Abstract
The supraoptic nucleus (SON) is part of the central osmotic circuitry that synthesises the hormone vasopressin (Avp) and transports it to terminals in the posterior lobe of the pituitary. Following osmotic stress such as dehydration, this tissue undergoes morphological, electrical and transcriptional changes to facilitate the appropriate regulation and release of Avp into the circulation where it conserves water at the level of the kidney. Here, the organisation of the whole transcriptome following dehydration is modelled to fit Zipf's law, a natural power law that holds true for all natural languages, that states if the frequency of word usage is plotted against its rank, then the log linear regression of this is -1. We have applied this model to our previously published euhydrated and dehydrated SON data to observe this trend and how it changes following dehydration. In accordance with other studies, our whole transcriptome data fit well with this model in the euhydrated SON microarrays, but interestingly, fit better in the dehydrated arrays. This trend was observed in a subset of differentially regulated genes and also following network reconstruction using a third-party database that mines public data. We make use of language as a metaphor that helps us philosophise about the role of the whole transcriptome in providing a suitable environment for the delivery of Avp following a survival threat like dehydration.
Keywords: Transcriptome, Supraoptic nucleus, Dehydration, Zipf, Microarray, Vasopressin
Introduction
“Minimum Effort, Maximum Grace” Brian Goodwin 1931-2009
The hypothalamo-neurohypophysial system (HNS) consists of the large peptidergic magnocellular neurons (MCNs) of the hypothalamic supraoptic nucleus (SON) and paraventricular nucleus (PVN), the axons of which course through the internal zone of the median eminence (ME) and terminate on blood capillaries of the posterior lobe of the pituitary gland (PP) (1). The HNS is the source of the major neuropeptide antidiuretic hormone vasopressin (VP) (2), which is synthesised from a prepropeptide precursor that is processed during anterograde axonal transportation to terminals in the PP (3). Here, biologically active VP is stored until mobilised for secretion into the circulation by MCN electrical activities evoked by physiological cues, resulting in the massive hormone release that is a characteristic of dehydration. VP is crucially involved in the maintenance of osmotic stability (4). Following dehydration, a rise in plasma osmolality is detected by intrinsic MCN osmoreceptor mechanisms (5-8) and by specialised osmoreceptive neurons in the circumventricular organs that project to the MCNs (5,6,9,10) that provide direct glutamatergic and GABAergic receptor-mediated inputs (11-13) that shape the firing activity of magnocellular neuroendocrine cells (14,15) for hormone secretion (16,17). Upon release, VP travels through the blood stream to specific receptor targets located in the kidney where it increases the permeability of the collecting ducts to water, reducing the renal excretion of water, thus promoting water conservation. Both dehydration stress and lactation evoke a remodeling of the HNS (18-20). A plethora of activity-dependent changes in the morphology have been documented (19-26). The response of the HNS to dehydration represents a unique and tractable model for understanding the processes whereby changes in gene expression mediate neuronal plasticity (18), but the molecular mechanics of these processes remain to be elucidated.
We have previously interrogated Affymetrix rat 230 2.0 Genome Chips, which consist of 31,000 oligonucleotide probe sets representing 30,000 transcripts encoded by 28,000 genes, with targets derived from the SON of euhydrated male rats and 3-day dehydrated male rats (27). We described the transcriptome of the SON and revealed lists of genes that are considered to be present in this tissue and how these genes responded to the physiological state. The challenge of making the most of such comprehensive data has been met through conventional molecular analysis (28), comparative studies (29,30), and using network reconstruction strategies (Jahans-Price T, Greenwood M, Campbell C, Murphy D, Hindmarch CCT,in preparation). As part of these analyses we hypothesised that the plasticity of this structure was dependent on global patterns in gene expression and we have tried to find ways to explore such patterns.
Zipf's law is a self-scaling natural power law that describes many types of data including word frequency in language (31) and city size (32). The frequency of the words in a text or the size of a city in a country is inversely proportional to its rank as a function of all words or cities. For example, there are very few words that are used with the greatest frequency: e.g., ‘the’, ‘of’ and ‘and’ and a great number of words used infrequently: e.g., ‘skedaddle’ and ‘crapulence’. With respect to city size, a country has only a handful of ‘mega’ cities, and a great number of villages and towns. Zipf's law also holds true for several types of data, including internet search patterns (33) and family names (34). Each of these data approximately scales to the same law when counted regardless of the number of data points plotted. When the frequencies of these data points are plotted against their rank in the frequency table, and a log-log plot is resolved, the slope of this relationship is at, or near -1. Transcriptome data from a variety of microarrays correspond to Zipf's law (35-38) and the distribution across human chromosomes has also been modeled (39).
Here, we take our previously published male (27) and female (29) SON microarray data from both euhydrated and dehydrated states and determine that when we rank each gene according to the magnitude of expression and plot the resulting log-expression against the log rank we see a Zipf-like distribution as previously reported. Importantly, when we compute the slope of the euhydrated data and compare this to the slope of the dehydrated data, we see that under physiological stress the slope is closer to Zipf's exponent of -1 than it is under the euhydrated conditions. Finally, we demonstrate that this Zipf-like distribution is self scaling, and fits to both subsets of these data and to a network assembled using publicly sourced data.
Methods and Results
Animals
All animal study complied with the Home Office Animals (Scientific Procedures) Act 1986. Full protocols for tissue extraction and experimental design and microarray hybridisation have been previously published (27,29). Briefly, following 72 h of total water deprivation, rats were sacrificed (between 11:00 and 13:00) and the tissue was isolated and processed as described below. Euhydrated controls were handled in the same manner. Rats were stunned and decapitated with a small animal guillotine (Harvard Apparatus, USA), the brain was removed and sections of ≅1 mm in thickness taken (using the optic chiasm as a landmark). The SON was carefully dissected with the visual aid of a dissecting microscope. Each Affymetrix 230 2.0 microarray represented 5 individual animals and 5 microarrays were used for each condition except for the female euhydrated rat, where n=4.
Data analysis
Raw data files from previously described microarray experiments (27,29) were imported into spreadsheets and filtered according to Present flags in all microarray chips in both the euhydrated and dehydrated groups. Male and female data were handled separately for lists of genes considered Present, and the arbitrary cutoff for differential regulation was 1.5-fold (Welch ANOVA, P<0.05, Benjamini and Hochberg multiple testing correction). The median expression value was then calculated for all samples and used to set the experiment rank that would help normalise the experiment. For each microarray and for the mean expression per group, the log expression (either Present or 1.5-fold regulated) was then plotted against the log median rank so that the slope, intercept, linear regression (least squares), and Pearson's correlation coefficients could be calculated per array (Table 1) and for the mean expression (Figure 1, all Present genes; Figure 2, 1.5-fold regulated genes). Finally, the top 200 upregulated and top 200 downregulated genes in the male dataset were filtered and the gene symbols extracted and presented to the free Internet-based program GeneMANIA (www.genemania.org). Of these 400 genes, GeneMANIA identified 19 unrecognised gene symbols and 16 duplicates, leaving 365 targets with which the algorithm could search interactions to generate a network (Figure 3A). The interaction table (Supplementary Table S1) lists pairwise interactions (with corroboratory information about the data from which the interaction was mined). We combined these two columns to identify the frequency with which each networked gene appeared as part of a rule and ranked those frequencies in order to identify whether Zipf's law held true. Figure 3B shows the network frequency regression against rank, which has an exponent close to -1.
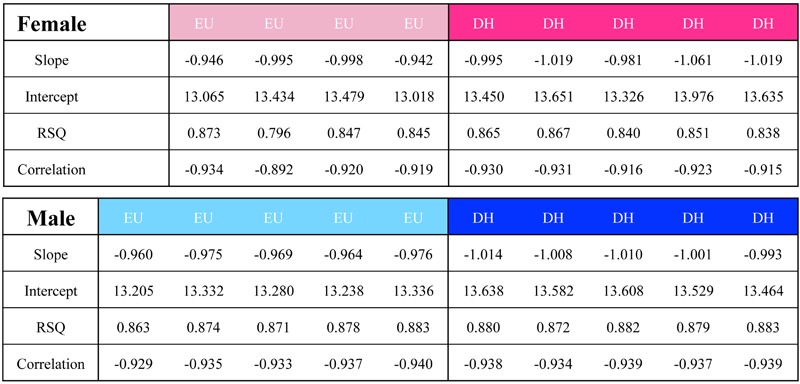
Figure 1. Raw microarray data from the supraoptic nucleus, considered Present in either the euhydrated (EU) or dehydrated (DH) state, were used. Male and female data were handled separately. The median expression value calculated for all samples set the experiment log rank against which the log expression per condition was then plotted. Slope, intercept and linear regression (least squares) were then calculated. The slope is closer to Zipf's exponent of -1 following DH compared to the EU arrays.
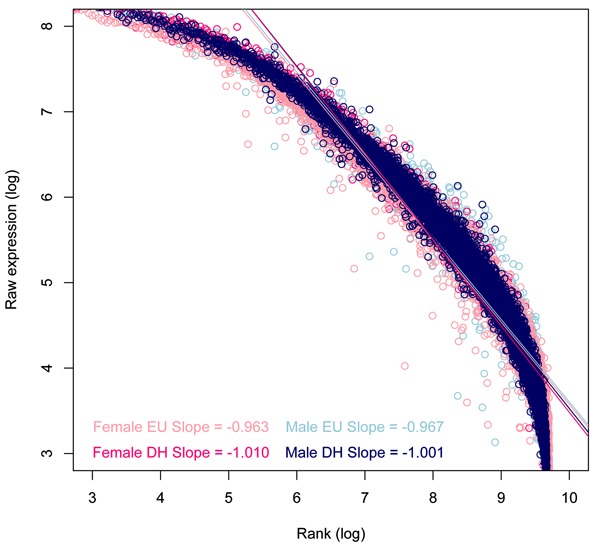
Figure 2. The list of probes used to construct the log rank-log expression was attenuated to only include those probes that were significantly regulated by dehydration. The slope is closer to Zipf's exponent of -1 following dehydration (DH) compared to the euhydrated (EU) arrays.
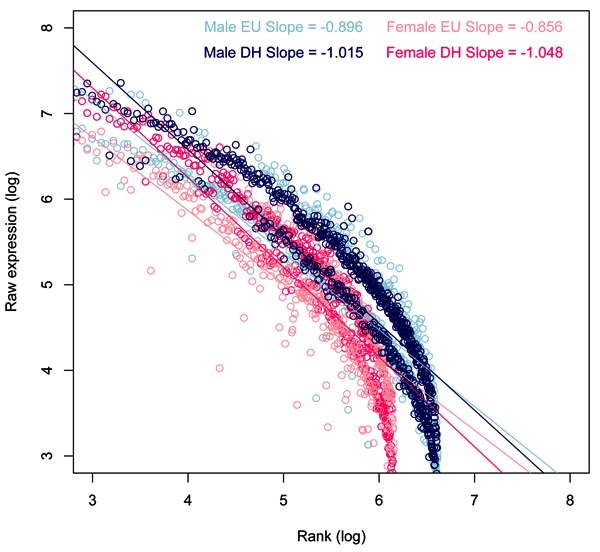
Figure 3. A, The top 200 upregulated and top 200 downregulated genes in the male dataset were filtered and the gene symbols extracted and presented to the free internet-based program GeneMANIA (www.genemania.org). B, When the frequency of gene appearance was ranked and used to construct the log rank-log expression graph, the exponent was close to -1.
Discussion
The SON is a structure whose central role in the osmotic feedback loop is well established (4-6,10,16). Circumventricular organs capable of detecting changes in plasma osmolality and volume communicate threats to water balance to the MCN of the PVN and SON. In turn, these nuclei respond with an increased production, transport and release of VP from the posterior lobe of the pituitary with the aim to conserve water at the level of the kidney. We have shown previously (27) that the transcriptome of both these nuclei is responsive to dehydration and hypothesised that whole transcriptome distribution might differ between these two physiological states and the response to a survival threat in this tissue is a global effort rather than the contribution of a handful of genes that directly relate to VP synthesis; e.g., the entire transcriptome favors a molecular environment within which the synthesis and release of vasopressin is appropriate.
We were concerned that the changes in slope represented some technical aspect of the experiments and feel this concern is worth discussion in this paper. Because the Affymetrix microarrays display remarkably low chip-chip variation as a consequence of their manufacturing and were hybridised at the same time, we would expect such variation to be random rather than specific to one group or other. In the absence of any global normalisation, the rank axis was calculated according to the median of each gene using the signal for all samples (euhydrated and dehydrated) ensuring that this axis was compatible; group-to-group variability was not responsible for the different slopes shown in our experiment. The second possibility for the change in slope may have arisen as a consequence of the hypertrophy of magnocellular cells and subsequent increase in transcription or the retraction of glial processes that characterises this nucleus following dehydration (19,20). While we cannot adequately control for this variable, it is definitive of the dehydrated state that is under scrutiny here.
Many efforts have been made by mathematicians to understand the mechanisms by which power laws hold true for so many kinds of uncorrelated data (34,37,40). The remainder of this manuscript will, however, discuss the data in philosophical rather than mathematical terms. We would like to refer to Zipf's original interpretation that dealt with language and explore whether language is a useful metaphor for the exploration or discussion of transcriptomic networks. We are critically aware that this metaphor is imperfect, not least because the relationship between word frequency and rank is completely unrelated to gene expression and rank. Nevertheless, we are interested in using this common relationship as a vehicle for the better understanding of whole transcriptome expression and how this distribution changes following a survival threat like dehydration.
To think of the transcriptome as a language should not stretch the imagination too far; the genome codes for a rich biological story that belies the relatively small number of gene words available for use. Words in a text are not in isolation; however, they are placed into context by the other words in a sentence, paragraph, chapter or book; the coordinated and appropriate choice of words gives greater meaning than the sum provided by the individual units. Like biological systems, natural languages are evolved and self-organising rather than designed, and therefore they have the capacity to resolve changes and new additions (often from other languages or from made up terms like brand names) without altering the underlying structure of the language (41). A network of words will resolve itself according to the relative emphasis that is put on the costs. If there is little importance in how a feature expresses itself, the information cost will be minimised and there will be a highly variable anarchic system in which the behaviour of elements pays no attention to other elements. If it is crucial how order is interpreted for other parts of the system, then the signification cost will be minimised and there will be a highly deterministic system in which each element has a fixed meaning. This is machine language that requires precision but allows no creative sensitivity to context. Zipf's law shows how language evolved as the result of these two pressures (42).
While no conversation exists in the data we present here, we do see that the same phenomenon that exists for natural languages also exists in the transcriptome (Table 1). Our examination of microarray data from the rat SON fits with the evidence from other authors (35-38) that transcriptome data exhibit a power-law distribution with an exponent close to -1 (i.e., obeys Zipf's law). We have also been interested to notice that the slope for the dehydrated microarrays is consistently closer to Zipf's ideal -1 slope than it is in the euhydrated microarrays, a pattern noticed in both male and female rats in data representing the entire catalogue of Present genes in the SON (Figure 1) and a sub-catalogue of regulated genes that were plotted as a function of expression against log (Figure 2).
Finally, we wanted to visualise the Zipfian power law in a manner independent of the numeric values of our data. We extracted the Gene symbols from the top 200 ‘UP’ and bottom 200 ‘DOWN’ regulated genes and presented them to GeneMANIA (www.genemania.org), a free internet software that looks for positive associations (using Pearson's correlation coefficients) of input gene lists within microarray datasets that have been presented to a public database such as the Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) to resolve a network of genes that putatively interact. When we ranked the frequency of these interactions and plotted the log-frequency against log-rank, we were satisfied to see a correlation that fitted to a Zipf distribution.
In the same way that the lexicon of language allows a coherent message to be resolved based on the appropriate choice of word assignments, the genomic lexicon allows a coherent biological message or outcome to be delivered based on the choice of gene assignments that are appropriate to the situation. For example, under non-stressed social circumstances, the conversationalist can adopt a more loose and relaxed vocabulary than would be chosen in a debate where the room for ambiguity would be low and the requirement for organised argument would be necessary. Following dehydration, the status of the animal represents one that has a tightly regulated and prioritised behavioural and hormonal agenda that is under transcriptomic control; the message is by vital necessity unambiguous and organised. The transcriptional environment within the SON is necessarily optimal for creative responses to a broad and overlapping range of environments and disturbances acting on a temporal scale; e.g., the capacity to mount a slightly different but nonetheless appropriate response to physiological stimulation by dehydration or salt-loading and to discriminate an appropriate response of either stimulus to 6- or 72-h dehydration.
We have applied the metaphor of language to biological organisation in an effort to help us to better understand how informational resources are applied dynamically to meet physiological circumstance with significant adaptive response. We have enjoyed modelling our ideas about how the whole transcriptome is capable of responding to a stimulus like dehydration in a modulated and plastic manner against that of how natural languages behave and are now intrigued as to how the organisation of the transcriptome might respond or change under pathological conditions or following gene manipulation. Following dehydration, we can conclude that an optimal and unambiguous message is delivered by the whole SON transcriptome rather than just vasopressin and genes directly linked to vasopressin.
Supplementary Material
Acknowledgments
Brian Goodwin, Professor of Biology at the Schumacher College, sadly passed away in July 2009. Brian was instrumental in forging the collaboration that led to this manuscript. He enjoyed a long and successful career in the field of evolutionary biology and was a pioneer of complexity science and theoretical biology. This paper is dedicated to Brian. The Intuitive Way of Knowing: a Tribute to Brian Goodwin (David Lambert, Chris Chetland and Craig Millar, Editors) was published by Floris Books in July 2013 (ISBN: 978-0863159657).
Footnotes
First published online December 2, 2013.
Presented at the Symposium on “Recent Advances in the Study of the Integrative Physiology with Emphasis on the Neuroendocrine Control of Energy Metabolism and Body Fluid Homeostasis”, Ribeirão Preto, SP, Brazil, August 29-31, 2012.
References
- 1.Bargmann W. Neurosecretion. Int Rev Cytol. 1966;19:183–201. doi: 10.1016/S0074-7696(08)60567-7. [DOI] [PubMed] [Google Scholar]
- 2.Burbach JP, Luckman SM, Murphy D, Gainer H. Gene regulation in the magnocellular hypothalamo-neurohypophysial system. Physiol Rev. 2001;81:1197–1267. doi: 10.1152/physrev.2001.81.3.1197. [DOI] [PubMed] [Google Scholar]
- 3.Brownstein MJ, Russell JT, Gainer H. Synthesis, transport, and release of posterior pituitary hormones. Science. 1980;207:373–378. doi: 10.1126/science.6153132. [DOI] [PubMed] [Google Scholar]
- 4.Antunes-Rodrigues J, de Castro M, Elias LL, Valenca MM, McCann SM. Neuroendocrine control of body fluid metabolism. Physiol Rev. 2004;84:169–208. doi: 10.1152/physrev.00017.2003. [DOI] [PubMed] [Google Scholar]
- 5.Bourque CW. Osmoregulation of vasopressin neurons: a synergy of intrinsic and synaptic processes. Prog Brain Res. 1998;119:59–76. doi: 10.1016/S0079-6123(08)61562-9. [DOI] [PubMed] [Google Scholar]
- 6.Bourque CW, Oliet SH, Richard D. Osmoreceptors, osmoreception, and osmoregulation. Front Neuroendocrinol. 1994;15:231–274. doi: 10.1006/frne.1994.1010. [DOI] [PubMed] [Google Scholar]
- 7.Bourque CW, Voisin DL, Chakfe Y. Stretch-inactivated cation channels: cellular targets for modulation of osmosensitivity in supraoptic neurons. Prog Brain Res. 2002;139:85–94. doi: 10.1016/S0079-6123(02)39009-5. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Z, Bourque CW. Osmometry in osmosensory neurons. Nat Neurosci. 2003;6:1021–1022. doi: 10.1038/nn1124. [DOI] [PubMed] [Google Scholar]
- 9.McKinley MJ, Gerstberger R, Mathai ML, Oldfield BJ, Schmid H. The lamina terminalis and its role in fluid and electrolyte homeostasis. J Clin Neurosci. 1999;6:289–301. doi: 10.1016/S0967-5868(99)90050-4. [DOI] [PubMed] [Google Scholar]
- 10.McKinley MJ, Mathai ML, McAllen RM, McClear RC, Miselis RR, Pennington GL, et al. Vasopressin secretion: osmotic and hormonal regulation by the lamina terminalis. J Neuroendocrinol. 2004;16:340–347. doi: 10.1111/j.0953-8194.2004.01184.x. [DOI] [PubMed] [Google Scholar]
- 11.Nissen R, Hu B, Renaud LP. N-methyl-D-aspartate receptor antagonist ketamine selectively attenuates spontaneous phasic activity of supraoptic vasopressin neurons in vivo . Neuroscience. 1994;59:115–120. doi: 10.1016/0306-4522(94)90103-1. [DOI] [PubMed] [Google Scholar]
- 12.Gies U, Theodosis DT. Synaptic plasticity in the rat supraoptic nucleus during lactation involves GABA innervation and oxytocin neurons: a quantitative immunocytochemical analysis. J Neurosci. 1994;14:2861–2869. doi: 10.1523/JNEUROSCI.14-05-02861.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Armstrong WE. Morphological and electrophysiological classification of hypothalamic supraoptic neurons. Prog Neurobiol. 1995;47:291–339. [PubMed] [Google Scholar]
- 14.Hu B, Bourque CW. NMDA receptor-mediated rhythmic bursting activity in rat supraoptic nucleus neurones in vitro . J Physiol. 1992;458:667–687. doi: 10.1113/jphysiol.1992.sp019440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Onaka T, Yagi K. Involvement of N-methyl-D-aspartic acid receptor activation in oxytocin and vasopressin release after osmotic stimuli in rats. J Neuroendocrinol. 2001;13:166–174. doi: 10.1046/j.1365-2826.2001.00607.x. [DOI] [PubMed] [Google Scholar]
- 16.Dyball RE, McKenzie DN, Thomas GP. Osmoresponsiveness of the rat supraoptic nucleus in vivo depends on glutamatergic inputs. Neurobiology. 1995;3:351–362. [PubMed] [Google Scholar]
- 17.Windle RJ, Forsling ML, Smith CP, Balment RJ. Patterns of neurohypophysial hormone release during dehydration in the rat. J Endocrinol. 1993;137:311–319. doi: 10.1677/joe.0.1370311. [DOI] [PubMed] [Google Scholar]
- 18.Sharman G, Ghorbel M, Leroux M, Beaucourt S, Wong LF, Murphy D. Deciphering the mechanisms of homeostatic plasticity in the hypothalamo-neurohypophyseal system - genomic and gene transfer strategies. Prog Biophys Mol Biol. 2004;84:151–182. doi: 10.1016/j.pbiomolbio.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 19.Hatton GI. Function-related plasticity in hypothalamus. Annu Rev Neurosci. 1997;20:375–397. doi: 10.1146/annurev.neuro.20.1.375. [DOI] [PubMed] [Google Scholar]
- 20.Theodosis DT, El Majdoubi M, Pierre K, Poulain DA. Factors governing activity-dependent structural plasticity of the hypothalamoneurohypophysial system. Cell Mol Neurobiol. 1998;18:285–298. doi: 10.1023/A:1022577105819. [DOI] [PubMed] [Google Scholar]
- 21.Catheline G, Touquet B, Lombard MC, Poulain DA, Theodosis DT. A study of the role of neuro-glial remodeling in the oxytocin system at lactation. Neuroscience. 2006;137:309–316. doi: 10.1016/j.neuroscience.2005.08.042. [DOI] [PubMed] [Google Scholar]
- 22.Miyata S, Hatton GI. Activity-related, dynamic neuron-glial interactions in the hypothalamo-neurohypophysial system. Microsc Res Tech. 2002;56:143–157. doi: 10.1002/jemt.10012. [DOI] [PubMed] [Google Scholar]
- 23.Miyata S, Takamatsu H, Maekawa S, Matsumoto N, Watanabe K, Kiyohara T, et al. Plasticity of neurohypophysial terminals with increased hormonal release during dehydration: ultrastructural and biochemical analyses. J Comp Neurol. 2001;434:413–427. doi: 10.1002/cne.1184. [DOI] [PubMed] [Google Scholar]
- 24.Tanaka M, Cummins TR, Ishikawa K, Black JA, Ibata Y, Waxman SG. Molecular and functional remodeling of electrogenic membrane of hypothalamic neurons in response to changes in their input. Proc Natl Acad Sci U S A. 1999;96:1088–1093. doi: 10.1073/pnas.96.3.1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tasker JG, Di S, Boudaba C. Functional synaptic plasticity in hypothalamic magnocellular neurons. Prog Brain Res. 2002;139:113–119. doi: 10.1016/S0079-6123(02)39011-3. [DOI] [PubMed] [Google Scholar]
- 26.Yang QZ, Hatton GI. Nitric oxide via cGMP-dependent mechanisms increases dye coupling and excitability of rat supraoptic nucleus neurons. J Neurosci. 1999;19:4270–4279. doi: 10.1523/JNEUROSCI.19-11-04270.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hindmarch C, Yao S, Beighton G, Paton J, Murphy D. A comprehensive description of the transcriptome of the hypothalamoneurohypophyseal system in euhydrated and dehydrated rats. Proc Natl Acad Sci U S A. 2006;103:1609–1614. doi: 10.1073/pnas.0507450103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qiu J, Yao S, Hindmarch C, Antunes V, Paton J, Murphy D. Transcription factor expression in the hypothalamo-neurohypophyseal system of the dehydrated rat: upregulation of gonadotrophin inducible transcription factor 1 mRNA is mediated by cAMP-dependent protein kinase A. J Neurosci. 2007;27:2196–2203. doi: 10.1523/JNEUROSCI.5420-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qiu J, Hindmarch CC, Yao ST, Tasker JG, Murphy D. Transcriptomic analysis of the osmotic and reproductive remodeling of the female rat supraoptic nucleus. Endocrinology. 2011;152:3483–3491. doi: 10.1210/en.2011-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stewart L, Hindmarch CC, Qiu J, Tung YC, Yeo GS, Murphy D. Hypothalamic transcriptome plasticity in two rodent species reveals divergent differential gene expression but conserved pathways. J Neuroendocrinol. 2011;23:177–185. doi: 10.1111/j.1365-2826.2010.02093.x. [DOI] [PubMed] [Google Scholar]
- 31.Zipf GK. Selected studies of the principle of relative frequency in language. Cambridge: Harvard University Press; 1932. [Google Scholar]
- 32.Zipf GK. Human behavior and the principle of least effort. Cambridge: Addison-Wesley; 1939. [Google Scholar]
- 33.Adamic LA, Huberman BA. Zipf's law and the Internet. Glottometrics. 2002;3:143–150. [Google Scholar]
- 34.Baek SK, Bernhardsson S, Minnhagen P. Zipf's law unzipped. New J Physics. 2011:13. [Google Scholar]
- 35.Furusawa C, Kaneko K. Zipf's law in gene expression. Phys Rev Lett. 2003;90: doi: 10.1103/PhysRevLett.90.088102. [DOI] [PubMed] [Google Scholar]
- 36.Hoyle DC, Rattray M, Jupp R, Brass A. Making sense of microarray data distributions. Bioinformatics. 2002;18:576–584. doi: 10.1093/bioinformatics/18.4.576. [DOI] [PubMed] [Google Scholar]
- 37.Ogasawara O, Kawamoto S, Okubo K. Zipf's law and human transcriptomes: an explanation with an evolutionary model. C R Biol. 2003;326:1097–1101. doi: 10.1016/j.crvi.2003.09.031. [DOI] [PubMed] [Google Scholar]
- 38.Ueda HR, Hayashi S, Matsuyama S, Yomo T, Hashimoto S, Kay SA, et al. Universality and flexibility in gene expression from bacteria to human. Proc Natl Acad Sci U S A. 2004;101:3765–3769. doi: 10.1073/pnas.0306244101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nebel J, Pezzulli S. Distribution of human genes observes Zipf's law . KURIR: Kinston Univ Res Innovation Rep. 2012;8:1–9. [Google Scholar]
- 40.Corominas-Murtra B, Sole RV. Universality of Zipf's law. Phys Rev E Stat Nonlin Soft Matter Phys. 2010;82: doi: 10.1103/PhysRevE.82.011102. [DOI] [PubMed] [Google Scholar]
- 41.Harremoes P, Topsoe F. Zipf's law, hyperbolic distributions and entropy loss. Isit: 2002 Ieee International Symposium on Information Theory, Proceedings. 2002:207–207. doi: 10.1109/ISIT.2002.1023479. [DOI] [Google Scholar]
- 42.Cancho RFI, Sole RV. Least effort and the origins of scaling in human language. Proceedings of the National Academy of Sciences of the United States of America. 2003:788–791. doi: 10.1073/pnas.0335980100. [DOI] [PMC free article] [PubMed] [Google Scholar]


