Abstract
The soil and moss dwelling oribatid mite family Scutoverticidae is considered to represent an assemblage of distantly related but morphologically similar genera. We used nucleotide sequences of one mitochondrial (COI) and two nuclear (28S rDNA, ef-1α) genes, and 79 morphological characters to elucidate the phylogenetic relationships among eleven nominal plus two undescribed European mite species of the family Scutoverticidae with a particular focus on the genus Scutovertex. Both molecular genetic and morphological data revealed a paraphyletic genus Scutovertex, with S. pictus probably representing a distinct genus, and Provertex kuehnelti was confirmed as member of the family Scutoverticidae. Molecular genetic data confirmed several recently described Scutovertex species and thus the high species diversity within this genus in Europe and suggest that S. sculptus represents a complex of several cryptic species exhibiting marked genetic, but hardly any morphological divergence.
Keywords: oribatid mites, Scutovertex, multidisciplinary approach, phylogeny, paraphylum, cryptic species
Introduction
In recent years, genetic studies have highlighted cryptic diversity in various groups of organisms, indicated by large genetic distances within traditionally recognized, sometimes even well-known, taxa (Edwards and Dimock, 1997; Hebert et al., 2004; Katongo et al., 2005; Kon et al., 2007; Mayer et al., 2007; Metzger et al., 2009). These cryptic species are so similar morphologically that they are almost or entirely indistinguishable based on morphological characters alone, albeit many cryptic species have been subsequently supported by subtle morphological differences found in post-hoc analyses of morphological data (Mathews et al., 2008; Padial and de la Riva, 2009). The potential for cryptic diversity seems particularly high in small-size and short generation time animals (Marzluff and Dial, 1991; Kon et al., 2007). Moreover, some biomes seem to home more cryptic species than others, and particularly in tropical and marine habitats it appears to be a widespread phenomenon (Baric and Sturmbauer, 1997; Wilcox et al., 1997; Bond and Sierwald, 2002; Hebert et al., 2004; Sáez and Lozano, 2005; Chan et al., 2007), whereas the number of cryptic species in temperate terrestrial biomes seems to be smaller (Schlick-Steiner et al., 2006; King et al., 2008; Murray et al,. 2008).
In mites, species identification is typically based on morphological character sets. There are many species which are morphologically very similar and thus hardly to distinguish. One example for such a group including morphologically very similar taxa is the oribatid mite family Scutoverticidae which is assigned to a subgroup of the Circumdehiscentiae (“Higher Oribatida”) at the base of the Poronota. This subgroup shows wrinkled nymphs and adults which bear sacculi on the notogaster homologous to the so called octotaxic system consisting of four pairs of porose areas characterizing the Poronota (Grandjean, 1953; 1969).
Despite their systematic position the Scutoverticidae are often considered as a conglomeration of distantly related but morphologically similar genera. Bernini (1976) already pointed out the urgent need for a comprehensive and detailed revision of this family to check the membership of the different genera to this taxon, whereby Scutovertex as the eponymous genus should serve as reference. Meanwhile, several new taxa were described and Shtanchaeva and Netuzhilin (2003) published a revision of the Scutoverticidae, describing some new species and summing up the knowledge without any attempt to include additional characters to eliminate the taxonomic uncertainties. Up to now the systematic classification within the Scutoverticidae has been suffering from two major problems: the short, fragmentary and often inaccurate descriptions of species and genera, and the limited knowledge of the amount of intraspecific variation and the diversity of this mite family. These two factors led to the description of new taxa (some of them may represent synonyms) and caused some taxonomical confusion, e.g. in the genus Provertex (Krisper and Schuster, 2009).
To date, this family comprises eight genera with about 60 species worldwide (Subías, 2004; resp. 2008) whereof only one-third occurs in Europe. Most species can be found in the very south-western (Spain) or eastern (Russia) European part. They are adapted to extreme environmental conditions such as regular desiccation, inundation and temperature fluctuation because their preferred habitats are mosses, lichens or tussocks on sun exposed rocks and roofs (Krisper et al., 2002; Smrž, 2006), as well as saline soils, salt marshes or inundation meadows (Schuster, 1958; Weigmann, 2004). Due to their adaptation to extreme environmental conditions, scutoverticid mites play an important ecological role as pioneer species at the first steps of succession (Skubala, 1995). General information on life span, population size etc. is more or less lacking but most members of the family reproduce sexually and generation times vary from two to six months (Ermilov, 2008; personal observations). Recent data on the genetic diversity in two Austrian species revealed marked inter-specific differences suggestive of differences in population size and dispersal ability (Schäffer et al., in press).
Especially in the genus Scutovertex species diversity is very high. Problems to distinguish between the two most widespread species S. minutus and S. sculptus (e.g. mentioned by Weigmann, 2006) were solved recently by detailed re-descriptions of both species (Schäffer and Krisper, 2007; Pfingstl et al., 2008) and their taxonomic discreteness and different population structure was also confirmed by molecular genetic data (Schäffer et al., 2008). In Central, Northern and Western Europe eight Scutovertex-species are known (S. alpinus, S. arenocolus, S. ianus, S. minutus, S. pictus, S. pileatus, S. pannonicus, and S. sculptus), some of which have been described only recently (Schäffer et al., 2008; Pfingstl et al., 2009, in press, submitted), plus five additional species representing three different genera (Lamellovertex caelatus; Exochocepheus hungaricus; Provertex delamarei, P. kuehnelti, P. mailloli).
In the present study we attempt to i) evaluate the taxonomic status of eleven nominal and two undescribed European species of the family Scutoverticidae ii) elucidate the phylogenetic relationships between the Scutovertex species and iii) uncover a potential cryptic diversity within the genus Scutovertex. To achieve our aims, we used three molecular markers (one mitochondrial and two nuclear genes) which allow us to resolve both ancient and recent nodes in a phylogenetic tree. Additionally 79 well defined characters and character states were used to obtain a morphology-based phylogeny for comparative purpose.
Materials and Methods
Sample collection
This study includes eleven nominal plus two undescribed species of the family Scutoverticidae, collected from different localities in Central, Northern and Western Europe between 2005 and 2009. Based on Grandjean (1969) we chose Unduloribates undulatus (Unduloribatidae) and three specimens of Cymbaeremaeus cymba (Cymbaeremaeidae) as outgroup taxa because both families also belong to the subgroup of Circumdehiscentiae with wrinkled nymphs. Information on sampling localities is given in Table 1 and Figs. 1a-b. Specimens were extracted from mosses and lichens collected on sun exposed rocks and roofs or salt marshes with Berlese-Tullgren funnels. Individuals for morphological analyses were preserved in 70% ethanol, those for molecular genetic analyses in absolute ethanol.
Table 1. Specimens, sample ID, sampling location and GenBank accession numbers for the samples analyzed in this study.
| GenBank Accession No. | |||||
|---|---|---|---|---|---|
| Species | Sample ID | COI | 28S | ef-1α | Sampling locality |
| Scutoverticidae | |||||
| Scutovertex | |||||
| S. alpinus | SalpHT1 | GU208673# | GU208524 | GU208619 | Fuscherkarkopf/Großglockner/Carinthia - A |
| SalpHT2 | GU208674# | GU208525 | GU208620 | Fuscherkarkopf/Großglockner/Carinthia - A | |
| SalpHT3 | GU208675# | GU208526 | GU208621 | Fuscherkarkopf /Großglockner/Carinthia - A | |
| S. arenocolus | SarCoast3 | GU208578 | GU208527 | GU208622 | Darss-Zingst/Baltic Coast - D |
| SarCoast7 | GU208579 | GU208528 | GU208623 | Darss-Zingst/Baltic Coast - D | |
| S. ianus | SianSt5 | GU208580 | GU208529 | GU208624 | Stiwoll/Styria - A |
| SianSch8 | GU208581 | GU208530 | GU208625 | Schladming/Styria - A | |
| SianAdm1 | GU208582 | GU208531 | GU208626 | Admont/Styria - A | |
| SianAu4 | GU208583 | GU208532 | GU208627 | Floodplain of Traun/Upper Astria - A | |
| SianAu5 | GU208584 | GU208533 | GU208628 | Floodplain of Traun/Upper Astria - A | |
| SianMos1 | GU208585 | GU208534 | GU208629 | Mosbach near Heidelberg - D | |
| S. minutus | SmBach3 | GQ890381* | GU208535 | GU208630 | Bachsdorf/Styria - A |
| SmPo3 | GQ890362* | GU208536 | GU208631 | Pogier/Styria - A | |
| SmKal3 | GQ890373* | GU208537 | GU208632 | Graz/Styria - A | |
| SmUsb4 | GQ890395* | GU208538 | GU208633 | Unterstinkenbrunn/Lower Austria - A | |
| S. pannonicus | SpaI_B8 | GQ890445* | GU208539 | GU208634 | Lake “ Zicklacke”/Burgenland - A |
| SpaI_C6 | GQ890444* | GU208540 | GU208635 | Lake “Oberer Stinker”/Burgenland - A | |
| S. pictus | SpKal6 | GU208586 | GU208541 | GU208636 | Graz/Styria - A |
| SpBH9 | GU208587 | GU208542 | GU208637 | Castle Hochosterwitz/Carinthia - A | |
| S. pileatus | SpilBH5 | GU208588 | GU208543 | GU208638 | Castle Hochosterwitz/Carinthia - A |
| SpilL3 | GU208589 | GU208544 | GU208639 | Laas/Carinthia - A | |
| S. species 1 | SspHu5 | GU208590 | GU208545 | GU208640 | Fülöpháza/Kiskunság National Park - H |
| SspHu6 | GU208591 | GU208546 | GU208641 | Fülöpháza/Kiskunság National Park - H | |
| S. species 2 | SspWa1 | GU208592 | GU208547 | GU208642 | Wangen am Ritten/South Tyrol - I |
| SspWa2 | GU208593 | GU208548 | GU208643 | Wangen am Ritten/South Tyrol - I | |
| S. sculptus | SsHlb2 | GQ890440* | GU208549 | GU208644 | Häuslberg/Styria - A |
| SsI_B6 | GQ890427* | GU208550 | GU208645 | Lake “Oberer Stinker”/Burgenland - A | |
| SsI_C13 | GQ890434* | GU208551 | GU208646 | Lake “Zicklacke”/Burgenland - A | |
| SsFliess3 | GQ890441* | GU208552 | GU208647 | Fliess/Tyrol - A | |
| SsRu1 | GU208594 | GU208553 | GU208648 | Nizhniy Novgorod - RUS | |
| SsRu2 | GU208595 | GU208554 | GU208649 | Nizhniy Novgorod - RUS | |
| SsRu3 | GU208596 | GU208555 | GU208650 | Nizhniy Novgorod - RUS | |
| SsSW1 | GU208597 | GU208556 | GU208651 | Endeby near Uppsala - S | |
| SsSW2 | GU208598 | GU208557 | GU208652 | Endeby near Uppsala - S | |
| SsMos2 | GU208599 | GU208558 | GU208653 | Mosbach near Heidelberg - D | |
| SsIRL1 | GU208600 | GU208559 | GU208654 | Ceide-Fields - IRL | |
| SsF1 | GU208601 | GU208560 | GU208655 | Seignosse/Les Bourdaines - F | |
| SsF3 | GU208602 | GU208561 | GU208656 | Seignosse/Les Bourdaines - F | |
| SsXa1 | GU208603 | GU208562 | GU208657 | Xanten - D | |
| SsXa2 | GU208604 | GU208563 | GU208658 | Xanten - D | |
| Lamellovertex | |||||
| L. caelatus | LcE3 | GU208605 | GU208564 | GU208659 | Ernstbrunn/Lower Austria - A |
| LcE6 | GU208606 | GU208565 | GU208660 | Ernstbrunn/Lower Austria - A | |
| Provertex | |||||
| P. kuehnelti | PkHT1 | GU208607 | GU208566 | GU208661 | F. Josef Höhe/Großglockner/Carinthia - A |
| PkGe1 | GU208608 | GU208567 | GU208662 | Gesäuse/Styria - A | |
| PkIRL1 | GU208609 | GU208568 | GU208663 | Galway - IRL | |
| PkRom2 | GU208610 | GU208569 | GU208664 | Braşov-Bucegi mountains - RO | |
| Exochocepheus | |||||
| E. hungaricus | EhungHu1 | GU208611 | GU208570 | GU208665 | Fülöpháza/Kiskunság National Park - H |
| EhungHu4 | GU208612 | GU208571 | GU208666 | Fülöpháza/Kiskunság National Park - H | |
| EhungHu5 | GU208613 | GU208572 | GU208667 | Fülöpháza/Kiskunság National Park - H | |
| Unduloribatidae | |||||
| Unduloribates | |||||
| U. undulatus | UuRom1 | GU208614 | GU208573 | GU208668 | Braşov-Bucegi mountains - RO |
| UuRom3 | GU208615 | GU208574 | GU208669 | Braşov-Bucegi mountains - RO | |
| Cymbaeremaeidae | |||||
| Cymbaeremaeus | |||||
| C. cymba | CcRoe3 | GU208616 | GU208575 | GU208670 | Röthelstein/Styria - A |
| CcRoe5 | GU208617 | GU208576 | GU208671 | Röthelstein/Styria - A | |
| CcPlatte1 | GU208618 | GU208577 | GU208672 | Graz/Styria - A | |
Sequences from COI region2 fragment alone.
Sequences not generated in the framework of this study were obtained from Schäffer et al. (in press).
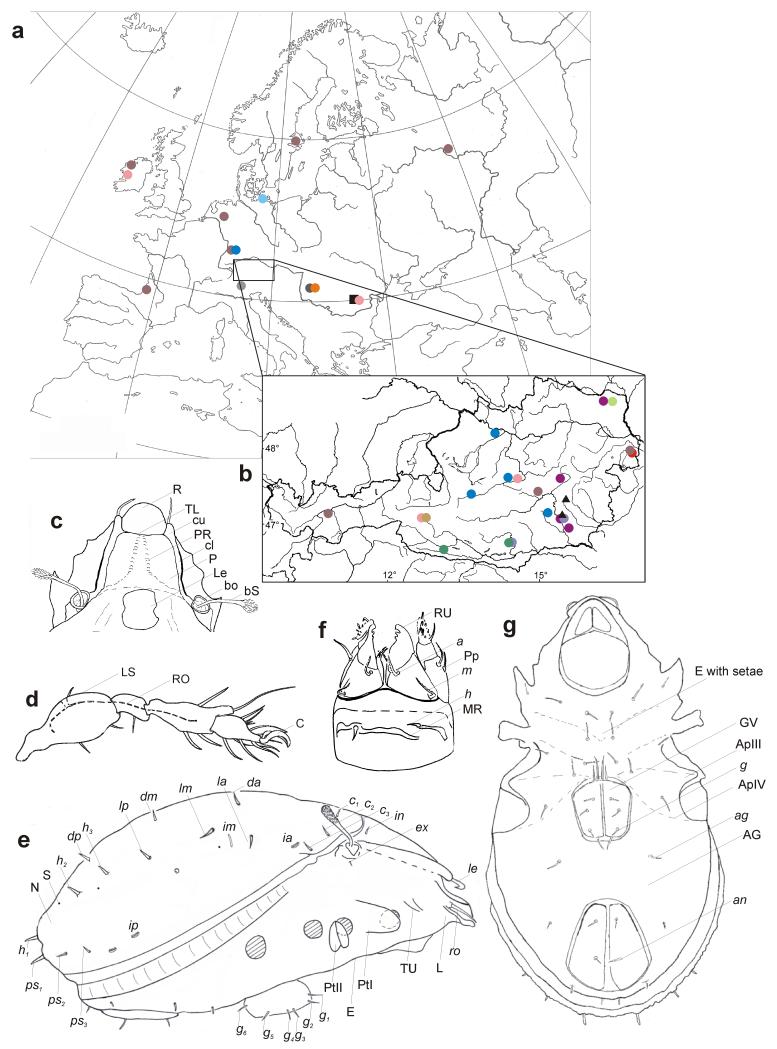
Fig. 1. (a-b) Sampling localities of the specimens used in this study: (a) Map of Europe. (b) Map of Austria.
Species are marked by different colors or symbols:  = Scutovertex alpinus,
= Scutovertex alpinus,  = S. arenocolus,
= S. arenocolus,  = S. ianus,
= S. ianus,  = S. minutus,
= S. minutus,  = S. pannonicus,
= S. pannonicus,  = S. pictus,
= S. pictus,  = S. pileatus,
= S. pileatus,  = S. sp.1,
= S. sp.1,  = S. sp.2,
= S. sp.2,  = S. sculptus,
= S. sculptus,  = Exochocepheus hungaricus,
= Exochocepheus hungaricus,  = Lamellovertex caelatus,
= Lamellovertex caelatus,  = Provertex kuehnelti, ▴ = Cymbaeremaeus cymba, ∎ = Unduloribates undulatus. (c-g) Schematic representation of morphological characters investigated in this study: (c) prodorsum (dorsal view); (d) leg with setae (lateral view); (e) lateral body view; (f) subcapitulum (ventral view); (g) ventral body view. Abbreviations: a, anterior subcapitular seta; AG, anogenital region; ag, aggenital seta; an, anal seta; Ap, apodem; bo, bothridium; bS, sensillus; C, claw; cl, lamella; cu, cusp; E, epimeral region; ex, exobothridial seta; GV, genital valve; g, genital seta; h, hysterostomatic seta; in, interlamellar seta; ia, im, ip, lyrifissures; le, lamellar seta; Le, lenticulus; LS, leg surface; m, median subcapitular seta; MR, rib on mentum; N, notogastral surface; P, prodorsal surface; PL, prodorsum lateral; Pp, pedipalp; PR, prodorsal ridges; PtI, PtII, pedotectum I/II; R, rostrum; RO, respiratory organ; ro, rostral seta; RU, rutellar teeth; S, saccule of the octotaxic system; TL, translamella; TU, tutorium; c1-3, da, dm, dp, la, lm, lp, h1-3, ps1-3, notogastral setae.
= Provertex kuehnelti, ▴ = Cymbaeremaeus cymba, ∎ = Unduloribates undulatus. (c-g) Schematic representation of morphological characters investigated in this study: (c) prodorsum (dorsal view); (d) leg with setae (lateral view); (e) lateral body view; (f) subcapitulum (ventral view); (g) ventral body view. Abbreviations: a, anterior subcapitular seta; AG, anogenital region; ag, aggenital seta; an, anal seta; Ap, apodem; bo, bothridium; bS, sensillus; C, claw; cl, lamella; cu, cusp; E, epimeral region; ex, exobothridial seta; GV, genital valve; g, genital seta; h, hysterostomatic seta; in, interlamellar seta; ia, im, ip, lyrifissures; le, lamellar seta; Le, lenticulus; LS, leg surface; m, median subcapitular seta; MR, rib on mentum; N, notogastral surface; P, prodorsal surface; PL, prodorsum lateral; Pp, pedipalp; PR, prodorsal ridges; PtI, PtII, pedotectum I/II; R, rostrum; RO, respiratory organ; ro, rostral seta; RU, rutellar teeth; S, saccule of the octotaxic system; TL, translamella; TU, tutorium; c1-3, da, dm, dp, la, lm, lp, h1-3, ps1-3, notogastral setae.
Morphological data
79 morphological characters or character states, respectively, (see Figs. 1c-g) were recorded for five individuals per species. Character coding was based on unordered multistate characters (Appendix A+B).
Phylogenetic reconstruction was carried out by MP using PAUP* (search options: heuristic search; random addition of taxa; TBR branch swapping with 1,000 replicates), using one specimen per species, since there were no intraspecific differences. Statistical support was assessed by bootstrapping (1,000 pseudo-replicates).
Molecular genetic analyses
Total genomic DNA was extracted from single individuals applying the CTAB (hexadecyltriethylammonium bromide) method described in Schäffer et al. (in press) or the DNeasy Blood & Tissue Kit (Qiagen, Vienna, Austria).
Fragments of COI, ef-1α and 28S rDNA genes were amplified by polymerase chain reaction (PCR) using the following primers: COI_1fwd (5′-GNTCAACAAWTCATWAAG-3′) and COI_2rev (5′-TAAACTTCNGGYTGNCCAAAAAATCA-3′) for COI region1 (modified after Heethoff et al., 2007), Mite COI-2F and Mite COI-2R (Otto and Wilson, 2001) for COI region 2, D3A and D3B (Litvaitis et al., 1994) for the D3 fragment of the 28S rDNA, and 40.71F and 52.RC (Regier and Shultz, 1997) for ef-1α. Since last-mentioned primer pair did not work well in all specimens we designed new ones: EF-SyFwd (5′-GGACAAACTGAAGGHW GAGMG-3′) and EF-SyRev (5′-RKNGGTCKTGAGGGCGGTTCC-3′). Purification of PCR products, and sequencing reaction followed the protocol described in Schäffer et al. (2008). DNA fragments were purified with SephadexTM G-50 (Amersham Biosciences) following the manufacturer’s instruction and visualized on a 3130xl capillary sequencer (Applied Biosystems). Sequences are available from GenBank under the accession numbers listed in Table 1.
We sequenced 1,259 bp of the mitochondrial COI gene, 316-324 bp of the D3 region of the nuclear 28S rDNA and 504 bp of the nuclear ef-1α gene in 54 specimens (in the three S. alpinus individuals the fragment of the COI-region1 could not be amplified). Sequences were verified by comparisons with known oribatid sequences from GenBank and aligned by eye in MEGA 3.1 (Kumar et al., 2004). One 18-26 bp fragment of the 28S D3 region could not be aligned unambiguously and was excluded (also base position 153 from SpKal6) from the analyses. For the further phylogenetic analyses we always used all available sequences for each gene fragment.
In a first step, for testing the performance of the single genes, we constructed separate phylogenies for the three genes plus a phylogeny for the COI region2 (because of S. alpinus, see above) using Bayesian inference (BI) as implemented in MrBayes 3.1.2 (Ronquist and Huelsenbeck, 2003). For all Bayesian analyses genes (except 28S rDNA) were partitioned by codon position. Rate heterogeneity was set according to a gamma distribution with six rate categories (GTR model) for each data partition. Posterior probabilities were obtained from a Metropolis-coupled Markov chain Monte Carlo simulation (2 independent runs; 4 chains with 3 million generations each; chain temperature: 0.2; trees sampled every 100 generations), with parameters estimated from the data set. Depending on the data set we applied different burn-ins to allow likelihood values to reach stationarity, so that the average standard deviation of split frequencies was <0.01.
In a second step, the fragments of all three genes were combined for further analyses (length of concatenated data set = 2,059 bp). We analyzed two data sets, one with all available sequences, entitled full set (FS) and one without COI region1, entitled reduced set (RS). The RS data set served to get information on the phylogenetic placement of S. alpinus. Phylogenetic reconstructions by neighbor joining (NJ) and maximum parsimony (MP) were conducted in PAUP* (Swofford, 2002), maximum likelihood (ML) in RAxML-7.0.3-WIN (Stamatakis, 2006) and BI in MrBayes. For NJ of FS and RS, the best-fit substitution model selected by the hierarchical likelihood ratio test (hLRT) implemented in Modeltest 3.06 (Posada and Crandall, 1998) was GTR+I+G (parameters of FS/RS: base frequencies: A = 0.3057/0.3112, C = 0.1742/0.1749, G = 0.1604/0.1790, T = 0.3597/0.3349; R-matrix: A↔C = 0.6075/0.9541; A↔G = 10.6077/8.1492; A↔T = 1.0908/1.1455; C↔G = 1.3283/1.3145; C↔T = 12.0869/12.9849; G↔T = 1.0000; proportion of invariable sites: I = 0.6165/0.5742; gamma shape parameter: α = 0.6548/0.3888). Heuristic tree searches under MP criteria applied random addition of taxa and TBR branch swapping (1,000 replicates). Statistical support for the resulting NJ and MP topologies was assessed by bootstrapping (1,000 pseudo-replicates). To find the best-scoring ML tree we used the default algorithm with 40 distinct rate categories, the GTR+I+G substitution model and COI and ef-1α were partitioned by gene and by codon position. Nodes were supported by bootstrapping (500 replicates). Settings for Bayesian Inference were same as mentioned above and the partitioning of COI and ef-1α was the same as for ML analysis.
To assess whether the topologies obtained by the different tree building algorithms differed significantly, we performed Kishino-Hasegawa (KH; Kishino and Hasegawa, 1989) and Shimodaira-Hasegawa (SH) tests (Shimodaira and Hasegawa, 1999) in PAUP*. Alternative phylogenetic hypotheses were compared to a strict consensus topology of the NJ, MP, ML and BI trees also by means of KH and SH tests.
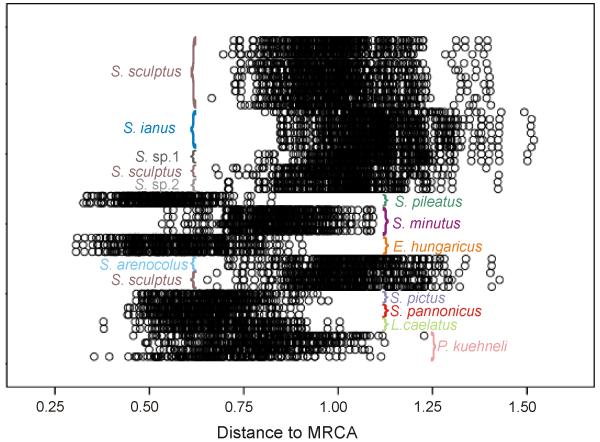
A Bayesian relative rates test according to the method describe by Wilcox et al. (2004) was conducted to test for significant differences in branch lengths and hence substitution rates at the COI gene, which is commonly used to estimated divergence times in arthropods (Brower, 1994; Juan et al., 1996; Quek et al., 2004), and also in oribatid mites (Salomone et al., 2002; Heethoff et al., 2007). The posterior probability distribution of branch lengths for all branches was obtained by saving branch lengths for every 100th sampled tree (after burn-in) of the MrBayes run. For each tree the distance from the most recent common ancestor (MRCA) of the ingroup to each of the terminal taxa was calculated with Cadence v.1.0.1 (Wilcox et al., 2004; available at http://www.biosci.utexas.edu/antisense/). Scutovertex alpinus was excluded from this analysis because of the lacking COI region1 sequence. The distribution of branch lengths was plotted in the program SPSS ver. 16.0. Because of considerable variations in relative rates among the ingroup taxa (Fig. 2), we refrained from applying a molecular clock to estimate divergence times (Wilcox et al. 2004). Given a lack of possible calibration points we were not able use relaxed clock models on our data to reliably estimate divergence times either.
Fig. 2. Results of the Bayesian relative rates test.
The distribution of branch lengths from the most recent common ancestor (MRCA) to the terminal taxa (outgroup was excluded) is shown.
Results
Pairwise sequence divergence (uncorrected p-distance) between scutoverticid species ranged from 15 to 24 % in the COI gene, from 0 to 7.4 % in the D3 fragment of 28S rDNA, and from 0.6 to 9.6 % in the ef-1α gene. In the combined data set, pairwise differences ranged from 8 to 19 %.
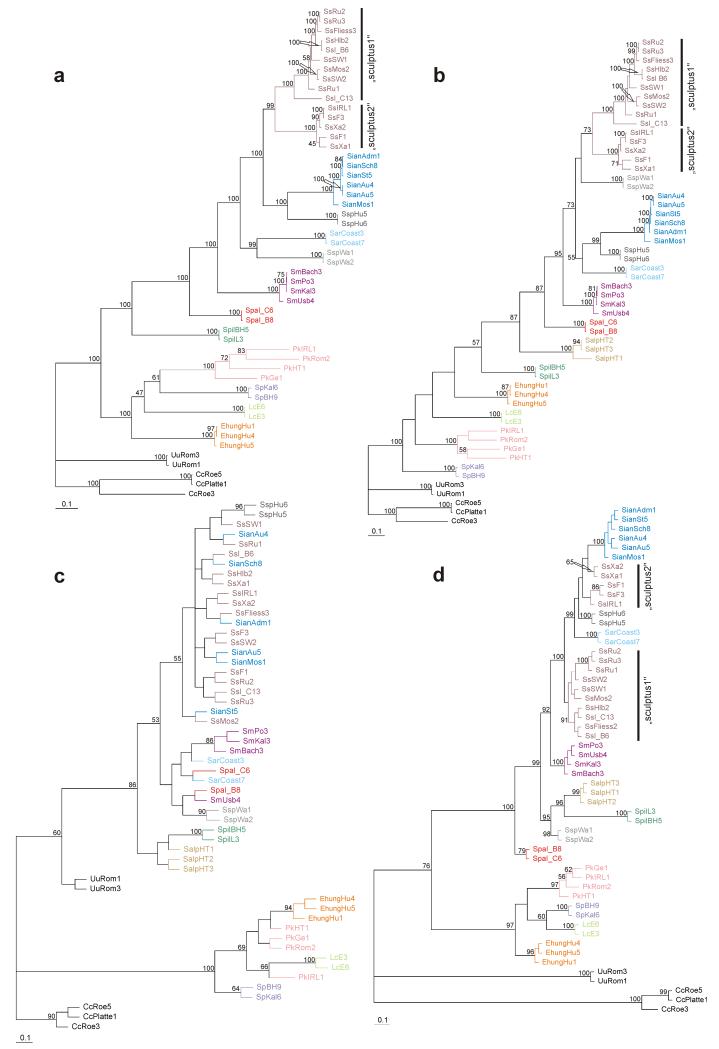
Phylogenetic analyses based on Bayesian inference of the whole COI gene, COI region 2 and ef-1α (Figs. 3a, 3b and 3d) yielded similar results and revealed well resolved topologies with high statistical support for the monophyly of the family Scutoverticidae and the monophyly of each species except S. sculptus, whose specimens clustered in two well supported clades: one included individuals from Russia, Sweden, Germany and Austria (“sculptus1”) and one comprised individuals from France, Ireland and Germany (“sculptus2”). By contrast, the 28S rDNA gene showed only a poorly resolved phylogeny (Fig. 3c). Only the monophyly of the genus Scutovertex and of the remaining genera was well supported. Single gene analyses resulted in different relative phylogenetic positions of the “sculptus1” and “sculptus2” clades.
Fig. 3. Phylogeny of eleven nominal plus two undescribed European species of the family Scutoverticidae based on single gene analyses.
Bayesian inference (BI) tree of three studied genes: (a) mitochondrial COI; (b) COI region2; (c) nuclear 28S rDNA; (d) nuclear ef-1α. Only posterior probabilities >50 are shown. Colors are the same as in Fig. 1.
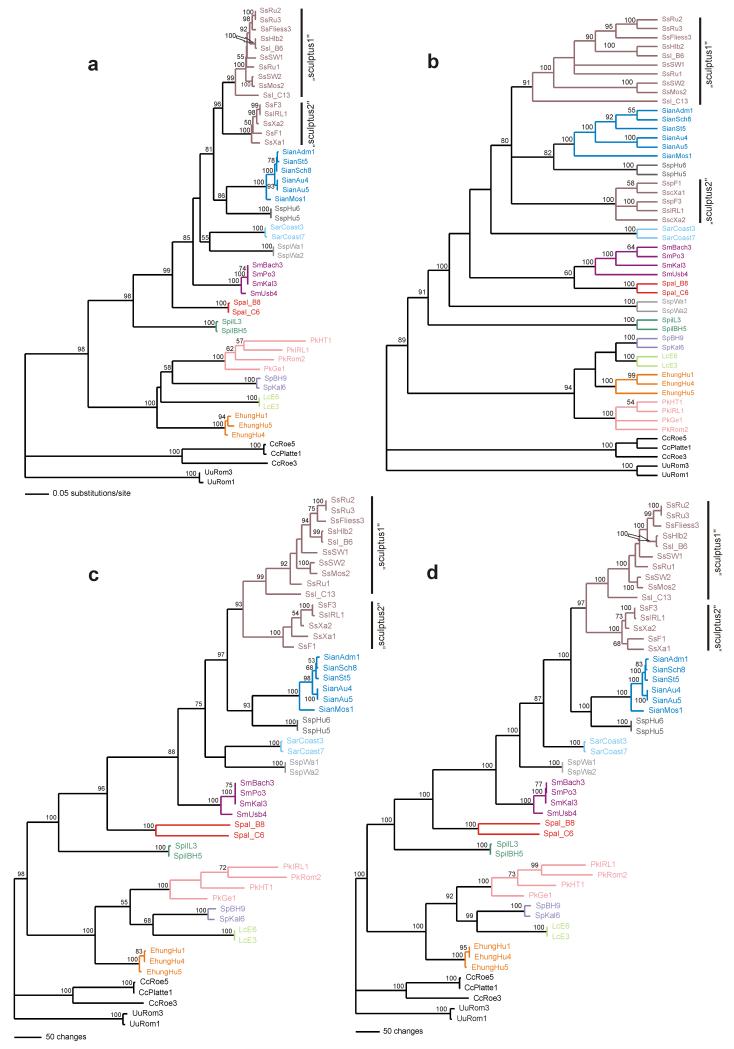
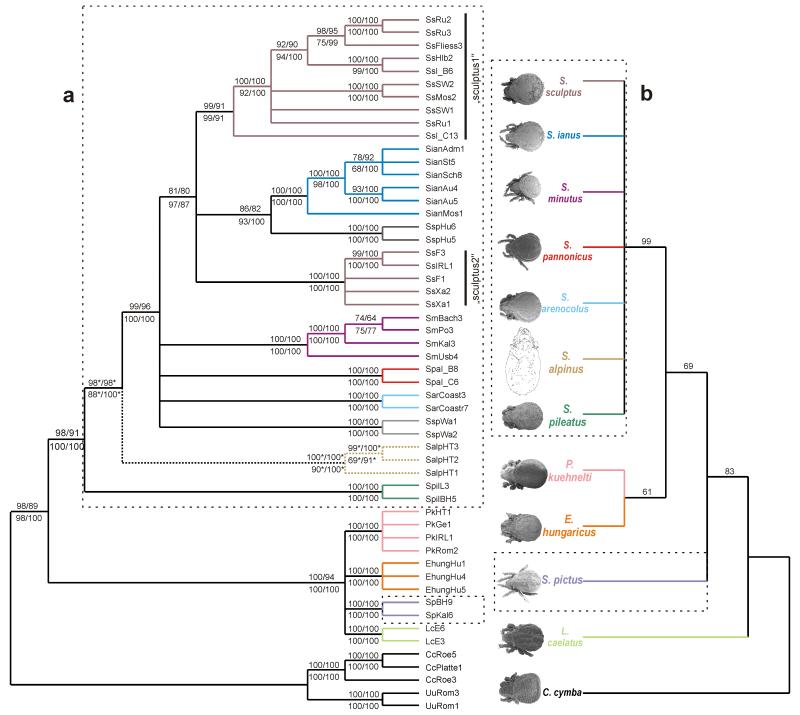
All analyses with the combined data sets revealed highly consistent topologies (Figs. 4a-d). Only slight differences were observed with respect to the tree building algorithm used. MP of the FS/RS yielded 12/2 most parsimonious trees with a length of 3,519/1,916 steps (CI excluding uninformative characters = 0.3224/0.3421; RI = 0.7372/0.7576; RC = 0.2413/0.2633). An evaluation of the phylogenetic hypotheses obtained from NJ, MP, ML and BI by means of KH and SH tests revealed no significant differences between the alternative topologies except for the MP tree in the SH test (Table 2). Compared to the other three tree topologies the MP tree showed a slightly different branching order (branching order among P. kuehnelti, L. caelatus, E. hungaricus and S. pictus; placement of S. pannonicus and S. sp. 2 (specimens from Wangen/South Tyrol) albeit with low bootstrap support. A strict consensus tree of NJ, MP, ML and Bayesian Inference is shown in Figure 5a. Scutovertex alpinus was added manually based on its position in the analyses of the RS data (data not shown). Within the family Scutoverticidae two main clusters became evident, one with species of the genus Scutovertex and one with the members of the three other genera P. kuehnelti, L. caelatus, E. hungaricus plus S. pictus, rendering the genus Scutovertex paraphyletic. This result conforms to the morphology-based phylogeny (21 constant, 16 parsimony-uninformative and 42 parsimony-informative characters; 34 most parsimonious trees; tree length = 139; CI excluding uninformative characters = 0.7080; RI = 0.7027; RC = 0.5359), in which S. pictus clusters between L. caelatus and E. hungaricus with P. kuehnelti as sister taxon (Fig. 5b). The test enforcing a monophyletic genus Scutovertex in the molecular phylogeny with S. pictus representing the most ancestral split, resulted in a significantly worse fit to the data (KH-test: tree length difference = 64 steps, s.d. = 10.77344, t = 5.9405, P = <0.0001; SH-test: Δ-lnL = 130.98973, P = 0.000). With the exception of S. sculptus, all species within the genus Scutovertex were recovered, with high statistical support, as monophyletic. The two “sculptus” clades resulted as sister taxa in all methods, except MP (Fig. 4b), which showed no resolution between S. sculptus, S. ianus and S. sp.1 specimens. Scutovertex pileatus and S. alpinus resulted as the most basal representatives of the genus Scutovertex. The phylogenetic relationship of the remaining species S. minutus, S. pannonicus, S. arenocolus and the undescribed species S. sp.2 with individuals from Wangen differs slightly depending on the method used. Unlike the molecular phylogeny, the morphological data (Fig. 5b) revealed, with good to high statistical support, the monophyly of all species but lacked resolution of the phylogenetic relationships among the Scutovertex species (with exception of S. pictus as already mentioned).
Fig. 4. Phylogeny of eleven nominal plus two undescribed European species of the family Scutoverticidae based on the concatenated data set of all available fragments of the COI, 28S rDNA and ef-1α genes.
(a) NJ tree using the GTR+I+G model; (b) strict consensus of 34 most parsimonious trees; (c) ML tree using the GTR+I+G; (d) Bayesian 50% majority rule consensus tree. Bootstrap values (for NJ, MP and ML), and posterior probabilities (for BI) are shown when >50. Colors are the same as in Fig. 1.
Table 2. Comparison of alternative phylogenetic hypotheses.
| KH test | SH test | ||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Tree | tree length diff. | s.d. (diff) | t | P | −lnL | Δ-lnL | P |
| NJ | 9 | 11.87557 | 0.7579 | 0.4486 | 16875.99005 | 19.70894 | 0.105 |
| BI | best | 16856.81429 | 0.53318 | 0.832 | |||
| MP | 20 | 13.48681 | 1.4829 | 0.1382 | 16886.10722 | 29.82611 | 0.026* |
| ML | 5 | 7.68222 | 0.6509 | 0.5152 | 16856.28111 | best | |
Kishino-Hasegawa (KH; Kishino and Hasegawa, 1989) and Shimodaira-Hasegawa tests (SH; Shimodaira and Hasegawa, 1998) were used to assess whether the topologies of NJ, MP, BI and ML differed significantly.
P<0.05.
Fig. 5. Phylogeny of eleven nominal plus two undescribed European species of the family Scutoverticidae.
(a) Strict consensus of NJ, MP, ML and BI trees of the concatenated data set of all available fragments of the COI, 28S rDNA and ef-1α genes. Bootstrap values of NJ and MP are shown above the branches, bootstrap values for ML and posterior probabilities for BI below (only values >50 are shown). (b) Strict consensus tree of 34 most parsimonious trees based on 79 external morphological characters or character states. Bootstrap values >50 are shown. Stippled lines highlight investigated members of the genus Scutovertex. * = Support values from analyses of the RS data. Colors are the same as in Fig. 1.
Discussion
Molecular genetic and morphological data revealed well resolved phylogenies, demonstrating the monophyly of all species, with the exception of Scutovertex sculptus. Despite only minor morphological differences to congeneric species, all recently described Scutovertex species included in this study appeared as genetically distinct. Thus, morphological differentiation in the studied European scutoverticid mites is accompanied by high degrees of genetic differentiation, which is not necessarily the case in other oribatid mite families (e.g., Avanzati et al., 1994).
The most important congruence between morphology and molecular genetic data concerned the phylogenetic reconstruction of the genus Scutovertex itself. Both trees revealed a paraphylum Scutovertex, supporting the morphology-based hypothesis of Sitnikova (1980) that S. pictus might not belong to the genus Scutovertex. Moreover, several morphological characteristics – e.g., the shape of the bothridium, the absence of the lenticulus and the type of respiratory organs in the legs - clearly separate S. pictus from all other members of the genus. Its definite position is still unclear because in the molecular tree it clusters, depending on the tree building algorithm used, with different members of the three other European scutoverticid genera. However, we hypothesize, that S. pictus does not belong to any other known genus of the family Scutoverticidae but rather constitutes a new genus pursuant to its distinct morphological characters mentioned above.
A further important finding concerns the phylogenetic placement of P. kuehnelti. In the molecular tree it was placed in a lineage together with E. hungaricus, L. caelatus and S. pictus, a grouping strongly supported by high bootstrap and posterior probability values. This result contradicts Woas’ statement (2002) that the genus Provertex would belong to the Cymbaeremaeidae because of sharing some morphological characters. With regard to our investigations, his argument seems not to be substantive as there is no close relationship between P. kuehnelti and Cymbaeremaeus cymba in any of the phylogenetic trees. Even in our morphology-based phylogeny P. kuehnelti does not occupy the most ancestral branch within the Scutoverticidae, further rejecting a close affinity to C. cymba.
Despite well resolved phylogenies and congruencies between both data sets, unexpected results emerged from the molecular genetic data. The most obvious one was the high genetic divergence among samples classified as S. sculptus: morphologically indistinguishable individuals were separated into two well supported clades, “sculptus1” and “sculptus2” (Figs. 5a-b). This separation became evident in both the mitochondrial COI gene and the nuclear ef-1α gene, whereas the 28S rDNA lacked resolution at this divergence level. These two clades are allopatrically distributed with “sculptus1” in Eastern and “sculptus2” in Western Europe (Fig. 1a), pointing to an ancient geographic separation of these two clades. Moreover, despite their well supported genetic distinctness, two unidentified specimens from Hungary (S. sp.1) showed close morphological resemblance to S. sculptus - e.g., cuticle and cerotegument structure of notogaster, shape of notogastral setae. Given the congruence among the different molecular markers, incomplete lineage sorting could be eliminated as possible cause for the patterns observed within S. sculptus. Instead, our findings are consistent with the possibility that S. sculptus actually represents a complex of cryptic species. Which one is representing the “real” S. sculptus can not be answered in this study since neither samples from the holo- or paratypes of this species (described by Michael, 1879) nor specimens from a location site in England (locus typicus) were available for our analyses.
There are several reasons why morphological characters might be not useful in discriminating species, but there appear to be two general and recurrent frames for cryptic species (Bickford et al., 2007): they are either differentiated by nonvisual mating signals (Byers and Struble, 1990; Henry, 1998; Feulner et al., 2006; Stuart et al., 2006) and/or appear to be under selection promoting morphological stasis (Vrijenhoek et al., 1994; Rothschild and Mancinelli, 2001; Lefébure et al., 2006; Finston et al., 2007). Regarding the first point, nonvisual mating signals could also be important in differentiating among the different S. sculptus lineages because within oribatid mites indirect sperm transfer occurs by means of spermatophores. For S. sculptus, the “completely dissociated transfer” after Proctor (1998) is applicable (Pfingstl, pers. observations), where males and females never meet, and chemical cues induce the uptake of spermatophores by the female. Moreover, since S. sculptus occurs in extreme environments such as saline soils, salt marshes and other very dry habitats, convergent evolution under harsh conditions in similar habitats likely produced similar morphologies in genetically distinct lineages (also see Vrijenhoek et al., 1994; Rothschild and Mancinelli, 2001; Lefébure et al., 2006; Finston et al., 2007). However, this raises the question ’what is really “extreme”‘? Therefore we want to conform to Rothschild and Mancinelli (2001) who stated ’all physical factors are on a continuum, and extremes in the conditions that make it difficult for organisms to function are ’extreme“ (p. 1093, lines 5-7). The main habitats of our investigated mite species are mosses and lichens on sun-exposed places. Considering that these habitats can both dry up and be flooded completely it is obvious that they are extreme for the specimens living in.
We emphasize that many European Scutovertex species are morphologically very similar and several species have been recognized only recently (Schäffer et al., 2008; Pfingstl et al., 2009; Weigmann, 2009). A good example is the new species S. ianus (Pfingstl et al., submitted) which exhibits morphological character states similar to either S. minutus or S. sculptus. Taking only a short look at S. ianus would certainly lead to wrong species identification. We note, that in the older literature species seem to have been mixed up, in particular S. minutus and S. sculptus, and clearly different morphological depictions have been referred to as one and the same species (Balogh, 1972; Giljarov and Krivolutsky, 1975; Pérez-Iñigo, 1993; Woas, 1998).
Furthermore, it should be noted that S. minutus is possibly not as common as it has been stated in literature. We received samples from many European countries but the “real” S. minutus could be identified hitherto only in Austria and in Germany (samples not included in this study). This suggests that the often-cited statement of the Palaearctic distribution of S. minutus (Subías, 2004, resp.2006; Weigmann, 2006) is not true. Scutovertex sculptus (or members of this cryptic species complex), on the other hand, seem to be very abundant throughout its Palaearctic distribution.
Conclusions
Molecular genetic and morphological data revealed a paraphyletic genus Scutovertex, with S. pictus likely representing a distinct genus, and confirmed the taxonomic placement of Provertex kuehnelti within the family Scutoverticidae. Furthermore, molecular genetic data confirmed several recently described Scutovertex species and thus the high species diversity within this genus in Europe and suggest that S. sculptus is a complex of several cryptic species showing marked genetic, but little (if any) morphological divergence.
Acknowledgements
Financial support was provided by the Austrian Science Fund (FWF, project number P19544-B16). We are grateful to K. Brandl, E. Ebermann, S. Ermilov, C. Hellig, P. Horak, J. Jagersbacher-Baumann, J. Knapp, I. Kulterer, E. McCullough and H. Schatz for providing moss samples for our study and we are indebted to the administration of the National Park “Neusiedler See-Seewinkel” for the permission to collect moss and soil samples. Furthermore, the authors thank Prof. Dr. F. Hofer and his team at the Research Institute for electron Microscopy (FELMI) for the cooperation in making SEM-micrographs.
Appendix A. Morphological characters and character states
1. Prodorsal surface (P): smooth, no foveae (0); smooth, foveae (1); granular, no foveae, no wrinkles (2); granular, no foveae, wrinkles (3); granular, foveae, no wrinkles (4); granular, foveae, wrinkles (5).
2. Bothridium (bo): closed border, roundly shaped (0); closed border, longish shaped (1); open border, roundly shaped (2); open border, longish shaped (3).
3. Sensillus (bS) dimension: short, slim (0); short, thick (1); long, slim (2); long, thick (3).
4. Sensillus shape: spinose, broad, clavate and flattened (0); spinose, broad, clavate and spherical (1); spinose, slender, clavate and flattened (2); spinose, slender, clavate and spherical (3).
5. Lamella (cl): absent (0); short, collateral (1); short, convergent (2); long, collateral (3), long, convergent (4); broad, laterally overhanging (5).
6. Lamellar seta (le): short, slim, smooth (0); short, slim, spinose (1); short, thick, smooth (2); short, thick, spinose (3); long, slim, smooth (4); long, slim, spinose (5); long, thick, smooth (6); long, thick, spinose (7).
7. Interlamellar seta (in): absent (0); short, slim (1); short, thick (2); long, slim (3); long, thick (4).
8. Rostral seta (ro) dimension: short, thick (0); short, slim (1); long, thick (2); long, slim (3).
9. Rostral seta shape: smooth, spiniform (0); smooth, lanceolate (1); spinose, spiniform (2); spinose, lanceolate (3).
10. Exobothridial seta (ex): absent (0); short (1); long (2).
11. Rostrum (R): with one ridge (0); with two ridges (1); with two clear projections (2).
12. Lenticulus (Le): absent (0); lateral borders bend inward (1); oval (2); rectangular (3).
13. Translamella (TL): absent (0); narrow, straight (1); narrow, bent (2); broad, straight (3); broad, bent (4).
14. Cusps (cu): absent (0); small (1); large (2); broad, overhanging (3).
15. Prodorsal ridges (PR): absent (0); collateral, reaching TL (1); collateral, not reaching TL (2); converging, not fused, reaching TL (3); converging, not fused, not reaching TL (4); converging, fused, reaching TL (5); converging, fused, not reaching TL (6).
16. Notogastral surface (N): foveae, no blocs, no granules, no bars, not netlike (0); foveae, blocs, no granules, no bars, not netlike (1); foveae, blocs, granules, no bars, not netlike (2); foveae, no blocs, granules, no bars, not netlike (3); no foveae, no blocs, granules, no bars, not netlike (4); no foveae, no blocs, no granules, bars, not netlike (5); no foveae, no blocs, no granules, bars, netlike (6); no foveae, no blocs, granules, bars, not netlike (7); almost smooth (8).
17. Foveae on notogaster: absent (0); indistinct borders (1); distinct borders (2).
18. Lyrifissure ia: inconspicuous, not on a nodule (0); on a nodule (1).
19. Lyrifissure im: inconspicuous (0); very long (1).
20. Lyrifissure ip: inconspicuous (0); on a protuberance (1).
21. Pairs of notogastral setae: 10 (0); 12 (1); 13 (2); 14 (3); 15 (4).
22. Saccules (S) of the octotaxic system: absent (0); 1 pair (1); 2 pairs (2); 3 pairs (3); 4 pairs (4).
23.-37. Notogastral setae c1-3, da, dm, dp, la, lm, lp, h1-3, ps1-3: absent (0); slim, not spinose (1); slim, spinose (2); broadened, not spinose (3); broadened, spinose (4); thick, not spinose (5); thick, spinose (6).
38. Lateral prodorsum (PL) surface: smooth (0); granular (1).
39. Tutorium (TU): absent (0); not V-shaped (1); V-shaped (2).
40. Pedotectum I (PtI): small, not triangular (0); small, triangular (1); large, not triangular (2); large, triangular (3).
41. Pedotectum II (PtII): small, Y-shaped (0); small, triangular (1); large, Y-shaped (2); large, triangular (3).
42. Dens tutorius: absent (0); small (1); large (2).
43. Subcapitulum (S) surface: smooth (0); granular (1).
44.-46. Subcapitular setae (m, a, h): smooth (0); spinose (1).
47. Rutellar teeth (RU): 2 (0); 3 (1); 4 (2).
48. Rib on mentum (MR): absent (0); slender, straight (1); slender, V-shaped (2); broad, straight (3); broad, V-shaped (4); massive, reaching anterior border (5).
49. Pedipalp (Pp) “corne double”: absent (0); incomplete (1); complete (2).
50. Chaetome pedipalp (solenidion excluded): 0-2-1-3-9 (0); alternative (1).
51. Apophysis on palptarsus: absent (0); present (1).
52. Epimeral region (E) surface: smooth (0); granular (1).
53. Apodemata (Ap) III + IV: both absent (0); III present, IV absent (1); both present (2).
54. Epimeral setal formula: 3-1-2-2 (0); 3-1-3-2 (1); 3-1-3-3 (2); 3-1-3-4 (3); 3-1-3-1 (4).
55. Anogenital region (AG) surface: smooth (0); granular (1).
56. Genital valves (GV) shape: rounded, anteriorly broadened (0); rounded, posteriorly broadened (1); rectangular, anteriorly broadened (2); rectangular, posteriorly broadened (3).
57. Genital setal (g) formula: 5+5 (0); 6+6 (1); 9+9 (2); >9<12 (3); >12 (4).
58. Aggenital setal (ag) formula: setae absent (0); 1+1 (1); 2+2 (2).
59. Anal setal (an) formula: 2+2 (0); 3+3 (1).
60. Placement from g1 to g2: in a row (0); side by side (1).
61. Placement from g3 to g6: in a row (0); displaced laterally (1).
62. Placement of anal setae: medially on anal valve (0); next to inner border of anal valve (1).
63. Leg surface (LS): smooth (0); granular, no ridges (1); granular, ridges (2).
64. Respiratory organs (RO) in legs: absent (0); planar areae porosae (1); saccules (2); platytracheae (3), brachytracheae (4); tracheae (5).
65. Claw (C) number: monodactylous (0); bidactylous (2); tridactylous (3)
66. Claw shape: homodactylous (0); heterodactylous (1).
67. Dorsal setae on legs coupled with solenidia: lost in adult stage (1); present in all stages (2); lost in all stages (3).
68. Apophysis on tibia I: absent (0); small (1); large (2).
69. Lateral setae on legs: slender, smooth (0); slender, dentate (1); broadened, smooth (2); broadened, dentate (3); extremely broadened (4).
70. Position of the respiratory organs in leg I and II: femur (0); femur, tibia (1); femur, tibia, tarsus (2); femur, tarsus (3).
71. Position of the respiratory organs in leg III and IV: trochanter, femur (0); trochanter, femur, tibia (1), trochanter, femur, tibia, tarsus (2); trochanter, femur, tarsus (3); femur, tarsus (4)
72. Chaetome leg I: 1-3-3-4-16 (0); 1-4-2-4-18 (1); 1-4-3-4-18 (2); 1-4-3-4-19 (3); 1-5-3-4-18 (4).
73. Chaetome leg II: 1-4-2-4-15 (0); 1-4-3-4-15 (1); 1-3-3-5-14 (2); 1-5-3-4-15 (3).
74. Chaetome leg III: 2-2-1-3-15 (0); 2-3-1-3-15 (1); 2-2-2-3-14 (2); 2-4-1-3-15 (3).
75. Chaetome leg IV: 1-2-2-3-12 (0); alternative (1).
76. Solenidia leg I: 1-2-2 (0); 1-2-3 (1).
77. Solenidia leg II: 1-1-2 (0); 1-1-1 (1).
78. Solenidia leg III: 0-1-0 (0); 1-1-0 (1).
79. Solenidia leg IV: 0-1-0 (0); alternative (1).
Appendix B. Matrix for morphological characters and character states
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||||||||
| S. alpinus | 22324 | 40200 | 11126 | 02101 | 04010 | 11010 | 11111 | 11123 | 20100 | 01310 | 01201 | 01101 | 00253 | 11230 | 02100 | 0010 |
| S. arenocolus | 22324 | 70200 | 11415 | 31100 | 03010 | 03010 | 33344 | 11123 | 20111 | 01410 | 01201 | 01101 | 10253 | 11230 | 02100 | 0010 |
| S. ianus | 22324 | 70200 | 11425 | 21100 | 03010 | 01010 | 11144 | 11123 | 20111 | 01310 | 01201 | 01101 | 10253 | 11230 | 02100 | 0010 |
| S. minutus | 22324 | 70200 | 11426 | 40100 | 13010 | 13310 | 33344 | 11123 | 20111 | 01410 | 01201 | 01101 | 10253 | 11230 | 02100 | 0010 |
| S. pannonicus | 22324 | 70200 | 11425 | 02100 | 03010 | 03010 | 33324 | 11123 | 20111 | 01010 | 01201 | 01101 | 10253 | 11230 | 02100 | 0010 |
| S. pictus | 32104 | 60200 | 00320 | 50000 | 00010 | 01010 | 11111 | 11103 | 20111 | 01400 | 11201 | 01100 | 01243 | 11231 | 13010 | 0010 |
| S. pileatus | 22323 | 00200 | 11324 | 70100 | 03010 | 33010 | 33331 | 11123 | 20111 | 01310 | 01201 | 01100 | 00253 | 11230 | 01000 | 0010 |
| S. sculptus | 22324 | 70200 | 11425 | 11100 | 03010 | 04010 | 66666 | 11123 | 20100 | 01410 | 01201 | 01101 | 10253 | 11230 | 02100 | 0010 |
| L. caelatus | 33304 | 70200 | 20120 | 60000 | 00101 | 11111 | 11111 | 10113 | 20100 | 01100 | 01201 | 01100 | 01240 | 01221 | 12110 | 0010 |
| P. kuehnelti | 22112 | 00220 | 00104 | 40000 | 30110 | 11111 | 11111 | 11101 | 20111 | 00010 | 01201 | 01100 | 01233 | 11131 | 12100 | 0010 |
| E. hungaricus | 22325 | 70220 | 00034 | 60001 | 01010 | 01010 | 11111 | 11113 | 20111 | 00010 | 01241 | 01100 | 01233 | 11241 | 12000 | 0010 |
| C. cymba | 30110 | 00101 | 00000 | 60000 | 20110 | 11111 | 11111 | 10103 | 30100 | 2300 | 11001 | 01100 | 01243 | 01202 | 20220 | 0110 |
References
- Avanzati AM, Baratti M, Bernini F. Molecular and morphological differentiation between steganacarid mites (Acari: Oribatida) from the Canary Islands. Bio. J. Linn. Soc. 1994;52:325–340. [Google Scholar]
- Balogh J. The oribatid genera of the world. Akdémiai Kiadó; Budapest: 1972. p. 188. 71 plates. [Google Scholar]
- Baric S, Sturmbauer C. Ecological parallelism and cryptic species in the genus Ophiothrix derived from mitochondrial DNA sequences. Mol. Phylogenet. Evol. 1999;11:157–162. doi: 10.1006/mpev.1998.0551. [DOI] [PubMed] [Google Scholar]
- Bernini F. Notulae oribatologicae XV. Lamellovertex, un nuovo genere per Scutovertex caelatus Berlese, 1895 (Acarida, Oribatei) Redia. 1976;59:311–321. [Google Scholar]
- Bickford D, Lohman DJ, Sodhi NS, Ng PKL, Meier R, Winker K, Ingram KK, Das I. Cryptic species as a window on diversity and conservation. Trends Ecol. Evol. 2007;22:148–155. doi: 10.1016/j.tree.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Bond JE, Sierwald P. Cryptic speciation in the Anadenobolus excisus millipede species complex on the Island of Jamaica. Evolution. 2002;56:1123–1135. doi: 10.1111/j.0014-3820.2002.tb01426.x. [DOI] [PubMed] [Google Scholar]
- Boyce TM, Zwick ME, Aquadro CF. Mitochondrial DNA in the bark weevils: size, structure and heteroplasmy. Genetics. 1989;123:825–836. doi: 10.1093/genetics/123.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brower AVZ. Rapid morphological radiation and convergence among races of the butterfly Heliconius erato inferred from patterns of mitochondrial DNA evolution. Proc. Natl. Acad. Sci. USA. 1994;91:6491–6495. doi: 10.1073/pnas.91.14.6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers JR, Struble DL. Identification of sex pheromones of two sibling species in dingy cutworm complex Feltia jaculifera (Lepidoptera: Noctuidae) J. Chem. Ecol. 1990;16:2981–2992. doi: 10.1007/BF00979489. [DOI] [PubMed] [Google Scholar]
- Chan BKK, Tsang LM, Chu KH. Cryptic diversity of the Tetraclita squamosa Complex (Crustacea: Cirripedia) in Asia: description of a new species from Singapore. Zool. Stud. 2007;46:46–56. [Google Scholar]
- Edwards DD, Dimock RV., Jr. Genetic differentiation between Unionicola formosa and U. foili (Acari: Unionicolidae): cryptic species of molluscan symbionts. Invertebr. Biol. 1997;116:124–133. [Google Scholar]
- Ermilov S, Łochyńska M, Olszanowski Z. The cultivation and morphology of juvenile stages of two species from genus Scutovertex (Acari: Oribatida: Scutoverticidae) Ann. Zool. 2008;58:433–443. [Google Scholar]
- Feulner PGD, Kirschbaum F, Schugardt C, Ketmaier V, Tiedemann R. Electrophysiological and molecular genetic evidence for sympatrically occuring cryptic species in African weakly electric fishes (Teleostei: Mormyridae: Campylomormyrus) Mol. Phylogenet. Evol. 2006;39:198–208. doi: 10.1016/j.ympev.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Finston TL, Johnson MS, Humphreys WF, Eberhard SM, Halse SA. Cryptic speciation in two widespread subterranean amphipod genera reflects historical drainage patterns. Mol. Ecol. 2007;16:355–365. doi: 10.1111/j.1365-294X.2006.03123.x. [DOI] [PubMed] [Google Scholar]
- Giljarov MS, Krivolutsky DA. Key to the soil-inhabiting mites of the Sarcoptiformes. Nauka, Moscow: 1975. p. 491. (in Russian) [Google Scholar]
- Grandjean F. Essai de classification des Oribates (Acariens) B. Soc. Zool. Fr. 1953;78:421–446. [Google Scholar]
- Grandjean F. Considérations sur le classement des Oribates. Leur division en 6 groupes majeurs. Acarologia. 1969;11:127–153. [Google Scholar]
- Hebert PDN, Penton EH, Burns JM, Janzen DH, Hallwachs W. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proc. Natl. Acad. Sci. USA. 2004;101:14812–14817. doi: 10.1073/pnas.0406166101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heethoff M, Domes K, Laumann M, Maraun M, Norton RA, Scheu S. High genetic divergences indicate ancient separation of parthenogenetic lineages of the oribatid mite Platynothrus peltifer (Acari, Oribatida) J. Evol. Biol. 2007;20:392–402. doi: 10.1111/j.1420-9101.2006.01183.x. [DOI] [PubMed] [Google Scholar]
- Henry CS. Singing and cryptic speciation in insects. Trends Ecol. Evol. 1994;9:388–392. doi: 10.1016/0169-5347(94)90061-2. [DOI] [PubMed] [Google Scholar]
- Juan C, Oromi P, Hewitt GM. Phylogeny of the genus Hegeter (Tenebrionidae, Coleoptera) and its colonisation of the Canary Islands deduced from cytochrome oxidase I mitochondrial DNA sequences. Heredity. 1996;76:392–403. doi: 10.1038/hdy.1996.57. [DOI] [PubMed] [Google Scholar]
- Katongo C, Koblmüller S, Duftner N, Makasa L, Sturmbauer C. Phylogeography and speciation in the Pseudocrenilabrus philander species complex in Zambian rivers. Hydrobiologia. 2005;542:221–233. [Google Scholar]
- King RA, Tibble AL, Symondson WOC. Opening a can of worms: unprecedented sympatric cryptic diversity within British lumbricid earthworms. Mol. Ecol. 2008;17:4684–4698. doi: 10.1111/j.1365-294X.2008.03931.x. [DOI] [PubMed] [Google Scholar]
- Kishino H, Hasegawa M. Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in hominoidea. J. Mol. Evol. 1989;29:170–179. doi: 10.1007/BF02100115. [DOI] [PubMed] [Google Scholar]
- Kon T, Yoshino T, Mukai T, Nishida M. DNA sequences identify numerous cryptic species of the vertebrate: A lesson from the gobioid fish Schindleria. Mol. Phylogenet. Evol. 2007;44:53–62. doi: 10.1016/j.ympev.2006.12.007. [DOI] [PubMed] [Google Scholar]
- Krisper G, Schmikl M, Ebermann E. Erstnachweis der felsbodenbewohnenden Hornmilben Scutovertex pictus Kunst, 1959 und Lamellovertex caelatus (Berlese, 1895) (Acari, Oribatida) für Österreich. Mitt. naturwiss. Ver. Steiermark. 2002;132:193–196. [Google Scholar]
- Krisper G, Schuster R. Morphological analysis of Provertex kuehnelti Mihelcic, 1959 – an oribatid mite of rocky habitats (Acari: Oribatida: Scutoverticidae) Contrib. Nat. Hist. 2009;81 (in press) [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Lefébure T, Douady CJ, Gouy M, Trontelj P, Briolay J, Gibert J. Phylogeography of a subterranean amphipod reveals cryptic diversity and dynamic evolution in extreme environments. Mol. Ecol. 2006;15:1797–1806. doi: 10.1111/j.1365-294X.2006.02888.x. [DOI] [PubMed] [Google Scholar]
- Litvaitis MK, Nunn G, Thomas WK, Kocher TD. A molecular approach for the identification of meiofaunal turbellarians (Platyhelminthes, Turbellaria) Mar. Biol. 1994;120:437–442. [Google Scholar]
- Marzluff JM, Dial KP. Life-history correlates of taxonomic diversity. Ecology. 1991;72:428–439. [Google Scholar]
- Mathews LM, Adams L, Anderson E, Basile M, Gottardi E, Buckholt MA. Genetic and morphological evidence for substantial hidden biodiversity in a freshwater crayfish species complex. Mol. Phylogenet. Evol. 2008;48:126–135. doi: 10.1016/j.ympev.2008.02.006. [DOI] [PubMed] [Google Scholar]
- Mayer F, Dietz C, Kiefer A. Molecular species identification boosts bat diversity. Front. Zool. 2007;4:4. doi: 10.1186/1742-9994-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger GA, Kraus F, Allison A, Parkison CL. Uncovering cryptic diversity in Aspidomorphus (Serpentes: Elapidae): Evidence from mitochondrial and nuclear markers. Mol. Phylogenet. Evol. 2009 doi: 10.1016/j.ympev.2009.07.027. (in press) doi:10.1016/j.ympev.2009.07.027. [DOI] [PubMed] [Google Scholar]
- Michael AD. A contribution to the knowledge of The British Oribatidae. J. Mic. Soc. London. 1879;2:225–251. [Google Scholar]
- Murray TE, Fitzpatrick Ú, Brown MJF, Paxton RJ. Cryptic species diversity in a widespread bumble bee complex revealed using mitochondrial DNA RFLPs. Conserv. Genet. 2008;9:653–666. [Google Scholar]
- Otto JC, Wilson KJ. Assessment of the usefulness of ribosomal 18S and mitochondrial COI sequences in Prostigmata phylogeny. In: Halliday RB, Walter DE, Proctor HC, Norton RA, Colloff J, editors. Acarology; Proceedings of the 10th International Congress; Melbourne: CSIRO Publishing; 2001. pp. 100–109. [Google Scholar]
- Padial JM, de la Riva I. Integrative taxonomy reveals cryptic Amazonian species of Pristimantis (Anura: Strabomantidae) Zool. J. Linn. Soc. 2009;155:97–122. [Google Scholar]
- Pérez-Iñigo C. Acari, Oribatei, Poronota. In: Ramos MA, et al., editors. Fauna Iberica. Vol. 3. Museo Nacional de Ciencias Naturales, CSIC; Madrid: 1993. p. 320. [Google Scholar]
- Pfingstl T, Schäffer S, Ebermann E, Krisper G. Intraspecific morphological variation of Scutovertex sculptus Michael (Acari: Oribatida: Scutoverticidae) and description of its juvenile stages. Zootaxa. 2008;1829:31–51. [Google Scholar]
- Pfingstl T, Schäffer S, Ebermann E, Krisper G. Differentiation between two epilittoral species, Scutovertex arenocolus spec. nov. and Scutovertex pilosetosus Polderman (Acari: Oribatida) from different European coasts. Zootaxa. 2009;2153:35–54. [Google Scholar]
- Pfingstl T, Schäffer S, Ebermann E, Krisper G. Scutovertex alpinus Willmann, 1953 – redescription and geographic distribution (Acari, Oribatida, Scutoverticidae) Journal of Natural History. (in press) [Google Scholar]
- Posada D, Crandall K. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Proctor HC. Indirect sperm transfer in arthropods: behavioral and evolutionary trends. Annu. Rev. Entomol. 1998;43:153–174. doi: 10.1146/annurev.ento.43.1.153. [DOI] [PubMed] [Google Scholar]
- Quek SP, Davies SJ, Itino T, Pierce N. Codiversification in an ant–plant mutualism: the phylogeny of host use in Crematogaster (Formicidae) associates of Macaranga (Euphorbiaceae) Evolution. 2004;58:554–570. [PubMed] [Google Scholar]
- Regier JC, Shultz JW. Molecular phylogeny of the major arthropod groups indicates polyphyly of crustaceans and a new hypothesis for the origin of hexapods. Mol. Biol. Evol. 1997;14:902–913. doi: 10.1093/oxfordjournals.molbev.a025833. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Rothschild LJ, Mancinelli RL. Life in extreme environments. Nature. 2001;409:1092–1101. doi: 10.1038/35059215. [DOI] [PubMed] [Google Scholar]
- Sáez AG, Lozano E. Body doubles. Cryptic species: as we discover more examples of species that are morphologically indistinguishable, we need to ask why and how they exist. Nature. 2005;433:111. doi: 10.1038/433111a. [DOI] [PubMed] [Google Scholar]
- Salomone N, Emerson BC, Hewitt GM, Bernini F. Phylogenetic relationships among the Canary Island Steganacaridae (Acari, Oribatida) inferred from mitochondrial DNA sequence data. Mol. Ecol. 2002:79–89. doi: 10.1046/j.0962-1083.2001.01421.x. [DOI] [PubMed] [Google Scholar]
- Schäffer S, Krisper G. Morphological analysis of the adult and juvenile instars of Scutovertex minutus (Acari, Oribatida, Scutoverticidae) Rev. Suisse Zool. 2007;114:663–683. [Google Scholar]
- Schäffer S, Krisper G, Pfingstl T, Sturmbauer C. Description of Scutovertex pileatus sp. nov. (Acari, Oribatida, Scutoverticidae) and molecular phylogenetic investigation of congeneric species in Austria. Zool. Anz. 2008;247:249–258. [Google Scholar]
- Schäffer S, Koblmüller S, Pfingstl T, Sturmbauer C, Krisper G. Contrasting mitochondrial DNA diversity estimates in Austrian Scutovertex minutus and S. sculptus (Acari, Oribatida, Brachypylina, Scutoverticidae) Pedobiologia. (in press) doi: 10.1016/j.pedobi.2009.09.004. [Google Scholar]
- Schlick-Steiner BC, Steiner FM, Moder K, Seifert B, Sanetra M, Dyreson E, Stauffer C, Christian E. A multidisciplinary approach reveals cryptic diversity in Western Palearctic Tetramorium ants (Hymenoptera: Formicidae) Mol. Phylogenet. Evol. 2006;40:259–273. doi: 10.1016/j.ympev.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Schuster R. Beitrag zur Kenntnis der Milbenfauna (Oribatei) in pannonischen Trockenböden. Sitzber. Österr. Akad. Wiss., Mathem.-naturw. Kl., Abt I. 1958;167:221–235. [Google Scholar]
- Shimodaira H, Hasegawa M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol. Biol. Evol. 1999;16:1114–1116. [Google Scholar]
- Shtanchaeva UY, Netuzhilin IA. A review of the world fauna of Scutoverticidae Oribatid mites (Acari, Oribatida) with description of new species. Zool. Zhurnal. 2003;82:781–803. [Google Scholar]
- Sitnikova LG. New species of mites, fam. Scutoverticidae (Acariformes, Oribatei) Parazitol. Sbornik. 1980;29:180–195. [Google Scholar]
- Skubala P. Moss mites (Acarina: Oribatida) on industrial dumps of different age. Pedobiologia. 1995;39:170–184. [Google Scholar]
- Smrž J. Microhabitat selection in the simple oribatid community dwelling in epilithic moss cover (Acari: Oribatida) Naturwissenschaften. 2006;93:570–576. doi: 10.1007/s00114-006-0141-y. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Stuart BL, Inger RF, Voris HK. High level of cryptic species diversity revealed by sympatric lineages of Southeast Asian forest frogs. Biol. Lett. 2006;2:470–474. doi: 10.1098/rsbl.2006.0505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subías LS. Listado sistemático, sinonímico y biogeográfico de los acaros oribátidos (Acariformes: Oribatida) del mundo. Graellsia. 2004;60:3–305. [Google Scholar]
- Swofford DL. PAUP*. Phylogenetic analysis using parsimony (* and other methods) Ver. 4 Sinauer Associates; Sunderland, MA: 2002. [Google Scholar]
- Vrijenhoek RC, Schutz SJ, Gustafson RG, Lutz RA. Cryptic species of deep sea clams (Mollusca: Bivalvia: Vesicomyidae) from hydrothermal vent and cold water seep environments. Deep-Sea Res. Part I. 1994;41:1171–1189. [Google Scholar]
- Weigmann G. Recovery of the oribatid mite community in a floodplain after decline due to long-term flooding. In: Weigmann G, Alberti G, Wohltmann A, Ragusa S, editors. Acarine Biodiversity in the Natural and Human Sphere; Phytophaga; Proc. Vth Symposium of EURAAC; Berlin. 2004.2004. pp. 201–207. [Google Scholar]
- Weigmann G. Hornmilben (Oribatida). Die Tierwelt Deutschlands, begründet 1925 von Friedrich Dahl. 76. Teil. Goecke & Evers, Keltern. 2006. p. 520.
- Weigmann G. Oribatid mites (Acari: Oribatida) from the coastal region of Portugal. III. New species of Scutoverticidae and Scheloribatidae. Soil Organisms. 2009;81 (in press) [Google Scholar]
- Wilcox TP, Hugg L, Zeh JA, Zeh DW. Mitochondrial DNA sequencing reveals extreme genetic differentiation in a cryptic species complex of neotropical pseudoscorpions. Mol. Phylogenet. Evol. 1997;7:208–216. doi: 10.1006/mpev.1996.0388. [DOI] [PubMed] [Google Scholar]
- Wilcox TP, García de León FJ, Hendrickson DA, Hillis DM. Convergence among cave catfishes: long-branch attraction and a Bayesian relative rate test. Mol. Phylogenet. Evol. 2004;31:1101–1113. doi: 10.1016/j.ympev.2003.11.006. [DOI] [PubMed] [Google Scholar]
- Woas . Mosaikverteilung der Merkmale basaler Höherer Oribatiden – Die Gattungen Passalozetes und Scutovertex (Acari, Oribatei) In: Ebermann E, editor. Arthropod Biology: Contributions to Morphology, Ecology and Systematics. Vol. 14. 1998. pp. 291–313. (Biosystematics and Ecology Series). [Google Scholar]
- Woas S. Acari: Oribatida. In: Adis J, editor. Amazonian Arachnida and Myriapoda. Pensoft Publishers; Sofia, Moscow: 2002. pp. 21–291. [Google Scholar]