Abstract
Longitudinal healthcare claims databases are frequently used for studying the comparative safety and effectiveness of medications, but results from these studies may be biased due to residual confounding. It is unclear whether methods for confounding adjustment that have been shown to perform well in small, simple nonrandomized studies are applicable to the large, complex pharmacoepidemiologic studies created from secondary healthcare data. Ordinary simulation approaches for evaluating the performance of statistical methods do not capture important features of healthcare claims. A statistical framework for creating replicated simulation datasets from an empirical cohort study in electronic healthcare claims data is developed and validated. The approach relies on resampling from the observed covariate and exposure data without modification in all simulated datasets to preserve the associations among these variables. Repeated outcomes are simulated using a true treatment effect of the investigator's choice and the baseline hazard function estimated from the empirical data. As an example, this framework is applied to a study of high versus low-intensity statin use and cardiovascular outcomes. Simulated data is based on real data drawn from Medicare Parts A and B linked with a prescription drug insurance claims database maintained by Caremark. Properties of the data simulated using this framework are compared with the empirical data on which the simulations were based. In addition, the simulated datasets are used to compare variable selection strategies for confounder adjustmentvia the propensity score, including high-dimensional approaches that could not be evaluated with ordinary simulation methods. The simulated datasets are found to closely resemble the observed complex data structure but have the advantage of an investigator-specified exposure effect.
Keywords: simulation, pharmacoepidemiology, propensity score, variable selection
1 Introduction
Longitudinal healthcare claims databases are frequently used for studying the comparative safety and effectiveness of medications. Administrative healthcare data generally provide a longitudinal record of medical services, procedures, diagnoses, and medications for large numbers of patients, and therefore provide a rich data source for conducting pharmacoepidemiologic research. Compared with randomized trials, the data available in healthcare claims better represent the full spectrum of patients that are exposed to a drug and the processes of care in routine practice (Schneeweiss and Avorn, 2005; Strom and Carson, 1990). However, drug studies in claims data may suffer from bias due to residual confounding (Brookhart et al., 2010), and it is unclear whether methods for confounding adjustment that have been shown to perform well in small, simple nonrandomized studies are applicable to the cohort studies created from complex healthcare claims data.
Monte Carlo simulation can be used to evaluate the performance of causal inference methods, but ordinary simulation approaches do not capture important features of healthcare claims. For example, healthcare claims databases often have hundreds, or even thousands, of measured covariates with complex covariance structures. These covariates, either singly or in combination, may serve as proxies for unmeasured confounders and be effectively used to remove bias (Schneeweiss et al., 2009). Further, the complexity of real-world data extends beyond confounding; patients’ follow-up time and censoring patterns are often associated with exposure and outcome via a path of underlying characteristics. These complexities cannot be replicated in fully synthetic simulated data, as they are generally not completely understood and vary by data source.
As an alternative to simulation, Vaughan et al. (Vaughan et al., 2009) suggested creating “plasmode” datasets. A plasmode is a real dataset that is created from natural processes but has some aspect of the data-generating model known, for example, a “spike-in” experiment in microarray analysis of gene expression where a known amount of genome transcript is added to each sample. Merging this concept with simulation techniques has led to several studies of methods performance that use real observed data augmented with simulated data (Elobeid et al., 2009; Gadbury et al., 2008). Other approaches utilize fully simulated data, but create associations among variables to match estimated associations from observed data (Chao et al., 2010; Erenay et al., 2011; McClure et al., 2008; Rolka et al., 2005; Schmidt et al., 2009), including one approach specifically designed for simulating an entire healthcare claims database (Murray et al., 2011). However, due to the massive size and complexity of the data in this approach, generally only one dataset is created for each set of simulation parameters, and the relative contributions ofbias and variance to estimation error cannot be judged. Furthermore, this data-generation process may produce spurious correlations among variables that are not present in the underlying empirical dataset.
In this paper, we outline a statistical and computational framework for creating replicated simulation datasets based on an empirical pharmacoepidemiologic cohort study in healthcare claims data. The objective of this work is to enable the evaluation of approaches to confounder adjustment in simulated data that preserve the complex features and information content of claims data but also have a known true treatment effect. As an example, we applied our framework to a study of high versus low-intensity statin use and cardiovascular outcomes. Simulated data was based on real data drawn from Medicare Parts A and B and eligibility files linked with Part D prescription drug insurance claims database maintained by Caremark. We compared properties of the data simulated using this framework with the empirical data on which the simulations were based. In addition, we used the simulated datasets to compare variable selection strategies for confounder adjustmentvia the propensity score (PS), including high-dimensional approaches that could not be evaluated with ordinary simulation methods since their performance depends on the information richness and complexity of the underlying empirical data source.
2 Methods
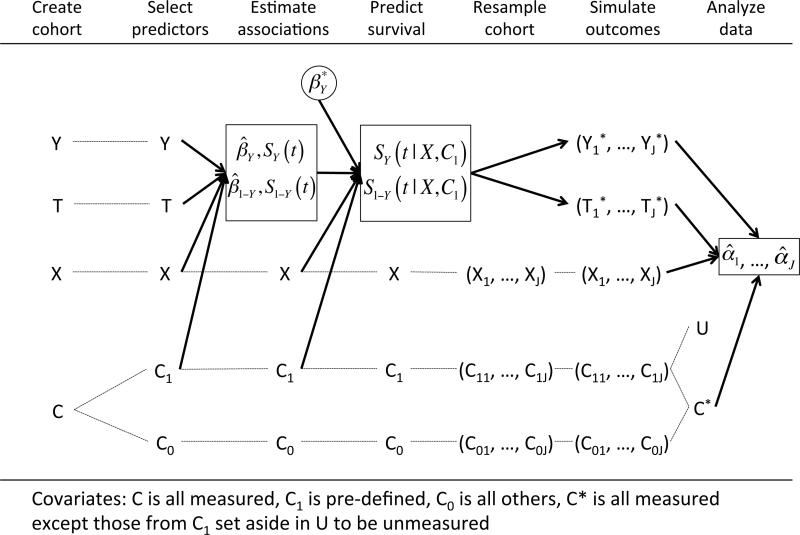
Our simulation approach relies on resampling from the observed covariate and exposure data without modification in all simulated datasets to preserve the empirical associations among these variables. Repeated outcomes are simulated using a true treatment effect of the investigator's choice and the baseline hazard function estimated from the empirical data (Figure 1). R code and documentation for the simulation setup are available in the Web Appendix.
Figure 1.
Diagram showing the steps in the simulation framework. Dashed lines represent reusing or resampling a data element without modification. Solid arrows represent using a data element to create new data structures. The end result is a sequence of exposure effect estimates (one for each of J simulated datasets) given by .
2.1 Construct the cohort
The first task in creating simulated datasets is to create the cohort on which the simulations will be based from the larger healthcare database. The specifics of the study design, including inclusion and exclusion criteria for the cohort, definitions of exposures and covariates, and determinations of follow-up and censoring for outcome events, are important in determining the performance of any statistical methods subsequently applied to the data. As these issues are not the focus of this paper, we refer the reader to the wide array of literature on the subject for specific information on these determinations. In general, we recommend a “new user design” with an active comparator (Ray, 2003; Schneeweiss, 2010), where two treatments with similar clinical indications are compared in patients initiating one treatment or the other with no history of use in the prior six months (or some other pre-specified period). Covariates (diagnoses, procedures, medications, and health system service use) are assessed in the period preceding initiation of treatment, and assessment of outcomes begins on or after the date of treatment initiation.
The result of this design is a dataset where each patient has information on exposure (X = 1 indicates initiating one treatment, X = 0 indicates initiating the reference treatment), presence of an outcome event (Y), and length of follow-up time (T). In addition, we assume that there is a large pool of potential covariates, C, that contains potentially hundreds or thousands of distinct codes for diagnoses, procedures, hospitalizations, medications and other health system service use in the period preceding treatment initiation (Schneeweiss et al., 2009). This dataset provides all of the information that we will use for constructingthe simulated datasets.
2.2 Select covariates for simulation basis
Within the hundreds or thousands of potential covariates in C, we identify a subset to be used for outcome generation. We recommend specifying a set of covariates that are believed to be associated with the outcome, including important demographic information such as age, gender, and race. We refer to this subset as C1, and we refer to the complement (everything in C not included in C1) as C0. The variables in C1 are used for simulating outcome variables. In general, including more covariates in C1 will result in more realistic simulated outcomes, as any associations between covariates and outcome present in the observed data will be lost if those covariates are not included in C1. However, including all potential covariates in C1 will generally be infeasible due to the model estimation required in subsequent steps. If any of the variables in C1 are associated with exposure, or if they are associated with other measured covariates that are correlates of exposure, then confounding will be present in the simulated datasets.
2.3 Estimate associations with outcome and censoring
In order to produce outcome and censoring times that have realistic associations with covariates, we estimate the empirical multivariate associations with two Cox proportional hazards models. In the first model, we estimate the hazard of the outcome event in the observed data. Investigators can specify this model as needed to capture important features of the relationships of covariates and exposure with outcome. For example, interaction terms beween covariates and exposure could be included to estimate (and in following steps, simulate) treatment effect heterogeneity. The second model is identical except that we estimate the hazard of censoring, represented in the model as the reverse of the outcome variable (1 – Y).
2.4 Predict survival and censoring
To translate the empirical associations into the simulated outcome data, we use the fitted models from the previous step to predict each patient's expected survival and censoring time given his C1 values. We extractthe Breslow estimates (Breslow, 1975) ofthe baseline event-free survival function, SY(t), and the baseline censor-free survival function, S1–Y(t). In addition, we extract the vector of estimated coefficients from each model ( and for the event and censoring models, respectively). The desired true effects are specified by selectively replacing the values in with desired values at this step. For example, an alternative true effect of exposure can be inserted by replacing the estimated coefficient on X with another value. In addition, one may increase the overall amount of confounding by replacing the covariate coefficients in with larger values. We denote the coefficient vector used for event time simulation as . This vector will define the true causal effects of X and C1 on the simulated outcomes.
A predicted event-free survival curve for each individual is then calculated as:
where Di is the row for patient i in the design matrix from the estimated time to event model. A predicted censor-free survival curve is calculated similarly using S1–Y(t) and with the exception that the predicted censor-free survival curve is set to zero on the date of administrative censoring if present.
2.4.1 Adjust baseline survival
If any values in are replaced in this step in , then the overall event rate in the subsequent simulated data will be different from that observed in the empirical data. In order to keep the overall event rate the same (or to specify another preferred event rate) in the simulated data, we adjust the baseline event-free survival function. Specifically, in order to guarantee that the probability ofhaving an event in the period defined by T=t is approximately p, we find the value of δ such that
This value of δ is then applied to the predicted survival function for each individual so that the adjusted survival curve is given by SY*(t|Xi, C1i) = SY(t|Xi, C1i)δ.
2.5 Resample and simulate
We now construct J simulated datasets of size n ≤ N, where N is the size of the full cohort. We describe the process for creating one simulated dataset, and the entire process is repeated J times. We first take a bootstrap resample of size n (sampled with replacement) from the complete set of covariate-exposure vectors (Ci, Xi). Because we do not modify or permute these variables, the systematic relationships among covariates and exposure remain intact in each sample. To simulate survival and censoring times for individuals in the sample, we use the fact that for any arbitrary distribution defined by the cumulative distribution function F, the distribution of F–1(R) is given by F, where R is a random uniform variable in (0,1) (Casella and Berger, 2001). Therefore, in order to simulate an event time from the patient-specific survival function SY(t|Xi, C1i), we simulate a random variate R~Unif(0,1). We then calculate the corresponding event times by inverting the patient-specific survival step function, Ei = mint{SY(t|Xi, C1i) < R}. Similarly, to simulate a censoring time for patient i, we generate another random uniform variate and calculate the corresponding censoring time, Fi = mint{S1–Y(t|Xi, C1i) < R}. For each patient in the sample, the simulated follow-up time is taken to be the minimum of the event and censoring times and outcome variables are created to reflect the simulated event status
2.6 Analyze simulated data
We now have J datasets of size n, each of which contains an exposure vector X, a large matrix of potential covariates C, and vectors containing event indicators, Y*, and follow-up times, T*, for all patients in the sample. Furthermore, we have created the outcomes in such a way that the complete data generating mechanism for the outcome is known, including the effect sizes for exposure and all covariates. The data generating mechanism for X and C remain unknown, as these data remain unaltered from their observed values, and any associations that exist among these variables in the observed data remain intact.
If desired, unobserved confounding can be created at this step by setting aside a subset of the predictors of outcome C1 to be the unobserved confounders U, so that the variables available for analysis are . By hiding the variables in U from the confounder adjustment methods applied to the simulated data, we may observe the performance of methods under unobserved confounding. By varying which covariates are set aside and their strength of association with outcome, we can vary the strength of the unobserved confounding in the simulations.
Analyzing these data, we return an estimate of exposure effect for each of the J simulated datasets, . Using these estimates, we may calculate features of the estimation procedure as in ordinary simulation studies, for example bias and variance.
3 Application
3.1 Source cohort and simulation
We applied our framework to a cohort study of high-intensity versus low-intensity statin medications for the prevention of cardiovascular events. These data come from a cohort of Medicare beneficiaries 65 years of age and older with prescription drug coverage through a Medicare Part D or employer-sponsored plan maintained by Caremark, a pharmacy benefits manager. Diagnostic, healthcare utilization, and demographic data from Medicare Parts A, B, and enrollment files were linked to prescription drug claims.
There were 236,314 eligible patients that initiated a statin between July 1, 2005 and December 31, 2008 and had continuous Medicare eligibility and demonstrated Medicare Parts A, B and prescription drug benefit use in the 6 months prior to statin initiation. The initiation date was defined as the first date a patient filled a prescription for a statin without a fill of any statin or statin combination drug in the prior 180 days. Exposure was classified according to the daily dose of statin dispensed as high-intensity or low-intensity therapy, as shown in the Web Appendix. We followed for outcomes for 180 days after treatment start, including hospitalization for myocardial infarction (MI) or acute coronary syndrome (ACS) with revascularization. Patients were censored before 180 days ifthey had an outcome event, died, lost eligibility in either Caremark or Medicare, or were hospitalized for more than 14 days.
We considered the full pool of covariates, C, to include all claims submitted through either Medicare or Caremark in the 180 days prior to statin initiation, including claims for diagnoses, procedures, hospitalizations, and medications. For simulating the outcome, we defined 61 covariates representing demographic information (age, sex, race), history of vascular conditions, history of other comorbid conditions, and overall use of the healthcare system, including use of preventive services, ordering of lipid tests, and frequency of physician visits. A complete list of the covariates in C1 with claims definitions is available in the Web Appendix.
In the survival and censoring model estimation step, we used penalized splines (Eilers and Marx, 1996; Hurvich et al., 1998) with a modest 2 degrees of freedom for estimating an independent effect for each continuous covariate (e.g., age, number of comorbid conditions) as a smooth nonlinear function. We also used ridge regression for estimating the effect of binary variables (including the exposure), so that extreme and imprecise parameter estimates on covariates with low prevalence are shrunk toward a more reasonable null value (Gray, 1992; Therneau and Grambsch, 2000; Therneau et al., 2003). We chose a penalty parameter of θ=1 (so that the penalty is one half the sum of squared coefficients). Each of these estimation techniques is implemented in the survival package in R (Therneau, 2011) and allow for precise estimation even when many predictors are included in the model.
Using these models, we simulated 500 datasets, each with 100,000 patients, a true high versus low-intensity treatment effect hazard ratio of 1.0, and the effects of all other predictors set at their estimated values (scenario 1). In order to keep the proportion of patients with an event approximately the same as in the observed data (1.7%), we adjusted the baseline hazard function as described in Section 2.4.1. We simulated a second set of 500 datasets with the same study size (100,000), but the true exposure hazard ratio set to 1.5 and the coefficients on all other predictors set at 2 times their estimated value (scenario 2). In this case, we adjusted the baseline hazard to ensure approximately 5% of patients would have an event within 180 days.
3.2 Comparison of simulated and observed data
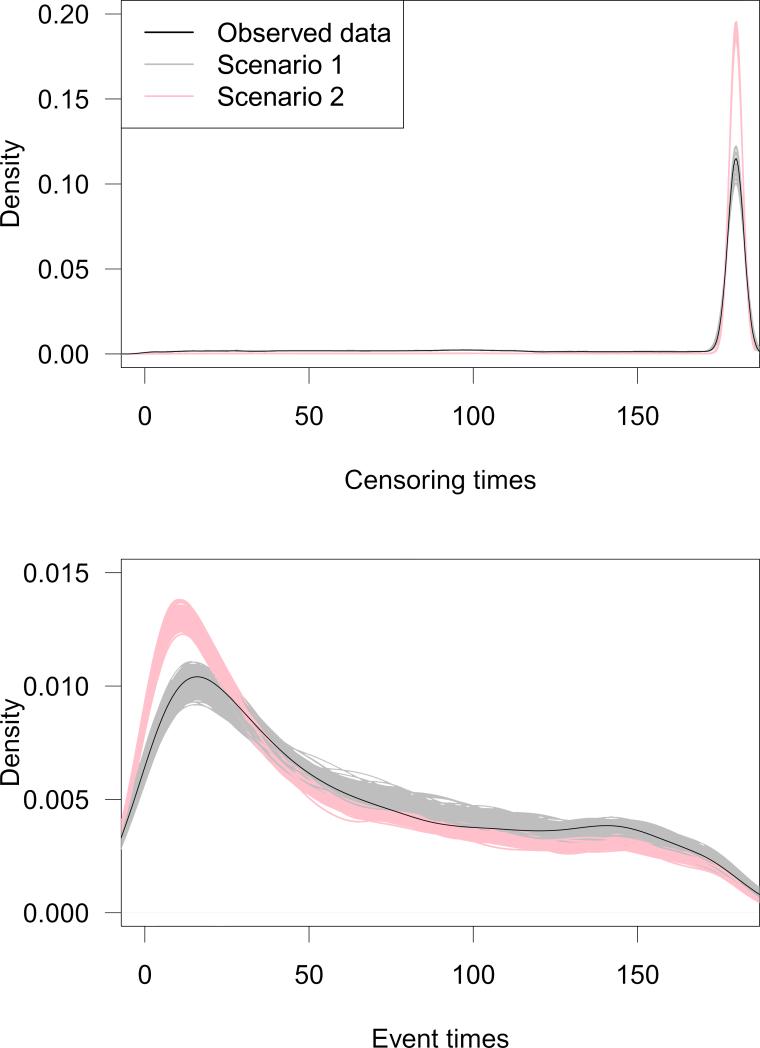
To ensure that our simulation strategy was producing realistic data that closely matched the observed data, we compared the observed and simulated data. Each simulated dataset contained data for 100,000 patients with an average prevalence of high-intensity therapy of 30.4%, matching the prevalence in the observed data. In Figure 2, we present density plots for censoring times (top panel) and event times (bottom panel). The densities from the observed data are plotted in black and the densities from each of the 500 simulated datasets are plotted in gray and red for scenarios 1 and 2, respectively. These plots show that the distribution of event times and censoring times were very similar for observed and simulated data, particularly in scenario 1. In scenario 2, the number of events was increased, causing a larger proportion of patients to have an event early during follow-up.
Figure 2.
Densities ofcensoringtimes (top) and eventtimes (bottom) for observed data (solid black curve) and simulated data (Scenario 1 is in gray; Scenario 2 is in red).
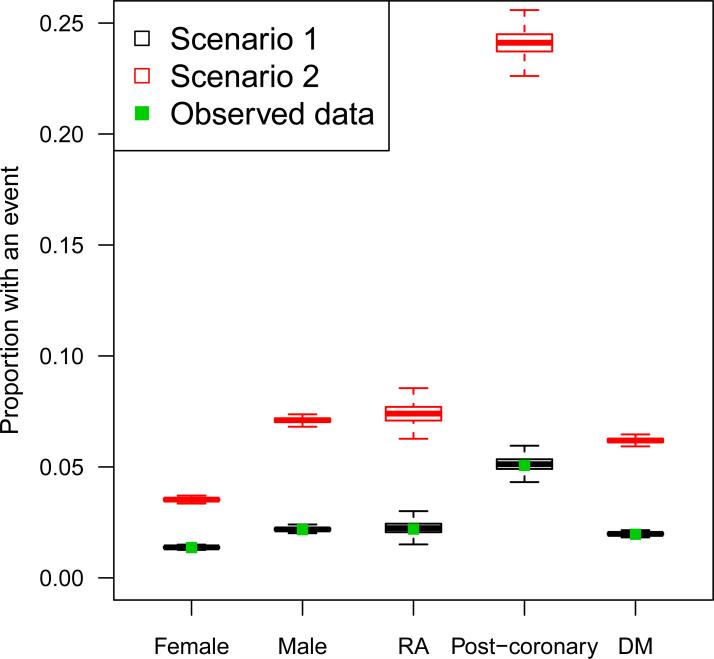
Figure 3 shows the proportion of patients with an event in each of five patient subgroups: 1) females, 2) males, 3) patients with a history of MI or ACS in the pre-exposure period (post-coronary), 4) patients with a history of diabetes mellitus (DM) in the preexposure period, and 5) patients with a history of rheumatoid arthritis (RA) in the preexposure period. These subgroups were chosen to show patient outcomes in the general population (males and females) as well as in subgroups with risk factors for cardiovascular events (post-coronary, DM, RA). The proportions in the observed data are shown with a solid green point and the distributions of proportions across the 500 simulated datasets are shown with black and red boxplots, separately for scenario 1 and scenario 2, respectively. Again, the data from scenario 1 closely resembled the observed data with some random variation. The proportions in scenario 2 were higher than the observed proportions as expected, but followed similar patterns.
Figure 3.
Proportion of patients with an event in observed data (green point) and simulated data, separately in patient subgroups. Black boxplots show the distribution of proportions across Scenario 1 datasets; red boxplots show Scenario 2.
3.3 Evaluating variable selection strategies in simulated data
To demonstrate the value of the proposed simulation framework, we used the simulated data from both scenarios to compare strategies for selecting variables to include in the PS. This example study is designed to evaluate the performance of the high-dimensional PS (hdPS) variable selection approaches frequently used in pharmacoepidemiology for identifying important confounders among the thousands of potential covariates in longitudinal claims data with little investigator input (Rassen et al., 2011; Schneeweiss et al., 2009). In “exposure-based hdPS,” variables are selected based on their association with exposure only. In “bias-based hdPS,” variables are selected based on their associations with both exposure and outcome. See the references above for additional details. In empirical studies with at least 100 outcome events in each group, hdPS algorithms have been shown to perform well by reproducing results observed in randomized trials, but they have not been studied via simulation. There were on average 573 and 2,136 outcome events in the high-intensity group in scenarios 1 and 2, respectively.
We compared hdPS algorithms with the correctly-specified outcome model and correctly-specified PS approach. Specifically, in each simulated dataset we estimated the effect of high versus low-intensity statins using several Cox proportional hazards models. We estimated 1) a “crude” model that only included a term for the exposure, 2) a model with terms for exposure, patient age, sex, and year of statin initiation (the A/S/Y model), and 3) a model that included exposure and linear terms for all covariates in C1 (the “all variables” model). In addition, we estimated 3 PS-adjusted models, including the PS models: 1) a PS model that included linear terms for all covariates in C1 (the all variables PS model), 2) exposure-based hdPS, and 3) bias-based hdPS. To estimate each PS-adjusted treatment effect, we used a Cox model with terms for exposure, age, sex, year, and indicators of PS decile. Therefore, we estimated 6 exposure effects in each dataset.
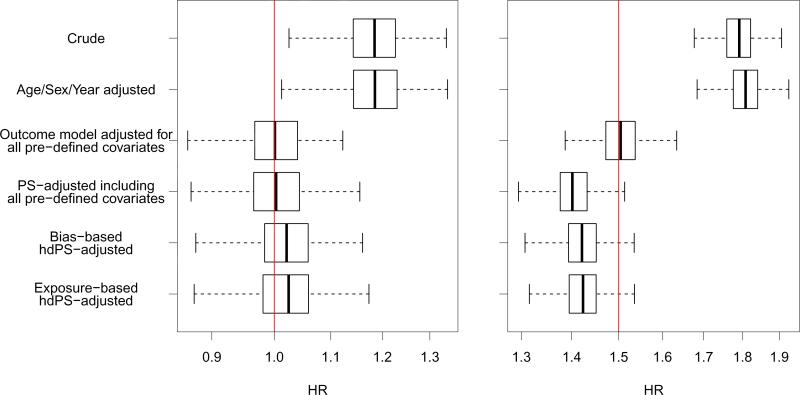
The results from this study are plotted in Figure 4. This figure displays boxplots for the distributions of exposure effect estimates across 500 simulated datasets from each of the 6 estimation approaches and 2 scenarios. As expected, the estimates from the crude model that did not adjust for confounding were positively biased for the true hazard ratio. Adjusting for age, sex, and year had little effect, but adjusting for all variables in the outcome model yielded a median hazard ratio estimate nearly identical to the truth in both simulation scenarios.
Figure 4.
Boxplots of exposure effect estimates across 500 simulated datasets. The true treatment effect is plotted with a dotted line, and the x-axis is plotted on the log-scale with axis values unlogged.
When adjusting for deciles of the all-variables PS, the exposure effect estimator was unbiased for the null treatment effect in scenario 1 (median OR [95% quantile interval]: 1.00 [0.90—1.11]) but negatively biased for the non-null effect in scenario 2 (1.40 [1.33—1.48]). This bias may be due to the non-collapsability of the hazard ratio; in the case of a non-null treatment effect, the conditional hazard ratio, as specified in the conditional proportional hazards data-generating model, will not match the marginal hazard ratio, even in the case of zero confounding (Austin et al., 2007). Furthermore, the hazard ratio conditional on PS deciles will not be equivalent to the hazard ratio conditional on covariates directly. Therefore, the PS approaches have a slightly different estimand from the specified conditional treatment effect. However, even with potential non-collapsability problems, the PS-adjusted hazard ratio resulted in a 62% reduction in bias over the crude estimate on the log scale.
The exposure effect estimates from both the bias-based and exposure-based hdPS algorithms were nearly identical and were slightly biased in both scenarios. These approaches resulted in a median reduction in bias from the crude analysis of 88% in scenariol and 70% in scenario 2. The variability of treatment effect estimates from the hdPS approaches was not increased over that of other methods considered, despite the fact that the hdPS algorithms selected covariates for inclusion in the PS in a data-driven way that resulted in a different set of variables selected in each simulation iteration.
These results indicate that in these scenarios hdPS algorithms, without any investigator input, performed nearly as well as an investigator-specified PS model that included all covariates used to generate the outcome. However, the hdPS algorithms were completely automated and required effectively no investigator input into the choice of covariates. By contrast, the correctly-specified PS model required detailed knowledge and investigator specification of all 61 covariates included. Given that complete knowledge of prognostic covariates for outcome is rarely (if ever) available, hdPS algorithms may be very useful for identifying covariates to include in the PS model in longitudinal claims data.
4 Discussion
In this paper, we have outlined steps for creating simulated cohort studies based on observed data from healthcare claims to evaluate the performance of analytic strategies in the specific data environment of interest to the researcher. We applied our simulation framework to a typical pharmacoepidemiologic study and compared the simulated and observed data. In addition, we provided an example of how this simulation framework can be used to evaluate statistical methods that were previously not able to be evaluated in simulated data. We found that our simulation framework created datasets that closely resembled the observed complex data structure, but had the advantage of an investigator-specified event rate, confounding strength, and exposure effect.
Although our framework was ideal for evaluating the hdPS algorithms, which rely on a large pool of pre-exposure healthcare claims from which to select covariates, the simulation procedure has some limitations that may lessen its utility in evaluating other methods. In our simulation framework, outcomes are generated based on a limited number of investigator-defined covariates, while in observed data, outcomes may be influenced by a much larger set of factors, both measured and unmeasured. However, because the covariate data and associations among covariates remain intact in our simulation framework, many covariates may influence outcome indirectly via their association with variables in C1, in addition to the variables that are used for directly generating outcome. Our simulation framework also provides the ability to set aside some covariates from C1 to be “unmeasured,” thereby mimicking this aspect of real-world claims data.
As with all simulation studies, the conclusions that may be drawn from a given simulation are limited to the specific data-generating scenarios explored. In particular, the performance of methods observed in simulations based on one observed cohort will likely not extend to all comparative studies in claims data. However, the approach introduced here greatly extends the types of data-generating scenarios that can be explored over ordinary simulation techniques. Therefore, despite these limitations, the simulation framework presented in this paper can be useful for evaluating methods for confounder adjustment in comparative safety and effectiveness analyses in data that mimic the complex structure of observed healthcare claims. The fact that this framework is data-based and cohort-specific will support those investigators who want to evaluate the performance of analytic strategies in data that is based on a specific motivating dataset.
Supplementary Material
Acknowledgements
The work presented in this paper was funded by a grant from the National Heart Lung and Blood Institute (RC4 HL106376).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Austin PC, Grootendorst P, Normand S-LT, Anderson GM. Conditioning on the propensity score can result in biased estimation of common measures of treatment effect: a Monte Carlo study. Statistics in Medicine. 2007;26:754–768. doi: 10.1002/sim.2618. [DOI] [PubMed] [Google Scholar]
- Breslow NE. Analysis of survival data under the proportional hazards model. International Statistical Review/Revue Internationale de Statistique. 1975:45–57. [Google Scholar]
- Brookhart MA, Sturmer T, Glynn RJ, Rassen J, Schneeweiss S. Confounding control in healthcare database research: challenges and potential approaches. Med Care. 2010;48:S114–S120. doi: 10.1097/MLR.0b013e3181dbebe3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casella G, Berger RL. Statistical Inference. 2nd ed. Duxbury Press; Pacific Grove, CA: 2001. [Google Scholar]
- Chao DL, Halloran ME, Obenchain VJ, Longini IM. FluTE, a publicly available stochastic influenza epidemic simulation model. PLoS Computational Biology. 2010;6:e1000656. doi: 10.1371/journal.pcbi.1000656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers PHC, Marx BD. Flexible smoothing with B-splines and penalties. Statistical Science. 1996:89–102. [Google Scholar]
- Elobeid MA, Padilla MA, McVie T, Thomas O, Brock DW, Musser B, Lu K, Coffey CS, Desmond RA, St-Onge MP. Missing data in randomized clinical trials for weight loss: scope of the problem, state of the field, and performance of statistical methods. PLoS One. 2009;4:e6624. doi: 10.1371/journal.pone.0006624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erenay FS, Alagoz O, Banerjee R, Cima RR. Estimatingthe unknown parameters of the natural history of metachronous colorectal cancer using discrete-event simulation. Medical Decision Making. 2011;31:611–624. doi: 10.1177/0272989X10391809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadbury GL, Xiang Q, Yang L, Barnes S, Page GP, Allison DB. Evaluating statistical methods using plasmode data sets in the age of massive public databases: an illustration using false discovery rates. PLoS Genetics. 2008;4:e1000098. doi: 10.1371/journal.pgen.1000098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray RJ. Flexible methods for analyzing survival data using splines, with applications to breast cancer prognosis. Journal of the American Statistical Association. 1992;87:942–951. [Google Scholar]
- Hurvich CM, Simonoff JS, Tsai CL. Smoothing parameter selection in nonparametric regression using an improved Akaike information criterion. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 1998;60:271–293. [Google Scholar]
- McClure DL, Glanz JM, Xu S, Hambidge SJ, Mullooly JP, Baggs J. Comparison of epidemiologic methods for active surveillance ofvaccine safety. Vaccine. 2008;26:3341–3345. doi: 10.1016/j.vaccine.2008.03.074. [DOI] [PubMed] [Google Scholar]
- Murray RE, Ryan PB, Reisinger SJ. Design and Validation of a Data Simulation Model for Longitudinal Healthcare Data. American Medical Informatics Association. 2011:1176. [PMC free article] [PubMed] [Google Scholar]
- Rassen JA, Glynn RJ, Brookhart MA, Schneeweiss S. Covariate selection in high-dimensional propensity score analyses of treatment effects in small samples. American Journal of Epidemiology. 2011;173:1404–1413. doi: 10.1093/aje/kwr001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray WA. Evaluating medication effects outside of clinical trials: new-user designs. American Journal of Epidemiology. 2003;158:915–920. doi: 10.1093/aje/kwg231. [DOI] [PubMed] [Google Scholar]
- Rolka H, Bracy D, Russell C, Fram D, Ball R. Using simulation to assess the sensitivity and specificity of a signal detection tool for multidimensional public health surveillance data. Statistics in Medicine. 2005;24:551–562. doi: 10.1002/sim.2035. [DOI] [PubMed] [Google Scholar]
- Schmidt WP, Genser B, Chalabi Z. A simulation model for diarrhoea and other common recurrent infections: a tool for exploring epidemiological methods. Epidemiology and Infection. 2009;137:644–653. doi: 10.1017/S095026880800143X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeweiss S. A basic study design for expedited safety signal evaluation based on electronic healthcare data. Pharmacoepidemiology and Drug Safety. 2010;19:858–868. doi: 10.1002/pds.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeweiss S, Avorn J. A review of uses of health care utilization databases for epidemiologic research on therapeutics. Journal of Clinical Epidemiology. 2005;58:323–337. doi: 10.1016/j.jclinepi.2004.10.012. [DOI] [PubMed] [Google Scholar]
- Schneeweiss S, Rassen JA, Glynn RJ, Avorn J, Mogun H, Brookhart MA. High-dimensional propensity score adjustment in studies of treatment effects using health care claims data. Epidemioloy (Cambridge, Mass.) 2009;20:512. doi: 10.1097/EDE.0b013e3181a663cc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom BL, Carson JL. Use of automated databases for pharmacoepidemiology research. Epidemiologic Reviews. 1990;12:87–107. doi: 10.1093/oxfordjournals.epirev.a036064. [DOI] [PubMed] [Google Scholar]
- Therneau T. survival: Survival analysis, including penalised likelihood, R package version 2.36-10. ed. 2011.
- Therneau TM, Grambsch PM. Modeling survival data: extending the Cox model. Springer Verlag; 2000. [Google Scholar]
- Therneau TM, Grambsch PM, Pankratz VS. Penalized survival models and frailty. Journal of Computational and Graphical Statistics. 2003;12:156–175. [Google Scholar]
- Vaughan LK, Divers J, Padilla MA, Redden DT, Tiwari HK, Pomp D, Allison DB. The use of plasmodes as a supplement to simulations: A simple example evaluating individual admixture estimation methodologies. Computational Statistics & Data Analysis. 2009;53:1755–1766. doi: 10.1016/j.csda.2008.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.