Abstract
Based on sequence data from cox1, cox2, ITS and LSU rDNA, it is shown that at least six species of Hyaloperonospora occur on the genus Cardamine, most of which were commonly classified under Peronospora dentariae. Based on sequences from their type hosts, Peronospora dentariae, Peronospora cardamines-laciniatae, Peronospora dentariae-macrophyllae, Peronospora malyi and Peronospora nasturtii-aquatici are combined into Hyaloperonospora, and their circumscription is clarified. Hyaloperonospora cardamines-enneaphyllos is described as a new species from Cardamine enneaphyllos. The host range of Hyaloperonospora nasturtii-aquatici, described from Nasturtium officinale, is shown to extend to various Cardamine species. Host range of species is shown to be highly diagnostic, with no overlap in their host range, but species commonly cannot be distinguished by morphology alone. Both cox1 and cox2 are confirmed to be good markers for phylogenetic species delimitation of closely related Hyaloperonospora species on Cardamine.
Keywords: Dentaria, Host range, Obligate parasites, Peronospora, Peronosporaceae
Introduction
Recent molecular phylogenetic studies showed that the genus Hyaloperonospora, established by Constantinescu and Fatehi (2002) for Peronospora species infecting Brassicaceae, Capparaceae, Cistaceae, Limnanthaceae, Resedaceae and Zygophyllaceae, comprises a high biodiversity, its species usually being highly host specific (e.g. Riethmüller et al. 2002; Choi et al. 2003; Voglmayr 2003; Göker et al. 2003, 2004, 2009a; Choi et al. 2011; Voglmayr and Göker 2011). Therefore, a narrow species circumscription as already advocated by Gäumann (1918, 1923) has been confirmed, disproving the widely applied concept of Yerkes and Shaw (1959), who classified all accessions from Brassicaceae under a single species, H. parasitica (see review in Voglmayr 2008).
Although in general the narrow species concept of Gäumann (1918, 1923) has been shown to be more appropriate, there are numerous problems in detail. Gäumann (1918, 1923) described numerous Peronospora species from Brassicaceae based on subtle morphological differences and on evidence of high host specificity obtained by cross-inoculation studies; however, as cross-inoculation studies could be performed only on a very limited number of hosts, most of his species were mainly based on host range in combination with often subtle differences in conidial size and shape. Therefore, he commonly classified accessions of various related host species under the same species if they were morphologically similar. As a result, he unfortunately did not select types for the species he described, but only a list of material examined, which especially becomes a problem if accessions from several hosts were classified under the same species. Recently, the extensive molecular phylogenetic study of Göker et al. (2009a) showed that several of his species were highly polyphyletic, as accessions from related hosts classified under the same species name by Gäumann (1918) are often not closely related, raising the problem of the correct naming of species. For this, nomenclatural and taxonomic decisions like lectotypification of heterogeneous entities are necessary. In addition, sequences have to be available for the species originating from the type host before appropriate reclassification can be achieved.
Five of these problematic taxa for which no nomenclatural decisions could yet be achieved due to reasons discussed above include Peronospora dentariae, P. cardamines-laciniatae, P. dentariae-macrophyllae, P. malyi and P. nasturtii-aquatici. Peronospora dentariae has already been described by Rabenhorst (1859) from an Italian collection from Cardamine (Dentaria) heptaphylla, a rather rare montane species confined to the Jura, south-western Alps and Apennines, but Gäumann (1918, 1923) also placed collections from various other Cardamine species in that species. Peronospora cardamines-laciniatae, P. dentariae-macrophyllae and P. nasturtii-aquatici have been described by Gäumann (1918), based on differences in conidial sizes. Peronospora cardamines-laciniatae was described for accessions from the North American Cardamine laciniata, P. dentariae-macrophyllae for accessions from the East Asian Cardamine leucantha (syn. C. macrophylla var. dasyloba), and P. nasturtii-aquatici for accessions from Nasturtium officinale. Based on his conidial measurements, Gäumann (1918) assumed that accessions from the European Cardamine bulbifera could also belong to P. cardamines-laciniatae, but later expressed strong doubts that they are conspecific (Gäumann 1923). However, Gustavsson (1959) found no significant differences in conidial size between accessions from various Cardamine species either classified as P. dentariae or P. cardamines-lacinatae and he considered them to be synonymous. Finally, Lindtner (1957) described a fifth species, Peronospora malyi, from Cardamine graeca, based on larger conidia and oospores.
The extensive investigation of Göker et al. (2009a) showed that Hyaloperonospora accessions from various Cardamine species were placed in five distinct subgroups, two of which were part of their clade 1, and three of their clade 6. However, as neither material from the type host of Peronospora dentariae, C. heptaphylla, nor from the type host of P. cardamines-laciniatae, C. laciniata, were available for study at that time, it remained unclear to which clades the names P. dentariae and P. cardamines-laciniatae should be applied. Therefore, the various Hyaloperonospora clades on Cardamine could not be properly classified. Due to these uncertainties, P. nasturtii-aquatici could also not be properly classified, as accessions from Nasturtium officinale were contained in a clade comprising accessions from various Cardamine species, e.g., C. amara, C. hirsuta and C. pratensis, which were all placed in the older P. dentariae by Gäumann (1918). No material from Cardamine graeca, the type host of P. malyi, was included in the investigation of Göker et al. (2009a), so the status of that species also remained unresolved.
To clarify these nomenclatural uncertainties, recent collections were obtained for the type hosts of all four species described from Cardamine. Four genes (ITS, LSU, cox1, cox2) were sequenced and analyzed for numerous accessions from Cardamine to evaluate the host ranges and species boundaries.
Materials and methods
Morphological analysis
Conidiophores and conidia were removed from the underneath of infected leaves, transferred to anhydrous lactic acid on a slide, carefully torn apart using forceps and needles, shortly heated using an alcohol burner and covered with a cover slip. Slides were examined and photographed using a Zeiss Axio Imager.A1 (Zeiss, Jena, Germany) microscope equipped with a Zeiss AxioCam ICc3 digital camera. Measurements are reported as maxima and minima in parentheses and the mean plus and minus the standard deviation of a number of measurements given in parentheses.
Sample sources
Information on the samples used for morphological analysis, sequencing and phylogenetic analyses is given Table 1.
Table 1.
Sources and GenBank accession numbers of Hyaloperonospora and Perofascia material used for molecular phylogenetic analyses. For institution codes of herbarium vouchers, see Thiers (2013); asterisks (*) denote sequences generated in the present study
| GenBank accession no. | ||||||||
|---|---|---|---|---|---|---|---|---|
| Taxon | Host | Geographic origins | Accession | Voucher | ITS | LSU | cox1 | cox2 |
| Hyaloperonospora arabidopsidis | Arabidopsis thaliana | Austria, Oberösterreich, St. Willibald | HV2091 | WU | EU049236 | EU054900 | – | – |
| H. barbareae | Barbarea vulgaris | Austria, Tirol, Schattwald | MG1862 | TUB 12260 | AY531395 | AY035499 | – | – |
| H. brassicae | Brassica napus | Austria, Oberösterreich, St. Willibald | HV2163 | WU | EU049248 | EU054911 | – | – |
| H. camelinae | Camelina sativa | Austria, Oberösterreich, St. Willibald | HV444 | WU | AY531456 | EU054841 | – | – |
| H. cardamines-laciniatae | Cardamine diphylla | USA, Tennessee, Knoxville | HV-PA10 | WU 32372 | KC494994* | KC494994* | KC494918* | KC494953* |
| H. cardamines-laciniatae | C. cf. diphylla | USA, Tennessee, Great Smoky Mts Natl. Park | HV2.4.P.P. | WU 32373 | KC494995* | KC494995* | KC494919* | KC494954* |
| H. cardamines-laciniatae | C. laciniata | USA, Maryland, Beltsville | HV2949 | WU 32371 | KC494996* | KC494996* | KC494920* | KC494955* |
| H. cardaminopsidis | Arabidopsis arenosa | Germany, Sachsen, Plattenthal | D23/7/97 | TUB 12446 | AY531435 | EU054829 | – | – |
| H. cheiranthi | Erysimum cheirei | Germany, Sachsen-Anhalt, Plossig | J3786/01 | TUB 12419 | AY531460 | EU054849 | – | – |
| H. dentariae | Cardamine bulbifera | Austria, Niederösterreich, Gießhübl | HV77 | WU 22896 | AY531399 | EU054837 | KC494921* | KC494956* |
| H. dentariae | C. bulbifera | Austria, Niederösterreich, Gießhübl | HV2029 | WU 32375 | KC494997* | KC494997* | – | KC494957* |
| H. dentariae | C. bulbifera | Austria, Niederösterreich, Mannersdorf/Leithageb. | HV2100 | WU 32376 | KC494998* | KC494998* | KC494922* | KC494958* |
| H. dentariae | C. bulbifera | Austria, Niederösterreich, Kaltenleutgeben | HV2106 | WU 32377 | KC494999* | KC494999* | KC494923* | KC494959* |
| H. dentariae | C. bulbifera | Austria, Niederösterreich, Tattendorf | HV2260 | WU 32378 | KC495000* | KC495000* | KC494924* | KC494960* |
| H. dentariae | C. bulbifera | Germany, Baden-Württemberg, Tübingen | MG2144 | TUB 12303 | EU049251 | EU054913 | – | – |
| H. dentariae | C. heptaphylla | Italy, Trentino, Storo, Val di Lorina | HV2927 | WU 32379 | KC495001* | KC495001* | KC494925* | KC494961* |
| H. dentariae | C. heptaphylla | Italy, Trentino, Storo, Val delle Communi | HV2928 | WU 32380 | KC495002* | KC495002* | KC494926* | KC494962* |
| H. dentariae | C. heptaphylla | Italy, Trentino, Val di Ampola | HV2930 | WU 32381 | KC495003* | KC495003* | KC494927* | KC494963* |
| H. dentariae | C. heptaphylla | Italy, Lombardia, Endine Gaiano, Valmaggiore | HV2932 | WU 32382 | KC495004* | KC495004* | KC494928* | KC494964* |
| H. dentariae | C. heptaphylla | Italy, Lombardia, Lecco, Canzo, Gajum | HV2935 | WU 32383 | KC495005* | KC495005* | KC494929* | KC494965* |
| H. dentariae | C. heptaphylla | Italy, Lombardia, Bellagio, Monte San Primo | HV2938 | WU 32384 | KC495006* | KC495006* | KC494930* | KC494966* |
| H. dentariae | C. heptaphylla | Italy, Lombardia, Valle Imagna, Valsecca | HV2941 | WU 32385 | KC495007* | KC495007* | KC494931* | KC494967* |
| H. dentariae | C. impatiens | Germany, Baden-Württemberg, Tübingen | HV855 | WU 32386 | KC495008* | KC495008* | - | KC494968* |
| H. dentariae | C. impatiens | Austria, Niederösterreich, Gießhübl | HV2174 | WU 32387 | – | – | – | KC494969* |
| H. dentariae | C. impatiens | Austria, Niederösterreich, Gießhübl | HV2289 | WU 32388 | KC495009* | KC495009* | KC494932* | KC494970* |
| H. dentariae | C. impatiens | Austria, Tirol, Steeg | GLM46902 | EU049231 | EU054896 | – | – | |
| H. dentariae | C. impatiens | Austria, Tirol, Schattwald | MG1840 | TUB 12261 | AY531400 | AY035500 | – | – |
| H. dentariae | C. impatiens | Germany, Baden-Württemberg, Tübingen | MG1939 | TUB 12282 | AY531397 | AY272000 | – | – |
| H. dentariae | C. pentaphyllos | Austria, Kärnten, Ferlach, Tscheppaschlucht | HV2334 | WU 32389 | KC495010* | KC495010* | KC494933* | KC494971* |
| H. dentariae | C. pentaphyllos | Italy, Lombardia, Endine Gaiano, Valmaggiore | HV2933 | WU 32390 | KC495011* | KC495011* | KC494934* | KC494972* |
| H. dentariae | C. pentaphyllos | Italy, Friuli, Val di Resia, Lischiazze | HV2944 | WU 32391 | KC495012* | KC495012* | KC494935* | KC494973* |
| H. dentariae-macrophyllae | C. leucantha | Korea, Chuncheon | SMK17273 | KUS-F 17273 | AY210990 | KC495013* | KC494936* | KC494974* |
| H. dentariae-macrophyllae | C. leucantha | Korea, Gangneung | SMK17298 | KUS-F 17298 | AY210991 | KC495014* | KC494937* | KC494975* |
| H. dentariae-macrophyllae | C. leucantha | Korea, Hongcheon | SMK17322 | KUS-F 17322 | AY210992 | KC495015* | KC494938* | KC494976* |
| H. dentariae-macrophyllae | C. leucantha | Korea, Gangneung | SMK17539 | KUS-F 17539 | AY210993 | KC495016* | KC494939* | KC494977* |
| H. cardamines-enneaphyllos | C. enneaphyllos | Austria, Niederösterreich, Gießhübl | HV2025 | WU 32392 | KC495017* | KC495017* | – | – |
| H. cardamines-enneaphyllos | C. enneaphyllos | Austria, Niederösterreich, Mannersdorf/Leithageb. | HV2099 | WU 32393 | KC495018* | KC495018* | KC494940* | KC494978* |
| H. cardamines-enneaphyllos | C. enneaphyllos | Austria, Niederösterreich, Gießhübl | HV2105 | WU 32394 | KC495019* | KC495019* | – | KC494979* |
| H. cardamines-enneaphyllos | C. enneaphyllos | Austria, Steiermark, Gröbming | HV2125 | WU 32395 | KC495020* | KC495020* | KC494941* | KC494980* |
| H. cardamines- enneaphyllos | C. enneaphyllos | Italy, Trentino, Storo, Val delle Communi | HV2929 | WU 32396 | KC495021* | KC495021* | KC494942* | KC494981* |
| H. hesperidis | Hesperis matronalis | Germany, Sachsen-Anhalt, Kloster Gröningen | J554/01 | TUB 12438 | EU049214 | EU054859 | – | – |
| H. isatidis | Isatis tinctoria | Germany, Sachsen-Anhalt, Rollsdorf | J928/01 | TUB 12429 | AY531443 | EU054851 | – | – |
| H. lobulariae | Lobularia maritima | Germany, Sachsen-Anhalt, Arendsee | J3454/01 | TUB 12414 | AY531410 | EU054856 | – | – |
| H. lunariae | Lunaria rediviva | Austria, Niederösterreich, Lilienfeld | HV362 | WU22867 | AY531401 | EU054891 | – | – |
| H. malyi | Cardamine graeca | Greece, Korfu, Episkepsis | HV2895 | WU 32397 | KC495022* | KC495022* | KC494943* | KC494982* |
| H. nasturtii-aquatici | C. amara | Austria, Oberösterreich, Raab | HV2094 | WU 32398 | KC495023* | KC495023* | – | KC494983* |
| H. nasturtii-aquatici | C. amara | Czech Republic, Krusne Hory, Krystofovy Hamry | D25/5/99 | TUB 12448 | AY531420 | EU054833 | – | – |
| H. nasturtii-aquatici | C. flexuosa | Germany, Baden-Württemberg, Tübingen, Lustnau | HV834 | WU 32399 | KC495024* | KC495024* | KC494944* | KC494984* |
| H. nasturtii-aquatici | C. flexuosa | Austria, Oberösterreich, St. Willibald | HV2018 | WU 32400 | KC495025* | KC495025* | KC494945* | KC494985* |
| H. nasturtii-aquatici | C. flexuosa | Austria, Tirol, St. Anton am Arlberg | GLM46918 | EU049230 | EU054895 | – | – | |
| H. nasturtii-aquatici | C. flexuosa | Germany, Sachsen-Anhalt, Koßweda | J121/01 | TUB 12434 | EU049212 | EU054850 | – | – |
| H. nasturtii-aquatici | C. hirsuta | Austria, Wien, Landstraße, Botanischal Garden | HV2030 | WU 32401 | KC495026* | KC495026* | KC494946* | KC494986* |
| H. nasturtii-aquatici | C. hirsuta | Austria, Oberösterreich, St. Willibald | HV2092 | WU 32402 | KC495027* | KC495027* | KC494947* | KC494987* |
| H. nasturtii-aquatici | C. hirsuta | USA, Maryland, Greenbelt | HV-PA3 | WU 32403 | KC495028* | KC495028* | KC494948* | KC494988* |
| H. nasturtii-aquatici | C. hirsuta | Germany, Sachsen-Anhalt, Tanne | J535/01 | TUB 12462 | EU049259 | EU054919 | – | – |
| H. nasturtii-aquatici | C. hirsuta | Germany, Nordrhein-Westfalen, Wuppertal | MG1821 | TUB 12225 | AY531421 | AY035505 | – | – |
| H. nasturtii-aquatici | C. pratensis | Germany, Baden-Württemberg, Tübingen, Lustnau | HV796 | WU 32404 | KC495029* | KC495029* | KC494949* | KC494989* |
| H. nasturtii-aquatici | C. pratensis | Germany, Baden-Württemberg, Tübingen | HV799 | WU 32405 | KC495030* | KC495030* | KC494950* | KC494990* |
| H. nasturtii-aquatici | C. pratensis | Germany, Baden-Württemberg, Tübingen, Lustnau | HV829 | WU 32406 | – | – | – | KC494991* |
| H. nasturtii-aquatici | C. pratensis | Austria, Niederösterreich, Mariensee | HV2400 | WU 32407 | KC495031* | KC495031* | KC494951* | KC494992* |
| H. nasturtii-aquatici | C. pratensis | Germany, Sachsen-Anhalt, Großpörthen | J145/01 | TUB 12456 | EU049258 | EU054918 | – | – |
| H. nasturtii-aquatici | C. pratensis | Germany, Baden-Württemberg, Tübingen | MG1820 | TUB 12221 | EU049205 | AY035504 | – | – |
| H. nasturtii-aquatici | C. pratensis | Germany, Baden-Württemberg, Niedernhall | MG1885 | TUB 12274 | AY531417 | EU054825 | – | – |
| H. nasturtii-aquatici | Nasturtium officinale | Germany, Sachsen-Anhalt, Sülldorf | J3493/01 | TUB 12415 | AY531419 | EU054870 | – | – |
| H. nasturtii-aquatici | N. officinale | Sweden, Gotland, Visby | GLM50769 | EU049233 | EU054897 | – | – | |
| H. nesliae | Neslia paniculata | Austria, Niederösterreich, Theresienfeld | HV203 | WU22913 | AY531458 | EU054892 | – | – |
| H. praecox | Draba verna | Austria, Wien, Botanical Garden | HV2144 | WU30285 | EU049239 | EU054903 | – | – |
| H. rorippae-islandicae | Rorippa palustris | Austria, Tirol, Steeg | GLM46904 | EU049235 | EU054899 | – | – | |
| H. sisymbrii-sophiae | Descurainia sophia | Austria, Niederösterreich, Hundsheim | HV276 | WU22928 | AY531430 | EU054910 | – | – |
| H. teesdaliae | Teedalia nudicaulis | Germany, Sachsen, Zschepa | J1186/01 | TUB 12406 | AY531415 | EU054860 | – | – |
| H. thlaspeos-arvensis | Thlaspi arvense | Austria, Oberösterreich, St. Willibald | HV762 | WU | AY531445 | EU054890 | – | – |
| H. thlaspeos-perfoliati | Noccaea caerulescens | Czech Republic, Krusne Hory, Vejprty | D24/4/99 | TUB 12444 | AY531433 | EU054831 | – | – |
| H. thlaspeos-perfoliati | N. caerulescens | Germany, Sachsen, Obercunnersdorf | J450/01 | TUB 12422 | AY531436 | EU054857 | – | – |
| Perofascia lepidii | Lepidium ruderale | Germany, Sachsen-Anhalt, Röden | J2068/01 | TUB 12409 | AY531467 | EU054855 | – | – |
| Perofascia lepidii | L. ruderale | Germany, Sachsen-Anhalt, Wendelstein | J3189/01 | TUB 1241 | AY531446 | EU054854 | – | – |
| Peronospora aubrietae | Aubrieta sp. | Sweden, Gotland, Visby | GLM50765 | EU049228 | EU054893 | – | – | |
| Peronospora crispula | Reseda lutea | Austria, Burgenland, Apetlon | HV1028 | WU | AY531437 | EU054847 | – | – |
| Peronospora lepidii-sativi | Lepidium draba | Austria, Niederösterreich, Guntramsdorf | HV246 | WU22908 | AY531463 | EU054889 | – | – |
| Peronospora rumicis | Rumex acetosa | Austria, Oberösterreich, Kopfing | HV312 | WU22925 | AY198287 | KC495032* | KC494952* | KC494993* |
DNA extraction, PCR and sequencing
For DNA extraction, infected dry host tissue was placed in 2-ml reaction tubes together with six sterile 2-mm glass beads and ground in a Retsch 200 mixer mill for 10 min at a frequency of 30 Hz. DNA was extracted using the modified CTAB protocol described in Riethmüller et al. (2002).
A ca. 2,200-bp-long fragment containing partial nuSSU-ITS-LSU rDNA was amplified using primers DC6 (Bonants et al. 1997) and LR6-O (Riethmüller et al. 2002) or LR6-O1 (designed here; 5′ CGCATCGCCAGACGAGC 3′). In cases where no product could be obtained, ITS and LSU were separately amplified using primers DC6 and ITS4 (White et al. 1990) and LR0R (Vilgalys and Hester 1990) and LR6-O1, respectively. For cycle sequencing, primers ITS5-P (designed here; 5′ GGAAGGTGAAGTCGTAACAAGG 3′), ITS4, LR0R and LR6-O were used. For the mitochondrial cytochrome c oxidase subunit I (cox1) sequences, primers Oom-CoxI-lev-up and Oom-CoxI-lev-lo (Robideau et al. 2011) were used for amplification and cycle sequencing; the cytochrome c oxidase subunit II (cox2) was amplified and cycle-sequenced with the forward and reverse primers of Hudspeth et al. (2000). The PCR products were purified using an enzymatic PCR cleanup (Werle et al. 1994) according to the protocol of Voglmayr and Jaklitsch (2008). DNA was cycle-sequenced using the ABI PRISM Big Dye Terminator Cycle Sequencing Ready Reaction Kit v.3.1 (Applied Biosystems, Warrington, UK) and an automated DNA sequencer (AB 3730xl Genetic Analyzer, Applied Biosystems).
Phylogenetic analysis
To reveal the phylogenetic position of the Hyaloperonospora clades on Cardamine, an ITS-LSU data matrix was used. For this, the new sequences generated during the current study were combined with those of accessions from Cardamine and Nasturtium included in Göker et al. (2009a); in addition, sequences of representative species from throughout the tree of Göker et al. (2009a) were added (Table 1), with Perofascia lepidii as outgroup to root the trees. All alignments were produced with Muscle v.3.6 (Edgar 2004).
For evaluation of species status, a multi-gene analysis of all four genes (ITS, LSU, cox1, cox2) was performed; due to lack of data available for other species, only accessions of all six clades from Cardamine were included, with Peronospora rumicis as outgroup to root the tree. Prior to phylogenetic analyses, the approach of Wiens (1998) was applied to test for significant levels of localized incongruence among the two gene partitions, using the level of bootstrap support (Sung et al. 2007). For this, the 70 % maximum parsimony (MP) bootstrap trees of the individual gene regions (ITS-LSU, cox1, cox2) were compared, which were calculated using the same parameters as for the combined analysis given below. No topological conflicts were observed between these bootstrap trees of genes, indicating the absence of significant incongruence and combinability of the matrices (Wiens 1998).
Maximum parsimony (MP) analysis was performed with PAUP* v.4.0 b10 (Swofford 2002), using 1,000 replicates of heuristic search with random addition of sequences and subsequent TBR branch swapping (MULTREES option in effect, COLLAPSE=MINBRLEN, steepest descent option not in effect), each replicate limited to 1 million rearrangements. All molecular characters were unordered and given equal weight; analyses were performed with gaps treated as missing data. Bootstrap analysis with 1,000 replicates was performed in the same way, but using 5 rounds of random sequence addition and subsequent branch swapping during each bootstrap replicate.
For maximum likelihood (ML) and Bayesian analyses, the well-known general time reversible model (GTR) was selected by Modeltest 3.6 (Posada and Crandall 1998) using the Akaike information criterion for all three genes; with a gamma distribution for the ITS-LSU (GTR+G) and with invariant sites and gamma distribution for the remaining sites for cox1 and cox2 (GTR+I+G). In the combined analyses of all gene regions, substitution parameters were estimated separately for each region. For ML analyses, 500 rounds of random addition of sequences as well as 500 fast bootstrap replicates were computed with RAxML (Stamatakis 2006) as implemented in raxmlGUI 0.95 (Silvestro and Michalak 2012) using the GTRGAMMA and GTRCAT substitution models, respectively. For Bayesian analyses using MrBayes v.3.1.2 (Huelsenbeck and Ronquist 2001), three parallel runs of four incrementally heated simultaneous Markov chains were performed over 1 million generations from which every 100th tree was sampled in each run. The first 500 trees were discarded, and a 90 % majority rule consensus of the remaining trees was computed to obtain posterior probabilities. The final matrix was deposited in TreeBASE (http://www.treebase.org) and is available under http://purl.org/phylo/treebase/phylows/study/TB2:S14017.
Results
After the exclusion of excessive leading and trailing gap regions and large insertions present in some single sequences, 2,172 characters were included in the ITS-LSU analyses, of which 428 were parsimony informative. MP analyses revealed 2,106 MP trees of 1,213 steps which were identical except for topologies within species, a polytomy containing H. lobulariae, H. brassicae and H. lunariae, a polytomy containing H. hesperidis and the H. cheiranthi–H. sisymbrii-sophiae clade, and a polytomy containing H. cardaminopsidis and the H. arabidopsidis–H. praecox clade. Backbone topology of the deeper unsupported nodes of the ML tree obtained with RAxML differed from the MP strict consensus tree (data not shown), but topologies of supported nodes were the same as in the MP analysis. One of the 2,106 MP trees is shown as phylogram in Fig. 1, with MP and ML bootstrap support above 60 % and posterior probabilities above 90 % given at first, second and third positions above/below the branches, respectively.
Fig. 1.
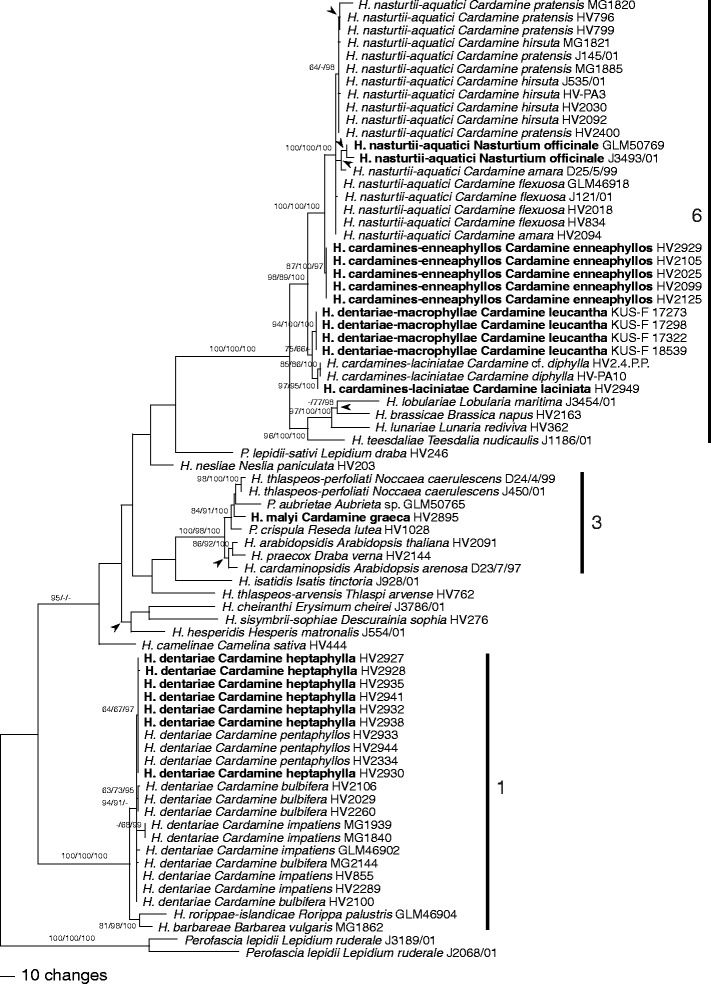
Phylogram showing 1 of 2,106 MP trees inferred from the complete ITS (ITS1, 5.8S rDNA, and ITS2)-LSU alignment with PAUP and rooted with Perofascia; arrowheads denoting branches/nodes collapsed in the strict consensus tree of all MP trees. MP and ML bootstrap support above 60 % and posterior probabilities above 90 % are given at first, second, and third positions, respectively, above/below the branches. Hyaloperonospora specimens from type hosts of the six species from Cardamine are shown in bold. Clade numbers correspond to those of Göker et al. (2009a)
The combined matrix contained 3,412 characters (2,150 from ITS-LSU, 681 from cox1, 581 from cox2), from which 374 were parsimony informative. MP analyses revealed 54 MP trees of 885 steps which were identical except for topologies within species. Tree topology of the ML tree obtained with RAxML was fully compatible with the MP strict consensus tree (data not shown). One of the 54 MP trees is shown as phylogram in Fig. 2, with MP and ML bootstrap support above 60 % and posterior probabilities above 90 % given at first, second, and third positions above/below the branches, respectively.
Fig. 2.
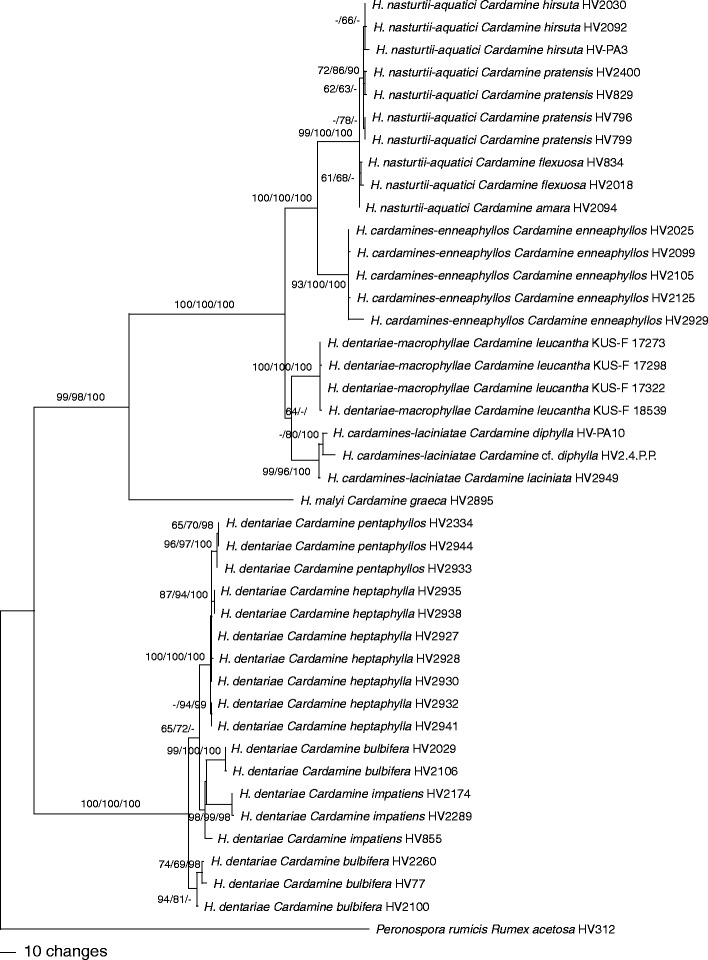
Phylogram showing 1 of 54 MP trees inferred from the combined ITS-LSU-cox1-cox2 sequence data matrix. MP and ML bootstrap support above 60 % and posterior probabilities above 90 % are given at first, second, and third positions, respectively, above/below the branches
In the ITS-LSU analyses (Fig. 1), the accessions from Cardamine were contained in three distinct clades of Göker et al. (2009a), one consisting of H. dentariae within clade 1, a second containing H. malyi within clade 3, and a third highly supported monophyletic clade containing H. cardamines-laciniatae, H. dentariae-macrophyllae, H. cardamines-enneaphyllos and H. nasturtii-aquatici within clade 6. In both ITS-LSU and combined analyses, all species were highly supported. Each Cardamine species harboured only a single Hyaloperonospora species. The accessions from the previously not sampled C. enneaphyllos formed a distinct clade representing a new species described as H. cardamines-enneaphyllos below. Hyaloperonospora cardamines-enneaphyllos, H. malyi and H. dentariae-macrophyllae each contained only accessions from a single host species, whereas within H. cardamines-laciniatae, H. dentariae and H. nasturtii-aquatici accessions from several host species were placed. Within H. dentariae and H. nasturtii-aquatici, some substructure was observed in the combined analyses according to the host species, indicating the presence of host-specific lineages within species. Within the H. dentariae clade, the accessions from C. heptaphylla and C. pentaphyllos formed a highly supported monophylum, the latter again forming a highly supported subclade; however, accessions from C. bulbifera and C. impatiens were not contained in monophyletic lineages. Within H. nasturtii-aquatici, the accessions from C. hirsuta and from C. flexuosa each formed weakly supported subclades, the latter being included in a moderately supported clade together with the accessions from C. pratensis.
Taxonomy
As a result of the molecular phylogenetic investigations, H. cardamines-enneaphyllos is described as a new species. In addition, P. cardamines-laciniatae, Peronospora dentariae, P. dentariae-macrophyllae, P. malyi and P. nasturtii-aquatici are combined into Hyaloperonospora.
Hyaloperonospora cardamines-enneaphyllos Voglmayr, sp. nov. Fig. 3
Fig. 3.
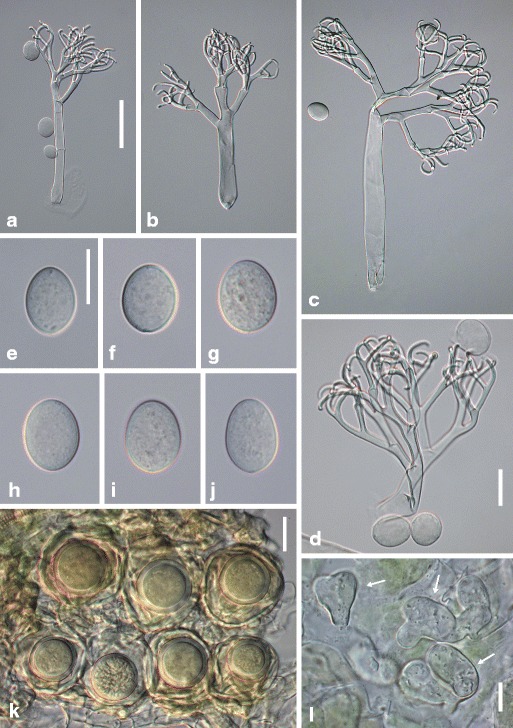
Hyaloperonospora cardamines-enneaphyllos. a–c conidiophores; d ultimate branchlets; e–j conidia; k oogonia and oospores in host tissue; l three vesicular-lobate haustoria (arrows). Sources: (a, d) WU 32395; (b, c, g–j) WU 32393; (e, f, k, l) holotype WU 32392. Scale bars (a–c) 50 μm, (d–l) 20 μm
Mycobank MB 803659
Etymology: Referring to its host, Cardamine enneaphyllos.
Infection commonly systemic, more rarely localized, when systemic whole plants or leaves stunted, chlorotic, dwarfed, when localized producing polyangular to confluent lesions without distinct margins. Down whitish, consisting of scattered conidiophores to dense and felt-like. Haustoria lobate, (9−)12−19(−21) μm long, (12−)15–26(−33) μm wide (n = 26), filling the host cell partly to almost completely. Conidiophores hyaline, stout, straight to slightly sinuous, (135−)200–325(−420) μm long; trunk straight, usually collapsed (flat), (70−)105–200(−290) μm long (n = 44), variable in width, 9–17 μm wide below the first branch, 13–25 μm wide above the base, with often somewhat swollen base; callose plugs absent; upper part monopodially or subdichotomously branched 4–5 times. Ultimate branchlets in pairs or rarely single, flexuous, sigmoid to strongly involuted, (9−)12−21(−30) μm long, 2–3 μm wide at the base (n = 66), apex obtuse. Conidia hyaline, subglobose, ellipsoidal to ovate, (17−)20.5–25(−29) μm long, (15−)17–20(−22.5) μm wide, mean 23.3 × 18.0 μm, l/w ratio (1.04−)1.16–1.32(−1.45) (n = 215), greatest width median or submedian, base and tip round; pedicel absent in most conidia but a scar visible at the point of attachment; producing germ tubes. Resting organs oogonia subglobose to irregular, pale yellowish, (41−)49–59(−64) μm diam., wall smooth, 1.5−4 μm thick (n = 43); oospores aplerotic, globose, (25−)28–33(−36) μm diam., wall 2.5–3.5 μm thick (n = 43), smooth.
Habitat: On living leaves and stems of Cardamine enneaphyllos (Brassicaceae).
Holotype: Austria, Niederösterreich, Distr. Mödling, Comm. Gießhübl, deciduous forest ca 700 m SW parking lot, 380 m, map grid 7963/1, 27 Apr. 2003, H. Voglmayr HV2025, WU 32392.
Additional specimens examined: Austria, Niederösterreich, Distr. Mödling, Comm. Gießhübl, deciduous forest ca 700 m SW parking lot, 380 m, map grid 7963/1, 18 Apr. 2004, H. Voglmayr HV2105, WU 32394. Distr. Bruck/Leitha, comm. Mannersdorf/Leitha, Schweinsgraben SE Mannersdorf, dediduous forest, 260 m, map grid 8063/3, 17 Apr. 2004, H. Voglmayr HV2099, WU 32393. Steiermark, Distr. Liezen, Comm. Gröbming, W Winkl, forest between Stickler and Sticklereck, 1200 m, map grid 8549/1, 10 June 2004, H. Voglmayr HV2125, WU 323935. Italy, Trentino, SE Storo, Val di Lorina, Val delle Communi, 25 May 2012, H. Voglmayr & I. Greilhuber HV2929, WU 32396.
Comments: Spore sizes of H. cardamines-enneaphyllos correspond to those recorded for the eastern Asian H. dentariae-macrophyllae (av. 23.3 × 18.0 μm, Gäumann 1918), under which name accessions from C. enneaphyllos have been classified by Săvulescu and Rayss (1930), Lindtner (1957) and Kochman and Majewski (1970). Molecular phylogenies reveal H. cardamines-enneaphyllos to be related to but clearly distinct from H. dentariae-macrophyllae, which is closest relative of the North American H. cardamines-laciniatae. The closest relative of H. cardamines-enneaphyllos, H. nasturtii-aquatici, differs in smaller spore sizes (av. lengths 17.3–20.5, av. widths 15–17.5; see Table 2).
Table 2.
Conidial measurements of Hyaloperonospora accessions from various Cardamine species and from Nasturtium officinale
| Species | Host | Length range (μm) | Width range (μm) | Mean length (μm) | Mean width (μm) | mean l/w ratio | Reference |
|---|---|---|---|---|---|---|---|
| H. cardamines-laciniatae | C. laciniata | (12-)18–22(−28) | (11-)14–18(−21) | 19.7 | 16.2 | 1.21 | Gäumann (1918) |
| C. diphylla | (18.5-)19.5–24(−29) (n = 54) | (16.5-)17.5–20(−22) (n = 54) | 21.8 | 18.7 | 1.16 | Present study (WU 32373) | |
| H. dentariae | C. heptaphylla | (11-)15–19(−23) | (9-)11–14(−18) | 16.5 | 13.4 | 1.23 | Gäumann (1918) |
| C. heptaphylla | 15–21 | 14–17 | – | – | – | Gustavsson (1959) | |
| C. heptaphylla | (15.5-)17–19(−21) (n = 55) | (13-)13,5–15,5(−17) (n = 55) | 18.0 | 14.5 | 1.24 | Present study (WU 32380) | |
| C. bulbifera | (14.5-)16–19(−21.5) (n = 73) | (13-)13.5–15.5(−18) (n = 73) | 17.6 | 14.6 | 1.20 | Present study (WU 32376) | |
| C. bulbifera | – | – | 19.3 | 16.0 | 1.20 | Gustavsson (1959) | |
| C. impatiens | (12.5-)14.5–17(−19.5) (n = 71) | (11-)12–13.5(−14.5) (n = 71) | 15.6 | 12.8 | 1.22 | Present study (WU 32388) | |
| C. pentaphyllos | (15.5-)18–21.5(−24.5) (n = 45) | (13.5-)15–17.5(−19.5) (n = 45) | 19.7 | 16.2 | 1.22 | Present study (WU 32391) | |
| H. dentariae-macrophyllae | C. leucantha | (16-)21–26(−29) | (12-)18–21(−24) | 23.1 | 18.8 | 1.23 | Gäumann (1918) |
| C. leucantha | (19-)22.5–25(−28) | (15-)17–21.5(−25) | 23.5 | 19.1 | 1.22 | Present study (KUS-F 17273) | |
| H. cardamines-enneaphyllos | C. enneaphyllos | (20.5-)22.5–24.5(−25) (n = 32) | (15,5-)16.5–19.5(−21.5) (n = 32) | 23.3 | 18.0 | 1.30 | Present study (WU 32392) |
| C. enneaphyllos | (19-)22–26.5(−29) (n = 44) | (16.5-)17.5–21(−22.5) (n = 44) | 24.2 | 19.3 | 1.26 | Present study (WU 32393) | |
| C. enneaphyllos | (17-)19.5–22.5(−25.5) (n = 73) | (16-)17–19(−20) (n = 73) | 21.2 | 17.9 | 1.18 | Present study (WU 32395) | |
| C. enneaphyllos | (19-)20.5–25(−28.5) (n = 66) | (15-)16.5–19.5(−22.5) (n = 66) | 22.7 | 18.0 | 1.26 | Present study (WU 32396) | |
| C. enneaphyllos | (16-)19–25(−28) | (12-)15–19(−24) | 22.0 | 18.3 | 1.20 | Săvulescu & Rayss (1930) | |
| C. enneaphyllos | – | – | 25.0 | 20.2 | 1.24 | Lindtner (1957) | |
| H. malyi | C. graeca | (16-)20–28(−33) | (14-)18–22(−25) | 26.4 | 21.2 | 1.24 | Lindtner (1957) |
| H. nasturtii-aquatici | N. officinale | ca. 16–27a | ca. 13–21a | 20.3 | 17.1 | 1.19 | Gäumann (1918) |
| N. officinale | 15–25 | 14–21 | – | – | – | Kochman and Majewski (1970) | |
| C. amara | (15.5-)18–22(−25) (n = 86) | (14-)16–19(−22) (n = 86) | 20.5 | 17.5 | 1.12 | Present study (WU 32398) | |
| C. amara | – | – | 19.1 | 15.6 | 1.23 | Gustavsson (1959) | |
| C. flexuosa | (14-)16–19(−20) (n = 39) | (12.5-)14–16 (−17) (n = 39) | 17.6 | 15.0 | 1.17 | Present study (WU 32400) | |
| C. hirsuta | (15-)16.5–19(−21.5) (n = 70) | (13-)14–16(−18) (n = 70) | 17.7 | 15.2 | 1.16 | Present study (WU 32402) | |
| C. hirsuta | – | – | 17.3 | 15.7 | 1.10 | Gustavsson (1959) | |
| C. pratensis | (15.5-)17.5–20(−21) (n = 59) | (14-)15–17(−18) (n = 59) | 18.8 | 16.0 | 1.17 | Present study (WU 32406) |
Hosts in bold denote type hosts. Note the variability of sporangial sizes commonly observed within collections from the same host, within the same Hyaloperonospora species, and the overlap in size range between most Hyaloperonospora species
arange measurements listed in Gäumann (1918: 528) erroneous; approximate range was estimated from spore size line graphs (Gäumann 1918: 466)
Hyaloperonospora cardamines-laciniatae (Gäum.) Voglmayr, comb. nov.
Basionym: Peronospora cardamines-laciniatae Gäum., Beih. bot. Zbl., Abt. 1 35(1): 523. 1918.
Mycobank MB 803660
Confirmed hosts: Cardamine laciniata, C. diphylla
Distribution: North America
Hyaloperonospora dentariae (Rabenh.) Voglmayr, comb. nov.
Basionym: Peronospora dentariae Rabenh., Fungi Eur. 86. 1859.
Mycobank MB 803661
Confirmed hosts: Cardamine heptaphylla, C. pentaphyllos, C. impatiens, C. bulbifera
Distribution: Europe
Hyaloperonospora dentariae-macrophyllae (Gäum.) Voglmayr, Y.J. Chin & H.D. Shin, comb. nov.
Basionym: Peronospora dentariae-macrophyllae Gäum., Beih. bot. Zbl., Abt. 1 35(1): 523. 1918.
Mycobank MB 803662
Confirmed hosts: Cardamine leucantha
Distribution: East Asia
Hyaloperonospora malyi (Lindtner) Voglmayr, comb. nov.
Basionym: Peronospora malyi Lindtner, Glasn. Muz. Srpsk. Zeml. (Bull. Mus. Hist. Nat. Pays Serbe), Ser. B, 9: 141. 1957.
Mycobank MB 803663
Confirmed hosts: Cardamine graeca
Distribution: Southern Europe
Hyaloperonospora nasturtii-aquatici (Gäum.) Voglmayr, comb. nov.
Basionym: Peronospora nasturtii-aquatici Gäum., Beih. bot. Zbl., Abt. 1 35(1): 528. 1918.
Mycobank MB 803664
Confirmed hosts: Nasturtium officinale, Cardamine amara, C. flexuosa, C. hirsuta, C. pratensis
Distribution: Europe, Asia, North America
Discussion
Phylogenetic analyses of the ITS-LSU data are largely congruent with the results of Göker et al. (2009a) concerning the placement of Hyaloperonospora accessions from Cardamine (Fig. 1) within their clades 1 and 6. However, there are differences in the deeper unsupported nodes of the tree which may be due to the more extensive taxon and accession sampling of Göker et al. (2009a). This is to be expected, as the deeper nodes of trees inferred from ITS-LSU data mostly lack support. Lack of support for deeper nodes in ITS as well as LSU phylogenies has also been reported from other downy mildew genera like Peronospora (e.g., Voglmayr 2003; Choi et al. 2007; García Blázquez et al. 2008; Göker et al. 2009b) or Plasmopara (e.g., Voglmayr et al. 2004; Voglmayr and Constantinescu 2008; Voglmayr and Thines 2007).
In the combined analyses, resolution as well as support within the main species clades is improved, showing that cox1 and cox2 add substantial resolution to the tree. cox1, chosen as barcoding locus for higher animals and considered to be the primary barcoding marker for organisms unless shown to be unsuitable (http://www.barcodeoflife.org), has also been shown to be an appropriate barcoding locus for oomycetes (Robideau et al. 2011), which is confirmed in the current study. cox2 shows similarly good resolution and may serve as an accessory barcoding marker; it also has some advantages over cox1, as it usually amplifies better especially in cases of low DNA quantity or older degraded samples (as also shown in Telle and Thines 2008), and thus cox2 sequences are available for many more species.
The results of the current study clearly show that the classification of Hyaloperonospora accessions from Cardamine proposed by Gäumann (1918, 1923) has to be substantially revised. In his classification, accessions from Cardamine heptaphylla, C. impatiens, C. amara, C. flexuosa, C. hirsuta and C. pratensis were placed in Peronospora dentariae. However, these are actually contained within two distinct clades in the molecular phylogenies, the first two being included in clade 1 as H. dentariae, whereas the latter four are part of clade 6 of Göker et al. (2009a), belonging to H. nasturtii-aquatici (Figs. 1, 2). Likewise, his P. cardamines-laciniatae contained accessions from C. bulbifera and C. laciniata which are phylogenetically also contained within clade 1 and 6, respectively, the former being placed in H. dentariae. This is understandable, as the conidial sizes and shapes, which were primary criteria for species classification, are similar between most of these phylogenetically distinct entities (Table 2), and other features like conidiophore morphology are highly variable and also unsuitable for morphological distinction (Gäumann 1918, own observations). In addition, conidial sizes show some variability between different collections of the same species (Table 2) and may be dependent on host, environmental conditions, and ontogenetic state of the collection, which has been recently confirmed for Pseudoperonospora cubensis (Runge et al. 2012). The inability to distinguish them morphologically led Gustavsson (1959) to synonymize P. cardamines-laciniatae with P. dentariae. On the other hand, Lindtner (1957) accepted the classification of Gäumann, and distinguished the various species by their deviating mean lengths and widths. Based on conidial sizes, Săvulescu and Rayss (1930) classified accessions from C. enneaphyllos under P. dentariae-macrophyllae, which was subsequently followed by Lindtner (1957) and Kochman and Majewski (1970).
The data on conidial sizes (Table 2) show that most phylogenetic species occurring on Cardamine cannot be distinguished by morphology alone, at least not by characters usually used for species identification; they should be considered cryptic species which is quite common amongst downy mildews (Voglmayr 2008). However, the hosts are diagnostic for the different Hyaloperonospora species investigated in the present study. Within Hyaloperonospora, usually only a single species is observed on a given host species. In exceptional cases where more than one Hyaloperonospora species has been observed to occur on a single host species, they were morphologically quite distinct (Voglmayr and Göker 2011). No such case of overlapping host range has yet been observed in Hyaloperonospora on Cardamine despite extensive sampling. However, reliable species identification is nowadays only possible by sequence data, especially in groups which are incompletely sampled.
Remarkably, H. nasturtii-aquatici contains accessions from two distinct genera, Cardamine and Nasturtium. Long considered to be closely related to Rorippa and sometimes even classified within that genus, it has been shown that Nasturtium actually is the closest relative of Cardamine (Al-Shehbaz and Price 1998; Franzke et al. 1998). The conidial size recorded for Hyaloperonospora accessions from Nasturtium officinale (range 15–27 × 13–21 μm, av. 20 × 17 μm; see Table 2) fits well the measurements recorded for accessions from Cardamine species placed within H. nasturtii-aquatici (range 14-25 × 12.5–22 μm, av. lengths 17.3–20.5 μm, av. widths 15–17.5 μm; see Table 2).
Some phylogenetic substructure was observed within both H. dentariae and H. nasturtii-aquatici, where subclades were formed according to the host species (Fig. 2), which indicates some host specificity also within species. This is evidence for active evolutionary radiation, which may result in speciation events, and some of these subclades may actually represent taxonomically separable entities on the subspecific level. Therefore, these species may be good candidates for population genetic investigations on a larger scale to investigate host specificity, gene flow, and speciation processes in detail. Remarkably, similar phylogenetic substructures were also observed in Albugo on various Cardamine species, which either formed phylogenetically distinct species or showed distinct substructures within a species according to their hosts (Ploch et al. 2010). This may imply that a common evolutionary pattern may be present in both Albugo and Hyaloperonospora on Cardamine, and a comparison of their divergence patterns could give some more general insights in the evolution of obligatory parasitic oomycetes in future studies.
Acknowledgment
Financial support by the Austrian Science Fund (FWF; project P22739-B20) to HV is gratefully acknowledged. YJC appreciates support by the Alexander-von-Humboldt Foundation.
References
- Al-Shehbaz IY, Price RA. Delimitation of the genus Nasturtium (Brassicaceae) Novon. 1998;8:124–126. doi: 10.2307/3391978. [DOI] [Google Scholar]
- Bonants P, van Hagenaar-de Weerdt M, Gent-Pelzer M, Lacourt I, Cooke D, Duncan J. Detection and identification of Phytophthora fragariae Hickman by the polymerase chain reaction. Europ J Plant Pathol. 1997;103:345–355. doi: 10.1023/A:1008640227432. [DOI] [Google Scholar]
- Choi YJ, Hong SB, Shin HD. Diversity of the Hyaloperonospora parasitica complex from core brassicaceous hosts based on ITS rDNA sequence. Mycol Res. 2003;107:1313–1321. doi: 10.1017/S0953756203008578. [DOI] [PubMed] [Google Scholar]
- Choi YJ, Hong SB, Shin HD. Re-consideration of Peronospora farinosa infecting Spinacia oleracea as distinct species, Peronospora effusa. Mycol Res. 2007;111:381–391. doi: 10.1016/j.mycres.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Choi YJ, Shin HD, Voglmayr H. Reclassification of two Peronospora species parasitic on Draba in Hyaloperonospora based on morphological and molecular phylogenetic data. Mycopathologia. 2011;171:151–159. doi: 10.1007/s11046-010-9340-3. [DOI] [PubMed] [Google Scholar]
- Constantinescu O, Fatehi J. Peronospora-like fungi (Chromista, Peronosporales) parasitic on Brassicaceae and related hosts. Nova Hedwigia. 2002;74:291–338. doi: 10.1127/0029-5035/2002/0074-0291. [DOI] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzke A, Pollmann K, Bleeker W, Kohrt R, Hurka H. Molecular systematics of Cardamine and allied genera (Brassicaceae): ITS and non-coding chloroplast DNA. Folia Geobot. 1998;33:225–240. doi: 10.1007/BF03216204. [DOI] [Google Scholar]
- García Blázquez G, Göker M, Voglmayr H, Martín MP, Tellería MT, Oberwinkler F. Phylogeny of Peronospora, parasitic on Fabaceae, based on ITS sequences. Mycol Res. 2008;112:502–512. doi: 10.1016/j.mycres.2007.10.007. [DOI] [PubMed] [Google Scholar]
- Gäumann E. Über die Formen der Peronospora parasitica (Pers.) Fries. Ein Beitrag zur Speziesfrage bei den parasitischen Pilzen. Beih Bot Centralbl. 1918;34(1):395–533. [Google Scholar]
- Gäumann E. Beiträge zu einer Monographie der Gattung Peronospora Corda. Beitr Kryptogamenfl Schweiz. 1923;5(4):1–360. [Google Scholar]
- Göker M, Voglmayr H, Riethmüller A, Weiß M, Oberwinkler F. Taxonomic aspects of Peronosporaceae inferred from Bayesian molecular phylogenetics. Can J Bot. 2003;81:672–683. doi: 10.1139/b03-066. [DOI] [Google Scholar]
- Göker M, Riethmüller A, Voglmayr H, Weiss M, Oberwinkler F. Phylogeny of Hyaloperonospora based on nuclear ribosomal internal transcribed spacer sequences. Mycol Prog. 2004;3:83–94. doi: 10.1007/s11557-006-0079-7. [DOI] [Google Scholar]
- Göker M, Voglmayr H, García-Blázquez G, Oberwinkler F. Species delimitation in downy mildews: the case of Hyaloperonospora in the light of nuclear ribosomal ITS and LSU sequences. Mycol Res. 2009;113:308–325. doi: 10.1016/j.mycres.2008.11.006. [DOI] [PubMed] [Google Scholar]
- Göker M, Voglmayr H, García Blázquez G, Tellería MT, Martín MP. Molecular taxonomy of phytopathogenic fungi: a case study in Peronospora. PLoS One. 2009;4:e6319. doi: 10.1371/journal.pone.0006319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustavsson A. Studies on nordic peronosporas. I. Taxonomic revision. Opera Bot Soc Bot Lundensi. 1959;3(1):1–271. [Google Scholar]
- Hudspeth DSS, Nadler SA, Hudspeth MES. A cox2 phylogeny of the Peronosporomycetes (Oomycetes) Mycologia. 2000;92:674–684. doi: 10.2307/3761425. [DOI] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Kochman J Majewski T (1970) Grzyby (Mycota). Tom IV. Glonowce (Phycomycetes), Wroslikowe (Peronosporales). Panstwowe Wydawnictwo Naukowe, Warzawa
- Lindtner V. Sur quelques nouvelles et rares Peronosporaces de Serbie. Godišnjak Biol Inst Sarajevu. 1957;9:123–130. [Google Scholar]
- Ploch S, Choi YJ, Rost C, Shin HD, Schilling E, Thines M. Evolution of diversity in Albugo is driven by high host specificity and multiple speciation events on closely related Brassicaceae. Mol Phylogenet Evol. 2010;57:812–820. doi: 10.1016/j.ympev.2010.06.026. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Rabenhorst GL (1859) Fungi Europaei exsiccati, Klotzschii herbarii vivi mycologici continuatio. Editio nova. Series secunda: no. 86
- Riethmüller A, Voglmayr H, Göker M, Weiβ M, Oberwinkler F. Phylogenetic relationships of the downy mildews (Peronosporales) and related groups based on nuclear large subunit ribosomal DNA sequences. Mycologia. 2002;94:834–849. doi: 10.2307/3761698. [DOI] [PubMed] [Google Scholar]
- Robideau GP, de Cock AWAM, Coffey MD, Voglmayr H, Brouwer H, Bala K, Chitty DW, Désaulniers N, Eggertson QA, Gachon CMM, Hu CH, Küpper FC, Rintoul TL, Sarhan E, Verstappen ECP, Zhang Y, Bonants PJM, Ristaino JB, Lévesque CA. DNA barcoding of oomycetes with cytochrome c oxidase subunit I and internal transcribed spacer. Mol Ecol Res. 2011;11:1002–1011. doi: 10.1111/j.1755-0998.2011.03041.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runge F, Ndambi B, Thines M. Which morphological characteristics are most influenced by the host matrix in downy mildews? A case study in Pseudoperonospora cubensis. PLoS One. 2012;7(11):e44863. doi: 10.1371/journal.pone.0044863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Săvulescu T, Rayss T. Contribution à la connaissance de Péronosporees de Roumanie. Ann Mycol. 1930;28:297–320. [Google Scholar]
- Silvestro D, Michalak I. raxmlGUI: a graphical front-end for RAxML. Org Div Evol. 2012;12:335–337. doi: 10.1007/s13127-011-0056-0. [DOI] [Google Scholar]
- Stamatakis E. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Sung GH, Sung JM, Hywel-Jones NL, Spatafora JW. A multi-gene phylogeny of Clavicipitaceae (Ascomycota, Fungi): identification of localized incongruence using a combinational bootstrap approach. Mol Phyl Evol. 2007;44:1204–1223. doi: 10.1016/j.ympev.2007.03.011. [DOI] [PubMed] [Google Scholar]
- Swofford DL (2002) PAUP*: phylogenetic analysis using parsimony (*and other methods) Version 4.0b10. Sinauer, Sunderland
- Telle S, Thines M. Amplification of cox2 ( 620 bp) from 2 mg of up to 129 years old herbarium specimens, comparing 19 extraction methods and 15 polymerases. PLoS One. 2008;3:e3584. doi: 10.1371/journal.pone.0003584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiers B (2013) Index herbariorum: a global directory of public herbaria and associated staff. http://sciweb.nybg.org/science2/IndexHerbariorum.asp
- Vilgalys R, Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol. 1990;172:4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voglmayr H. Phylogenetic relationships of Peronospora and related genera based on nuclear ribosomal ITS sequences. Mycol Res. 2003;107:1132–1142. doi: 10.1017/S0953756203008438. [DOI] [PubMed] [Google Scholar]
- Voglmayr H. Progress and challenges in systematics of downy mildews and white blister rusts: new insights from genes and morphology. Eur J Pl Pathol. 2008;122:3–18. doi: 10.1007/s10658-008-9341-y. [DOI] [Google Scholar]
- Voglmayr H, Constantinescu O. Revision and reclassification of three Plasmopara species based on morphological and molecular phylogenetic data. Mycol Res. 2008;112:487–501. doi: 10.1016/j.mycres.2007.10.009. [DOI] [PubMed] [Google Scholar]
- Voglmayr, Göker Morphology and phylogeny of Hyaloperonospora erophilae and H. praecox sp. nov., two downy mildew species co-occurring on Draba verna sensu lato. Mycol Prog. 2011;10:283–292. doi: 10.1007/s11557-010-0699-9. [DOI] [Google Scholar]
- Voglmayr H, Jaklitsch W. Prosthecium species with Stegonsporium anamorphs on Acer. Mycol Res. 2008;112:885–905. doi: 10.1016/j.mycres.2008.01.020. [DOI] [PubMed] [Google Scholar]
- Voglmayr H, Thines M. Phylogenetic relationships and nomenclature of Bremiella sphaerosperma (Chromista, Peronosporales) Mycotaxon. 2007;100:11–20. [Google Scholar]
- Voglmayr H, Riethmüller A, Göker M, Weiß M, Oberwinkler F. Phylogenetic relationships of Plasmopara, Bremia and other genera of downy mildews with pyriform haustoria based on Bayesian analysis of partial LSU rDNA sequence data. Mycol Res. 2004;108:1011–1024. doi: 10.1017/S0953756204000954. [DOI] [PubMed] [Google Scholar]
- Werle E, Schneider C, Renner M, Völker M, Fiehn W. Convenient single-step, one tube purification of PCR products for direct sequencing. Nucleic Acids Res. 1994;22:4354–4355. doi: 10.1093/nar/22.20.4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor JW. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetic. In: Innis MA Gelfand DH, Sninsky JJ, White TJ, editors. PCR Protocols, a Guide to Methods and Application. London: Academic; 1990. pp. 315–322. [Google Scholar]
- Wiens JJ. Combining data sets with different phylogenetic histories. Syst Biol. 1998;47:568–581. doi: 10.1080/106351598260581. [DOI] [PubMed] [Google Scholar]
- Yerkes WD, Shaw CG. Taxonomy of the Peronospora species on Cruciferae and Chenopodiaceae. Phytopathology. 1959;49:499–507. [Google Scholar]


