first-line chemotherapy in patients with advanced/metastatic non-small-cell lung cancer. Br J Cancer 104, 316–23 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boukovinas I. et al. Tumor BRCA1, RRM1 and RRM2 mRNA expression levels and clinical response to first-line gemcitabine plus docetaxel in non- small-cell lung cancer patients. PLoS One 3, e3695 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosell R. et al. BRCA1: a novel prognostic factor in resected non-small-cell lung cancer. PLoS One 2, e1129 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Font A. et al. BRCA1 mRNA expression and outcome to neoadjuvant cisplatin-based chemotherapy in bladder cancer. Ann Oncol 22, 139–44 (2011). [DOI] [PubMed] [Google Scholar]
- Weberpals J. et al. The DNA repair proteins BRCA1 and ERCC1 as predictive markers in sporadic ovarian cancer. Int J Cancer 124, 806–15 (2009). [DOI] [PubMed] [Google Scholar]
- Dabholkar M. et al. ERCC1 and ERCC2 expression in malignant tissues from ovarian cancer patients. J Natl Cancer Inst 84, 1512–7 (1992). [DOI] [PubMed] [Google Scholar]
- Middleton M. R. & Margison G. P. Improvement of chemotherapy efficacy by inactivation of a DNA-repair pathway. Lancet Oncol 4, 37–44 (2003). [DOI] [PubMed] [Google Scholar]
- Esteller M. et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med 343, 1350–4 (2000). [DOI] [PubMed] [Google Scholar]
- Helleday T., Petermann E., Lundin C., Hodgson B. & Sharma R. A. DNA repair pathways as targets for cancer therapy. Nat Rev Cancer 8, 193–204 (2008). [DOI] [PubMed] [Google Scholar]
- Mikeska T., Bock C., Do H. & Dobrovic A. DNA methylation biomarkers in cancer: progress towards clinical implementation. Expert Rev Mol Diagn 12, 473–87 (2012). [DOI] [PubMed] [Google Scholar]
- Curtin N. J. DNA repair dysregulation from cancer driver to therapeutic target. Nat Rev Cancer 12, 801–17 (2012). [DOI] [PubMed] [Google Scholar]
- Mikeska T., Candiloro I. & Dobrovic A. The implications of heterogeneous DNA methylation for the accurate quantification of methylation. Epigenomics 2, 561–573 (2010). [DOI] [PubMed] [Google Scholar]
- Cottrell S. E. & Laird P. W. Sensitive detection of DNA methylation. Ann N Y Acad Sci 983, 120–30 (2003). [DOI] [PubMed] [Google Scholar]
- Brandes J. C., Carraway H. & Herman J. G. Optimal primer design using the novel primer design program: MSPprimer provides accurate methylation analysis of the ATM promoter. Oncogene 26, 6229–37 (2007). [DOI] [PubMed] [Google Scholar]
- Wojdacz T. K. & Dobrovic A. Methylation-sensitive high resolution melting (MS-HRM): a new approach for sensitive and high-throughput assessment of methylation. Nucleic Acids Res 35, e41 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord R. V. et al. Low ERCC1 expression correlates with prolonged survival after cisplatin plus gemcitabine chemotherapy in non-small cell lung cancer. Clin Cancer Res 8, 2286–91 (2002). [PubMed] [Google Scholar]
- Huang K. T. et al. DNA methylation profiling of phyllodes and fibroadenoma tumours of the breast. Breast Cancer Res Treat 124, 555–65 (2010). [DOI] [PubMed] [Google Scholar]
- Wong E. M. et al. Constitutional methylation of the BRCA1 promoter is specifically associated with BRCA1 mutation-associated pathology in early- onset breast cancer. Cancer Prev Res (Phila) 4, 23–33 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikeska T., Carney D. A., Seymour J. F. & Dobrovic A. No evidence for DNA methylation of the ATM promoter CpG island in chronic lymphocytic leukemia. Leuk Lymphoma 53, 1420–2 (2012). [DOI] [PubMed] [Google Scholar]
- Wu Y. H. et al. Xeroderma pigmentosum group C gene expression is predominantly regulated by promoter hypermethylation and contributes to p53 mutation in lung cancers. Oncogene 26, 4761–73 (2007). [DOI] [PubMed] [Google Scholar]
- Chaisaingmongkol J. et al. Epigenetic screen of human DNA repair genes identifies aberrant promoter methylation of NEIL1 in head and neck squamous cell carcinoma. Oncogene 31, 5108–16 (2012). [DOI] [PubMed] [Google Scholar]
- Chen H. Y., Shao C. J., Chen F. R., Kwan A. L. & Chen Z. P. Role of ERCC1 promoter hypermethylation in drug resistance to cisplatin in human gliomas. Int J Cancer 126, 1944–54 (2010). [DOI] [PubMed] [Google Scholar]
- Peng B. et al. Epigenetic silencing of the human nucleotide excision repair gene, hHR23B, in interleukin-6-responsive multiple myeloma KAS-6/1 cells. J Biol Chem 280, 4182–7 (2005). [DOI] [PubMed] [Google Scholar]
- Virmani A. K. et al. Aberrant methylation of the adenomatous polyposis coli (APC) gene promoter 1A in breast and lung carcinomas. Clin Cancer Res 7, 1998–2004 (2001). [PubMed] [Google Scholar]
- Brock M. V. et al. DNA methylation markers and early recurrence in stage I lung cancer. N Engl J Med 358, 1118–28 (2008). [DOI] [PubMed] [Google Scholar]
- Vaissiere T. et al. Quantitative analysis of DNA methylation profiles in lung cancer identifies aberrant DNA methylation of specific genes and its association with gender and cancer risk factors. Cancer Res 69, 243–52 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegi M. E. et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med 352, 997–1003 (2005). [DOI] [PubMed] [Google Scholar]
- Hegi M. E., Sciuscio D., Murat A., Levivier M. & Stupp R. Epigenetic deregulation of DNA repair and its potential for therapy. Clin Cancer Res 15, 5026–31 (2009). [DOI] [PubMed] [Google Scholar]
- Tomii K. et al. Aberrant promoter methylation of insulin-like growth factor binding protein-3 gene in human cancers. Int J Cancer 120, 566–73 (2007). [DOI] [PubMed] [Google Scholar]
- Das P. M. et al. Methylation mediated silencing of TMS1/ASC gene in prostate cancer. Mol Cancer 5, 28 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enokida H. et al. Ethnic group-related differences in CpG hypermethylation of the GSTP1 gene promoter among African-American, Caucasian and Asian patients with prostate cancer. Int J Cancer 116, 174–81 (2005). [DOI] [PubMed] [Google Scholar]
- Herman J. G., Graff J. R., Myohanen S., Nelkin B. D. & Baylin S. B. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A 93, 9821–6 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang J. W., Karlan B. Y., Cass L. & Baldwin R. L. BRCA1 promoter methylation predicts adverse ovarian cancer prognosis. Gynecol Oncol 101, 403–10 (2006). [DOI] [PubMed] [Google Scholar]
- Dobrovic A. & Simpfendorfer D. Methylation of the BRCA1 gene in sporadic breast cancer. Cancer Res 57, 3347–50 (1997). [PubMed] [Google Scholar]
- Marsit C. J. et al. Inactivation of the Fanconi anemia/BRCA pathway in lung and oral cancers: implications for treatment and survival. Oncogene 23, 1000–4 (2004). [DOI] [PubMed] [Google Scholar]
- Lee M. N. et al. Epigenetic inactivation of the chromosomal stability control genes BRCA1, BRCA2, and XRCC5 in non-small cell lung cancer. Clin Cancer Res 13, 832–8 (2007). [DOI] [PubMed] [Google Scholar]
- van Dijk J. P. et al. A novel, essential control for clonality analysis with human androgen receptor gene polymerase chain reaction. Am J Pathol 161, 807–12 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupper D., Reuter M., Meisel A. & Kruger D. H. Reliable detection of DNA CpG methylation profiles by the isoschizomers MspI/HpaII using oligonucleotide stimulators. Biotechniques 23, 843–7 (1997). [DOI] [PubMed] [Google Scholar]
- Nouspikel T. DNA repair in mammalian cells: Nucleotide excision repair: variations on versatility. Cell Mol Life Sci 66, 994–1009 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan O. et al. HR23B is a biomarker for tumor sensitivity to HDAC inhibitor- based therapy. Proc Natl Acad Sci U S A 107, 6532–7 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyanaga A. et al. Antitumor activity of histone deacetylase inhibitors in non- small cell lung cancer cells: development of a molecular predictive model. Mol Cancer Ther 7, 1923–30 (2008). [DOI] [PubMed] [Google Scholar]
- Bandaru V., Sunkara S., Wallace S. S. & Bond J. P. A novel human DNA glycosylase that removes oxidative DNA damage and is homologous to Escherichia coli endonuclease VIII. DNA Repair (Amst) 1, 517–29 (2002). [DOI] [PubMed] [Google Scholar]
- Rosenquist T. A. et al. The novel DNA glycosylase, NEIL1, protects mammalian cells from radiation-mediated cell death. DNA Repair (Amst) 2, 581–91 (2003). [DOI] [PubMed] [Google Scholar]
- Virmani A. K. et al. Hierarchical clustering of lung cancer cell lines using DNA methylation markers. Cancer Epidemiol Biomarkers Prev 11, 291–7 (2002). [PubMed] [Google Scholar]
- Cancer Genome Atlas Research N. Comprehensive genomic characterization of squamous cell lung cancers. Nature 489, 519–25 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M. et al. Genome-wide DNA methylation profiling using Infinium(R) assay. Epigenomics 1, 177–200 (2009). [DOI] [PubMed] [Google Scholar]
- Safar A. M. et al. Methylation profiling of archived non-small cell lung cancer: a promising prognostic system. Clin Cancer Res 11, 4400–5 (2005). [DOI] [PubMed] [Google Scholar]
- Safar A. M. et al. Promoter hypermethylation for molecular nodal staging in non-small cell lung cancer. Arch Pathol Lab Med 131, 936–41 (2007). [DOI] [PubMed] [Google Scholar]
- Wang Y. et al. Multiple gene methylation of nonsmall cell lung cancers evaluated with 3-dimensional microarray. Cancer 112, 1325–36 (2008). [DOI] [PubMed] [Google Scholar]
- Wu J. Y. et al. Association of O6-methylguanine-DNA methyltransferase (MGMT) promoter methylation with p53 mutation occurrence in non-small cell lung cancer with different histology, gender, and smoking status. Ann Surg Oncol 15, 3272–7 (2008). [DOI] [PubMed] [Google Scholar]
- Feng Q. et al. DNA methylation in tumor and matched normal tissues from non-small cell lung cancer patients. Cancer Epidemiol Biomarkers Prev 17, 645–54 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Lan Q., Siegfried J. M., Luketich J. D. & Keohavong P. Aberrant promoter methylation of p16 and MGMT genes in lung tumors from smoking and never-smoking lung cancer patients. Neoplasia 8, 46–51 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. T. et al. Prognostic implication of aberrant promoter hypermethylation of CpG islands in adenocarcinoma of the lung. J Thorac Cardiovasc Surg 130, 1378 (2005). [DOI] [PubMed] [Google Scholar]
- Tang M., Torres-Lanzas J., Lopez-Rios F., Esteller M. & Sanchez-Cespedes M. Wnt signaling promoter hypermethylation distinguishes lung primary adenocarcinomas from colorectal metastasis to the lung. Int J Cancer 119, 2603–6 (2006). [DOI] [PubMed] [Google Scholar]
- Seng T. J. et al. DLEC1 and MLH1 promoter methylation are associated with poor prognosis in non-small cell lung carcinoma. Br J Cancer 99, 375–82 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. C. et al. Inactivation of hMLH1 and hMSH2 by promoter methylation in primary non-small cell lung tumors and matched sputum samples. J Clin Invest 111, 887–95 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng X., Wang F., Zhang L. & Zhang W. M. Loss of heterozygosity combined with promoter hypermethylation, the main mechanism of human MutL Homolog (hMLH1) gene inactivation in non-small cell lung cancer in a Chinese population. Tumori 95, 488–94 (2009). [DOI] [PubMed] [Google Scholar]