Summary
The cyanobacterial circadian clock generates genome-wide transcriptional oscillations and regulates cell division, but the underlying mechanisms are not well understood. Here we show that the response regulator RpaA serves as the master regulator of these clock outputs. Deletion of rpaA abrogates gene expression rhythms globally and arrests cells in a dawn-like expression state. Although rpaA deletion causes core oscillator failure by perturbing clock gene expression, rescuing oscillator function does not restore global expression rhythms. We show that phosphorylated RpaA regulates the expression of not only clock components, generating feedback on the core oscillator, but also a small set of circadian effectors that in turn orchestrate genome-wide transcriptional rhythms. Expression of constitutively active RpaA is sufficient to switch cells from a dawn-like to a dusk-like expression state as well as to block cell division. Hence, complex global circadian phenotypes can be generated by controlling the phosphorylation of a single transcription factor.
Introduction
The circadian clock of the cyanobacterium Synechococcus elongatus PCC7942 drives daily genome-wide oscillations in mRNA expression levels, controls genome compaction and supercoiling, and modulates cell division (Johnson et al., 2011). The clock contains a core oscillator consisting of the proteins KaiA, KaiB, and KaiC, which together generate circadian (i.e., approximately 24-hour) oscillations in KaiC phosphorylation (Markson and O’Shea, 2009). Remarkably, the KaiC phosphorylation oscillations observed in vivo can be reconstituted in vitro simply by mixing the three Kai proteins and ATP (Nakajima et al., 2005). In vivo, this proteinaceous post-translational oscillator (PTO) is embedded in a transcription-translation feedback loop (TTL) that regulates expression of the kaiBC operon, enhancing the precision of the clock by stabilizing its phase (Johnson et al., 2011; Qin et al., 2010; Teng et al., 2013; Zwicker et al., 2010).
Although much is known about the mechanism by which the PTO keeps time, less is understood about how the clock uses the time information encoded in the PTO to generate outputs like global gene expression rhythms and modulation of cell division. Most genes show circadian expression oscillations in constant light, displaying a variety of amplitudes, phases, and waveforms (Ito et al., 2009; Johnson et al., 2011; Vijayan et al., 2009). The distribution of phases is biphasic, with expression of one population peaking around subjective dusk (class 1) and the other peaking around subjective dawn (class 2). (“Subjective dusk” and “subjective dawn” refer to the times at which light-to-dark or dark-to-light transitions would occur in a 12 h light-12 h dark environmental cycle. The term “subjective” is used when, as here, environmental conditions are held constant in order to isolate clock-driven from environmentally-driven processes.) Genome compaction and DNA supercoiling also oscillate in a circadian manner (Smith and Williams, 2006; Woelfle et al., 2007), and supercoiling oscillations contributes to the generation of gene expression oscillations (Vijayan et al., 2009). Finally, the clock gates cell division, prohibiting it during the subjective night (Dong et al., 2010; Mori et al., 1996; Yang et al., 2010).
Genetic and biochemical approaches have revealed key players in the output pathway that connects the PTO to these output responses. The response regulator RpaA has been implicated genetically in circadian gene expression control (Takai et al., 2006; Taniguchi et al., 2007). The phase of the PTO is transmitted to RpaA via the histidine kinases SasA and CikA (Gutu and O’Shea, 2013; Takai et al., 2006; Taniguchi et al., 2010): in vivo, SasA phosphorylates and CikA dephosphorylates RpaA via their respective kinase and phosphatase activities. The PTO generates temporal separation of SasA and CikA activities, producing circadian oscillations in RpaA phosphorylation levels (Gutu and O’Shea, 2013). The response regulator RpaB, a paralog of RpaA, also may play a role in circadian clock output: it binds in vitro and in vivo to the promoters of several circadian genes, and this binding is antagonized by RpaA in vitro (Hanaoka et al., 2012).
Deletion of rpaA eliminates oscillations in the activity of the ~10 circadian promoters that were assayed by bioluminescence reporters (Takai et al., 2006), but the role of RpaA in regulating circadian expression genome-wide is not known. RpaA is predicted to be a transcription factor (Takai et al., 2006), but recent studies failed to detect binding of RpaA to candidate target promoters (Hanaoka et al., 2012; Takai et al., 2006). Similarly, cell division gating by the clock requires RpaA (Dong et al., 2010), but the mechanism is not understood.
Here we employ a multifaceted approach to elucidate the molecular and functional roles of RpaA in circadian clock output. We find that circadian gene expression oscillations are absent genome-wide in an rpaA mutant, with cells being arrested in a subjective dawn-like transcriptional state. Through chromatin immunoprecipitation with high-throughput sequencing (ChIP-Seq) and in vitro DNase I footprinting, we show that phosphorylated RpaA binds directly to over one hundred locations in the genome, including the promoter of kaiBC. Finally, we show that overexpression of a phosphomimetic mutant of RpaA is sufficient to drive cells from the subjective dawn to the subjective dusk gene expression state and also to close the cell division gate, demonstrating that RpaA is the global regulator of circadian output in this organism.
Results
rpaA is required for global circadian gene expression and its deletion arrests cells in a subjective dawn-like state
To determine whether RpaA is required for global gene expression rhythms, we measured gene expression over 48 hours by microarray in an rpaA mutant strain (Figure 1A). We found that circadian oscillations were abolished genome-wide (Figure 1A), even for the genes that oscillate with the highest amplitude in the wild-type (Figure S1). Hence, RpaA is required for the generation of global circadian gene expression rhythms.
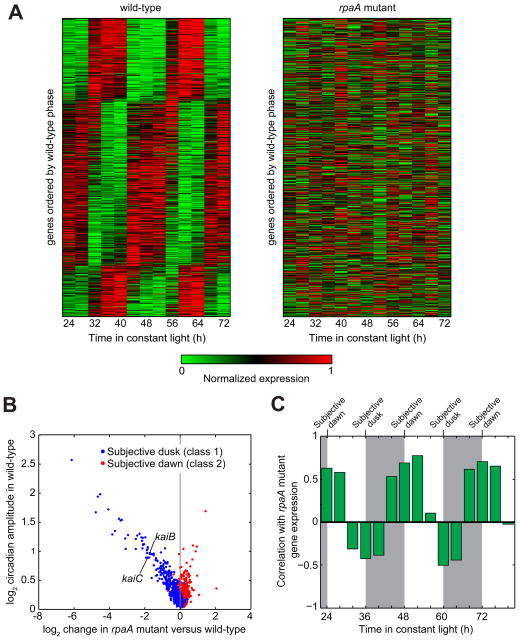
Figure 1. Gene expression is globally perturbed by deletion of rpaA.
A. Global gene expression timecourse in the wild-type and rpaA mutant. (Left) Circadian gene expression in the wild-type strain (data from Vijayan et al, 2009). Expression timecourses of genes reproducibly oscillating with a circadian period (n = 856; see Extended Experimental Procedures) were normalized to the interval [0 1] and sorted by phase. (Right) Gene expression in the rpaA mutant. Expression timecourse of the same set of genes as in wild-type, displayed in the same order.
B. Comparison of gene expression change in the rpaA mutant with circadian amplitude in the wild-type strain. We computed the expression change in the rpaA mutant by comparing the average rpaA mutant expression over one day with the average wild-type expression over one day (see Experimental Procedures). Only genes that oscillate with circadian periodicity in the wild-type strain are shown (n = 856). kaiB and kaiC are indicated; kaiA is not classified as circadian and hence is not shown.
C. Correlation of global gene expression in the rpaA mutant with each timepoint in the wild-type timecourse. Expression of all genes (both circadian and non-circadian) in the rpaA mutant was time-averaged as in Figure 1B, and the correlation between this time-averaged expression and the expression in the wild-type strain at each timepoint over a 60-h timespan was computed. Wild-type data are from Vijayan et al., 2009. Subjective night is shaded. Subjective dawn occurs at 24, 48, and 72 h, while subjective dusk falls at 36 and 60 h.
To gain insight into the role of RpaA in circadian gene regulation, we searched for genes which showed the greatest magnitude of transcript level difference between the wild-type and rpaA mutant strains (Table S1). Consistent with previous results (Takai 2006), kaiBC expression decreased approximately 3.5-fold, while kaiA expression was not affected substantially; this disparate effect on kai gene expression perturbs Kai protein stoichiometry, likely situating it in a regime that does support PTO function (Nakajima et al., 2010). Overall, we found that the expression of 67 genes decreased more than two-fold, while the expression of 16 genes increased by at least that amount. Strongly downregulated genes included four sigma factors, two transcription factors, and several genes encoding proteins involved in energy production and metabolism (particularly carbohydrate metabolism). Circadian genes downregulated in the rpaA mutant were highly enriched for subjective dusk phasing, whereas upregulated genes were enriched for subjective dawn phasing (Figure 1B). Interestingly, the decrease in expression of subjective dusk genes in the rpaA mutant is directly proportional to the gene’s circadian amplitude in the wild-type strain (Figure 1B). These observations suggest that RpaA is responsible for promoting subjective dusk gene expression and repressing subjective dawn gene expression. Consistent with this scenario, global gene expression in the rpaA mutant is most positively correlated with wild-type subjective-dawn expression and most negatively correlated with wild-type subjective dusk expression (Figure 1C). Hence, deletion of rpaA arrests cells in a subjective dawn-like state.
Rescue of PTO function in the rpaA mutant reveals that RpaA is directly responsible for the orchestration of circadian gene expression
Interpretation of the role of RpaA is confounded by the loss of PTO function in the rpaA mutant (Takai et al., 2006): the absence of gene expression oscillations in the rpaA mutant could merely be a secondary effect of the loss of PTO function rather than an indication of a master regulator role for RpaA. Indeed, it is possible that the primary role of RpaA is simply to sustain Kai post-translational oscillations by, for example, modulating kaiBC expression to maintain permissive Kai protein stoichiometry.
To distinguish between direct and indirect contributions of RpaA to circadian gene expression, we asked whether rescuing the PTO in an rpaA mutant background would restore global gene expression oscillations. We rescued the PTO by ectopically expressing kaiBC from the IPTG-inducible Ptrc promoter (Murayama et al., 2008) in a ΔrpaA ΔkaiBC background (ΔrpaA ΔkaiBC Ptrc::kaiBC), and, as a control, in a ΔkaiBC background (ΔkaiBC Ptrc::kaiBC). We refer to these strains as the ΔrpaA clock rescue and the control clock rescue, respectively. In both strains, we obtained strong KaiC phosphorylation rhythms of similar amplitude in the presence of 6 μM IPTG (Figures 2 and S2A). Note that ectopic expression of KaiA was not necessary because its levels are not substantially perturbed by deletion of rpaA (Takai et al., 2006).
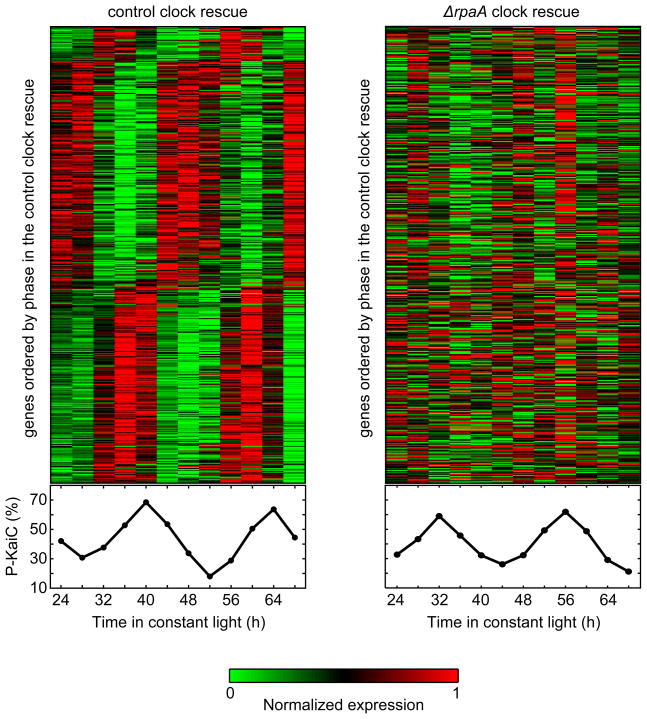
Figure 2. RpaA is required for control of global gene expression by the PTO.
(Left) Gene expression timecourse upon rescue of Kai oscillator function in a ΔkaiBC background via ectopic expression of kaiBC (ΔkaiBC Ptrc::kaiBC, termed the “control clock rescue”). The heatmap shows the expression timecourse of the 471 genes that oscillate with a circadian period in the control clock rescue in each of two biological replicates, with genes normalized individually and ordered by phase. KaiC phosphorylation levels during the timecourse are shown below the heatmap.
(Right) Gene expression timecourse upon rescue of Kai oscillator function in the absence of rpaA via ectopic expression of kaiBC (ΔrpaA ΔkaiBC Ptrc::kaiBC, termed the “ΔrpaA clock rescue”). The heatmap shows the expression timecourse of the same set of genes as in (A), displayed in the same order. KaiC phosphorylation levels during the timecourse are shown below the heatmap.
We used microarrays to examine global gene expression dynamics in the two rescue strains (Figure 2 and Table S2). In the control clock rescue, circadian gene expression oscillations were restored robustly for both subjective dusk and subjective dawn genes (Figures 2, S2B, and S2C). In contrast, no strong oscillations were observed in either class of genes in the ΔrpaA clock rescue (Figures 2, S2B, and S2C). We conclude that RpaA is required for the orchestration of robust circadian gene expression by the PTO, ruling out the scenario in which RpaA’s role is limited to supporting oscillator function.
Phosphorylated RpaA binds to the kaiBC promoter and upregulates kaiBC expression
While the rescue experiments demonstrate that RpaA plays a critical and central role in producing global gene expression rhythms, they do not indicate the means by which it does so. As previous studies have failed to identify direct RpaA binding to DNA, it has been proposed that RpaA acts indirectly via displacement of RpaB from circadian promoters (Hanaoka et al., 2012). However, these studies have assayed only a few regions for RpaA binding, and it is possible that RpaA acts via rhythmic binding in conditions or at locations that have not been examined.
To determine whether and where RpaA associates with genome, we generated an anti-RpaA antibody (Figure S3A) and used it to perform chromatin immunoprecipitation (ChIP) over a 36 h circadian timecourse. We found that RpaA localized to the kaiBC promoter in an oscillatory manner, in phase with both RpaA phosphorylation (RpaA~P) and expression of the kaiBC transcript (Figures 3A and S3B). These results are not an artifact of off-target binding of the anti-RpaA antibody, as ChIP-qPCR analysis of a strain expressing epitope-tagged RpaA (HA-RpaA) using an anti-HA antibody demonstrated that it localized to the kaiBC promoter with a circadian period and in phase with HA-RpaA phosphorylation (Figure S3C).
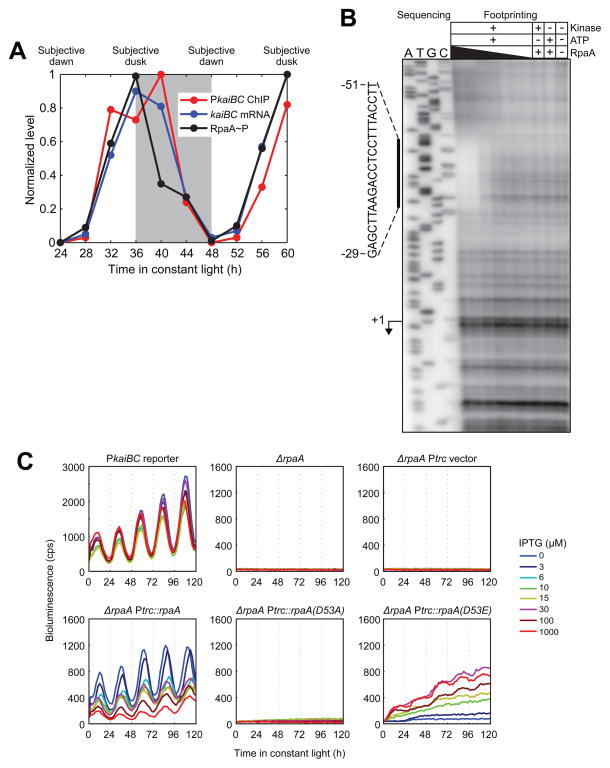
Figure 3. RpaA binds to the kaiBC promoter in vivo and in vitro and promoteskaiBC expression in a phosphorylation-dependent manner.
A. Correlation between RpaA phosphorylation, RpaA enrichment at the kaiBC promoter, and abundance of the kaiBC transcript. Subjective night is shaded. RpaA phosphorylation was measured by Phos-tag Western blot (see Figure S3B for gel image), association with PkaiBC by ChIP-qPCR, and kaiBC expression by RT-qPCR. Note that while the apex of the ChIP-qPCR enrichment is at 40 h (four hours after subjective dusk) in this experiment, the precise phase of RpaA (as well as that of global gene expression oscillations) varies between experiments, with RpaA binding typically peaking at or a few hours before subjective dusk.
B. In vitro DNase I footprinting of RpaA on the kaiBC promoter as a function of recombinant RpaA phosphorylation and concentration. Sanger sequencing reactions used to identify the location of the footprint are shown on the left; footprinting reactions are shown on the right. The region protected from digestion by RpaA~P is indicated by the vertical bar. The kaiBC transcription start site (Kutsuna et al., 2005) is indicated with an arrow. RpaA pre-treatment and concentration are indicated above each footprinting lane. RpaA was at least 50% phosphorylated in the presence of both CikA and ATP but was unphosphorylated otherwise (Figure S3D). RpaA was added to a final concentration of 6.0, 3.0, 0.6, 0.3, 0.06, or 0.03 μM as indicated by the thickness of the wedge.
C. Activity of the kaiBC promoter was assayed using a PkaiBC::luxAB luciferase reporter in various genetic backgrounds: wild-type, ΔrpaA, and ΔrpaA expressing wild-type RpaA, unphosphorylatable RpaA (D53A), or an RpaA phosphomimetic (D53E) from the IPTG-inducible Ptrc promoter. IPTG was added at the indicated concentration prior to entrainment with two 12-h dark pulses.
See also Figure S3.
To determine whether localization of RpaA to the kaiBC promoter results from direct association with DNA, we assayed for physical interaction of RpaA with promoter DNA in vitro using DNase I footprinting. We observed a clear footprint, strictly dependent on RpaA phosphorylation, between 29 and 51 bp upstream of the transcription start site (Figure 3B). The strict phosphorylation dependence may explain the inability to detect RpaA binding to the kaiBC promoter in previous in vitro studies (Hanaoka et al., 2012; Takai et al., 2006): these studies used either unphosphorylated RpaA (Takai et al., 2006) or RpaA putatively phosphorylated by treatment with acetyl phosphate (Hanaoka et al., 2012), which we found does not actually result in RpaA phosphorylation (Figure S3D). Intriguingly, the footprint of RpaA~P in the kaiBC promoter coincides with a region in which mutations substantially reduce promoter activity in vivo (Kutsuna et al., 2005). We tested three of those mutations in the footprinting assay, finding that all of them reduced or eliminated RpaA~P binding (Figure S3E).
To test whether RpaA~P drives expression of the kaiBC transcript in vivo, we expressed phosphorylation site mutants mimicking phosphorylated or unphosphorylated RpaA in an rpaA mutant background and used a PkaiBC-driven luciferase reporter to assess the effect of these mimetics on PkaiBC activity (Figure 3C). Aspartate 53 (D53) of RpaA is predicted by sequence homology with the well-studied E. coli OmpR protein to be the site of phosphorylation by SasA, and we therefore used a glutamate mutation at residue 53 (D53E) to mimic RpaA~P and an alanine mutation (D53A) to mimic unphosphorylated RpaA. All of the RpaA variants were expressed in the presence of IPTG (Figure S3F). We observed no expression of the luciferase reporter in a control strain containing the Ptrc promoter without RpaA. Leaky expression of wild-type RpaA from the Ptrc promoter (Figure S3F) rescued circadian activity of the kaiBC reporter (Figure 3C). Consistent with a previous report (Taniguchi et al., 2007), increasing levels of wild-type RpaA with IPTG induction progressively repressed promoter activity, likely due to an increase in the ratio of unphosphorylated RpaA to RpaA~P (Figure S3G). In contrast, kaiBC promoter activity was absent at all doses of RpaA(D53A). Expression of RpaA(D53E), however, restored activity from the promoter in a dose-dependent manner (Figure 3C), consistent with positive regulation of promoter activity by RpaA~P. No aspartate phosphorylation was present in RpaA(D53A) or RpaA(D53E) (Figure S3G). Collectively, our observations suggest that rhythmic association of RpaA~P with the kaiBC promoter drives circadian expression of this operon. Therefore, the circadian TTL is directly mediated by RpaA.
ChIP-Seq reveals the landscape of RpaA binding
To identify RpaA binding sites genome-wide, we used our anti-RpaA antibody to perform circadian timecourse ChIP analyzed by high throughput sequencing (ChIP-Seq). We identified 110 binding sites (peaks), all located on the main chromosome (see Experimental Procedures) (Figure 4A and Table S3A). A well-defined binding site was identified upstream of the kaiBC locus, the occupancy of which varied with circadian time (Figure 4B). RpaA binds in a circadian manner and with a similar phase at all 110 binding sites (Figure 4C).
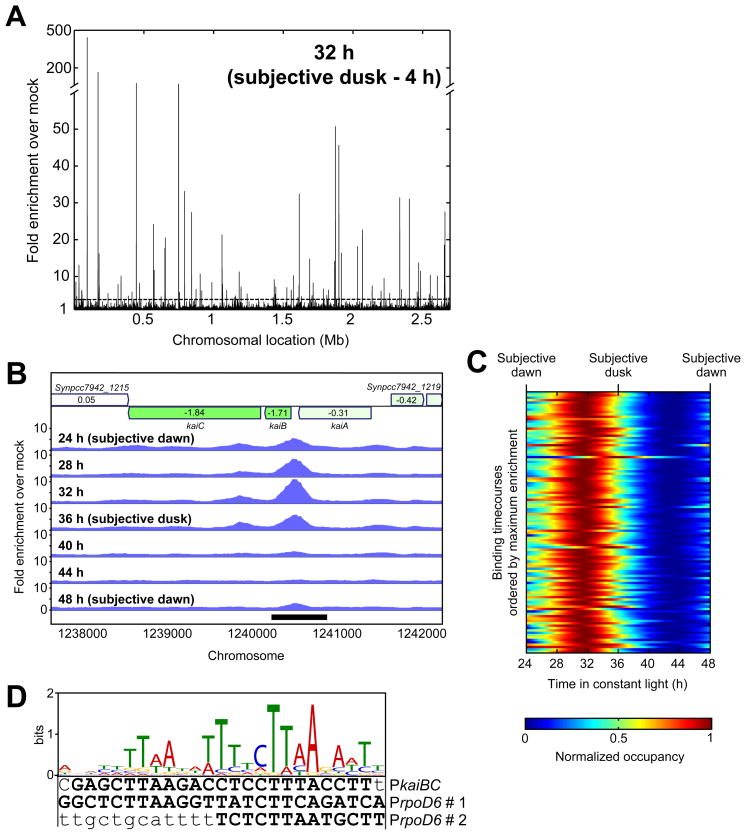
Figure 4. Identification of RpaA binding sites by ChIP-Seq.
A. Genome-wide binding profile of RpaA by ChIP-Seq. The enrichment of read density in the RpaA ChIP-Seq (anti-RpaA antibody on the wild-type strain) at four hours prior to subjective dusk (32 h), relative to the mock ChIP-Seq (anti-RpaA antibody on the rpaA mutant), is plotted as a function of position on the chromosome. The dotted line indicates the 3-fold enrichment cutoff for identification of RpaA binding sites.
B. Genome browser view of ChIP-Seq enrichment profiles in the vicinity of the kaiBC locus over one day in the wild-type strain. Genes are shown at the top, with the log2 of their expression change in the rpaA mutant represented by shading (green is decreased expression and red is increased expression) and indicated by the text inside the gene.
C. Timecourse of RpaA enrichment at the 110 RpaA binding sites. ChIP-Seq was performed every four hours for 24 h, and enrichment relative to the mock IP was calculated at the location of maximum wild-type ChIP-Seq read density within each binding site. Enrichment at intermediate timepoints was computed by interpolation with cubic splines. Each row in the heatmap represents the binding timecourse for one binding site; the rows are sorted by the maximum enrichment observed during the timecourse, which ranged from 411-fold (top) to 3.1-fold (bottom). The dynamic range (maximum enrichment divided by minimum enrichment for each binding site) varied from 38-fold to 1.4-fold.
D. A 25-basepair motif is overrepresented near RpaA binding sites (E-value, 1.7 × 10−36; Table S4A) and is found within the RpaA~P footprint in the kaiBC promoter (Figure 3B) and in both footprints in the rpoD6 promoter (Figure S4E). Bases protected by RpaA~P binding in the DNase I footprinting assays are capitalized and boldfaced.
See also Figure S4 and Tables S3 and S4.
To ensure that the enrichment we observed did not reflect off-target binding of the anti-RpaA antibody, we performed a ChIP-Seq experiment using the HA-RpaA strain and the anti-HA antibody (Table S3B). The genome-wide HA-RpaA binding profile (Figure S4A) resembled that of wild-type RpaA, although enrichments were generally substantially lower (Figure S4B and Table S3B), consistent with the lower overall phosphorylation of HA-RpaA (Figure S4C). We found that 66 out of the 110 wild-type binding sites (60%) were also present in the HA-RpaA ChIP experiment at a minimum of 2-fold enrichment (Figure S4D and Table S3A).
To determine whether RpaA can bind directly to DNA at locations other than the kaiBC promoter, we performed in vitro DNase I footprinting on a strongly-enriched region upstream of the gene encoding the sigma factor RpoD6. RpaA bound to the rpoD6 promoter in a phosphorylation- and concentration-dependent manner (Figure S4E).
We identified an A/T-rich motif overrepresented within the RpaA binding sites (Figure 4D and Table S4A). This motif is present in the footprints of RpaA~P on the kaiBC and rpoD6 promoters (Figures 3B, 4D, and S4E), and over half (55%) of the RpaA binding sites contain one or more instances of this motif with p-values less than 0.001 (Tables S3A and S4B). We conclude that the peaks observed in the ChIP-Seq data primarily result from direct binding of RpaA to DNA.
The RpaA regulon contains genes mediating a variety of cellular processes
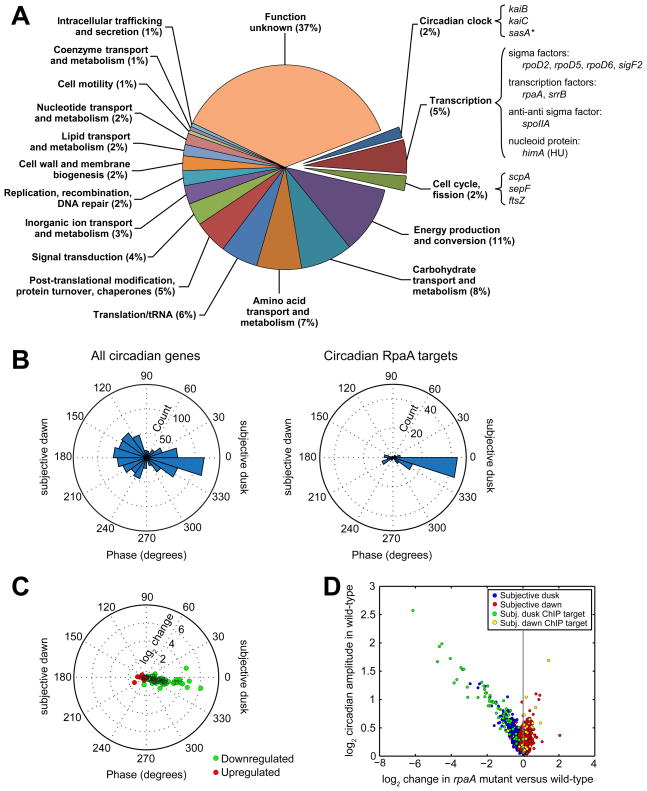
We systematically identified targets of RpaA genome-wide by searching for genic (annotated mRNA, tRNA, or rRNA) transcripts and high-confidence non-coding transcripts (Vijayan et al., 2011) with 5′ ends near the RpaA binding sites (see Extended Experimental Procedures). We found 134 such target transcripts; together, these comprise the RpaA regulon. Ninety-three of these transcripts collectively encode 170 genes (many being co-expressed in operons) (Figure 5A), while 41 of the target transcripts are high-confidence non-coding RNAs (Tables S1 and S5). Three peaks were located too far from any transcripts to be associated with a target.
Figure 5. The RpaA regulon.
A. Functional characterization of protein and tRNA ChIP targets of RpaA. We identified 134 transcripts closest to the 110 binding sites (see Extended Experimental Procedures). Of those transcripts, 93 encode proteins or tRNAs (corresponding to 170 genes, some of which are co-expressed in operons), while the other 41 are classified as non-coding RNAs (Vijayan et al., 2011). Because the function of the non-coding RNAs is not known, we restrict our functional analysis to the 170 protein-coding or tRNA genes (“RpaA ChIP target genes”). RpaA ChIP target genes were categorized as described in the Extended Experimental Procedures. Some genes of particular interest are highlighted. The asterisk (*) indicates that the gene’s classification as an RpaA ChIP target is artifactual because of assignment to an incorrectly-demarcated operon containing a bona fide target (Vijayan et al., 2011).
B. Comparison of the distribution of phases of all circadian genes (left, n = 856, from Figure 1A) and of the ChIP target genes whose expression oscillates with circadian periodicity (right, n = 95).
C. Change in expression of circadian ChIP target genes downregulated (green, n = 72) or upregulated (red, n = 23) in the rpaA mutant plotted as a function of their phase in the wild-type strain.
D. Comparison of gene expression change in the rpaA mutant with circadian amplitude in the wild-type strain (from Figure 1B). Only genes that oscillate with circadian periodicity in the wild-type strain are shown (n = 856). Circadianly-expressed RpaA ChIP target genes (n = 95) are highlighted.
See also Tables S1 and S6.
Expression of most of the genic RpaA ChIP targets oscillates with circadian periodicity, and these targets are strongly enriched for subjective dusk expression (Figure 5B). Expression of a small number of targets peaks at subjective dawn, including, quite interestingly, the canonical subjective dawn gene purF (Paddock et al., 2013). In the rpaA mutant, subjective dusk RpaA ChIP targets decrease in expression while subjective dawn targets increase in expression (Figure 5C), suggesting that RpaA functions as an activator of subjective dusk targets and a repressor of subjective dawn targets. This helps to explain the dawn phase arrest of the rpaA mutant (Figures 1B and 1C). We also found that most strongly downregulated genes in the rpaA mutant are RpaA ChIP targets, while more weakly downregulated genes typically are not targets (Figure 5D). This implies that RpaA acts as a master regulator by directly modulating the expression of a subset of high-amplitude circadian genes, whose products in turn effect the fine global pattern of circadian gene expression.
The RpaA regulon (Figure 5A and Tables S1 and S5) contains targets involved in transcription regulation, including four sigma factors. Interestingly, RpaA also targets the himA gene (synpcc7942_2248) encoding the nucleoid protein HU, which is downregulated 1.7-fold in the rpaA mutant. Because both DNA compaction (Smith and Williams, 2006) and himA expression are regulated as a function of circadian time (Vijayan et al., 2009), himA could link oscillatory RpaA activity to circadian genome compaction, providing another route for RpaA to influence gene expression globally.
Several RpaA targets are enzymes of the glycolysis, glycogen, and pentose phosphate metabolic pathways, suggesting a direct link between the circadian clock and energy production and storage. RpaA also directly targets the translation initiation factor IF-3 (infC), the protein chaperone trigger factor (tig), and a ClpXP protease system (clpX and clpP2), implying that the clock may modulate translation and protein homeostasis directly, connections that have not been reported previously in this organism. In addition, the rpaA promoter itself is an RpaA ChIP target, suggesting the presence of autoregulatory feedback. We note that some 37% of the RpaA ChIP targets have no known function; among these targets there may be unforeseen control nodes with roles in global gene expression regulation.
Cells mutated for rpaA display increased efficiency of energy transfer from the light-harvesting phycobilisomes to photosystem II relative to photosystem I, and for this reason the gene was named regulator of phycobilisome association A (Ashby and Mullineaux, 1999). We find that RpaA directly targets the rpaC gene, which encodes an integral membrane protein implicated in controlling the stability of the photosystem II–phycobilisome interaction (Joshua and Mullineaux, 2005). rpaC is circadianly expressed and is downregulated approximately twofold in the rpaA mutant, consistent with positive regulation by RpaA.
Finally, we found that the RpaA regulon is enriched for genes whose expression increases in darkness in a kaiABC-dependent manner (Hosokawa et al., 2011) (p = 4.4 × 10−6, Fisher’s exact test), suggesting that RpaA is involved in clock modulation of the gene expression response to darkness.
Active RpaA is sufficient to switch cells between the two major gene expression states produced by the circadian clock
To test directly whether RpaA serves as the master regulator governing circadian gene expression, we asked whether the RpaA~P phosphomimetic RpaA(D53E) is sufficient to induce global changes in expression similar to those that occur over the course of a circadian cycle. We hypothesized that overexpression of RpaA(D53E) in an rpaA mutant would switch cells from the subjective dawn state in which the mutant resides to the subjective dusk state that coincides with the time of maximal RpaA activity in the wild-type strain. To isolate transcriptional changes resulting directly from activity of RpaA(D53E) from potentially confounding Kai oscillator-dependent processes, we introduced the Ptrc::rpaA(D53E) construct into a strain lacking kaiBC as well as rpaA, producing a ΔrpaA ΔkaiBC Ptrc::rpaA(D53E) strain that we refer to as “OX-D53E.”
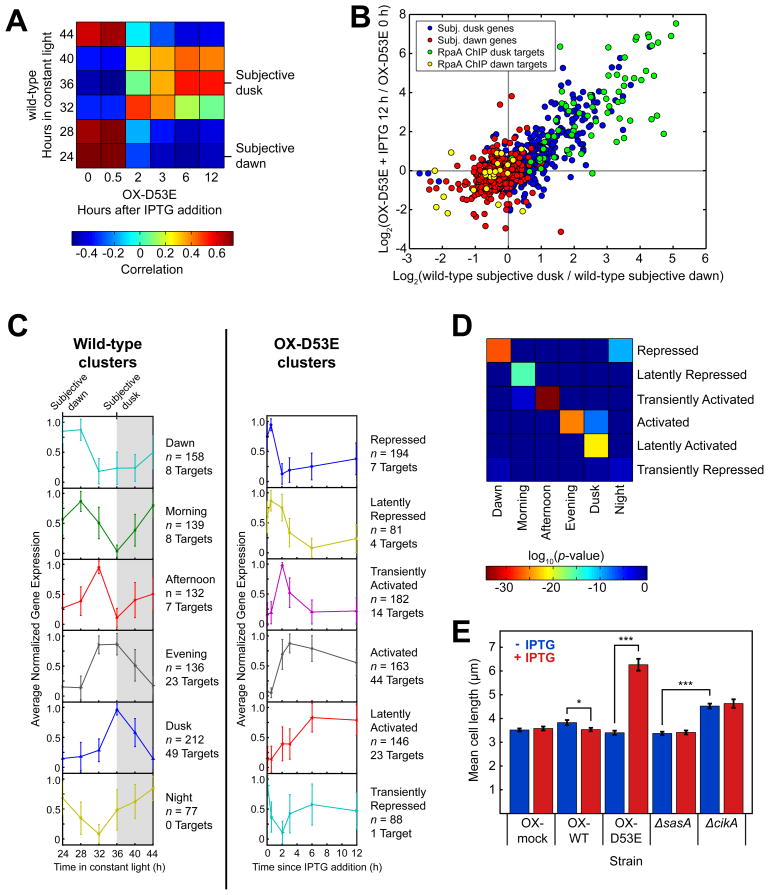
We used high-throughput RNA sequencing (RNA-Seq) to compare gene expression changes caused by induction of RpaA(D53E) expression with IPTG (Figure S5A) to those experienced during the course of a day in constant light in the wild-type strain (Table S6). First we calculated the correlation between expression of circadian genes during a timecourse of OX-D53E induction and during a wild-type circadian timecourse (Figure 6A). Consistent with the gene expression state of the rpaA mutant (Figure 1C), pre-induction OX-D53E is most correlated with wild-type at 24 h (subjective dawn) and most anti-correlated with wild-type at 36 h (subjective dusk). Over 12 h of RpaA(D53E) induction with IPTG, the correlations reverse (Figure 6A): OX-D53E becomes most similar to the wild-type 36 h timepoint and most anti-correlated with the wild-type 24 h timepoint. Importantly, these shifts are not observed when IPTG is added to a control strain (OX-mock) in which no gene is inserted downstream of the Ptrc promoter or when IPTG is not added to the OX-D53E strain (Figures S5B and S5C).
Figure 6. RpaA orchestrates global circadian gene expression and controls the cell division gate.
A. Correlation between the expression of circadian genes (n = 856) in the wild-type strain over the course of one day and the expression of those genes in the ΔrpaA ΔkaiBC Ptrc::rpaA(D53E) (OX-D53E) strain before (t = 0 h) and after induction with IPTG. Gene expression was measured by RNA-Seq.
B. Correlation between the change in expression of circadian genes (n = 856) caused by induction of RpaA(D53E) in the OX-D53E strain (y-axis) with the change in expression between subjective dusk and dawn in the wild-type strain (x-axis). RpaA ChIP target genes are highlighted (n = 95; 71 subjective dusk and 24 subjective dawn). Gene expression was measured by RNA-Seq.
C. K-means identification of gene expression clusters in the wild-type and OX-D53E strains. Gene expression was measured by RNA-Seq. With K = 6, wild-type circadian genes (n = 856) were separated into six clusters with distinct expression phases (left), consistent with previous microarray observations (Vijayan et al., 2009). Timecourses of the same set of genes in the OX-D53E strain were also clustered using K = 6 (right). The traces show the average normalized timecourse of genes within each cluster; error bars show standard deviation. The numbers of all genes (n) and RpaA ChIP target genes in each cluster are indicated. Non-coding RNAs were not included in this analysis.
D. Mapping between clusters in the wild-type (x-axis) and OX-D53E (y-axis) strains. Each element of the heatmap shows the log10 of the statistical significance (Fisher’s exact test) of the overlap between the corresponding clusters on each axis.
E. Mean cell lengths in ΔrpaA ΔkaiBC strains containing a Ptrc promoter driving expression of wild-type RpaA (OX-WT), RpaA(D53E) (OX-D53E), or an empty multi-cloning site (OX-mock) grown in the presence (red) or absence (blue) of the inducer IPTG (100 μM). At least 80 cells were analyzed for each strain. Error bars show standard error of the mean (SEM). *, p < 0.05; ***, p < 10−13 (one-way ANOVA).
The flip of the dawn-to-dusk gene expression switch is illustrated by plotting each circadian gene’s expression change upon IPTG induction in OX-D53E against its change between subjective dawn and dusk in the wild-type (Figure 6B). After induction, subjective dusk gene expression increases in proportion to its change from subjective dawn to dusk in the wild-type, while subjective dawn gene expression decreases in proportion to its change in the wild-type (correlation = 0.8). Furthermore, expression of 85% of circadian genes (725 of 856) differed from baseline by greater than 1.5-fold in at least one timepoint after IPTG induction. The strong correlation between the dawn-to-dusk expression change in the wild-type and the expression change upon RpaA(D53E) induction in OX-D53E shows that active RpaA suffices to switch cells between dawn and dusk expression states.
Consistent with our analysis of the ChIP-Seq data (Figures 5B–5D), constitutively active RpaA strongly induced expression of ChIP dusk targets, while it had weaker repressive activity toward a minority of its subjective ChIP dawn targets (Figure 6B). Interestingly, RpaA ChIP targets comprise only a subset of significantly affected genes; the remaining genes must be activated or repressed by one of RpaA’s direct targets. Collective these results demonstrate that RpaA is the master regulator of circadian gene expression, acting as the most upstream node in a network of circadian regulators that together orchestrate global gene expression rhythms.
The RpaA regulon produces complex gene expression dynamics that parallel those observed during a circadian period
We examined whether the dynamics of the subjective dawn to dusk transition induced by active RpaA mirror those observed in the wild-type strain. We used K-means clustering to identify patterns in expression of circadian genes in the wild-type strain and of those same genes in the OX-D53E strain (Figures 6C and S5D). In the wild-type strain, clustering separated genes according to their time of maximum expression: Dawn, Morning, Afternoon, Evening, Dusk, and Night (Vijayan et al., 2009). We named the clusters obtained for the OX-D53E strain according to their dynamics following induction: Repressed, Latently Repressed, Transiently Activated, Activated, Latently Activated, and Transiently Repressed. Some genes in the Repressed and Latently Repressed category were slightly induced at 30 minutes after IPTG addition but were subsequently repressed as RpaA(D53E) levels further increased (Figure S5A). This suggests that low levels of RpaA promote the expression of these genes, while higher levels have a much stronger repressive effect on their expression. Notably, the timescale of response to induction varies amongst the clusters, with some clusters responding in concert with RpaA(D53E) accumulation (Activated and Repressed) and others more slowly (Latently Activated and Latently Repressed). The Transiently Activated and Transiently Repressed clusters display markedly non-monotonic responses to RpaA(D53E) induction. Hence, the network of regulators downstream of RpaA encodes a variety of responses to continuous accumulation of active RpaA. Note that RpaA ChIP targets never comprise more than one-quarter of the genes in a cluster. Therefore, the majority of genes in each cluster are controlled by the RpaA regulon rather than by RpaA itself.
If the gene expression dynamics observed in the OX-D53E induction timecourse resemble those in the wild-type circadian timecourse, there should be an overlap between the clusters in each strain. Indeed, we find a nearly one-to-one mapping between the two sets of clusters (Figure 6D). The Dawn cluster in the wild-type strain maps to the Repressed cluster in OX-D53E, Morning maps to Latently Repressed, Afternoon maps to Transiently Activated, Evening maps to Activated, Dusk maps to Latently Activated, and Night maps to Repressed (p ≤ 0.001 for each pair). Remarkably, this mapping arises despite the difference in the timing of RpaA activation in the two strains. In the wild-type strain, RpaA~P abundance varies sinusoidally, increasing over a ~12 hour timespan and then decreasing over the same timespan (Figure 3A). In contrast, RpaA(D53E) accumulates rapidly, plateauing after 3 h of induction and remaining high for the remainder of the timecourse (Figure S5A). Nonetheless, genes respond to active RpaA in a stereotyped manner in both strains, suggesting that the dynamics of global circadian gene expression are hard-wired into the RpaA regulon.
Active RpaA closes the cell division gate
During the subjective night cells elongate but do not divide, a phenomenon referred to as cell division gating (Dong et al., 2010; Mori et al., 1996; Yang et al., 2010). When the gate is closed, cells form elongating rods, allowing the status of the gate in a given strain to be inferred from the cell length distribution (Dong et al., 2010). The observation that deletions of sasA and cikA have opposite effects on cell cycle gating (Dong et al., 2010) suggests that RpaA~P could be responsible for closing the gate, as SasA and CikA have opposite effects on RpaA phosphorylation (Gutu and O’Shea, 2013).
To test this hypothesis, we examined the effect of overexpression of wild-type and phosphomimetic RpaA on cell length. We characterized cell length in strains ectopically expressing wild-type RpaA (OX-WT), RpaA(D53E) (OX-D53E), or an empty multi-cloning site (OX-mock) from the Ptrc promoter with and without IPTG treatment (Figures 6E, S5E, and S5F). Consistent with our hypothesis, cells showed pronounced elongation in the OX-D53E strain after induction. Conversely, induction of OX-WT shortens the median cell length, consistent with the reduction in RpaA~P levels (Figure S5F) and the repression of kaiBC gene expression in this condition (Figure 3C). Induction of OX-mock had no effect on cell length. These data are consistent with a causative role for RpaA~P in clock-mediated cell division gating.
Several RpaA ChIP targets are involved in cell division and so might mediate gating. The most prominent of these are the bacterial tubulin homolog ftsZ and the FtsZ regulator sepF (Marbouty et al., 2009). Intriguingly, FtsZ is mislocalized in ΔcikA strains (Dong et al., 2010). However, mean ftsZ and sepF expression is unchanged by deletion of rpaA (Table S1), so the functional relevance of these targets is unclear. Other ChIP targets of interest are genes involved in the peptidoglycan biosynthetic pathway (synpcc7042_0482, synpcc7042_1740/murB, synpcc7042_1741/murC), the last of which has been shown to interact with several Fts cell division proteins (Munshi et al., 2013).
Discussion
RpaA is the hub through which the circadian clock controls cellular physiology
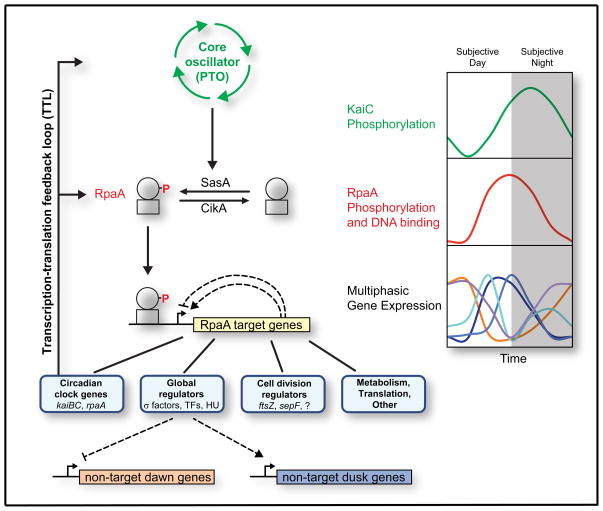
RpaA phosphorylation links the core Kai oscillator to two of the most striking physiological outputs of the clock: global transcriptome oscillations and gating of cell division (Figure 7). Time information encoded in the PTO is read out through the histidine kinases SasA and CikA, which antagonistically regulate RpaA phosphorylation to generate oscillations of RpaA~P that peak at or immediately preceding subjective dusk (Gutu and O’Shea, 2013) (Figures 3A and 7). The accumulation of RpaA~P switches the cell’s gene expression program from subjective dawn to subjective dusk (Figures 6A and 6B) via transcriptional programs hardwired into the RpaA regulon (Figures 6C and 6D). RpaA~P accumulation also initiates closure of the cell division gate (Figure 6E). RpaA~P decreases during the subjective night (Gutu and O’Shea, 2013) (Figure 3A), allowing gene expression to revert to its default dawn-like state (Figures 1B and C) and also opening the cell division gate.
Figure 7. Model for RpaA and the cyanobacterial circadian program.
Model for control of clock output by RpaA. Time encoded in the PTO is transduced into RpaA phosphorylation via SasA and CikA, producing oscillations in RpaA~P (red) that phase-lead those of phosphorylated KaiC (green) by approximately 4 hours (Gutu and O’Shea, 2013). RpaA~P binds to DNA and controls the expression of the RpaA regulon, which consists of global regulators, cell division regulators, certain clock genes (kaiBC and rpaA), and genes involved in metabolism and translation. The global regulators are at the top of a transcriptional cascade that orchestrates multiphasic circadian gene expression, repressing subjective dawn genes while activating subjective dusk genes. Fine patterns within the dusk and dawn categories could be generated by a network of interactions amongst RpaA ChIP targets (hypothetical positive and negative feedbacks are shown as dotted lines). RpaA control of kaiBC and rpaA expression forms the clock TTL.
Previous reports have implicated circadianly-regulated DNA supercoiling in driving gene expression oscillations (Vijayan et al., 2009; Woelfle et al., 2007). We sought to investigate the relationship between RpaA binding and supercoiling by measuring supercoiling in the rpaA mutant, ΔrpaA clock rescue, and OX-D53E strains, but had difficulty obtaining reproducible results and so do not report any here. Nonetheless, we can gain insight into to the relative importance of supercoiling and RpaA binding in generating global gene expression oscillations by comparing the magnitudes of expression changes in response to perturbations in supercoiling and RpaA activity. When supercoiling is rapidly relaxed by treatment with a pharmacological inhibitor of DNA gyrase (Vijayan et al., 2009), global gene expression changes in the same manner as it does upon induction of RpaA(D53E) (Figure 6B). However, the magnitude of change is substantially larger for induction of RpaA(D53E) than for relaxation of supercoiling (compare Figure 6B here to Figure 4 in (Vijayan et al., 2009)). Moreover, induction of RpaA(D53E) almost quantitatively reproduces the magnitude of gene expression change observed over circadian time in the wild-type strain (Figure 6B). On this basis, we suggest that circadian oscillations in RpaA activity play a dominant role in driving global circadian gene expression oscillations. Perhaps one or more members of the RpaA regulon induces oscillation in supercoiling, which acts in concert with RpaA via a feedforward loop to actuate circadian gene expression. Alternatively, RpaA binding affinity could be positively influenced by supercoiling, in which case relaxation of supercoiling by pharmacological inhibition of gyrase would release RpaA from the chromosome, leading to observed effects on global gene expression state (Vijayan et al., 2009). Future studies will be required to establish the molecular connections between RpaA activity and supercoiling. Our observation that RpaA targets the himA gene encoding the nucleoid protein HU could provide a starting point for such studies.
Also meriting future investigation are possible roles for RpaA in the integration of environmental and time information. Environmental cues may influence RpaA phosphorylation directly, as the activity of its phosphatase CikA is regulated not only by the PTO but also by the cellular redox state, which in turn reflects environmental conditions such as light availability (Ivleva et al., 2006; Kim et al., 2012). Hence, information about the environment and circadian time may be integrated at the level of RpaA phosphorylation. RpaB also may play a role: RpaB phosphorylation is regulated by a number of environmental cues (Moronta-Barrios et al., 2012), and RpaB binds to the promoters of kaiBC and rpoD6 (Hanaoka et al., 2012), both of which are RpaA targets. In fact, RpaB binding to the kaiBC promoter is antagonized by RpaA (Hanaoka et al., 2012), consistent with the overlap of the RpaA footprint (Figure 3B) with the HLR1 motif bound by RpaB. A systematic exploration of the interaction of RpaA and RpaB at promoters and its effect on gene expression will be required.
A recent study found that activity of the canonical subjective dawn promoter PpurF is affected by the presence of the kaiC gene in an rpaA mutant background, suggesting the presence of an RpaA-independent output pathway (Paddock et al., 2013). The authors proposed a model for PpurF control in which PTO-modulated RpaA~P levels repress the activity of a “predominant” RpaA-independent output pathway, which in turn links serine-phosphorylated KaiC to activation of the promoter. This model predicts that purF transcript abundance will oscillate in our ΔrpaA mutant clock rescue experiment, in which KaiC phosphorylation oscillates with high amplitude (Figures 2 and S2). Instead, we found purF levels to be constant, locked at the level of maximum expression in the control clock rescue (Figure S2C).
While our data do not rule out the existence of an RpaA-independent clock output pathway, they do suggest that any such pathway is weak relative to the RpaA-dependent output pathway we describe here. Moreover, our ability to switch the genome-wide transcriptional program from subjective dawn to dusk by expression of an RpaA phosphomimetic in a strain lacking both kaiB and kaiC (Figures 6 and S5) shows that RpaA phosphorylation is sufficient to control the circadian gene expression program. Notably, purF expression is switched from a high to a low state upon phosphomimetic induction (Figure S5D); this is straightforwardly explained by the fact that purF is a direct ChIP target of RpaA (Tables S1 and S5). We found that RpaA~P binds to PpurF in vitro and that point mutations affecting its expression phase (Vijayan and O’Shea, 2013) fall within the RpaA~P footprint; at least two of these mutations impair RpaA~P binding (Figure S4F).
Generation of complex gene expression patterns with a one-dimensional signal
The complex gene expression dynamics observed upon induction of OX-D53E (Figure 6C) demonstrates that a smooth, univariate signal like RpaA phosphorylation (Figure 3A and (Gutu and O’Shea, 2013)) can generate a diverse array of dynamic responses. We suggest that the smooth RpaA~P signal is converted into a mosaic of dynamic patterns through network motifs composed of the gene expression regulator targets of RpaA (e.g., sigma factors) and their own downstream targets.
Interestingly, the RpaA~P level does not uniquely specify the time of day, as intermediate RpaA~P levels are experienced during both the subjective day and subjective night (Figure 3A). The single-bit encoding of time in RpaA phosphorylation contrasts sharply with its encoding in the PTO, in which differential phosphorylation at two residues of KaiC specifies two bits of information (Nishiwaki et al., 2007; Rust et al., 2007). The four phosphoforms of KaiC appear in an ordered pattern during each circadian cycle in a manner that uniquely maps the time of day to a particular phosphoform distribution (Nishiwaki et al., 2007; Rust et al., 2007). It seems paradoxical that time encoded in the PTO would be read out through a single-bit channel that cannot uniquely encode it, causing a loss of information. However, the network motifs that likely generate the complex circadian gene expression dynamics (Figure 6C and 6D) could make the functional effect of a given RpaA~P level history-dependent (hysteretic). The current level of RpaA~P and its history together would suffice to fully specify time.
Relevance to eukaryotic circadian clock output pathways
In directly controlling a large number of transcriptional regulators, RpaA resembles eukaryotic circadian effectors like White Collar Complex (WCC) in Neurospora crassa (Smith et al., 2010), CLOCK and CYCLE in Drosophila melanogaster (Abruzzi et al., 2011), CLOCK and BMAL1 in mice (Koike et al., 2012; Rey et al., 2011), and PRR5 in Arabidopsis thaliana (Nakamichi et al., 2012). Transcription factors in these eukaryotic clock output pathways have been proposed to initiate hierarchical transcriptional cascades that effect widespread circadian rhythms in gene expression (Edery, 2011), similar to what we propose occurs with RpaA. However, the output pathways in these eukaryotic clocks are inseparable from the core circadian oscillators: the core oscillators are built from the output pathways themselves, which form time-delayed TTLs that drive oscillations. Multiple interlocking positive and negative transcriptional feedback loops exist in these clocks, with transcription factors serving simultaneously as clock outputs and TTL components. In contrast, the cyanobacterial clock is fundamentally a PTO with a subsidiary and dispensable TTL (Qin et al., 2010; Teng et al., 2013), with the output pathway serving mainly as a conduit for transmitting time information from the PTO to the genome in order to control circadian gene expression and cell division gating. More work is required to determine if these are indeed bona fide differences in the topology of these systems, or if the eukaryotic and prokaryotic clocks share more similarities than currently appreciated.
Synthetic biology applications
Our work has implications for synthetic biology. Identification of the binding motif and binding dynamics of RpaA (Figures 3A and 4) provide the molecular details required to rationally design connections between the PTO and synthetic transcriptional outputs, enabling reconstitution of the PTO with a transcription-based reporter in orthogonal organisms. Industrial applications arise, as cyanobacteria present an attractive platform for the engineered production of chemicals directly from the abundant fuel source of sunlight (Ducat et al., 2011). Synthetic pathways could be coupled to the circadian clock via RpaA; as the clock enhances the fitness of the wild-type organism in oscillating environmental conditions (Ouyang et al., 1998), so too might it enhance the productivity of synthetic pathways.
Experimental Procedures
Gene expression analysis
Microarray analysis of gene expression timecourses was conducted as described previously (Vijayan et al., 2009). To compare gene expression in the wild-type strain to that in the rpaA mutant (Figures 1B, 1C, and S1), we prepared time-averaged, pooled cDNA samples for each strain (constructed by combining equal mass quantities of cDNA from each timepoint), labeled them with different dyes, and hybridized them against one another. Dye ratios were normalized by lowess regression. To reduce dye bias, we analyzed a second microarray in which the dyes were swapped and averaged the lowess-normalized values from the two arrays.
RNA-Seq library preparation is described in the Extended Experimental Procedures. Expression levels were median- and z-score normalized across samples. K-means clustering (with K = 6) was performed in Matlab with squared Euclidean distance as the distance metric.
ChIP-qPCR and ChIP-Seq
ChIP was performed as described previously (Hanaoka and Tanaka, 2008; Vijayan et al., 2011) with modifications described in the Extended Experimental Procedures. ChIP was performed using affinity-purified anti-RpaA antibody (see Extended Experimental Procedures) or anti-HA agarose beads (Pierce). Mock ChIP samples were prepared using the anti-RpaA antibody with crosslinked rpaA mutant cells or the anti-HA antibody with crosslinked wild-type cells.
For ChIP-qPCR, enrichment was computed by normalizing the abundance of the promoter of interest to the abundance of the coding region of synpcc7942_0612, which shows no enrichment for RpaA occupancy by ChIP-Seq. See Extended Experimental Procedures for more details.
Preparation and sequencing of ChIP-Seq libraries and ChIP-seq data analysis are described in the Extended Experimental Procedures.
Data availability
High-throughput data are available from Gene Expression Omnibus under accession number GSE50922.
Supplementary Material
Highlights.
RpaA is required for transmission of time from the core oscillator to clock outputs
rpaA deletion arrests cells in a dawn-like state, with no circadian gene expression
RpaA binds to 110 sites and activates global regulators such as sigma factors
Overexpression of activated RpaA switches cells from dawn to dusk state
Acknowledgments
We thank Vikram Vijayan for the 48-h wild-type gene expression timecourse and for code, technical assistance, and discussion; Andrian Gutu for Phos-tag Western blots; and Andrian Gutu, Xu Zhou, Abbas Rizvi, Brian Zid, Guogang Dong (UCSD) and Susan Golden (UCSD) for discussion and advice. We thank Richard Losick, Bodo Stern, and Andrian Gutu for comments on the manuscript and Takao Kondo (Nagoya University) and Susan Golden for plasmids and strains. This work was supported by the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abruzzi KC, Rodriguez J, Menet JS, Desrochers J, Zadina A, Luo W, Tkachev S, Rosbash M. Drosophila CLOCK target gene characterization: implications for circadian tissue-specific gene expression. Genes Dev. 2011;25:2374–2386. doi: 10.1101/gad.178079.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashby MK, Mullineaux CW. Cyanobacterial ycf27 gene products regulate energy transfer from phycobilisomes to photosystems I and II. FEMS Microbiol Lett. 1999;181:253–260. doi: 10.1111/j.1574-6968.1999.tb08852.x. [DOI] [PubMed] [Google Scholar]
- Dong G, Yang Q, Wang Q, Kim YI, Wood TL, Osteryoung KW, van Oudenaarden A, Golden SS. Elevated ATPase activity of KaiC applies a circadian checkpoint on cell division in Synechococcus elongatus. Cell. 2010;140:529–539. doi: 10.1016/j.cell.2009.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducat DC, Way JC, Silver PA. Engineering cyanobacteria to generate high-value products. Trends Biotechnol. 2011;29:95–103. doi: 10.1016/j.tibtech.2010.12.003. [DOI] [PubMed] [Google Scholar]
- Edery I. A master CLOCK hard at work brings rhythm to the transcriptome. Genes Dev. 2011;25:2321–2326. doi: 10.1101/gad.180984.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutu A, O’Shea EK. Two antagonistic clock-regulated histidine kinases time the activation of circadian gene expression. Mol Cell. 2013;50:288–294. doi: 10.1016/j.molcel.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanaoka M, Takai N, Hosokawa N, Fujiwara M, Akimoto Y, Kobori N, Iwasaki H, Kondo T, Tanaka K. RpaB, another response regulator operating circadian clock-dependent transcriptional regulation in Synechococcus elongatus PCC 7942. J Biol Chem. 2012;287:26321–26327. doi: 10.1074/jbc.M111.338251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanaoka M, Tanaka K. Dynamics of RpaB-promoter interaction during high light stress, revealed by chromatin immunoprecipitation (ChIP) analysis in Synechococcus elongatus PCC 7942. Plant J. 2008;56:327–335. doi: 10.1111/j.1365-313X.2008.03600.x. [DOI] [PubMed] [Google Scholar]
- Hosokawa N, Hatakeyama TS, Kojima T, Kikuchi Y, Ito H, Iwasaki H. Circadian transcriptional regulation by the posttranslational oscillator without de novo clock gene expression in Synechococcus. Proc Natl Acad Sci U S A. 2011;108:15396–15401. doi: 10.1073/pnas.1019612108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Mutsuda M, Murayama Y, Tomita J, Hosokawa N, Terauchi K, Sugita C, Sugita M, Kondo T, Iwasaki H. Cyanobacterial daily life with Kai-based circadian and diurnal genome-wide transcriptional control in Synechococcus elongatus. Proc Natl Acad Sci U S A. 2009;106:14168–14173. doi: 10.1073/pnas.0902587106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivleva NB, Gao T, LiWang AC, Golden SS. Quinone sensing by the circadian input kinase of the cyanobacterial circadian clock. Proc Natl Acad Sci U S A. 2006;103:17468–17473. doi: 10.1073/pnas.0606639103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CH, Stewart PL, Egli M. The cyanobacterial circadian system: from biophysics to bioevolution. Annu Rev Biophys. 2011;40:143–167. doi: 10.1146/annurev-biophys-042910-155317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshua S, Mullineaux CW. The rpaC gene product regulates phycobilisome-photosystem II interaction in cyanobacteria. Biochimica et biophysica acta. 2005;1709:58–68. doi: 10.1016/j.bbabio.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Kim YI, Vinyard DJ, Ananyev GM, Dismukes GC, Golden SS. Oxidized quinones signal onset of darkness directly to the cyanobacterial circadian oscillator. Proc Natl Acad Sci U S A. 2012;109:17765–17769. doi: 10.1073/pnas.1216401109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, Takahashi JS. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338:349–354. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutsuna S, Nakahira Y, Katayama M, Ishiura M, Kondo T. Transcriptional regulation of the circadian clock operon kaiBC by upstream regions in cyanobacteria. Mol Microbiol. 2005;57:1474–1484. doi: 10.1111/j.1365-2958.2005.04781.x. [DOI] [PubMed] [Google Scholar]
- Marbouty M, Saguez C, Cassier-Chauvat C, Chauvat F. Characterization of the FtsZ-interacting septal proteins SepF and Ftn6 in the spherical-celled cyanobacterium Synechocystis strain PCC 6803. J Bacteriol. 2009;191:6178–6185. doi: 10.1128/JB.00723-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markson JS, O’Shea EK. The molecular clockwork of a protein-based circadian oscillator. FEBS Lett. 2009;583:3938–3947. doi: 10.1016/j.febslet.2009.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T, Binder B, Johnson CH. Circadian gating of cell division in cyanobacteria growing with average doubling times of less than 24 hours. Proc Natl Acad Sci U S A. 1996;93:10183–10188. doi: 10.1073/pnas.93.19.10183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moronta-Barrios F, Espinosa J, Contreras A. In vivo features of signal transduction by the essential response regulator RpaB from Synechococcus elongatus PCC 7942. Microbiology. 2012;158:1229–1237. doi: 10.1099/mic.0.057679-0. [DOI] [PubMed] [Google Scholar]
- Munshi T, Gupta A, Evangelopoulos D, Guzman JD, Gibbons S, Keep NH, Bhakta S. Characterisation of ATP-Dependent Mur Ligases Involved in the Biogenesis of Cell Wall Peptidoglycan in Mycobacterium tuberculosis. PLoS One. 2013;8:e60143. doi: 10.1371/journal.pone.0060143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama Y, Oyama T, Kondo T. Regulation of circadian clock gene expression by phosphorylation states of KaiC in cyanobacteria. J Bacteriol. 2008;190:1691–1698. doi: 10.1128/JB.01693-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima M, Imai K, Ito H, Nishiwaki T, Murayama Y, Iwasaki H, Oyama T, Kondo T. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. 2005;308:414–415. doi: 10.1126/science.1108451. [DOI] [PubMed] [Google Scholar]
- Nakajima M, Ito H, Kondo T. In vitro regulation of circadian phosphorylation rhythm of cyanobacterial clock protein KaiC by KaiA and KaiB. FEBS Lett. 2010;584:898–902. doi: 10.1016/j.febslet.2010.01.016. [DOI] [PubMed] [Google Scholar]
- Nakamichi N, Kiba T, Kamioka M, Suzuki T, Yamashino T, Higashiyama T, Sakakibara H, Mizuno T. Transcriptional repressor PRR5 directly regulates clock-output pathways. Proc Natl Acad Sci U S A. 2012;109:17123–17128. doi: 10.1073/pnas.1205156109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiwaki T, Satomi Y, Kitayama Y, Terauchi K, Kiyohara R, Takao T, Kondo T. A sequential program of dual phosphorylation of KaiC as a basis for circadian rhythm in cyanobacteria. Embo J. 2007;26:4029–4037. doi: 10.1038/sj.emboj.7601832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Y, Andersson CR, Kondo T, Golden SS, Johnson CH. Resonating circadian clocks enhance fitness in cyanobacteria. Proc Natl Acad Sci U S A. 1998;95:8660–8664. doi: 10.1073/pnas.95.15.8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddock ML, Boyd JS, Adin DM, Golden SS. Active output state of the Synechococcus Kai circadian oscillator. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1315170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X, Byrne M, Xu Y, Mori T, Johnson CH. Coupling of a core post-translational pacemaker to a slave transcription/translation feedback loop in a circadian system. PLoS Biol. 2010;8:e1000394. doi: 10.1371/journal.pbio.1000394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey G, Cesbron F, Rougemont J, Reinke H, Brunner M, Naef F. Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol. 2011;9:e1000595. doi: 10.1371/journal.pbio.1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust MJ, Markson JS, Lane WS, Fisher DS, O’Shea EK. Ordered phosphorylation governs oscillation of a three-protein circadian clock. Science. 2007;318:809–812. doi: 10.1126/science.1148596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KM, Sancar G, Dekhang R, Sullivan CM, Li S, Tag AG, Sancar C, Bredeweg EL, Priest HD, McCormick RF, et al. Transcription factors in light and circadian clock signaling networks revealed by genomewide mapping of direct targets for neurospora white collar complex. Eukaryot Cell. 2010;9:1549–1556. doi: 10.1128/EC.00154-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RM, Williams SB. Circadian rhythms in gene transcription imparted by chromosome compaction in the cyanobacterium Synechococcus elongatus. Proc Natl Acad Sci U S A. 2006;103:8564–8569. doi: 10.1073/pnas.0508696103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai N, Nakajima M, Oyama T, Kito R, Sugita C, Sugita M, Kondo T, Iwasaki H. A KaiC-associating SasA-RpaA two-component regulatory system as a major circadian timing mediator in cyanobacteria. Proc Natl Acad Sci U S A. 2006;103:12109–12114. doi: 10.1073/pnas.0602955103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi Y, Katayama M, Ito R, Takai N, Kondo T, Oyama T. labA: a novel gene required for negative feedback regulation of the cyanobacterial circadian clock protein KaiC. Genes Dev. 2007;21:60–70. doi: 10.1101/gad.1488107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi Y, Takai N, Katayama M, Kondo T, Oyama T. Three major output pathways from the KaiABC-based oscillator cooperate to generate robust circadian kaiBC expression in cyanobacteria. Proc Natl Acad Sci U S A. 2010;107:3263–3268. doi: 10.1073/pnas.0909924107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng SW, Mukherji S, Moffitt JR, de Buyl S, O’Shea EK. Robust circadian oscillations in growing cyanobacteria require transcriptional feedback. Science. 2013;340:737–740. doi: 10.1126/science.1230996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayan V, Jain IH, O’Shea EK. A high resolution map of a cyanobacterial transcriptome. Genome Biol. 2011;12:R47. doi: 10.1186/gb-2011-12-5-r47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayan V, O’Shea EK. Sequence determinants of circadian gene expression phase in cyanobacteria. J Bacteriol. 2013;195:665–671. doi: 10.1128/JB.02012-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayan V, Zuzow R, O’Shea EK. Oscillations in supercoiling drive circadian gene expression in cyanobacteria. Proc Natl Acad Sci U S A. 2009;106:22564–22568. doi: 10.1073/pnas.0912673106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woelfle MA, Xu Y, Qin X, Johnson CH. Circadian rhythms of superhelical status of DNA in cyanobacteria. Proc Natl Acad Sci U S A. 2007;104:18819–18824. doi: 10.1073/pnas.0706069104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Pando BF, Dong G, Golden SS, van Oudenaarden A. Circadian gating of the cell cycle revealed in single cyanobacterial cells. Science. 2010;327:1522–1526. doi: 10.1126/science.1181759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwicker D, Lubensky DK, ten Wolde PR. Robust circadian clocks from coupled protein-modification and transcription-translation cycles. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:22540–22545. doi: 10.1073/pnas.1007613107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
High-throughput data are available from Gene Expression Omnibus under accession number GSE50922.