Abstract
Objectives
To test the hypothesis that changes in DNA methylation are involved in vitamin D deficiency–related immune cell regulation using an unbiased genome-wide approach combined with a genomic and epigenomic integrative approach.
Study design
We performed a genome-wide methylation scan using the Illumina HumanMethylation 27 BeadChip on leukocytes DNAs of 11 cases of vitamin D deficiency (serum 25(OH)D≤ 25 nmol/L) and 11 age-matched controls (serum 25(OH)D>75 nmol/L); the subjects were African American normal-weight (BMI<85th percentile) males aged 14-19 years. The Limma package was used to analyze each CpG site for differential methylation between cases and controls. To correct for multiple testing, the set of raw p values were converted to false discovery rates (FDR). We also compared our findings with the recent data from GWAS of circulating 25(OH) D levels and then performed a permutation test to examine whether the “double hit” genes were randomly enriched.
Results
A total of 79 CpG sites achieved raw p<0.001. Of the 79 CpG sites, 2 CpG sites survived multiple testing: cg16317961 (raw p=3.5 × 10-6, FDR=0.078, in MAPRE2) and cg04623955 (raw p=5.9 × 10-6, FDR=0.078, in DIO3). Furthermore, 3 out of the 4 genes previously identified in the two GWAS studies were also significant at the methylation level (DHCR7: cg07487535, p=0.015 & cg10763288, p=0.017; CYP2R1: cg25454890, p=0.040; CYP24A1: cg18956481, p=0.022), reflecting significant enrichment (p=0.0098).
Conclusion
Severe vitamin D deficiency is associated with methylation changes in leukocyte DNA. The genomic and epigenomic approach reinforce the crucial roles played by the DHCR7, CYP2R1 and CYP24A1 genes in vitamin D metabolism.
Keywords: Vitamin D deficiency, DNA methylation, genome-wide association study, African Americans, extreme phenotypes
Vitamin D is considered to play an important role in a broad range of bodily functions beyond bone health 1-4. The latest assay-adjusted National Health and Nutrition Examination Survey (NHANES) 2001-2006 in 5,867 adolescents aged 12-19 years demonstrates that low 25-hydroxyvitamin D [25(OH)D] is associated with several cardiometabolic risk factors such as metabolic syndrome and insulin resistance5.
The prevalence of 25(OH)D levels of < 25 nmol/L increased from 2% during NHANES 1988-1994 to 6% during NHANES 2001-2004. The prevalence of 25(OH)D levels of < 25 nmol/L in African Americans rose from 9% to 29%6. We recently reported that African American adolescents living in a year-round sunny climate area like Augusta, Georgia (∼33° North latitude), circulating 25(OH)D concentrations were less than half those of Caucasian adolescents in every season7. In addition, 6.2% African American males and 17.2% African America females suffered from severe vitamin D deficiency (<25 nmol/L)7.
Vitamin D has known immunomodulatory effects on a wide range of immune cells including activated CD4 and CD8 T lymphocytes, B lymphocytes, macrophages as well as dendritic cells1,8. Serum 25(OH)D levels are inversely associated with levels of C-reactive protein (CRP), interleukin-6 (IL-6) and tumor necrosis factor -α (TNF-α)9. Vitamin D supplementation has been shown to down-regulate nuclear factor-kB activity, increase anti-inflammatory IL-10 levels and decrease the pro-inflammatory cytokines IL-6, IL-12, interferon-γ and TNF-α10-11. Recent hierarchical and model-based clustering analysis of 200 vitamin-related proteins revealed that vitamin D-related proteins were strongly represented among network hubs, highlighting the pervasive effects of this nutrient primarily related to immune system and cancer process12.
Epigenetic factors are at the interface between environmental stimuli and long-lasting molecular, cellular and behavioral phenotypes13. Epigenetics defines all meiotically and mitotically heritable changes in chromosome function/gene expression that are not coded in the DNA sequence itself. Recently, there has been a greater appreciation of the role of histone modification in vitamin D-mediated immune-regulatory and inflammation response14. However, whether DNA methylation is also involved in vitamin D-mediated immune regulation remains unknown.
Genome-wide association studies (GWAS) of circulating 25 (OH)D, identified 4 genes, which are involved in cholesterol synthesis, hydroxylation, and vitamin D transport15-16. Genetic variation at these loci identifies individuals who have substantially elevated risk of vitamin D insufficiency. Therefore, the goal of this study was to characterize DNA methylation profile in peripheral blood leukocytes in African American adolescents with and without severe vitamin D deficiency using an unbiased genome-wide approach combined with a genomic and epigenomic integrative approach comparing our methylation profiling data with the candidate genes identified from GWAS on circulating vitamin D and vitamin D insufficiency
Methods
The genome-wide methylation analysis was conducted in 11 children with vitamin D deficiency (plasma 25(OH)D≤ 25 nmol/L) and 11 age-matched controls (plasma 25(OH)D> 75 nmol/L) selected at the two extreme ends of the 25(OH)D distribution (extreme phenotypes) from previous studies7,17. All the subjects from the two studies were apparently healthy and were not taking any medications or vitamin supplements. The 22 subjects aged 14-19 were selected based on the following criteria: (1) African American ancestry by self-report; (2) male; (3) apparently healthy (normotensive, non-diabetic) (4) body mass index (BMI) < 85th percentile for age and sex; (5) having leukocyte DNA available; (6) at the lowest and the highest ends of the 25(OH)D distribution. The institutional Review Board at the Georgia Health Sciences University approved the studies. Informed consent was obtained from all subjects and by parents if subjects were less than 18 years of age.
Height and weight were measured by standard methods using a wall-mounted stadiometer and a scale, respectively. BMI was calculated as weight/height2. After participants quietly rested for 10 minutes, systolic blood pressure and diastolic blood pressure were measured in a supine position with a Dinamap monitor (Critikon, Tampa, FL). Blood samples were collected. The plasma and buffy coat were separated and stored at -80°C.
Plasma 25(OH)D measurement
Liquid chromatography tandem mass spectroscopy (LC-MS/MS) was used to measure circulating plasma levels of 25(OH)D2 and 25(OH)D3 as described previously18. LC-MS/MS procedures is one of the most sensitive methods of measuring 25 (OH)D19. The detection limits for 25(OH)D2 and 25(OH)D3 were 4.6 and 1.2 nmol/L. The detection range is 1-250 nmol/L18. The intra-assay (inter-assay) coefficients of variation for 25(OH)D2 and 25(OH)D3 were 6-9% (9-12%) and 7-11% (8-13%) respectively. Regarding the assay specificity, LC–MS/MS has the advantage of being able to measure both 25(OH)D2 and 25(OH)D3 independently19. The levels of total 25(OH)D including 25(OH)D2 and 25(OH)D3 were used in the selection process.
Genome-wide methylation chip
The HumanMethylation27 BeadChip from Illumina (Illumina, San Diego, CA, USA) was used. This chip can quantitatively measure 27, 000 CpG sites, covering more than 14,000 well-annotated genes at single-CpG resolution. After bisulfite treatment, 200ng of the converted DNA was whole genome amplified (WGA) and enzymatically fragmented. The bisulfite-converted WGA-DNA samples were purified and applied to the BeadChips. Image processing and intensity data extraction were performed according to Illumina's instructions (http://www.illumina.com/products/infinium_humanmethylation27_beadchip_kits.ilmn). Each methylation data point is represented by fluorescent signals from the methylated and unmethylated alleles. DNA methylation beta values are continuous variables between 0 (completely unmethylated) and 1 (completely methylated), representing the ratio of the intensity of the methylated bead type to the combined locus intensity. Initial array processing and quality control were performed with BeadStudio software.
Statistical Analyses
The general characteristics of the subjects are presented as mean ± SD. Independent t-tests were conducted to examine the differences of covariates between cases and controls using SPSS17.0 (SPSS Inc., Chicago, IL). For the genome-wide methylation analysis, the Limma package20 was used to analyze each CpG site for differential methylation between cases and controls under the design matrix of a paired test. CpG sites on the X and Y chromosomes were excluded from the analysis. Each CpG site was assigned a raw p value based on a moderated t statistic. To correct for multiple testing, the set of raw p values were converted to false discovery rates (FDR) according to Benjamini and Hochberg21. Rather than expressing the probability of a single false-positive result among all tests, the FDR value estimates the proportion of results declared interesting that are actually false. We used a FDR value threshold of 0.2022, meaning that one should expect at most 20% of declared discovery to be false.
Gene Ontology (GO) analysis was conducted with the FatiGO tool 23. FatiGO takes two lists of genes and converts them into two lists of GO terms. Then a Fisher's exact test for 2 × 2 contingency tables was used to check for significant over-presentation of GO terms in one of the sets with respect to the other one. Multiple testing correction (indexed by adjusted p-values) to account for the multiple hypotheses tested (one for each GO term) was applied to reduce false positives. Because at least two CpG sites were included for the majority of genes in this genome wide chip, for each gene we only selected the CpG sites with the lowest p value.
Additionally, we applied an integrative approach compared our methylation profiling data with the candidate SNPs identified from GWAS on circulating vitamin D and vitamin D insufficiency15-16 to identiy the “double hit” genes. We then performed a permutation test to examine whether the “double hit” genes were randomly enriched (STATA 8, StataCorp, College station, TX).
Results
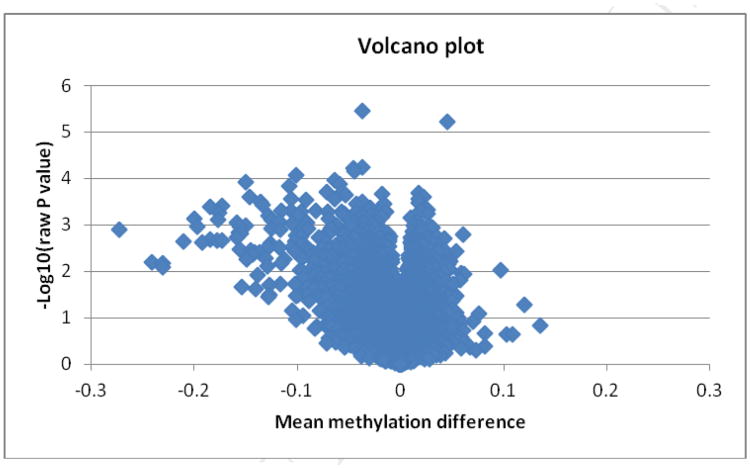
Table I displays the general characteristics of the study sample. Except for the plasma 25(OH)D concentrations, all other variables were similar for the case and control groups. The Figure is a volcano plot showing the raw p-values for all CpG sites versus mean methylation difference between the severe vitamin D deficiency group and the control group. In general, participants with severe vitamin D deficiency showed a pattern of reduced methylation compared with controls. Table II lists the top 10 most differently methylated CpG sites. Again, 9 out of the top 10 CpG sites showed reduced methylation levels compared with controls. The top two significan CpG sites survived multiple testing using an FDR of 10% (cg16317961, raw p = 3.5 × 10-6, FDR=0.078, in MAPRE2; cg04623955, raw p= 5.92 × 10-6, FDR=0.07, in DIO3).
Table 1. General characteristics of the African American male participants.
| Cases 25(OH)D ≤ 25 nmol/L & BMI < 85th percentile | Controls 25(OH)D > 75 nmol/L & BMI < 85th percentile | p | |
|---|---|---|---|
| N | 11 | 11 | - |
| Age, years | 16.2 ± 1.2 | 16.1 ± 1.3 | 0.791 |
| Age range, years | 14.8-18 | 14.2-18.6 | - |
| Height, cm | 174.6 ± 6.1 | 174.3 ± 8.8 | 0.930 |
| Weight, kg | 63.2 ± 7.4 | 62.5 ± 8.5 | 0.841 |
| BMI, kg/m2 | 20.7 ± 2.5 | 20.6 ± 2.4 | 0.897 |
| BMI percentile (%)* | 43.6 ± 24.5 | 46.2 ± 25.8 | 0.739 |
| Waist circumference, c | m 71.2 ± 4.7 | 70.9 ± 4.3 | 0.878 |
| SBP, mmHg | 113.0 ± 5.7 | 112.6 ± 4.7 | 0.855 |
| DBP, mmHg* | 58.5 ± 6.1 | 58.2 ± 4.5 | 0.940 |
| 25(OH)D, nmol/L | 19.8 ± 3.8 | 92.0 ± 12.4 | <0.0001 |
Values are mean ± SD. Tests of significance between groups were based on independent t test. SBP: systolic blood pressure. DBP: diastolic blood pressure.
BMI% and DBP were log-transformed prior to the analysis.
Figure 1.
Legend: Volcano plot showing raw p-values versus mean methylation differences between severe vitamin D deficient cases and matched controls.
Table 2. Top 10 differentially methylated CpG sites.
| Gene | Illumina ID | Distance to TSS | Methylation | P | FDR | |
|---|---|---|---|---|---|---|
|
| ||||||
| Case/Control (%) | Difference (%) | |||||
| MAPRE2 | cg16317961 | 357 | 8.0/11.7 | -3.7 | 0.0000035 | 0.078 |
| DIO3 | cg04623955 | 1312 | 21.3/16.7 | 4.6 | 0.0000059 | 0.078 |
| ASTN | cg23492043 | 242 | 14.1/17.8 | -3.7 | 0.0000588 | 0.317 |
| PAWR | cg00864867 | 478 | 6.5/11.1 | -4.6 | 0.0000592 | 0.317 |
| MEPCE | cg11979382 | 510 | 9.5/14.1 | -4.6 | 0.0000693 | 0.317 |
| FLI23191 | cg24646710 | 402 | 15.9/26.0 | -10.1 | 0.0000866 | 0.317 |
| ZBTB38 | cg17718515 | 479 | 6.6/13.0 | -6.4 | 0.000107 | 0.317 |
| MLLT6 | cg04133652 | 904 | 20.1/35.1 | -15.0 | 0.000118 | 0.317 |
| PARL | cg24355091 | 482 | 28.7/34.6 | -5.9 | 0.000132 | 0.317 |
| HRK | cg16826718 | 573 | 32.4/43.2 | -10.8 | 0.000148 | 0.317 |
TSS, transcription starting site.
Gene Ontology analysis was performed to test whether some common functional trends in molecular functions and biological processes were associated with the genes exhibiting differences between cases of vitamin D deficiency and controls in the genome-wide chip. We assigned those genes with a raw p <0.001 to the first list (n=79) and included all other genes in the second list (n=13,811). As expected from a study in only 22 subjects, we did not observe any GO categories that survived multiple testing. Overall, we observed enriched functional processes that are potentially relevant for regulation of: metabolic processes; cellular development; cell adhesion, motility and differentiation; cellular defense; transcriptional factor binding; sterol binding; and Wnt-protein binding. Table III (available at www.jpeds.com) lists the GO categories with raw p values < 0.05.
Table III. Gene-Ontology analysis.
| GO. ID | Term | P value | FDR | |
|---|---|---|---|---|
| Biological process | GO:0006139 | Nucleotide and nucleic acid metabolic process | 0.0081 | 0.936 |
| GO:0019222 | Regulation of metabolic process | 0.014 | 0.936 | |
| GO:0050794 | Regulation of cellular process | 0.0141 | 0.936 | |
| GO:0048518 | Positive regulation of biological process | 0.0182 | 0.936 | |
| GO:0040029 | Regulation of gene expression, epigenetic | 0.0221 | 0.936 | |
| GO:0040012 | Regulation of locomotion | 0.0328 | 0.936 | |
| GO:0006928 | Cell motility | 0.036 | 0.936 | |
| GO:0030154 | Cell differentiation | 0.036 | 0.936 | |
| GO:0006968 | Cellular defense response | 0.0409 | 0.936 | |
| GO:0050790 | Regulation of catalytic activity | 0.0469 | 0.936 | |
| GO:0043283 | Biopolymer metabolic process | 0.0481 | 0.936 | |
| Molecular function | GO:0008134 | Transcriptional factor binding | 0.00058 | 0.11 |
| GO:0032934 | Sterol binding | 0.00079 | 0.11 | |
| GO:0030020 | Extracellular matrix structural constituent conferring tensile strength | 0.0063 | 0.58 | |
| GO:0030023 | Extracellular matrix constituent conferring elasticity | 0.0125 | 0.78 | |
| GO:0003704 | Specific RNA polymerase II transcription factor activity | 0.0141 | 0.78 | |
| GO:0017147 | Wnt-protein binding | 0.0187 | 0.87 | |
| GO:0015248 | Sterol transporter activity | 0.0431 | 1 |
Furthermore, our genomic and epigenomic integration approach showed that of the 3 loci (within or near GC, DHCR7/NADSYN1 and CYP2R1 genes) independently discovered in these two recent GWAS studies15-16, the methylation differences in the promoter CpG sites of the two genes (DHCR7 and CYP2R1) were statistically significant between the severe vitamin D deficiency group and the control group (DHCR7: cg07487535, p=0.015 & cg10763288, p=0.017; CYP2R1: cg25454890, p=0.040). Moreover, a SNP near CYP24A1 showed an overall p value of 6.0 × 10-10 in one of the GWAS studies16. The methylation differences in the CpG site of the CYP24A1 gene were also statistically significant between the severe vitamin D deficiency group and the control group (cg18956481, p=0.022, Table IV).
Table 4. Differentially methylated CpG sites in 3 genes identified in GWAS of vitamin D level and insufficency.
| Gene | Illumina ID | Distance to TSS | Methylation | P | FDR | |
|---|---|---|---|---|---|---|
|
| ||||||
| Case/Contrl (%) | Difference (%) | |||||
| DHCR7 | cg07487535 | 506 | 11.5/10.1 | 1.4 | 0.015 | 0.543 |
| cg10763288 | 474 | 39.3/42.6 | -3.3 | 0.017 | 0.554 | |
| CYP2R1 | cg25454890 | 131 | 16.7/14.8 | 1.9 | 0.040 | 0.635 |
| CYP24A1 | cg18956481 | 385 | 8.3/9.8 | -1.5 | 0.022 | 0.586 |
TSS, transcription starting site.
To test whether the above finding (ie, 3 of the 4 candidates with GWA genes were significantly associated with severe vitamin D deficiency) was randomly enriched, we performed the permutation test. Of total 26,486 CpG sites, 26,460 were mapped on 13,872 genes. For each gene, we assigned the highest statistic value (i.e. smallest p value) among the multiple CpG sites mapped to the gene as the statistic value of the gene. Four genes were randomly selected from these 13,872 genes. The number of genes with p value ≤0.05 was counted. This procedure was repeated 10000 times, and a distribution of count number of significant genes (p≤0.05) out of 4 genes was obtained. In these 10000 repetitions, only 98 had a count number equal or larger than 3. In other words, these 3 genes (DHCR7, CYP2R1 and CYP24A1) were significantly enriched with respect to severe vitamin D deficiency (p=0.0098), indicative of not a chance finding.
Discussion
In this study, we identified methylation differences in leukocyte DNA in relation to vitamin D deficiency. Moreover, the genomic and epigenomic integrative approach confirmed the DHCR7, CYP2R1 and CYP24A1 genes, which may play crucial roles in vitamin D metabolism.
MAPRE2 (also known as EB2, RP1) has the most significant differently methylated promoter CpG site. The methylation level was 37% lower (3.7%/10%) in the group with vitamin D deficiency compared with the controls. Moreover, the other CpG site of the MAPRE2 gene was also significantly differentially methylated (p=0.016). The MAPRE gene family consists of three members (MAPRE1, 2 and 3) that code for three microtubule associated proteins. They are important for many cellular processes i.e. cell division, cytoplasmatic organization, maintenance of cell polarity, chromosome segregation and cell migration24-26. These microtubule associated proteins regulate microtubule functions and dynamics, which is fundamentally important to the way cells response to their environment24, 27. MAPRE2 is expressed in T cells and lymphocyte tumor cell lines and activated by cell surface antigens and/or cytokines28. MAPRE2 over expression induced nuclear accumulation of β-catenin and promoted the transcriptional activity of β-catenin/T-cell factor29. Because the expression is rapidly up-regulated in activated T cells, MAPRE2 is considered a member of the immediate-early T-cell regulatory gene family28. The proteins encoded by the immediate-early gene can act as pleiotropic regulators of cellular activation, such as transcription factors and proteins involved in signal transduction cascades30. MAPRE2 can also be found in rapidly proliferating tumor cells and its expression correlates with the cell cycle activity indicates that its protein (RP1) might be critically involved in processes controlling cell proliferation28, 31.
DIO3, the second most significant site identified, is located in the imprinted region in chromosome 14q32 and plays a critical role in maturation and function of the thyroid axis which is also critical to the development of the human central nervous system32. Meta-analysis of genes modulating intracellular T3 bio-availability has revealed a possible role for DIO3 gene in osteoarthritis33. Recent data have shown that this gene is hypermethylated in hematological tumors34-35. In the present study, the promoter CpG site methylation was 24.2% (4.6%/19%) higher in the group with vitamin D deficiency as compared with the controls.
As demonstrated by previous studies, 3-10% of the human genome is regulated directly and indirectly by the vitamin D endocrine system36-37. Genome-wide VDR has a few thousand chromosomal target sites that regulate several hundred 1, 25(OH)2D3 target genes in a cell-specific fashion38-39. Methylation/demethylation reflects vitamin D-mediated transcriptional control via vitamin D response element (VDRE)40. VDRE can directly bind to DNA methyltransferases 1 and 3B (DNMT1 and DNMT3B)41. A global transcriptomic study showed that 1, 25 (OH)2D3 significantly changed the RNA expression profile in human colon cancer cells involving many different cell functions, including transcription, cell adhesion, DNA synthesis, apoptosis and intracellular signaling42. Recent hierarchical and model-based clustering analysis of the protein-protein interaction network of 200 vitamin-related proteins revealed that a subgroup of 22 proteins that are highly central, most of these proteins are key immune regulators such as cytokines and transcription factors such as SMAD3 and VDR. Moreover, 17 out of the 22 central proteins in the protein-protein interaction network are vitamin D-related proteins, highlighting the pervasive effects of this nutrient primarily related to immune system and cancer process12. We also showed that many functional pathways are involved in severe vitamin D deficiency-related methylation changes in leukocytes including regulation of metabolic processes, cellular development, cell adhesion, motility and differentiation, cellular defense response as well as transcriptional factor binding, sterol binding, and Wnt-protein binding (Table III). The finding that vitamin D receptor deficiency enhances Wnt/β-Catenin signaling in human color cancer cells43 parallels our finding that Wnt-protein binding was an enriched molecular function for differentially methylated genes in leukocytes of individuals with vitamin D deficiency.
Our epigenomic findings confirmed 3 genes out of the 4 genes in GWAS findings, which reinforce the crucial roles played by those 3 genes in vitamin D metabolism. DHCR7 encodes the enzyme 7-dehydrocholesterol (7-DHC) reductase, which converts 7-DHC to cholesterol, thereby removing the substrate from the synthetic pathway of vitamin D3, a precursor of 25OHD3. DHCR7 is a novel gene for association with vitamin D levels identified in the two recent GWAS studies15-16. CYP2R1 (cytochrome P450, family 2, subfamily R, polypeptide 1) encodes a hepatic microsomal enzyme 25-hydroxylase that converts vitamin D3 to an active vitamin D receptor ligand. Previous candidate44-45 and GWAS15-16 studies suggested that this gene is associated with vitamin D metabolism. CYP24A1, which encodes 24-hydroxylase, has been identified as a candidate gene for vitamin D insufficiency in one GWAS16, not the other15. This mitochondrial protein initiates the degradation of 1,25-dihydroxyvitamin D3 and plays a role in calcium homeostasis and vitamin D metabolism. Our findings suggested that subjects with vitamin D deficiency are more likely to have reduced synthesis and increased metabolism of active vitamin D. Our epigenetic data provide the additional body of evidence that those 3 enzymes are the key enzymes underlying the crucial steps in vitamin D metabolism.
The observed DNA methylation differences between cases with vitamin D deficiency and controls were relatively small (<5%), which is in agreement with the two recent genome-wide methylation studies in leukocyte DNA on obesity and type 2 diabetes46,47. Toperoff et al demonstrated that the odd of belonging to the type 2 diabetes group increased by 6% for every 1% decrease in methylation in whole blood DNA47. It is possible that the methylation differences identified in leukocyte DNA could be attributed to differences in the leukocyte profile, rather than epigenetic changes induced within specific subtypes of leukocytes. However, Toperoff et al further analyzed the potential contribution of differential case-control composition of blood cells to the observed methylation differences. No significant differences in blood counts appeared between cases and controls, and adjustment for blood lineage count did not affect the association of methylation with the incidence of type 2 diabetes47.
Our use of a hypothesis-free approach avoided the limitations imposed by candidate gene studies and allowed us to search the whole genome in an unbiased manner. Genomic and epigenomic data integration provides greater understanding of physiology and etiology of the complex traits. The use of extreme phenotypes based on plasma vitamin D values maximized the power to make discoveries. The selection of normal weight cases and controls eliminated the confounding effect of obesity on methylation differences. The focus on adolescents prior to the development of target organ damages optimized the chances to unmask important etiologic relationships.
Several limitations are noteworthy. First, we were unable to determine whether the between group methylation differences we observed were associated with differences in transcription level between the two groups because cellular RNA samples were unavailable. Second, although we hypothesize that severe vitamin D deficiency will lead to methylation changes in leukocyte DNA, our case-control study design cannot determine whether the identified methylation differences are the cause or the consequence of severe vitamin D deficiency. Third, the methylation profiling was done in whole blood leukocytes. However, the blood lineage data was not collected at the time of sampling. Forth, the Infinium HumanMethylation27 Beadchip was used because of the availability at that time. The limited coverage of this genome-wide chip will restrict the findings to certain CpG sites within certain genes. Genome chips with more complete coverage such as the recently released 450K Infinium Methylation BeadChip from Illumina should be used in future studies. Last, the study is limited by lack of cases with severe vitamin D deficiency and matched controls for further validation; future studies are needed to replicate and validate the findings by independent methods in independent cohorts including female populations.
Acknowledgments
The participants in this study were previously recruited and funded by National Institutes of Health (NIH; HL64157 and HL77230). H.Z., G.H., and Y.D. are supported by NIH (HL69999).
Abbreviations
- 25(OH)D
25-hydroxyvitamin D
- BMI
Body mass index
- DBP
Diastolic blood pressure
- FDR
False discovery rate
- GO
Gene Ontology
- GWAS
Genome-wide association study
- IL
Interleukin
- SBP
Systolic blood pressure
- WGA
whole genome amplification
Footnotes
The authors declare no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Veldman CM, Cantorna MT, DeLuca HF. Expression of 1,25-dihydroxyvitamin D(3) receptor in the immune system. Arch Biochem Biophys. 2000;374:334–8. doi: 10.1006/abbi.1999.1605. [DOI] [PubMed] [Google Scholar]
- 2.Holick MF. Vitamin D deficiency. N Engl J Med. 2007;357:266–81. doi: 10.1056/NEJMra070553. [DOI] [PubMed] [Google Scholar]
- 3.Lee JH, O'Keefe JH, Bell D, Hensrud DD, Holick MF. Vitamin D deficiency an important, common, and easily treatable cardiovascular risk factor? J Am Coll Cardiol. 2008;52:1949–56. doi: 10.1016/j.jacc.2008.08.050. [DOI] [PubMed] [Google Scholar]
- 4.Woloszynska-Read A, Johnson CS, Trump DL. Vitamin D and cancer: Clinical aspects. Best Pract Res Clin Endocrinol Metab. 2011;25:605–15. doi: 10.1016/j.beem.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ganji V, Zhang X, Shaikh N, Tangpricha V. Serum 25-hydroxyvitamin D concentrations are associated with prevalence of metabolic syndrome and various cardiometabolic risk factors in US children and adolescents based on assay-adjusted serum 25-hydroxyvitamin D data from NHANES 2001-2006. Am J Clin Nutr. 2011;94:225–33. doi: 10.3945/ajcn.111.013516. [DOI] [PubMed] [Google Scholar]
- 6.Ginde AA, Liu MC, Camargo CA., Jr Demographic differences and trends of vitamin D insufficiency in the US population, 1988-2004. Arch Intern Med. 2009;169:626–32. doi: 10.1001/archinternmed.2008.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dong Y, Pollock N, Stallmann-Jorgensen IS, Gutin B, Lan L, Chen TC, et al. Low 25-hydroxyvitamin D levels in adolescents: race, season, adiposity, physical activity, and fitness. Pediatrics. 2010;125:1104–11. doi: 10.1542/peds.2009-2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Provvedini DM, Tsoukas CD, Deftos LJ, Manolagas SC. 1,25-dihydroxyvitamin D3 receptors in human leukocytes. Science. 1983;221:1181–3. doi: 10.1126/science.6310748. [DOI] [PubMed] [Google Scholar]
- 9.Bellia A, Garcovich C, D'Adamo M, Lombardo M, Tesauro M, Donadel G, et al. Serum 25-hydroxyvitamin D levels are inversely associated with systemic inflammation in severe obese subjects. Intern Emerg Med. 2011 Mar 25; doi: 10.1007/s11739-011-0559-x. [DOI] [PubMed] [Google Scholar]
- 10.Schleithoff SS, Zittermann A, Tenderich G, Berthold HK, Stehle P, Koerfer R. Vitamin D supplementation improves cytokine profiles in patients with congestive heart failure: a double-blind, randomized, placebo-controlled trial. Am J Clin Nutr. 2006;83:754–9. doi: 10.1093/ajcn/83.4.754. [DOI] [PubMed] [Google Scholar]
- 11.Mathieu C, Adorini L. The coming of age of 1,25-dihydroxyvitamin D(3) analogs as immunomodulatory agents. Trends Mol Med. 2002;8:174–9. doi: 10.1016/s1471-4914(02)02294-3. [DOI] [PubMed] [Google Scholar]
- 12.Nguyen TP, Scotti M, Morine MJ, Priami C. Model-based clustering reveals vitamin D dependent multi-centrality hubs in a network of vitamin-related proteins. BMC Syst Biol. 2011;5:195. doi: 10.1186/1752-0509-5-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petronis A. Epigenetics as a unifying principle in the aetiology of complex traits and diseases. Nature. 2010;465:721–7. doi: 10.1038/nature09230. [DOI] [PubMed] [Google Scholar]
- 14.Gynther P, Toropainen S, Matilainen JM, Seuter S, Carlberg C, Vaisanen S. Mechanism of 1alpha,25-dihydroxyvitamin D(3)-dependent repression of interleukin-12B. Biochim Biophys Acta. 2011;1813:810–8. doi: 10.1016/j.bbamcr.2011.01.037. [DOI] [PubMed] [Google Scholar]
- 15.Ahn J, Yu K, Stolzenberg-Solomon R, Simon KC, McCullough ML, Gallicchio L, et al. Genome-wide association study of circulating vitamin D levels. Hum Mol Genet. 2010;19:2739–45. doi: 10.1093/hmg/ddq155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang TJ, Zhang F, Richards JB, Kestenbaum B, van Meurs JB, Berry D, et al. Common genetic determinants of vitamin D insufficiency: a genome-wide association study. Lancet. 2010;376:180–8. doi: 10.1016/S0140-6736(10)60588-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petty KH, Li K, Dong Y, Fortenberry J, Stallmann-Jorgensen I, Guo D, et al. Sex dimorphisms in inflammatory markers and adiposity in African-American youth. Int J Pediatr Obes. 2010;5:327–33. doi: 10.3109/17477160903497019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen TC, Turner AK, Holick MF. Methods for the determination of the circulating concentration of 25-hydroxyvitamin D. J Nutr Biochem. 1990;1:315–9. doi: 10.1016/0955-2863(90)90067-u. [DOI] [PubMed] [Google Scholar]
- 19.Wallace AM, Gibson S, de la Hunty A, Lamberg-Allardt C, Ashwell M. Measurement of 25-hydroxyvitamin D in the clinical laboratory: current procedures, performance characteristics and limitations. Steroids. 2010;75:477–88. doi: 10.1016/j.steroids.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 20.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Statistical applications in genetics and molecular biology. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 21.Klipper-Aurbach Y, Wasserman M, Braunspiegel-Weintrob N, Borstein D, Peleg S, Assa S, et al. Mathematical formulae for the prediction of the residual beta cell function during the first two years of disease in children and adolescents with insulin-dependent diabetes mellitus. Medical hypotheses. 1995;45:486–90. doi: 10.1016/0306-9877(95)90228-7. [DOI] [PubMed] [Google Scholar]
- 22.Smith NL, Hindorff LA, Heckbert SR, Lemaitre RN, Marciante KD, Rice K, et al. Association of genetic variations with nonfatal venous thrombosis in postmenopausal women. JAMA. 2007;297:489–98. doi: 10.1001/jama.297.5.489. [DOI] [PubMed] [Google Scholar]
- 23.Al-Shahrour F, Diaz-Uriarte R, Dopazo J. FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics. 2004;20:578–80. doi: 10.1093/bioinformatics/btg455. [DOI] [PubMed] [Google Scholar]
- 24.Morrison EE. Action and interactions at microtubule ends. Cell Mol Life Sci. 2007;64:307–17. doi: 10.1007/s00018-007-6360-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Musch A. Microtubule organization and function in epithelial cells. Traffic. 2004;5:1–9. doi: 10.1111/j.1600-0854.2003.00149.x. [DOI] [PubMed] [Google Scholar]
- 26.Valiron O, Caudron N, Job D. Microtubule dynamics. Cell Mol Life Sci. 2001;58:2069–84. doi: 10.1007/PL00000837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carvalho P, Tirnauer JS, Pellman D. Surfing on microtubule ends. Trends Cell Biol. 2003;13:229–37. doi: 10.1016/s0962-8924(03)00074-6. [DOI] [PubMed] [Google Scholar]
- 28.Renner C, Pfitzenmeier JP, Gerlach K, Held G, Ohnesorge S, Sahin U, et al. RP1, a new member of the adenomatous polyposis coli-binding EB1-like gene family, is differentially expressed in activated T cells. J Immunol. 1997;159:1276–83. [PubMed] [Google Scholar]
- 29.Wang Y, Zhou X, Zhu H, Liu S, Zhou C, Zhang G, et al. Overexpression of EB1 in human esophageal squamous cell carcinoma (ESCC) may promote cellular growth by activating beta-catenin/TCF pathway. Oncogene. 2005;24:6637–45. doi: 10.1038/sj.onc.1208819. [DOI] [PubMed] [Google Scholar]
- 30.Kelly K, Siebenlist U. Immediate-early genes induced by antigen receptor stimulation. Curr Opin Immunol. 1995;7:327–32. doi: 10.1016/0952-7915(95)80106-5. [DOI] [PubMed] [Google Scholar]
- 31.Abiatari I, Gillen S, DeOliveira T, Klose T, Bo K, Giese NA, et al. The microtubule-associated protein MAPRE2 is involved in perineural invasion of pancreatic cancer cells. Int J Oncol. 2009;35:1111–6. doi: 10.3892/ijo_00000426. [DOI] [PubMed] [Google Scholar]
- 32.Hernandez A, Martinez ME, Fiering S, Galton VA, St Germain D. Type 3 deiodinase is critical for the maturation and function of the thyroid axis. J Clin Invest. 2006;116:476–84. doi: 10.1172/JCI26240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meulenbelt I, Bos SD, Chapman K, van der Breggen R, Houwing-Duistermaat JJ, Kremer D, et al. Meta-analyses of genes modulating intracellular T3 bio-availability reveal a possible role for the DIO3 gene in osteoarthritis susceptibility. Ann Rheum Dis. 2011;70:164–7. doi: 10.1136/ard.2010.133660. [DOI] [PubMed] [Google Scholar]
- 34.Martin-Subero JI, Ammerpohl O, Bibikova M, Wickham-Garcia E, Agirre X, Alvarez S, et al. A comprehensive microarray-based DNA methylation study of 367 hematological neoplasms. PLoS One. 2009;4:e6986. doi: 10.1371/journal.pone.0006986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Figueroa ME, Wouters BJ, Skrabanek L, Glass J, Li Y, Erpelinck-Verschueren CA, et al. Genome-wide epigenetic analysis delineates a biologically distinct immature acute leukemia with myeloid/T-lymphoid features. Blood. 2009;113:2795–804. doi: 10.1182/blood-2008-08-172387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bouillon R, Carmeliet G, Verlinden L, van Etten E, Verstuyf A, Luderer HF, et al. Vitamin D and human health: lessons from vitamin D receptor null mice. Endocr Rev. 2008 Oct 29;6:726–76. doi: 10.1210/er.2008-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manson JE. Vitamin D and the heart: why we need large-scale clinical trials. Cleve Clin J Med. 2010;77:903–10. doi: 10.3949/ccjm.77gr.10004. [DOI] [PubMed] [Google Scholar]
- 38.Carlberg C, Seuter S. Dynamics of nuclear receptor target gene regulation. Chromosoma. 2010;119:479–84. doi: 10.1007/s00412-010-0283-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pike JW. Genome-wide principles of gene regulation by the vitamin D receptor and its activating ligand. Mol Cell Endocrinol. 2011;347:3–10. doi: 10.1016/j.mce.2011.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim MS, Kondo T, Takada I, Youn MY, Yamamoto Y, Takahashi S, et al. DNA demethylation in hormone-induced transcriptional derepression. Nature. 2009;461:1007–12. doi: 10.1038/nature08456. [DOI] [PubMed] [Google Scholar]
- 41.Rhee I, Bachman KE, Park BH, Jair KW, Yen RW, Schuebel KE, et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002;416:552–6. doi: 10.1038/416552a. [DOI] [PubMed] [Google Scholar]
- 42.Palmer HG, Sanchez-Carbayo M, Ordonez-Moran P, Larriba MJ, Cordon-Cardo C, Munoz A. Genetic signatures of differentiation induced by 1alpha,25-dihydroxyvitamin D3 in human colon cancer cells. Cancer Res. 2003;63:7799–806. [PubMed] [Google Scholar]
- 43.Larriba MJ, Ordonez-Moran P, Chicote I, Martin-Fernandez G, Puig I, Munoz A, et al. Vitamin D Receptor Deficiency Enhances Wnt/beta-Catenin Signaling and Tumor Burden in Colon Cancer. PLoS One. 2011;6:e23524. doi: 10.1371/journal.pone.0023524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramos-Lopez E, Bruck P, Jansen T, Herwig J, Badenhoop K. CYP2R1 (vitamin D 25-hydroxylase) gene is associated with susceptibility to type 1 diabetes and vitamin D levels in Germans. Diabetes Metab Res Rev. 2007;23:631–6. doi: 10.1002/dmrr.719. [DOI] [PubMed] [Google Scholar]
- 45.Cheng JB, Levine MA, Bell NH, Mangelsdorf DJ, Russell DW. Genetic evidence that the human CYP2R1 enzyme is a key vitamin D 25-hydroxylase. Proc Natl Acad Sci U S A. 2004;101:7711–5. doi: 10.1073/pnas.0402490101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang X, Zhu H, Snieder H, Su S, Munn D, Harshfield G, et al. Obesity related methylation changes in DNA of peripheral blood leukocytes. BMC Med. 2010;8:87. doi: 10.1186/1741-7015-8-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Toperoff G, Aran D, Kark JD, Rosenberg M, Dubnikov T, Nissan B, et al. Genome-wide survey reveals predisposing diabetes type 2-related DNA methylation variations in human peripheral blood. Hum Mol Genet. 2012;21:371–83. doi: 10.1093/hmg/ddr472. [DOI] [PMC free article] [PubMed] [Google Scholar]