Abstract
Sensitivity to temozolomide (TMZ) is restricted to a subset of glioblastoma patients, with the major determinant of resistance being a lack of promoter methylation of the gene encoding the repair protein DNA methyltransferase MGMT, although other mechanisms are thought to be active. There are, however, limited preclinical data in model systems derived from paediatric glioma patients. We have screened a series of cell lines for TMZ efficacy in vitro, and have investigated the differential mechanisms of resistance involved. In the majority of cell lines, a lack of MGMT promoter methylation, and subsequent protein overexpression, was linked to TMZ resistance. An exception was the paediatric glioblastoma line KNS42. Expression profiling data revealed a co-ordinated upregulation of HOX gene expression in resistant lines, especially KNS42, which was reversed by PI3-kinase pathway inhibition. High levels of HOXA9/HOXA10 gene expression were associated with a shorter survival in paediatric high grade glioma patient samples. Combination treatment in vitro of pathway inhibition and TMZ resulted in a highly synergistic interaction in KNS42 cells. The resistance gene signature further included contiguous genes within the 12q13-q14 amplicon including the Akt enhancer PIKE, significantly overexpressed in the KNS42 line. These cells were also highly enriched for CD133 and other stem cell markers. We have thus demonstrated an in vitro link between PI3-kinase-mediated HOXA9/HOXA10 expression, and a drug-resistant, progenitor cell phenotype in MGMT-independent paediatric glioblastoma.
Keywords: glioma, temozolomide, paediatric, MGMT, PI3-kinase, HOX, CD133
INTRODUCTION
Glioblastoma is the most common tumour of the central nervous system, affecting patients of all ages, and being essentially refractory to treatment - the clinical outcome remains dismal regardless of age at diagnosis. The median survival of a patient with glioblastoma is 15 months, and this has improved little in the last four decades(1). The mainstays of treatment during this time have been surgical resection and radiotherapy, often with nitrosurea-based chemotherapy. The use of adjuvant temozolomide (TMZ) has more recently emerged as a new standard of care in glioblastoma, with concurrent and sequential treatment in the initial therapy of patients resulting in a modest improvement in median survival(2).
TMZ is a DNA methylating agent which is orally bioavailable, crosses the blood:brain barrier and exhibits schedule-dependent anti-tumour activity(3). The improved survival benefits in glioblastoma are largely restricted to the subset of patients lacking expression of the DNA repair enzyme O6-methylguanine-DNA-methyl-transferase (MGMT)(4). TMZ induces cytotoxic O6-guanine methyl adducts which are removed directly by functional MGMT, thereby producing drug resistance. Downregulation of MGMT usually occurs in tumours by gene promoter hypermethylation, in which greater than 50% methylation has been shown to silence gene expression(5).
Deficiencies in DNA mismatch repair (MMR) are also linked with resistance to alkylating agents such as TMZ(6), as are elevated levels of Ape1/Ref-1, a major component of the base excision repair (BER)(7) system, with attempts to enhance TMZ-induced cytotoxicity by disrupting BER by means of inhibition of poly-[ADP-ribose]-polymerase (PARP) proving effective in vitro and in vivo(8).
The vast majority of the above work has taken place in adult glioblastoma and preclinical models derived from adult patients. In the paediatric setting, MGMT promoter hypermethylation predicts for response to alkylating agents(9); however, the survival of children treated with adjuvant TMZ does not appear to be improved when compared with historical controls(10-14). The mechanisms of drug resistance in paediatric high grade glioma are poorly understood, in part due to the lack of availability of suitable models of the disease. We have screened a series of paediatric and adult glioma cell lines for TMZ efficacy in vitro, and have investigated the differential mechanisms of resistance involved, highlighting the involvement in paediatric cells of processes outside of the usual MGMT/MMR/BER axis.
MATERIALS AND METHODS
Cell culture
Adult glioblastoma cell lines A172, LN229, SF268, U87MG, U118MG and U138MG, and paediatric glioma cell lines SF188, KNS42, UW479, Res259 and Res186 were obtained and cultured as previously described(15). For the spheroid formation assay, cells were grown in Neurosphere medium, which consisted of NDiff RHB□A Medium (Stem Cell Sciences, Cambridge, UK) supplemented with growth factors EGF and FGF-2, each at 20ng/ml.
Growth inhibition studies
TMZ was obtained from Apin Chemicals, Abingdon, UK; O6-benzylguanine from Calbiochem, San Diego, CA, USA; and PI-103 from Piramed Pharma, Slough, UK or synthesized in-house. Growth inhibition was determined using the sulforhodamine B(16) or MTS(17) assays as previously described. To attempt reversion of resistance to TMZ, O6-benzylguanine was added at the highest non-toxic concentration (10-15% of cell growth inhibition, 20 μM). For the assessment of combination effects, cells were treated with increasing concentrations of drugs either alone or concurrently at their equipotent molar ratio and combination indices calculated by the method of Chou and Talalay(18). All values are given as mean ± SD of at least three independent experiments.
Promoter methylation analysis
Cell line DNA was treated with sodium bisulphite using the Epitect kit (Qiagen, Crawley, UK) according to the manufacturer’s instructions. Methylation-specific (MS)-PCR for the MGMT promoter was performed as described previously(19). MS-MLPA was carried out as previously reported(15) according to manufacturer’s instructions (MRC-Holland, Amsterdam, Netherlands)(20). HOXA9/HOXA10 methylation was assessed by comparing expression profiles of 5-Aza-2′-deoxycytidine-treated cells with vehicle-treated controls on Illumina Human-6 v2 Expression BeadChips (Illumina Inc, San Diego, CA, USA), ArrayExpress accession number E-TABM-858.
Western blot analysis
Immunodetection was performed as previously described(15) using antibodies against MGMT (1:500, Zymed, Carlsbad, CA, USA), MLH1 (1:500, Pharmingen, San Diego, CA, USA), MLH3 (1:500, Santa Cruz Biotechnologies, Santa Cruz, CA, USA), MSH2 (1:500, Calbiochem), MSH3 (1:250, BD Bioscience, San Diego, CA, USA), MSH6, PMS2 (both 1:500, BD Bioscience), PARP1/2 (1:1000, Cell Signaling), XRCC1 (1:500, Cell Signaling), APE1 (Novus Biochemicals, Littleton, CO, USA), p85, p110α (Cell Signalling), p110β, p110δ (Santa Cruz), PIKE-A/PIKE-L (all 1:1000, Abcam, Cambridge, UK), phospho-AktSer473, Akt (both 1:1000, Cell Signaling) and GAPDH (1:2000, Chemicon, Hampshire, UK).
mRNA expression profiling analysis
Cell line expression profiling by Affymetrix U133 oligonucleotide arrays has been previously published(15) (ArrayExpress accession number E-TABM-579). Supervised analysis was performed using an absolute signal to noise metric of greater than 1.5 in GenePattern software (http://www.broad.mit.edu/cancer/software/genepattern/). Co-ordinate gene regulation was identified using Gene Set Enrichment Analysis (GSEA, www.broad.mit.edu/gsea/), with a nominal p value cut-off of 0.001. “Core enriched” genes are defined as belonging to the leading-edge subset within the gene set, and thus contribute the most to the enrichment result. Assessment of HOX gene expression after 24h treatment with PI-103 at 5xIC50 was carried out using Illumina HT-12 BeadChips (ArrayExpress accession number E-TABM-890). Affymetrix U133 expression data from The Cancer Genome Atlas glioblastoma study(21) was assessed for cross-correlations of probesets corresponding to HOXA9 by calculating Pearson’s correlation coefficients in R. GSEA and clinical correlations were further carried out on a published dataset(22) of Affymetrix U133 expression array profiling of 78 paediatric high grade gliomas (Gene Expression Omnibus accession number GSE19578; http://www.ncbi.nlm.nih.gov/geo/).
Immunofluorescence and flow cytometry
CD133 protein expression was measured by both flow cytometry using a BD FACS Vantage SEDiVa system (BD Biosciences, San Jose, CA, USA), and immunofluorescence on cytospin preparations, using anti-CD133 antibody (AC133/1, Miltenyi Biotec, Bergisch Gladbach, Germany) at 1:50 and 1:100 dilution respectively. Cells were co-stained with nestin (196908, R & D Systems, Minneapolis, MN, USA) and visualised with DAPI (Vector Laboratories Inc., Burlingame, CA, USA). Cell cycle analysis was also carried out by FACS.
RESULTS
Sensitivity of glioma cell lines to TMZ in vitro is largely but not exclusively dependent on MGMT promoter methylation and lack of protein expression
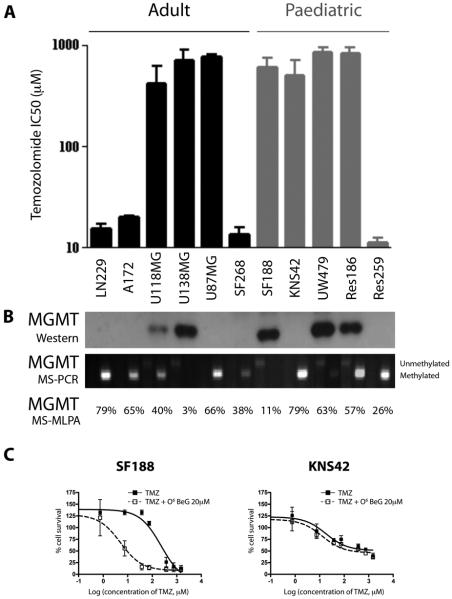
We first determined the response to TMZ in vitro of our panel of five paediatric (SF188, KNS42, UW479, Res259, Res186) and six adult (A172, LN229, SF268, U87MG, U118MG, U138MG) glioma cell lines. Four lines (A172, LN229, SF268, Res259) were classed as TMZ-sensitive, with IC50 values of between 10-20 μM (Figure 1A). The remaining cells were resistant to treatment with the alkylating agent, with IC50s of greater than 500 μM.
Figure 1. Sensitivity of paediatric and adult glioma cell lines to TMZ and relationship to MGMT status.
(A) Adult (LN229, A172, U118MG, U138MG, U87MG, SF268) and paediatric (SF188, KNS42, UW479, Res186, Res259) glioma cells were treated with TMZ, and cytotoxicity assessed by the SRB assay. IC50 values are plotted on a log10 scale. (B) Western blot for MGMT protein expression correlated with extent of promoter methylation as assessed by methylation-specific PCR and MLPA. In most cases expression correlates with TMZ resistance, with the exception of U87MG and KNS42 cells, which are hypermethylated, do not express the protein, and are resistant to TMZ. (C) SF188 and KNS42 cells were treated with MGMT substrate analogue O6-benzylguanine, demonstrating the MGMT-dependent nature of TMZ resistance in SF188, but not KNS42 cells. Growth inhibition was determined by the SRB assay. Concentration of TMZ is on a log10 scale.
We assessed MGMT promoter methylation by methylation-specific PCR and MLPA, and protein expression by Western blot (Figure 1B). Extensive methylation resulted in an absence of MGMT protein expression in LN229, A172, U87MG, SF268, Res259 and KNS42 cells. There was, for the most part, a direct correlation between MGMT methylation/lack of expression and TMZ sensitivity. An exception to this were the paediatric glioblastoma KNS42 cells, which displayed insensitivity in the absence of MGMT protein, implying that alternate mechanisms of resistance must be operative. To confirm this, we treated paediatric glioblastoma SF188 and KNS42 cells with TMZ in the presence of the substrate analogue O6-benzyl guanine (O6BeG), which depletes the enzyme and increases cytotoxicity(23) (Figure 1C). Treatment with 20μM O6BeG increased the efficacy of TMZ in SF188 cells nearly 40-fold (IC50 without O6BeG = 194 μM, IC50 in presence of O6BeG = 5 μM), thus confirming the dependence of these cells on MGMT in conferring TMZ resistance. By contrast, no such effect was seen in KNS42 cells, demonstrating the MGMT-independent nature of the insensitivity.
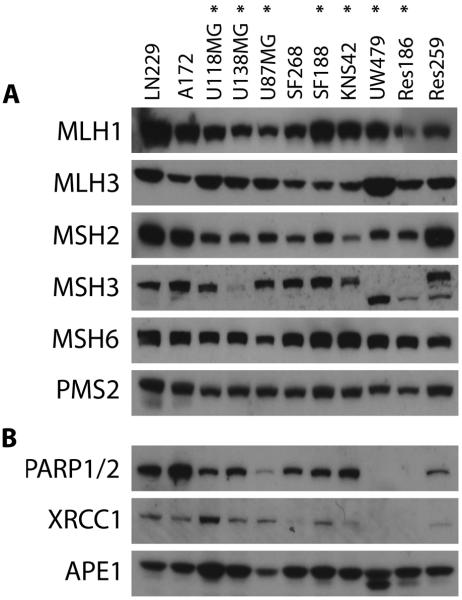
Dysregulation of mismatch repair and base excision repair proteins do not explain resistance to TMZ in MGMT deficient KNS42 cells
Enzymes involved in DNA mismatch repair (MLH1, MLH3, MSH2, MSH3, MSH6, PMS2) were evaluated by Western blot (Figure 2A). Although we identified three TMZ resistant lines with abrogated expression of MSH3 (U138MG, UW479 and Res186), there was no deficiency in KNS42 or U87MG cells. These data correlated well with levels of promoter methylation assessed by MS-MLPA(15). We further investigated components of the base excision repair pathway, including PARP1/2, XRCC1 and APE1. Although TMZ resistant UW479 and Res186 appeared to lack PARP1/2 and XRCC1 expression, there were no apparent alterations in other glioma lines, including KNS42 or U87MG (Figure 2B).
Figure 2. Assessment of DNA mismatch repair and base excision repair pathways in paediatric and adult glioma cell lines.
Western blot for proteins involved in (A) mismatch repair, and (B) base excision repair. Although deficiencies in MSH3 are noted in U138MG, UW479 and Res186, and there is some variability in PARP and XRCC1 expression, U87MG and KNS42 have normal expression of proteins involved in both pathways. TMZ-resistant cell lines are highlighted (*).
Identification of a HOX/stem cell gene expression signature associated with TMZ resistance in paediatric glioblastoma cells
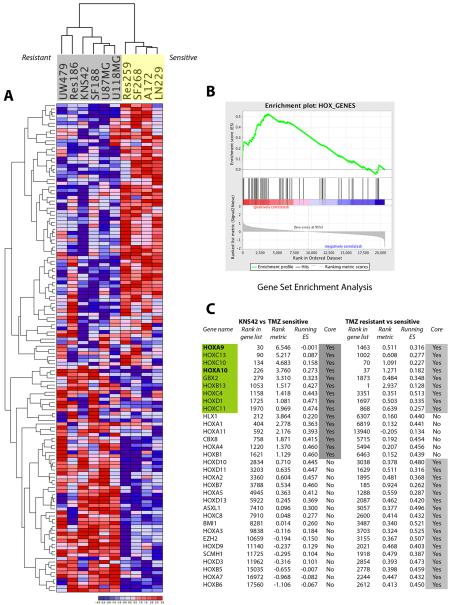
Using expression microarrays, we identified 135 genes differentially expressed between sensitive and resistant glioma cell lines (Figure 3A). Included in this list were MGMT and PARP2, despite these enzymes not explaining the resistance in all cell lines. Also included were several kinases including MAPK9, and CDK6, which may prove suitable targets for pharmacological modulation; PIK3C3 (Vps34), suggesting a possible link to the autophagic response (24); and genes encoding elements of the immune response such as IL10 and IL16.
Figure 3. Expression profiling reveals a HOX / stem cell signature associated TMZ resistance in glioma cell lines.
(A) Heatmap demonstrating hierarchical clustering of 135 differentially expressed genes between resistant (grey - UW479, Res186, KNS42, SF188, U87MG, U118MG) and sensitive (yellow – Res259, SF268, A172, LN229) high grade glioma cell lines. (B) Gene Set Enrichment Analysis highlighting co-ordinated differential expression of gene sets defined a priori. Enriched in TMZ resistant cell lines were the HOX_GENES set. (Enrichment Score = 0.54, nominal p = 0.01; FDR q = 0.403). (C) Ranked genes derived from GSEA of KNS42 versus TMZ sensitive cell lines, and all TMZ resistant versus sensitive lines. Genes are provided along with their rank in the total gene list, rank enrichment metric, and running enrichment score are provided for both analyses. Genes present within the core enrichment signature are highlighted in grey, with those genes present in both analyses highlighted in green. These genes which are also present in the HOX gene signature in Murat et al. (26), HOXA9 and HOXA10, are highlighted in bold.
When we applied Gene Set Enrichment Analysis (GSEA) to our data, we observed co-ordinated differential expression of the HOX_GENES set (MSigDB C2:curated gene sets) in resistant versus sensitive cell lines (Figure 3B), with an Enrichment Score of 0.54 (nominal p = 0.01; FDR q = 0.403). Furthermore, the HOX_GENES list was also identified as significant using a GSEA “Preranked” analysis based on differentially expressed genes between KNS42 alone and TMZ-sensitive cell lines (Figure 3C). In these analyses, the genes in both “core enriched” lists, which contribute to the leading-edge subset within the gene set(25), included HOXA9, HOXA10, HOXB13, HOXC4, HOXC10, HOXC11, HOXC13, HOXD1 and GBX2.
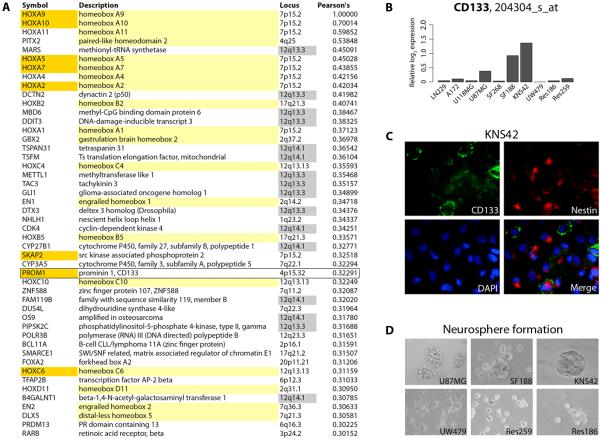
As co-ordinated expression of HOX genes had recently been noted in glioblastoma clinical samples(26), and was reported as evidence of a ‘self-renewal’ signature as it included the stem cell marker PROM1 (CD133), we sought further evidence for this in a published glioblastoma dataset and our cell line models. When we investigated The Cancer Genome Atlas (TCGA) expression profiles of 163 glioblastomas for genes which correlated with the top-ranking HOX gene in our KNS42 GSEA list, HOXA9, we noted a remarkable parallel expression of numerous other homeobox genes (Figure 4A). Of the top 47 genes by this analysis, 18 were homeobox genes found at nine distinct genomic loci, and seven were included in the ‘self-renewal’ signature of Murat et al.(26). These latter genes included PROM1. Intriguingly, the vast majority of the non-homeobox genes identified by this analysis are contiguous genes found commonly amplified in glioblastoma at the genomic locus 12q13-q14.
Figure 4. Co-ordinated upregulation of HOX genes in primary glioblastomas and a striking degree of CD133 positivity on KNS42 cells.
(A) Genes co-ordinately expressed with HOXA9 from the TCGA dataset (21) were determined by calculating Pearson’s correlation coefficients. All genes with values greater than 0.3 are listed, in rank order. Those highlighted in yellow are homeobox genes, genes in orange were also present in the HOX gene/self-renewal signature of Murat et al. (26), including PROM1 (CD133, box); genes highlighted in grey are found within the 12q13-q14 amplicon. (B) Affymetrix expression analysis of PROM1(CD133) mRNA levels in the panel of glioma cell lines, plotted as relative log2 expression. (C) Immunofluorescence assay demonstrating extensive expression of stem cell markers CD133 (green) and nestin (red) in KNS42 cells grown as a monolayer. (D) Neurosphere formation assay highlighting the tight three-dimensional spheroids formed by KNS42 cells in contrast to the other cell lines studied.
To determine whether an enrichment of the stem cell marker CD133 may be playing a role in the resistance of KNS42 cells to TMZ, we assessed the levels of mRNA expression relative to the other cell lines (Figure 4B), and noted considerably higher levels of PROM1 in KNS42 cells than any other line in our panel. Of note, the only other two cell lines to express PROM1 at above background levels were the similarly TMZ-resistant U87MG and SF188. This was visualised by immunofluorescent staining for CD133, co-labelled with nestin (Figure 4C). We had previously reported the relatively high levels of stem cell markers in SF188 and KNS42 by immunocytochemistry(15), and in order to more accurately quantify this, we used the more sensitive flow cytometry analysis to reveal an usually high degree of expression in KNS42, with 17.0% of cells positive for CD133. There were also high levels of CD133-positive cells in the SF188 line (4.8%) compared with 0.0 - 0.02% in other cell lines. KNS42 cells grown as monolayers also expressed by far the greatest levels of other stem cell markers such as nestin, SOX2 and musashi-1 (Affymetrix U133, data not shown). Consistent with the cancer stem/progenitor cell-associated gene expression profile, neurosphere formation assays demonstrated that KNS42 cells form tight three-dimensional spheroids, which may be serially passaged and undergo self-renewal. In contrast, SF188 cells form smaller, more loosely packed spheres, whilst U87MG grow as cell aggregates rather than neurospheres per se. Neither UW479, Res259 nor Res186 formed spheres under these conditions. Taken together, KNS42 cells appear to have a significant cancer stem/progenitor cell-associated gene expression signature and biological phenotype.
Expression of HOXA9/HOXA10 is a result of PI3-kinase-mediated demethylation, inhibition of which synergistically interacts with TMZ in KNS42 cells
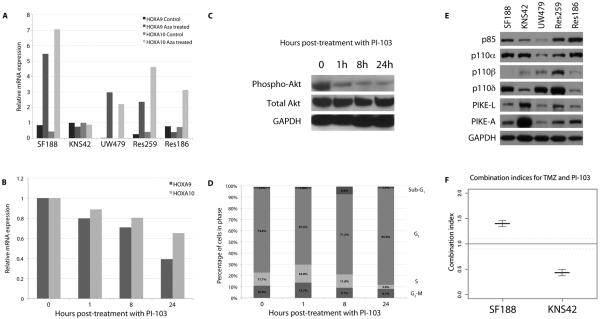
Treatment with the demethylating agent 5-aza-2′-deoxycytidine resulted in highly differential levels of expression of HOXA9 and HOXA10 in all paediatric cell lines with the exception of KNS42, in which no changes were observed, indicative of a lack of methylation in the untreated cells (Figure 5A). As a recent study has proposed a mechanism for this observation whereby transcriptional activation of the HOXA cluster is reversible by a PI3-kinase inhibitor through an epigenetic mechanism involving histone H3K27 trimethylation(27), we sought to investigate whether this mechanism was active in our system. Treatment for 1, 8 and 24 hours with the dual PI3-kinase/mTOR inhibitor PI-103(28-30) at 5x IC50 resulted in significantly reduced expression of both HOXA9 and HOXA10 in KNS42 cells in a time-dependent manner (Figure 5B). These effects are not due to fluctuations in HOX gene expression with the cell cycle, as diminished HOXA9/HOXA10 was observed as early as 1 hour post-treatment, at which time there is no evidence of G1 arrest (Figure 5C), despite inhibition of PI3K as seen by reduced phospho-Akt levels (Figure 5D).
Figure 5. HOXA9/HOXA10 expression in KNS42 cells is driven by a lack of promoter methylation in a PI3-kinase-dependent manner.
(A) Relative mRNA expression levels of HOXA9 and HOXA10 before and after treatment with 5-Aza-2′-deoxycytidine in paediatric glioma cell lines. An absence of expression changes after 5-Aza-2′-deoxycytidine treatment in KNS42 cells is indicative of an absence of constitutive promoter methylation. (B) HOXA9/HOXA10 expression is reduced by treatment with PI-103 in KNS42 cells in a time-dependent manner. Treatment with the dual PI3-kinase/mTOR inhibitor PI-103 at 5xIC50 for 1, 8 and 24 hours led to a reduction in expression of both HOXA9 and HOXA10 in KNS42 cells as early as 1 hour post-treatment. (C) Western blot analysis of phospho- and total Akt after treatment of KNS42 cells with PI-103 at 5xIC50 for 1, 8 and 24 hours. Inhibition of PI3K signalling is observed at the earliest timepoint (D) Cell cycle analysis of KNS42 cells after treatment with PI-103 at 5xIC50 for 1, 8 and 24 hours. There was no G1 arrest evident at the early timepoints at which reduced HOXA9/HOXA10 expression was observed. (E) Western blot analysis of PI3-kinase regulatory and catalytic subunits and enhancers in paediatric glioma cells. Expression of the specific Akt enhancer proteins PIKE-A and PIKE-L were considerably elevated in KNS42 cells compared with other paediatric glioma cells. (F) Synergistic interaction of PI-103 and TMZ. Co-treatment with PI-103 and TMZ resulted in a high degree of synergy in MGMT-independent KNS42 cells (combination index = 0.43) as calculated by the median effect analysis. By contrast, SF188 cells showed an antagonistic interaction (combination index = 1.401).
Next we sought to determine whether there was any specific dysregulation of the PI3-kinase/PTEN system in KNS42 cells that may be responsible for the HOX gene overexpression. Mutation screening for PTEN, PIK3CA, PIK3R1 and PIK3R3 did not identify any sequence variations (data not shown), and Western blot analysis confirmed a lack of overexpression of PI3-kinase regulatory and catalytic subunits (Figure 5E). By contrast, there were significantly elevated levels of the enhancer proteins PIKE-A and especially PIKE-L in KNS42 cells in comparison with the other lines. Both PIKE proteins are encoded by the CENTG1 (AGAP2) gene found within the 12q13-q14 amplicon co-ordinately upregulated in association with the HOX cluster, and likely represent a significant target for this genomic event in human glioblastoma.
Finally, we investigated the efficacy of targeting PI3-kinase as a strategy for overcoming TMZ resistance in our paediatric glioma cells. Combination treatment in vitro of TMZ with the dual PI3K/mTOR inhibitor PI-103 resulted in a highly synergistic interaction in KNS42 as measured by median effect analysis (combination index = 0.43) (Figure 5F). By contrast, in SF188 cells an antagonistic response was observed with the same combination (combination index = 1.40).
HOXA9/HOXA10 expression is associated with shorter survival in paediatric high grade glioma patients
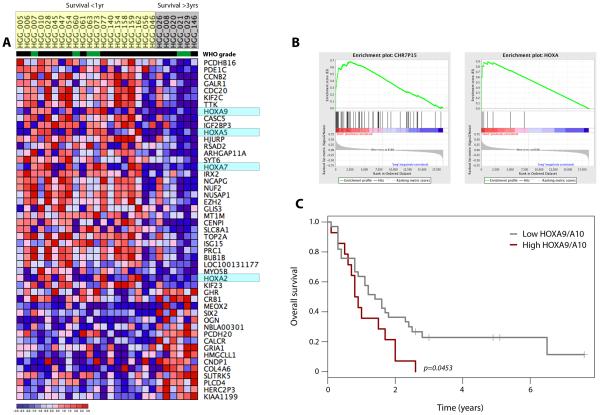
In order to assess the translational relevance of the HOX gene expression in paediatric high grade glioma patient samples, we examined published data detailing expression profiles of 78 tumours arising in childhood(22). Although the number of long-term (>3 years) survivors was small, we identified 49 genes that were differentially expressed between patients with long and short (<1 year) overall survival (Figure 6A). Included in this list were HOXA2, HOXA5, HOX7 and HOXA9. By applying GSEA to the dataset, we identified co-ordinated upregulation in the short-term survivors of genes at the chromosome 7p15 cytoband, with an Enrichment Score of 0.68 (nominal p < 0.001) albeit with a high false discovery rate value (FDR q = 0.930). With the HOXA cluster found at this locus, running GSEA on the HOXA gene list itself gave a highly significant Enrichment Score of 0.90 (nominal p < 0.001, FDR q < 0.001) (Figure 6B). There was considerable correlation between gene expression of all members of the HOXA family in the paediatric samples (Pearson’s correlation coefficients 0.03-0.90). Taking the values of HOXA9 and HOXA10, and segregating samples into ‘high’ and ‘low’ expressers (combined expression greater or less than the 75th percentile of all values, respectively), we demonstrated a significantly reduced overall survival of paediatric high grade glioma patients with high HOXA9/HOXA10 expression (log-rank test, p=0.0453) (Figure 6C) independent of the WHO grade of the tumour (p=0.635, Fishers exact test).
Figure 6. High levels of HOXA gene expression are associated with shorter survival in paediatric high grade glioma patients.
(A) Heatmap representing differentially expressed genes between short (<1 year, yellow) and long-term (>3 years, grey) survivors. WHO grade IV (black) and III (green) tumours are indicated. HOXA genes are highlighted in light blue. (B) GSEA analysis demonstrating significant enrichment of genes at the chromosome 7p15 locus (Enrichment Score = 0.68, nominal p < 0.001, FDR q = 0.930); and of HOXA genes in particular (Enrichment Score = 0.90, nominal p < 0.001, FDR q < 0.001), upregulated in short term survivors. (C) Kaplan-Meier plot demonstrating a significantly shorter survival of patients with high levels of HOXA9/HOXA10 gene expression (p=0.0453, log-rank test).
DISCUSSION
Promoter methylation of the MGMT gene is generally accepted as the major determinant of sensitivity to the alkylating agent TMZ in glioblastoma cells, and as such has major significance in the treatment of these patients. We have identified the paediatric glioblastoma cell line KNS42 to be resistant to TMZ in vitro despite an absence of MGMT expression, a competent MMR system, and an intact double strand break repair pathway. Clues as to the mechanism of resistance in these cells may help to identify factors contributing to childhood glioblastoma patients who remain refractory to TMZ treatment.
Gene expression profiling of a panel of paediatric and adult glioma cell lines highlighted co-ordinated expression of numerous HOX genes in the resistant cell lines, and most especially KNS42, and provided in vitro model system evidence in support of data from TMZ-treated adult glioblastoma patients(26). Using a similar expression profiling and GSEA approach, Murat and co-workers identified a HOX-dominated gene cluster as an independent predictive factor of resistance. Integrating the core gene lists of the present study to that dataset highlights HOXA9 and HOXA10 as the key effectors in both systems. This converges with a recent study identifying the HOXA cluster, and HOXA9 in particular, to be an independent negative prognostic marker in adult glioblastoma(27). Herein we provide evidence for an additional prognostic role in paediatric high grade glioma.
HOX genes are essential in axis determination during embryonic development and are known to be involved in cancer, including glioblastomas(31, 32); however, it is not immediately apparent what role they may play in resistance to alkylating agents. Costa et al. propose that HOXA9 exerts anti-apoptotic and pro-proliferative effects after upregulation via an epigenetic mechanism controlled by PI3-kinase, and independent of mTOR(27). Our findings confirm this observation, and add to previous evidence demonstrating the synergistic interactions of TMZ and PI3-kinase pathway inhibitors in in vitro and in vivo models of adult glioblastoma(33); this combination may therefore also be beneficial in glioblastoma patients with an MGMT-independent, HOX gene signature-associated mechanism of resistance to TMZ.
Murat et al. suggested strong HOXA10 expression in glioblastoma-derived neurospheres to be in line with a role of HOX genes in the glioma stem-like cell compartment, and showed that the resistance signature as a whole is evocative of self-renewal(26). Of note is the presence of a high proportion (17.0%) of CD133-positive cells present in our KNS42 cell monolayer cultures, with additional expression of other stem cell markers in co-ordination with the HOX gene signature. Such a HOX/stem cell signature was also found to be tightly regulated in an analysis of the TCGA glioblastoma expression data(21). The paediatric glioblastoma cell line KNS42 may be an excellent experimental model for investigating such interactions with MGMT-independent treatment failure.
We also found a remarkable link between the HOX/stem cell signature and co-ordinated overexpression of genes within the CDK4 amplicon at 12q13-q14 in glioblastoma patient samples. This association was also present, and correlated with poor response to TMZ chemoradiotherapy, in the Murat et al. dataset(26). Here we provide a possible mechanism for this co-expression of HOX and 12q13-q14 genes in the form of overexpression of the Akt enhancer PIKE (CENTG1, AGAP2), also present at very high levels in the unamplified KNS42 cells, which may drive the changes in H3K27 methylation of the HOXA cluster mediated via PI3-kinase pathway signalling(27).
It is apparent that a variety of processes, not all involving repair or tolerance of alkyl lesions, may promote alkylator resistance(23). As well as providing therapeutic guidance for those patients whose tumours are intrinsically resistant to treatment, characterisation of additional determinants of resistance is necessary in order to develop new targets for therapy in tumours that acquire resistance to TMZ in vivo in the presence of hypermethylated MGMT.
ACKNOWLEDGEMENTS
We would like to thank Dr Daphne Haas-Kogan (UCSF) and Dr Michael Bobola (University of Washington) for provision of the paediatric glioma cell lines, and Dr Michael Hubank (Institute of Child Health, University College London) for assistance with the expression profiling. This research is supported by Cancer Research UK (C1178/A10294, C309/A2187, C309/A8274), the Oak Foundation (LM) and La Fondation de France (NG). We acknowledge NHS funding to the NIHR Biomedical Research Centre. Paul Workman is a Cancer Research UK Life Fellow.
Footnotes
Disclosure of Potential Conflicts of Interest
Nathalie Gaspar, Lynley Marshall, Lara Perryman, Dorine Bax, Suzanne Little, Marta Viana-Pereira, Swee Sharp, Andrew Pearson, Paul Workman and Chris Jones are or were employees of The Institute of Cancer Research which has a commercial interest in the development of PI3K inhibitors and operates a rewards-to-inventors scheme. Paul Workman and his team have been involved in a commercial collaboration with Yamanouchi (now Astellas Pharma) and with Piramed Pharma, and intellectual property arising from the programme has been licensed to Genentech. Paul Workman was a founder of, consultant to, Scientific Advisory Board member of, and stockholder in Piramed Pharma, which was acquired by Roche.
REFERENCES
- 1.Mrugala MM, Chamberlain MC. Mechanisms of disease: temozolomide and glioblastoma--look to the future. Nat Clin Pract Oncol. 2008;5:476–86. doi: 10.1038/ncponc1155. [DOI] [PubMed] [Google Scholar]
- 2.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–96. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 3.Newlands ES, Blackledge GR, Slack JA, Rustin GJ, Smith DB, Stuart NS, et al. Phase I trial of temozolomide (CCRG 81045: M&B 39831: NSC 362856) Br J Cancer. 1992;65:287–91. doi: 10.1038/bjc.1992.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 5.Esteller M, Garcia-Foncillas J, Andion E, Goodman SN, Hidalgo OF, Vanaclocha V, et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343:1350–4. doi: 10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]
- 6.Middlemas DS, Stewart CF, Kirstein MN, Poquette C, Friedman HS, Houghton PJ, et al. Biochemical correlates of temozolomide sensitivity in pediatric solid tumor xenograft models. Clin Cancer Res. 2000;6:998–1007. [PubMed] [Google Scholar]
- 7.Silber JR, Bobola MS, Blank A, Schoeler KD, Haroldson PD, Huynh MB, et al. The apurinic/apyrimidinic endonuclease activity of Ape1/Ref-1 contributes to human glioma cell resistance to alkylating agents and is elevated by oxidative stress. Clin Cancer Res. 2002;8:3008–18. [PubMed] [Google Scholar]
- 8.Cheng CL, Johnson SP, Keir ST, Quinn JA, Ali-Osman F, Szabo C, et al. Poly(ADP-ribose) polymerase-1 inhibition reverses temozolomide resistance in a DNA mismatch repair-deficient malignant glioma xenograft. Mol Cancer Ther. 2005;4:1364–8. doi: 10.1158/1535-7163.MCT-05-0128. [DOI] [PubMed] [Google Scholar]
- 9.Pollack IF, Hamilton RL, Sobol RW, Burnham J, Yates AJ, Holmes EJ, et al. O6-methylguanine-DNA methyltransferase expression strongly correlates with outcome in childhood malignant gliomas: results from the CCG-945 Cohort. J Clin Oncol. 2006;24:3431–7. doi: 10.1200/JCO.2006.05.7265. [DOI] [PubMed] [Google Scholar]
- 10.Broniscer A, Chintagumpala M, Fouladi M, Krasin MJ, Kocak M, Bowers DC, et al. Temozolomide after radiotherapy for newly diagnosed high-grade glioma and unfavorable low-grade glioma in children. J Neurooncol. 2006;76:313–9. doi: 10.1007/s11060-005-7409-5. [DOI] [PubMed] [Google Scholar]
- 11.Nicholson HS, Kretschmar CS, Krailo M, Bernstein M, Kadota R, Fort D, et al. Phase 2 study of temozolomide in children and adolescents with recurrent central nervous system tumors: a report from the Children’s Oncology Group. Cancer. 2007;110:1542–50. doi: 10.1002/cncr.22961. [DOI] [PubMed] [Google Scholar]
- 12.Ruggiero A, Cefalo G, Garre ML, Massimino M, Colosimo C, Attina G, et al. Phase II trial of temozolomide in children with recurrent high-grade glioma. J Neurooncol. 2006;77:89–94. doi: 10.1007/s11060-005-9011-2. [DOI] [PubMed] [Google Scholar]
- 13.Hargrave DR, Zacharoulis S. Pediatric CNS tumors: current treatment and future directions. Expert Rev Neurother. 2007;7:1029–42. doi: 10.1586/14737175.7.8.1029. [DOI] [PubMed] [Google Scholar]
- 14.Lashford LS, Thiesse P, Jouvet A, Jaspan T, Couanet D, Griffiths PD, et al. Temozolomide in malignant gliomas of childhood: a United Kingdom Children’s Cancer Study Group and French Society for Pediatric Oncology Intergroup Study. J Clin Oncol. 2002;20:4684–91. doi: 10.1200/JCO.2002.08.141. [DOI] [PubMed] [Google Scholar]
- 15.Bax DA, Little SE, Gaspar N, Perryman L, Marshall L, Viana-Pereira M, et al. Molecular and phenotypic characterisation of paediatric glioma cell lines as models for preclinical drug development. PLoS ONE. 2009:4. doi: 10.1371/journal.pone.0005209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skehan P, Storeng R, Scudiero D, Monks A, McMahon J, Vistica D, et al. New colorimetric cytotoxicity assay for anticancer-drug screening. J Natl Cancer Inst. 1990;82:1107–12. doi: 10.1093/jnci/82.13.1107. [DOI] [PubMed] [Google Scholar]
- 17.Cory AH, Owen TC, Barltrop JA, Cory JG. Use of an aqueous soluble tetrazolium/formazan assay for cell growth assays in culture. Cancer Commun. 1991;3:207–12. doi: 10.3727/095535491820873191. [DOI] [PubMed] [Google Scholar]
- 18.Chou TC, Talaly P. A simple generalized equation for the analysis of multiple inhibitions of Michaelis-Menten kinetic systems. J Biol Chem. 1977;252:6438–42. [PubMed] [Google Scholar]
- 19.Esteller M, Hamilton SR, Burger PC, Baylin SB, Herman JG. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res. 1999;59:793–7. [PubMed] [Google Scholar]
- 20.Nygren AO, Ameziane N, Duarte HM, Vijzelaar RN, Waisfisz Q, Hess CJ, et al. Methylation-specific MLPA (MS-MLPA): simultaneous detection of CpG methylation and copy number changes of up to 40 sequences. Nucleic Acids Res. 2005;33:e128. doi: 10.1093/nar/gni127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McLendon R, Friedman A, Bigner D, Van Meir EG, Brat DJ, Mastrogianakis M, et al. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paugh BS, Qu C, Jones C, Liu Z, Adamowicz-Brice M, J Z, et al. Integrated molecular profiling of pediatric high grade gliomas reveals key differences with the adult disease. J Clin Oncol. doi: 10.1200/JCO.2009.26.7252. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bobola MS, Silber JR, Ellenbogen RG, Geyer JR, Blank A, Goff RD. O6-methylguanine-DNA methyltransferase, O6-benzylguanine, and resistance to clinical alkylators in pediatric primary brain tumor cell lines. Clin Cancer Res. 2005;11:2747–55. doi: 10.1158/1078-0432.CCR-04-2045. [DOI] [PubMed] [Google Scholar]
- 24.Workman P, van Montfort RL. Unveiling the secrets of the ancestral PI3 kinase Vps34. Cancer Cell. 2010;17:421–3. doi: 10.1016/j.ccr.2010.04.016. [DOI] [PubMed] [Google Scholar]
- 25.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murat A, Migliavacca E, Gorlia T, Lambiv WL, Shay T, Hamou MF, et al. Stem cell-related “self-renewal” signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J Clin Oncol. 2008;26:3015–24. doi: 10.1200/JCO.2007.15.7164. [DOI] [PubMed] [Google Scholar]
- 27.Costa BM, Smith JS, Chen Y, Chen J, Phillips HS, Aldape KD, et al. Reversing HOXA9 Oncogene Activation by PI3K Inhibition: Epigenetic Mechanism and Prognostic Significance in Human Glioblastoma. Cancer Res. 2010:70. doi: 10.1158/0008-5472.CAN-09-2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hayakawa M, Kaizawa H, Moritomo H, Koizumi T, Ohishi T, Yamano M, et al. Synthesis and biological evaluation of pyrido[3′,2′:4,5]furo[3,2-d]pyrimidine derivatives as novel PI3 kinase p110alpha inhibitors. Bioorg Med Chem Lett. 2007;17:2438–42. doi: 10.1016/j.bmcl.2007.02.032. [DOI] [PubMed] [Google Scholar]
- 29.Raynaud FI, Eccles S, Clarke PA, Hayes A, Nutley B, Alix S, et al. Pharmacologic characterization of a potent inhibitor of class I phosphatidylinositide 3-kinases. Cancer Res. 2007;67:5840–50. doi: 10.1158/0008-5472.CAN-06-4615. [DOI] [PubMed] [Google Scholar]
- 30.Workman P, Collins I. Probing the probes: fitness factors for small molecule tools. Chem Biol. 2010;17:561–77. doi: 10.1016/j.chembiol.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abate-Shen C. Deregulated homeobox gene expression in cancer: cause or consequence? Nat Rev Cancer. 2002;2:777–85. doi: 10.1038/nrc907. [DOI] [PubMed] [Google Scholar]
- 32.Abdel-Fattah R, Xiao A, Bomgardner D, Pease CS, Lopes MB, Hussaini IM. Differential expression of HOX genes in neoplastic and non-neoplastic human astrocytes. J Pathol. 2006;209:15–24. doi: 10.1002/path.1939. [DOI] [PubMed] [Google Scholar]
- 33.Guillard S, Clarke PA, Te Poele R, Mohri Z, Bjerke L, Valenti M, et al. Molecular pharmacology of phosphatidylinositol 3-kinase inhibition in human glioma. Cell Cycle. 2009;8:443–53. doi: 10.4161/cc.8.3.7643. [DOI] [PubMed] [Google Scholar]