SUMMARY
The tetrahydrofolate (THF) riboswitch regulates folate transport and metabolism in a number of Firmicutes by cooperatively binding two molecules of THF. To further understand this riboswitch’s specificity for THF, binding and regulatory activity of a series of THF analogs and antifolates was examined. Our data reveal that while binding is dominated by the RNA’s interactions with the pterin moiety, the para-aminobenzoic acid (pABA) moiety plays a significant role in transcriptional regulation. Further, we find that adenine and several other analogs bind with high affinity by an alternative binding mechanism. Despite a similar affinity to THF, adenine is a poor regulator of transcriptional attenuation. These results demonstrate that binding alone does not determine a compound’s effectiveness in regulating the activity of the riboswitch—a complication in current efforts to develop antimicrobials that target these RNAs.
Keywords: riboswitch, tetrahydrofolate, RNA structure, transcriptional regulation, x-ray crystallography, calorimetry
INTRODUCTION
Folate and its reduced derivatives are essential for normal cellular metabolism in all life (Jabrin et al., 2003). Reduced folates such as tetrahydrofolate (THF) are critically important for single carbon unit transfer reactions in the synthesis of methionine, thymine, and purines (Scott and Weir, 1998). This has made enzymes that synthesize and utilize folates major targets of therapeutics including sulfonamides directed against folate biosynthesis in bacteria (Bermingham and Derrick, 2002) and methotrexate and related compounds that act as anticancer agents (Gonen and Assaraf, 2012). Research into how folates and their inhibitors act in the biological context has shown to be one of the few fruitful endeavors for the development of antibiotics (Bermingham and Derrick, 2002). Further studies into how folates regulate gene expression in bacteria may offer a new avenue to apply this proven set of molecules for the discovery of novel antibiotics as well as further our understanding of a unique RNA base regulatory system.
In many Firmicutes, genes involved in folate transport and synthesis are regulated by an element located within their mRNA leader called a riboswitch (Ames et al., 2010). These non-protein coding elements consist of an aptamer domain that directly interacts with an effector molecule and a regulatory domain that generally contains a secondary structural switch directing the expression machinery (Barrick and Breaker, 2007) (Breaker, 2012). Members of the THF riboswitch family (Rfam RF01831) have a highly conserved aptamer domain with a secondary structure containing a three-way junction motif and a pseudoknot (Figure 1A, 1B). Upon THF binding the aptamer domain, the regulatory domain will either terminate transcription or inhibit translation. This RNA was found to bind a variety of reduced folates including tetrahydrofolate (THF), dihydrofolate (DHF), and tetrahydrobiopterin (BH4) with high affinity while strongly discriminating against folate (Ames et al., 2010). This suggested that a key element of recognition between the RNA and folates is the presence of a hydrogen bond donor at position N8 established by the reduction of the pterin ring (Figure 1C) (Ames et al., 2010).
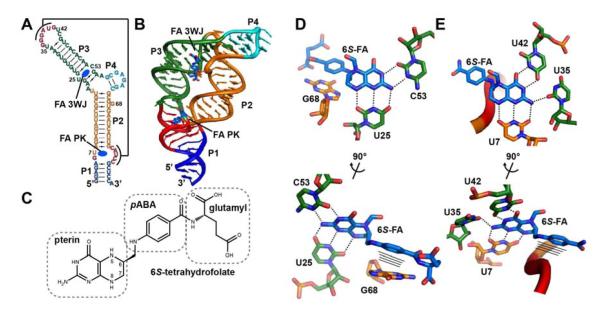
Figure 1.
Structure of the THF riboswitch. (A) Representation of the secondary structure of the THF riboswitch (THF in black, P1 in blue, P2 in orange, P3 in green, P4 in cyan, pseudoknot in red). (B) X-ray crystal structure of the THF riboswitch bound to 6S-folinic acid (4LVV). (C) Chemical structure of 6S-tetrahydrofolate (6S-THF), emphasizing the pterin, para-aminobenzoic acid (pABA) and glutamyl moieties. (D) The three-way junction (3WJ) binding site of the THF riboswitch bound to 6S-folinic acid shown from two angles to emphasize the hydrogen bonding to the pterin ring (top) and interactions of the pABA moiety (below). (E) The pseudoknot (PK) binding site of the THF riboswitch bound to 6S-folinic acid shown from two angles.
To reveal the structural basis for reduced folate recognition, two groups independently solved the structure of a member of the THF riboswitch family in complex with a THF derivative, folinic acid (FA). The structure solved by the Patel group revealed a single THF binding site in the three-way junction, suggesting to the authors that the THF induces a long-range pseudoknot whose formation directs the downstream regulatory switch (Huang et al., 2011). In contrast, the structure presented by the Batey laboratory revealed two THF binding sites, one in the three-way junction that was identical to that observed in the Patel structure, and another in the pseudoknot (Trausch et al., 2011). Notably, in the Patel structure, the pseudoknot was a site of an artifact of crystallization involving strand exchange between two neighboring molecules within the crystalline array of molecules, but was dismissed as not relevant to the biological form of the RNA (Huang et al., 2011). Conversely, it can be argued that the second site observed by the Batey group is an artifact of the high folinic acid concentration used in the crystallization conditions. Thus, it remains an open question as to which structure is the most biologically relevant representative of the THF family (Serganov and Patel, 2012).
Both structures reveal that reduced folates are primarily recognized by the RNA through their pterin moiety (Huang et al., 2011; Trausch et al., 2011). In the three-way junction (3WJ) site, two bases within the P3 helical element form a base triple-like interaction with the pterin, while the pABA moiety forms a stacking interaction with a flipped out conserved purine (Figure 1B, 1D). The pseudoknot (PK) binding site is formed through a Watson-Crick base pairing interaction between L3 and J2/1. Similar to the 3WJ site, the PK site primarily interacts with the pterin ring of THF, while the pABA moiety makes van der Waals’ interactions with the backbone of the RNA (Figure 1B, 1E). The observation that recognition is primarily mediated through the reduced pterin was reinforced by calorimetric data showing that guanine, whose ring system has an analogous set of hydrogen bond acceptors and donors as reduced pterin, can bind the THF riboswitch with nearly identical affinity at 10 mM magnesium ion concentration (Trausch et al., 2011).
Despite the two THF sites having nearly identical affinities for the effector, they appear to have very different roles in transcriptional attenuation. While the wild type Streptococcus mutans (Smu) riboswitch did not function in a single-turnover in vitro transcriptional termination assay, a common method for assessing riboswitch activity, a chimera of the Smu aptamer and Bacillus subtilis metE regulatory switch is strongly THF-responsive (Trausch et al., 2011). Chimeric riboswitches have been validated with numerous aptamer/expression platform pairings and found to be an accurate reporter ligand-dependent regulatory activity (Ceres et al., 2013a; Ceres et al., 2013b). In the context of the chimera, loss of the 3WJ site has a moderate impact on regulatory activity while a similar mutation in the PK site conferred a dramatic loss in THF-dependent transcriptional attenuation. These data reveal that the PK site is the primary regulatory site with the 3WJ playing a supporting role (Trausch et al., 2011).
These findings suggest that binding affinity is not perfectly correlated to regulation— there are features of the THF-RNA interaction that do not contribute to binding but are critical for regulatory activity. To investigate this phenomenon, we probed the THF riboswitch with a series of THF, pterin, purine, and pyrimidine analogs chosen to interrogate different features of the effector. We were also able to take advantage of the wealth of antifolate therapeutics (Gonen and Assaraf, 2012) to further this analysis. Using a combination of structural, calorimetric and activity measurements, we clearly reveal that the pABA moiety plays a significant role in promoting regulation. Surprisingly, an alternative small molecule binding mechanism is also possible in which adenine and its analogs 2,6-diaminpurine (DAP) and 2-aminopurine (2AP) bind to the PK site in a reverse orientation relative to pterin and guanine. While this enables DAP to bind to the RNA with 26-fold higher affinity than THF, this compound is particularly ineffective at promoting regulatory activity. Since riboswitches have been suggested to be potentially important target for novel antimicrobials, understanding the disconnect between the effector’s affinity for the aptamer domain of the riboswitch and regulatory activity mediated by the downstream secondary structural switch should highlight challenges and yield new insights into how to approach this problem.
RESULTS
The majority of THF riboswitches bind two ligands at distinct sites
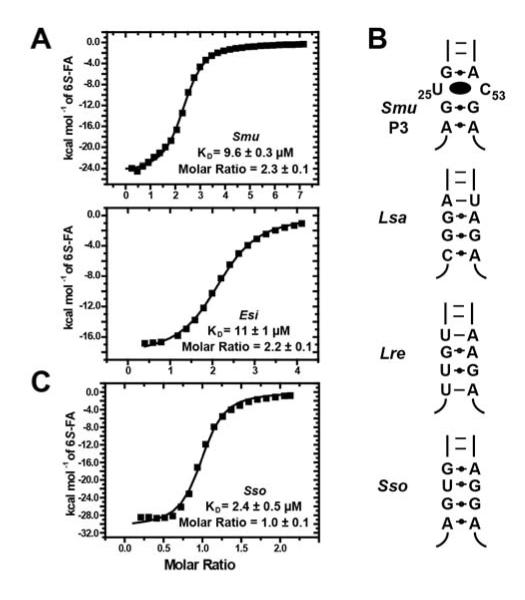
It has been proposed that binding two molecules of the aptamer domain is not a universal feature of the THF family of riboswitches (Serganov and Patel, 2012). Instead, the single site observed in the structure of the Eubacterium siraeum (Esi) THF riboswitch (Huang et al., 2011) might represent a subset of this family that only binds the effector to the three-way junction site (Serganov and Patel, 2012). This is in contrast to the observation that the conservation of bases in direct contact with THF at the PK site is nearly universal--including the Esi variant. Further, the authors of the structural study of the Esi variant did not present any solution data in support of the claim of a single binding site. Therefore, we titrated the EsiTHF aptamer with the THF analog folinic acid by ITC to verify its binding stoichiometry. Consistent with the SmuTHF receptor structure, the EsiTHF receptor binds two equivalents of ligand, with nearly the same apparent KD (Figure 2A). These data strongly suggest that the lack of the PK site in the Patel crystal structure is artifactual.
Figure 2.
The majority, but not all, of THF riboswitches contain two sites. (A) ITC titrations of Streptococcus mutans (Smu) and Eubacterium siraeum (Esi) THF riboswitch binding to 6S-folinic acid. Data generated from three independent titrations. Error represents standard deviation. (B) A few phylogenetic representatives of P3, which contains the three-way junction binding (3WJ) site. Smu is an example of a P3 that represents the majority of THF riboswitches containing a consensus THF binding site. Lactobacillus saliva (Lsa), Lactobacillus reuteri (Lre) Streptococcus sobrinus (Sso) all contain sequence variations in P3 stems inconsistent with binding at the 3WJ site. (C) ITC titration of Streptococcus sobrinus (Sso) THF riboswitch binding 6S-folinic acid, revealing binding to a single site.
Despite the above finding, there may exist a subset of THF riboswitches with only a single site within the pseudoknot. Recent expansion of THF riboswitch sequences available in Rfam 11.0 revealed a subset of aptamers with sequence variation at the 3WJ site that is expected to disrupt binding. These variants do not have the equivalent of the U25/C53 juxtaposition at the 3′-side of the P3 helix that forms the 3WJ site (Figure 2B). To investigate if these variants only bind a single THF, we tested Streptococcus sobrinus, a sequence containing a representative C53G variation. This RNA binds THF with a KD of 2.4 ± 0.5 M and an n-value of 1.0 ± 0.1 (Figure 2C). This demonstrates that although the majority of THF riboswitches exhibit a sequence consistent with binding two ligands, there is a subset of sequences that bind only one THF molecule at the critical regulatory PK site. Thus, the 3WJ site is not an essential feature of the THF family--only the PK site. Still, the conclusions that binding and regulation of this receptor based upon the two-site model are more representative of the family and all further analysis of the THF riboswitch is considered in light of the two-site structure that represents the vast majority of RNAs in this family (Trausch et al., 2011).
The reduced pterin moiety is the primary determinant of high-affinity binding and regulatory activity
The central binding feature at both sites is that RNA-THF contacts are mediated primarily through the pterin moiety with little apparent contribution from either the pABA or glutamyl groups (Figure 1D, 1E). To further define the role of functional groups, we interrogated the structure and activity of the riboswitch with a series of THF, pterin, and purine analogs. In contrast to previous studies, we took care to use the same, or very similar, buffer and temperature conditions in our single-turnover in vitro transcription assay and ITC conditions (70 mM buffer, pH 8.0, 70 mM NaCl, 2.5 mM MgCl2 at 37 °C) to correlate ligand binding and regulatory activities.
The moderate importance of the pABA and glutamyl moieties is exhibited through calorimetric measurements of the THF-RNA interaction. In our original study, we obtained THF as a mixture of the 6S and 6R epimers, whereas in biology THF exists exclusively in the 6S conformation (Illarionova et al., 2002). This mixture yielded a 2:1 THF:RNA binding stoichiometry with no indications that the RNA was discriminating between the two epimers. However, modeling of 6R-THF into the crystal structure of the bound state suggests that this epimer must adopt an extreme pucker in the pterin ring to yield a conformation that enables pABA-RNA contacts at either site (Figure S1 of the Supplemental Information). To clarify this issue, we obtained purified 6S-THF along with 6R-THF and measured the binding and regulatory activity (Table 1). 6S- and 6R-THF bind the receptor with similar affinities: 13 ± 1 M and 19 ± 2 M, respectively. However, in a single-turnover in vitro transcription assay under the same buffer and temperature conditions, these compounds display a greater difference in the concentration required to elicit the half-maximal regulatory response (referred to as T50). 6S-THF yielded a T50 of 32 ± 1 M while the 6R-epimer showed a moderately, but significantly, higher T50 (110 ± 10 M). This demonstrates that interactions between the pABA moiety and the RNA contribute to stabilizing the receptor domain in a fashion that prevents the competing antiterminator stem-loop structure from forming. Attempts to crystallize the receptor with purified 6S- and 6R-THF in high concentrations of reducing agent (100 mM DTT) and under inert atmosphere (argon) resulted in crystals, but analysis of the diffraction data revealed that in both cases oxidation led to the cleavage of the pABA/glutamyl moieties (Reed and Archer, 1980).
Table 1.
Affinity and Transcriptional Regulation by THF and THF analogs
| Ligand | KD (μM) | n | T50 (μM) | Relative T50/KD |
|---|---|---|---|---|
| 6S-tetrahydrofolate | 13 ± 1 | 1.9 ± 0.1 | 32 ± 1 | 2.5 |
| 6R-tetrahydrofolate | 19 ± 2 | 2.1 ± 0.1 | 110 ± 10 | 5.8 |
| 6S-folinic Acid | 9.6 ± 0.3 | 2.3 ± 0.1 | 19 ± 1 | 2.0 |
| dihydrofolate | 23 ± 1 | 2.0 ± 0.1 | 89 ± 6 | 3.9 |
| tetrahydrobiopterin | 18 ± 3 | 1.9 ± 0.1 | 190 ± 20 | 10 |
| 6,7-dimethyl-tetrahydrobiopterin | 57 ± 3 | 2.1 ± 0.2 | 590 ± 30 | 10 |
| 7-deazaguanine | 66 ± 1 | 2.0 ± 0.1 | 690 ± 20 | 10 |
| Pemetrexed | 110 ± 10 | n.d. | >10000 | n.a. |
| Lometrexol | 37 ± 2 | 1.9 ± 0.2 | 190 ± 20 | 5.1 |
| 2,6-diaminopurine | 0.5 ± 0.1 | 1.0 ± 0.1 | 130 ± 10 | 260 |
| adenine | 8 ± 1 | 1.2 ± 0.1 | 2500 ± 800 | 310 |
| 2-aminopurine | 26 ± 3 | 0.9 ± 0.1 | >10000 | n.a. |
| 8-aminoadenine | 160 ± 20 | 1.1 ± 0.1 | >10000 | n.a. |
| 6-hydroxy-2,4,5-triaminopyrimidine | 84 ± 5 | 1.8 ± 0.1 | 1400 ± 200 | 780 |
n.d. = not determined due to solubility limitations
n.a. = not applicable
Error in KD and n represents the standard deviation of three independent experiments. Error in T50 represents the error in the fit of the average of three independent experiments.
To address this issue, we looked to a more oxidation resistant form of THF, folinic acid (5-formyl-THF or FA, Figure 3). This compound is a naturally occurring intermediate of THF (Kariluoto et al., 2010) and also a drug (leucovorin) used to ease the side effects of the antifolate chemotherapeutic methotrexate (Keshava et al., 1998). We have previously shown that folinic acid binds the THF riboswitch at both sites with high affinity (Trausch et al., 2011). In an effort to advance our understanding, we obtained purified 6S-folinic acid (6S-FA, a kind gift from Merck). We were able to obtain high quality crystals of 6S-FA bound to the riboswitch. The structure was very similar to the one solved using a mixture of epimers but showed better density for the pABA and glutamic acid of FA (Figures 1D, 1E, and S2 of the Supplemental Information). We observed an affinity for 6S-FA by ITC of 9.6 ± 0.3 M (Table 1). The data showed a 2:1 THF:RNA stoichiometry but a single apparent binding curve reflecting near equivalent binding sites. In vitro transcription revealed a T50 of 19 ± 1 M (Figure 4), making it a more effective regulator than 6S-THF. It is not surprising that a modification at the N5 position is easily tolerated within the binding pocket. In the cell, THF exists in a variety of forms such as 5-methyl-THF, 5-formyl-THF (FA), and 5,10-methenyl-THF and likely the THF riboswitch recognizes all these forms (Kariluoto et al., 2010). The ability of 6S-folinic acid to terminate transcription was also tested with mutants that knockout either binding site (U7C for PK site and U25C for 3WJ site). These data are consistent with previous results using a mixture of (6S,6R)-folinic acid epimers, in that knocking out the PK side is much more deleterious to the switch than knocking out the 3WJ (Figure 4). However, we do note that the U7C mutant was previously observed to be ~100-fold lower activity than wildtype (Trausch et al., 2011), whereas in the current study it is completely inactive. This difference may be attributable to the use of the purified 6S-FA epimer in these experiments rather than the mixture of epimers used previously as well as minor changes to the transcription conditions (higher NTP concentration in the current study). Notably, the Hill coefficient (nH) is much higher with both sites compared to the U25C mutant containing only the PK site, indicating a strongly cooperative regulatory response.
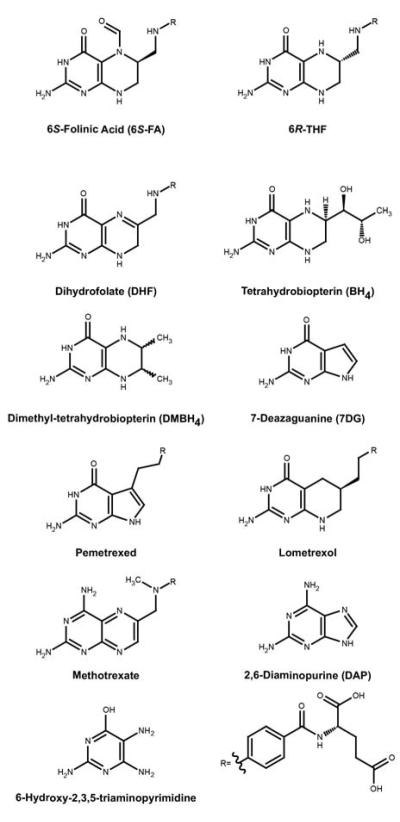
Figure 3.
Ligands used in binding and transcription termination studies. The R group in select compounds represents the pABA attached to L-glutamic acid (lower right) found in folate and its natural derivatives.
Figure 4.
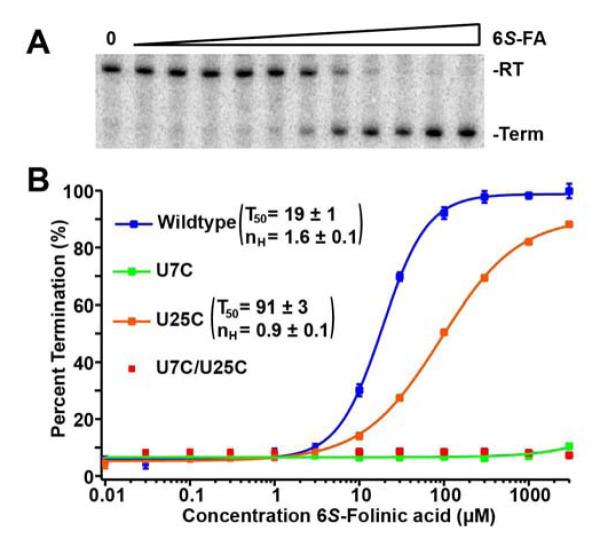
The role of the two binding sites in transcription regulation. (A) Example of an in vitro transcription titration using 6S-folinic acid (0 to 3 mM) and the quantification of this data. Read through (RT) and termination (Term) products were separated using a 6% denaturing PAGE. (B) Graph of quantified titrations done with wild type (WT), U7C, U25C, and U7C/U25C. Error bars represent the standard deviation based on three independent experiments.
The finding that the pABA moiety plays a role in regulation is further supported by comparable measurements with dihydrofolate (DHF), the partially reduced form of THF. In DHF, the N8 of the pterin ring is reduced such that it still forms a critical hydrogen bond with residue U7. However, N5 is not reduced such that C6 remains sp2 hybridized (Figure 3), likely disallowing full Van der Waals interactions between the pABA moiety and RNA. Consistent with this, we find that DHF binds the RNA with about two-fold lower affinity than 6S-THF and a higher T50; these values are comparable to 6R-THF (Table 1).
To further assess the role of the pABA/glutamyl moieties in binding and regulation, we examined a set of compounds that remove part or wholly these groups. Tetrahydrobiopterin (BH4) is a common reduced pterin in biology in which the pABA and glutamyl moieties are replaced by a propylene glycol moiety at the C6 position (Figure 3). BH4 binds with a KD only slightly higher than 6S-THF, strongly supporting that the pABA/glutamyl moieties contribute marginally to binding (Ames et al., 2010). This is rather remarkable considering the pABA/glutamyl makes up over 50% of the molecular weight of THF, and is in stark contrast to the majority of riboswitches in which the majority, if not the entirety, of the effector is directly recognized (Serganov and Nudler, 2013). However, BH4 has a T50 of 190 ± 20 M, or about 10-fold higher than its KD. The structure of the THF riboswitch bound to BH4 shows that the placement of the pterin ring in BH4 is identical to that of THF, but the propylene glycol does not interact with the RNA in either binding site, as indicated by the very weak electron density in this region (Figure 5A, 5B).
Figure 5.
Structure of the THF riboswitch bound to a set of analogs. (A, B) Structure of tetrahydrobiopterin (BH4) bound to the three-way junction binding site and the pseudoknot binding site, respectively. (C, D) Structure of 7-deazaguanine (7DG) bound to the three-way junction binding site and the pseudoknot binding site, respectively. (E, F) Structure of Pemetrexed (Pem) bound to the three-way junction binding site and the pseudoknot binding site, respectively. For all structures, the electron density from a 2Fo-Fc simulated annealing omit map (calculated using a model in which all of the illustrated nucleotides and ligand were omitted from the model) contoured at 1σ is shown in grey.
A synthetic ligand similar to BH4 is 6,7-dimethyl-tetrahydropterin (DMPH4). It should be noted that this compound is available only as a mixture of the four diastereomers at the 6- and 7-positions (Figure 3). DMPH4 shows a KD of 57 ± 3 M and a stoichiometry of 2:1 DMPH4:RNA (Table 1). The observed affinity is weaker than 6S-THF or BH4, which might be due to this compound possessing two chiral centers that have no specific distinction. It is possible that some of the diastereomers in this mixture bind more poorly than others and that an averaged affinity is observed by ITC. Similar to BH4, the T50 for DMPH4 was ~10-fold higher than the KD. Although the crystal structure showed density in both the PK and 3WJ sites corresponding to the ring system, only poor density was observed for the methyl groups, likely resulting from the bound ligand at both sites being a mixture of all diastereomers (data not shown).
In our original study, we noted that guanine has a similar pattern of hydrogen bond donors and acceptors as the reduced pterin ring of THF (Trausch et al., 2011). In exponentially growing E. coli guanine is found at a relatively high concentration of 190 M, (Bennett et al., 2009) suggesting that it could act as an effector of the THF riboswitch within the cell. For example, the glmS riboswitch integrates signal from two distinct metabolites, one as an activator of mRNA cleavage and several as inhibitors to elicit the appropriate regulatory response to the global metabolic state of the cell (Watson and Fedor, 2011). To investigate this hypothesis, we previously tested 7-deazaguanine (7DG, a water soluble analog of guanine) for affinity and transcriptional regulation (Trausch et al., 2011). Data showed that 7DG binds with high affinity, but poorly regulates transcription (Trausch et al., 2011). Unfortunately, in the previous study, the KD and T50 measurements were not taken under buffer/temperature matched conditions, complicating the comparison. Under the new conditions of this study, 7DG binds with a KD of 66 ± 1 M and exhibits a T50 of 690 ± 20 M (Table 1). Like BH4 and DMPH4, 7DG has a T50 ~10-fold higher than the KD, again reflecting the loss of interactions mediated by the pABA/glutamyl moieties. The structure of the THF riboswitch was solved in complex with 7DG (Figure 5C, 5D). 7DG is observed bound to both the 3WJ and PK sites, making a set of hydrogen bonding interactions to the RNA analogous to reduced pterin. This structure confirms the previous hypothesis that guanine and certain derivatives like 7DG are able to engage the riboswitch, but do not regulate under biologically relevant guanine concentrations. Instead, under physiological conditions, intracellular guanine may act as a competitive inhibitor of regulation such that higher THF concentrations are required to downregulate gene expression.
A pyrimidine compound is also able to productively bind the THF riboswitch. The purine riboswitch, which uses a similar set of ligand-RNA contacts for recognition as the THF riboswitch, is capable of binding several pyrimidine analogs (Dixon et al., 2010; Gilbert et al., 2006a; Mulhbacher et al., 2010). Within a series of pyrimidine analogs tested, 2,4,6-triaminopyrimidine, 2,4,5,6-tetraaminopyrimidine, 2,4-diamiopyrimidine, and a 5-member ring compound 5-aminoimidazole-4-carboxamide all failed to bind (data not shown). Conversely, 6-hydroxy-2,4,5-triaminopyrimidine (Figure 3) binds the THF aptamer with a modest KD of 84 ± 5 M and a 2:1 stoichiometry, similar to the values observed for 7DG (Table 1). The T50 is of 1400 ± 200 M, ~16-fold higher than the KD, comparable to that of BH4 and DMPH4 (Table 1). While we were able to obtain crystals of this complex, they were of extremely poor quality and not suitable for diffraction studies. Thus, while the orientation of 6-hydroxy-2,4,5-triaminopyrimidine in the binding sites is unknown, it is possible that the hydrogen bonding is similar to interactions made by 7DG.
Antifolate drugs are able to bind and regulate the THF riboswitch
A number of drugs have been developed over the last half-century that target enzymes involved in THF synthesis or THF related pathways (Gonen and Assaraf, 2012). These compounds are mainly anticancer agents that target proteins such as dihydrofolate reductase, thymidylate synthase and glycineamide ribonucleotide formyltransferase (Gonen and Assaraf, 2012). Many of these compounds are designed to closely mimic THF and therefore might represent a rich pool of potential effectors of the THF riboswitch.
Lometrexol (DDATHF), a compound developed for the treatment of cancer, (Gonen and Assaraf, 2012) has a structure with the greatest similarity to THF and folinic acid. This compound replaces N5 and N10 with carbons and the lack of N5 prevents the oxidative loss of the N8 hydrogen (Figure 3), which, like folinic acid, might improve its performance. Lometrexol binds to the THF riboswitch with an affinity of 37 ± 2 M and regulates transcriptional termination with a T50 of 190 ± 20 M (Table 1). The ~5-fold higher T50 relative to KD is similar to 6R-THF, suggesting that the C5 and/or C10 substitutions have a deleterious effect. One possibility is that conjugation of N10 with the benzyl ring in THF restricts conformational flexibility of the pABA/glutamyl moiety enabling it to position the pABA moiety optimally for interacting with the RNA at each site. The additional conformational freedom in Lometrexol has the same effect of the 6R epimer of THF that prohibits these interactions.
Pemetrexed (Alimta®), a rationally designed drug used in the treatment of lung cancer, (Gonen and Assaraf, 2012) replaces the pterin moiety with 7DG and N10 with a methylene group (Figure 3). While 7DG is a poor regulator of transcriptional attenuation, the presence of the pABA/glutamyl moiety might improve activity. However, Pemetrexed exhibits one of the poorest affinities tested (KD = 110 ± 10 μM) and is almost completely unable to terminate transcription (T50 ≥ 10 mM). The ~100-fold difference between KD and T50 is far greater than that observed for 7DG, indicating that the pABA/glutamyl moiety in the context of a guanine scaffold is deleterious. A crystal structure of the Pemetrexed-RNA complex revealed that in the 3WJ site, Pemetrexed is bound almost identically as THF (Figure 5E, 5F); the purine ring is correctly positioned and there is weak density to support pABA stacking on G68. In the PK site the purine ring is also correctly positioned, but there is no density to support the pABA in any conformation. The observed disorder of the pABA/glutamyl moiety likely reflects C9 being coplanar with the purine ring along with the greater conformational flexibility imparted by C10. This observation may suggest that the key interaction for regulation is between the pABA moiety and A80 in the PK site; the analogous stack with G68 in the 3WJ site is not as important.
Methotrexate is a commonly used antifolate in the treatment of many cancers and autoimmune disorders (Gonen and Assaraf, 2012). Methotrexate, and many antifolates therapeutics exhibit modifications to the pterin ring and the pABA/glutamyl moiety relative to THF. In the case of methotrexate, these modifications come in the form of an amino group replacing the carbonyl oxygen of the pterin, a methyl group on N10, and the pterin ring being fully oxidized (Figure 3). These deviations from THF make the pattern of hydrogen bond donors and acceptors in this compound incompatible with either binding pocket. Not surprisingly, methotrexate does not bind to the THF riboswitch or terminate transcription, and it is very likely that similar antifolates such as pralatrexate, raltitrexed, trimetrexate, piritrexim, and talotrexin also do not interact with the THF riboswitch.
Retro-binding of adenine and derivatives to the THF riboswitch
Given the above result with methotrexate, it was a great surprise to find that adenine and one of its derivatives, 2,6-diaminopurine (DAP), bind the THF riboswitch more tightly than 6S-folinic acid. Like methotrexate, the loss of hydrogen bonding to U42 in the PK site (Fig. 1C) or C53 in the 3WJ in adenine and its derivatives would be expected to be a significant challenge for high affinity binding. To the contrary, DAP, 2-aminopurine (2AP), and adenine (Ade) all bind with affinities either higher or comparable to 6S-THF (Table 1). The affinity of DAP is most striking (KD = 0.5 ± 0.1 μM) in that it is ~20-fold higher than 6S-folinic acid. However, ITC also revealed that these compounds bind the THF aptamer with a 1:1 ligand:RNA stoichiometry, indicating that they productively bind only one site. Comparing the regulatory activity of 1 mM DAP for the wild type aptamer against point mutants that abrogate THF binding to the PK site (U7C), the 3WJ site (U25C), or both (U7C, U25C) revealed that DAP only interacts with the PK site (Figure 6A).
Figure 6.
Diaminopurine (DAP) as a binder and effector of the THF riboswitch. (A) PAGE separation of the terminated (Term) and read-through (RT) products from in vitro transcription done in the absence of ligand as well as 1 mM of folinic acid (FA), 2,6-diaminopurne (DAP), adenine (Ade), and 2-aminpurine (2AP) in the context of the WT, U7C ( PK), U25C ( 3WJ), U7C/U25C ( PK+ 3WJ) variants. (B, C) The structure of the pseudoknot binding site bound to DAP and adenine, respectively. Simulated annealing omit map is shown in grey, calculated in the same fashion as those in Figure 5. (D) The overlay of two structures of dihydrofolate reductase bound to folate (Bystroff et al., 1990) (red) and methotrexate (Bolin et al., 1982) (blue).
The finding that DAP productively binds to the riboswitch appears to be inconsistent with the lack of binding seen with methotrexate, whose pterin moiety has a similar pattern of hydrogen bond donors and acceptors. To understand this discrepancy, we solved the structure of the THF riboswitch bound to DAP. No electron density consistent with ligand was observed in the three-way junction site. Conversely, strong electron density was observed adjacent to U7, confirming binding to the PK site. More strikingly, the orientation of the DAP in the PK site is rotated ~180° within the binding pocket relative to the ring systems of folinic acid and 7DG (Figure 6B). This orients the Watson-Crick face of DAP away from U7/U35/U42, such that it makes no hydrogen bonds with the RNA. Instead, it employs the Hoogsteen and sugar edges of the nucleobase to interact with the RNA. The crystal structure of Ade bound reveals the same reverse binding mechanism as DAP (Figure 6C). Superimposition of the DAP-bound structure on that of folinic acid or 7DG reveals that the RNA does not undergo significant conformational change to accommodate binding in the reverse orientation. This suggests that the PK site is rigid and may not be able to adopt alternative conformations to accommodate small molecules with different patterns of hydrogen bond acceptors and donors.
DAP binds the THF riboswitch with remarkable affinity but fails to act as an equally efficient effector of transcriptional regulation. DAP terminates transcription with a T50 of 130 ± 10 M, approximately 250-fold higher than the KD. DAP has two disadvantages in acting as an effector for the THF riboswitch. First, it does not have a pABA moiety, which in the case of BH4, DMPH4, and 7DG comes at a ~10-fold reduction in T50 relative to KD. Second, DAP only binds at one site, which results in an additional ~5-fold reduction in T50 in the SmuTHF aptamer. These effects added together may contribute to the drastic difference in KD and T50 seen with DAP, adenine, and 2AP. Like guanine (see above), it is conceivable that adenine could act as a competitive inhibitor to FA-dependent transcriptional termination. It is estimated that the intracellular adenine concentration in rapidly dividing E. coli is 1.5 μM (Bennett et al., 2009), which is not substantially below the observed affinity for adenine binding to the PK site.
DISCUSSION
The structure of the THF riboswitch revealed several unusual aspects of the aptamer distinguishing it from other riboswitches. Uniquely, this aptamer contains two binding sites (Trausch et al., 2011). By calorimetry, the RNAs used in both crystallographic analyses bind two equivalents of THF; the observation of only a single site in one structure (Huang et al., 2011) is an artifact of strand swapping within the crystal. However, this is not a universal feature of the THF riboswitch family. Inspection of an expanded phylogenetic alignment of almost 600 sequences available in Rfam 11.0 reveals that some Lactobacilli, Streptococcus, and Clostridium species harbor THF riboswitches that have sequence variations that ablate the 3WJ site. Unexpectedly, the SsoTHF aptamer that only binds one molecule of THF has a higher affinity for THF than the SmuTHF aptamer. This could be a result from closing P3 with a G·U base pair, instead of the C and U that normally binds THF, which orders the 3WJ as if there were ligand bound. Thus, the 3WJ site is not essential for biological function. One explanation of why an aptamer would evolve two binding sites is a selection pressure for a more digital regulatory response. Positive cooperative systems are ubiquitous in protein-based regulatory mechanisms, and it should be no surprise that RNA has evolved a similar mechanism. A second hypothesis relates to the co-transcriptional nature of regulation. In this model, during transcription of the aptamer domain, the 3WJ site is formed first, allowing it to bind THF first and facilitate organization of the PK and increasing the rate of association of THF to the regulatory site. Thus, the 3WJ site acts as a folding center that makes the kinetics of ligand binding and the coupled regulatory response more favorable.
The other novel feature of the THF riboswitch is that it primarily recognizes only one of the three main moieties—the pterin—in reduced folates. In contrast, many riboswitches almost completely encapsulate their ligand, usually through a conformational change coupled to the binding event, and as a consequence recognize almost all features of their effector (Batey et al., 2004). In this study we examined THF in greater detail to reveal potential minor contributions made by the pABA/glutamyl moieties to binding and regulation. Our data clearly show that while these groups do not contribute substantially to binding energy, the pABA group significantly contributes to regulatory activity. Differences between binding and regulatory efficiency have also been observed in the lysine riboswitch (Garst et al., 2012). For example, the natural effector lysine has a T50 to KD ratio of 1.0 (under transcription conditions of 50 μM NTPs) such that binding and regulation occur at the same concentration. The close derivative L-lysine methyl ester displays near equivalent binding affinity to lysine, but has a T50 to KD ratio of ~6. These observations can be explained in several ways. First is that differences in the kinetics of association and dissociation between the natural effector and their close chemical analogs result in differences in their efficiency in eliciting a regulatory response. Since many riboswitches are proposed to be under “kinetic control”, (Haller et al., 2011; Wickiser et al., 2005) parameters such as kon and koff of the ligand are expected to have significant effect on the observed T50. We should note that while the in vitro transcription assay cannot completely mirror cellular conditions for a variety of reasons, the behavior of riboswitches between the two sets of conditions are consistent (Ceres et al., 2013a; Lemay et al., 2011; Tomsic et al., 2008).
An alternative hypothesis is that these analogs disrupt a set of interactions between the ligand and RNA critical for the regulatory switch. In most riboswitches, the ligand binding site lies adjacent to nucleotides at the 3′-end of the aptamer domain that are involved in the regulatory switch (Garst and Batey, 2009). It may be that certain contacts are crucial for stabilizing the incorporation of the switching sequence (a sequence element that is partitioned into either the aptamer domain or the expression platform) into the aptamer domain, thereby preventing formation of one of the alternative secondary structures in the expression platform. In the case of THF, the pABA moiety forms van der Waals’ contacts on the ribose-phosphate backbone around A80. This interaction may be sufficient to reduce invasion of the expression platform into the aptamer that would be required to form an alternative secondary structure. In the case of the lysine riboswitch, the carboxyl moiety mediates binding of a potassium ion that interacts with nucleotides on the 3′-side of the P1 helix that are also involved in alternative secondary structure formation (Garst et al., 2008; Serganov et al., 2008).
These data illustrate an important consideration in the development of compounds that target riboswitches. Since the discovery that these RNA elements are widely distributed in bacteria and control expression of genes critical for survival and virulence in some medically important pathogens, it has been proposed that they might be important new targets for antimicrobial therapeutics (Blount and Breaker, 2006). In fact, it has been demonstrated that the natural antimicrobial compounds roseoflavin and pyrophosphate pyrithiamine acquire their antibacterial properties from interactions with the FMN and TPP riboswitches, respectively (Lee et al., 2009; Sudarsan et al., 2005). A number of studies looking for compounds that could be potential riboswitch effectors have often focused on looking for high affinity binders (Deigan and Ferre-D’Amare, 2011; Guan and Disney, 2012). However, this approach has yielded a number of hits that bind but do not efficiently regulate the switch (Cressina et al., 2011). This study would suggest that certain aspects of the effector’s interaction with the RNA must be preserved for a chemical analog to be able to also regulate the riboswitch.
Another surprise in this study is the observation of an alternative binding mode for adenine and two related compounds. In binding studies exploring chemical analogs of the native effector to riboswitches, all of the productive binders appear to bind to the RNA in the orientation as the biological effector. For example, every lysine, (Garst et al., 2012; Wilson-Mitchell et al., 2012) purine, (Gilbert et al., 2006b) and thiamine pyrophosphate (Thore et al., 2008) analog tested to date has shown that the compound lies in the binding pocket in the same orientation and conformation as the native ligand. However, in the search for compounds that target a particular riboswitch family, it is highly desirable to discover chemical scaffolds from which to build novel classes of compounds. In screening purine compounds that bind to the THF riboswitch, we uncovered DAP. This molecule binds 26-fold more tightly than 6S-THF and in an opposite orientation within the pseudoknot binding pocket. Unfortunately, DAP regulates extremely poorly, likely due to the fact that it is unable to form interactions outside of the ring system like the pABA (10-fold effect) and that DAP only binds to one site (~5-fold effect). Nonetheless, the Watson-Crick face (e.g. N1) provides a starting point for the addition of chemical features that could mimic the pABA moiety and restore efficient regulatory activity. The reverse binding mechanism of DAP is not unprecedented amongst proteins and is even seen in proteins recognizing folates (Saraste and Musacchio, 1994). Dihydrofolate reductase naturally binds folate and is inhibited by methotrexate (Saraste and Musacchio, 1994). Although these compounds sterically have a similar pterin ring system, the hydrogen bonding pattern that the ring systems present are quite different (analogous to THF/7DG and DAP). As a consequence, methotrexate is flipped within the binding pocket with respect to folate (Figure 5D)(Bolin et al., 1982; Bystroff et al., 1990). The observation of “retro-binding” by the similar DAP/methotrexate ring systems to protein and RNA reinforces that in the search for small molecules that target RNA, many of the same principles and tools that have been developed for proteins can be applied (Daldrop et al., 2011).
SIGNIFICANCE
Riboswitches have been advanced as a potentially new class of targets for antimicrobial therapeutics as they control genes essential for virulence and survival in medically important pathogens. Further, riboswitches, by virtue of containing a known small molecule binding site, are important model systems for establishing the approaches and methodologies for targeting RNAs with novel compounds. In this work, we clearly establish that a series of pterin, purine and pyrimidine compounds can efficiently bind the THF riboswitch, but are not able to fully recapitulate the regulatory properties of the natural effector. Importantly, several established antifolates that are used to target folate-dependent mechanisms in humans are able to bind and regulate riboswitch activity. This suggests that there exists a pool of known protein-binding therapeutics that might be employed to target RNA. Further, the finding that adenine and several of its analogs bind the switch in an alternative binding mechanism demonstrate that an issue that complicates drug discovery in proteins also must be considered in efforts to target RNA. Together, these results highlight important issues in the development of RNA targeting compounds.
Experimental Procedures
RNA and ligand preparation
Transcription and purification of RNA was preformed using methods described previously (Reyes et al., 2009). Briefly, RNA was transcribed from a DNA template generated by PCR using T7 RNA polymerase and the resulting RNA was purified with a 12% denaturing acrylamide gel. RNA was visualized by UV shadowing, the band corresponding to full-length product excised and eluted from the gel using the “crush-and-soak” method overnight in 0.5X T.E. buffer (5 mM Tris-Cl, pH 8.0, 0.5 mM EDTA, pH 8.0) at 4 °C. The supernatant was concentrated with a 10,000 molecular weight cutoff centrifugal concentrator, exchanged three times into 0.5X T.E. to remove urea, and passed through a 0.2 μm filter. RNA was stored at -80 °C until use. All sequences used in this study are listed in Table S1 of the Supplemental Information. Oxygen-sensitive ligands (6S-THF, 6R-THF, DHF) were brought up in buffer and used immediately to limit loss in activity,
Isothermal Titration Calorimetry (ITC)
Calorimetric measurements of the equilibrium dissociation constant (KD) were performed as previously described (Trausch et al., 2011). Prepared RNA was dialyzed overnight against ITC buffer (70 mM Na-HEPES, pH 8.0, 70 mM NaCl, 2.5 mM MgCl2, 1 mM DTT). Ligand was resuspended from solid stocks with ITC buffer. The concentration of RNA and ligand used in a titration was determined by attempting to achieve a c-value greater than 10 but less then 100 (Gilbert and Batey, 2009). All titrations using a MicroCal iTC200 (GE Healthcare) were run at 37 °C to match both temperature and buffer conditions of the single-turnover transcription assay. Data were processed using the MicroCal processing software Origin 7. High purity samples of 6S-THF, 6R-THF, DHF, and 6S-folinic acid used in experiments were provided as a generous gift by Merck & Cie.
Single-turnover transcription assays
The chimeric riboswitch between the S. mutans folT THF aptamer domain and B. subtilis metE expression platform used previously (Trausch et al., 2011) was cloned into pUC19 and sequence verified. Transcription of the riboswitch is driven by a strong T7A1 promoter to allow for efficient initiation with E. coli RNA polymerase. Template DNA was synthesized by PCR amplification from this plasmid and purified using the Qiagen PCR purification kit.
Single turnover in vitro transcription was adopted from methods previously described (Dann et al., 2007). 17.5 μL volume reactions were prepared containing 50 ng of DNA template, 12.5 l of 2X transcription buffer (140 mM Tris-Cl, pH 8.0, 140 mM NaCl, 0.2 mM EDTA, pH 8.0, 28 mM β-mercaptoethanol, and 70 g/ml BSA), 2.5 l of 25 mM MgCl2, 0.5 Ci of [α-32P]ATP, and 0.25 units of E. coli RNA polymerase σ70 holoenzyme (Epicentre). These solutions were incubated at 37 °C for 10 minutes. Transcription wa s initiated by adding 7.5 l of NTP mix (75 M final concentration in each NTP, 0.2 mg/ml heparin and ligand at 3.33X concentration) and further incubated at 37 °C for 20 minutes. Transcri ption was quenched by adding 25 l RNA loading dye (95% (v/v) deionized formamide, 10 mM EDTA, pH 8.0, 0.25% (w/v) bromophenol blue, 0.25% (w/v) xylene cyanol) followed by raising the temperature to 65 °C for 3 minutes. A 6% 29:1 acrylamide:bisacrylamide denaturing polyacrylamide gel was used to separate terminated and read through products. The gel was imaged with a phosphor screen (Molecular Dynamics) and scanned with a Typhoon 9400. The bands were quantified using ImageQuant 5.2 (Molecular Dynamics). Data was fit to a standard two-state binding model with a Hill coefficient parameter using IGOR Pro 6 (WaveMetrics).
X-ray crystallography, structure solution and refinement
The Smu THF riboswitch aptamer domain in complex with various ligands was crystallized using the hanging drop/vapor diffusion method. One microliter of RNA mix (500 M RNA and 1-2 mM ligand) was mixed with two microliters of crystallization buffer comprised of 6-8% 2-methyl-2,4-pentanediol (MPD), 8-12 mM spermine HCl, 40 mM sodium cacodylate, pH 7.0, 80 mM sodium chloride and 1 mM dithiothreitol. Hanging drops were incubated at 30 °C for 3-5 days over 500 l of 35% MPD. The resulting needle-like crystals were mounted on a nylon loop and flash frozen in liquid nitrogen.
Data sets were collected at the Advanced Light Source (5.0.2). Data were indexed, integrated and scaled using iMosflm (Battye et al., 2011). Initial electron density maps were generated with molecular replacement using PHENIX(Adams et al., 2010) with model 3SD1 (Trausch et al., 2011). All atoms corresponding to solvent, metal ions, and ligand were removed. Bases in the hydrogen-bonding plane of the ligands, as well as those involved in base stacking to the pABA, were also removed (U7, U35, U42, U25, C53, G68). Model refinement was performed using a combination of energy minimization, simulated annealing, and atomic B-factor refinement, all done in PHENIX (Adams et al., 2010). At each stage, high temperature simulated annealing was employed to reduce model bias (Brunger and Rice, 1997). Following initial refinement of the model used for molecular replacement, the deleted nucleotides were rebuilt using Coot (Emsley et al., 2010). Solvent was subsequently built automatically and edited manually. Finally, the ligand was modeled with the aid of 2Fo-Fc and Fo-Fc maps; energy restraints for ligands were generated using PHENIX eLBOW. Crystallographic data and refinement statistics are given in Table S2 of the Supplemental Information. To validate that the crystallographic data supported the placement of each ligand in the fully refined model, 2Fo-Fc simulated annealing omit maps were generated by removing the ligand as well as bases involved in ligand binding. All figures of models and maps were completed with PyMol. The model and structure factors were submitted to the Protein Data Bank (PDB) under the accession codes 4LVV (6S-folinic Acid), 4LVX (tetrahydrobiopterin), 4LVW (7-deazaguanine), 4LVY (Pemetrexed), 4LVZ (2,6-diaminopurine), and 4LW0 (adenine).
Supplementary Material
HIGHLIGHTS.
While most THF riboswitches contain two binding sites, one is not vital for function
The pABA moiety of THF greatly enhances regulatory activity but not binding affinity
Some well-established antifolates can the regulate the riboswitch’s activity
Adenine and derivatives exhibit “retro-binding” to the THF riboswitch
Acknowledgements
We gratefully acknowledge Merck & Cie for generously providing folate derivatives for this study.
Funding Source Statement This work was supported by a grant from the National Institutes of Health (R01 GM073850) to R.T.B. and a predoctoral Creative Training in Molecular Biology Grant (NIH 5-T32-GM07135) to J.J.T.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica Section D, Biological crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames TD, Rodionov DA, Weinberg Z, Breaker RR. A eubacterial riboswitch class that senses the coenzyme tetrahydrofolate. Chemistry & biology. 2010;17:681–685. doi: 10.1016/j.chembiol.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick JE, Breaker RR. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome biology. 2007;8:R239. doi: 10.1186/gb-2007-8-11-r239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batey RT, Gilbert SD, Montange RK. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature. 2004;432:411–415. doi: 10.1038/nature03037. [DOI] [PubMed] [Google Scholar]
- Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta crystallographica Section D, Biological crystallography. 2011;67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett BD, Kimball EH, Gao M, Osterhout R, Van Dien SJ, Rabinowitz JD. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nature chemical biology. 2009;5:593–599. doi: 10.1038/nchembio.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermingham A, Derrick JP. The folic acid biosynthesis pathway in bacteria: evaluation of potential for antibacterial drug discovery. BioEssays : news and reviews in molecular, cellular and developmental biology. 2002;24:637–648. doi: 10.1002/bies.10114. [DOI] [PubMed] [Google Scholar]
- Blount KF, Breaker RR. Riboswitches as antibacterial drug targets. Nature biotechnology. 2006;24:1558–1564. doi: 10.1038/nbt1268. [DOI] [PubMed] [Google Scholar]
- Bolin JT, Filman DJ, Matthews DA, Hamlin RC, Kraut J. Crystal structures of Escherichia coli and Lactobacillus casei dihydrofolate reductase refined at 1.7 A resolution. I. General features and binding of methotrexate. The Journal of biological chemistry. 1982;257:13650–13662. [PubMed] [Google Scholar]
- Breaker RR. Riboswitches and the RNA world. Cold Spring Harbor perspectives in biology. 2012;4 doi: 10.1101/cshperspect.a003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Rice LM. Crystallographic refinement by simulated annealing: methods and applications. Methods in enzymology. 1997;277:243–269. doi: 10.1016/s0076-6879(97)77015-0. [DOI] [PubMed] [Google Scholar]
- Bystroff C, Oatley SJ, Kraut J. Crystal structures of Escherichia coli dihydrofolate reductase: the NADP+ holoenzyme and the folate.NADP+ ternary complex. Substrate binding and a model for the transition state. Biochemistry. 1990;29:3263–3277. doi: 10.1021/bi00465a018. [DOI] [PubMed] [Google Scholar]
- Ceres P, Garst AD, Marcano-Velazquez JG, Batey RT. Modularity of Select Riboswitch Expression Platforms Enables Facile Engineering of Novel Genetic Regulatory Devices. ACS synthetic biology. 2013a doi: 10.1021/sb4000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceres P, Trausch JJ, Batey RT. Engineering modular ‘ON’ RNA switches using biological components. Nucleic Acids Research, (accepted) 2013b doi: 10.1093/nar/gkt787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cressina E, Chen LH, Abell C, Leeper FJ, Smith AG. Fragment screening against the thiamine pyrophosphate riboswitch thiM. Chem Sci. 2011;2:157–165. [Google Scholar]
- Daldrop P, Reyes FE, Robinson DA, Hammond CM, Lilley DM, Batey RT, Brenk R. Novel ligands for a purine riboswitch discovered by RNA-ligand docking. Chemistry & biology. 2011;18:324–335. doi: 10.1016/j.chembiol.2010.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dann CE, 3rd, Wakeman CA, Sieling CL, Baker SC, Irnov I, Winkler WC. Structure and mechanism of a metal-sensing regulatory RNA. Cell. 2007;130:878–892. doi: 10.1016/j.cell.2007.06.051. [DOI] [PubMed] [Google Scholar]
- Deigan KE, Ferre-D’Amare AR. Riboswitches: discovery of drugs that target bacterial gene-regulatory RNAs. Accounts of chemical research. 2011;44:1329–1338. doi: 10.1021/ar200039b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon N, Duncan JN, Geerlings T, Dunstan MS, McCarthy JE, Leys D, Micklefield J. Reengineering orthogonally selective riboswitches. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:2830–2835. doi: 10.1073/pnas.0911209107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta crystallographica Section D, Biological crystallography. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garst AD, Batey RT. A switch in time: detailing the life of a riboswitch. Biochimica et biophysica acta. 2009;1789:584–591. doi: 10.1016/j.bbagrm.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garst AD, Heroux A, Rambo RP, Batey RT. Crystal structure of the lysine riboswitch regulatory mRNA element. The Journal of biological chemistry. 2008;283:22347–22351. doi: 10.1074/jbc.C800120200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garst AD, Porter EB, Batey RT. Insights into the regulatory landscape of the lysine riboswitch. Journal of molecular biology. 2012;423:17–33. doi: 10.1016/j.jmb.2012.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert SD, Batey RT. Monitoring RNA-ligand interactions using isothermal titration calorimetry. Methods Mol Biol. 2009;540:97–114. doi: 10.1007/978-1-59745-558-9_8. [DOI] [PubMed] [Google Scholar]
- Gilbert SD, Mediatore SJ, Batey RT. Modified pyrimidines specifically bind the purine riboswitch. Journal of the American Chemical Society. 2006a;128:14214–14215. doi: 10.1021/ja063645t. [DOI] [PubMed] [Google Scholar]
- Gilbert SD, Stoddard CD, Wise SJ, Batey RT. Thermodynamic and kinetic characterization of ligand binding to the purine riboswitch aptamer domain. Journal of molecular biology. 2006b;359:754–768. doi: 10.1016/j.jmb.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Gonen N, Assaraf YG. Antifolates in cancer therapy: structure, activity and mechanisms of drug resistance. Drug resistance updates : reviews and commentaries in antimicrobial and anticancer chemotherapy. 2012;15:183–210. doi: 10.1016/j.drup.2012.07.002. [DOI] [PubMed] [Google Scholar]
- Guan L, Disney MD. Recent advances in developing small molecules targeting RNA. ACS chemical biology. 2012;7:73–86. doi: 10.1021/cb200447r. [DOI] [PubMed] [Google Scholar]
- Haller A, Souliere MF, Micura R. The dynamic nature of RNA as key to understanding riboswitch mechanisms. Accounts of chemical research. 2011;44:1339–1348. doi: 10.1021/ar200035g. [DOI] [PubMed] [Google Scholar]
- Huang L, Ishibe-Murakami S, Patel DJ, Serganov A. Long-range pseudoknot interactions dictate the regulatory response in the tetrahydrofolate riboswitch. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:14801–14806. doi: 10.1073/pnas.1111701108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illarionova V, Eisenreich W, Fischer M, Haussmann C, Romisch W, Richter G, Bacher A. Biosynthesis of tetrahydrofolate. Stereochemistry of dihydroneopterin aldolase. The Journal of biological chemistry. 2002;277:28841–28847. doi: 10.1074/jbc.M204046200. [DOI] [PubMed] [Google Scholar]
- Jabrin S, Ravanel S, Gambonnet B, Douce R, Rebeille F. One-carbon metabolism in plants. Regulation of tetrahydrofolate synthesis during germination and seedling development. Plant physiology. 2003;131:1431–1439. doi: 10.1104/pp.016915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariluoto S, Edelmann M, Herranen M, Lampi AM, Shmelev A, Salovaara H, Korhola M, Piironen V. Production of folate by bacteria isolated from oat bran. International journal of food microbiology. 2010;143:41–47. doi: 10.1016/j.ijfoodmicro.2010.07.026. [DOI] [PubMed] [Google Scholar]
- Keshava C, Keshava N, Whong WZ, Nath J, Ong TM. Inhibition of methotrexate-induced chromosomal damage by folinic acid in V79 cells. Mutation research. 1998;397:221–228. doi: 10.1016/s0027-5107(97)00216-9. [DOI] [PubMed] [Google Scholar]
- Lee ER, Blount KF, Breaker RR. Roseoflavin is a natural antibacterial compound that binds to FMN riboswitches and regulates gene expression. RNA biology. 2009;6:187–194. doi: 10.4161/rna.6.2.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemay JF, Desnoyers G, Blouin S, Heppell B, Bastet L, St-Pierre P, Masse E, Lafontaine DA. Comparative study between transcriptionally- and translationally-acting adenine riboswitches reveals key differences in riboswitch regulatory mechanisms. PLoS genetics. 2011;7:e1001278. doi: 10.1371/journal.pgen.1001278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulhbacher J, Brouillette E, Allard M, Fortier LC, Malouin F, Lafontaine DA. Novel riboswitch ligand analogs as selective inhibitors of guanine-related metabolic pathways. PLoS pathogens. 2010;6:e1000865. doi: 10.1371/journal.ppat.1000865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed LS, Archer MC. Oxidation of Tetrahydrofolic Acid by Air. J Agr Food Chem. 1980;28:801–805. [Google Scholar]
- Reyes FE, Garst AD, Batey RT. Strategies in RNA crystallography. Methods in enzymology. 2009;469:119–139. doi: 10.1016/S0076-6879(09)69006-6. [DOI] [PubMed] [Google Scholar]
- Saraste M, Musacchio A. Backwards and forwards binding. Nature structural biology. 1994;1:835–837. doi: 10.1038/nsb1294-835. [DOI] [PubMed] [Google Scholar]
- Scott JM, Weir DG. Folic acid, homocysteine and one-carbon metabolism: a review of the essential biochemistry. Journal of cardiovascular risk. 1998;5:223–227. [PubMed] [Google Scholar]
- Serganov A, Huang L, Patel DJ. Structural insights into amino acid binding and gene control by a lysine riboswitch. Nature. 2008;455:1263–1267. doi: 10.1038/nature07326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Nudler E. A decade of riboswitches. Cell. 2013;152:17–24. doi: 10.1016/j.cell.2012.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Patel DJ. Metabolite recognition principles and molecular mechanisms underlying riboswitch function. Annual review of biophysics. 2012;41:343–370. doi: 10.1146/annurev-biophys-101211-113224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsan N, Cohen-Chalamish S, Nakamura S, Emilsson GM, Breaker RR. Thiamine pyrophosphate riboswitches are targets for the antimicrobial compound pyrithiamine. Chemistry & biology. 2005;12:1325–1335. doi: 10.1016/j.chembiol.2005.10.007. [DOI] [PubMed] [Google Scholar]
- Thore S, Frick C, Ban N. Structural basis of thiamine pyrophosphate analogues binding to the eukaryotic riboswitch. Journal of the American Chemical Society. 2008;130:8116–8117. doi: 10.1021/ja801708e. [DOI] [PubMed] [Google Scholar]
- Tomsic J, McDaniel BA, Grundy FJ, Henkin TM. Natural variability in S-adenosylmethionine (SAM)-dependent riboswitches: S-box elements in bacillus subtilis exhibit differential sensitivity to SAM In vivo and in vitro. Journal of bacteriology. 2008;190:823–833. doi: 10.1128/JB.01034-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trausch JJ, Ceres P, Reyes FE, Batey RT. The structure of a tetrahydrofolate-sensing riboswitch reveals two ligand binding sites in a single aptamer. Structure. 2011;19:1413–1423. doi: 10.1016/j.str.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson PY, Fedor MJ. The glmS riboswitch integrates signals from activating and inhibitory metabolites in vivo. Nature structural & molecular biology. 2011;18:359–363. doi: 10.1038/nsmb.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickiser JK, Winkler WC, Breaker RR, Crothers DM. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Molecular cell. 2005;18:49–60. doi: 10.1016/j.molcel.2005.02.032. [DOI] [PubMed] [Google Scholar]
- Wilson-Mitchell SN, Grundy FJ, Henkin TM. Analysis of lysine recognition and specificity of the Bacillus subtilis L box riboswitch. Nucleic Acids Res. 2012;40:5706–5717. doi: 10.1093/nar/gks212. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.