Abstract
Background
Previous studies have demonstrated that genital infection with high-risk types of human papillomavirus (HPV), most often HPV16, is the most significant risk factor for the development of cervical cancer. However, serologic assays that have been developed to identify high-risk HPV infection have either failed to associate serum reactivity with other indicators of HPV infection or have identified only a minority of HPV-infected individuals.
Purpose
Our purpose was to determine whether a specifically developed enzyme-linked immunosorbent assay (ELISA) could detect IgG anti-HPV16 virion antibodies in the sera of women who had tested positive for genital HPV16 infection by DNA-based methods.
Methods
An ELISA was developed using newly developed HPV16 virus-like particles as antigens to detect anti-HPV16 virion IgG antibodies. These particles are comprised of HPV16 structural proteins that are self-assembled in insect cells after expression by recombinant baculo-viruses. The sera of 122 women, whose HPV status had been previously evaluated by nucleic acid-based methods, were tested by this ELISA.
Results
The sera of 59% of women (32 of 54) positive for genital HPV16 DNA by polymerase chain reaction (PCR) were positive in the ELISA assay compared with sera from women who had tested negative for HPV DNA (P<.0005). In contrast, 6% of HPV DNA-negative women (two of 31) and 9% of women positive for low-risk HPV6/11 DNA (one of 11) were ELISA positive by this criterion. The sera of women who were DNA positive for two additional high-risk HPV types were evaluated; the sera of 31% of HPV18-positive (four of 13) and 38% of HPV31-positive women (five of 13) were positive in the HPV16 particle ELISA. The sera of 75% of HPV16 DNA-positive women with severe dysplasias (12 of 16) gave positive ELISA results. The sera of 67% of women (28 of 42) who tested positive for HPV16 DNA by both PCR and the less sensitive ViraType assay tested positive in the ELISA compared with 33% of women (four of 12) who were positive by PCR but negative by ViraType (P<.05).
Conclusion
The majority of women with cervical HPV16 infection generate an IgG antibody response to conformationally dependent epitopes of HPV16 L1 that can be detected by ELISA.
Implication
This particular ELISA, or a similar one incorporating virus-like particles of additional HPV types, may be useful in determining the natural history of high-risk HPV infection and perhaps help to identify women at risk for developing cervical cancer.
Genital human papillomaviruses (HPV) are common sexually transmitted agents that induce a variety of proliferative genital lesions, including condylomata acuminata and benign cervical atypia. Infection by high-risk HPV types (most often HPV16 and HPV18) is by far the most significant risk factor for the development of cervical intraepithelial neoplasia (increased risk of 10-fold to 40-fold), which can progress to invasive cervical carcinomas (increased risk of 200-fold to 300-fold) (1-3). Infection by low-risk types (most often HPV6 and HPV11) is common, but lesions induced by these types are rarely associated with carcinogenic progression.
The identification of women who are at risk for developing cervical cancer has depended primarily on the use of the Papanicolaou (Pap) test, which detects cytologic abnormalities in cells from cervico-vaginal scrapes. In recent years, it has become evident that most of the cytologic changes represent manifestations of genital HPV infection. Although the Pap test is a very useful screening assay, it has a relatively high proportion of misdiagnosis and requires highly trained personnel to obtain and analyze the samples (4). Therefore, attempts have been made to devise assays to complement or partially replace Pap screening. Most of the ancillary assays are based on the direct detection of HPV nucleic acids in exfoliated cervico-vaginal cells and biopsy specimens. Two major shortcomings of the DNA-based assays are that they are beyond the scope of procedures currently in use in most clinical laboratories and, since the viral lesions are focal, they are subject to the same sampling errors as Pap smears (5).
A serologic assay for genital HPV infection may offer advantages over currently available HPV diagnostic methods, since it would measure systemic responses to a significant viral infection and could be performed by most clinical laboratories. In addition, an antibody test might be useful for evaluating past exposure to the viruses. Attempts to develop serologic assays for high-risk HPV infections, however, have thus far been relatively unsuccessful. Responses to nonstructural viral proteins appear to be low and show substantial variability, with no single protein eliciting a detectable response in more than a minority of HPV-infected people (6,7). Assays using bacterially produced LI major or L2 minor virion structural proteins have not shown a good association between serum reactivity and other measures of HPV infection (8). The latter finding may indicate that HPV infections that are localized to the genital mucosa do not regularly elicit a significant systemic immune response to the virion proteins. Alternatively, most of the immunogenic epitopes of HPV virions recognized during natural infection may not be displayed by the bacterially derived virion proteins, which are generally isolated as denatured proteins and lack eukaryotic specific modifications.
Humoral responses to native genital HPV virions have only been studied using virions of the low-risk HPV11 virions propagated in human tissues grown in immunologically impaired mice (9,10) because no efficient in vitro system exists for generating preparative amounts of native virions. However, we have reported (11) that the L1 virion proteins of bovine papillomavirus type 1 (BPV1) and HPV16 have the intrinsic capacity to self-assemble into virus-like particles when expressed in insect cells via recombinant baculoviruses. We have now used purified HPV16 L1 plus L2 virus-like particles (12) as the antigen to develop an enzyme-linked immunosorbent assay (ELISA). We have tested whether this assay can detect IgG anti-HPV16 virion antibodies in the sera of women who test positive for genital HPV16 infection by DNA-based methods.
Patients and Methods
Patients
From participants of an epidemiologic study of cervical HPV infection at the University of New Mexico, Albuquerque (13,14), a sample of 122 women was selected to participate in this study. These women, aged 18-42 years, were attendees at either gynecology or student health clinics that primarily serve a population of Hispanic women; however, other white women also attend. The study sample was selected on the basis of known genital HPV infection and cervical pathology status and included 91 women infected with one of the following low- or high-risk HPV types: HPV6/11, HPV16, HPV18, or HPV31 (Table 1). A previously reported (15) consensus polymerase chain reaction (PCR) method that accurately and reproducibly detects the presence of a broad spectrum of HPV types in clinical specimens followed by type-specific PCR was used to determine the cervico-vaginal HPV status in the study population (13,14). Additionally, the ViraType hybridization test (Digene Diagnostics, Silver Spring, Md., Inc.) was used.
Table 1. ELISA reactivity of women’s sera to HPV16 L1/L2 particles.
| No. of serum samples tested |
No. ELISA positive |
% positive | |
|---|---|---|---|
| A. By PCR type | |||
| Negative | 31 | 2 | 6 |
| HPV6/11 | 11 | 1 | 9 |
| HPV16 | 54 | 32 | 59 |
| HVP18 | 13 | 4 | 31 |
| HPV31 | 13 | 5 | 38 |
| B. By ViraType* | |||
| Negative | 12 | 4 | 33 |
| Positive | 42 | 28 | 67 |
| C. By cytology* | |||
| Normal | 15 | 8 | 53 |
| Atypical | 4 | 2 | 50 |
| Mild dysplasia | 7 | 5 | 71 |
| Moderate dysplasia | 11 | 5 | 45 |
| Severe dysplasia | 16 | 12 | 75 |
HPV16 positive by PCR. Pap test results were not available for one woman.
To facilitate interpretation of results, women infected with two or more HPV types were excluded. As a normal control population, we selected 31 women who were either virgins and had once tested negative for HPV DNA or women who had tested negative for HPV DNA by PCR in 10 consecutive assays. This population was selected to minimize the confounding effects of false negatives (i.e., women with previous or current but undetected HPV infection) in the analysis of the ELISA results. The women were subjected to routine pelvic examination and Pap testing, and colposcopy and direct cervical biopsies were carried out on those women with positive Pap smears (13,14). Sera were drawn and stored at −70 °C prior to testing.
ELISA
Recombinant baculoviruses for simultaneous expression of the L1 gene of HPV16 strain 114/B or the L1 gene of the prototype strain (16) and the L2 gene of the prototype HPV16 were generated (12) and used to infect Sf-9 insect cells. In addition, we used single-expression baculoviruses that express either of the L1 proteins alone (11). Insect cells were infected for 72 hours. The cells were lysed, and L1/L2 or L1 particles were purified by cesium chloride gradient centrifugation as previously described (11). Purified particle preparation (1.0 μg) (as determined by quantitation of total L1 protein after polyacrylamide gel electrophoresis) in 50 μL phosphate-buffered saline (PBS) was added to each well of a 96-well Immulon II microtiter plate (Dynatech Laboratories, Inc., Chantilly, Va.). This amount was saturating in that increasing the amount of particles did not increase serum reactivity. The plate was covered and incubated at 37 °C for 1.5 hours and then washed three times.
These and subsequent washes were done with PBS without calcium and magnesium, using an ELISA plate washer (Corning Glass Inc., Corning, N.Y.). The wells were blocked for 2 hours in PBS containing 0.5% nonfat dry milk (Giant Foods, Washington, D.C.) plus 0.1% newborn calf serum (GIBCO-BRL, Grand Island, N.Y.) at room temperature and then washed three times. Human serum (50 μL) was diluted in 200 μL of 0.5% dry milk in PBS and added to the wells immediately after removing the last wash. The plates were covered and incubated for 2.5 hours with gentle rocking. After five washes, 50 μL of horseradish peroxidase conjugated goat F(ab)2 antihuman IgG (Tago, Inc., Burlingame, Calif.), diluted 1:10 000 in 0.5% milk–PBS prior to use, was added to the wells. The plate was then incubated at room temperature for 1 hour with gentle rocking and washed three times. The peroxidase substrate ABTS (50 μL) (prepared according to the manufacturer’s instructions; Boehringer Mannheim, Indianapolis, Ind.) was added to the plate, which was then incubated at room temperature for 45 minutes. The optical densities (ODs) were read at 405 nm in a Thermo Max microplate reader (Molecular Devices Corp., Menlo Park, Calif.). To test the reactivity to denatured particles, concentrated particle preparations were diluted to 20 μg/mL in 0.2 M NaHCO3 (pH 10.6) containing 10 mM dithiothreitol, and 50 μL (1 μg) was added to each well. After air drying, the wells were washed three times with PBS and the ELISA was conducted as described above.
Statistical Analysis
The data were analyzed with SAS Version 6 (SAS Institute Inc., Cary, N.C.), using simple procedures (FREQ, UNIVARIATE, and NPARIWAY). Chi-square tests were used to determine differences between ELISA test results. Exact methods were used when sample sizes were small (less than five). Age was examined as a continuous variable or stratified by 5-year intervals, with similar findings in both analyses. Test reproducibility and interassay variability were evaluated on a representative subset of 74 patients.
Results
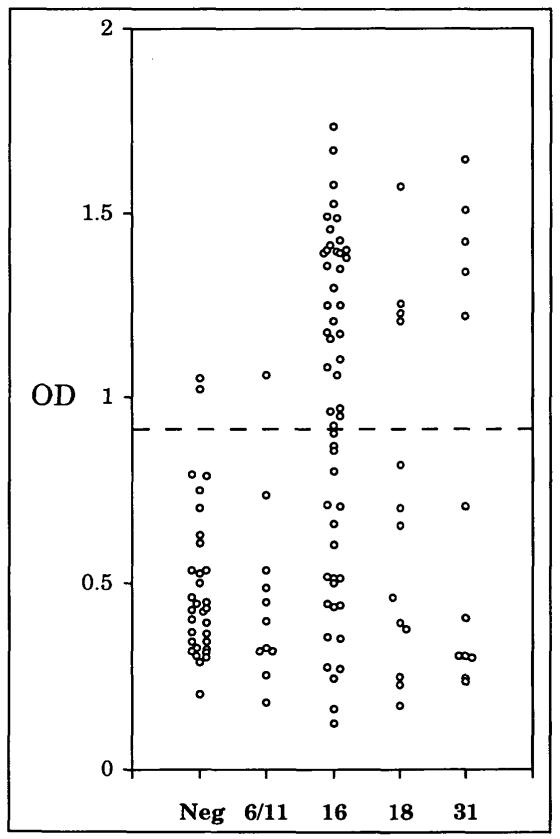
Sera from 122 women attending the University of New Mexico Women’s and Student Health Clinics were examined for IgG reactivity to native HPV16 virion-like particles in an ELISA. To promote maximum sensitivity, a saturating amount of particles was added to each well. To determine if the particle ELISA could be used to detect genital HPV infection, a sample of 31 women who were cytologically normal and had tested negative for genital HPV DNA by PCR were designated as “normal” controls. The sera from 54 women who had tested positive for cervical HPV16 DNA by PCR were considered positive for HPV16 infection. All sera were tested in duplicate against HPV16 L1/L2 particles purified from recombinant baculovirus-infected insect cells, and the mean ODs of duplicate samples tested on separate plates were calculated (Fig. 1).
Fig. 1. Reactivity of sera to native HPV16 virus-like particles by HPV type.
OD = optical density at 405 nm. Neg = negative for HPV DNA, 6/11 = positive for HPV6 or HPV11 DNA, 16 = positive for HPV16 DNA, 18 = positive for HPV18 DNA, 31 = positive for HPV31 DNA. Each circle represents the average reading of two assays done on separate plates. The dashed line at OD = 0.89 indicates the cut-off used to distinguish positive from negative results.
The test results obtained with sera of 31 HPV DNA-negative women were used to calculate the means plus 2.0 standard deviations of the ODs, and the value obtained (0.89; dotted lines in Fig. 1) was selected as the cutoff for designating ELISA reactivities as positive. By these criteria, the sera of 94% (29 of 31) of women who were negative for HPV DNA were negative in the ELISA assay. A second serum sample, drawn approximately 2 months later from each of the two HPV DNA-negative women who tested positive in the first assay, was also positive. In contrast, a second serum from each of 15 women whose first serum tested negative gave a negative result. It is therefore unlikely that the two positive results were due to handling errors. Neither of the positive sera came from a virgin, raising the possibility that these women had previously been infected with HPV16.
In contrast to the results obtained with sera from women who tested negative for cervical HPV16 DNA, the sera from 59% (32 of 54) of the women who tested positive for HPV16 by PCR were ELISA positive (Fig. 1 and Table 1, A; P<.0005 by 2 × 2 chi-squared test). The serum reactivity in the L1/L2 particle ELISA was primarily directed against L1, since assays using virus-like particles containing L1 alone gave similar results (data not shown). The assay distinguished women with high-risk HPV16 infection from those infected with low-risk types HPV6 and HPV11; the sera from only 9% (one of 11) of the women who tested positive for HPV6 or HPV11 DNA by PCR were positive in the HPV16 ELISA (P<.002). In addition, when sera from women who tested PCR-positive for either of two other high-risk HPV types, HPV18 or HPV31, were examined, 31% (four of 13) of women positive for HPV18 and 38% (five of 13) of women positive for HPV31 were positive by HPV16 ELISA (P<.01 when both groups were combined and compared with HPV DNA-negative women).
A higher percentage of women who tested positive for HPV16 DNA by both PCR and the less sensitive ViraType assay tested positive in the ELISA (67%; 28 of 42) than women who were positive by PCR but negative by ViraType (33%; four of 12), a result that was statistically significant (P<.05; Table 1, B). For all study participants, age was not significantly different between those testing negative (26.2 years) and those testing positive (27.3 years) in the ELISA (P>.2 by Wilcoxon test). For women positive for HPV16 DNA by PCR, the mean age of women who were ELISA positive was 27.6 years; for those who were ELISA negative, it was 28.7 years (P>.6).
The percentage of HPV16 DNA-positive women with cervical abnormalities who had positive ELISA results was 63% (24 of 38) compared with 53% (eight of 15) of cytologically normal women (Table 1, C), but this difference was not statistically significant (P>.6). For HPV16 DNA-positive women with benign reactive cervical atypia or mild dysplasia, 64% (seven of 11) were positive by ELISA; for those with moderate dysplasia, 45% (five of 11) were positive by ELISA; and for those with severe dysplasia, 75% (12 of 16) were positive by ELISA. We detected serum antibodies in 67% (six of nine) of cytologically normal women who were both ViraType and PCR positive for HPV16 but only in 33% (two of six) of cytologically normal women who were positive only by PCR. There was a strong association between cervical abnormalities and ELISA positivity in the HPV31-positive women, although a limited number of samples were tested. Five (71%) of seven HPV31 DNA-positive women who had dysplastic lesions were ELISA positive, while 0% (zero of six) of cytologically normal women with this HPV type were ELISA positive.
ELISA test reproducibility was evaluated on the sera of a representative subset of 74 patients. Two tests, performed on different days, were compared. Overall, the sera of 95% (70 of 74) of the women gave the same result in both assays. The correlation for the sera from HPV16-positive women was 96% (25 of 26), for women positive for other HPV types it was 94% (32 of 34), and for HPV-negative women it was 93% (13 of 14). Un-weighted kappa coefficients of 0.88 and 0.92 were statistically significant (P<.05) for the sera of all 74 women and for the sera from women testing positive for HPV16, respectively.
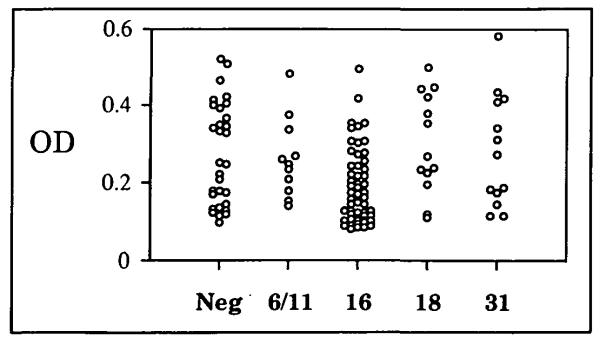
Since antibodies raised in animals after experimental inoculation with native BPV1 virions have been shown to primarily recognize conformationally dependent epitopes (17), we wished to determine if this were also true for the human anti-virion antibodies to natural HPV infection detected in the ELISA. Therefore, the HPV16 L1/L2 particles were subjected to denaturing conditions (pH 10.6 carbonate buffer) prior to their addition to the wells. Essentially, all of the specific reactivity to the particles was lost after this treatment (i.e., no more reactivity was detected in the sera from HPV-positive women than in the sera from HPV-negative women) (Fig. 2). The lack of reactivity to the denatured particles was specific to the women’s sera, since strong reactivity was detected using rabbit sera raised against denatured HPV 16 L1 protein (data not shown). We conclude that during natural infection, the majority of human serum IgG antibodies against genital papillomavirus virions are to L1 epitopes that have secondary structure.
Fig. 2. Reactivity of sera to denatured HPV16 virus-like panicles by HPV type.
The designations are the same as in the legend of Fig. 1.
We also tested a subset of the sera in an HPV16 L1/L2 ELISA using L1 protein derived from the prototype HPV16 strain (16). The L1 from the prototype HPV16 strain has three amino acid differences and self-assembles into virion-like particles in the insect cells at least 1000-fold less efficiently than does the L1 protein used in the above assays. None of the sera from women who were positive for HPV16 by PCR demonstrated reactivity to the prototype L1 that differed significantly from that of the sera of 10 negative controls (data not shown). The mean ODs (± SD) for the prototype L1/L2 were 0.20 (± 0.12) and 0.20 (± 0.08), respectively. In contrast, the mean OD to the efficiently assembled L1/L2 particles was 1.25 (± 0.26) for the sera from HPV16 DNA-positive women compared with 0.35 (± 0.12) for the sera from HPV DNA-negative women. These results imply either that the majority of the prototype L1 is improperly folded so that it does not present the conformationally dependent epitopes or, less likely, that the amino acid differences in the prototype L1 are in the primary antigenic sites of the virion.
Discussion
In this article, we have examined the reactivity of human sera to immobilized HPV16 virus-like particles in an ELISA. It had not previously been determined if women infected with high-risk genital HPV mount a significant humoral immune response to native virion structural proteins, since preparative amounts of HPV16 virions or virus-like particles have not been available for testing. Our findings indicate that the majority of infected women do mount a systemic anti-virion antibody response. This response appears to be primarily directed against conformationally dependent epitopes found on L1 of intact virions, since L1/L2 and L1-only particles gave similar results. Significant reactivities were not detected to denatured HPV16 particles or to preparations of the prototype L1 protein, which contains an alteration in amino acid sequence that prevented efficient particle assembly. Since previous HPV16 L1 serologic assays used denatured and/or prototype L1, this finding could explain the negative results. Human antibodies to nongenital HPV1 virions also appear to react principally with conformationally dependent epitopes (18,19). Previous research has shown that approximately 30% of patients with HPV6- or HPV11-induced condylomata acuminata react in an ELISA to HPV11 virions derived from the mouse xenograft Kreider system (10). It was unclear if the modest sensitivity of the HPV11 assay reflected a limited response to infection by these low-risk HPV types or to technical limitations imposed by the low concentration of available virions.
There are several reasons to suspect that the potential sensitivity and specificity of our ELISA for the detection of clinically significant HPV16 infection may be greater than is indicated from the results presented here. First, the DNA-based detection methods used to determine HPV infection status are subject to errors that can lead to misclassification. Second, our serologic assay, unlike DNA-based assays, might detect past as well as current HPV16 infections. Therefore, the few women whose sera appeared to give false-positive results might have had HPV16 infections that had become undetectable or eliminated prior to sampling. Third, there is no reason to suspect that the sera were drawn during the peak of the antivirion IgG immune response, since only one serum sample from each woman was analyzed. Recently infected women may not yet have mounted a significant immune response, and the antibody titers in women with persistent progressed lesions may have waned, since moderate and severely dysplastic lesions may express few if any virion proteins (20).
A sensitive and specific virus-like particle ELISA would be useful in determining the natural history of genital HPV infections and their relationship to cervical neoplasia. Possible associations between infection, anti-virion humoral response, and the status of HPV-induced lesions could be evaluated. In prospective studies, it may be possible to determine how long after initial infection antibodies appear and if the lesions of individuals with high antibody titers are more or less likely to undergo regression or malignant progression. The assay might also serve as a primary screen to measure the prevalence of genital HPV infections in men. High-risk HPV infections in males are often asymptomatic and, because of the difficulties in obtaining specimens for DNA typing, the demographics of men with genital HPV infections are not well understood (21).
Establishing the clinical usefulness of an antivirion ELISA test of the type described here will require the analysis of sera from larger and more varied sets of individuals, and further refinements in the assay are likely to be made. Nevertheless, several encouraging trends emerged from the current study. First, our assay appears to identify HPV infections in a majority of HPV16 DNA-positive women with cervical abnormalities (75% of women with severe dysplasia were ELISA positive). Detection of cervical disease in this group is clinically important, and, in some studies, false-negative results were obtained in over 20% of Pap smears of such women (22). The clinical relevance of PCR-based HPV16 positivity in women without cytologic abnormalities is less clear. A lower percentage of women in this group tested positive in our assay (33% of PCR-positive, ViraType-negative women were ELISA positive), presumably because the ELISA is less likely to detect transient or subclinical infection. Second, while approximately 60% of women positive for HPV16 by PCR analysis had positive sera, less than 10% of women who tested positive for low-risk viruses were positive by ELISA, a result that was not statistically different from that found with the HPV DNA-negative women (P>.7). It would be useful for a serologic assay to discriminate between infections by high- and low-risk types, since infection by low-risk viruses does not seem to be an independent risk factor for cervical cancer (2). If it were clinically advantageous to specifically identify low-risk genital HPV infections, it is likely that an assay based on insect cell-derived HPV6 and HPV11 particles could also be developed [(23); our unpublished results].
The assay using HPV 16 particles detected approximately 35% of women who were positive by PCR for two other high-risk types, HPV18 and HPV31. It is not clear if the positive reactions seen with these sera indicate a cross-reactivity among the virions of high-risk types or if they detected infections by HPV16, either prior or concurrent, that were undetected by the DNA assays. The latter reason is possible because HPV16 is the most common type detected, and infection by more than one genital HPV type frequently occurs in the population tested (22%) (13,14). It is anticipated that an ELISA designed to maximize detection of infection by high-risk types would use a mixed antigen consisting of virus-like particles of HPV16, HPV18, HPV31, HPV45, and perhaps other less common high-risk types. Such an assay, coupled with type-specific assays, might aid in determining the natural history of high-risk HPV infection and perhaps help to identify women at risk for developing cervical cancer.
Acknowledgments
Supported in part by Public Health Service grant R0132917-03 (C. M. Wheeler) from the National Institute of Arthritis and Musculoskeletal and Skin Diseases and CA48003 (T. M. Becker) from the National Cancer Institute, National Institutes of Health, Department of Health and Human Services.
C. M. Wheeler is a recipient of an American Social Health/Pfizer fellowship.
T. M. Becker is a recipient of a Junior Faculty Research Award from the American Cancer Society.
We thank Kathy Altobelli for her help with the statistical analysis.
Contributor Information
Reinhard Kirnbauer, Laboratory of Cellular Oncology, National Cancer Institute, Bethesda, Md..
Nancy L. Hubbert, Laboratory of Cellular Oncology, National Cancer Institute, Bethesda, Md.
Cosette M. Wheeler, New Mexico Cancer Center, Department of Cell Biology and the New Mexico Tumor Registry, Albuquerque, N.M.
Thomas M. Becker, New Mexico Tumor Registry and University of New Mexico, Department of Medicine and Department of Family and Community Medicine, Albuquerque.
Douglas R. Lowy, Laboratory of Cellular Oncology, National Cancer Institute, Bethesda, Md.
John T. Schiller, Laboratory of Cellular Oncology, National Cancer Institute, Bethesda, Md..
References
- (1).Koutsky LA, Holmes KK, Critchlow CW, et al. A cohort study of the risk of cervical intraepithelial neoplasia grade 2 or 3 in relation to papillomavirus infection. N Engl J Med. 1992;327:1272–1278. doi: 10.1056/NEJM199210293271804. [DOI] [PubMed] [Google Scholar]
- (2).Lorincz AT, Reid R, Jenson AB, et al. Human papillomavirus infection of the cervix: relative risk associations of 15 common anogenital types. Obstet Gynecol. 1992;79:328–337. doi: 10.1097/00006250-199203000-00002. [DOI] [PubMed] [Google Scholar]
- (3).Schiffman MH, Bauer HM, Hoover RN, et al. Epidemiologic evidence showing that human papillomavirus infection causes most cervical intraepithelial neoplasia. J Natl Cancer Inst. 1993;85:958–964. doi: 10.1093/jnci/85.12.958. [DOI] [PubMed] [Google Scholar]
- (4).Koss LG. The Papanicolaou test for cervical cancer detection. A triumph and a tragedy. JAMA. 1989;261:737–743. [PubMed] [Google Scholar]
- (5).Schiffman MH. Validation of hybridization assays: correlation of filter in situ, dot blot and PCR with Southern blot. In: Muñoz N, Bosch FX, Shah KV, et al., editors. The Epidemiology of Cervical Cancer and Human Papillomavirus. International Agency for Research on Cancer; Lyon: 1992. pp. 169–179. (Scientific Publication No. 119). [PubMed] [Google Scholar]
- (6).Jha PK, Beral V, Peto J, et al. Antibodies to human papillomavirus and to other genital infectious agents and invasive cervical cancer risk. Lancet. 1993;341:1116–1118. doi: 10.1016/0140-6736(93)93128-n. [DOI] [PubMed] [Google Scholar]
- (7).Jochmus-Kudielka I, Schneider A, Braun R, et al. Antibodies against the human papillomavirus type 16 early proteins in human sera: correlation of anti-E7 reactivity with cervical cancer. J Natl Cancer Inst. 1989;81:1698–1704. doi: 10.1093/jnci/81.22.1698. [DOI] [PubMed] [Google Scholar]
- (8).Galloway DA. Serological assays for the detection of HPV antibodies. IARC Sci Publ. 1992;119:147–161. [PubMed] [Google Scholar]
- (9).Christensen ND, Kreider JW, Shah KV, et al. Detection of human serum antibodies that neutralize infectious human papillomavirus type 11 virions. J Gen Virol. 1992;73:1261–1267. doi: 10.1099/0022-1317-73-5-1261. [DOI] [PubMed] [Google Scholar]
- (10).Bonnez W, Da Rin C, Rose RC, et al. Use of human papillomavirus type 11 virions in an ELISA to detect specific antibodies in humans with condylomata acuminata. J Gen Virol. 1991;72:1343–1347. doi: 10.1099/0022-1317-72-6-1343. [DOI] [PubMed] [Google Scholar]
- (11).Kirnbauer R, Booy F, Cheng N, et al. Papillomavirus L1 major capsid protein self-assembles into virus-like particles that are highly immunogenic. Proc Natl Acad Sci U S A. 1992;89:12180–12184. doi: 10.1073/pnas.89.24.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Kirnbauer R, Taub J, Greenstone H, et al. Efficient self-assembly of human papillomavirus type 16 L1 and L1-L2 into virus-like particles. J Virol. 1993;67:6929–6936. doi: 10.1128/jvi.67.12.6929-6936.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Becker TM, Wheeler CM, McGough N, et al. Cervical papillomavirus infection and cervical dysplasia in Hispanic, Native American, and non-Hispanic white women in New Mexico. Am J Public Health. 1991;81:582–586. doi: 10.2105/ajph.81.5.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Wheeler CM, Parmenter CA, Hunt WC, et al. Determinants of genital human papillomavirus infection among cytologically normal women attending the University of New Mexico student health center. Sex Transm Dis. 1993;20:286–289. doi: 10.1097/00007435-199309000-00009. [DOI] [PubMed] [Google Scholar]
- (15).Bauer HM, Ting Y, Greer CE, et al. Genital human papillomavirus infection in female university students as determined by a PCR-based method. JAMA. 1991;265:472–477. [PubMed] [Google Scholar]
- (16).Dürst M, Gissmann L, Ikenberg H, et al. A papillomavirus DNA from a cervical carcinoma and its prevalence in cancer biopsy samples from different geographic regions. Proc Natl Acad Sci U S A. 1983;80:3812–3815. doi: 10.1073/pnas.80.12.3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Ghim S, Christensen ND, Kreider JW, et al. Comparison of neutralization of BPV-1 infection of C127 cells and bovine fetal skin xenografts. Int J Cancer. 1991;49:285–289. doi: 10.1002/ijc.2910490224. [DOI] [PubMed] [Google Scholar]
- (18).Steele JC, Gallimore PH. Humoral assays of human sera to disrupted and nondisrupted epitopes of human papillomavirus type 1. Virology. 1990;174:388–398. doi: 10.1016/0042-6822(90)90092-6. [DOI] [PubMed] [Google Scholar]
- (19).Carter JJ, Hagensee M, Taflin MC, et al. HPV-1 capsids expressed in vitro detect human serum antibodies associated with foot warts. Virology. 1993;195:456–462. doi: 10.1006/viro.1993.1396. [DOI] [PubMed] [Google Scholar]
- (20).Shirasawa H, Tomita Y, Kubota K, et al. Transcriptional differences of the human papillomavirus type 16 genome between precancerous lesions and invasive carcinomas. J Virol. 1988;62:1022–1027. doi: 10.1128/jvi.62.3.1022-1027.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Barrasso R, De Brux J, Croissant O, et al. High prevalence of papillomavirus-associated penile intraepithelial neoplasia in sexual partners of women with cervical intraepithelial neoplasia. N Engl J Med. 1987;317:916–923. doi: 10.1056/NEJM198710083171502. [DOI] [PubMed] [Google Scholar]
- (22).van der Graaf Y, Vooijs GP, Gaillard HL, et al. Screening errors in cervical cytology screening. Acta Cytol. 1987;31:434–438. [PubMed] [Google Scholar]
- (23).Rose RC, Bonnez W, Reichman RC, et al. Expression of human papillomavirus type 11 L1 protein in insect cells: in vivo and in vitro assembly of viruslike particles. J Virol. 1993;67:1936–1944. doi: 10.1128/jvi.67.4.1936-1944.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]