Summary
Strains of Pseudomonas aeruginosa (PA) isolated from the airways of cystic fibrosis patients constitutively add palmitate to lipid A, the membrane anchor of lipopolysaccharide. The PhoPQ regulated enzyme PagP is responsible for the transfer of palmitate from outer membrane phospholipids to lipid A. This enzyme had previously been identified in many pathogenic Gram-negative bacteria, but in PA had remained elusive, despite abundant evidence that its lipid A contains palmitate. Using a combined genetic and biochemical approach, we identified PA1343 as the PA gene encoding PagP. Although PA1343 lacks obvious primary structural similarity with known PagP enzymes, the β-barrel tertiary structure with an interior hydrocarbon ruler appears to be conserved. PA PagP transfers palmitate to the 3’ position of lipid A, in contrast to the 2 position seen with the enterobacterial PagP. Palmitoylated PA lipid A alters host innate immune responses, including increased resistance to some antimicrobial peptides and an elevated pro-inflammatory response, consistent with the synthesis of a hexa-acylated structure preferentially recognized by the TLR4/MD2 complex. Palmitoylation commonly confers resistance to cationic antimicrobial peptides, however, increased cytokine production resulting in inflammation is not seen with other palmitoylated lipid A, indicating a unique role for this modification in PA pathogenesis.
Keywords: Lipopolysaccharide, lipid A, Pseudomonas aeruginosa, PagP, innate immunity, palmitate
Introduction
Gram-negative bacteria have double membranes; an inner membrane composed of a phospholipid bilayer partitioning the cytoplasm from the periplasm, and a complex outer membrane with an inner leaflet composed of phospholipids and an outer leaflet dominated by lipopolysaccharide (LPS). This structuring of membranes allows the bacteria to rely on the outer membrane to defend against environmental stressors and host derived factors through alterations in the composition of porin/proteins, lipids, and LPS (Nikaido & Vaara, 1987, Raetz et al., 2007, Needham & Trent, 2013).
LPS can be divided into three major constituents: O-antigen projecting from the cell surface, a conserved core oligosaccharide, and lipid A that anchors LPS to the bacterial outer membrane. O-antigen is a repeating sugar unit of variable length and among the first bacterial cellular components to come into contact with the host upon infection; as such, it is critical for evading host defenses, including phagocytosis and activated complement (Dasgupta et al., 1994). The core oligosaccharide is composed of non-repeating sugar moieties linking O-antigen to lipid A. Core modifications contribute to the release of bacterial toxins (Horstman et al., 2004), as well as porin incorporation within the membrane (Diedrich et al., 1990). The hydrophobic portion of LPS, lipid A, plays a distinct role in innate immune system signaling and modulation (Fujimoto et al., 2013, Curtis et al., 2011, Galanos et al., 1985) as well as influencing outer membrane fluidity and permeability (Raetz et al., 2007).
Lipid A remains a focus for study due in large part to its endotoxic properties leading to endotoxemia, localized tissue destruction, and septic shock. While the general structure of lipid A is conserved between Gram-negative bacteria, variations in the acyl chain number (4 to 7) and carbon length (10 to 18) are observed between species (Nikaido & Vaara, 1987, Caroff et al., 2002); such variation contributes to the diversity in virulence seen within bacterial populations (Wilkinson, 1996, Needham & Trent, 2013). The ability to control lipid A variation in response to environmental cues further enhances pathogenesis.
Many Gram-negative bacteria modify their lipid A through palmitoylation (addition of a saturated C16 fatty acid), which has the effect of altering host responses. Palmitoylation results in increased resistance to cationic antimicrobial peptides (CAMPs) (Trent, 2004, Trent et al., 2006, Ernst et al., 2001) and alterations in the ability to stimulate innate immunity through toll-like receptor (TLR) 4 mediated reponses (Alexander & Rietschel, 2001, Kawasaki et al., 2004, Kong et al., 2011).
The enzyme, PagP, was first shown to be responsible for palmitoylation of lipid A in enteric bacteria. The pagP gene was identified in Salmonella (Guo et al., 1998) and its homologue in E. coli was later purified from the outer membrane and shown to be a phospholipid::lipid A palmitoyltransferase enzyme (Bishop et al., 2000, Bishop, 2005) Homologues of PagP are found in Bordetellae (Preston et al., 2004), Yersinia (Rebeil et al., 2004), and Legionella (Robey et al., 2001) and have been implicated in the pathogenesis of successful infection. Unlike most lipid A biosynthetic enzymes that require access to cytosolic substrates, PagP is located in the outer membrane where it transfers palmitate from the sn-1 position of a phospholipid to the 2 position of lipid A (Bishop et al., 2000).
Cystic fibrosis (CF) is the most common lethal autosomal recessive disease of Caucasians. In this disorder, chronic lung infections with Pseudomonas aeruginosa (PA) are strongly associated with disease progression and premature mortality (Davis et al., 1996, Folkesson et al., 2012). Our previous studies have demonstrated that one of the earliest known PA adaptations to the CF lung is the synthesis of a characteristic lipid A structure distinct from lipid A isolated from acute infection (blood, ear, eye and urinary tract), bronchiectasis (non-CF lung infections), or environmental isolates (Ernst et al., 1999). The CF specific lipid A structure differs from other lipid A species in the constitutive addition of palmitate (Miller et al., 2011, Moskowitz et al., 2004, Ernst et al., 1999, Ernst et al., 2007).
The constitutive expression of palmitoylated lipid A by PA isolated from CF patients with chronic lung infections indicates that this modification is critical to adaptation and survival within the CF lung (Moskowitz & Ernst, 2010). In this disorder, chronic lung infections with PA are strongly associated with disease progression and premature mortality (Davis et al., 1996, Folkesson et al., 2012). Although the enzyme responsible for this CF specific modification has remained elusive, its characterization will be essential in understanding PA early adaptation to the CF airway. Here, we report the identification of PA1343 (www.pseudomonas.com) as the gene encoding the palmitoyltransferase enzyme, PagP. We hypothesize that PagP dependent palmitoylation of lipid A mediates important adaptations of PA to host microenvironments, including increased resistance to antimicrobial peptides and alterations of host innate immune responses.
Results
Screening for the PA lipid A palmitoyltransferase gene, pagP
To identify candidate genes with palmitoyltransferase activity, we utilized a variety of in silico analysis strategies including fold and function assignment system (FFAS) profiling, multiple sequence alignments, and transmembrane prediction. Initially, we used FFAS03 (Jaroszewski et al., 2005) to identify genes of interest. For further analysis, profiles were created for all PA genes encoding ORFs shorter than 300 amino acids, as PagP in other bacteria were approximately 160–180 amino acids in length. A profile for the 1THQ structure of E. coli PagP crystallized from LDAO was searched against each PA protein (e.g. using BLOSUM substitution matrixes), FFAS profiles were then constructed from 1THQ and this profile was searched against the PA profiles, generating 23 potential hits. Based on this information, we further identified the putative transmembrane spanning regions of these proteins. Utilizing the multiple sequence alignments algorithm (DAS-http://www.sbc.su.se/~miklos/DAS/), a unique pagP profile having the S. Typhimurium, and E. coli subspecies enzymes as representative examples, produced a pattern with two increases in DAS profile score at either end of the sequence and three smaller increases in the middle (Supplementary Fig. S1) Curves are obtained by pairwise comparison of the proteins in the test set in “each against the rest” fashion. Two cutoffs are indicated on the plots, a strict one at 2.2 DAS score, and a loose one at 1.7. The horizontal solid line (strict cutoff) and dashed line (loose cutoff) indicates the number of matching segments and the actual location of the transmembrane segment respectively. Using this information, twenty-three transposon mutants from the University of Washington two-allele transposon mutant library, (www.genome.washington.edu/UWGC/pseudomonas/index.cfm) were obtained and screened for loss of palmitoyltransferase activity.
Matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) lipid A analysis reveals loss of palmitoyltransferase activity in strains with mutations in PA1343
Individual transposon insertion mutants were grown under PhoPQ-activating conditions shown (low magnesium - 8 µM) and screened by MALDI-TOF mass spectrometry (MS) for lipid A lacking palmitate, as compared to the wild-type (WT) strain, PAO-1. A single mutant that contained a transposon insertion in the gene PA1343 lacked the characteristic ion peak seen upon PagP induction by magnesium limitation.
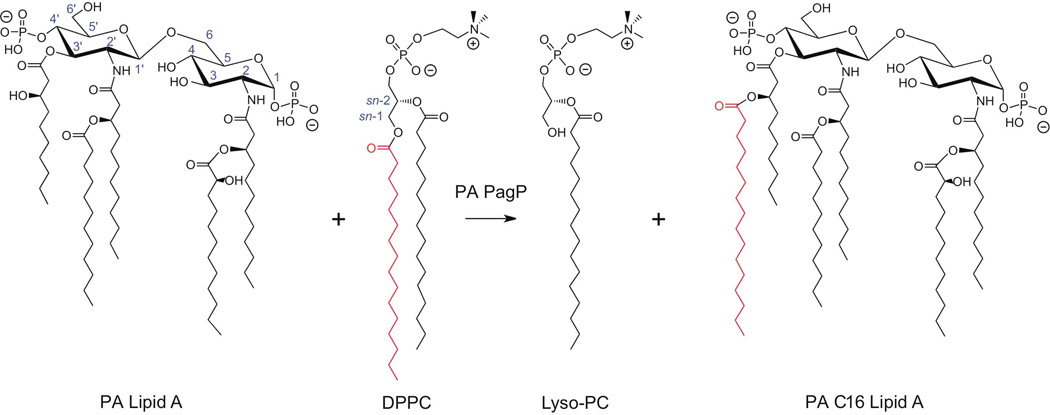
PA lipid A is a β-(1’,6)-linked glucosamine disaccharide substituted with phosphate groups at the 1 and 4’ positions (Fig. 1). Primary fatty acyl chains include amide-linked R-3-hydroxylaurate at the 2 and 2’ positions and ester-linked R-3-hydroxydecanoate at the 3 and 3’ positions. Secondary fatty acyl chains include the acyloxyacyl additions of laurate to the 2’ position (m/z 1418), and of S-2-hydroxylaurate to the 2 position (m/z 1616). Deacylation of the R-3-hydroxydecanoate at the 3 position is also seen in the majority of PA strains (m/z 1446). PA isolated from individuals with CF pulmonary disease synthesize lipid A palmitoylated at the 3’ position, resulting in a hexa-acylated structure (m/z 1684)(Ernst et al., 2007).
Fig. 1.
Pseudomonas lipid A palmitoylation by PagP. Predominant lipid A species isolated from WT under non-inducing conditions. PA PagP transfers palmitate from the sn-1 position of a phospholipid to position 3’ of lipid A.
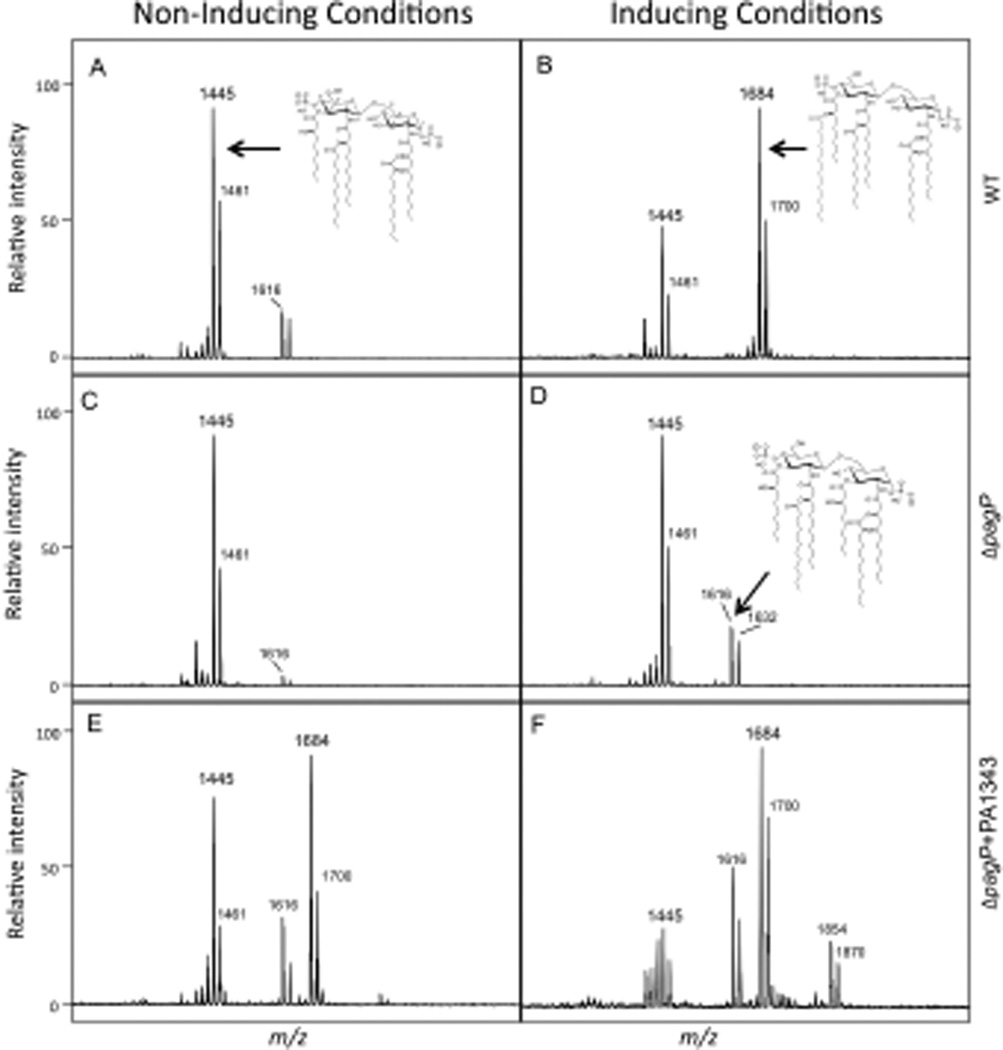
To confirm PA1343 was the PagP enzyme in PA, a clean deletion of the PA1343 was constructed using standard allelic exchange techniques (Invitrogen Gateway Technology) and confirmed by sequencing. WT (PAO-1), ΔpagP and ΔpagP+PA1343 driven off the pUCP19 plasmid were grown in inducing (low magnesium) and non-inducing conditions (high magnesium – 1mM) followed by lipid A isolation and analysis by MALDI-TOF MS. All strains were able to synthesize the lipid A species corresponding to the major peak m/z 1446 (penta-acylated) and the minor species at m/z 1616 (non-palmitoylated hexa-acylated). Each of these peaks shows a neighboring peak with a m/z difference of +16, indicating an additional hydroxylation of the laurate at the 2’ position. Upon magnesium-limited growth of WT, palmitoylation observed at the 3’ hydroxydecanoate of the penta-acylated lipid A results in additional peaks m/z 1684 (one hydroxylaurate) and m/z 1700 (two hydroxylaurates) (Fig. 2A and 2B). The ΔpagP mutant strain did not show any palmitoylated lipid A species under either inducing or non-inducing conditions (Fig. 2C and 2D). Overexpression of pagP in the ΔpagP+PA1343 strain resulted in palmitoylated species under both inducing and non-inducing conditions. Additional species m/z 1854 and m/z 1870 under magnesium limited conditions indicate the retention of the 3-hydroxydecanoate at the 3 position (Fig. 2E and 2F) resulting in the synthesis of a hepta-acylated lipid A species. Electrospray ionization (ESI) tandem MS was performed to confirm the addition of palmitate at the 3’ position. A summary of all structures and corresponding peaks can be found in Supplementary Table S1.
Fig. 2.
PA1343 is required for palmitoylation. (A and B) Lipid A was isolated from WT under inducing and non-inducing conditions. Inducing conditions resulted in hexa-acylated lipid A containing palmitate m/z 1684. (C and D) ΔpagP did not produce palmitoylated lipid A under non-inducing or inducing conditions. (E and F) PagP activity was restored in the ΔpagP+PA1343 strain when grown under both inducing and non-inducing conditions. Peaks present at m/z 1684 and m/z 1854 correspond to the palmitoylated hexa and hepta-acyl lipid A respectively.
Gas chromatography (GC) analysis of lipid A confirms PA1343 is necessary for addition of palmitate
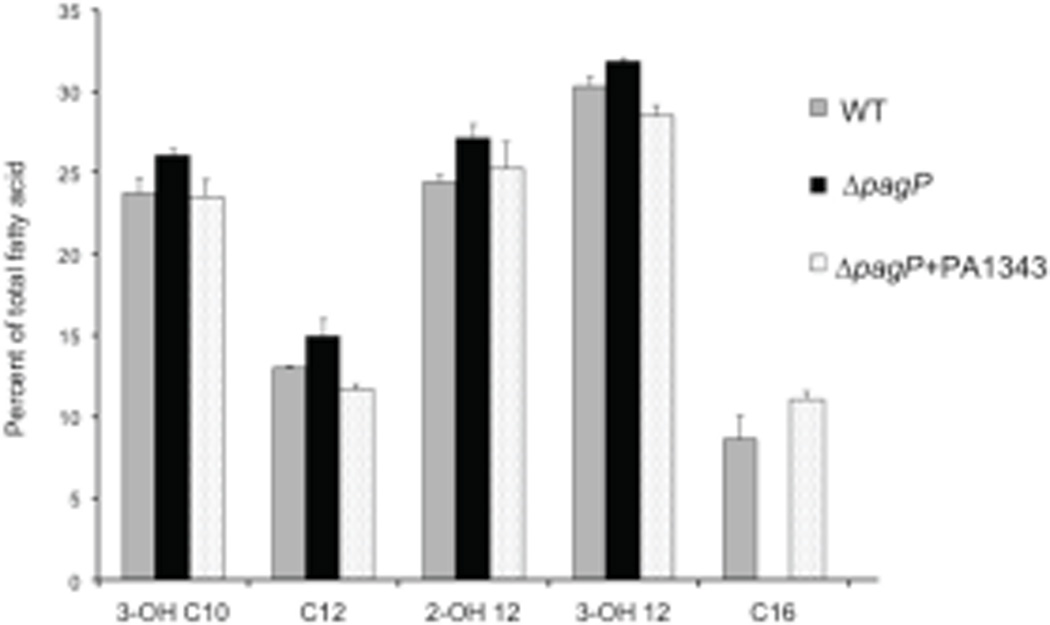
GC analysis of lipid A was performed to confirm that PA1343 is required for the addition of palmitate in PA. WT, ΔpagP, and ΔpagP+PA1343 strains were grown in inducing conditions and analyzed by GC with flame ionization detection. Total fatty acids were isolated and quantified against a C15 internal standard. Evaluation of the total fatty acid composition of WT lipid A showed an incorporation of 8% palmitate. No incorporation of palmitate was seen in the ΔpagP strain, as was indicated by MS results. Palmitoyltransferase activity was restored after complementation of the gene in trans with palmitate making up 11% of the total fatty acids of lipid A (Fig. 3). Taken together the data strongly indicates PA1343 acts as PagP in PA.
Fig. 3.
GC analysis of total fatty acid content of lipid A isolated from WT (grey), ΔpagP (black bars) and ΔpagP+PA1343 (dotted bars). Complete loss of C16 is seen in ΔpagP. C16 incorporation was restored in the complemented strain. Results represent three biological replicates.
Sequence analysis of the PA pagP gene and protein indicates PA PagP is unique among PagP enzymes
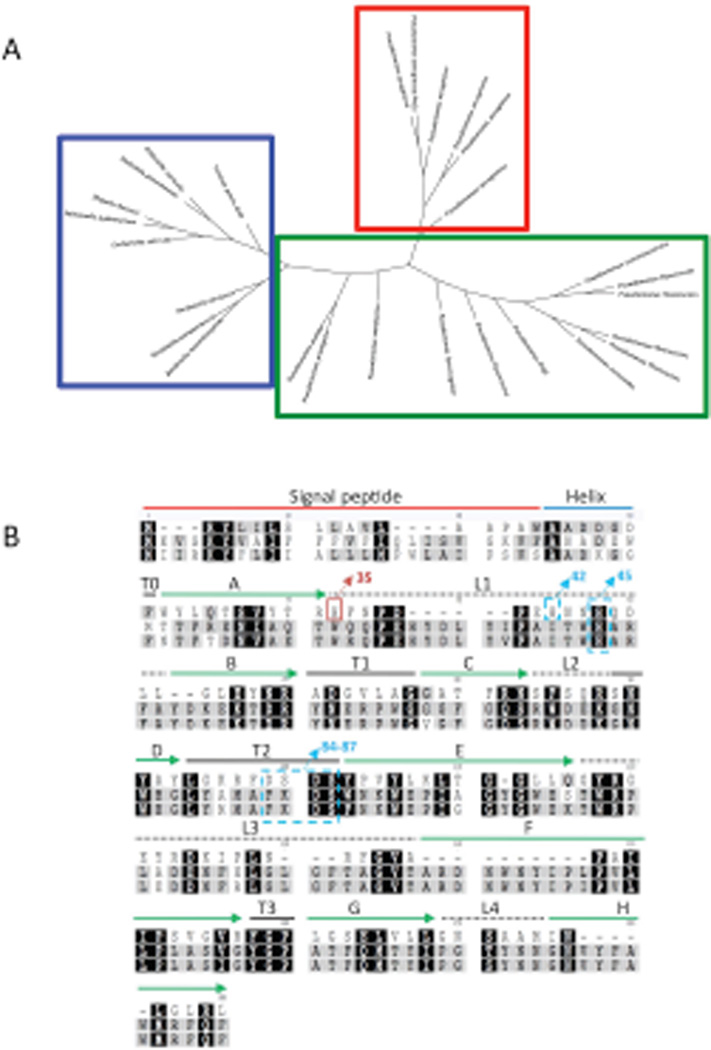
After successful identification of the PA pagP gene, a phylogenetic tree was constructed to determine the evolutionary relatedness between PA pagP and pagP genes and homologues in other Gram-negative pathogens. These results demonstrate that the pagP sequences from E. coli, Yersinia, Salmonella, and Bordetella subspecies, grouped together indicating these pagP genes derived from a common ancestor (Fig. 4A blue box). In contrast, pagP genes identified in the Pseudomonads clustered loosely together with a separate and distinct cluster containing the PA pagP genes (Fig. 4A red box). This separation indicates PA pagP has dramatically diverged to the point of no longer being comparable to the known PagP sequences.
Fig. 4.
Tree diagram of the palmitoyltransferase enzymes of gram-negative bacteria and Pseudomonas species generated by using Geneious. (A) Clustering patterns show PA PagP (red box) is distinct and separate from other non-aeruginosa Pseudomonas PagP (green box), as well as other known PagP enzymes (blue box). (B) Alignment of PagP from three different pathogenic Gram-negative bacterial genera PA, E. coli and Salmonella typhimurium using ClustalW (www.geneious.com). Absolutely conserved residues are marked with black background. Residues indicated in grey denote weakly conserved residues, white indicates non-conserved residues. PA PagP domains are predicted based on the E. coli PagP sequence of the mature protein. Residues from 1-18AA represents the signal sequence, the predicted α-helix extends from 19AA to 37AA. The putative catalytic residues (H-42 and 45/D-84 and 86/S- 85 and 87) are indicated in blue dashed boxes. His35 in the red box indicates the catalytically necessary residue. Extracellular loops are indicated by dashed lines, solid green arrows indicate β-sheet.
A previous structural alignment of PagP enzymes in E. coli, S. Typhimurium, Erwinia chrysanthemi, Yersinia pestis, Photorhabdus luminescens, Legionella pneumophila, and B. pertussis resulted in strong sequence alignment between all species (Ahn et al., 2004). Alignment of PA PagP with Salmonella and E. coli PagP enzymes showed little indication that these proteins were similar at the protein level (Fig. 4B). The E. coli PagP protein aligned with the translated PA PagP sequence shows an identity of (BLAST). PA PagP aligned with 24% S. Typhimurium PagP showed 21% identity, while E. coli compared to Salmonella resulted in 75% identity (Fig. 4B).
PA PagP purification and detergent dependence of its enzymatic activity
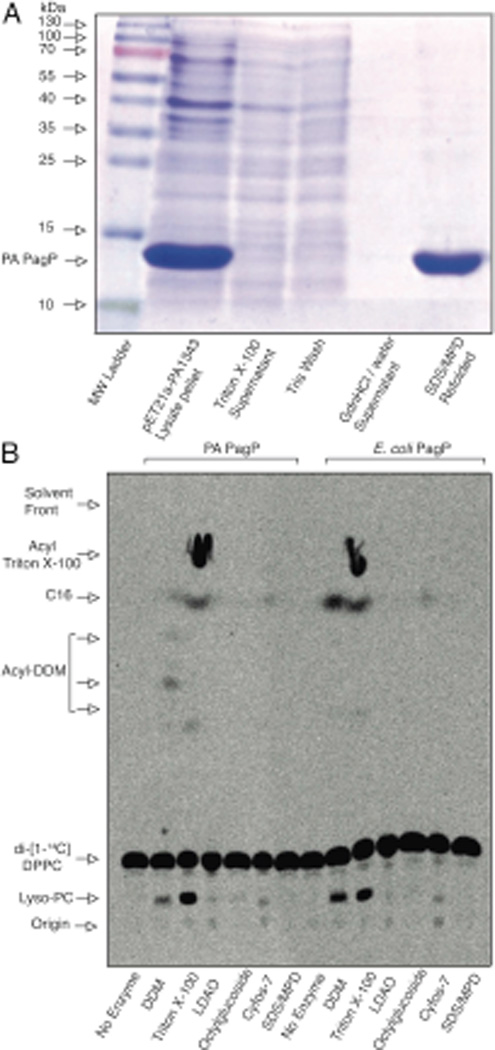
Experimental evidence provides support for the presence in PA PagP of a hydrocarbon ruler to incorporate only a C16 fatty acid into PA lipid A because the enzyme must exclude acyl chains longer and shorter than palmitate, which are abundantly present in the cellular phospholipid pool. Despite the low overall sequence similarity, a procedure for purification and folding of E. coli PagP (Cuesta-Seijo et al.) served to produce purified and enzymatically active PA PagP (Fig. 5A). A strikingly similar profile of detergents that either support or inhibit enzymatic activity of both PA PagP and E. coli PagP was observed (Fig. 5B). Since detergents that effectively mimic the structures of fatty acyl chains are E. coli PagP competitive inhibitors, only detergents with bulky substituents that sterically preclude their binding within the hydrocarbon ruler are able to support the lipid A palmitoyltransferase reaction (Khan et al., 2010a, Khan et al., 2010b). As shown in Fig. 5B, phospholipase activity for both PA PagP and E. coli PagP depends on Triton X-100, DDM, or Cyfos-7 in decreasing order of specific activity. In each case, Triton X-100 participates as a substrate in the palmitoyltransferase reaction. Although DDM is the preferred detergent for E. coli PagP because it readily supports activity without participating as a substrate (Khan et al., 2007), trace amounts of DDM appear to be acylated by PA PagP. Both enzymes appear to slowly utilize Cyfos-7 as a supporting detergent without it participating as a substrate. LDAO, octylglucoside, and SDS/MPD are all detergent systems that support PagP folding, but without supporting activity due to competitive detergent inhibition. The overall similarity in the detergent-activity relationships strongly supports common structure-function relationships between PA PagP and E. coli PagP.
Fig. 5.
Detergent dependence and sensitivity of PA and E. coli PagP. (A) SDS-PAGE assessment of PA PagP purification by selective precipitation. 40 µg of protein from cell lysate pellet and refolded PA PagP samples were resolved with similar volumes of wash material and stained with Coomassie Blue dye. (B) Thin layer chromatogram showing phospholipase activity in the absence of lipid A acceptors. Phospholipase activity is associated with production of free palmitate (C16) and Lyso-PC from DPPC in the presence of dodecylmaltoside (DDM), Triton X-100, and Cyfos-7. LDAO (lauryldimethylamine-N-oxide); SDS/MPD (mixture of SDS and 2,4-methyl-2-pentanediol)
Regiospecificity of lipid A palmitoylation by PA PagP
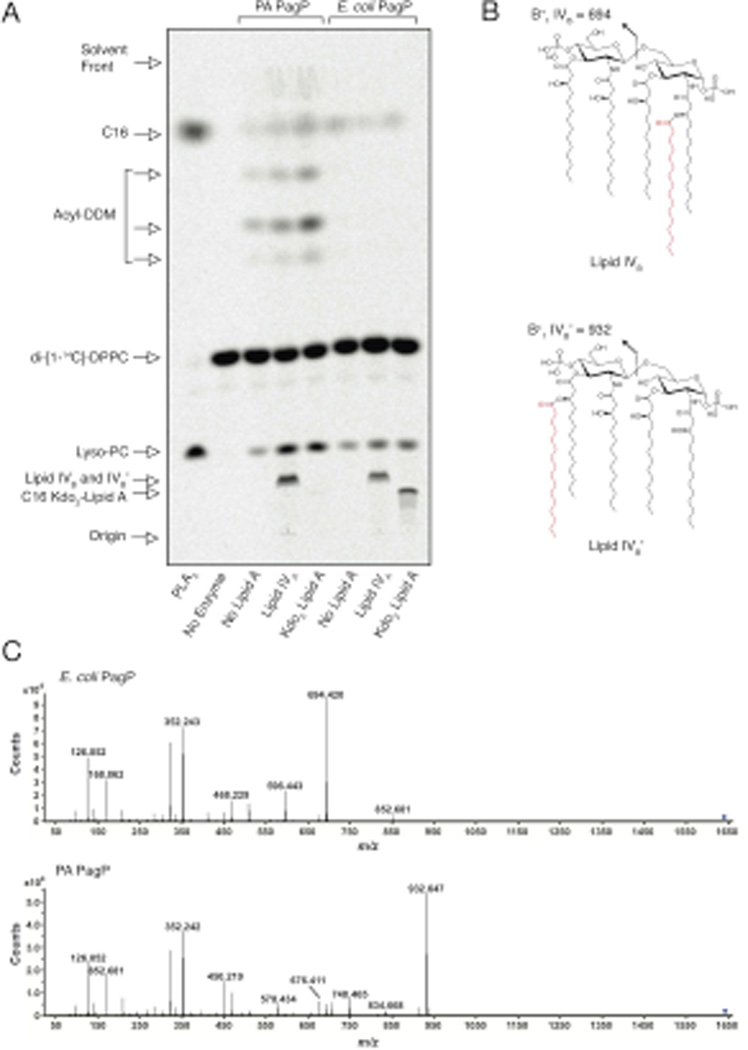
To compare the phospholipid::lipid A palmitoyltransferase activity of PA PagP and E. coli PagP, we monitored palmitoylation of lipid IVA and E. coli Kdo2-lipid A (Fig. 6). Whereas E. coli PagP could palmitoylate both substrates, PA PagP could only palmitoylate lipid IVA, consistent with the available regiospecificities for palmitoylation at the 3’ position in PA PagP and for the 2 position in E. coli PagP; both these positions are free in lipid IVA while only the latter is free in Kdo2-lipid A. In order to determine the location of the palmitate added to lipid IVA, the in vitro lipid IVB and lipid IVB’ products (Fig. 6B) generated by E. coli PagP and PA PagP, respectively, were analyzed using liquid chromatography ESI quadrupole TOF (LC-ESI-Q-TOF) MS. The expected product was readily detected in both the negative ([M–H]−, m/z 1642.076) and positive mode ([M+H+]+, m/z 1644.091), and eluted at ∼50.5 minutes. Collision induced dissociation MS (MS/MS) was performed on the [M+H+]+ ion to determine the location of the added palmitate. MS/MS analysis of the [M+H+]+ generated from E. coli PagP yielded an oxonium product ion (referred to as the B+1 ion) (Raetz et al., 1985, Qureshi et al., 1983, Costello & Vath, 1990, Que et al., 2000) with a m/z of 694.420 consistent with the palmitate being added to an acyl chain of the distal glucosamine (Fig. 6C). The ion at m/z 596.443 corresponds to the B+1 ion having lost the 1-phosphate. In contrast MS/MS analysis of the [M+H+]+ generated from the PA PagP yielded a B+1 ion with a m/z of 932.647 consistent with the palmitate being added to an acyl chain of the proximal glucosamine (Fig. 6C). The ion at m/z 834.668 corresponds to the B+1 ion having additionally lost the 1-phosphate. In addition, product-ions corresponding to the loss of the palmitate from B+1 (with or without the 1-phosphate) at m/z 676.411 and 578.434 respectively are readily detected in the in vitro product generated using the PA PagP. Taken together, these results verify that the purified E. coli and PA PagP enzymes palmitoylate lipid IVA on the proximal and distal glucosamine units, respectively.
Fig. 6.
Regiospecificity of phospholipid::lipid A palmitoyltransferase activity. (A) Thin layer chromatogram as described in Fig. 6, but showing palmitoylation of 100 µM lipid IVA and Kdo2-lipid A to produce lipid IVB, lipid IVB’, and C16-Kdo2-lipid A. (B) Structure of lipid IVB and lipid IVB’ (C) MS/MS performed on the [M+H+]+ lipid IVB and lipid IVB’ ions. PA and E. coli PagP reveal distinctly different B+1 ions (exact masses 932.6586 and 694.4290 daltons), characteristic of palmitoylation on the distal and proximal lipid A glucosamine units, respectively.
His35 is necessary for PA PagP palmitoyltransferase activity
Once the PagP activity and regiospecificity of PA PagP had been established, we focused on identifying the amino acid residues critical for activity. His33, Asp76 and Ser77 are highly conserved among characterized PagP enzymes (E. coli and Salmonella) and are thought to make up the enzymatic catalytic site (Hwang et al., 2002). These residues are seemingly duplicated at residues His42 and His45, Asp84 and Asp86, and Ser85 and Ser87 in PA PagP. An additional histidine, His35, was also found in PA PagP and may also be important for enzyme function. In order to pinpoint which of the indicated amino acids were critical for activity, each of the sites were mutated to uncharged and hydrophobic amino acids. Mutations were made by introduction of the suicide plasmid pEXGWD resulting in the following amino acid substitutions: H35F, H35N, H42F, H42N, H45F, H45L, H45N, D84A, D84N, S85A, S85G, D86A, D86N, S87A, S87G and DSDS-AGNA (D84A, S85G, D86N, S87A). All mutants were grown under inducing conditions and LPS was prepared for MALDI-TOF analysis (Caroff et al., 1988, El Hamidi et al., 2005). Alterations of His45, Asp84, Asp86, Ser85 or Ser87 alone did not alter protein function as evaluated by the presence of a peak at m/z 1684. Function was retained even after replacement of all four aspartic acid and serine residues. In contrast, mutation of the His35 rendered PA PagP non-functional (Table 1). Based on this analysis, we demonstrated that catalytic activity of PA PagP is dependent on His35 without the additional residues implicated in E. coli PagP function, suggesting that the distinctly different regiospecificities of these two enzymes might affect distinctive arrangements of catalytic residues.
Table 1.
Mutations of the proposed catalytic residues.
| Mutation | Presence of C16 (m/z 1684) |
|---|---|
| WT | + |
| ΔpagP | − |
| H35F | − |
| H35N | − |
| H42F | + |
| H42N | + |
| H45F | + |
| H45L | + |
| H45N | + |
| D84A | + |
| D84N | + |
| S85A | + |
| S85G | + |
| D86A | + |
| D86N | + |
| S87A | + |
| S87G | + |
| DSDS-AGNA | + |
PA PagP structural model and hydrocarbon ruler
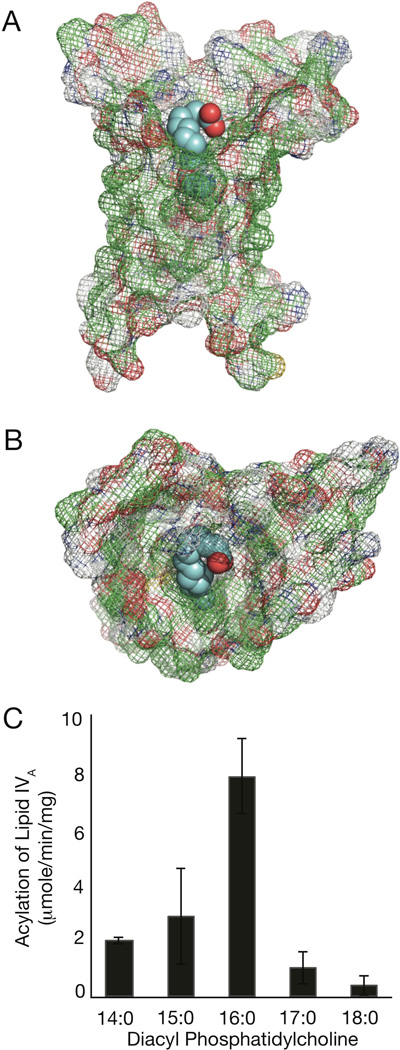
In order to visualize the PA PagP protein structure, its sequence was uploaded to the I-TASSER online protein structure and function predictions tool. This system uses threading, where the sequence and predicted secondary structure of the protein are matched with solved structures in the Protein Data Bank (PDB) library to identify the best templates. The I-TASSER program then splits the sequence into fragments based on the template alignments, which are then reassembled to full-length models. Finally, hydrogen bonding networks are optimized and steric overlaps are removed (Roy et al., 2011). Interestingly, five of the top ten template structures used by I-TASSER from the PDB were of PagP, and the top template was PagP crystallized in SDS/MPD (Cuesta-Seijo et al., 2010). We then used the Autodock/Autodock Vina (Seeliger & de Groot, 2010, Trott & Olson, 2010) PyMOL plugins, to run a docking simulation with palmitate (Seeliger & de Groot, 2010). The results of the simulation in Fig. 7A and B show the mesh representation of PA PagP with palmitate docked in the interior hydrophobic pocket reminiscent of the E. coli hydrocarbon ruler. Hydrophobic residues on the exterior of the protein are located where they would be embedded in the membrane, while the hydrophilic portions are at the expected solvent-exposed membrane interface regions. The terminal six methylene units of the palmitate chain maintain van der Waals contacts with the E. coli hydrocarbon ruler (Khan et al., 2010b) and a similar interaction is apparent in PA PagP (Fig. 7A and B). To validate the presence of a hydrocarbon ruler in PA PagP, we prepared [32P]-lipid IVA from strain BKT09 (Emptage et al., 2012) for use as an acceptor with non-radioactive phosphatidylcholines of defined acyl chain composition as donors (Fig. 7C). The results demonstrate that PA PagP clearly shares E. coli PagP’s unique ability to discriminate acyl chains that differ from palmitate by only a single methylene unit (Khan et al., 2010a, and 2010b). Based on the PA PagP homology model, His35, like His33 in E. coli PagP, maps to the cell surface region adjacent to the hydrocarbon ruler
Fig. 7.
Homology model and hydrocarbon ruler of PA PagP. Mesh representations of PA PagP viewed parallel (A) and above (B) the membrane plane with bound palmitate colored in cyan. Panel (C) shows the specific activity for the conversion by PA PagP of [32P]-lipid IVA to lipid IVB’ using symmetrical phosphatidylcholine donors of saturated acyl chain compositions varying from C14 to C18 in methylene unit increments.
PA PagP confers resistance to C18G, but does not influence resistance to polymyxins
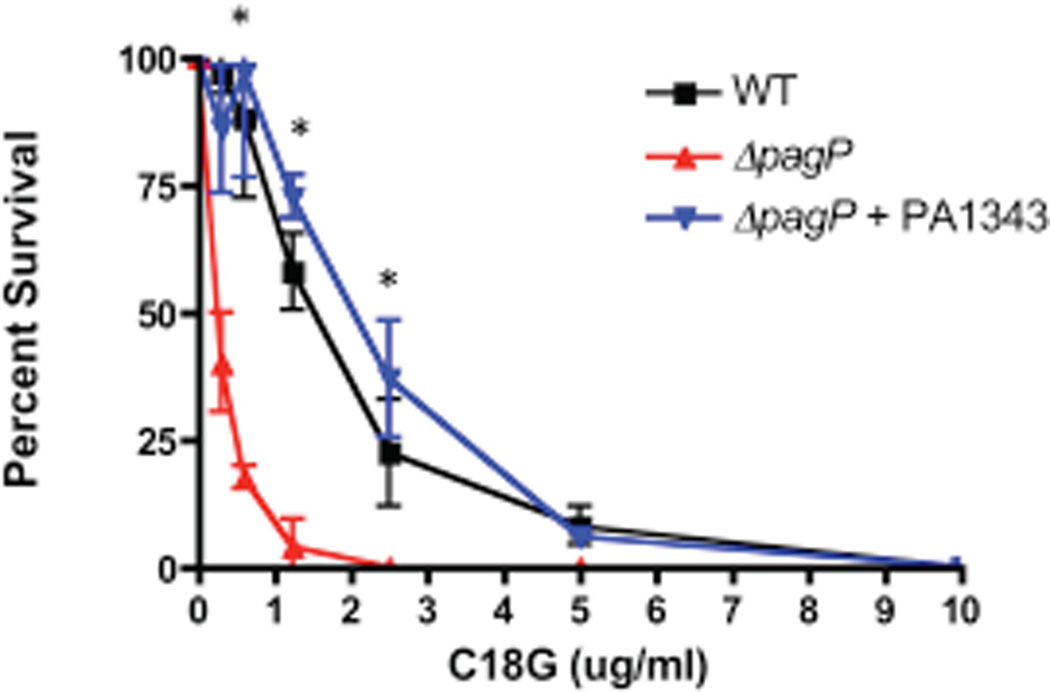
After identification and functional analysis of PA PagP was established, we focused on identifying its role within the context of the host innate immune system. Increased lipid A acylation by palmitoylation has been reported to promote resistance to CAMPs, possibly via increase in the outer membrane permeability barrier and altered lipid packing leading to repulsion of hydrophobic portions on CAMPs (Ernst et al., 2001). Killing assays were performed using C18G, a synthetic α-helical CAMP derived from human platelet factor IV, to determine the role PA PagP may play in resistance to CAMPs. WT, ΔpagP, and ΔpagP+PA1343 strains were grown under magnesium limited conditions to promote palmitoylation and strains were challenged with serial dilutions of C18G followed by plating to assess survival. Colonies were enumerated and compared to non-treated samples for survival counts. C18G resistance in these backgrounds (WT, ΔpagP, ΔpagP+PA1343) was dependent of PagP enzymatic activity, because the concentration of peptide at which 50% of bacteria were killed was approximately six times greater for WT and complemented strain (1.8 and 2.0 µg/ml, respectively) than for the ΔpagP null mutant (0.31 µg/ml) (Fig. 8).
Fig. 8.
Palmitoylated lipid A imparts resistance to C18G. WT, ΔpagP and ΔpagP+PA1343 were incubated with increasing amounts of the CAMP C18G and plated to assess susceptibility. Expression of PA1343 imparted a significant survival advantage against C18G.
In contrast, we did not detect an effect of pagP deletion when constitutively polymyxin-resistant pmrB12 and ΔphoQ strains (Miller et al., 2011, Moskowitz et al., 2004) were grown under magnesium-replete conditions and tested by conventional colistin agar dilution and alternative polymyxin B sulfate plate assays (Table 2). The lack of effect on the pmrB12 strain phenotype is not surprising, given the demonstrated inhibitory effect of this system on lipid A palmitoylation (Moskowitz et al., 2012). However, the ΔphoQ strain is known to add palmitate to its lipid A constitutively (Miller et al., 2011), thus the lack of pagP deletion effect in this strain background indicates that lipid A palmitoylation does not influence PhoPQ-dependent polymyxin resistance in PA.
Table 2.
Deletion of pagP does not affect the polymyxin resistance of relevant PA mutants.
| Strain | Colistin agar dilution MIC | Polymyxin B plate 50% CFU kill |
|---|---|---|
| (mg/L) | (mg/L) | |
| PAK | 1 | 1 |
| PAK ΔphoQ | 16 | 10 |
| PAK ΔphoQ ΔpagP | 16 | 10 |
| PAK pmrB12 | 256 | 80 |
| PAK pmrB12 ΔpagP | 256 | 80 |
Palmitoylation of PA lipid A increases IL-8 production
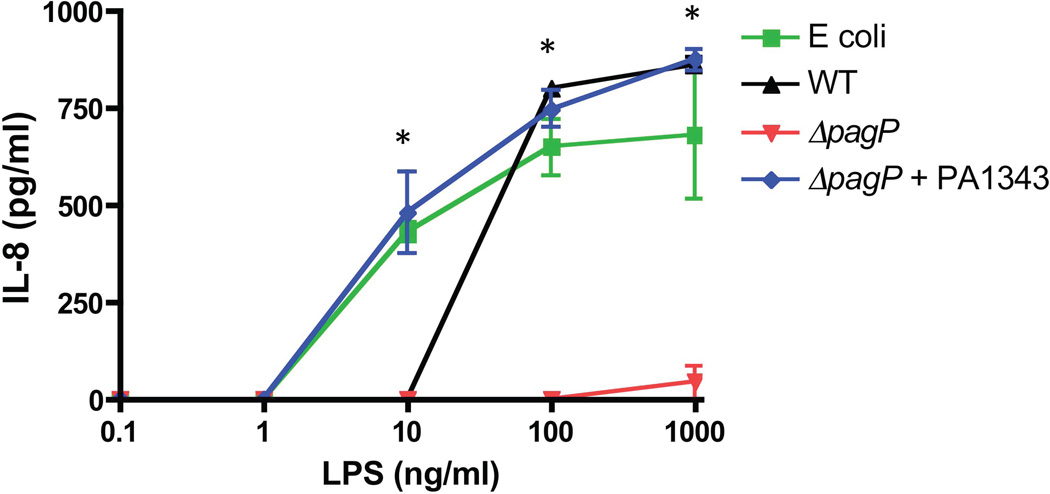
Previous studies have reported changes in cytokine levels resulting from TLR4 stimulation with palmitoylated lipid A in addition to the effects seen on CAMP resistance. Palmitoylation in other Gram-negative pathogens have been described as imparting antagonistic effects when used to stimulate TLR4 (Tanamoto & Azumi, 2000, Loppnow et al., 1986, Feist et al., 1989). We have previously shown that CF-specific lipid A containing palmitate was associated with increased inflammatory responses (Ernst et al., 1999) indicating that this modification is likely involved in progression of airway disease. Activation of TLR4 by LPS up-regulates the expression of pro-inflammatory cytokines via the NFκB pathway. The pro-inflammatory cytokine IL-8 was used as a readout for TLR4 activity. Immune response effects of PA PagP activity were tested by isolating LPS from PAO1, ΔpagP, and the ΔpagP+PA1343 after growth in PagP inducing conditions followed by stimulation of Vitamin D3 differentiated human-derived monocytic THP-1 cells for 16 hours. Supernatants were harvested and tested by ELISA for TLR activation by IL-8 expression using E. coli LPS as a positive control. Overall, IL-8 expression by cells stimulated with ΔpagP LPS was significantly lower than WT and ΔpagP+PA1343 expressing a functional PagP (Fig. 9). The complemented strain induced significantly higher levels of IL-8 than WT and ΔpagP when LPS was added at a concentration of 10 ng/ml (p=0.01). Both WT and ΔpagP+PA1343 induced significantly higher levels of IL-8 at 100 ng/ml (p=0.001) and 1ug/ml (p=0.0001) than the deletion strain.
Fig. 9.
THP-1 alveolar macrophages were stimulated with LPS prepared from E. coli, WT, ΔpagP, and ΔpagP+PA1343 grown under magnesium-limiting conditions for 16 hrs. After incubation supernatants were harvested and used for ELISA measuring IL-8 production. WT and ΔpagP+PA1343 were able to stimulate significantly higher levels of IL-8 than the ΔpagP mutant. All experiments were performed in three biological replicates.
Discussion
Gram-negative bacteria modify their LPS in response to external stimuli. Within the host, bacteria must defend against fluctuations in pH, temperature, ion limitation, as well as protect against assault by immune cells and toxic reactive products. Addition of palmitate to lipid A by PagP is seen frequently in Gram-negative pathogens in response to host factors. This additional fatty acyl chain aids in resistance to CAMPs, dampens TLR signaling, and contributes to the overall remodeling and stabilization of the membrane. Lipid A isolated from PA infecting the CF lung shows constitutive addition of palmitate making identification and characterization of the PagP enzyme in PA of particular interest. Understanding the role palmitoylated lipid A plays in this unique infection might help explain long term PA survival and target this modification as a potential treatment to block disease progression.
Upon in silico identification of candidate pagP genes, individual transposon mutants representing them were screened for loss of palmitoyltransferase activity by MALDI-TOF MS after induction by magnesium limitation. One mutant, PA1343, showed loss of PagP activity. Deletion of this gene resulted in the inability to produce the palmitolylated hexa-acylated lipid A species (m/z 1684) upon growth in inducing conditions. Further analysis of fatty acids isolated from the ΔpagP mutant lipid A showed no incorporation of palmitate, whereas WT and in trans complemented strains were able to produce hexa-acylated lipid A containing palmitate.
Although PA PagP imparts a similar function to that of previously identified PagP enzymes, phylogenetic tree analysis, as well as amino acid sequence alignment showed little similarity to known PagP proteins. This analysis at first indicated PA PagP might not have evolved from the common PagP ancestor and was potentially a distinctly different enzyme, albeit one similar both in size and in being targeted for secretion by an N-terminal signal peptide. However, most lipid A acylation enzymes possess active sites that are exposed to the cytosol in order to utilize thioester substrates, yet PagP uses a phospholipid as the palmitoyl donor and is uniquely capable of functioning within the outer membrane. If PA palmitoylates its lipid A using a phospholipid donor, then it could possess a true homolog of E. coli PagP among the pool of small β-barrel proteins in the PA outer membrane. This, in fact, appears to be the case, and our finding that purified PA PagP recapitulates the enzymology of E. coli PagP is certainly consistent with an orthologous relationship.
What sets E. coli PagP apart from all known outer membrane β-barrel structures is its interior hydrophobic pocket lined by a detergent molecule, which demarcates the hydrocarbon ruler for measuring the selected sn-1 palmitoyl group within the phospholipid donor (Khan et al., 2010b). The evidence that PA PagP and E. coli PagP are true homologs comes from the observations that the PA PagP homology model recreates a similar interior hydrophobic pocket for binding the terminal 6 methylene units of hydrocarbon chains as does E. coli PagP, and that the predicted existence of a hydrocarbon ruler in PA PagP could be confirmed by the observed enzymatic exclusion of acyl chains longer or shorter than palmitate. To our knowledge, PagP from E. coli and PA are the only integral membrane enzymes of lipid metabolism known to possess a hydrocarbon ruler capable of distinguishing acyl chains with methylene unit precision. Additionally, PA PagP can be purified as an active enzyme using a procedure that was developed for purification of active E. coli PagP, the purified PA PagP enzyme displays inhibition by the same detergents that bind within the E. coli PagP hydrocarbon ruler, and both enzyme activities depend on the same narrow spectrum of detergents that do not bind within the hydrocarbon ruler. Structural details of a PagP:phospholipid:lipid A ternary complex remain to be elucidated, but this structure will likely reveal an ordering of residues in the dynamic cell surface loops and evince the details of the PagP catalytic mechanism. Our finding that the organization of catalytic His residues in the cell surface loops is not perfectly conserved likely reflects the distinctly different regiospecificities for palmitoylation of lipid A at the 2 and 3’ positions, respectively, in E. coli and PA PagP. Perhaps these differences, combined with known functional differences between the outer membrane barriers for enterobacteria and the pseudomonads, will eventually explain how these two distinct families of PagP homologs have diverged beyond the level of detection at the amino acid sequence level. Nevertheless, the common elements of surface catalytic His residues flanking a conserved β-barrel interior hydrophobic pocket seem to be rudimentary for enzymatic palmitoylation of lipid A in bacterial outer membranes.
Resistance to CAMPs is an important evasion mechanism for pathogens. Many infections resistant to antibiotics are treated with polymyxins as a last resort. In addition, immune cells, such as neutrophils, release CAMPs to aid in bacterial clearance. Palmitate addition leads to resistance to these CAMPs in E. coli and Salmonella. Susceptibility experiments carried out here using PA expressing palmitoylated lipid A showed protection was limited to the α-helical peptide C18G, while resistance to polymyxins, typically mediated by lipid A phosphate modification, did not change. This limited resistance pattern indicates protection against these peptides is not the primary function of palmitoylation, leading our focus to other means of immune modulation.
PA lipid A isolated from patients with CF has been shown previously to produce a pro-inflammatory response. Here we were able to show modification of lipid A by PA PagP contributes to this pro-inflammatory property. Multiple factors influence agonistic properties of lipid A including terminal phosphorylation, chain length and position, unsaturation and overall acylation quantity of the lipid A molecule. These factors play a key role in optimal binding of MD2-TLR4-LPS leading to dimerization and downstream immune signaling. TLR4 antagonists, while able to bind to the complex, are not capable of inducing dimerization events. Strong agonists typically have fatty acid chains of 12 and 14 carbons in length, PA lipid A is comprised of fatty acids of 10 and 12 carbon chains contributing to a suboptimal fit within the MD2 binding pocket. Addition of the longer palmitoyl chain to PA lipid A may alter binding by seating the molecule higher in the pocket exposing regions of lipid A necessary for MD2/TLR4 dimerization. Additionally, the change from a penta-acylated to hexa-acylated lipid A is ideal for shifting of the glucosamine backbone necessary for phosphate interaction with the positively charged residues on TLR4 (Park et al., 2009).
Modifications to lipid A resulting in a proinflammatory molecule may be advantageous within the CF lung where an influx of immune cells can provide nutrients and growth factors but are not able to clear infection. Inflammation increases the trafficking of cells, such as neutrophils to sites of infection leading to bacterial clearance. Within the CF lung, responding immune cells have a decreased ability to penetrate into the host mucus layer and clear bacteria. Additionally, PA evades neutrophils through release of secreted factors leading to cell death. Increasing inflammation and thus increasing cellular trafficking leads to lysis of trapped neutrophils and release of cellular contents containing nutrients into the mucus layer depositing elements necessary for growth and survival within this unique environment.
The ability of PA to adapt to and persist within the CF lung depends on tight control of lipid A modifications in response to environmental conditions. The detailed understanding of the PA PagP enzyme that has been gained through these studies will support the development of novel strategies for combating a pathogen with emerging resistance to many treatment options. Modulation of this enzyme may help control levels of inflammation that already contribute to disease progression as well as disrupting a mechanism for metabolite acquisition critical to survival within the CF lung environment.
Experimental Procedures
Bacterial Strains and Growth Conditions
PA and other bacterial strains used during these studies are listed in Supplementary Table S2. PA PAO1 strain was used for mutagenesis and subsequently served as the WT control. Bacterial cells for LPS or lipid A analysis were obtained after overnight growth with aeration in N-minimal medium (5 mM KCl, 7.5 mM (NH4)2SO4, 0.5 mM K2SO4, 1 mM KH2PO4, 0.1 M Tris HCl, 0.1% w/v Casamino acids, 38 mM Glycerol) supplemented with either 1 mM (high or repressing) or 8 μM (low or inducing) MgCl2, or in LB medium supplemented with 1 mM MgCl2.
Recombinant DNA Techniques
Restriction enzyme digestions, ligations, transformations, and DNA electrophoresis were performed as described by manufacturer’s instructions. The oligonucleotide primers used for DNA sequencing and PCR gene amplification were manufactured by Invitrogen and are listed in Supplementary Table S3. Purification of genomic DNA and plasmids, PCR products, and restriction fragments were performed with the Qiagen kits, according to the manufacturer’s instructions. DNA sequencing was performed at the University of Maryland core facility. Plasmids used in these studies are shown in Supplementary Table S2. We cloned PA1343 on pUCP19-USER vector using USER™ Friendly Cloning Kit (New England BioLab Cat. #E5500S). The resulting plasmids were electroporated into Pseudomonas. The transformants were then selected by 200 µg ml−1 carbenicillin for Pseudomonas. The recombinant PA1343 was expressed by induction with isopropyl β-D-1-thiogalactopyranoside (IPTG). For the clean knockout of PA1343, a deleted fragment of PA1343 was amplified from genomic DNA by PCR with Pfu Turbo DNA polymerase (Invitrogen). The PCR fragment was then cloned in counter-selectable plasmid pEXGWD using Gateway Cloning system (Invitrogen). Briefly, the PCR fragment was first cloned on the Gateway-compatible vector pDONR201 using BP Clonase II (Invitrogen Cat. #11789-020). The resulting product was then introduced into E. coli DH5α cells by heat shock, and the bacterial transformants were selected on plates containing kanamycin (50 µg ml−1). The PCR fragment was subsequently introduced into the broad-host-range plasmid pEXGW-D using LR Clonase II (Invitrogen Cat. #11791-020). The plasmid was then introduced into PAO1 or PAK strain derivatives by 10-minute electroporation protocol (Choi et al., 2006). The resulting merodiploids were formed via the integration of suicide plasmid by a single crossover event. The merodiploid state was then resolved via sucrose selection in the presence of gentamicin, resulting in deletion of the WT gene. For the generation of point mutations in PA1343, a full-length PA1343 including 1 kb flanking sequence on both sides was amplified and cloned on pEXGW-D as described above. The plasmid then underwent site-directed mutagenesis by using QuikChange® XL Site-Directed Mutagenesis Kit (Stratagene Cat. #200516). The mutated plasmids were then transformed into PAO1 and counter selected as described above. The final genomic point mutations were confirmed by sequencing.
Lipid A Extraction
For MS analysis, lipid A was extracted using Caroff method (El Hamidi et al., 2005) Briefly, bacteria were suspended in isobutyric acid and ammonium hydroxide (1M) solution ratio 5:3 (vol./vol.). The samples were incubated at 100°C for 1 hour and centrifuged. The supernatant was collected in new tubes and diluted with equal volume water and lyophilized. The dry pellets were then washed with 100% methanol and finally extracted in chloroform-methanol mixtures.
MALDI TOF MS Analysis
Lipid A structures were assessed by negative-ion MALDI-TOF MS. Lyophilized lipid A was extracted in chloroform/methanol and then 1 µl was mixed with 1µl of Norharmane MALDI matrix. All MALDI-TOF experiments were performed using a Bruker Autoflex Speed MALDI-TOF mass spectrometer (Bruker Daltonics Inc., Billerica, MA). Each spectrum was an average of 300 shots. ES tuning mix (Aligent, Palo Alto, CA) was used for calibration.
LPS Extraction for GC
The rapid extraction of LPS was performed as described before (Somerville et al., 1996). Briefly, 10 mg bacterial cell pellet was suspended in 500 µl of toxin free water and the equal amount of 90% phenol and incubated at 70°C for 1 hour with constant shaking. The mixture was cooled on ice for 5 minutes and centrifuged at 9,391 x g for 10 minutes. The supernatant was collected in a new glass tube. The collected fraction was washed with diethyl ether two times by centrifugation at 845 x g for 5 minutes. Finally, the lower phase containing LPS was collected and then lyophilized. For the GC, fatty acids were converted into methyl esters by methanolysis (2M HCl in methanol; Alltech, Lexington, KY) of dried LPS at 90°C for 18 hours. The mixture was extracted twice with hexane before subjecting to GC using an HP 5890 series II with a 7673 autoinjector. Fatty acid methyl esters (Matreya bacterial acid methyl esters, CP mix no.1114) and pentadecanoic acid (Sigma, St. Louis, MO) were used as standard of a known fatty acid mixture and internal concentration control, respectively.
Phylogenetic Tree Analysis
Phylogenetic tree analysis was done using the Geneious tree builder. The Cost Matrix Blosum62 was used with a Gap open penalty of 10 and a Gap extension penalty of 1. The global alignment was performed with free end gaps. Jukes-Cantor was used as the genetic distance model with the UPGMA tree build method and no outgroup.
PA PagP Structure and Palmitate Docking Simulation
The PA1343 sequence was uploaded to the I-TASSER (Roy et al., 2010) (Zhang, 2008) database which predicts secondary structure and models a 3D structure of the protein. The results of the simulation were visualized in PyMOL (PyMOL, Schrödinger, LLC). Docking simulations were performed using the Autodock/Autodock Vina (Seeliger & de Groot, 2010) (Trott & Olson, 2010) plugins for PyMOL.
PA PagP Enzymology
Plasmid pET21a-PA1343 was constructed by cloning the 402 bp fragment carrying PA1343 without signal sequence from plasmid pUCP19-PA1343 into the IPTG-inducible T7-promoter expression vector pET21a(+), which was opened by XhoI-NdeI digestion. PCR gene amplification was performed with 5 U of Taq polymerase in a volume of 50 µl of the supplied buffer with 150 ng of pUCP19-PA1343 as template, 0.2 µM of the appropriate primers (Forward: 5’-TATACATATGGCCGACGGCGACTT-3’ and Reverse: 5’-TATACTCGAGTCAGAGACGCAGGCCGA-3’) and 10 µM dNTPs. After initial denaturation for 1 min at 94°C, 33 cycles of 30 s at 94°C, 1 min at 53°C and 1.5 min at 72°C were performed, followed by 5 min at 72°C. The XhoI-NdeI-digested PCR product that was amplified from pUCP19-PA1343 was cloned into pET21a+ digested with the same enzymes to create plasmid pET21a-PA1343. All cloned PCR products were subjected to double strand DNA sequencing at the McMaster University MOBIX Lab DNA Sequencing and Oligo Synthesis Facility to confirm the absence of any spurious mutations.
E. coli BL21(DE3) cells were transformed with pET21a-PA1343 and plated on LB agar plates with 100 µg/ml ampicillin. The plates were incubated at 37°C for 16–20 hours and then stored at 4°C. A colony was selected from the plate and added to an overnight culture (10 ml LB with 0.1 mg/ml ampicillin). The cultures were incubated for 16–20 hours, and then added to a new 1 L culture (LB with 0.1 mg/ml ampicillin). Cultures were grown at 37°C with shaking at 200 rpm and induced with 1 mM IPTG at an OD600 of 0.4 to 0.6. The cultures continued to grow for another 4 hours at 37°C. The cells were centrifuged at 7,500 g for 10 minutes, the LB was poured off, and pellet was stored at −80°C. The cell pellet was then resuspended in 40 mL 50 mM Tris-HCl (pH 8) and 5 mM EDTA. The cells were then lysed in a Thermo French Press Cell Disrupter and centrifuged at 34,000 g for 20 min at 4°C. The supernatant was poured off and the pellet was resuspended in 24 mL 2% Triton X-100 and 50 mM Tris-HCl (pH 8) and then centrifuged at 34,000 g for 20 min at 4°C. The pellet was resuspended in 32 mL 50 mM Tris-HCl (pH 8) and centrifuged at 7,600 g for 20 min at 4°C. The final pellet was resuspended in 20 mL 6M guanidine-HCl with 50 mM Tris-HCl (pH 8) and centrifuged at 7,600 g for 10 min at 4°C. The supernatant was collected and stored at 4°C. Samples were collected at each stage of the isolation and concentrations were determined using the BCA method (Smith et al., 1985). Samples in 6M Guanidine-HCl and 10 mM Tris-HCl (pH 8) were dialyzed against 4 L water with stirring for 4 hrs, then the water changed and stirred overnight. The precipitated samples were centrifuged at 7,600 g for 15 minutes and the water was poured off. The pellets were dissolved in 20 ml of 1% SDS, 1M MPD (Sigma), and 10 mM Tris-HCl (pH 8) (Cuesta-Seijo et al., 2010). The solutions were heated in a boiling water bath for 2 minutes, removed from the water bath and allowed to return to room temp on the bench overnight. The samples were centrifuged at 7,600 g for 20 minutes to remove any remaining precipitate. The liquid samples were collected and stored at 4°C. The concentrations were determined by measuring the absorbance using a molar extinction coefficient (ε) of 17210 cm−1 M−1.
The palmitoyltransferase in vitro assays are adapted from assays described by (Cuesta-Seijo et al., 2010). Phosphatidylcholines and Kdo2-lipid A were obtained from Avanti Polar Lipids, lipid IVA was obtained from Peptides International, Inc. (Louisville, KY), and dipalmitoyl-1-14C -DPPC was obtained from Perkin Elmer. Briefly, the reactions were carried out in a final volume of 25 µl, with enough 14C-DPPC to obtain a final concentration of 20 µM. 14C-DPPC was dried by leaving the microcentrifuge tubes open to the air for 30 minutes. Lipid A was dried down under a stream of N2(g) and dissolved in 22.5 µl reaction buffer (0.25% DDM, 100 mM Tris-HCl (pH 8), and 10 mM EDTA. The activity buffer with lipid A was added to the microcentrifuge tubes and equilibrated for 30 minutes. Reactions were started by addition of 2.5 µl of PA PagP (25 ng total). As a control, phospholipase A2 (PLA2) was added to the first lane, but substituting CaCl2 for EDTA in the reaction buffer. Reactions were run at 30°C for 1 hour, at which point they were stopped by adding 12.5 µl of the reaction to 22.5 µl of 1:1 CHCl3/MeOH. The bottom layer was then spotted (5 µl) on a silica gel 60 TLC plate. The TLC plates were developed for 2.5 hours in CHCl3:MeOH:H2O (65:25:4 v/v) solvent system in a sealed glass tank. The plates were then exposed overnight to a PhosphorImager screen and developed the next day with a Molecular Dynamics Typhoon 9200 PhosphorImager. Non-radioactive dried lipid films were resuspended in CHCl3:CH3OH, (2:1 v/v) and analyzed by normal phase LC-ESI-Q-TOF and MS/MS as described previously (Garrett et al., 2011).
[32P]-lipid IVA was prepared from E. coli BKT09 (provided by Pei Zhou, Duke University) (Supplementary Table S2) by mild acid hydrolysis as described (Zhou et al., 1999) followed by extraction from TLC plates. The dried 32P-labeled lipids were dissolved in 100uL of CHCl3/MeOHl (4:1 v/v) and spotted onto a TLC plate and developed in the solvent system CHCl3/pyridine/88% formic acid/H2O (50:50:16:5 v/v). After drying, the plate was exposed to an x-ray film to locate the relevant lipid IVA species. The desired compound was scraped off the plate and the silica chips were extracted with 5mL of a single phase Bligh/Dyer mixture of CHCl3:MeOH:H2O (1:2:0.8, v/v) at room temperature for 1 hour. The suspension was centrifuged and the supernatant was passed through a glass Pasteur pipette fitted with a small glass wool plug. The flow-through was collected and converted into a two-phase Bligh/Dyer mixture by adding 1.3 mL CHCl3 and 1.3 mL MeOH, mixed thoroughly and partitioned by centrifugation. The lower phase containing [1,4’ 32P]-lipid IVA was collected and dried under a stream of nitrogen. Assays were performed with [32P]-lipid IVA at ∼100 cpm/µL, cold lipid IVA at 100 µM, and phosphatidylcholines at 1mM in 0.25% DDM, 100 mM Tris-HCl (pH 8), and 10 mM EDTA as described above.
Cytokine Stimulation Assay
96-well plate (Corning Costar, Acton, MA) was seeded with 200 µl of 8×104 cells/ml of human monocytic THP-1 cells in Gibco RPMI 1640 (Invitrogen) supplemented with 10% Gibco heat-inactivated fetal bovine serum (Invitrogen), 100 units/ml penicillin/streptomycin and 50nM vitamin D3 (Sigma, St. Louis, MO). The cells were incubated at 37°C in the humid air with 5% CO2 for 72 hours for cells differentiation. For stimulation, purified LPS was resuspended in sterile water and followed by sonication. The LPS was diluted as needed in RPMI medium supplemented with 2% human serum and then added to the cell suspension. After 16-hour incubation, the supernatant was harvested for IL-8 level measurement using Human CXCL8/IL-8 DuoSet ELISA kit (R&D systems, Minneapolis, MN Cat#DY208) according to the manufacturer’s instruction. The highly purified LPS were extracted for this assay by the phenol-water method of Westphal and Jann was used (Westphal, 1965).
Susceptibility Assays
A CAMP killing assay was used to test mutant strains of PAO1 for resistance to C18G, an α-helical CAMP derived from the COOH-terminus of the human platelet factor IV. Briefly, the overnight culture in N-minimal broth supplemented with 8µM MgCl2 was diluted 1:40 and grown to an OD600 of 0.07. Then the culture was further diluted 1:50, and 100µl was inoculated into a microtiter plate. The bacterial cultures were challenged with serial dilutions of C18G. After 2 hours, the culture was diluted and plated on LB plate for counting the CFU and calculating survival percentage. All experiments were done in triplicate.
Conventional colistin agar dilution and alternative polymyxin B sulfate plate assays, used to test mutant strains of PAK for resistance to the polymyxins, were performed as described (CLSI, 2006, Miller et al., 2011).
Supplementary Material
Acknowledgements
This study was supported by the Canadian Institutes of Health Research operating grant MOP-125979 (REB), the National Science Foundation-Major Research Instrumentation Award 1029659 (TAG) and the National Institutes of Health grants R01AI067653 (SMM), RO1AI030479 (SIM), and R01AI047938 (RKE),
Footnotes
Supporting information
Additional supporting information may be found in the online version of this article at the publisher’s web-site.
References
- Ahn VE, Lo EI, Engel CK, Chen L, Hwang PM, Kay LE, Bishop RE, Prive GG. A hydrocarbon ruler measures palmitate in the enzymatic acylation of endotoxin. EMBO J. 2004;23:2931–2941. doi: 10.1038/sj.emboj.7600320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander C, Rietschel ET. Bacterial lipopolysaccharides and innate immunity. Journal of Endotoxin Research. 2001;7:167–202. [PubMed] [Google Scholar]
- Bishop RE. The lipid A palmitoyltransferase PagP: molecular mechanisms and role in bacterial pathogenesis. Molecular Microbiology. 2005;57:900–912. doi: 10.1111/j.1365-2958.2005.04711.x. [DOI] [PubMed] [Google Scholar]
- Bishop RE, Gibbons HS, Guina T, Trent MS, Miller SI, Raetz CR. Transfer of palmitate from phospholipids to lipid A in outer membranes of gram-negative bacteria. EMBO J. 2000;19:5071–5080. doi: 10.1093/emboj/cdd507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caroff M, Karibian D, Cavaillon JM, Haeffner-Cavaillon N. Structural and functional analyses of bacterial lipopolysaccharides. Microbes Infect. 2002;4:915–926. doi: 10.1016/s1286-4579(02)01612-x. [DOI] [PubMed] [Google Scholar]
- Caroff M, Tacken A, Szabo L. Detergent-accelerated hydrolysis of bacterial endotoxins and determination of the anomeric configuration of the glycosyl phosphate present in the “isolated lipid A” fragment of the Bordetella pertussis endotoxin. Carbohydr Res. 1988;175:273–282. doi: 10.1016/0008-6215(88)84149-1. [DOI] [PubMed] [Google Scholar]
- Choi KH, Kumar A, Schweizer HP. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J Microbiol Methods. 2006;64:391–397. doi: 10.1016/j.mimet.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Clinical Laboratory Standards Institute. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; aproved standard M7-A7. seventh ed. Wayne, PA: Clinical Laboratory Standards Institute; 2006. [Google Scholar]
- Costello CE, Vath JE. Tandem mass spectrometry of glycolipids. Methods Enzymol. 1990;193:738–768. doi: 10.1016/0076-6879(90)93448-t. [DOI] [PubMed] [Google Scholar]
- Cuesta-Seijo JA, Neale C, Khan MA, Moktar J, Tran CD, Bishop RE, Pomes R, Prive GG. PagP crystallized from SDS/cosolvent reveals the route for phospholipid access to the hydrocarbon ruler. Structure. 2010;18:1210–1219. doi: 10.1016/j.str.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MA, Percival RS, Devine D, Darveau RP, Coats SR, Rangarajan M, Tarelli E, Marsh PD. Temperature-dependent modulation of Porphyromonas gingivalis lipid A structure and interaction with the innate host defenses. Infection and immunity. 2011;79:1187–1193. doi: 10.1128/IAI.00900-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta T, Kievit TRde, Masoud H, Altman E, Richards JC, Sadovskaya I, Speert DP, Lam JS. Characterization of lipopolysaccharide-deficient mutants of Pseudomonas aeruginosa derived from serotypes O3, O5, and O6. Infection and Immunity. 1994;62:809–817. doi: 10.1128/iai.62.3.809-817.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis PB, Drumm M, Konstan MW. Cystic fibrosis. Am J Respir Crit Care Med. 1996;154:1229–1256. doi: 10.1164/ajrccm.154.5.8912731. [DOI] [PubMed] [Google Scholar]
- Diedrich DL, Stein MA, Schnaitman CA. Associations of Escherichia coli K-12 OmpF trimers with rough and smooth lipopolysaccharides. Journal of Bacteriology. 1990;172:5307–5311. doi: 10.1128/jb.172.9.5307-5311.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Hamidi A, Tirsoaga A, Novikov A, Hussein A, Caroff M. Microextraction of bacterial lipid A: easy and rapid method for mass spectrometric characterization. J Lipid Res. 2005;46:1773–1778. doi: 10.1194/jlr.D500014-JLR200. [DOI] [PubMed] [Google Scholar]
- Emptage RP, Daughtry KD, Pemble CWt, Raetz CR. Crystal structure of LpxK, the 4’-kinase of lipid A biosynthesis and atypical P-loop kinase functioning at the membrane interface. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:12956–12961. doi: 10.1073/pnas.1206072109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst RK, Guina T, Miller SI. Salmonella typhimurium outer membrane remodeling: role in resistance to host innate immunity. Microbes Infect. 2001;3:1327–1334. doi: 10.1016/s1286-4579(01)01494-0. [DOI] [PubMed] [Google Scholar]
- Ernst RK, Moskowitz SM, Emerson JC, Kraig GM, Adams KN, Harvey MD, Ramsey B, Speert DP, Burns JL, Miller SI. Unique lipid a modifications in Pseudomonas aeruginosa isolated from the airways of patients with cystic fibrosis. The Journal of Infectious Diseases. 2007;196:1088–1092. doi: 10.1086/521367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst RK, Yi EC, Guo L, Lim KB, Burns JL, Hackett M, Miller SI. Specific lipopolysaccharide found in cystic fibrosis airway Pseudomonas aeruginosa . Science. 1999;286:1561–1565. doi: 10.1126/science.286.5444.1561. [DOI] [PubMed] [Google Scholar]
- Feist W, Ulmer AJ, Musehold J, Brade H, Kusumoto S, Flad HD. Induction of tumor necrosis factor-alpha release by lipopolysaccharide and defined lipopolysaccharide partial structures. Immunobiology. 1989;179:293–307. doi: 10.1016/S0171-2985(89)80036-1. [DOI] [PubMed] [Google Scholar]
- Folkesson A, Jelsbak L, Yang L, Johansen HK, Ciofu O, Hoiby N, Molin S. Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: an evolutionary perspective. Nat Rev Microbiol. 2012;10:841–851. doi: 10.1038/nrmicro2907. [DOI] [PubMed] [Google Scholar]
- Fujimoto Y, Shimoyama A, Saeki A, Kitayama N, Kasamatsu C, Tsutsui H, Fukase K. Innate immunomodulation by lipophilic termini of lipopolysaccharide; synthesis of lipid As from Porphyromonas gingivalis and other bacteria and their immunomodulative responses. Mol Biosyst. 2013;9:987–996. doi: 10.1039/c3mb25477a. [DOI] [PubMed] [Google Scholar]
- Galanos C, Hansen-Hagge T, Lehmann V, Luderitz O. Comparison of the capacity of two lipid A precursor molecules to express the local Shwartzman phenomenon. Infection and Immunity. 1985;48:355–358. doi: 10.1128/iai.48.2.355-358.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett TJ, Merves M, Yost RA. Characterization of protonated phospholipids as fragile ions in quadrupole ion trap mass spectrometry. Int J Mass Spectrom. 2011;308:299–306. doi: 10.1016/j.ijms.2011.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Lim KB, Poduje CM, Daniel M, Gunn JS, Hackett M, Miller SI. Lipid A acylation and bacterial resistance against vertebrate antimicrobial peptides. Cell. 1998;95:189–198. doi: 10.1016/s0092-8674(00)81750-x. [DOI] [PubMed] [Google Scholar]
- Horstman AL, Bauman SJ, Kuehn MJ. Lipopolysaccharide 3-deoxy-D-manno-octulosonic acid (Kdo) core determines bacterial association of secreted toxins. J of Biol Chem. 2004;279:8070–8075. doi: 10.1074/jbc.M308633200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang PM, Choy W-Y, Lo EI, Chen L, Forman-Kay JD, Raetz CRH, Privé GG, Bishop RE, Kay LE. Solution structure and dynamics of the outer membrane enzyme PagP by NMR. Proc Natl Acad Sci USA. 2002;99:13560–13565. doi: 10.1073/pnas.212344499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaroszewski L, Rychlewski L, Li Z, Li W, Godzik A. FFAS03: a server for profile--profile sequence alignments. Nucleic Acids Res. 2005;33:W284–288. doi: 10.1093/nar/gki418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki K, Ernst RK, Miller SI. Deacylation and palmitoylation of lipid A by Salmonellae outer membrane enzymes modulate host signaling through Toll-like receptor 4. Journal of Endotoxin Research. 2004;10:439–444. doi: 10.1179/096805104225006264. [DOI] [PubMed] [Google Scholar]
- Khan MA, Moktar J, Mott PJ, Bishop RE. A thiolate anion buried within the hydrocarbon ruler perturbs PagP lipid acyl chain selection. Biochemistry. 2010a;49:2368–2379. doi: 10.1021/bi901669q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MA, Moktar J, Mott PJ, Vu M, McKie AH, Pinter T, Hof F, Bishop RE. Inscribing the perimeter of the PagP hydrocarbon ruler by site-specific chemical alkylation. Biochemistry. 2010b;49:9046–9057. doi: 10.1021/bi1011496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MA, Neale C, Michaux C, Pomes R, Prive GG, Woody RW, Bishop RE. Gauging a hydrocarbon ruler by an intrinsic exciton probe. Biochemistry. 2007;46:4565–4579. doi: 10.1021/bi602526k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Q, Six DA, Liu Q, Gu L, Roland KL, Raetz CR, Curtiss R., 3rd Palmitoylation state impacts induction of innate and acquired immunity by the Salmonella enterica serovar Typhimurium msbB mutant. Infection and Immunity. 2011;79:5027–5038. doi: 10.1128/IAI.05524-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loppnow H, Brade L, Brade H, Rietschel ET, Kusumoto S, Shiba T, Flad HD. Induction of human interleukin 1 by bacterial and synthetic lipid A. Eur J Immunol. 1986;16:1263–1267. doi: 10.1002/eji.1830161013. [DOI] [PubMed] [Google Scholar]
- Miller AK, Brannon MK, Stevens L, Johansen HK, Selgrade SE, Miller SI, Hoiby N, Moskowitz SM. PhoQ mutations promote lipid A modification and polymyxin resistance of Pseudomonas aeruginosa found in colistin-treated cystic fibrosis patients. Antimicrobial agents and chemotherapy. 2011;55:5761–5769. doi: 10.1128/AAC.05391-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskowitz S, Ernst R. The role of Pseudomonas lipopolysaccharide in cystic fibrosis airway infection. Subcell Biochem. 2010;53:241–253. doi: 10.1007/978-90-481-9078-2_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskowitz SM, Brannon MK, Dasgupta N, Pier M, Sgambati N, Miller AK, Selgrade SE, Miller SI, Denton M, Conway SP, Johansen HK, Hoiby N. PmrB mutations promote polymyxin resistance of Pseudomonas aeruginosa isolated from colistin-treated cystic fibrosis patients. Antimicrobial Agents and Chemotherapy. 2012;56:1019–1030. doi: 10.1128/AAC.05829-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskowitz SM, Ernst RK, Miller SI. PmrAB, a two-component regulatory system of Pseudomonas aeruginosa that modulates resistance to cationic antimicrobial peptides and addition of aminoarabinose to lipid A. Journal of Bacteriology. 2004;186:575–579. doi: 10.1128/JB.186.2.575-579.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needham BD, Trent MS. Fortifying the barrier: the impact of lipid A remodelling on bacterial pathogenesis. Nat Rev Microbiol. 2013;11:467–481. doi: 10.1038/nrmicro3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H, Vaara M. Outer membrane. In: Neidhardt F, Ingraham J, Low KB, Magasanik B, Schaechter M, Umbarger H, editors. Escherichia coli and Salmonella: Cellular and Molecular Biology. Washington D. C: American Society for Microbiology; 1987. pp. 7–22. [Google Scholar]
- Park BS, Song DH, Kim HM, Choi BS, Lee H, Lee JO. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature. 2009;458:1191–1195. doi: 10.1038/nature07830. [DOI] [PubMed] [Google Scholar]
- Preston A, Parkhill J, Maskell DJ. The bordetellae: lessons from genomics. Nat Rev Microbiol. 2004;2:379–390. doi: 10.1038/nrmicro886. [DOI] [PubMed] [Google Scholar]
- Que NL, Lin S, Cotter RJ, Raetz CR. Purification and mass spectrometry of six lipid A species from the bacterial endosymbiont Rhizobium etli. Demonstration of a conserved distal unit and a variable proximal portion. J Biol Chem. 2000;275:28006–28016. doi: 10.1074/jbc.M004008200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi N, Takayama K, Heller D, Fenselau C. Position of ester groups in the lipid A backbone of lipopolysaccharides obtained from Salmonella typhimurium . J Biol Chem. 1983;258:12947–12951. [PubMed] [Google Scholar]
- Raetz CR, Purcell S, Meyer MV, Qureshi N, Takayama K. Isolation and characterization of eight lipid A precursors from a 3-deoxy-D-manno-octylosonic acid-deficient mutant of Salmonella typhimurium . J Biol Chem. 1985;260:16080–16088. [PubMed] [Google Scholar]
- Raetz CR, Reynolds CM, Trent MS, Bishop RE. Lipid A modification systems in gram-negative bacteria. Annu Rev Biochem. 2007;76:295–329. doi: 10.1146/annurev.biochem.76.010307.145803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebeil R, Ernst RK, Gowen BB, Miller SI, Hinnebusch BJ. Variation in lipid A structure in the pathogenic yersiniae. Mol Microbiol. 2004;52:1363–1373. doi: 10.1111/j.1365-2958.2004.04059.x. [DOI] [PubMed] [Google Scholar]
- Robey M, O’Connell W, Cianciotto NP. Identification of Legionella pneumophila rcp, a pagP-like gene that confers resistance to cationic antimicrobial peptides and promotes intracellular infection. Infection and Immunity. 2001;69:4276–4286. doi: 10.1128/IAI.69.7.4276-4286.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Xu D, Poisson J, Zhang Y. A protocol for computer-based protein structure and function prediction. J Vis Exp. 2011:e3259. doi: 10.3791/3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeliger D, de Groot BL. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J Comput Aided Mol Des. 2010;24:417–422. doi: 10.1007/s10822-010-9352-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- Somerville JE, Jr., Cassiano L, Bainbridge B, Cunningham MD, Darveau RP. A novel Escherichia coli lipid A mutant that produces an antiinflammatory lipopolysaccharide. Journal of Clinical Investigation. 1996;97:359–365. doi: 10.1172/JCI118423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanamoto K, Azumi S. Salmonella-type heptaacylated lipid A is inactive and acts as an antagonist of lipopolysaccharide action on human line cells. J Immunol. 2000;164:3149–3156. doi: 10.4049/jimmunol.164.6.3149. [DOI] [PubMed] [Google Scholar]
- Trent MS. Biosynthesis, transport, and modification of lipid A. Biochem Cell Biol. 2004;82:71–86. doi: 10.1139/o03-070. [DOI] [PubMed] [Google Scholar]
- Trent MS, Stead CM, Tran AX, Hankins JV. Diversity of endotoxin and its impact on pathogenesis. Journal of Endotoxin Research. 2006;12:205–223. doi: 10.1179/096805106X118825. [DOI] [PubMed] [Google Scholar]
- Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphal OaJK. Bacterial Lipopolysaccharides: extraction with phenol-water and further applications of the procedure. In: RL W, editor. Methods in Carbohydrate Chemistry. New York: Academic Press, Inc; 1965. pp. 83–91. [Google Scholar]
- Wilkinson SG. Bacterial lipopolysaccharides--themes and variations. Prog Lipid Res. 1996;35:283–343. doi: 10.1016/s0163-7827(96)00004-5. [DOI] [PubMed] [Google Scholar]
- Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Lin S, Cotter RJ, Raetz CR. Lipid A modifications characteristic of Salmonella typhimurium are induced by NH4VO3 in Escherichia coli K12. Detection of 4-amino-4-deoxy-L-arabinose, phosphoethanolamine and palmitate. The J of Biol Chem. 1999;274:18503–18514. doi: 10.1074/jbc.274.26.18503. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.