Abstract
Chromatin, composed of DNA wrapped around an octamer of histones, is the relevant substrate for all genetic processes in eukaryotic nuclei. Changes in chromatin structure are associated with the activation and silencing of gene transcription and reversible post-translational modifications of histones are now known to direct chromatin structure transitions. Recent studies in several fungal species have identified a chromatin-based regulation of secondary metabolism (SM) gene clusters representing an upper-hierarchical level for the coordinated control of large chromosomal elements. Regulation by chromatin transition processes provides a mechanistic model to explain how different SM clusters located at dispersed genomic regions can be simultaneously silenced during primary metabolism. Activation of SM clusters has been shown to be associated with increased acetylation of histones H3 and H4 and, consequently, inhibition of histone de-acetylase activities also leads to increased production of secondary metabolites. New findings suggest that SM clusters are silenced by heterochromatic histone marks and that the “closed” heterochromatic structures are reversed during SM activation. This process is mediated by the conserved activator of SM, LaeA. Despite the increase in knowledge about these processes, much remains to be learned from chromatin-level regulation of SM. For example, which proteins “position” the chromatin restructuring signal onto SM clusters or how exactly LaeA works to mediate the low level of heterochromatic marks inside different clusters remain open questions. Answers to these and other chromatin-related questions would certainly complete our understanding of SM gene regulation and signaling and, because for many predicted SM clusters corresponding products have not been identified so far, anti-silencing strategies would open new ways for the identification of novel bioactive substances.
Keywords: Chromatin, Epigenetics, Heterochromatin, Secondary metabolism, Aspergillus, Neurospora
1. Introduction
Production of secondary metabolites by filamentous asco- and basidiomycetes is a specialized process that occurs only under specific environmental conditions or at specific points in their life cycle (Bennett, 1987; Hoffmeister and Keller, 2007). The term “secondary metabolism” (SM) is not well defined but it is generally accepted that it denotes a stage in the fungal life history at which “primary metabolism”, i.e. metabolic processes connected with active growth (cell expansion and cell division), is phased out. It has been shown that the transition from primary to secondary metabolism is often correlated with depletion of nutrients, absence of light, development of reproductive structures, or changes in ambient pH (Calvo et al., 2002; Yu and Keller, 2005). Secondary metabolism is intimately connected to sexual fruiting body formation in Aspergillus nidulans or the production of sclerotia in Aspergillus flavus (Calvo et al., 2002; Chang et al., 2002). Also, conidiation (asexual spore formation) is coordinated with SM-gene expression through the production of spore-related products such as spore pigments by polyketide synthetases (Adams and Yu, 1998). Despite the enormous progress made in understanding the regulation and signal transduction of secondary metabolite gene expression, the plethora of these diverse environmental signals until now results in a quite fuzzy picture of regulatory keypoints in gene regulation of secondary metabolite genes.
Aspergilli represent a highly diverse group of filamentous fungi with a high capacity to produce secondary metabolites and the aflatoxin, sterigmatocystin (ST) and penicillin (PN) production pathways have served as instructive models to understand, at the molecular level, the regulation of the involved genes. Studies in A. nidulans, mainly on PN and ST gene clusters, have paved the way for a general understanding of secondary metabolic gene regulation. Results obtained from this model provided a constructive guidance for similar studies in economically important species such as A. flavus, Aspergillus parasiticus, Penicillium chrysogenum or Fusarium graminearum (Goswami et al., 2006; Kosalkova et al., 2009; Payne and Yu, 2010). Secondary metabolite (SM) signaling and regulation in A. nidulans has been dealt with in detail in a large number of reviews (Brakhage et al., 2004; Calvo, 2008; Hoffmeister and Keller, 2007; Keller et al., 2005). Recent results from a number of laboratories added a novel aspect of a higher-hierarchical level of regulation in this model fungus, i.e. the role of chromatin structure and nucleosome modifications in expression of secondary metabolite genes. Chromatin structure and function is currently one of the most actively studied topics in molecular biology and progress in the field is constantly reviewed from different perspectives. Numerous reviews have extensively dealt with the function and modifications of chromatin for basic cellular processes such as transcription, DNA replication and repair, RNA interference, and the role chromatin plays in mitotic and meiotic chromosome separation (Holbert and Marmorstein, 2005 and references therein).
Here we focus our view on chromatin processes related to fungal secondary metabolism and refer the reader to recent overview articles for more detailed information on mechanisms regulating chromatin-related processes.
2. Chromatin modifications and transcription
Chromatin is the complex of nuclear DNA with proteins in which the genetic material of eukaryotes is packed. The fundamental subunit of chromatin is the nucleosome. It consists of roughly 165 bp of DNA wrapped in two superhelical turns around an octamer of four different core histone proteins (two each of H2A, H2B, H3 and H4). This physical arrangement of DNA on nucleosomes results in a roughly 10-fold compaction which is additionally increased by interactions of neighboring nucleosomes to form a 30 nm chromatin fiber (Tremethick, 2007). Additional interactions with non-histone chromatin proteins and RNAs are believed to mediate higher-order chromatin structures, such as condensed mitotic or meiotic chromosomes (Felsenfeld and Groudine, 2003; Luger and Hansen, 2005; Woodcock and Dimitrov, 2001).
However, chromatin is both the natural substrate and a barrier for the molecular machinery that needs to contact and move along DNA during transcription or DNA replication. Thus, chromatin structure needs to be dynamic in order to adapt to different cellular requirements. Flexibility of the chromatin structure is achieved by ATP-dependent chromatin remodeling (Cairns, 2009) and by decorating specific residues of histone proteins with an array of different post-translational modifications (PTMs). This reversible marking includes positioning or removal of acetyl and methyl groups on lysines or arginines, but also phosphorylation of serines or threonines and ubiquitination of lysines (Kouzarides, 2007). An additional level of complexity is introduced by “trans-histone” regulation in which a given set of histone modifications regulates the positioning or removal of other modifications on the same histone or on neighboring histones in the nucleosome (Fischle et al., 2003; Suganuma and Workman, 2008; Wang et al., 2001).
The specific pattern of covalent modifications on a given genomic region is proposed to generate a “histone code” (Jenuwein and Allis, 2001) where the modified histone residues serve as docking stations for proteins which promote either an open (euchromatic) or a closed (heterochromatic) chromatin conformation. Euchromatic regions are rich in coding sequences, and their high transcriptional activity correlates with hyper-acetylated nucleosomal histones. Methylated lysines in H3 and H4 can have different impacts on chromatin structure. Whereas euchromatin is usually enriched in methylated H3K4, typical heterochromatic marks are characterized by H3K9, H3K27 and H4K20 methlyation. Pericentromeric and (sub)telomeric segments represent typical gene-poor “heterochromatic” regions which exhibit low levels of transcription (Grewal and Jia, 2007). Essential to the understanding of heterochromatin formation was the identification of the direct link between tri-methylation of H3K9 and binding of HP-1, a central protein functioning in the formation of repressive, highly condensed heterochromatin (Bannister et al., 2001; Horn and Peterson, 2006; Jacobs and Khorasanizadeh, 2002). Surprisingly, certain histone modifications can display dual functions as they can have different effects depending on the physical location of the chromatin stretch. For example, in Saccharomyces cerevisiae, H3K4 tri-methylation is a permissive mark for gene transcription in euchromatic regions but it is also required for gene silencing at mating type loci and sub-telomeric regions (Bryk et al., 2002; Mueller et al., 2006).
In filamentous fungi chromatin structure and function has only been studied in a few model systems (summarized in Fig. 1). Pioneering work has revealed an essential function of heterochromatic marks and HP-1 for DNA methylation and telomere silencing in Neurospora crassa (Honda and Selker, 2008; Kouzminova and Selker, 2001; Lewis et al., 2009; Selker et al., 2002, 2003; Smith et al., 2008; Tamaru and Selker, 2001; Tamaru et al., 2003). In the same organism histone acetylation and chromation remodeling has been studied in relation to transcriptional activation and repression processes during light regulation (Belden et al., 2007; Grimaldi et al., 2006). Experimental chromatin work in A. nidulans initially focussed on nucleosomal organization (Morris, 1976) and the connection of chromatin to regulation of the mitotic cell cycle (Osmani et al., 1988). The genomic inventory of the basic chromatin components in different filamentous fungi has been recently reviewed and chromatin regulation of several catabolic genes was summarized (Brosch et al., 2008; Scazzocchio and Ramon, 2008).
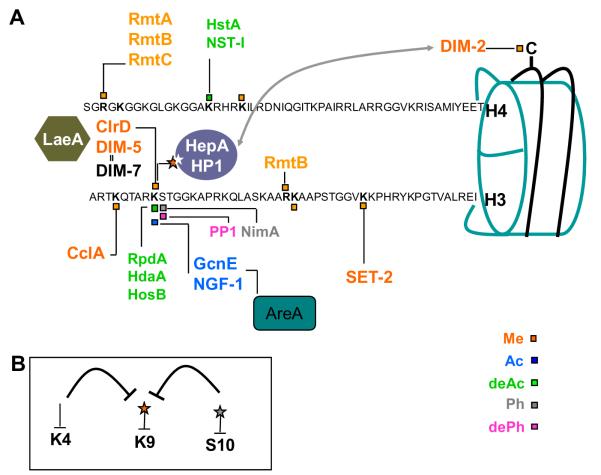
Fig. 1.
(A) Representation of histones H3 and H4 N terminal sequences for which post-translational covalent modifications (epigenetic marks) have been shown to influence the function of filamentous fungal chromatin. Squares indicate the type of modification carried out by the corresponding enzyme. Me, methylation; Ac, acetylation; deAc, deacetylation; Ph, phosphorylation; dePh, dephosphorylation. In A. nidulans H3K9 is subject to methylation by ClrD and recognition by HepA, the presence of these marks are associated with the repression of several SM gene clusters in A. nidulans during primary metabolism (Reyes-Dominguez et al., 2010). LaeA is a positive global regulator of SM in fungi (Bok and Keller, 2004; Perrin et al., 2007; Wiemann et al., in press), that counteracts the methylation of H3K9 and HepA binding at SM clusters by an unknown mechanism. H3K4 is subject to methylation by a putative COMPASS complex harboring CclA. In Neurospora crassa H3K9 is methylated by DIM-5 and recognized by HP1, this mark directs the methylation of cytosine (C) on the DNA and leads to silencing. A direct interaction between HP1 and DIM-2 is required for DNA methylation (gray line) (Honda and Selker, 2008; Tamaru and Selker, 2001; Tamaru et al., 2003). Dim-7 is essential for the recruitment of Dim-5 and the formation of heterochromatin, these proteins interact in vivo (double line) (Lewis et al., 2009). SET-2 is a H3K36 methyltransferase essential for normal growth and development in N. crassa. H3K36me and H3K4me are enriched in actively transcribed genes (Adhvaryu et al., 2005). The recombinant A. nidulans arginine methyltransferases, RmtA, RmtB and RmtC methylate H4R3 in vitro. RmtB methylates preferentially H4 and to a lesser extent H3 and H2A. H4R3 and H3R26 are methylated in vivo (Trojer et al., 2004). In Ascobolus, the methylation of the downstream portion of a gene promotes its stable silencing and triggers the production of truncated transcripts, in this regions there is an increase of H3K9me2 and a decrease of H3K14me2 (Barra et al., 2005). In A. nidulans, GcnE (HAT catalytic subunit) and AdaB (an architectural protein) of the putative SAGA complex contribute to the increase of H3K9K14ac in a crucial nucleosome of the prnD–prnB bidirectional promoter. In this case, acetylation is required for strict nucleosomal positioning and for the repression of the prnB–prnD cluster (Reyes-Dominguez et al., 2008). In contrast, H3 acetylation is correlated with activation of the nitrate cluster and the broad domain GATA factor AreA mediates this effect (Berger et al., 2008). NGF-1 is homologous to GcnE and required for induction of the light responsive al-3 gene in N. crassa (Grimaldi et al., 2006). The activity of histone de-acetylases HdaA, HosB and HstA was shown to be involved in germination and SM production in A. fumigatus and in A. nidulans HdaA is responsible for the majority of HDAC activities (Lee et al., 2009; Shwab et al., 2007; Tribus et al., 2005). In N. crassa, NST-I is a H4 HDAC required for silencing at telomeric regions. The subtelomeric regions of this organism are enriched with H3K9me3, H3K27me3, and H4K20me3 (Smith et al., 2008). NimA is a H3S10 kinase required for entry into mitosis and chromatin condensation in A. nidulans (De Souza et al., 2000). (B) Cross-talk between residue modifications. In Neurospora crassa, loss of PP1 phosphatase function results in the absence of H3K9me3 and the abolishment of DNA methylation at several parts of the genome (Adhvaryu and Selker, 2008). The lack of di and trimethylation on H3K4 inside the A. nidulans monodictyphenone cluster negatively influences the amount of H3K9me3 (Bok et al., 2009).
3. Regulation of fungal secondary metabolism by histone acetylation
The biosynthesis of secondary metabolites is a multi-enzyme process, and in most of the cases, the genes coding for the enzymes in a given biosynthetic pathway are clustered in the fungal genome (Keller and Hohn, 1997; Khaldi et al., 2010). In several pathways the specific transcription factor is also embedded within the cluster; such as aflR regulating sterigmatocystin (ST) production in A. nidulans (Hoffmeister and Keller, 2007), aflatoxin production in A. flavus and A. parasiticus (Chang et al., 1993; Smith et al., 2007). Also the F. graminearum (teleomorph Giberella zeae) genome has been found to contain biosynthetic gene clusters harboring pathway activators, e.g. the aurofusarin and trichotecene clusters (Kimura and Horikoshi, 2004; Malz et al., 2005).
It was intuitive to propose that this genomic arrangement may facilitate a coordinated regulation of secondary metabolite gene clusters. First evidence of chromatin involvement was obtained in studies using histone de-acetylase (HDAC) deletion mutants in A. nidulans. Keller, Grässle and colleagues demonstrated that genetic or chemical inactivation of HDACs in a range of SM-producing filamentous fungi leads to up-regulation of SM genes and to an elevated level of several metabolites (ST, PN and terrequinone A) (Shwab et al., 2007). This broad effect on the chemical diversity in one organism and a similar function in unrelated fungi (e.g. Alternaria, Penicillium) suggested a conserved mechanism for SM-gene expression by concomitant chromatin acetylation (Williams et al., 2008). Direct biochemical evidence (by chromatin immunoprecipitation-ChIP assays) for the involvement of histone acetylation in the regulation of the A. parasiticus aflatoxin (AF) cluster (Roze et al., 2007) and the A. nidulans ST cluster (Reyes-Dominguez et al., 2010) has been subsequently obtained. Which histone acetyltransferases (HATs) mediate the increase in histone acetylation of ST genes in A. nidulans remains to be determined. HAT-components of the conserved SAGA/ADA complex (Timmers and Tora, 2005) are probably not involved in this process because mutants lacking the catalytic enzyme (GcnE) or an adaptor (AdaB) of the putative A. nidulans SAGA complex (Reyes-Dominguez et al., 2008) showed no difference in ST production capacities on solid media (Reyes and Strauss, unpublished). This indicates that other acetylation activities (e.g. NuA4/p300/Esa1-type complexes acting mainly on histone H4) may be more relevant for the ST gene activation process.
Interestingly, the specific order of histone H4 acetylation pattern in chromatin domains of the A. parasiticus aflatoxin gene cluster roughly paralleled the pattern of gene activation events. Although AflR binding coincided with the spread of H4 acetylation along the cluster, it was surprising to find AflR function not being required for these chromatin rearrangement processes. Based on previous work (Roze et al., 2004) and the function of cAMP-response element binding proteins (CREB) in recruitment of chromatin remodeling factors (Blobel, 2002) the authors hypothesized that a CREB homolog, instead of AflR, might recruit the HAT complexes to the AF cluster. Subsequent binding of and activation by AflR would turn on this SM gene cluster. Future work in A. parasiticus probably will provide additional experimental evidence.
4. LaeA, a conserved fungal protein involved in defining the chromatin landscape
The isolation of a laeA (stands for “loss of aflR expression-A”) mutant by the Keller lab certainly was an important contribution to better understand the global aspect of SM gene regulation. Besides A. nidulans, a crucial function for LaeA in secondary metabolism has so far been documented for A. flavus (Amaike and Keller, 2009; Kale et al., 2008), Aspergillus fumigatus (Bok et al., 2005; Perrin et al., 2007), P. chrysogenum (Kosalkova et al., 2009) and recently also in a Fusarium species (Wiemann et al., in press). LaeA does not show homology to proteins with known function and has no typical transcription factor signature (e.g. DNA-binding motif), but instead carries an S-adenosyl-l-methionine (SAM) binding domain. SAM domains are identifying features of methyltransferases, transferring a methyl group from the ubiquitous SAM to either nitrogen, oxygen or carbon atoms. This reaction is frequently employed in organisms from all phyla and modifies not only proteins but also DNA, RNA and small molecules. Thus, the regulatory role executed by LaeA could be diverse but the presence of such domains in histone methyltransferases has nourished speculations that LaeA is a transcription factor involved in histone methylation thereby changing the chromatin landscape of a larger genomic region.
As mentioned above, the first hint for LaeA involvement in chromatin modification came from studies which demonstrated that the block of the HdaA-HDAC activity resulted in a partial bypass of laeA requirement for ST production. To fully bypass ST production in laeAΔ, the simultaneous deletion of classical HDACs (HdaA, HosB) as well as the sirtiun-type HstA (Shwab et al., 2007) is necessary. Sirtiuns are known to cooperate with classical HDACs to silence sub-telomeric regions in S. pombe (Wiren et al., 2005) and N. crassa sirtuin NST-1 has been shown to be necessary to silence a reporter gene inserted near a telomere (Smith et al., 2008).
However, in hdaAΔ, the tested SM genes are not de-repressed “by default” during primary metabolism suggesting that the secondary metabolite signal (SMS) is still necessary for SM-gene expression in HDAC mutant strains. In addition, ChIP experiments in the promoter region of A. nidulans aflR showed that the acetylation pattern in a laeAΔ mutant is not significantly different from wild type (Reyes-Dominguez et al., 2010) which indicates that LaeA and HDACs function in different pathways.
5. LaeA counteracts heterochromatin formation
According to the proposed “trans-histone” cross-talk (Fischle, 2008) hypoacetylation can influence the establishment of heterochromatic marks, such as H3K9me3 and subsequent binding of heterochromatin protein (Schotta et al., 2002). In a preliminary transcriptome analysis of heterochromatin mutants we noticed higher expression of several genes involved in SM biosynthesis in a strain deleted for the A. nidulans HP-1 homolog, HepA (Y. Reyes-Dominguez, C. Scazzocchio and J. Strauss, unpublished observation). Following this initial finding, we have reported recently that deletion of the A. nidulans H3K9 methyltransferase ClrD and the HP-1 homolog HepA, results in over-expression of several genes belonging to characterized SM clusters (Reyes-Dominguez et al., 2010). Subsequent analysis of histone marks by ChIP demonstrated that heterochromatic marks were enriched in the ST cluster under conditions of primary metabolism, but were reduced when secondary metabolism conditions were applied. Importantly, ST production was fully restored to wild-type levels on solid media when either hepAΔ or clrDΔ were combined with laeAΔ indicating that LaeA and heterochromatin formation function in the same pathway but with opposite roles. ChIP analysis of heterochromatic marks in laeAΔ strains revealed that H3K9me3 and HepA binding to the tested ST locus genes are dramatically increased. These results for the first time directly link the function of the SAM-domain containing protein LaeA to histone H3 methylation (and heterochromatin formation). This study also revealed functional “border” elements separating the ST cluster from immediately neighboring genomic regions. Both expression studies and ChIP experiments demonstrated that reversal of the heterochromatin status by the LaeA-mediated process is confined to the cluster region and sequences outside the cluster remained heterochromatic under all tested metabolic conditions. The functional elements creating an active border are not known at the moment but the presence of repetitive sequences flanking the ST cluster could play a role in border definition, similar to the transposable element sequences shown to be required for efficient expression of the PN cluster (Shaaban et al., 2010).
Regulation of SM clusters by facultative heterochromatin would be an efficient way to silence larger genomic regions under conditions where their expression would not be beneficial for the organism, i.e. during favorable environmental conditions, active growth and plenty of available nutrients. When nutrients become limited and environmental factors unfavorable for growth, a LaeA-mediated process would trigger in such a scenario the removal of heterochromatic marks, allowing the positioning of activation marks and assembly of the transcriptional machinery.
6. Does position matter?
Whether a positional bias exists for LaeA-regulated SM clusters towards chromosome ends (sub-telomeric regions) has been discussed but experimental data from A. nidulans and A. fumigatus have shown LaeA target gene clusters positioned at different chromosomal locations, from 30 kb sub-telomeric to central positions on different chromosome arms (Perrin et al., 2007; Shwab et al., 2007). This genomic arrangement makes it more likely that specific features also present in sub-telomeric or pericentromeric regions, such as multiple sequence repeats or relics of transposon insertions, are associated with SM clusters. The preferential location of SM genes in highly recombinogenic regions, whether sub-telomeric or not, might reflect the evolution of SM clusters and represent relics of horizontal gene transfers from other organisms. The sub-telomeric location and the presence of paralogues for penicillin biosynthetic genes in A. nidulans (Brakhage et al., 2009; Walton, 2000) provides an example for such specific genomic arrangement that would allow facile acquisition and coordinated regulation of gene clusters. Consistent with the latter hypothesis are also results which show that ectopic expression of SM activators lead to gene expression of SM genes out of context of the usual signaling pathways. For example, addition of an extra copy of the specific transcriptional regulator aflR outside the A. nidulans ST cluster remediates ST production in a strain lacking LaeA (Bok et al., 2006). Using the strategy of over-expression of a candidate activator residing within a predicted SM cluster without a known metabolite, the Brakhage lab successfully activated a silent cluster (aspyridone) for which the natural conditions of expressions are not known (Bergmann et al., 2007).
7. Does silencing of SM clusters need a COMPASS?
As outlined above, histone methylation marks active and inactive chromatin, depending on the residue and the type of methylation. Actively transcribed genes in euchromatic regions are usually enriched in di- and tri-methylated lysine 4, a mark deposited by COMPASS, a macromolecular complex containing the Set1 (MLL in human) methyltransferase as catalytic subunit (Shilatifard, 2006; Sims and Reinberg, 2006). H3K4me3 is mainly found at 5′ regions of transcribed genes, coinciding with high levels of Set1 (Ng et al., 2003) and several chromatin remodeling factors such as CHD1, TAF1 or NurF have been shown to co-localize with this activating mark (Bhaumik et al., 2007; Kouzarides, 2007). In addition, H3K4me2/3 displays a context-dependent readout, i.e. it can trigger not only activating, but also repressing events. This is the case in budding yeast, where COMPASS is required for silencing of the mating type loci and sub-telomeric regions (Bryk et al., 2002; Meijsing and Ehrenhofer-Murray, 2001; Mueller et al., 2006). A repressive downstream event is also known in mammals, where ING2, a member of the human HDAC1 complex, specifically binds to H3K4me2/3 via its PHD domain and thereby mediates gene repression and growth inhibition upon DNA damage (Pena et al., 2006; Shi et al., 2006).
In N. crassa, H3K4 methylation has also been found to co-localize with active genes (Adhvaryu et al., 2005; Lewis et al., 2009). In an effort to test H3K4me involvement in SM-gene expression the Keller lab created mutants in putative A. nidulans COMPASS complex genes which, with the exception of putative Set1, do not seem to be essential in individual deletions (Bok and Keller, personal communication). Deletion of cclA, the yeast Bre2 homolog, showed a remarkable shift in the secondary metabolite profile and the deletion strain produced at least eight additional metabolites, among them monodictyphenone (MDP), emodin and four emodin analogs not seen before in this fungus (Bok et al., 2009). Subsequent ChIP analysis showed a strong reduction in H3K4 di- and tri-methylation in promoters and genes inside and outside the proposed MDP cluster. These data established the functionality of an A. nidulans Bre2-homolog in H3K4me2/3 marking. However, the biological readout of H3K4 hypo-methylation was different to what could be expected from COMPASS mutants as an “anti-silencing” effect was observed for some cryptic SM clusters. Strikingly, only genes inside the proposed cluster were upregulated, although a reduction in H3K4me2/3 was also observed in the flanking regions. Specific to the genes inside the cluster, however, was a simultaneous reduction of the canonical heterochromatic mark H3K9me3. This is reminiscent of the situation found within the A. nidulans ST cluster in which reduction of H3K9me3 marks lead to over-expression of ST genes (Reyes-Dominguez et al., 2010).
A mechanistic model for a cross-talk between H3K4me and H3K9me exists so far only for the activation event. This involves a H3K9 demethylase (JMJD2A) which directly binds to H3K4me2/3 via its TUDOR domain and concomitantly removes the neighboring repressive H3K9me3 mark, paving the way for gene activation (Huang et al., 2006). In the MDP cluster H3K4me2/3 seems to be required for the opposite effect, i.e. mediating the deposition of the repressive H3K9me3 mark and silencing. This would be more similar to the situation in budding yeast, where H3K4 methylation is associated with sub-telomeric and rDNA silencing (Mueller et al., 2006). It is worth noting, however, that the function of COMPASS and heterochromatic marks seem to be conserved in regulating fungal SM, as deletion of hepA and cclA-homologs from the F. graminearum genome also results in an altered chemical profile (Y. Reyes-Dominguez, S. Boedi, G. Adam and J. Strauss, unpublished).
Curiously, an overlapping set of genes (MDP cluster) has been found upregulated using a totally different approach: co-cultivation of A. nidulans with some specific Streptomycetes strains in controlled fermenters led to the same effect of MDP accumulation as identified in the cclA mutant (Schroeckh et al., 2009). If this coincidence is not mere serendipity, these results imply that the co-cultivation resulted in inactivation of CclA and/or reduction in H3K9 methylation. These astounding findings point towards an important ecological function of SM regulation by chromatin modification and it will be interesting to see which characteristics of the bacterial strains define whether silent fungal SM clusters become activated or not.
8. Summary and perspectives
Arguably, one of the most thoroughly studied SM pathways in fungi are sterigmatocystin (ST) and aflatoxin (AF) formation in aspergilli. Fig. 2 shows a simplified model of the current knowledge in SM gene regulation. This sketch should illustrate the degree of integration that is necessary to coordinate the activity of transcriptional regulators for SM production. Ultimately, signaling culminates in the activation of two known main transcriptional regulators, AflR and LaeA. AflR is transcriptionally and post-transcriptionally activated by SMS (secondary metabolite signals), binds to target sequences in the promoters of ST or AF genes and, most likely in cooperation with co-activators such as AflS, the pH-regulator PacC, or a proposed CREB-P (cAMP-response-element-binding-protein) initiates efficient transcription (Payne and Yu, 2010). However, as the aflR gene as well as many other cluster-specific SM transcriptional regulators resides inside the SM cluster, SMS must first activate the expression of the regulator before cluster activation can move forward. This activation requires the reversal of repressive epigenetic marks mediated by LaeA.
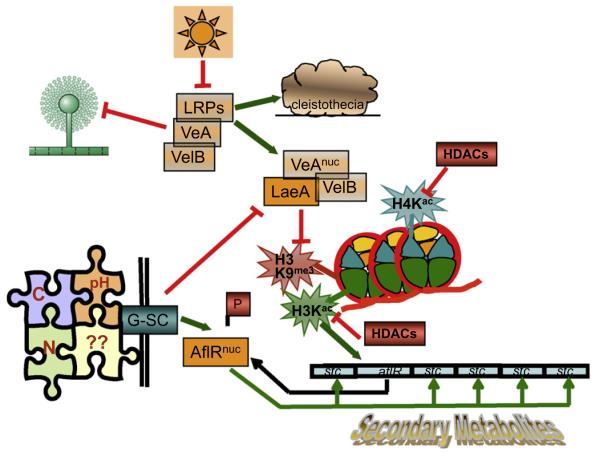
Fig. 2.
Simplified representation of an integrated pathway connecting environmental signals to SM production. The environmental signals can be divided into two main categories: physical conditions (light, temperature, humidity, oxygen) and nutritional-chemical conditions (nutrient quality and quantity, pH and other, less studied factors, represented by question marks). Light-responsive proteins (LRPs) regulate nuclear accumulation of the velvet complex (VeAnuc–VelB) which acts as an inhibitor of conidiation and stimulator of sexual reproduction (cleistothecia) and SM. For activating SM, the velvet complex associates with LaeA and mediates removal of heterochromatic marks (H3K9me3) which are, during primary metabolism, established in SM clusters with the help of histone de-acetylases (HDACs). Hyperacetylation of H3 and H4N-termini (H3Kac, H4Kac) parallels the activation of SM genes (aflR, stc) in the cluster and generates a euchromatic landscape competent for transcriptional activation. The nutritional conditions are integrated into the signaling pathway by a cAMP/PKA pathway which employs G-protein mediated signaling cascades (G-SC) to activate both LaeA and the pathway-specific regulator AflR. De-phosphorylated AflR preferentially translocates to the nucleus and the euchromatic structure now allows AflR (in cooperation with putative co-activators) to bind to exposed recognition sites in biosynthetic gene promoters eventually leading to transcription of all SM genes in the cluster. Arrows represent activation whereas inhibition is symbolized by blocked lines.
From present data it is clear that reversal of silencing is necessary but not sufficient for turning on SM clusters. The activation process can be more efficient when silencing is attenuated (e.g. in cclAΔ, HDACΔ, clrDΔ/hepAΔ strains) and also when the pathway regulator is ectopically over-expressed or when only broad-domain regulators are required for activation (e.g. in the penicillin cluster). This is in line with published data and indicates two sequential activation steps as mentioned above. Furthermore, the model postulates that deacetylation of histones is a prerequisite for establishment of silenced SM clusters, which is in good agreement with the known functions of HDACs in gene repression, recruitment of H3K9 methylases and heterochromatin formation in other organisms (Grewal and Jia, 2007).
Silencing of SM genes during active growth by the higher-order facultative heterochromatin structure represents an appealing hypothesis to explain coordinated regulation of physically linked gene clusters. To get a more complete picture of epigenetic regulation of SM clusters, at least three open questions should be answered. One is the exact mechanism of LaeA function in mediating removal of heterochromatic marks. Because LaeA is required for H3K9 de-methylation a simple model would place the function of LaeA in context of directly inhibiting H3K9 methyltransferase ClrD or activating potential H3K9 de-methylases. The genetic and/or physical interaction of LaeA with the velvet light regulation complex consisting of VeA, VelB, LreA, LreB and FphA (Bayram et al., 2008; Calvo, 2008; Purschwitz et al., 2009), and the dependence on the cdc2-like ImeB kinase (Bayram et al., 2009) might bring in additional players.
The second question of general relevance is how the heterochromatic borders of SM clusters are defined. All chromatin studies on SM clusters so far have shown that chromatin modifications are not reaching beyond the cluster genes. Chromatin domain barriers represent fixed or flexible “walls” that inhibit the spread of heterochromatin into transcriptionally active euchromatic regions and have been extensively studied in many systems (Kimura and Horikoshi, 2004; Sun and Elgin, 1999; West et al., 2002). It is far beyond the limits of this review to discuss possible mechanisms that might govern border establishment of SM genes but basic building blocks, such as insulator elements and histone variant exchange, known to define heterochromatin–euchromatin borders in other organisms (Sarma and Reinberg, 2005) might play a role in defining filamentous fungal SM cluster borders.
Finally, it will be relevant to understand how the SMS pathway(s) transmits the signal to the chromatin restructuring machinery. How the LaeA/VeA/VelB complex finds their target SM cluster regions to mediate heterochromatin reversal remains totally obscure; although binding to DNA cannot be excluded, none of the velvet factors possess a known DNA-binding domain. A possibility would involve the cAMP-response regulator CREB-P in targeting the LaeA-dependent system for chromatin opening, because experimental data in A. flavus have clearly shown that promoter sequences in SM genes match to putative binding sites for such regulator and the cAMP levels have been shown in several fungi to play crucial roles in the expression of SM-related genes (Roze et al., 2004; Shimizu et al., 2003). The pathway-specific transcription factors, e.g. AflR in ST and AF, are unlikely candidates for such targeting, because their corresponding genes are also under heterochromatin control and still silenced thus limiting the active protein concentration. Moreover, AflR functions through a LaeA-independent pathway as ectopic over-expression of aflR bypasses the requirement for LaeA in ST gene expression (Bok et al., 2006). It remains to be determined which activity mediates reversal of HC marks in the latter case, when LaeA is absent.
9. Conclusion
This review summarizes our actual knowledge on chromatin-level regulation of SM cluster genes. A solid body of evidence is starting to accumulate that heterochromatic histone marks such as hypoacetylation of H3 and H4 as well as methylation of H3K9 and binding of heterochromatin protein-1 are associated with SM-cluster silencing during active growth. SM signals lead to LaeA-mediated removal of these repressive marks and replacement by activating marks on histones inside the SM cluster (but not outside). These modifications generate a chromatin landscape required subsequently for the function of downstream activators. In our view this higher-order regulation, although still fragmentary, represents a good starting point to better understand the molecular physiology of toxin and antibiotic production. A solid knowledge of chromatin regulatory events might be relevant to develop technologies for the production of novel bioactive compounds (Chiang et al., 2009; Cichewicz, 2009; Williams et al., 2008).
Acknowledgments
We thank Nancy Keller for providing unpublished data and for stimulating discussions and one anonymous reviewer for many valuable suggestions to improve the manuscript. We are grateful to Bradley Matthews and Sotiris Amillis for critical reading of the manuscript. Work on chromatin-level regulation of fungal gene expression is supported by Austrian Science Fund Grants P19731-B11 and F3703-B11 and by the Vienna Science and Technology Fund (WWTF) Grant LS 09-042 to JS.
References
- Adams TH, Yu JH. Coordinate control of secondary metabolite production and asexual sporulation in Aspergillus nidulans. Curr. Opin. Microbiol. 1998;1:674–677. doi: 10.1016/s1369-5274(98)80114-8. [DOI] [PubMed] [Google Scholar]
- Adhvaryu KK, Morris SA, Strahl BD, Selker EU. Methylation of histone H3 lysine 36 is required for normal development in Neurospora crassa. Eukaryot. Cell. 2005;4:1455–1464. doi: 10.1128/EC.4.8.1455-1464.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhvaryu KK, Selker EU. Protein phosphatase PP1 is required for normal DNA methylation in Neurospora. Genes Dev. 2008;22:3391–3396. doi: 10.1101/gad.1738008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaike S, Keller NP. Distinct roles for VeA and LaeA in development and pathogenesis of Aspergillus flavus. Eukaryot. Cell. 2009;8:1051–1060. doi: 10.1128/EC.00088-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Barra JL, Holmes AM, Gregoire A, Rossignol JL, Faugeron G. Novel relationships among DNA methylation, histone modifications and gene expression in Ascobolus. Mol. Microbiol. 2005;57:180–195. doi: 10.1111/j.1365-2958.2005.04665.x. [DOI] [PubMed] [Google Scholar]
- Bayram O, Krappmann S, Ni M, Bok JW, Helmstaedt K, Valerius O, Braus-Stromeyer S, Kwon NJ, Keller NP, Yu JH, Braus GH. VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science. 2008;320:1504–1506. doi: 10.1126/science.1155888. [DOI] [PubMed] [Google Scholar]
- Bayram O, Sari F, Braus GH, Irniger S. The protein kinase ImeB is required for light-mediated inhibition of sexual development and for mycotoxin production in Aspergillus nidulans. Mol. Microbiol. 2009;71:1278–1295. doi: 10.1111/j.1365-2958.2009.06606.x. [DOI] [PubMed] [Google Scholar]
- Belden WJ, Loros JJ, Dunlap JC. Execution of the circadian negative feedback loop in Neurospora requires the ATP-dependent chromatin-remodeling enzyme CLOCKSWITCH. Mol. Cell. 2007;25:587–600. doi: 10.1016/j.molcel.2007.01.010. [DOI] [PubMed] [Google Scholar]
- Bennett JW. Mycotoxins, mycotoxicoses, mycotoxicology and mycopathologia. Mycopathologia. 1987;100:3–5. doi: 10.1007/BF00769561. [DOI] [PubMed] [Google Scholar]
- Berger H, Basheer A, Bock S, Reyes-Dominguez Y, Dalik T, Altmann F, Strauss J. Dissecting individual steps of nitrogen transcription factor cooperation in the Aspergillus nidulans nitrate cluster. Mol. Microbiol. 2008;69:1385–1398. doi: 10.1111/j.1365-2958.2008.06359.x. [DOI] [PubMed] [Google Scholar]
- Bergmann S, Schumann J, Scherlach K, Lange C, Brakhage AA, Hertweck C. Genomics-driven discovery of PKS-NRPS hybrid metabolites from Aspergillus nidulans. Nat. Chem. Biol. 2007;3:213–217. doi: 10.1038/nchembio869. [DOI] [PubMed] [Google Scholar]
- Bhaumik SR, Smith E, Shilatifard A. Covalent modifications of histones during development and disease pathogenesis. Nat. Struct. Mol. Biol. 2007;14:1008–1016. doi: 10.1038/nsmb1337. [DOI] [PubMed] [Google Scholar]
- Blobel GA. CBP and p300: versatile coregulators with important roles in hematopoietic gene expression. J. Leukocyte Biol. 2002;71:545–556. [PubMed] [Google Scholar]
- Bok JW, Keller NP. LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot. Cell. 2004;3:527–535. doi: 10.1128/EC.3.2.527-535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bok JW, Balajee SA, Marr KA, Andes D, Nielsen KF, Frisvad JC, Keller NP. LaeA, a regulator of morphogenetic fungal virulence factors. Eukaryot. Cell. 2005;4:1574–1582. doi: 10.1128/EC.4.9.1574-1582.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bok JW, Noordermeer D, Kale SP, Keller NP. Secondary metabolic gene cluster silencing in Aspergillus nidulans. Mol. Microbiol. 2006;61:1636–1645. doi: 10.1111/j.1365-2958.2006.05330.x. [DOI] [PubMed] [Google Scholar]
- Bok JW, Chiang YM, Szewczyk E, Reyes-Dominguez Y, Davidson AD, Sanchez JF, Lo HC, Watanabe K, Strauss J, Oakley BR, Wang CC, Keller NP. Chromatin-level regulation of biosynthetic gene clusters. Nat. Chem. Biol. 2009;5:462–464. doi: 10.1038/nchembio.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brakhage AA, Sprote P, Al-Abdallah Q, Gehrke A, Plattner H, Tuncher A. Regulation of penicillin biosynthesis in filamentous fungi. Adv. Biochem. Eng. Biotechnol. 2004;88:45–90. doi: 10.1007/b99257. [DOI] [PubMed] [Google Scholar]
- Brakhage AA, Thon M, Sprote P, Scharf DH, Al-Abdallah Q, Wolke SM, Hortschansky P. Aspects on evolution of fungal beta-lactam biosynthesis gene clusters and recruitment of trans-acting factors. Phytochemistry. 2009;70:1801–1811. doi: 10.1016/j.phytochem.2009.09.011. [DOI] [PubMed] [Google Scholar]
- Brosch G, Loidl P, Graessle S. Histone modifications and chromatin dynamics: a focus on filamentous fungi. FEMS Microbiol. Rev. 2008;32:409–439. doi: 10.1111/j.1574-6976.2007.00100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryk M, Briggs SD, Strahl BD, Curcio MJ, Allis CD, Winston F. Evidence that Set1, a factor required for methylation of histone H3, regulates rDNA silencing in S. cerevisiae by a Sir2-independent mechanism. Curr. Biol. 2002;12:165–170. doi: 10.1016/s0960-9822(01)00652-2. [DOI] [PubMed] [Google Scholar]
- Cairns BR. The logic of chromatin architecture and remodelling at promoters. Nature. 2009;461:193–198. doi: 10.1038/nature08450. [DOI] [PubMed] [Google Scholar]
- Calvo AM. The VeA regulatory system and its role in morphological and chemical development in fungi. Fungal Genet. Biol. 2008;45:1053–1061. doi: 10.1016/j.fgb.2008.03.014. [DOI] [PubMed] [Google Scholar]
- Calvo AM, Wilson RA, Bok JW, Keller NP. Relationship between secondary metabolism and fungal development. Microbiol. Mol. Biol. Rev. 2002;66:447–459. doi: 10.1128/MMBR.66.3.447-459.2002. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang PK, Cary JW, Bhatnagar D, Cleveland TE, Bennett JW, Linz JE, Woloshuk CP, Payne GA. Cloning of the Aspergillus parasiticus apa-2 gene associated with the regulation of aflatoxin biosynthesis. Appl. Environ. Microbiol. 1993;59:3273–3279. doi: 10.1128/aem.59.10.3273-3279.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang PK, Bennett JW, Cotty PJ. Association of aflatoxin biosynthesis and sclerotial development in Aspergillus parasiticus. Mycopathologia. 2002;153:41–48. doi: 10.1023/a:1015211915310. [DOI] [PubMed] [Google Scholar]
- Chiang YM, Lee KH, Sanchez JF, Keller NP, Wang CC. Unlocking fungal cryptic natural products. Nat. Prod. Commun. 2009;4:1505–1510. [PMC free article] [PubMed] [Google Scholar]
- Cichewicz RH. Epigenome manipulation as a pathway to new natural product scaffolds and their congeners. Nat. Prod. Rep. 2009;27:11–22. doi: 10.1039/b920860g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza CP, Osmani AH, Wu LP, Spotts JL, Osmani SA. Mitotic histone H3 phosphorylation by the NIMA kinase in Aspergillus nidulans. Cell. 2000;102:293–302. doi: 10.1016/s0092-8674(00)00035-0. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G, Groudine M. Controlling the double helix. Nature. 2003;421:448–453. doi: 10.1038/nature01411. [DOI] [PubMed] [Google Scholar]
- Fischle W. Talk is cheap – cross-talk in establishment, maintenance, and readout of chromatin modifications. Genes Dev. 2008;22:3375–3382. doi: 10.1101/gad.1759708. [DOI] [PubMed] [Google Scholar]
- Fischle W, Wang Y, Allis CD. Histone and chromatin cross-talk. Curr. Opin. Cell Biol. 2003;15:172–183. doi: 10.1016/s0955-0674(03)00013-9. [DOI] [PubMed] [Google Scholar]
- Goswami RS, Xu JR, Trail F, Hilburn K, Kistler HC. Genomic analysis of host-pathogen interaction between Fusarium graminearum and wheat during early stages of disease development. Microbiology. 2006;152:1877–1890. doi: 10.1099/mic.0.28750-0. [DOI] [PubMed] [Google Scholar]
- Grewal SI, Jia S. Heterochromatin revisited. Nat. Rev. Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- Grimaldi B, Coiro P, Filetici P, Berge E, Dobosy JR, Freitag M, Selker EU, Ballario P. The Neurospora crassa White Collar-1 dependent blue light response requires acetylation of histone H3 lysine 14 by NGF-1. Mol. Biol. Cell. 2006;17:4576–4583. doi: 10.1091/mbc.E06-03-0232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmeister D, Keller NP. Natural products of filamentous fungi: enzymes, genes, and their regulation. Nat. Prod. Rep. 2007;24:393–416. doi: 10.1039/b603084j. [DOI] [PubMed] [Google Scholar]
- Holbert MA, Marmorstein R. Structure and activity of enzymes that remove histone modifications. Curr. Opin. Struct. Biol. 2005;15:673–680. doi: 10.1016/j.sbi.2005.10.006. [DOI] [PubMed] [Google Scholar]
- Honda S, Selker EU. Direct interaction between DNA methyltransferase DIM-2 and HP1 is required for DNA methylation in Neurospora crassa. Mol. Cell Biol. 2008;28:6044–6055. doi: 10.1128/MCB.00823-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn PJ, Peterson CL. Heterochromatin assembly: a new twist on an old model. Chromosome Res. 2006;14:83–94. doi: 10.1007/s10577-005-1018-1. [DOI] [PubMed] [Google Scholar]
- Huang Y, Fang J, Bedford MT, Zhang Y, Xu RM. Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A. Science. 2006;312:748–751. doi: 10.1126/science.1125162. [DOI] [PubMed] [Google Scholar]
- Jacobs SA, Khorasanizadeh S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science. 2002;295:2080–2083. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Kale SP, Milde L, Trapp MK, Frisvad JC, Keller NP, Bok JW. Requirement of LaeA for secondary metabolism and sclerotial production in Aspergillus flavus. Fungal Genet. Biol. 2008;45:1422–1429. doi: 10.1016/j.fgb.2008.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller NP, Hohn TM. Metabolic pathway gene clusters in filamentous fungi. Fungal Genet. Biol. 1997;21:17–29. [PubMed] [Google Scholar]
- Keller NP, Turner G, Bennett JW. Fungal secondary metabolism – from biochemistry to genomics. Nat. Rev. Microbiol. 2005;3:937–947. doi: 10.1038/nrmicro1286. [DOI] [PubMed] [Google Scholar]
- Khaldi N, Seifuddin FT, Turner G, Haft D, Nierman WC, Wolfe KH, Fedorova ND. SMURF: genomic mapping of fungal secondary metabolite clusters. Fungal Genet. Biol. 2010;47:736–741. doi: 10.1016/j.fgb.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura A, Horikoshi M. Partition of distinct chromosomal regions: negotiable border and fixed border. Genes Cells. 2004;9:499–508. doi: 10.1111/j.1356-9597.2004.00740.x. [DOI] [PubMed] [Google Scholar]
- Kosalkova K, Garcia-Estrada C, Ullan RV, Godio RP, Feltrer R, Teijeira F, Mauriz E, Martin JF. The global regulator LaeA controls penicillin biosynthesis, pigmentation and sporulation, but not roquefortine C synthesis in Penicillium chrysogenum. Biochimie. 2009;91:214–225. doi: 10.1016/j.biochi.2008.09.004. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Kouzminova E, Selker EU. Dim-2 encodes a DNA methyltransferase responsible for all known cytosine methylation in Neurospora. EMBO J. 2001;20:4309–4323. doi: 10.1093/emboj/20.15.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Oh JH, Shwab EK, Dagenais TR, Andes D, Keller NP. HdaA, a class 2 histone deacetylase of Aspergillus fumigatus, affects germination and secondary metabolite production. Fungal Genet. Biol. 2009;46:782–790. doi: 10.1016/j.fgb.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis ZA, Honda S, Khlafallah TK, Jeffress JK, Freitag M, Mohn F, Schubeler D, Selker EU. Relics of repeat-induced point mutation direct heterochromatin formation in Neurospora crassa. Genome Res. 2009;19:427–437. doi: 10.1101/gr.086231.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Hansen JC. Nucleosome and chromatin fiber dynamics. Curr. Opin. Struct. Biol. 2005;15:188–196. doi: 10.1016/j.sbi.2005.03.006. [DOI] [PubMed] [Google Scholar]
- Malz S, Grell MN, Thrane C, Maier FJ, Rosager P, Felk A, Albertsen KS, Salomon S, Bohn L, Schafer W, Giese H. Identification of a gene cluster responsible for the biosynthesis of aurofusarin in the Fusarium graminearum species complex. Fungal Genet. Biol. 2005;42:420–433. doi: 10.1016/j.fgb.2005.01.010. [DOI] [PubMed] [Google Scholar]
- Meijsing SH, Ehrenhofer-Murray AE. The silencing complex SAS-I links histone acetylation to the assembly of repressed chromatin by CAF-I and Asf1 in Saccharomyces cerevisiae. Genes Dev. 2001;15:3169–3182. doi: 10.1101/gad.929001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris NR. Nucleosome structure in Aspergillus nidulans. Cell. 1976;8:357–363. doi: 10.1016/0092-8674(76)90147-1. [DOI] [PubMed] [Google Scholar]
- Mueller JE, Canze M, Bryk M. The requirements for COMPASS and Paf1 in transcriptional silencing and methylation of histone H3 in Saccharomyces cerevisiae. Genetics. 2006;173:557–567. doi: 10.1534/genetics.106.055400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell. 2003;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- Osmani SA, Pu RT, Morris NR. Mitotic induction and maintenance by overexpression of a G2-specific gene that encodes a potential protein kinase. Cell. 1988;53:237–244. doi: 10.1016/0092-8674(88)90385-6. [DOI] [PubMed] [Google Scholar]
- Payne GA, Yu J. Ecology, development and gene regulation in Aspergillus flavus. In: Masayuki M, Katsuya G, editors. Aspergillus: Molecular Biology and Genomics. Caister Academic Press; Norfolk, UK: 2010. [Google Scholar]
- Pena PV, Davrazou F, Shi X, Walter KL, Verkhusha VV, Gozani O, Zhao R, Kutateladze TG. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature. 2006;442:100–103. doi: 10.1038/nature04814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin RM, Fedorova ND, Bok JW, Cramer RA, Wortman JR, Kim HS, Nierman WC, Keller NP. Transcriptional regulation of chemical diversity in Aspergillus fumigatus by LaeA. PLoS Pathog. 2007;3:e50. doi: 10.1371/journal.ppat.0030050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purschwitz J, Muller S, Fischer R. Mapping the interaction sites of Aspergillus nidulans phytochrome FphA with the global regulator VeA and the White Collar protein LreB. Mol. Genet. Genomics. 2009;281:35–42. doi: 10.1007/s00438-008-0390-x. [DOI] [PubMed] [Google Scholar]
- Reyes-Dominguez Y, Narendja F, Berger H, Gallmetzer A, Fernandez-Martin R, Garcia I, Scazzocchio C, Strauss J. Nucleosome positioning and histone H3 acetylation are independent processes in the Aspergillus nidulans prnD–prnB bidirectional promoter. Eukaryot. Cell. 2008;7:656–663. doi: 10.1128/EC.00184-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Dominguez Y, Bok JW, Berger H, Shwab EK, Basheer A, Gallmetzer A, Scazzocchio C, Keller N, Strauss J. Heterochromatic marks are associated with the repression of secondary metabolism clusters in Aspergillus nidulans. Mol. Microbiol. 2010;76:1376–1386. doi: 10.1111/j.1365-2958.2010.07051.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roze LV, Arthur AE, Hong SY, Chanda A, Linz JE. The initiation and pattern of spread of histone H4 acetylation parallel the order of transcriptional activation of genes in the aflatoxin cluster. Mol. Microbiol. 2007;66:713–726. doi: 10.1111/j.1365-2958.2007.05952.x. [DOI] [PubMed] [Google Scholar]
- Roze LV, Miller MJ, Rarick M, Mahanti N, Linz JE. A novel cAMP-response element, CRE1, modulates expression of nor-1 in Aspergillus parasiticus. J. Biol. Chem. 2004;279:27428–27439. doi: 10.1074/jbc.M400075200. [DOI] [PubMed] [Google Scholar]
- Sarma K, Reinberg D. Histone variants meet their match. Nat. Rev. Mol. Cell Biol. 2005;6:139–149. doi: 10.1038/nrm1567. [DOI] [PubMed] [Google Scholar]
- Scazzocchio C, Ramon A. Chromatin in the genus Aspergillus. In: Goldman G, Osmani SA, editors. The Aspergilli: Genomics, Medical Aspects, Biotechnology, and Research Methods. CRC Press; Boca Raton, FL: 2008. pp. 321–342. [Google Scholar]
- Schotta G, Ebert A, Krauss V, Fischer A, Hoffmann J, Rea S, Jenuwein T, Dorn R, Reuter G. Central role of Drosophila SU(VAR)3-9 in histone H3-K9 methylation and heterochromatic gene silencing. EMBO J. 2002;21:1121–1131. doi: 10.1093/emboj/21.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeckh V, Scherlach K, Nutzmann HW, Shelest E, Schmidt-Heck W, Schuemann J, Martin K, Hertweck C, Brakhage AA. Intimate bacterial-fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans. Proc. Natl. Acad. Sci. USA. 2009;106:14558–14563. doi: 10.1073/pnas.0901870106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selker EU, Freitag M, Kothe GO, Margolin BS, Rountree MR, Allis CD, Tamaru H. Induction and maintenance of nonsymmetrical DNA methylation in Neurospora. Proc. Natl. Acad. Sci. USA. 2002;99(Suppl. 4):16485–16490. doi: 10.1073/pnas.182427299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selker EU, Tountas NA, Cross SH, Margolin BS, Murphy JG, Bird AP, Freitag M. The methylated component of the Neurospora crassa genome. Nature. 2003;422:893–897. doi: 10.1038/nature01564. [DOI] [PubMed] [Google Scholar]
- Shaaban M, Palmer JM, El-Naggar WA, El-Sokkary MA, Habib ES, Keller NP. Involvement of transposon-like elements in penicillin gene cluster regulation. Fungal Genet. Biol. 2010;47:423–432. doi: 10.1016/j.fgb.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, Carney D, Pena P, Lan F, Kaadige MR, Lacoste N, Cayrou C, Davrazou F, Saha A, Cairns BR, Ayer DE, Kutateladze TG, Shi Y, Cote J, Chua KF, Gozani O. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu. Rev. Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- Shimizu K, Hicks JK, Huang TP, Keller NP. Pka, Ras and RGS protein interactions regulate activity of AflR, a Zn(II)2Cys6 transcription factor in Aspergillus nidulans. Genetics. 2003;165:1095–1104. doi: 10.1093/genetics/165.3.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shwab EK, Bok JW, Tribus M, Galehr J, Graessle S, Keller NP. Histone deacetylase activity regulates chemical diversity in Aspergillus. Eukaryot. Cell. 2007;6:1656–1664. doi: 10.1128/EC.00186-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ, 3rd, Reinberg D. Histone H3 Lys 4 methylation: caught in a bind? Genes Dev. 2006;20:2779–2786. doi: 10.1101/gad.1468206. [DOI] [PubMed] [Google Scholar]
- Smith CA, Woloshuk CP, Robertson D, Payne GA. Silencing of the aflatoxin gene cluster in a diploid strain of Aspergillus flavus is suppressed by ectopic aflR expression. Genetics. 2007;176:2077–2086. doi: 10.1534/genetics.107.073460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KM, Kothe GO, Matsen CB, Khlafallah TK, Adhvaryu KK, Hemphill M, Freitag M, Motamedi MR, Selker EU. The fungus Neurospora crassa displays telomeric silencing mediated by multiple sirtuins and by methylation of histone H3 lysine 9. Epigenetics Chromatin. 2008;1:5. doi: 10.1186/1756-8935-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suganuma T, Workman JL. Crosstalk among histone modifications. Cell. 2008;135:604–607. doi: 10.1016/j.cell.2008.10.036. [DOI] [PubMed] [Google Scholar]
- Sun FL, Elgin SC. Putting boundaries on silence. Cell. 1999;99:459–462. doi: 10.1016/s0092-8674(00)81534-2. [DOI] [PubMed] [Google Scholar]
- Tamaru H, Selker EU. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature. 2001;414:277–283. doi: 10.1038/35104508. [DOI] [PubMed] [Google Scholar]
- Tamaru H, Zhang X, McMillen D, Singh PB, Nakayama J, Grewal SI, Allis CD, Cheng X, Selker EU. Trimethylated lysine 9 of histone H3 is a mark for DNA methylation in Neurospora crassa. Nat. Genet. 2003;34:75–79. doi: 10.1038/ng1143. [DOI] [PubMed] [Google Scholar]
- Timmers HT, Tora L. SAGA unveiled. Trends Biochem. Sci. 2005;30:7–10. doi: 10.1016/j.tibs.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Tremethick DJ. Higher-order structures of chromatin: the elusive 30 nm fiber. Cell. 2007;128:651–654. doi: 10.1016/j.cell.2007.02.008. [DOI] [PubMed] [Google Scholar]
- Tribus M, Galehr J, Trojer P, Brosch G, Loidl P, Marx F, Haas H, Graessle S. HdaA, a major class 2 histone deacetylase of Aspergillus nidulans, affects growth under conditions of oxidative stress. Eukaryot. Cell. 2005;4:1736–1745. doi: 10.1128/EC.4.10.1736-1745.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trojer P, Dangl M, Bauer I, Graessle S, Loidl P, Brosch G. Histone methyltransferases in Aspergillus nidulans: evidence for a novel enzyme with a unique substrate specificity. Biochemistry. 2004;43:10834–10843. doi: 10.1021/bi049626i. [DOI] [PubMed] [Google Scholar]
- Walton JD. Horizontal gene transfer and the evolution of secondary metabolite gene clusters in fungi: an hypothesis. Fungal Genet. Biol. 2000;30:167–171. doi: 10.1006/fgbi.2000.1224. [DOI] [PubMed] [Google Scholar]
- Wang H, Cao R, Xia L, Erdjument-Bromage H, Borchers C, Tempst P, Zhang Y. Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol. Cell. 2001;8:1207–1217. doi: 10.1016/s1097-2765(01)00405-1. [DOI] [PubMed] [Google Scholar]
- West AG, Gaszner M, Felsenfeld G. Insulators: many functions, many mechanisms. Genes Dev. 2002;16:271–288. doi: 10.1101/gad.954702. [DOI] [PubMed] [Google Scholar]
- Wiemann P, Brown DW, Kleigrewe K, Bok JW, Keller NP, Humpf HU, Tudzynski B. FfVel1 and FfLae1, components of a velvet-like complex in Fusarium fujikuroi, affect differentiation, secondary metabolism and virulence. Mol. Microbiol. doi: 10.1111/j.1365-2958.2010.07263.x. in press. doi:10.1111j.1365-2958.2010.07263.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RB, Henrikson JC, Hoover AR, Lee AE, Cichewicz RH. Epigenetic remodeling of the fungal secondary metabolome. Org. Biomol. Chem. 2008;6:1895–1897. doi: 10.1039/b804701d. [DOI] [PubMed] [Google Scholar]
- Wiren M, Silverstein RA, Sinha I, Walfridsson J, Lee HM, Laurenson P, Pillus L, Robyr D, Grunstein M, Ekwall K. Genomewide analysis of nucleosome density histone acetylation and HDAC function in fission yeast. Embo J. 2005;24:2906–2918. doi: 10.1038/sj.emboj.7600758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock CL, Dimitrov S. Higher-order structure of chromatin and chromosomes. Curr. Opin. Genet. Dev. 2001;11:130–135. doi: 10.1016/s0959-437x(00)00169-6. [DOI] [PubMed] [Google Scholar]
- Yu JH, Keller NP. Regulation of secondary metabolism in filamentous fungi. Annu. Rev. Phytopathol. 2005 doi: 10.1146/annurev.phyto.43.040204.140214. [DOI] [PubMed] [Google Scholar]