Abstract
Glioblastoma multiforme (GBM) is an aggressive, treatment-refractory type of brain tumor for which effective therapeutic targets remain important to identify. Here we report that cyclophilin B (CypB), a prolyl isomerase residing in the endoplasmic reticulum (ER), provides an essential survival signal in GBM cells. Analysis of gene expression databases revealed that CypB is upregulated in many cases of malignant glioma. We found that suppression of CypB reduced cell proliferation and survival in human GBM cells in vitro and in vivo. We also found that treatment with small molecule inhibitors of cyclophilins, including the approved drug cyclosporine, greatly reduced the viability of GBM cells. Mechanistically, depletion or pharmacologic inhibition of CypB caused hyperactivation of the oncogenic RAS-MAPK pathway, induction of cellular senescence signals, and death resulting from loss of MYC, mutant p53, Chk1 and JAK/STAT3 signaling. Elevated reactive oxygen species, ER expansion and abnormal unfolded protein responses in CypB-depleted GBM cells indicated that CypB alleviates oxidative and ER stresses and coordinates stress adaptation responses. Enhanced cell survival and sustained expression of multiple oncogenic proteins downstream of CypB may thus contribute to the poor outcome of GBM tumors. Our findings link chaperone-mediated protein folding in the ER to mechanisms underlying oncogenic transformation, and they make CypB an attractive and immediately targetable molecule for GBM therapy.
Keywords: glioblastoma multiforme, cyclophilin B, cellular senescence, MYC oncogene and mutant p53, endoplasmic reticulum stress
Introduction
Malignant gliomas have an extremely poor prognosis (1, 2), and require improved understanding of the molecular mechanisms supporting tumor cell survival. Cyclophilins, intracellular receptors for Cyclosporin A (CsA) (3), have peptidyl-prolyl isomerase activity, which accelerates the folding of proteins (4). The first cyclophilin identified, cyclophilin A, mediates the immunosuppressive effect of CsA by binding to calcineurin (5, 6). Cyclophilin B (CypB) is a highly related cyclophilin in the endoplasmic reticulum (ER) (7) and nucleus (8). Previous research suggested that CypB participates in multiple functions, including hepatitis virus replication (9), immunosuppression (10), chemotaxis (11), and prolactin signaling (12). To determine its role in vivo, we generated CypB knockout mice (13), and found that, other than moderate osteoporosis at older ages, CypB is not required for viability, and is well tolerated.
Recent studies revealed several signaling pathways frequently activated in GBM: the RTK pathway, the RB pathway and the p53 pathway (1). Most GBM tumors have mutations in all three to enhance cell proliferation and survival, while allowing the tumor cells to evade cell-cycle arrest, senescence, and death. Discovering ways to modulate these survival signals will improve therapeutic approaches for GBM. Because they are “druggable”, cyclophilins are considered good therapeutic targets. Recently, reports have implicated CypB in Stat3 activation and in generation of reactive oxygen specises (ROS) in other cancer cells (14, 15). Here, we identified CypB, for the first time, as a key regulator of several signals that fuel oncogenesis in GBM, including mutant p53, c-Myc (MYC), and Chk1. These findings offer proof-of-principle that CypB inhibitors may be effective as novel therapeutics for GBM.
Materials and Methods
Cell Culture and Reagents
GBM cell lines, primary human GBM xenograft cells (Mayo Brain Tumor Core), and human astrocytes (ScienCell Research) were grown in DMEM, 10%FBS, Pen/Strep. Xenografts using U251 cells in nude mice were performed as described (16). Brain tumor xenografts were obtained from the Mayo Brain Tumor Core Facility. Cell lines were obtained from the ATCC, and their identities verified by the ATCC (except for U251, which is no longer tested by the ATCC). Reagents were purchased from Sigma (cyclosporine, chloroquine, 10058-F4, H2O2, 17-AAG, AICAR), Tocris (thapsigargin, tunicamycin, eeyarestatin), Enzo (etoposide, daunorubicin), or Calbiochem (WP1066, Stattic, MG132, ALLN). CSA-dimer and compound 41 were from the Mayo Chemical Core.
shRNA-mediated silencing
TRC1.5 and TRC2 Lentiviral shRNA vectors (Sigma) were prepared using the manufacturer instructions, and transduced into GBM cells for 14 hours, followed by puromycin selection 24 hours later. Specific sequences are listed in Supplementary Materials.
Western blotting
Cell lysates were prepared at indicated times by lysis with 1%NP-40, and fractionated on SDS-PAGE. Westernblotting was conducted as described (13), using the indicated antibodies (sources listed in Supplementary Methods).
Gene expression analysis
Trizol-isolated total RNA was transcribed into cDNA using polydT primers and amplified to generate biotin-labeled cRNA. Gene expression microarray utilized the Illumina HumanHT-12v4 BeadChip, visualized by staining with Streptavidin-Cy3 after hybridization. Expression values were calculated using Illumina GenomeStudio® Data analysis software (GEO accession number GSE50756). Real-time PCR reactions utilized total RNA, reverse transcribed into cDNA, and were run in triplicate. mRNA levels were normalized to Actin or Gapdh, expressed as ΔΔCt values.
Cell viability, cell death, colony formation assays
Cells seeded in triplicate were cultured for the indicated times, and then directly counted using trypan blue exclusion for proliferation assays. Cell viability tests used AlamarBlue® (Invitrogen) according to the manufacturer instructions. Cell death was determined 3–5 days after lentiviral transduction by FACS analysis of PI/annexin-V-Cy5 staining. For colony formation, cells suspended in complete medium containing 0.3% agar deposited onto solidified 0.6% agar in 6-well plates were quantified for colonies after 17 days.
Senescence-associated β-galactosidase (SA-β-gal) asaay
Cells plated in triplicate at 104 cells per well in 6-well plates were assessed for SA-β-gal activity after 4 days (Cell Signaling SA-β-gal kit) following the manufacturers instructions. Percentages were assessed by counting at least 300 cells for each.
NFAT activation assay
NFAT activation was quantified by FACS analysis of a CD8 reporter under the control of a trimeric NFAT binding site in Jurkat cells stimulated with PMA/ionomycin in the presence or absence of cyclophilin inhibitors.
PDI staining
Cells were cultured on glass cover slides, fixed with 2.5% paraformaldehyde in PBS and permeabilized using 0.2% TritonX-100 in PBS. Cells were blocked with PBS containing 5% goat serum and stained with anti-PDI (Stressgen, 1D3).
Statistical analyses
Two-tailed Student’s t tests were used to measure the significance of a difference deviation between two means. Data are shown as mean±SD, and significant difference is represented by *P<0.05.
Results
CypB is overexpressed in brain tumors
Given the potential that CypB regulates Stat3-dependent cell proliferation (15), we asked whether CypB was expressed in neoplasms. Oncomine database (www.oncomine.org) inspection revealed that CypB was upregulated in 98/398 analyses of tumors (p<0.05). The highest upregulation was in brain tumors, with 14 studies comparing normal tissue to GBM, astrocytomas, or oligodendrogliomas (Fig. 1A,S1A). Rembrandt Repository (http://rembrandt.nci.nih.gov) analysis indicated highest CypB expression in GBM tumors compared to all others, while lowest levels were in non-tumor brain (Fig. S1B). Measurement of CypB abundance by western blot in primary human GBM cells revealed high amounts of CypB compared to primary human astrocytes (Fig. 1B,S1C).
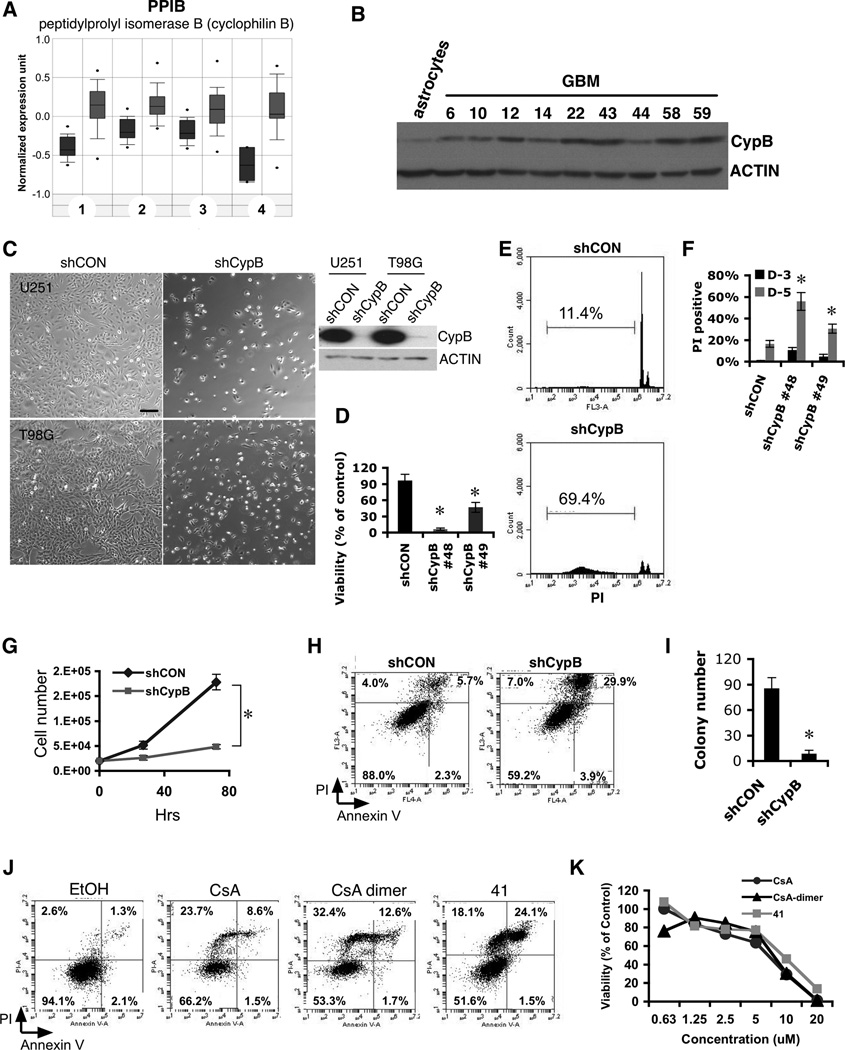
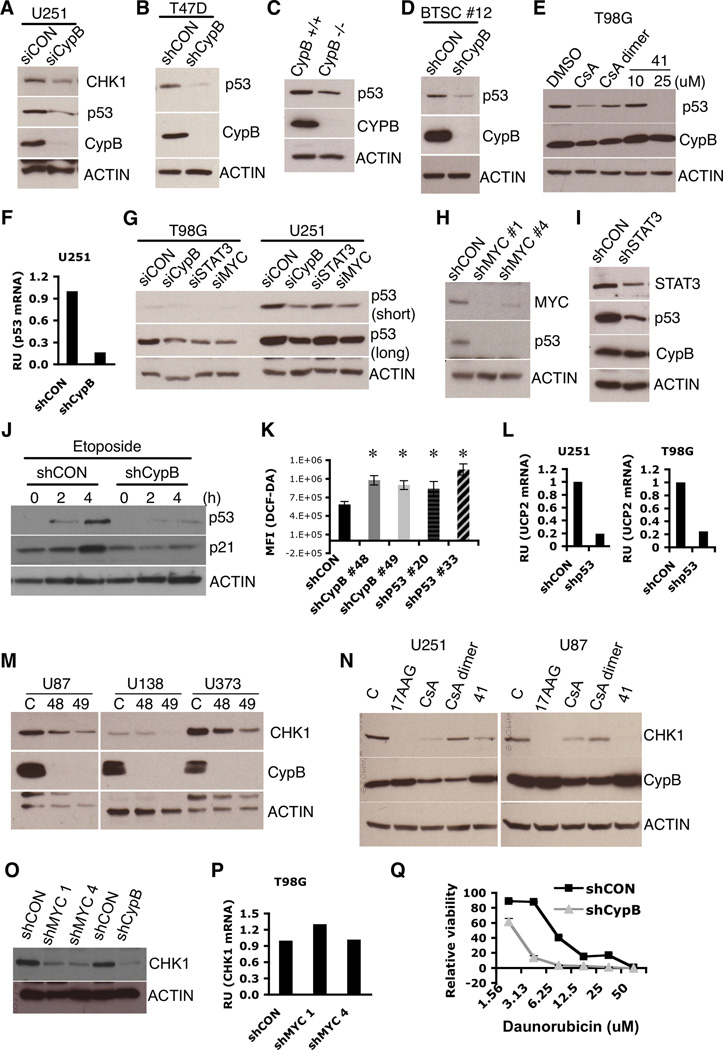
Figure 1. Knockdown or inhibition of Cyclophilin B reduces glioblastoma cell survival.
(A) Increased expression of CypB in biopsy samples (in gray) from patients with glioblastoma multiforme (p=7.9×10−20), astrocytoma (p=1×10−8), oligodendroglioma (p=4.6×10−8), and another astrocytoma study (p=2.5×10−5), compared to normal brain tissue (dark gray) (www.Oncomine.org). (B) Increased CypB protein expression in primary human GBM cells. (C) Cell death induced by CypB knockdown (shCypB) in U251 or T98G cells compared to control lentivirus (shCON). Bar=200µm. (D) AlamarBlue assay of cell viability in control U251 or two different CypB-knockdown U251 cultures (D-5 after knockdown). (E and F) CypB silencing increases sub-G1 fraction and PI positivity of U251 cells (D-3 or D-5). (G) Growth rates measured by counting control or CypB-knockdown U251 cells. (H) Cell death due to CypB knockdown detected by annexin-V/PI staining (D-4). (I) Decreased anchorage-independent tumor growth in CypB-depleted T98G cells analyzed in a soft agar growth assay. (J) Cell death induced in U251 cells treated for 48h with cyclosporine (CsA), cyclosporine dimer (CsA-dimer) or compound 41 (41). (K) Cyclophilin inhibitors decreased the survival of U251 cells, as measured by AlamarBlue assay. Errorbars show ±SD. *p<0.05.
Genetic ablation of CypB reduces glioma cell survival and proliferation
We depleted CypB in human GBM cells using lentiviral-delivered shRNA. Knockdown of CypB significantly decreased the viability of GBM cells (Fig. 1C,1G,1D). Increased death in CypB-depleted U251 cells was confirmed by increased subG1 fractions (Fig. 1E,1F) and annexin-V/PI staining (Fig. 1H). CypB knockdown also reduced anchorage-independent colony growth in soft agar (Fig. 1I).
Inhibition of CypB kills GBM cells
CypB Inhibition by cyclosporin A (CsA) also reduced GBM cell viability and proliferation (Fig. 1J). Immunosuppression by CsA is an obstacle to its use as a cancer therapeutic, so we synthesized a non-immunosuppressive derivative of CsA (CsA-dimer), which cannot inhibit calcineurin, yet retains cyclophilin-binding activity (17). CsA-dimer had equal effectiveness in killing U251 cells (Fig. 1J), yet was not immunosuppressive, indicating that its cytotoxic effect results from cyclophilin inhibition, rather than calcineurin inhibition (Fig. S1D). Also, compound 41, an unrelated small molecule shown previously to inhibit cyclophilin (18) killed glioblastoma cells (Fig. 1J,S1E) yet demonstrated little immunosuppression (Fig. S1D). These inhibitors also killed two medulloblastoma cell lines (Fig. 1K,S1F,S1G,S1H).
CypB regulates Stat3 mediated GBM survival by supporting Jak2 expression
Stat3 nuclear localization in myeloma was reported to be dependent upon CypB (15). We tested whether CypB knockdown similarly altered Stat3 nuclear accumulation in GBM cells. Unexpectedly, knockdown of CypB instead strongly decreased Stat3 phosphorylation upon Oncostatin-M (OSM) stimulation (Fig. 2A), without affecting cell surface expression of its receptor (Fig. 2B). CypB-knockdown T98G cells had reduced amounts of the Stat3-specific kinase Jak2 (Fig. 2C,2D), potentially explaining decreased Stat3 phosphorylation. Jak2 mRNA was not reduced (rather increased) by CypB silencing (Fig. 2E,2F), suggesting that CypB regulates Jak2 expression post-transcriptionally. Consistent with this, Stat3 inhibitors (19) increased cell death upon CypB knockdown, but had little effect on control GBM cells (Fig. 2G,2H). Combined treatment with CsA-dimer and Stat3 inhibitors also caused synergistic killing of tumor cells (Fig. 2I).
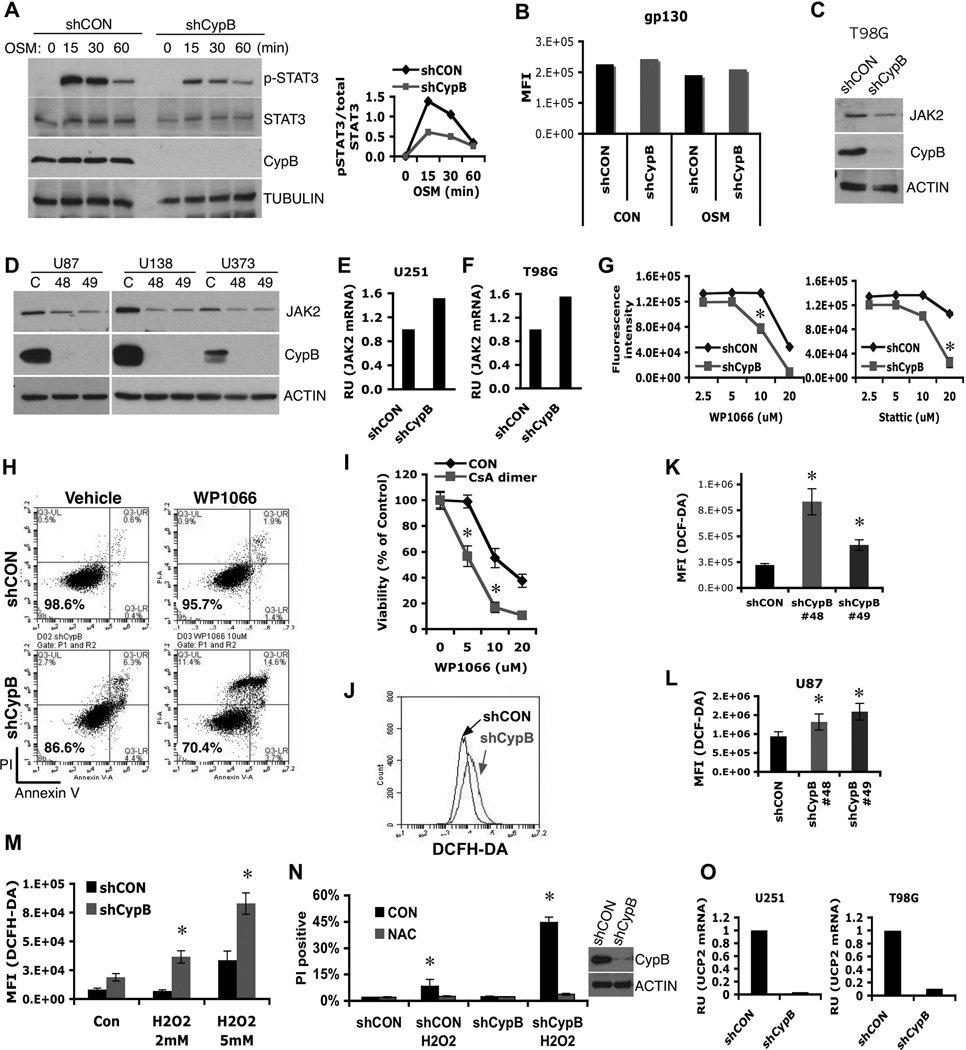
Figure 2. Cyclophilin B controls Jak2-Stat3 signals and ROS generation.
(A) Control or CypB-knockdown U251 cells were treated with Oncostatin M (OSM) for the indicated times and analyzed by westernblotting with the indicated antibodies (left). The results of phospho-Stat3 (pStat3) and total Stat3 westernblots were quantified using ImageJ. Relative intensity values of pStat3 to total Stat3 were plotted (right). (B) Normal cell surface expression of gp130 in CypB-depleted U251 cells, as measured by flow cytometry. (C and D) CypB depletion reduced Jak2 protein abundance in T98G, U87, U138, and U373 GBM cells. (E and F) Knockdown of CypB did not reduce Jak2 mRNA levels in U251 and T98G cells, measured by real-time PCR. (G and H) Decreased cell viability in CypB-depleted U251 cells in response to treatment with Stat3 inhibitors. Control or CypB-depleted U251 cells were treated with the indicated doses of Stat3 inhibitor, WP1066 (left) or Stattic (right) for 48h (G). U251 cells were treated with WP1066 (10µM) for 24h and cell viability was quantified by annexin-V/PI staining (H). (I) Increased cell killing effects with the indicated compounds for 24hr. Cell viability was observed by AlamarBlue assay. (J–L) Knockdown of CypB increased ROS generation in T98G (J), U251 (K), and U87 cells (L). Cellular ROS was measured using flow cytometry after staining with DCFH-DA (0.5mM for 30min). (M) CypB-depleted T98G cells had higher ROS production in response to H2O2 treatment. Cells were treated with H2O2 for 2hr and cellular ROS was measured by DCFH-DA staining. (N) CypB-depleted T98G cells showed increased sensitivity to oxidative stress-induced cell death. Cells were pretreated with 5mM N-acetyl cysteine (NAC) or vehicle, and subsequently with H2O2 for 24h. Cellular viability was measured by PI stain exclusion. (O) CypB silencing decreased UCP2 mRNA levels in U251 (left) or T98G (right) cells. Errorbars show ±SD. *p<0.05.
CypB suppresses generation of ROS
CypB-depleted GBM cells showed higher ROS levels than control cells (Fig. 2J,2K,2L), and developed higher increases in ROS following H2O2 exposure (Fig. 2M), indicating reduced ability to handle oxidative stress. CypB-knockdown GBM cells died more following H2O2 treatment (Fig. 2N), which was reversed by the antioxidant, N-acetyl cysteine (Fig. 2N).
CypB loss induces cellular senescence
Microarray analysis of RNA from control or CypB-knockdown U251 cells revealed approximately 130 genes affected more than 2-fold by CypB depletion (Fig. 3A,Table S1,Table S2). CypB-knockdown cells had a 3.4-fold reduction in the transcript for uncoupling protein-2 (UCP2), which suppresses ROS production during oxidative phosphorylation (20). These microarray results were similar to expression profiles of oncogenic Ras-induced senescent cells (Fig. 3D) (21), including increased levels of members of the senescence-associated secretome, such as IL-11, IL-6, IL-8, PLAU and SERPINE1. Several of these microarray results were validated by real-time PCR (Fig. 3B,2O) and westernblot (Fig. 3C).
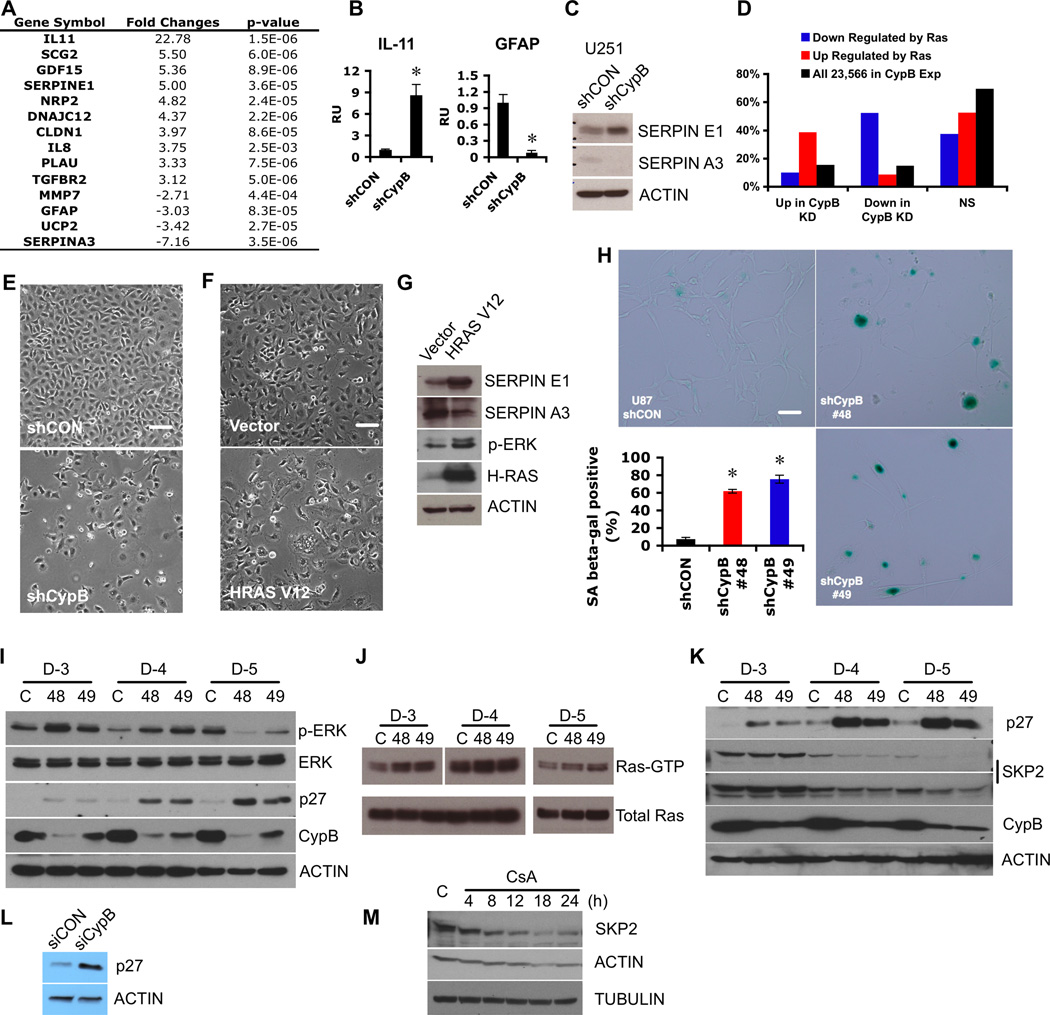
Figure 3. Cyclophilin B suppresses cellular senescence.
(A) Representative gene expression changes in CypB-depleted U251 cells. (B) Increased IL-11 (left) or decreased GFAP (right) mRNA levels in CypB-depleted U251 cells were confirmed by real-time PCR. (C) Increased SERPINE1 and decreased SERPINA3 expression were confirmed by westernblotting in CypB-depleted U251 cells. (D) Cross match analysis between microarray results from CypB silencing and those from oncogenic H-Ras-activated senescent cells. (E) Morphological changes in CypB-depleted U251 cells. CypB-knockdown cells had cytoplasmic vacuolization and large, flat appearance. Bar=200µm. (F and G) Morphological features (F) and molecular changes (G) of oncogenic H-Ras-expressed GBM cells. U251 cells were transduced with a lentivirus expressing oncogenic H-Ras (HRASV12) or control lentivirus (Vector) and cell morphology was examined by microscopy (F). Ras expression, subsequent Erk activation, and expression changes in SERPINE1 and SERPINA3 were measured by westernblotting (G). Bar=200µm. (H) Increased positive staining of the senescence-associated β-galactosidase marker in CypB-silenced U87 cells. Bar=200µm. (I) Increased Erk activation and p27 accumulation in CypB-depleted U251 cells. Cells were transduced with shCypB (48 or 49) or control lentivirus (C), and analyzed at days 3, 4, and 5. (J) Increased Ras activation in CypB-depleted U251 cells. Active form of Ras (RAS-GTP) was pulled down with Raf-1 RBD (Millipore) and detected by westernblot. (K and L) Increased expression of p27 in CypB-depleted T98G (K) or U87 cells (L). (M) CsA treatment (20 µM) reduced Skp2 expression in U251 cells. Errorbars show ±SD. *p<0.05.
As suggested by these findings, although the majority of U251 cells died upon CypB knockdown, surviving cells showed microscopic changes characteristic of senescence, including large and flat morphology, binuclear cells, and massive vacuolization (Fig. 3E). Furthermore, CypB knockdown in U87 cells markedly increased SA-β-gal staining (Fig. 3H). Overexpression of oncogenic Ras (HRASV12) in GBM cells evoked similar senescence-associated morphological and molecular changes (Fig. 3F,3G). Oncogenic Ras activates the MAPK/ERK cascades in senescent cells (21). CypB knockdown indeed increased Erk phosphorylation in U251 cells on day 3 and 4 following shRNA expression (Fig. 3I), and caused Ras activation (Fig. 3J). MEK inhibitor (U0126) treatment reduced this early hyperactivation of Erk and blocked cellular senescence in CypB-depleted GBM cells (Fig. S2). Ras hyperactivation causes suppression of its own downstream signals, due to Ras-dependent induction of the negative feedback inhibitors Sprouty, RasGap120, and DUSP proteins (22, 23). Consistent with this, by day 5 following CypB-depletion, phospho-Erk was reduced below the starting baseline (Fig. 3I). Furthermore, DUSP5 and Sprouty2 were among the top 35 upregulated genes in CypB-knockdown GBM cells (Table S1). CypB knockdown or inhibition also increased levels of the cell cycle inhibitor p27/Kip1 (24) (Fig. 3I,3K,3L) and strongly reduced the p27-specific E3 ubiquitin ligase Skp2 (Fig. 3M). These results suggest that aberrant activation of the RAS-MAPK pathway occurs following CypB suppression, and induces cellular senescence in GBM cells, possibly via increased p27.
CypB regulates MYC stability
We examined cells for known Ras targets previously implicated in cancer, such as MYC (25, 26). CypB depletion in GBM cells reduced steady-state levels of MYC (Fig. 4A). Induction of MYC by OSM or AICAR (27) was also blocked by CypB-knockdown (Fig. 4B,4C). CypB regulation of MYC is post-transcriptional since its silencing did not reduce MYC mRNA (Fig. 4D).
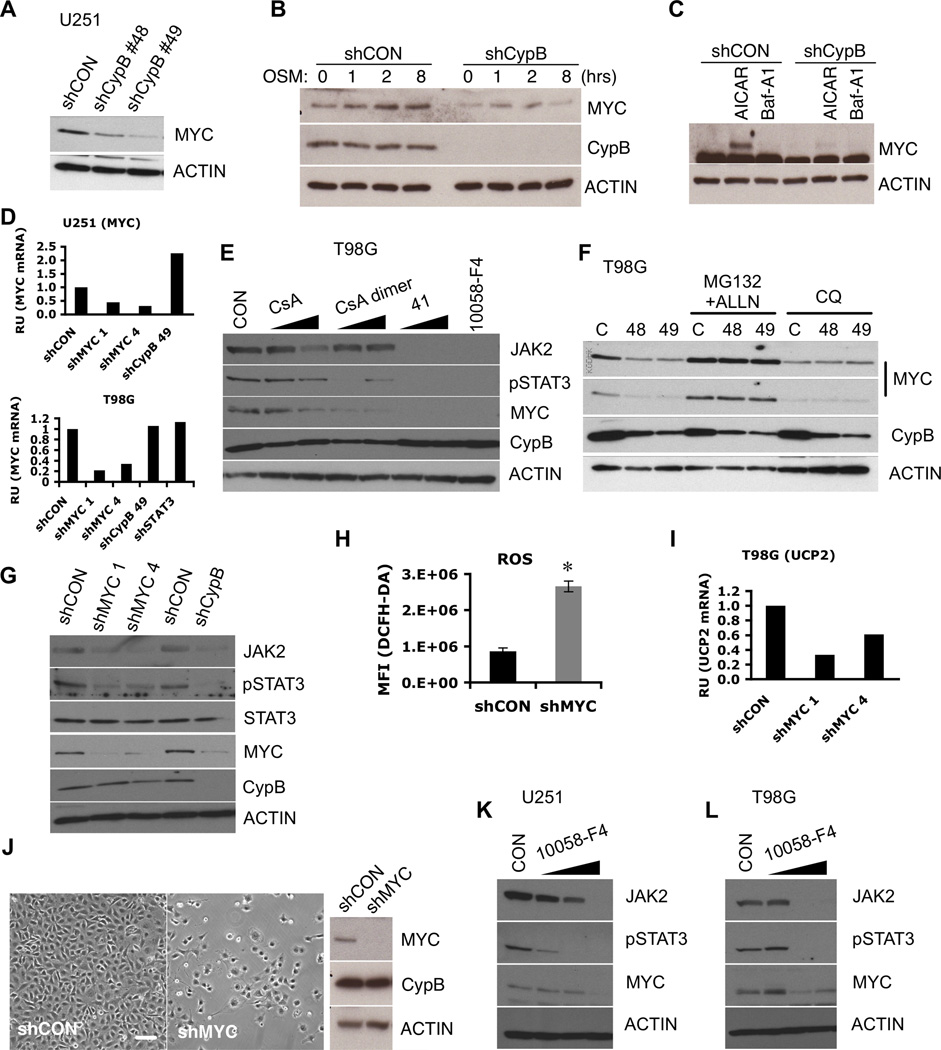
Figure 4. Cyclophilin B controls MYC stability.
(A) Reduced MYC protein in U251 cells transduced with shCypB (48 or 49). (B) MYC induction by OSM was reduced by CypB knockdown in U251 cells (C) Control or CypB-depleted U251 cells were treated with AICAR or bafilomycin A1 (Baf-A1) for 12hr. MYC levels were measured by westernblotting. (D) CypB silencing did not reduce MYC mRNA levels, as measured by real-time PCR. (E) Pharmacological inhibition of CypB reduced the levels of MYC, Jak2, and phospho-Stat3. T98G cells were treated with cyclophilin inhibitors (CsA, CsA-dimer, or 41) for 24h. (F) Treatment with proteasome inhibitors (MG132 and ALLN for 4h) rescued MYC expression in CypB silenced GBM cells; chloroquine had no effect. (G) MYC depletion reduced levels of Jak2 and phospho-Stat3 in U251 cells. (H and I) Increased levels of ROS and decreased UCP2 transcription in MYC depleted T98G cells. ROS levels were monitored by DCF-DA staining (H) and UCP2 mRNA levels were measured by real-time PCR (I). (J) shRNA-mediated MYC depletion reduced U251 cell survival. U251 cells were transduced with shMYC or control lentivirus and observed by phase microscopy. Bar=200µm. (K and L) Pharmacological inhibition of MYC reduced Jak2 expression and Stat3 phosphorylation. U251 (K) or T98G cells (L) were treated with the MYC inhibitor, 10058-F4, for 24h. Errorbars show ±SD. *p<0.05.
Cyclophilin inhibitors also decreased MYC expression (Fig. 4E), which was reversed by the proteasome inhibitors MG132 and ALLN (Fig. 4F). Jak2 and phospho-Stat3 were similarly reduced in MYC-knockdown cells, suggesting that Stat3 activation is partially dependent upon MYC (Fig. 4G). Furthermore, MYC-knockdown cells demonstrated higher levels of ROS and decreased UCP2 mRNA (Fig. 4H,4I), implicating MYC as a mediator of CypB knockdown effects. MYC knockdown also blocked proliferation and survival of GBM cells and induced morphological changes similar to CypB knockdown (Fig. 4J,S3D). The MYC inhibitor 10058-F4 (28) also reduced the survival and proliferation of GBM cells, and reduced Jak2 expression and Stat3 phosphorylation (Fig. 4K,4L). We conclude that CypB regulates MYC expression through enhancing its stability, an effect that is critical for GBM cell viability.
CypB regulates p53 and Chk1 expression
ROS are regulated downstream of several transcription factors including NRF2 and p53 (29). Although NRF2 levels were normal in CypB-depleted GBM cells (data not shown), p53 expression was remarkably reduced (Fig. 5A,5B,5D). Immortalized CypB−/ − fibroblast cells also had decreased p53 (Fig. 5C). CsA, CsA-dimer, and compound 41 also reduced p53 expression in GBM cells (Fig. 5E).
Figure 5. Cyclophilin B regulates p53 and Chk1 expression.
(A–D) Knockdown of CypB reduced mutant p53 and Chk1 levels in U251 cells (A), mutant p53 in T47D breast cancer cells (B), wildtype p53 in CypB deficient fibroblasts (C), and wildtype p53 in glioma stem cells (D). (E) Treatment with cyclophilin inhibitors (CsA, CsA-dimer, and 41) reduced mutant p53 levels in T98G cells. (F) Knockdown of CypB reduced p53 mRNA levels in U251 cells. (G) Knockdown of MYC or Stat3 by siRNA decreased mutant p53 expression in T98G or U251 cells. (H and I) shRNA-mediated MYC (H) or Stat3 (I) silencing reduced mutant p53 expression in U251 cells. (J) CypB silencing in U87 cells ablated (wildtype) p53 induction after treatment with DNA damaging drug, etoposide. (K) Increased ROS in p53-depleted U251 cells. (L) p53 silencing reduced UCP2 mRNA levels in U251 (left) and T98G cells (right). (M) Reduced Chk1 expression in CypB-depleted GBM cells. Cells were transduced with two different shCypB (48 and 49) or shCON lentiviruses. (N) Treatment with cyclophilin inhibitors (CsA, CsA-dimer, or 41) reduced Chk1 expression in U251 or U87 cells. Hsp90 inhibitor, 17-AAG was used as a positive control. (O) MYC silencing decreased Chk1 expression in U251 cells. (P) MYC depletion did not decrease Chk1 mRNA levels. (Q) Reduced cell viability in response to 24-hour treatment with daunorubicin in CypB-depleted U251 cells. Errorbars show ±SD. *p<0.05.
CypB-silencing decreased the amount of p53 transcript by approximately 5-fold (Fig. 5F). Mutant p53 expression in U251 cells was dependent upon endogenous MYC, because knockdown of MYC suppressed p53 expression (Fig. 5G,5H). Mutant p53 was also suppressed by knockdown of Stat3 (Fig. 5G,5I).
In U87 and MCF-7 cells (with wildtype p53), CypB knockdown suppressed induction of p53 and its transcriptional target p21 by etoposide or daunorubicin (Fig. 5J,S3A,S3B). Silencing of endogenous mutant p53 in GBM cells reduced survival, repressed UCP2 expression, and induced ROS (Fig. S3C,5K,5L).
Chk1 contributes to the DNA damage response and is induced by MYC (30, 31). CypB knockdown or inhibition reduced Chk1 expression (Fig. 5A,5M,5N), as did knockdown of MYC itself (Fig. 5O,5P). Consistent with this, CypB-knockdown cells were more sensitive to DNA damage induced by daunorubicin (Fig. 5Q). These data suggest that depletion or inhibition of CypB in GBM cells causes multiple deleterious effects due to loss of MYC, mutant p53, Chk1, and JAK/STAT3 signaling.
CypB loss causes ER stress and an abnormal Unfolded Protein Response (UPR)
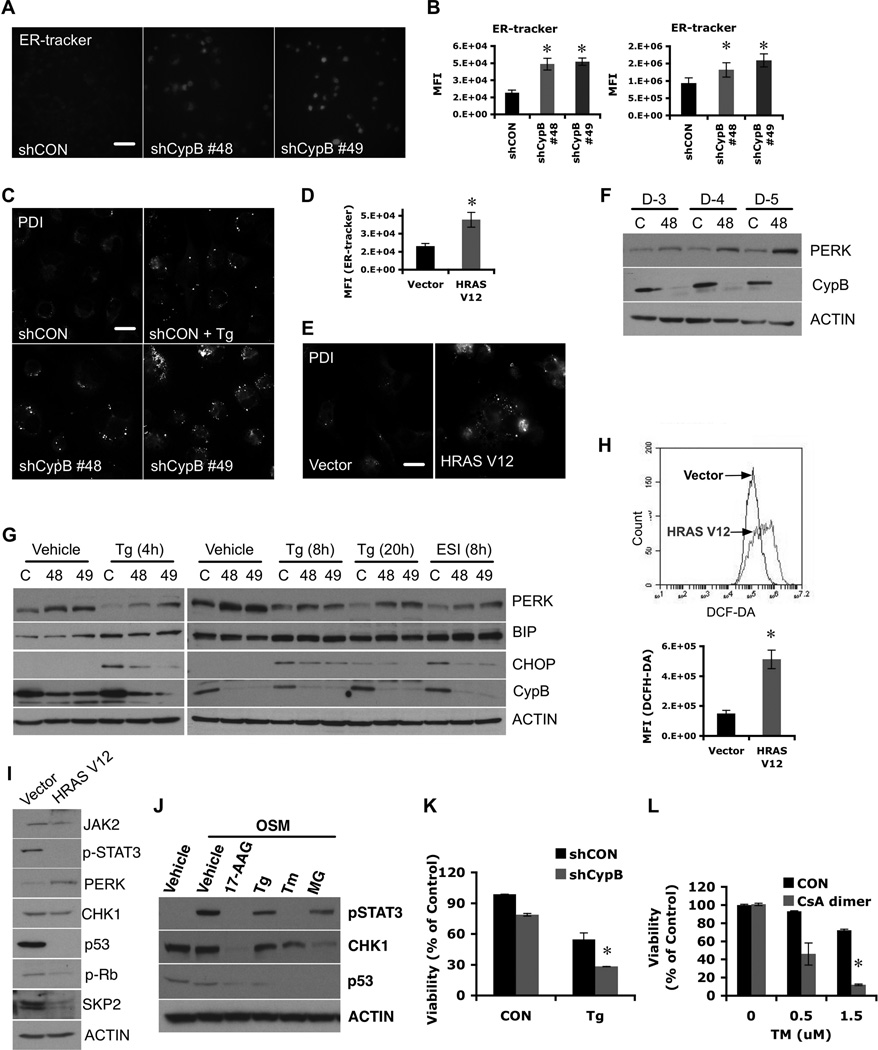
Because CypB chaperones some ER proteins (13), we asked whether CypB-knockdown might impact cells through the UPR (32). ER-tracker® (Molecular Probes, 250nM for 30 min) staining showed that CypB-depleted cells have increased ER content (Fig. 6A,6B). CypB silencing also induced a dot-like protein disulfide isomerase (PDI) staining pattern (Fig. 6C), similar to thapsigargin (Tg) treatment (Fig. 6C) (33). HRASV12 expression in GBM cells also increased ER content (Fig. 6D) and induced similar PDI dots (Fig. 6E). The ER stress sensor PERK (32) was also increased in CypB-depleted cells (Fig. 6F), and in HRASV12-expressing cells (Fig. 6H,6I) and by treatment with tunicamycin (Tm) or Tg (Fig. 6J). Induction of CHOP (but not BiP) by Tg or eeyarestatin was impaired following CypB knockdown (Fig. 6G).
Figure 6. Suppression of Cyclophilin B induces abnormal ER stress.
(A) Increased ER content in CypB-depleted U251 cells. ER content was measured using ER-tracker staining. Bar=40µM. (B) Quantification of ER-tracker signal by flow cytometry in U251 (left) or U87 cells (right). (C) Increased PDI staining in ER stress-induced (thapsigarin treated) or CypB-depleted U251 cells. Bar=40µM. (D and E) Increased staining for ER tracker (D) or PDI (E) in HRASV12-transduced U251 cells. Bar=40µM. (F) Increased expression of PERK in CypB-depleted U251 cells. (G) ER stress-mediated CHOP induction was decreased in CypB-depleted GBM cells. Contol or CypB-depleted cells were treated with thapsigargin (Tg; 4, 8 and 20h) or eeyarestatin Ι (ESI; 8h). Induction of ER chaperones, PERK, BiP, or CHOP, was measured by westernblotting. (H) Increased ROS levels in HRASV12-transduced U251 cells (HRASV12). (I) Activation of oncogenic Ras induced molecular changes similar to CypB silencing, including increased PERK expression and reduced levels of mutant p53, Skp2, and phospho-Stat3. (J) ER stress suppressed Chk1 and mutant p53 expression and Stat3 activation. Cells were pretreated with 17AAG, Tg, tunicamycin (Tm) or MG132 (MG) and stimulated with OSM. (K) Reduced cell viability in response to ER stress in CypB-depleted cells. Control or CypB-depleted U251 cells (D-1) were treated with Tg for 48h. Cell viability was measured by AlamarBlue assay. (L) Combined treatments with CsA-dimer and Tm increased tumor cell killing effects in T98G cells, as determined by AlamarBlue assay. Bar=200µm. Errorbars show ±SD. *p<0.05.
CypB-knockdown cells were more easily killed by Tg (Fig. 6K). Tm also increased susceptibility of GBM cells to CsA-dimer cytotoxicity (Fig. 6L). Calculation of the combination index and isobologram analysis indicated a true synergistic interaction of CsA-dimer with Tg or Tm (Fig. S4).
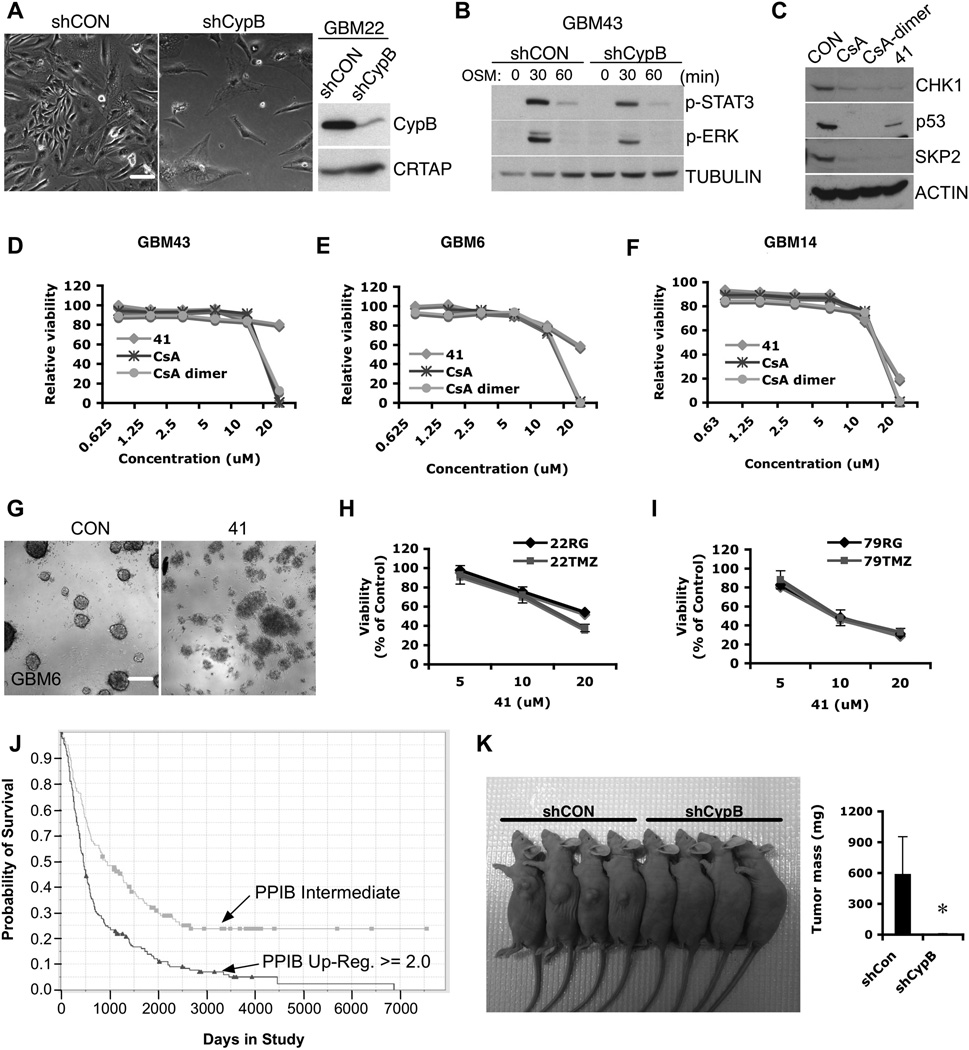
CypB targeting in primary human GBM cells
Knockdown or inhibition of CypB decreased the viability of primary human GBM cells from freshly isolated human GBM xenografts passaged in nude mice (Fig. 7A,7D–7F,7G), and reduced Stat3 and Erk activation in response to OSM (Fig. 7B). Cyclophilin inhibitors reduced Chk1, mutant p53, and Skp2 expression (Fig. 7C). Inhibition of CypB killed both primary (22RG, 79RG) as well as temozolomide (TMZ)-resistant recurrent GBM cells (22TMZ, 79TMZ) (Fig. 7H,7I).
Figure 7. Targeting of Cyclophilin B in primary human GBM cells.
(A) GBM22 cells were transduced with shCypB or control lentivirus. Bar=40µm. (B) Control or CypB knockdown GBM43 cells (D-5) were treated with OSM for the indicated times, harvested and analyzed by westernblotting with the phospho-Stat3 or phospho-Erk antibody. (C) Pharmacological inhibition of CypB reduced levels of Chk1, mutant p53, and Skp2. GBM43 cells were treated cyclophilin inhibitors (CsA, CsA-dimer, or 41) for 24h. (D–F) Treatment with cyclophilin inhibitors decreased the survival of primary human GBM cells. Cells were treated with indicated doses of CsA, CsA-dimer, or 41 for 72h. Cell survival was measured using AlamarBlue assay in GBM43 cells (D), GBM6 cells (E), and GBM14 cells (F). (G) GBM cells were treated with vehicle or compound 41. Neurosphere formation was observed by microscopy. Bar=200µm. (H–I) Treatment with cyclophilin inhibitors decreased the viability and survival of both TMZ sensitive (22RG and 79RG) and resistant (22TMZ and 79TMZ) matched sets of primary human GBM cells. Cells were treated with indicated doses of compound 41 for 72h. (J) Kaplan-Meier curves showing that overall survival of patients with brain tumors having higher PPIB gene expression (>2.0; 167cases) was significantly lower than those with a gene expression of 2.0 or below (176cases). Data were obtained from the Rembrandt database of the National Cancer Institute. (K) shRNA-mediated CypB depletion inhibited in vivo tumor cell growth. U251 cells expressing shRNAs targeting CypB or control shRNAs were implanted into the flanks of nude mice. The resulting tumor sizes were visualized and tumor volumes quantified. Genetic ablation of CypB significantly reduced in vivo tumor formation. Errorbars show ±SD. *p<0.05.
Rembrandt database inspection revealed that overall survival was better in glioma patients with lower CypB expression in their tumors (Fig. 7J), in part due to highest levels in GBMs, which have the worst prognosis. Among astrocytoma cases, higher CypB expression was associated with shorter overall survival (Fig. S6). Also, patients whose tumors had deletions of PPIB gene encoding CypB did better than those with PPIB amplification (Fig. S7, p=0.0492). To determine the effect of CypB inhibition on tumor cell growth in vivo, we implanted control or CypB-depleted U251 cells into nude mice and measured the rates of tumor formation. Ablation of CypB strongly suppressed tumor formation in this xenograft model (Fig. 7K).
Discussion
Gene expression studies revealed that CypB is highly upregulated in malignancies, suggesting a widespread role in folding of ER proteins to reduce ER stress, a known problem for cancers (34, 35). Previous studies found that CypB may support the survival of transformed cells through suppression of ROS or by enhancing the nuclear localization of Stat3. However, the mechanisms underlying these effects were unclear. We report here that ablation of CypB expression in GBM cells suppresses several canonical oncogenic signaling pathways, which cause the dramatic induction of cellular senescence and loss of tumor cell survival (Fig. S8).
A key feature of CypB is its ability to sustain expression of MYC (Fig. 4), which is essential for many cancer cells (26). We found that MYC induces Jak2 expression and subsequently, STAT3 activation (Fig. 4G). MYC knockdown increased ROS in GBM cells, thus explaining several of the CypB-dependent phenotypes (Fig. 4H). ROS generation downstream of CypB loss was likely due to reduced UCP2 (Fig. 2J–2O). Although MYC was not known previously to regulate UCP2, we found that knockdown of MYC decreased UCP2 mRNA by 80% (Fig. 4I).
Chk1 loss downstream of CypB depletion (Fig. 5A, 5M) or inhibition (Fig. 5N) may also be mediated via MYC, since MYC-knockdown similarly extinguished its expression (Fig. 5O). Chk1 has been proposed to be a potential target for therapy of MYC-driven lymphomas (36). GBM cells lacking CypB were significantly more sensitive to death caused by the DNA damage drug daunorubicin (Fig. 5Q).
Most importantly, knockdown of MYC recapped the dramatic killing of U251 cells that we observed following CypB knockdown (Fig. 4J). CypB knockdown affected MYC posttranscriptionally (Fig. 4D), and MYC protein was significantly rescued by proteasome inhibition (Fig. 4F). Clinically useful inhibitors of MYC have been difficult to develop, so our finding that the druggable protein CypB supplies a critical level of support for MYC in GBM cells provides an attractive approach for targeting it therapeutically.
Mutant p53 drives malignancy, as shown here (Fig. S3C) and elsewhere (37), and we found that its expression depends upon CypB. Although wildtype p53 and its target p21 also required CypB for induction after DNA damage, we note that U87 and primary GBM cells that have wildtype p53 were effectively killed by CypB knockdown.
Gene expression pattern changes in CypB-knockdown cells were most consistent with a Ras-induced senescence signature (Fig. 3D) (21), as further demonstrated by activated Ras expression in GBM cells. Although knockdown of CypB caused a transient increase in Ras activation, not all of its effects could be induced by exogenous expression of activated Ras. In particular, destabilization of MYC protein was not seen in Ras-overexpressing cells, and we attribute this to an alternative effect of CypB loss.
CypB knockdown induced dilation of the ER and PDI aggregation, indicators of ER stress and altered redox status (33, 38). Although CypB suppression did not evoke BiP and CHOP induction, it did upregulate PERK. Alteration of UPR sensors in CypB-knockdown cells may underlie the defective UPR response to ER stress, as evidenced by reduced CHOP in response to ER stress. Moreover, CypB knockdown rendered cells more vulnerable to ER stress-related death (Fig. 6K–L). CypB has a crucial role in ER protein quality control through the removal from the ER of ERAD-LS substrates (39), consistent with our finding of a dramatic defect in eeyarestatin-induced CHOP. Though our results suggest that CypB regulates ER stress and UPR signaling, further studies will be required to define the precise molecular mechanism of regulation of the UPR by CypB, which may involve its known interactions with other ER chaperones, including BiP, Grp94, calnexin, and calreticulin (14).
Cellular senescence is emerging as a fundamental mechanism of tumor suppression (40). Here, we showed that senescence is induced by disruption of the stress response machinery by targeting CypB. ER stress chaperones, including CypB, are apparently induced during GBM development, and we suspect that the increased buffering capacity of the ER serves to counterbalance cellular stress in the transformed cells. CypB silencing leads to suppression of several inhibitors of senescence, including MYC and Skp2, as well as the hyperactivation of the oncogenic RAS-MAPK cascade. The fact that CypB silencing drives cellular senescence in the absence of p53 or p16 suggests that additional tumor-suppressive mechanisms may be involved. Could targeting of other ER chaperones induce similar pro-senescent effects? Several secreted proteins, including interleukins and PAI1, can contribute to senescence in different cell types (41). Because CypB silencing induces upregulation of these secreted proteins, evaluation of their impact on the senescence response to inhibition of CypB will be important to study in the future.
An additional impact of CypB silencing in GBM was development of increased ROS and oxidative stress sensitivity. ROS are byproducts of protein oxidation in the ER. Since perturbation of ER stress chaperones induces oxidative stress, persistent ROS elevation, particularly in the context of a compromised ER stress response, may initiate a vicious cycle, leading to ER collapse and cell death (42). In addition to oxidative stress from the increased demand of protein folding in the ER, altered metabolism in rapidly proliferating cancer cells produces abnormally high levels of mitochondrial ROS. Cells typically relieve these effects of ROS by increasing production of cellular antioxidants, including reduced glutathione, thioredoxin, and NADPH. These antioxidant programs are activated by multiple intracellular signaling pathways, including MYC and mutant p53 (29). CypB likely constrains the mitochondrial redox state by controlling the transcription of UCP2, which is at least partially due to the maintained expression of MYC and mutant p53. We conclude that CypB regulates mitochondrial redox homeostasis in GBM cells.
Our proof-of-principle study suggests that pharmacological inhibition of CypB induces several tumor suppressive responses, including cell death and senescence in GBM cells. Although CypB is ubiquitously expressed, because inhibition of CypB lacks serious systemic toxicity (13), it is attractive as a novel molecular target for therapy of GBM. The elucidation of molecular mechanisms downstream of CypB, including modulation of p27, mutant p53, MYC, Chk1, ROS, and Stat3, further suggest combinatorial approaches that will have synergistic effects on tumor cell killing, as we found when testing Stat3 inhibitors with CypB-knockdown cells. However, all cyclophilin inhibitors tested in this study inhibit multiple isoforms of cyclophilin encoded by different genes. It should be considered that other closely related cyclophilins serve different functions in different cellular compartments and, more importantly, have distinct roles in cell proliferation, apoptosis, and necrosis (15, 43). We anticipate that development of compounds that more specifically inhibit CypB will yield better treatment of multiple cancer types, including brain tumors such as GBM.
Supplementary Material
Acknowledgements
We appreciate Abdul Fauq and Robert Chapman for synthesizing CsA-dimer and 41 compound, Lonn Lindquist for mouse experiments, and the Mayo Gene Expression Core for gene expression analysis. We thank all members of Bram Lab for invaluable discussions.
Grant Support
This work was supported by the NIH Grant R01-NS77555, Mayo Brain Tumor SPORE CA108961, and a grant from the Minnesota partnership for Biotechnology and Medical Genomics (to R.J.B.).
Footnotes
Conflicts of Interest: None.
Supplementary Data
Supplemental materials and methods, figures, and tables are included.
References
- 1.Chen J, McKay RM, Parada LF. Malignant glioma: lessons from genomics, mouse models, and stem cells. Cell. 2012;149:36–47. doi: 10.1016/j.cell.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Handschumacher RE, Harding MW, Rice J, Drugge RJ, Speicher DW. Cyclophilin: a specific cytosolic binding protein for cyclosporin A. Science. 1984;226:544–547. doi: 10.1126/science.6238408. [DOI] [PubMed] [Google Scholar]
- 4.Liu J, Albers MW, Chen CM, Schreiber SL, Walsh CT. Cloning, expression, and purification of human cyclophilin in Escherichia coli and assessment of the catalytic role of cysteines by site-directed mutagenesis. Proc Natl Acad Sci U S A. 1990;87:2304–2308. doi: 10.1073/pnas.87.6.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Friedman J, Weissman I. Two cytoplasmic candidates for immunophilin action are revealed by affinity for a new cyclophilin: one in the presence and one in the absence of CsA. Cell. 1991;66:799–806. doi: 10.1016/0092-8674(91)90123-g. [DOI] [PubMed] [Google Scholar]
- 6.Liu J, Farmer JD, Jr, Lane WS, Friedman J, Weissman I, Schreiber SL. Calcineurin is a common target of cyclophilin-cyclosporin A and FKBP-FK506 complexes. Cell. 1991;66:807–815. doi: 10.1016/0092-8674(91)90124-h. [DOI] [PubMed] [Google Scholar]
- 7.Price ER, Zydowsky LD, Jin MJ, Baker CH, McKeon FD, Walsh CT. Human cyclophilin B: a second cyclophilin gene encodes a peptidyl-prolyl isomerase with a signal sequence. Proc Natl Acad Sci U S A. 1991;88:1903–1907. doi: 10.1073/pnas.88.5.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim Y, Jang M, Lim S, Won H, Yoon KS, Park JH, Kim HJ, Kim BH, Park WS, Ha J, Kim SS. Role of cyclophilin B in tumorigenesis and cisplatin resistance in hepatocellular carcinoma in humans. Hepatology. 2011;54:1661–1678. doi: 10.1002/hep.24539. [DOI] [PubMed] [Google Scholar]
- 9.Watashi K, Ishii N, Hijikata M, Inoue D, Murata T, Miyanari Y, Shimotohno K. Cyclophilin B is a functional regulator of hepatitis C virus RNA polymerase. Mol Cell. 2005;19:111–122. doi: 10.1016/j.molcel.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 10.Swanson SK, Born T, Zydowsky LD, Cho H, Chang HY, Walsh CT, Rusnak F. Cyclosporin-mediated inhibition of bovine calcineurin by cyclophilins A and B. Proc Natl Acad Sci U S A. 1992;89:3741–3745. doi: 10.1073/pnas.89.9.3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bukrinsky MI. Cyclophilins: unexpected messengers in intercellular communications. Trends Immunol. 2002;23:323–325. doi: 10.1016/s1471-4906(02)02237-8. [DOI] [PubMed] [Google Scholar]
- 12.Rycyzyn MA, Reilly SC, O'Malley K, Clevenger CV. Role of cyclophilin B in prolactin signal transduction and nuclear retrotranslocation. Mol Endocrinol. 2000;14:1175–1186. doi: 10.1210/mend.14.8.0508. [DOI] [PubMed] [Google Scholar]
- 13.Choi JW, Sutor SL, Lindquist L, Evans GL, Madden BJ, Bergen HR, 3rd, Hefferan TE, Yaszemski MJ, Bram RJ. Severe osteogenesis imperfecta in cyclophilin B-deficient mice. PLoS Genet. 2009;5:e1000750. doi: 10.1371/journal.pgen.1000750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim J, Choi TG, Ding Y, Kim Y, Ha KS, Lee KH, Kang I, Ha J, Kaufman RJ, Lee J, Choe W, Kim SS. Overexpressed cyclophilin B suppresses apoptosis associated with ROS and Ca2+ homeostasis after ER stress. J Cell Sci. 2008;121:3636–3648. doi: 10.1242/jcs.028654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bauer K, Kretzschmar AK, Cvijic H, Blumert C, Loffler D, Brocke-Heidrich K, Schiene-Fischer C, Fischer G, Sinz A, Clevenger CV, Horn F. Cyclophilins contribute to Stat3 signaling and survival of multiple myeloma cells. Oncogene. 2009;28:2784–2795. doi: 10.1038/onc.2009.142. [DOI] [PubMed] [Google Scholar]
- 16.Carlson BL, Pokorny JL, Schroeder MA, Sarkaria JN. Establishment, maintenance and in vitro and in vivo applications of primary human glioblastoma multiforme (GBM) xenograft models for translational biology studies and drug discovery. Curr Protoc Pharmacol. 2011;Chapter 14(Unit 14):16. doi: 10.1002/0471141755.ph1416s52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Belshaw PJ, Ho SN, Crabtree GR, Schreiber SL. Controlling protein association and subcellular localization with a synthetic ligand that induces heterodimerization of proteins. Proc Natl Acad Sci U S A. 1996;93:4604–4607. doi: 10.1073/pnas.93.10.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu YQ, Belyakov S, Choi C, Limburg D, Thomas IB, Vaal M, Wei L, Wilkinson DE, Holmes A, Fuller M, McCormick J, Connolly M, Moeller T, Steiner J, Hamilton GS. Synthesis and biological evaluation of non-peptidic cyclophilin ligands. J Med Chem. 2003;46:1112–1115. doi: 10.1021/jm020409u. [DOI] [PubMed] [Google Scholar]
- 19.Iwamaru A, Szymanski S, Iwado E, Aoki H, Yokoyama T, Fokt I, Hess K, Conrad C, Madden T, Sawaya R, Kondo S, Priebe W, Kondo Y. A novel inhibitor of the STAT3 pathway induces apoptosis in malignant glioma cells both in vitro and in vivo. Oncogene. 2007;26:2435–2444. doi: 10.1038/sj.onc.1210031. [DOI] [PubMed] [Google Scholar]
- 20.Baffy G, Derdak Z, Robson SC. Mitochondrial recoupling: a novel therapeutic strategy for cancer? Br J Cancer. 2011;105:469–474. doi: 10.1038/bjc.2011.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mason DX, Jackson TJ, Lin AW. Molecular signature of oncogenic ras-induced senescence. Oncogene. 2004;23:9238–9246. doi: 10.1038/sj.onc.1208172. [DOI] [PubMed] [Google Scholar]
- 22.Hanafusa H, Torii S, Yasunaga T, Nishida E. Sprouty1 and Sprouty2 provide a control mechanism for the Ras/MAPK signalling pathway. Nat Cell Biol. 2002;4:850–858. doi: 10.1038/ncb867. [DOI] [PubMed] [Google Scholar]
- 23.Britson JS, Barton F, Balko JM, Black EP. Deregulation of DUSP activity in EGFR-mutant lung cancer cell lines contributes to sustained ERK1/2 signaling. Biochem Biophys Res Commun. 2009;390:849–854. doi: 10.1016/j.bbrc.2009.10.061. [DOI] [PubMed] [Google Scholar]
- 24.Sa G, Stacey DW. P27 expression is regulated by separate signaling pathways, downstream of Ras, in each cell cycle phase. Exp Cell Res. 2004;300:427–439. doi: 10.1016/j.yexcr.2004.07.032. [DOI] [PubMed] [Google Scholar]
- 25.Bachireddy P, Bendapudi PK, Felsher DW. Getting at MYC through RAS. Clin Cancer Res. 2005;11:4278–4281. doi: 10.1158/1078-0432.CCR-05-0534. [DOI] [PubMed] [Google Scholar]
- 26.Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adamo L, Zhang Y, Garcia-Cardena G. AICAR activates the pluripotency transcriptional network in embryonic stem cells and induces KLF4 and KLF2 expression in fibroblasts. BMC Pharmacol. 2009;9:2. doi: 10.1186/1471-2210-9-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang MJ, Cheng YC, Liu CR, Lin S, Liu HE. A small-molecule c-Myc inhibitor, 10058-F4, induces cell-cycle arrest, apoptosis, and myeloid differentiation of human acute myeloid leukemia. Exp Hematol. 2006;34:1480–1489. doi: 10.1016/j.exphem.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 29.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 30.Bartek J, Bartkova J, Lukas J. DNA damage signalling guards against activated oncogenes and tumour progression. Oncogene. 2007;26:7773–7779. doi: 10.1038/sj.onc.1210881. [DOI] [PubMed] [Google Scholar]
- 31.Dai Y, Grant S. New insights into checkpoint kinase 1 in the DNA damage response signaling network. Clin Cancer Res. 2010;16:376–383. doi: 10.1158/1078-0432.CCR-09-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 33.Salazar M, Carracedo A, Salanueva IJ, Hernandez-Tiedra S, Lorente M, Egia A, Vazquez P, Blazquez C, Torres S, Garcia S, Nowak J, Fimia GM, Piacentini M, Cecconi F, Pandolfi PP, Gonzalez-Feria L, Iovanna JL, Guzman M, Boya P, Velasco G. Cannabinoid action induces autophagy-mediated cell death through stimulation of ER stress in human glioma cells. J Clin Invest. 2009;119:1359–1372. doi: 10.1172/JCI37948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fang F, Zheng J, Galbaugh TL, Fiorillo AA, Hjort EE, Zeng X, Clevenger CV. Cyclophilin B as a co-regulator of prolactin-induced gene expression and function in breast cancer cells. J Mol Endocrinol. 2010;44:319–329. doi: 10.1677/JME-09-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams PD, Owens CR, Dziegielewski J, Moskaluk CA, Read PW, Larner JM, Story MD, Brock WA, Amundson SA, Lee JK, Theodorescu D. Cyclophilin B expression is associated with in vitro radioresistance and clinical outcome after radiotherapy. Neoplasia. 2011;13:1122–1131. doi: 10.1593/neo.111398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ferrao PT, Bukczynska EP, Johnstone RW, McArthur GA. Efficacy of CHK inhibitors as single agents in MYC-driven lymphoma cells. Oncogene. 2012;31:1661–1672. doi: 10.1038/onc.2011.358. [DOI] [PubMed] [Google Scholar]
- 37.Freed-Pastor WA, Prives C. Mutant p53: one name, many proteins. Genes Dev. 2012;26:1268–1286. doi: 10.1101/gad.190678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Merksamer PI, Trusina A, Papa FR. Real-time redox measurements during endoplasmic reticulum stress reveal interlinked protein folding functions. Cell. 2008;135:933–947. doi: 10.1016/j.cell.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bernasconi R, Solda T, Galli C, Pertel T, Luban J, Molinari M. Cyclosporine A-sensitive, cyclophilin B-dependent endoplasmic reticulum-associated degradation. PLoS One. 2010;5 doi: 10.1371/journal.pone.0013008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nardella C, Clohessy JG, Alimonti A, Pandolfi PP. Pro-senescence therapy for cancer treatment. Nat Rev Cancer. 2011;11:503–511. doi: 10.1038/nrc3057. [DOI] [PubMed] [Google Scholar]
- 41.Kuilman T, Peeper DS. Senescence-messaging secretome: SMS-ing cellular stress. Nat Rev Cancer. 2009;9:81–94. doi: 10.1038/nrc2560. [DOI] [PubMed] [Google Scholar]
- 42.Xu W, Trepel J, Neckers L. Ras, ROS and proteotoxic stress: a delicate balance. Cancer Cell. 2011;20:281–282. doi: 10.1016/j.ccr.2011.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schubert A, Grimm S. Cyclophilin D, a component of the permeability transition-pore, is an apoptosis repressor. Cancer Res. 2004;64:85–93. doi: 10.1158/0008-5472.can-03-0476. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.