Abstract
Pancreatic cancer is the fifth most aggressive malignancy and urgently requires new biomarkers to facilitate early detection. For providing impetus to the biomarker discovery, we have developed Pancreatic Cancer Methylation Database (PCMDB, http://crdd.osdd.net/raghava/pcmdb/), a comprehensive resource dedicated to methylation of genes in pancreatic cancer. Data was collected and compiled manually from published literature. PCMdb has 65907 entries for methylation status of 4342 unique genes. In PCMdb, data was compiled for both cancer cell lines (53565 entries for 88 cell lines) and cancer tissues (12342 entries for 3078 tissue samples). Among these entries, 47.22% entries reported a high level of methylation for the corresponding genes while 10.87% entries reported low level of methylation. PCMdb covers five major subtypes of pancreatic cancer; however, most of the entries were compiled for adenocarcinomas (88.38%) and mucinous neoplasms (5.76%). A user-friendly interface has been developed for data browsing, searching and analysis. We anticipate that PCMdb will be helpful for pancreatic cancer biomarker discovery.
Pancreatic cancer remains the fifth leading cause of cancer-related deaths with an overall 5-year survival rate less than 4%1. Both developed and developing countries are in the grip of this deadly disease. Despite the considerable progress in the fight against other cancers in recent years, the prognosis for patients diagnosed with pancreatic cancer has remained extremely poor. One of the major reasons for this poor prognosis is the unavailability of appropriate biomarkers for early diagnosis2. If this cancer could be caught before overt metastasis to other parts of the body, patients could be more effectively treated with surgery. Thus, the identification of adequate biomarkers in pancreatic cancer is of utmost importance.
In the past, considerable efforts have been carried out to identify potential biomarkers that include aberrantly expressed genes, proteins, miRNA detectable through non-invasive techniques in cancerous tissue and body fluids3,4. In addition, mutations in few genes have also been identified to be associated with the progression of pancreatic cancer5,6. The involvement of DNA methylation, an epigenetic process, in carcinogenesis has been well established7. Loss of gene expression due to methylation of promoter CpG island that is otherwise unmethylated in a normal cell has been the most widely investigated epigenetic event in cancer8,9 and thus has drawn significant attention as a biomarker candidate. Knowledge of DNA methylation in pancreatic cancer is rapidly increasing owing to the development of genome-wide techniques for their identification. Though promoter CpG island hypermethylation has been realized to be an efficient tumor biomarker since 1990s, it was only in the subsequent decade that the marker genes displaying change in methylation status found a place in clinical practice of cancer detection, diagnosis and prognosis7. Another significant development is the finding that DNA methylation is an efficient predictor of the response to chemotherapeutic drugs. For example, promoter hypermethylation of MGMT gene confers enhanced drug sensitivity to alkylating agent drugs like carmustine and temozolomide in patients with gliomas10,11. Biomarker genes for sensitivity to the drugs Gemcitabine and Docetaxel in pancreatic cancer cell lines and xenografts have been recently confirmed12,13. All such studies accentuate the need for consolidation of methylation studies in pancreatic cancer to have a holistic view of the methylation status at the genome level such that a conclusive panel of biomarkers with clinical utility is achieved.
Tremendous efforts have been made in the past for providing comprehensive databases containing methylation status of genes, facilitating the user to associate such data with various diseases. MethyCancer is one such resource, hosting methylation data of CpG island clones derived from large scale sequencing14. Most recent database is MENT (Methylation and Expression database of Normal and Tumor tissues), presenting correlation of DNA methylation and gene expression in paired normal and tumor samples15. Another recent database is DiseaseMeth that houses gene centric methylation data for 72 diseases encompassing various experimental techniques and platforms, the ultimate purpose being assistance in biomarker discovery16. Although all these databases are comprehensive, very little information against pancreatic cancer is available. The inaugural release of MENT database reports altered methylation of genes from a single high throughput study for pancreatic cancer. Moreover, some of these are even confined to only high throughput data that further needs locus specific validation. Considering the grim situation of pancreatic cancer and lack of appropriate biomarkers for early detection, we have developed a comprehensive repository named pancreatic cancer methylation database (PCMdb) that provides comprehensive information of methylation status of genes in pancreatic cancer.
PCMdb includes methylation data for pancreatic cancer circumscribing various experimental platforms, both high throughput and gene specific studies. In PCMdb, data from both tumor tissue and pancreatic cancer cell lines were compiled systematically. The cell line data in particular is highly useful in pancreatic cancer research, as it has been externally linked to available drug sensitivity data, actuating the search for new therapeutic options in pancreatic cancer. We hope that PCMdb will be helpful to expedite the process of materializing methylation data into methods for disease detection, diagnosis, prognosis and even deciding therapeutic regimen.
Results
Data statistics
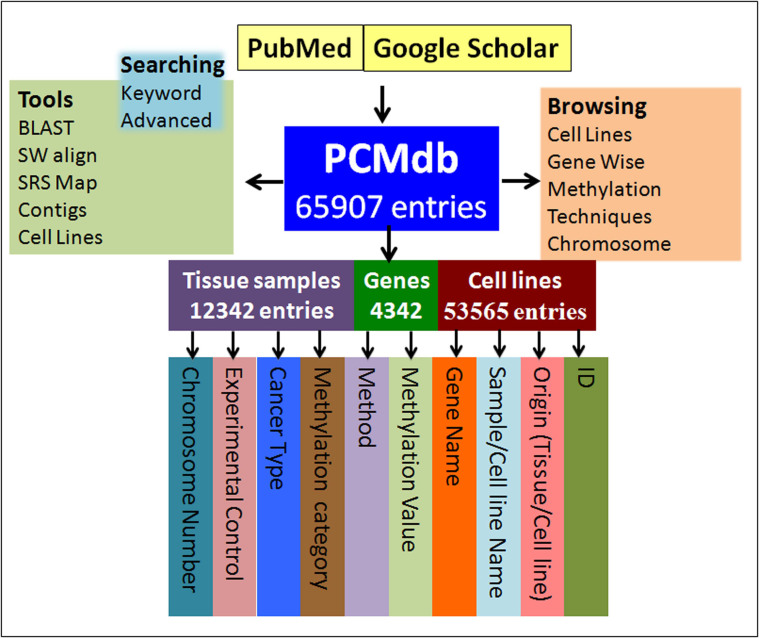
Being a compilation of methylation status of genes, PCMdb is directed towards methylation biomarker discovery for pancreatic cancer. All data, for both cell lines and tissue samples, have been compiled from 109 research articles. The inaugural release of PCMdb has 65907 entries for 4342 unique genes (Figure 1). We have made multiple entries for a single gene, if methylation status of a particular gene has been reported from more than one study or if the methylation status of the same gene has been reported from more than one cell lines. Most of the entries are from the cell line data (88 cell lines, 53565 entries, 81%) while about 19% (3078 tissue samples, 12342 entries) data have been occupied by the tissue sample data.
Figure 1. Architecture of PCMdb.
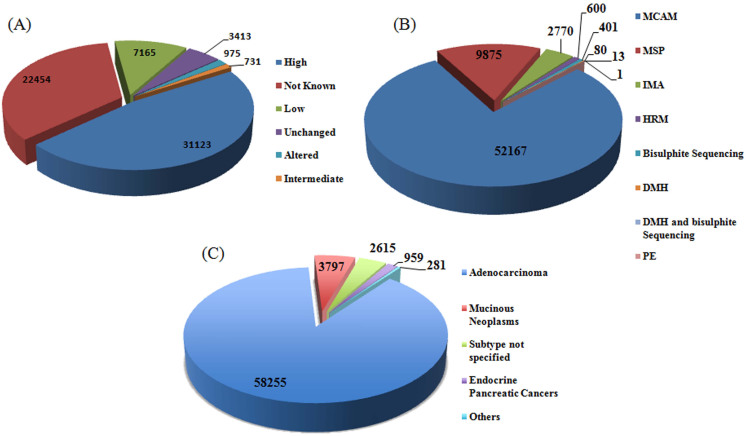
In literature, different terminologies (e.g., dense methylation, partial methylation, less frequent methylation, etc.) have been used for reporting methylation status of genes. To make it convenient, we have categorized the methylation status into five categories: high, intermediate, low, altered and not known. This classification is based on the level of DNA methylation reported compared to the control. In many cases, the source research article reported methylation status of the gene as methylated as compared to the control rather than explaining whether the methylation is more, less or altered with respect to the control. All such type of entries have been compiled in status ‘Not Known’ category. In PCMdb, almost half of the entries (47.22%) reported a high level of methylation for the corresponding genes (Figure 2A). Only 10.87% of the entries reported low level of methylation. Approximately 34% entries have been covered under the term ‘Not Known’ in the field of methylation status.
Figure 2. Data statistics based on (A) Methylation category, (B) Techniques used to detect methylation, and (C) types of pancreatic cancer subtypes.
Due to advancements in technologies, nowadays numerous methods are available for evaluating the locus specific methylation status yet the majority of the data (79%) housed by PCMdb comes from the experiments that used Methylated CpG Island Amplification and Microarray (MCAM) as the technique for evaluating methylation status (Figure 2B). This is expected, as MCAM is a high throughput technique. But such high throughput techniques do not undermine the reliability of locus specific techniques like Methylation Specific PCR (MSP) (14.98% of the entries in PCMdb) that need to be performed in the final confirmation of conferring the biomarker status to the methylation of a gene. Since pancreatic cancer has many subtypes, this information was also included for each entry. Majority of the entries (~88%) have been compiled for adenocarcinomas, which is the most common type of pancreatic tumors. In addition, entries were also made for Intraductal papillary mucinous neoplasms (IPMN), which are precursors to invasive pancreatic cancer6. Despite the fact that IPMN too represent an opportunity to cure pancreatic neoplasia before an invasive cancer develops, only a few studies characterizing methylation pattern of IPMN were carried out17,18. We have compiled data from these studies and a total of 5.76% entries were compiled for IPMN (Figure 2C). In the rest ~4% of the total entries, the pancreatic cancer subtype has not been specified in the source research article.
We have also integrated the methylation data of cell lines with the drug sensitivity data of pancreatic cancer cell lines. Among the 88 cell lines covered by PCMdb, drug resistance data for 33 out of 88 cell lines is available in CancerDR19. Methylation and drug resistance data for these cell lines have been incorporated as a separate submenu under the ‘Drug Resistance’ section of PCMdb. Apart from this, several genes have been reported in literature to be the source of nucleic acid oligomers circulating in the body fluids, especially in case of cancer. For 77 of these genes, the methylation data is available in the literature. Hence, these have been consolidated as the submenu ‘Biomarker’ in the ‘Summary’ menu.
Discussion
Pancreatic cancer is one of the most aggressive cancers with the mortality rate almost equal to the incidence rate. The only way to win the battle against this deadly disease is to catch it at an early stage when the tumor could be removed by surgery. Over the past decade, considerable efforts have been made to develop efficient biomarkers3, which can be useful to detect this disease before it becomes incurable. Development of PCMdb is also a step in this direction for the identification of efficient DNA methylation-based biomarkers for pancreatic cancer. The main aim of PCMdb is to provide comprehensive and quality data on DNA methylation in pancreatic cancer. However, a few repositories have been developed so far, which provide information of DNA methylation. But most of these databases have either covered many diseases or many types of cancers with less information for pancreatic cancer14,16. In our attempt to compile gene centric methylation data in pancreatic cancer, the primary emphasis is on facilitating biomarker discovery for pancreatic cancer detection, diagnosis and prognosis.
A unique feature of PCMdb is that it includes methylation data from both, cancerous tissue samples, as well as from cancerous cell lines. Cell line data is helpful in integrating gene methylation data with mutational and drug sensitivity data available for pancreatic cancer. On the other hand, tissue sample data is clinically more relevant representing more realistic status of cancer pathogenesis.
Apart from being suitable biomarker for cancer detection, diagnosis and prognosis, DNA methylation of genes has also been realized to be an effective predictor of response to chemotherapeutic cancer drugs20. This is the primary reason for integrating methylation data of PCMdb with drug sensitivity data of CancerDR. A total of 33 pancreatic cancer cell lines were selected for which the methylation status for genes has been included in PCMdb, and their half maximal inhibitory concentration (IC50) is available in CancerDR for selective drugs. These two types of data for 33 pancreatic cancer cell lines are systematically compiled in the Cell Lines sub-menu of ‘Drug Resistance’ section of PCMdb. This is extremely useful for making the relationship between methylation and drug sensitivity in these cancer cell lines.
DNA methylation on the CpG palindromes is recognized by DNA binding proteins most of them being transcription factors thus leading to gene silencing. Hence, investigations into the cellular pathways associated with gene expression that get affected due to DNA methylation in cancer could open new avenues for therapeutic intervention. With this objective in consideration, the ‘Function’ sub-menu of Summary section of PCMdb enlists the major cellular functions associated with gene transcription, translation and regulation, all of these ultimately affecting expression and the corresponding genes in PCMdb that perform these functions.
Methods
Data collection and compilation
The main aim of our PCMdb database is to collect and compile high quality DNA methylation data of pancreatic cancer. Therefore, the gene centric methylation data have been manually collected from the published research articles. These articles were searched using keywords like DNA methylation in pancreatic cancer from a simple search in PubMed that resulted into 414 articles as hits. Further, an advanced search with keywords ‘methylation’ and ‘pancreatic cancer’ was performed in PubMed, which ended into 436 abstracts. All these research papers from both searches were downloaded and compiled systematically for data curation. After careful reading of these research papers, comprehensive information related to genes, their methylation status, techniques used for examining the methylation, the experimental control and the type of sample or cell line, etc. were extracted and compiled. For associating methylation with drug sensitivity, the latter data available in the database CancerDR was integrated in PCMdb with external links to CancerDR. Cancer Cell Line Encyclopedia17 or the CCLE database contains the names of genes that are cancer drug targets. For some of these genes, the methylation status in pancreatic cancer has been included in PCMdb.
Database framework and web interface
PCMDB is developed on apache server and is based on MySQL relational database system. Front end is developed using HTML, PHP and javascript while the back end is supported by PHP and PERL programming languages. Apache and MySQL are preferred as these are efficient and open source software.
Data organization
The entries in PCMdb are broadly compiled into two categories – the cell line data and the tissue data (Figure 1). Consequently, two types of unique IDs have been assigned to each entry. First, there is a unique PCMdb ID for each entry, which provides a kind of global address within the database. The second ID is for convenience of locating the entry within one of the two categories of data origin – cell line or tissue. The primary data of PCMdb consists of following major fields. (i) Source (e.g., cell line or tissue), (ii) Gene name (e.g., MUC17) (iii) Control: It represents the source of experimental control DNA used for comparison of the methylation level of DNA from pancreatic cancer tissue sample or cell line, (iv) Cancer types: It gives information about sub-types of pancreatic cancer (e.g., Adenocarcinoma), (v) Methylation status: It represents the status of methylation (e.g., high, low or intermediate), (vii) Methods: It gives information about methods/assays used in the experiments (e.g., Bisulphite Sequencing).
The methylation data, being the primary data, has been manually curated from research articles. In addition, the methylation data has been integrated with the drug sensitivity data available from CancerDR and CCLE databases.
Implementation of tools
Data searching
PCMdb provides extensive cross-references and user-friendly interface. Various search and browsing tools have been integrated, which make data retrieval convenient. The following two search tools have been integrated.
Keyword search This search option allows the users to search PCMdb in a very simple way using various keywords. In order to search extensively, various fields have been provided, which can be selected by the user.
Advanced search This is a provision for systematic search that allows the user to build a query using individual keywords for each field where the search is desired. A complex query can be built where a combination of keywords can be defined to be included together or searched alternatively or excluded. This is possible using the conditionals = , <, > and the logical operators OR, LIKE and AND.
Data browsing
Various browsing tools have been integrated, which will facilitate various types of data retrieval. The current version of PCMdb contains information of 4342 methylated genes in pancreatic cancer. Considering the large number of objects in a single field of genes and to make browse page user-friendly, the gene wise browse page has been organized to search gene names alphabetically. Browsing based on gene name helps the user to know the methylation status of a particular gene across multiple studies. In addition, various external links to various resources, which provide comprehensive (e.g., gene symbol, gene name, chromosomal location, etc.) information related to a particular gene, has been linked to each gene name entry. PCMdb provides information of methylation data obtained from 88 pancreatic cancer cell lines. We have developed a robust browsing facility to extract maximum information related to cell lines. The cell line browse option allows users to retrieve all the methylation data for a particular cell line.
The user can fetch all the entries of PCMdb with the same category of methylation status in one go using the Methylation Browse page of PCMdb. Very often in methylation studies, the user would like to know the genes showing increased or decreased DNA methylation pattern in pancreatic cancer to corroborate a newly performed experiment. The Methylation Status Browse page in PCMdb would serve this purpose. There are many methods reported in the literature for identification of methylation events. Apart from the methylation status, users may want to know the techniques used to identify methylation. Therefore, we have compiled such experimental techniques under the heading ‘Techniques’. An experimentalist working on methylation would be facilitated by the techniques-based browse option in PCMdb.
For looking at the genes undergoing change of methylation level in pancreatic cancer located on the same chromosome, the Chromosome browse page has been provided in PCMdb. The browse page of Chromosome redirects the user to a table that displays genes lying on the particular chromosome number chosen by the user for which the methylation has been evaluated in the literature of pancreatic cancer. This table also has fields of the cancer type, methylation category, PMID, techniques used for investigating methylation and the expression of that gene in selective cell lines (data taken from CCLE) in the form of bar plot.
Data analysis
To assist the cancer biology community interested in analyzing the methylation data in the form of sequences and short reads generated from NGS, various web tools have been integrated to PCMdb. A short description of these tools is as follows:
BLAST BLAST search tool21 has been integrated to PCMdb that assists users to align query sequences against sets of genes available in PCMdb.
SW align In order to have an optimal local alignment of the methylated fragment on the gene sequence, Smith-Watermann algorithm22 has been integrated. User can submit sequences in FASTA format.
SRS map Owing to advances in NGS sequencing technologies, nowadays sequencing whole transcriptome, exome, and genome of a cancer patient is possible. Therefore, we have integrated a tool, where users can align NGS short read data directly to the reference genes in PCMdb. The alignment can be visualized for any variation in the query sequences.
Contigs Genomic fragments (i.e., contigs) can be submitted to PCMdb for gene prediction. The predicted genes in the cotings could be aligned to methylation target genes.
Limitations and future prospects
Although a rigorous search for methylation information in pancreatic cancer has been made in literature for compiling it in PCMdb, new hits of research articles in PubMed with different combinations of keywords cannot be ruled out. Furthermore, the initial release of PCMdb has been confined to the concluded data available in literature excluding raw methylation data for pancreatic cancer, for example, that available from TCGA. Such information first needs to be mapped on the human genome and then analyzed to arrive at the gene loci with altered methylation status as compared to the control. Apart from this the detailed information at sequence level could be added to PCMdb in the future to make it more informative and more helpful in biomarker discovery.
Update of PCMdb
We have included the most recent data from literature in PCMdb. We will try to incorporate the new data as soon as it will be available and update the database on a regular basis.
Author Contributions
G.N. and M.S. collected and compiled the data. G.N., S.K., S.G. and K.C. organized the data. G.N., S.K., S.G. and K.C. developed the web interface and integrated the tools. G.N., S.K., S.G., K.C. and A.G. analyzed the data. G.N., A.G. and G.P.S.R. wrote the manuscript. G.P.S.R. conceived the idea and coordinated the project.
Acknowledgments
Authors are thankful to funding agencies Council of Scientific and Industrial Research (project Open Source Drug Discovery and GENESIS BSC0121) and Department of Biotechnology (project BTISNET), Govt. of India for financial support.
References
- Siegel R., Naishadham D. & Jemal A. Cancer statistics, 2012. CA Cancer J Clin 62, 10–29 (2012). [DOI] [PubMed] [Google Scholar]
- Costello E., Greenhalf W. & Neoptolemos J. P. New biomarkers and targets in pancreatic cancer and their application to treatment. Nat Rev Gastroenterol Hepatol 9, 435–444 (2012). [DOI] [PubMed] [Google Scholar]
- Harsha H. C. et al. A compendium of potential biomarkers of pancreatic cancer. PLoS Med 6, e1000046 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer A. S. et al. Diagnosis of pancreatic ductal adenocarcinoma and chronic pancreatitis by measurement of microRNA abundance in blood and tissue. PLoS One 7, e34151 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yachida S. & Iacobuzio-Donahue C. A. Evolution and dynamics of pancreatic cancer progression. Oncogene 32, 5253–5260 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacobuzio-Donahue C. A., Velculescu V. E., Wolfgang C. L. & Hruban R. H. Genetic basis of pancreas cancer development and progression: insights from whole-exome and whole-genome sequencing. Clin Cancer Res 18, 4257–4265 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylin S. B. & Jones P. A. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer 11, 726–734 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P. A. & Baylin S. B. The fundamental role of epigenetic events in cancer. Nat Rev Genet 3, 415–428 (2002). [DOI] [PubMed] [Google Scholar]
- Jones P. A. & Baylin S. B. The epigenomics of cancer. Cell 128, 683–692 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M. et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med 343, 1350–1354 (2000). [DOI] [PubMed] [Google Scholar]
- Hegi M. E. et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med 352, 997–1003 (2005). [DOI] [PubMed] [Google Scholar]
- Tan A. C. et al. Characterizing DNA methylation patterns in pancreatic cancer genome. Mol Oncol 3, 425–438 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran K., Miller H., Gordian E., Rocha-Lima C. & Singal R. Methylation-mediated silencing of TMS1 in pancreatic cancer and its potential contribution to chemosensitivity. Anticancer Res 30, 3919–3925 (2010). [PubMed] [Google Scholar]
- He X. et al. MethyCancer: the database of human DNA methylation and cancer. Nucleic Acids Res 36, D836–841 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek S. J. et al. MENT: methylation and expression database of normal and tumor tissues. Gene 518, 194–200 (2013). [DOI] [PubMed] [Google Scholar]
- Lv J. et al. DiseaseMeth: a human disease methylation database. Nucleic Acids Res 40, D1030–1035 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- House M. G., Guo M., Iacobuzio-Donahue C. & Herman J. G. Molecular progression of promoter methylation in intraductal papillary mucinous neoplasms (IPMN) of the pancreas. Carcinogenesis 24, 193–198 (2003). [DOI] [PubMed] [Google Scholar]
- Hong S. M. et al. Multiple genes are hypermethylated in intraductal papillary mucinous neoplasms of the pancreas. Mod Pathol 21, 1499–1507 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R. et al. CancerDR: Cancer Drug Resistance Database. Sci Rep 3, 1445; 10.1038/srep01445 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyn H. & Esteller M. DNA methylation profiling in the clinic: applications and challenges. Nat Rev Genet 13, 679–692 (2012). [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W. & Lipman D. J. Basic local alignment search tool. J Mol Biol 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- Pearson W. R. Flexible sequence similarity searching with the FASTA3 program package. Methods Mol Biol 132, 185–219 (2000). [DOI] [PubMed] [Google Scholar]