Abstract
Self-incompatibility (SI) of the Brassicaceae family can be overcome by CO2 gas treatment. This method has been used for decades as an effective means to obtain a large amount of inbred seeds which can then be used for F1 hybrid seed production; however, the molecular mechanism by which CO2 alters the SI pathway has not been elucidated. In this study, to obtain new insights into the mechanism of CO2-induced SI breakdown, the focus was on two inbred lines of Brassica rapa (syn. campestris) with different CO2 sensitivity. Physiological examination using X-ray microanalysis suggested that SI breakdown in the CO2-sensitive line was accompanied by a significant accumulation of calcium at the pollen–stigma interface. Pre-treatment of pollen or pistil with CO2 gas before pollination showed no effect on the SI reaction, suggesting that some physiological process after pollination is necessary for SI to be overcome. Genetic analyses using F1 progeny of a CO2-sensitive×CO2-insensitive cross suggested that CO2 sensitivity is a semi-dominant trait in these lines. Analysis of F2 progeny suggested that CO2 sensitivity could be a quantitative trait, which is controlled by more than one gene. Quantitative trait locus (QTL) analyses identified two major loci, BrSIO1 and BrSIO2, which work additively in overcoming SI during CO2 treatment. No QTL was detected at the loci previously shown to affect SI stability, suggesting that CO2 sensitivity is determined by novel genes. The QTL data presented here should be useful for determining the responsible genes, and for the marker-assisted selection of desirable parental lines with stable but CO2-sensitive SI in F1 hybrid breeding.
Key words: Brassica rapa, calcium, CO2, F1 hybrid, QTL, self-incompatibility.
Introduction
Self-incompatibility (SI) is a widespread genetic system in many flowering plants which serves to prevent self-fertilization and maintain genetic diversity. It is based on self/non-self pollen–pistil recognition interactions followed by inhibition of self-pollen hydration, germination, or pollen tube growth.
In the Brassicaceae, SI is sporophytically controlled by a multiallelic locus termed the S locus (Bateman, 1955). Male and female determinants have been identified as SP11/SCR (S-locus protein 11/S-locus cysteine-rich) (Schopfer et al., 1999; Takayama et al., 2000) and SRK (S receptor kinase) (Takasaki et al., 2000), respectively. When a compatible pollen grain lands on the stigma, it swells and a pollen tube is allowed to grow, whereas when self-pollen attaches to the stigma, SP11/SCR binds specifically to the extracellular domain of SRK of the same S-haplotype (Takayama et al., 2001), which triggers an SI signalling pathway to reject self-pollen. Another stigmatically expressed gene located at the S locus, S locus glycoprotein (SLG) (Nasrallah et al., 1987; Takayama et al., 1987), has been shown to enhance the recognition process between self-pollen and stigma (Takasaki et al., 2000); however, this function of SLG remains controversial (Silva et al., 2001). A recent study suggested that the plants in the Brassicaceae genus Levenworthia use paralogous SRK and SP11/SCR genes, Lal2 (Leavenworthia alabamica SRK-related 2) and SCRl (SCR-like), for self/non-self recognition in SI, but the function of their orthologues in other Brassicaceae genera also remains unknown (Chantha et al., 2013).
Many studies focusing on the downstream components involved in this type of SI signalling pathway have been performed and, thus far, two components have been identified as positive effectors. ARC1 (arm repeat containing 1) was identified by a yeast two-hybrid screen using the kinase domain of SRK as the bait (Gu et al., 1998; Stone et al., 1999). ARC1 is a U-box protein with E3 ubiquitin ligase activity (Stone et al., 2003), and has been shown to interact with Exo70A1, a putative component of the exocyst complex required for compatible pollination (Samuel et al., 2009). MLPK (M-locus protein kinase) was identified by positional cloning as the gene responsible for the self-compatible mutation of Brassica rapa var. Yellow sarson (Murase et al., 2004). MLPK is a membrane-anchored cytoplasmic protein kinase and interacts directly with SRK to transduce SI signalling (Kakita et al., 2007). However, the importance of these two components in the Brassicaceae SI mechanism remains controversial (Kitashiba et al., 2011; Indriolo et al., 2012).
In the Brassicaceae, it has been known that SI can be overcome under some physiological and environmental conditions such as plant age (Ockendon, 1978; Horisaki and Niikura, 2008), stigmatic chemical treatments (e.g. ether, KOH, and NaCl) (Tatebe, 1968; Tao and Yong, 1986; Monterio and Gabelman, 1988), and high temperature (Matsubara, 1980; Okazaki and Hinata, 1987). CO2 gas (3–5%) treatment (Nakanishi et al., 1969) is the most effective way to overcome SI. Today, most cultivated lines of crucifer vegetables, such as cabbage, broccoli, Chinese cabbage, and radish are F1 hybrids whose seeds are produced by mix-planting two self-incompatible inbred parental lines. In this economical F1 hybrid breeding system, CO2 gas treatment has been used to suppress SI and allow self-fertilization, thereby providing large-scale seed propagation of parental lines. This method has been used all over the world for many years, but the molecular mechanism leading to SI breakdown by CO2 gas treatment is entirely unknown.
Previous studies suggested that not all lines respond equally to CO2, and there are variations in SI response to CO2 (CO2 sensitivity) in the Brassicaceae (Nakanishi and Hinata, 1973; Niikura and Matsuura, 2000). Preliminary genetic analysis using lines with different CO2 sensitivity in radish (Raphanus sativus) suggested that high CO2 sensitivity was controlled by a recessive gene independent of the S-locus (Niikura and Matsuura, 2000). In another genetic analysis using CO2-sensitive and CO2-insensitive lines of Chinese cabbage (B. rapa), high CO2 sensitivity was suggested to be controlled by a dominant gene (Hyun et al., 2007); however, no responsible genes have been identified from these studies so far.
In this study, new inbred lines of B. rapa with different CO2 sensitivity, a CO2-sensitive line (HA-11621) and a CO2-insensitive line (HA-11623), were selected and analysed. X-ray microanalysis suggested that SI breakdown in the CO2-sensitive line was accompanied by significant calcium accumulation at the pollen–stigma interface. Independent pre-treatment of pollen or pistil with CO2 gas before pollination showed no effect on the SI reaction, suggesting that some physiological process that occurs after pollination is necessary for SI to be overcome. Genetic analyses using F1 and F2 progeny of a CO2-sensitive×CO2-insensitive cross suggested that CO2 sensitivity is a semi-dominant and quantitative trait. Furthermore, quantitative trait locus (QTL) analyses identified two major responsible loci, BrSIO1 and BrSIO2, which function additively in overcoming SI during CO2 treatment.
Materials and methods
Plant materials
Two inbred lines of B. rapa (2n=20), a CO2-sensitive line (HA-11621) and a CO2-insensitive line (HA-11623), were established at Tohoku Seed Co., Ltd, and grown in the greenhouse with 16h light and 8h dark conditions at 20 °C. Both lines show stable SI under normal (open-air) condition but have different CO2 sensitivity; SI in HA-11621 breaks down following treatment with 4.5% CO2 whereas SI in HA-11623 is unaffected. HA-11621 and HA-11623 are reciprocally compatible, and their F1 progeny were obtained under normal conditions. Buds (1–2 d before flowering) from a randomly chosen F1 were used for F2 production. Young petals and stamens were removed from the bud, and the immature pistil was pollinated with pollen grains from mature flowers of the same plant (bud pollination). Pollinated pistil was then covered with a paper bag for 3 d and seeds from the pistil were harvested. More than 20 pistils were pollinated, and harvested seeds were used as the F2 population. A total of 110 F2 plants were used for phenotypic and genetic analysis.
Cryo-scanning electron microscopy and energy-dispersive X-ray analysis
Flowers were self- or cross-pollinated and incubated for 1.5h with or without 4.5% CO2 gas. These pollinated and non-pollinated pistils were submersed in liquid nitrogen slush and frozen under vacuum. While under vacuum, the sample was transferred to the microscope cryo stage (ALTO 1000, Gatan), and then the stage temperature was increased to –95 °C to remove frost that had settled on top of the specimen as a result of condensation. When all surface frost had been removed by sublimation, as verified by electron microscopy, the temperature was reduced to –140 °C. Imaging was performed using an ETD (Everhart–Thornley detector) by Quant 250 scanning electron microscopy (FEI). The chamber pressure was 30 Pa and the accelerating voltage was 15kV. EDX (energy-dispersive X-ray spectroscopy) analysis of the element assay was performed on selected papilla cells using INCA X-ray analysis software (Oxford Instruments, http://www.oxinst.com/Pages/home.aspx, last accessed 14 December 2013), with the detector’s processing time set at 2. X-ray data were collected with 4.5 nA probe current for 2min. Each 2–3 pistils were used in one experiment and three individual experimental sets were performed.
Evaluation of reaction level of SI to CO2 (RLSICO2)
Three to five flowers were cut at the peduncle and stood on a 1% (w/v) solid agar plate. Flowers were self-pollinated, placed into a CO2 incubator, and treated with 4.5% CO2 for 4h at 23 °C. After 1 d at room temperature, pistils were fixed in ethanol:acetic acid (3:1) overnight, softened in 1 N NaOH at 60 °C for 2h, then stained with 0.01% (w/v) decolorized aniline blue in 2% K3PO4 for 6h. Pollen tube behaviour was observed under a fluorescent microscope (Axiophot 2, Zeiss). CO2 sensitivity was measured using the RLSICO2 index. RLSICO2 was classified into five categories, based on the number of pollen tubes penetrating into the stigma: 1, 0 pollen tubes; 2, 1–5 pollen tubes; 3, 6–15 pollen tubes; 4, 16–30 pollen tubes; and 5, >30 pollen tubes. Three replicates were performed on each plant on different days. Non-CO2-treated self-pollinated flowers were used as controls. In all cases, no pollen tubes penetrated into the control stigmas.
Genotyping of S-haplotypes
S-haplotypes of B. rapa were identified using primers PS5 (5′-ATGAAAGGCGTAAGAAAAACCTA-3′) and PS15 (5′-CCG TGTTTTATTTTAAGAGAAAGAGCT-3′) (Nishio et al., 1996) to amplify a fragment of the SLG gene. PCR-RFLP (restriction fragment length polymorphism) was used to distinguish the two S-haplotypes based on differential digest with the restriction enzyme KpnI (TaKaRa, Japan). Digested DNA was electrophoresed on a 1.5% agarose gel.
Molecular markers and detection of DNA polymorphism
To screen for markers that show polymorphism between B. rapa lines, primers specific for simple sequence repeat (SSR) markers from different sources [UK, prefixes Ra, Na, Ol, and ENA (Lowe et al., 2004; http://brassica.bbsrc.ac.uk); Japan, prefixes BRMS, KBr, and EST (Suwabe et al., 2002, 2004, 2006; http://vegmarks.nivot.affrc.go.jp, NIVTS); China, prefix sau_um (Ge et al., 2011); and Korea, prefix AMCP (Ramchiary et al., 2011)] were used. SSR, RFLP, and insertion/deletion (InDel) markers (prefixes XT and Bra) were also designed based on the Brassica database (BRAD) (http://brassicadb.org/brad/, last accessed 14 December 2013) (Supplementary Table S1 available at JXB online).
Total genomic DNA was extracted from young leaves of two parental lines and F2 progeny using the cetyltrimethylammonium bromide (CTAB) method (Murray and Thompson, 1980). DNA polymorphism analysis with SSRs was carried out using PCR with fluorescent dyes, performed according to Suwabe et al. (2008) with some modifications. The M13 (–21) universal primer sequence (18bp) was fused to the 5′ end of the original forward primer, and the M13 (–21) universal primer was labelled with 6-FAM, NED, VIC, or PET fluorescent dye (Applied Biosystems, CA, USA). PCRs were performed in a 10 μl reaction volume containing 10ng of template DNA, 4.7 μM of labelled M13 (–21) universal primer and reverse primer, 0.3 μM of forward primer, 1× PCR buffer, 1× dNTP, 1× MgCl2, and 0.5U of rTaq (TOYOBO, Japan). Conditions for PCR were as follows: initial denaturation was carried out at 94 °C for 3min followed by 37 cycles at 94 °C for 30 s, 55 °C (slope of 0.5 °C s–1) for 30 s, 72 °C (slope of 0.5 °C s–1) for 30 s, and a final extension at 72 °C for 4min. A 1 μl aliquot of 50-fold diluted PCR product was added to 8.9 μl of Hi-Di™ Formamide and 0.1 μl of GeneScan™ 600 LIZ™ Size Standard (Applied Biosystems, USA) and applied to an ABI 3730 DNA Analyzer (Applied Biosystems). Data were analysed using ABI GeneMapper® software.
For polymorphism analysis with RFLP and InDel markers, PCR was carried out in a 10 μl reaction volume with 5 pmol of forward and reverse primers instead of fluorescent dyes. For RFLP markers, amplified fragments were digested using restriction enzymes for 1h. Fragments of digested DNA were separated on a 2–4% agarose gel.
Linkage map construction and QTL analysis
A genetic map was constructed using JoinMap® version 4 (Van Ooijen, 2006) utilizing the double pseudo-testcross strategy with a log10 of odds (LOD) threshold of 6.0 for linkage group identification. The best marker order was calculated with the regression mapping algorithm, and marker order was retained from the first round only. Map distance units in centiMorgans (cM) were converted from recombination frequencies using the Kosambi mapping function (Kosambi, 1944). Interval mapping (IM) was performed to identify putative QTLs using the established linkage map and the observed phenotypic traits. This method was run using MapQTL® version 6 (Van Ooijen, 2009). With this software, a P < 0.05 LOD score significance threshold was calculated by creating a group-wide distribution of the data based on a 1000 permutation test. LOD peaks were used to estimate the position of QTLs on the map.
Statistical analysis
Box plots were prepared by Ekuseru-Toukei 2012 software (Social Survey Research Information Co., Ltd, Japan) to compare the phenotypic difference, as this plot type gives a good sense of environmental data distribution (Upton and Cook, 1996). Kruskal–Wallis analysis of variance (ANOVA) by ranks was used between paired comparisons of markers to examine marker association.
Results
Phenotypic analysis of B. rapa lines in response to CO2 treatment
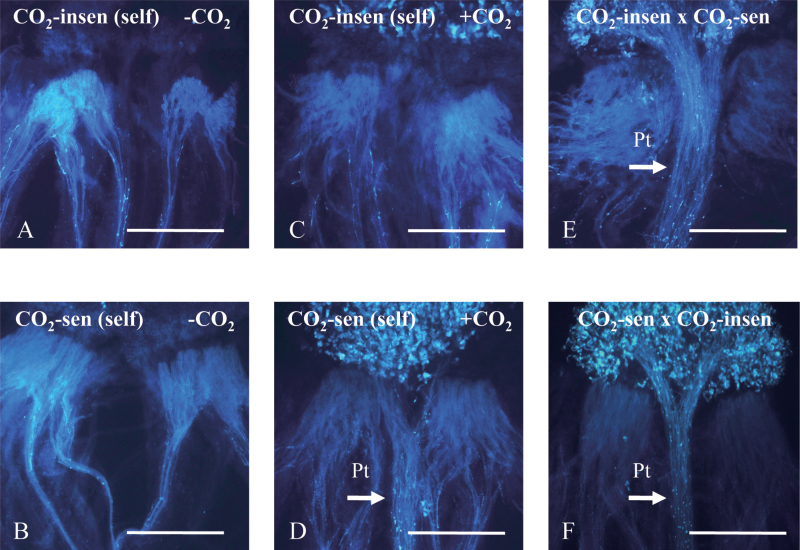
Two inbred lines of B. rapa with different CO2 sensitivity, a CO2-sensitive line (HA-11621) and a CO2-insensitive line (HA-11623), were used in this study. Flowers were self-pollinated by hand pollination and incubated in a CO2 incubator (4.5% CO2) for 4h. Both lines were self-incompatible under normal conditions (control) (Fig. 1A, B), whereas they showed significantly different responses to CO2 gas treatment (Fig. 1C, D). Specifically, in the CO2-sensitive line, many pollen tubes were seen to penetrate into papilla cells after CO2 treatment. This pollination test confirmed that the CO2-sensitive line had high sensitivity to CO2 while the CO2-insensitive line hardly responded to 4.5% CO2. Cross-pollination was performed as a positive control (Fig. 1E, F).
Fig. 1.
Phenotype of inbred Brassica rapa lines used in this study. (A, B) Pollen tube behaviour after self-pollination of CO2-sensitive (HA-11621) and CO2-insensitive (HA-11623) lines under normal conditions (without CO2 treatment). No penetrated or elongated pollen tubes are observed in either line. (C, D) Pollen tube behaviour after self-pollination of CO2-sensitive and CO2-insensitive lines under 4.5% CO2 gas treatment. Pollen tubes could penetrate into the stigma and elongate through the style only in the CO2-sensitive line. (E, F) Cross-pollination as a positive control; the arrow shows pollen tubes which have penetrated. Pt, pollen tubes. Bar=1000 μm.
Physiological changes in papilla cells after CO2 treatment
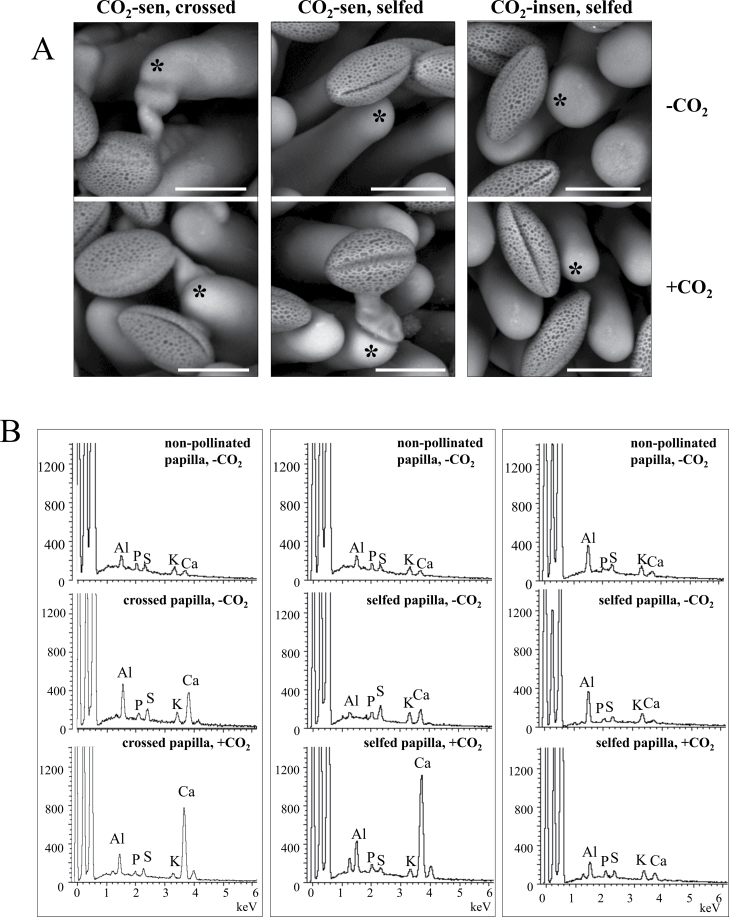
Previous work using X-ray microanalysis has revealed the accumulation of calcium at the stigmatic surface following compatible cross-pollination in Brassica oleracea (Elleman and Dickinson, 1999). X-ray mapping strongly indicated that a high concentration of calcium was localized at the points where the pollen grain made contact with the surface of the stigmatic papilla cell. The calcium accumulation was also observed in B. rapa, especially in compatible pollination (Iwano et al., 1999). To examine the physiological effect of CO2 on pollination reactions, this calcium accumulation was examined by using cryo-scanning electron microscopy fitted with an X-ray microanalysis system. When the CO2-sensitive line was cross-pollinated with the CO2-insensitive line, pollen grain hydrated and the pollen tube penetrated into the papilla cell, but when CO2-sensitive and CO2-insensitive lines were self-pollinated under normal conditions (without CO2 treatment), few pollen grains hydrated, and no pollen tube germination was observed in either line (Fig. 2A, upper panel). After CO2 treatment, the cross-pollinated pollen grains did not show a significant difference, neither did the self-pollinated CO2-insensitive line. However, the self-pollinated CO2-sensitive line showed obvious changes: pollen hydration and germination were observed under CO2 treatment (Fig. 2A, lower panel). The emission of elements (Kα) in the tip of the papilla cells was then analysed after selecting the pollinated papilla cells which faced nearly the same direction in relation to the X-ray detector. Emissions of P-Kα, S-Kα, K-Kα, and Ca-Kα were detected together with C and O, which are constituent elements of biological materials. The detected emission of Al-Kα originates from the stub that held the samples (Fig. 2B). There was no significant difference in the elemental emissions between CO2-sensitive and CO2-insensitive lines before pollination. Ca2+ accumulation was observed after cross-pollination as previously reported (Iwano et al., 1999), and Ca-Kα emission was increased with CO2 treatment. After self-pollination, Ca-Kα emission was slightly increased in the CO2-sensitive line in the normal CO2 condition. This Ca-Kα increase became more pronounced (~6-fold) in the CO2-sensitive line when it was self-pollinated in the high CO2 condition. In the CO2-insensitive line, no significant Ca-Kα increase was observed after self-pollination in normal or high CO2 conditions (Fig. 2B). Although the biological significance of the accumulation of calcium at the pollen–stigma interface is not clear, it has been suggested that calcium plays some role in the successful development of the pollen tube tip into the region of expanded stigmatic wall (Elleman and Dickinson, 1999). The present results suggest that, in the CO2-sensitive line, high CO2 activates a compatible pollination pathway or disturbs an SI signalling pathway leading to Ca2+ accumulation at the pollen–stigma interface.
Fig. 2.
Electron micrographs and X-ray microanalysis of B. rapa. (A) Cryo-scanning electron micrographs of pollinated papilla cells were taken 1.5h after pollination. Representative examples of the cross-pollinated (left column) and self-pollinated (middle column) CO2-sensitive line, and the self-pollinated (right column) CO2-insensitive line are shown. Without CO2 treatment (upper panels), only cross-pollen is accepted, and self-pollen grains maintain a spheroid shape without swelling in both lines. With 4.5% CO2 gas treatment (lower panels), pollen grains swell and germinate in the CO2-sensitive line (lower middle) but not in the CO2-insensitive line (lower right). Bar=25 μm. (B) Representative examples of energy-dispersive X-ray spectra of non-pollinated and pollinated papilla cell surfaces. Scanning positions for X-ray analyses are indicated by asterisks in (A). The emissions of Al-Kα, P-Kα, S-Kα, K-Kα, and Ca-Kα were detected at the papilla cell surfaces, and the intensity of Ca emission was increased after cross-pollination (left column). The increase of Ca emission was also observed after self-pollination with 4.5% CO2 gas treatment in the CO2-sensitive line (middle column) but not in the CO2-insensitive line (right column). These spectrum patterns are reproducible in three individual experiment sets. The emission of Al-Kα is mostly derived from the stub that held the samples.
The efficiency of CO2 treatment
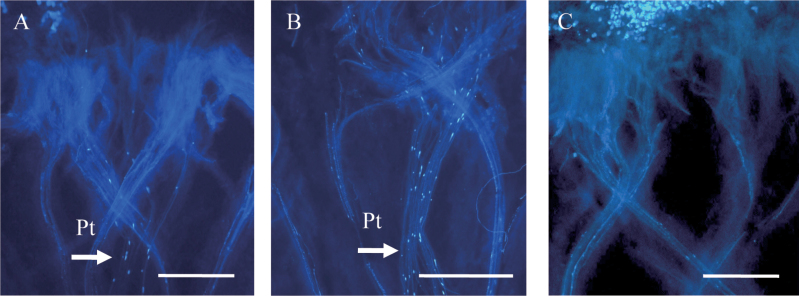
A previous study suggested that the effect of CO2 on SI breakdown depends on the timing of treatment (Nakanishi and Hinata, 1973). In the CO2-sensitive line, when self-pollinated flowers were immediately treated with 4.5% CO2 for 4h, SI could be overcome, and typically >10 pollen tubes penetrated into the stigma (Fig. 3A). When CO2 treatment started 3h after self-pollination, SI could still be overcome (Fig. 3B). However, when CO2 treatment started 6h after self-pollination, the number of penetrating pollen tubes was decreased (Fig. 3C). These results indicate that self-pollen inhibition in SI is biostatic, as previously suggested (Sarker et al., 1988), and can be reversed at least at 3h after pollination. However, at 6h after pollination, SI inhibition enters an irreversible phase that cannot be overcome by CO2 treatment.
Fig. 3.
The efficiency of CO2 treatment of the CO2-sensitive line. (A) Self-pollinated stigmas were treated with 4.5% CO2 immediately after self-pollination, and pollen tube penetration and elongation were observed. (B) Self-pollinated stigmas were treated with 4.5% CO2 3h after self-pollination. The pollen tube can still penetrate into papilla cells. (C) When CO2 treatment began 6h after self-pollination, the number of penetrating pollen tubes was decreased. Pt, pollen tubes. Bar=1000 μm.
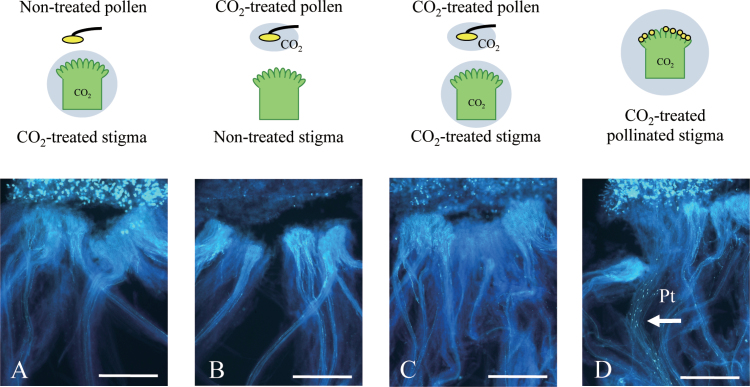
Next, in order to narrow down the stage of SI affected by CO2 treatment, experiments with high CO2-pre-treated pollen or pistil from the CO2-sensitive line were performed (Fig. 4). SI could not be overcome by pre-treatment of either tissue, and no pollen tube penetration could be observed even when both pollen and pistil were treated separately prior to pollination (Fig. 4). SI could be overcome only when the self-pollinated pistil was treated with high CO2. These results suggest that some post-pollination physiological process is affected by high CO2, in the process of SI breakdown.
Fig. 4.
Pollination assay using CO2-treated non-pollinated flowers of the CO2-sensitive line as either CO2-treated pistil or pollen. Pollen tube behaviour is observed after a 4h CO2 treatment immediately after self-pollination. (A) A pistil from a non-pollinated flower treated with CO2 for 4h was pollinated with pollen from the same plant which was not treated with CO2. (B) A pistil from a non-treated non-pollinated flower was pollinated with pollen from the same plant which was treated with CO2 for 4h. (C) A pistil from a CO2-treated non-pollinated flower (CO2-treated) was pollinated with pollen from the same plant which was treated with CO2 for 4h. (D) Self-pollinated flower treated with CO2 after pollination, positive control. Pt, pollen tubes. Bar=1000 μm.
S-allele characterization and phenotype of CO2 sensitivity in F1 and F2
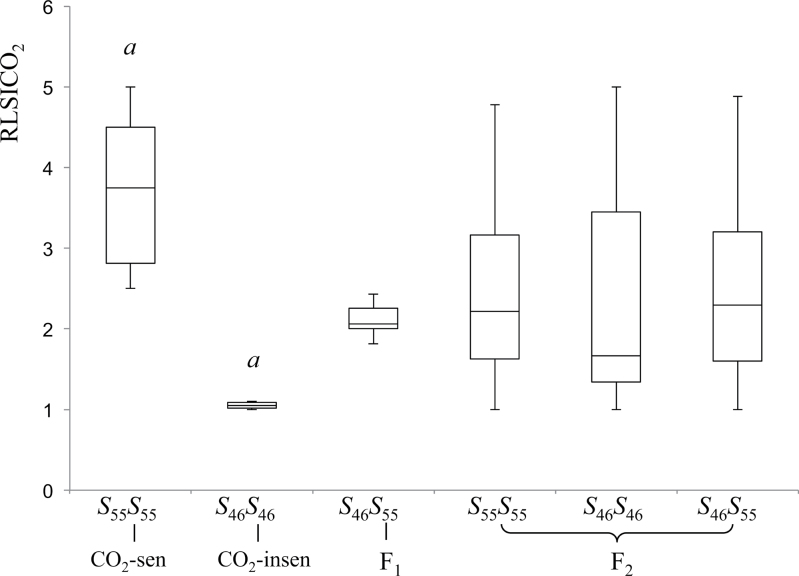
The S-haplotypes of the two parental inbred lines were first determined by amplifying their SLG genes (Nishio et al., 1996). The sequence data suggested that the S-haplotypes of the CO2-sensitive and CO2-insensitive lines were S 55 S 55 and S 46 S 46, respectively. To dissect genetically the gene(s) that determines sensitivity to CO2 treatment, six F1 plants (S 46 S 55) were produced by crossing CO2-sensitive and CO2-insensitive lines. These F1 plants exhibited an intermediate CO2 sensitivity phenotype where self-pollen tubes penetrated into the stigma under high CO2 treatment but there were fewer penetrating pollen tubes than observed in a self-pollination of the CO2-sensitive parent. An F2 population of 110 individuals derived from a bud-pollinated F1 plant was made and used for further genetic analyses of the CO2 sensitivity. F2 individuals were genotyped using PCR-RFLP to distinguish SLG alleles (Supplementary Fig. S1 at JXB online). S 55- and S 46-haplotypes were segregated in the F2 population according to Mendelian transmission (Supplementary Table S2). Pollen tube behaviour after CO2 treatment varied among individuals and, in order to quantify the strength of CO2 sensitivity, the modified RLSICO2 (reaction level of SI to CO2) index was employed (Niikura and Matsuura, 2000), which calculates CO2 sensitivity based on the number of penetrating pollen tubes after self-pollination under high CO2 conditions (see the Materials and methods). The RLSICO2 of 110 F2 individuals is presented in Supplementary Fig. S2, and the summarized box-plot data are shown in Fig. 5, together with the RLSICO2 of F1 and the parental inbred lines. F1 had an RLSICO2 score intermediate to the two parental lines, suggesting that the high CO2 sensitivity is a semi-dominant (incompletely dominant) trait in these inbred lines. Furthermore, the RLSICO2 of F2 individuals was continuously distributed and did not follow a simple one-locus biallelic Mendelian distribution (Supplementary Fig. S2). These results suggest that CO2 sensitivity in the inbred lines used here could be a quantitative trait which is controlled by more than one gene.
Fig. 5.
Box plots of CO2 sensitivity phenotypes. Data show the distribution of RLSICO2 with 25th, 50th, and 75th percentiles (horizontal bars), interquartile ranges (columns), and 1.5 interquartile ranges (error bars) of RLSICO2 from six CO2-sensitive (S 55 S 55) and six CO2-insensitive (S 46 S 46), individuals, six F1 individuals (S 46 S 55), and 110 F2 individuals (22 S 46 S 46, 64 S 46 S 55, and 24 S 55 S 55). a indicates a significant difference (P < 0.01) between CO2-sensitive and CO2-insensitive lines.
Relationship between S-alleles and CO2 sensitivity
To investigate whether CO2 sensitivity is related to S-haplotypes, the 110 F2 individuals were grouped into three genotypes (S 55 S 55, S 46 S 46, and S 46 S 55). The RLSICO2 of each group is shown in Fig. 5. In the three F2 groups, RLSICO2 scores were distributed from 1 to 5, and interquartile ranges overlapped, indicating that CO2 sensitivity is not linked to the S-locus in these two lines.
Reproductive tissue controlling CO2 sensitivity
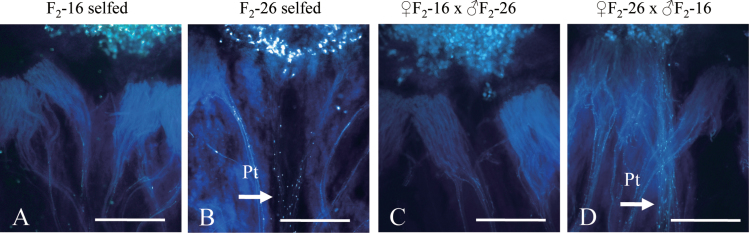
From the F2 population, two S 46 S 46 homozygotes with different RLSICO2 were selected: F2-16, a CO2-insensitive line (RLSICO2=1±0); and F2-26, a CO2-sensitive line (RLSICO2=4.15±0.53). These two lines were used to examine the reproductive tissue controlling CO2 sensitivity. Reciprocal crosses were performed between these two SI lines with or without high CO2 gas treatment. Because all crosses under normal conditions (without CO2 treatment) were incompatible, only data from crosses performed in the high CO2 condition are shown in Fig. 6. The cross between CO2-sensitive F2-26 pistil and CO2-insensitive F2-16 pollen was CO2 sensitive, showing many penetrating pollen tubes under high CO2. On the other hand, the cross between CO2-insensitive F2-16 pistil and CO2-sensitive F2-26 pollen was CO2 insensitive, showing no penetrating pollen tubes even under high CO2. These results suggest that CO2 sensitivity is controlled by genes expressed in the female tissue (pistil).
Fig. 6.
Reciprocal crosses with CO2 treatment between two S 46 homozygous individuals from the F2 population with different RLSICO2. (A) CO2-sensitive F2 self-pollination. (B) CO2-insensitive F2 self-pollination. (C) A CO2-sensitive F2 pistil pollinated with pollen from a CO2-insensitive F2. (D) A CO2-insensitive F2 pistil pollinated with pollen from a CO2-sensitive F2. Pt, pollen tubes. Bar=1000 μm. n=3.
Marker analysis and construction of a linkage map
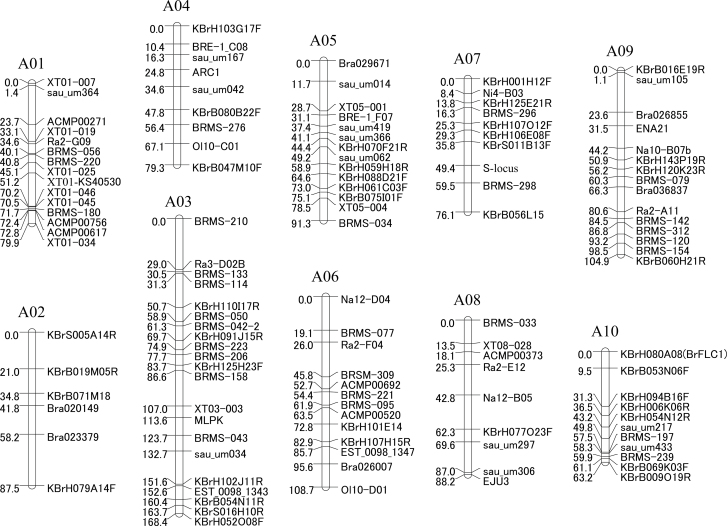
In order to map QTLs that determine CO2 sensitivity, a linkage map was constructed for this F2 population. A total of 911 different genetic markers were examined in the two parental lines. To clarify the relationship between previously identified SI-related genes and CO2 sensitivity, SLG (an S-locus marker), MLPK, and ARC1 were also selected. Though a very low level of polymorphism (14.7%) was detected for all types of markers, 123 polymorphic markers were selected, which include 113 SSRs, five RFLPs, and five InDel markers. These 123 markers were used for linkage mapping, and generated 10 linkage groups (A01–A10) at a LOD threshold value of 6.0 (Fig. 7). The total length of the map was 947.5 cM, and the length of the linkage groups ranged from 63.2 cM (A10) to 168.4 cM (A03). The distance between markers varied from 0 to 29.3 cM, with an average interval of 7.7 cM. SLG, MLPK, and ARC1 were mapped to A07, A03, and A04, respectively, which is consistent with previous reports (Ajisaka et al., 2001; Hatakeyama et al., 2010).
Fig. 7.
A linkage map of selected DNA markers from the F2 population. Map distances are shown to the left of vertical lines of the linkage group in cM, and marker names are shown to the right.
QTL analysis and association of markers with high CO2 sensitivity
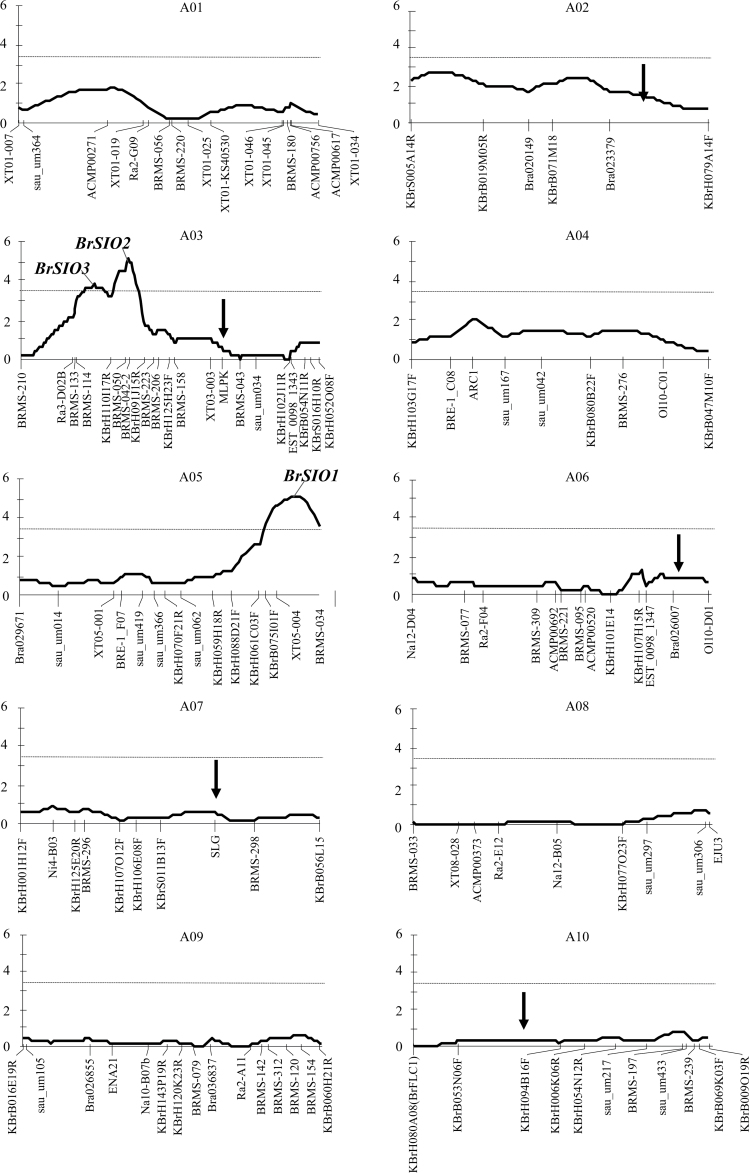
Using the constructed linkage map, QTLs responsible for high CO2 sensitivity were analysed. Three QTLs were identified on linkage groups A03 and A05 based on a LOD threshold of 3.40 (1000 permutation test, P < 0.05) (Fig. 8, Table 1). These QTLs were tentatively named Brassica rapa SI Overcome (BrSIO) 1–3, and these results further supported the prediction that CO2 sensitivity of SI is controlled by a polygenic system. BrSIO1 on A05 and BrSIO2 on A03 are two major QTLs that explained 19.3% and 19.0% of phenotypic variation, respectively. BrSIO3, located near BrSIO2, accounted for 14.5% of the variance (Table 1).
Fig. 8.
QTL analysis results. The solid line indicates the LOD score and the dotted line indicates the QTL threshold (LOD=3.4) determined using a 1000 permutation test (P < 0.05). The x-axis represents each linkage group (cM) and the y-axis indicates the QTL score. Two QTLs (BrSIO2 and 3) are detected in A03 and one in A05 (BrSIO1). Arrows show loci involved in SI stability reported by Hatakeyama et al. (2010).
Table 1.
Summary of CO2 sensitivity QTLs
| QTL | LG | Closest marker | QTL peak (cM)a | LOD | R 2b | Additive effectc |
|---|---|---|---|---|---|---|
| BrSIO1 | A05 | XT05-004 | 83.50 | 5.17 | 19.30 | 0.72 |
| BrSIO2 | A03 | BRMS-042-2 | 60.87 | 4.46 | 19.00 | 0.69 |
| BrSIO3 | A03 | KBrH110I17R | 41.25 | 3.76 | 14.50 | 0.65 |
a QTL peak position, detected by interval mapping, between two markers.
b Amount of phenotypic variation explained by the QTL.
c Additive effect of the CO2-sensitive HA-11621 allele on RLSICO2.
To examine the significance of these QTLs, F2 progeny were classified into groups based on the genotypes of the linkage markers nearest these three newly identified loci, and the relationship of the loci to RLSICO2 in individual plants was analysed using Kruskal–Wallis ANOVA by ranks (Table 2). Alleles from CO2-sensitive (HA-11621) and CO2-insensitive (HA-11623) lines are presented as S and I, respectively. Almost all classifications using the closest linkage markers showed a higher RLSICO2 index in the SS group with significance at P < 0.01, except marker BRMS-114, which showed significance at P < 0.05.
Table 2.
Statistical analysis of QTL effect
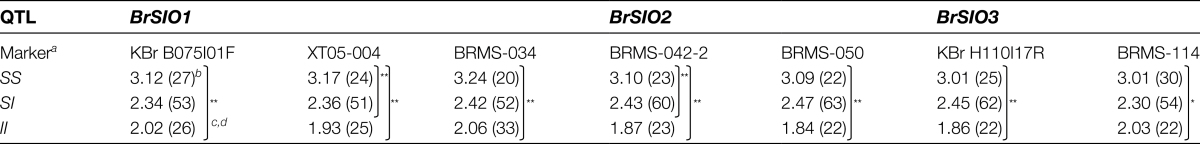
a S, CO2-sensitive HA-11621 allele; I, CO2-insensitive HA-11623 allele.
b Individuals whose genotype was unidentified are excluded.
c Kruskal–Wallis analysis comparing phenotype between genotype groups with individuals in the same groups.
d Significance level: **P <0.01; *P <0.05.
Marker association was further examined with combinations of BrSIO1 and BrSIO2 since BrSIO3 is a minor QTL closely linked to BrSIO2, making it difficult to identify it as an independent QTL. F2 progeny were classified into nine groups based on the genotypes of their closest linkage markers (Table 3). According to this classification, for example, the two above analysed S 46 S 46 homozygous lines with different RLSICO2, F2-16 (CO2-insensitive line) and F2-26 (CO2-sensitive line), were classified into group 8 and group 1, respectively. When groups with the same BrSIO1 genotype were compared, the BrSIO2 SS group showed a higher RLSICO2 index compared with the BrSIO2 II group. Likewise, when the groups with the same BrSIO2 genotype were compared, the BrSIO1 SS group showed a higher RLSICO2 compared with the BrSIO1 II group. Although the numbers of F2 individuals in each group were rather low, significance (P < 0.05) was detected between groups 1 and 6, 2 and 6, and 2 and 8. These data suggest that BrSIO1 and BrSIO2 work additively in overcoming SI during CO2 treatment in the CO2-sensitive (HA-11621) line. No QTL was detected at genes known to affect SI stability (MLPK, ARC1, or the S-locus), indicating that CO2 sensitivity is determined by novel genes in the experimental lines used here.
Table 3.
QTL association for CO2 sensitivity
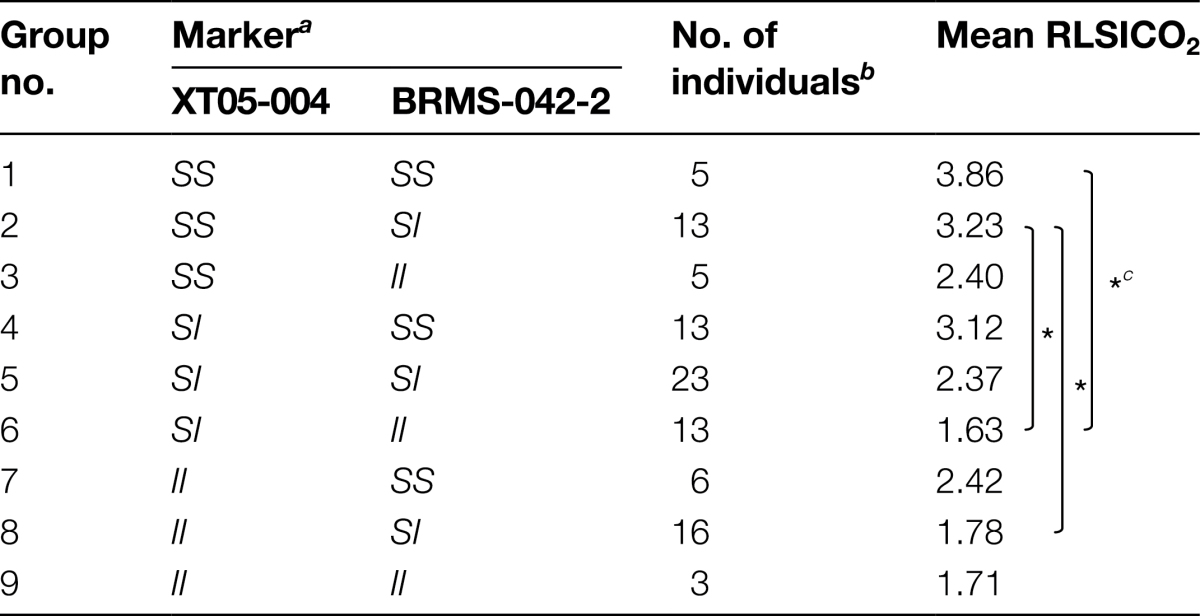
a S, CO2-sensitive HA-11621 allele; I, CO2-insensitive HA-11623 allele.
b Individuals whose genotype was unidentified are excluded.
c Significance level, *P <0.05.
Associated gene prediction by in silico comparative mapping
Using the B. rapa genome sequence (Cheng et al., 2011), BrSIO1 could be mapped to a 569kb region flanked by InDel marker XT05-004 and SSR marker BRMS-034, and BrSIO2 to a 1469kb region flanked by SSR markers BRMS-042-2 and KBrH110I17R. These two regions include 121 and 280 genes annotated in the Brassica database (BRAD), respectively (Supplementary Tables S3, S4 at JXB online). Comparison of the A. thaliana genome with the Brassicaceae genome (reviewed by Schranz et al., 2006) suggests that BrSIO1 has synteny on A. thaliana chromosome 2 and BrSIO2 has synteny on both chromosomes 3 and 4. It is assumed that these two QTLs do not have the same genetic origin and could be two independent regions controlling high CO2 sensitivity. Based on reciprocal cross results, the CO2 sensitivity trait may be controlled by genes expressed in the female organ (Fig. 8). A total of 121 and 280 annotated genes in BrSIO1 and BrSIO2 have 103 and 243 homologues in A. thaliana, respectively, and 54 and 141 of these genes are expressed in A. thaliana pistil (microarray data of carpel at stage 12, http://affymetrix.arabidopsis.info/narrays/search.pl?f1=1&s1=ATGE_37, last accessed 14 December 2013, Supplementary Tables S3, S4).
Genes involved in related biological processes are often expressed cooperatively and their co-expression information is important for understanding biological systems (Eisen et al., 1998). ATTED-II (http://atted.jp/, last accessed 14 December 2013) is a gene co-expression database useful for identifying the potential partners working in the same biological processes (Obayashi et al., 2007). Co-expression analysis was performed using ATTED-II with these 195 genes and it was found that MAP kinase 6 (At2g43790 in BrSIO1) and ethylene overproducer 1 (At3g511770 in BrSIO2) showed the strongest co-expression and calmodulin-like 41 (At3g50770 in BrSIO2) has weak co-expression with cytochrome c oxidase 10 (At2g44520in BrSIO1) and beta glucosidase 28 (At2g44460 in BrSIO1). In addition to these co-expressed genes, these two regions encode highly homologous family member proteins, for example matrixin proteins (At2g45040 in BrSIO1 and At4g16640 in BrSIO2) and senescence-associated proteins (At2g44670 in BrSIO1 and At4g17670 in BrSIO2). All these can be candidate responsible genes, although the biological functions of these genes are mostly unknown.
Discussion
It has been >40 years since Nakanishi et al. (1969) first reported that SI could be overcome by CO2 and Nakanishi and Hinata (1973) demonstrated the applicability of this technique to commercial use. Nowadays, seed companies have adopted this method to obtain inbred parental seeds of crucifer vegetables for large-scale commercial F1 hybrid seed production. However, there is still very limited understanding of the mechanism by which SI is overcome.
Lee et al. (2001) showed a shrunken and distorted papilla cell surface in the CO2-sensitive B. rapa cv. Hiratsuka, and suggested that these structural changes could cause SI to be overcome. The cryo-scanning electron microscopy data reported here did not show any structural changes in CO2-sensitive or CO2-sensitive lines (Fig. 2A). Additionally, pre-treatment of non-pollinated pistils with high CO2 gas did not cause SI breakdown (Fig. 4). Therefore, a completely different SI breakdown mechanism must be present, at least in the CO2-sensitive line. In contrast, massive Ca accumulation was observed at the pollen–stigma interface specifically in CO2-sensitive plants under high CO2 conditions (Fig. 2B). Brewbaker and Kwack (1963) were the first to describe the need for a high concentration of Ca2+ for pollen germination and pollen tube growth. The high concentration of Ca2+ could be needed for activating pectinase to loosen the papilla cell wall, allowing the pollen tube to penetrate (Black and Charlwood, 1995), or for keeping the pollen tube cell wall rigid enough not to burst (Hepler and Winship, 2010). Although causal relationships remain unclear, the data suggest that CO2 treatment induces a certain compatible reaction leading to Ca2+ accumulation at the pollen–stigma interface.
To date, several genetic studies have been performed to understand the mechanism of SI breakdown for breeding purposes. Niikura and Matsuura (2002) reported that in Japanese radish high CO2 sensitivity is controlled by a recessive gene that governs the construction and/or metabolism of the stigma, which reacts to CO2 without any changes in gene expression. In contrast, Hyun et al. (2007) reported a dominant, S-haplotype-linked high CO2 sensitivity phenotype in B. rapa. In contrast to these previous reports, F1 plants had an intermediate CO2 sensitivity and the F2 population had a continuous frequency distribution of RLSICO2 in the present study (Fig. 6). These results suggest that in the lines used for this study, CO2 sensitivity is a quantitative trait which is controlled by more than one gene.
Genetic linkage maps based upon frequency of recombination in segregating populations are fundamental and powerful tools for associating phenotypic trait-specific genetic regions. Linkage mapping can be used to understand the biological basis of complex traits and to dissect genetic determinants underlying the expression of agronomically important breeding traits (Paran and Zamir, 2003). Using QTL analysis, two major QTLs for high CO2 sensitivity were successfully identified (Fig. 8). BrSIO1 and BrSIO2 had similar LOD scores and explained similar amounts of phenotypic variation (19.3% and 19%), and these could be two major factors controlling high CO2 sensitivity. Very recently, five QTLs associated with stability of SI in B. rapa have been identified. Two of them co-localized with SLG (A07) and MLPK (A03) and the other three were on A02, A06, and A10 (Hatakeyama et al., 2010). CO2 sensitivity did not link with the S-locus in the present study (Fig. 5) and none of the other reported loci co-localized with QTLs detected here (Fig. 8), indicating that CO2 sensitivity of the lines in this study is determined by novel genes different from those known to affect SI stability. Genes in BrSIO1 and BrSIO2 regions have 103 and 243 homologues in A. thaliana, respectively, and 54 and 141 of these genes are expressed in A. thaliana pistil. In silico comparative analyses identified several co-expressing genes and highly homologous genes encoded in these two regions. All these can be candidate responsible genes; however, to dentify the genes responsible for high CO2 sensitivity in the QTL regions in B. rapa more accurately, it would be necessary to narrow down the regions by developing near-isogenic lines (NILs).
To maintain F1 seed quality, inbred lines with strong but CO2-sensitive SI are ideal for F1 hybrid breeding, and it is very important to understand the genetic relationships between SI-related genes and CO2 sensitivity phenotypes. These results could be useful for the marker-assisted selection of parental lines with both stable SI and high CO2 sensitivity.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. S-haplotype analysis of F2 plants by PCR-RFLP.
Figure S2. RLSICO2 in CO2-sensitive and CO2-insensitive lines, and F1 and F2 progeny based on the number of penetrating pollen tubes after self-pollination under high CO2 conditions.
Table S1. Genetic markers and their primers used for linkage analysis.
Table S2. S-haplotype segregation in the F2 population.
Table S3. Annotated genes and Arabidopsis homologues in BrSIO1.
Table S4. Annotated genes and Arabidopsis homologues in BrSIO2.
Acknowledgements
The authors thank Ms Rina Nagai, Ms Hitomi Ishikawa, and Ms Yuko Yoshimura for their excellent technical assistance. This work was supported by a Grant-in-Aid for the Scientific Research on Innovative Areas (21112003, 23113002) and by Grants in Aid for Scientific Research (23570056, 21248014, 25252021) from the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT) and by the Japan Advanced Plant Science Network.
Glossary
Abbreviations:
- InDel
insertion/deletion
- LG
linkage group
- LOD
log of odds
- QTL
quantitative trait locus
- RFLP
restriction fragment length polymorphism
- RLSICO2
reaction level of SI to CO2
- SI
self-incompatibility
- SSR
simple sequence repeat.
References
- Ajisaka H, Kuginuki Y, Yui S, Enomoto S, Hirai M. 2001. Identification and mapping of a quantitative trait locus controlling extreme late bolting in Chinese cabbage (Brassica rapa L. ssp. pekinensis syn. campestris L.) using bulked segregant analysis: a QTL controlling extreme late bolting in Chinese cabbage. Euphytica 118, 75–81 [Google Scholar]
- Bateman AJ. 1955. Self-incompatibility systems in angiosperms. III. Cruciferae. Heredity 9, 52–68 [Google Scholar]
- Black M, Charlwood B. 1995. The physiology and biochemistry of plant cell walls. London: Chapman & Hall [Google Scholar]
- Brewbaker JL, Kwack BH. 1963. The essential role of calcium ion in pollen germination and pollen tube growth. American Journal of Botany 50, 859–865 [Google Scholar]
- Chantha S-C, Herman AC, Platts AE, Vekemans X, Shoen DJ. 2013. Secondary evolution of a self-incompatibility locus in the Brassicaceae genus Leavenworthia. PLoS Biology 11, e1001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F, Liu S, Wu J, Fang L, Sun S, Liu B, Li P, Hua W, Wang X. 2011. BRAD, the genetics and genomics database for Brassica plants. BMC Plant Biology 11, 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. 1998. Cluster analysis and display of genome-wide expression patterns. Proceedings of the National Academy of Sciences, USA 95, 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elleman CJ, Dickinson HG. 1999. Commonalities between pollen/stigma and host/pathogen interactions: calcium accumulation during stigmatic penetration by Brassica oleracea pollen tubes. Sexual Plant Reproduction 12, 194–202 [Google Scholar]
- Ge Y, Ramchiary N, Wang T, Liang C, Wang N, Wang Z, Choi SR, Lim YP, Piao ZY. 2011. Development and linkage mapping of unigene-derived microsatellite markers in Brassica rapa L. Breeding Science 61, 160–167 [Google Scholar]
- Gu T, Mazzurco M, Sulaman W, Matias DD, Goring DR. 1998. Binding of an arm repeat protein to the kinase domain of the S-locus receptor kinase. Proceedings of the National Academy of Sciences, USA 95, 382–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama K, Horisaki A, Niikura S, Narusaka Y, Abe H, Yoshiaki H, Ishida M, Fukuoka H, Matsumoto S. 2010. Mapping of quantitative trait loci for high level of self-incompatibility in Brassica rapa L. Genome 53, 257–265 [DOI] [PubMed] [Google Scholar]
- Hepler PK, Winship LJ. 2010. Calcium at the cell wall–cytoplast interface. Journal of Integrative Plant Biology 52, 147–160 [DOI] [PubMed] [Google Scholar]
- Horisaki A, Niikura S. 2008. Developmental and environmental factors affecting level of self-incompatibility response in Brassica rapa L. Sexual Plant Reproduction 21, 123–132 [Google Scholar]
- Hyun JY, Gothandam KM, Baek NK, Wang G, Chung YY. 2007. Dominance relationship between two self-incompatible Brassica campestris haplotypes in response to CO2 gas. Journal of Plant Biology 50, 161–166 [Google Scholar]
- Indriolo E, Tharmapalan P, Wright SI, Goring DR. 2012. The ARC1 E3 ligase gene is frequenctly deleted in self-compatible Brassicaceae species and has a conserved role in Arabidopsis lyrata self-pollen rejection. The Plant Cell 24, 4607–4620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwano M, Wada M, Morita Y, Shiba H, Takayama S, Isoga A. 1999. X-ray microanalysis of papillar cells and pollen grains in the pollination process in Brassica using a variable-pressure scanning electron microscope. Journal of Electron Microscopy 48, 909–917 [Google Scholar]
- Kakita M, Murase K, Iwano M, Matsumoto T, Watanabe M, Shiba H, Isogai A, Takayama S. 2007. Two distinct forms of M-locus protein kinase localize to the plasma membrane and interact directly with S-locus receptor kinase to transduce self-incompatibility signalling in Brassica rapa . The Plant Cell 19, 3961–3973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitashiba H, Liu P, Nishio T, Nasrallah JB, Nasrallah ME. 2011. Functional test of Brassica self-incompatibility modifiers in Arabidopsis thaliana . Proceedings of the National Academy of Sciences, USA 108, 16173–16178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosambi DD. 1944. The estimation of map distance from recombination values. Annals of Eugenics 12, 172–175 [Google Scholar]
- Lee SH, Hong MY, Kim S, Lee JS, Kim BD, Min BH, Baek NK, Chung YY. 2001. Controlling self-incompatibility by CO2 gas treatment in Brassica campestris: structural alteration of papillae cell and differential gene expression by increased CO2 gas. Molecules and Cells 11, 186–191 [PubMed] [Google Scholar]
- Lowe AJ, Moule C, Trick M, Edwards KJ. 2004. Efficient large-scale development of microsatellites for marker and mapping applications in Brassica crop species. Theoretical and Applied Genetics 108, 1103–1112 [DOI] [PubMed] [Google Scholar]
- Matsubara S. 1980. Overcoming self-incompatibility in Raphanus sativus L. with high temperature. Journal of the American Society for Horticultural Science 105, 842–846 [Google Scholar]
- Monterio AA, Gabelman WH. 1988. Use of sodium chloride solution to overcome self-incompatibility in Brassica campestris . HortScience 23, 876–877 [Google Scholar]
- Murase K, Shiba H, Iwano M, Che FS, Watanabe M, Isogai A, Takayama S. 2004. A membrane-anchored protein kinase involved in Brassica self-incompatibility signaling. Science 303, 1516–1519 [DOI] [PubMed] [Google Scholar]
- Murray HG, Thompson WF. 1980. Rapid isolation of high molecular weight DNA. Nucleic Acids Research 8, 4321–4325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi T, Esashi Y, Hinata K. 1969. Control of self-incompatibility by CO2 gas in Brassica . Plant and Cell Physiology 10, 925–927 [Google Scholar]
- Nakanishi T, Hinata K. 1973. An effective time for CO2 gas treatment in overcoming self- incompatibility in Brassica . Plant and Cell Physiology 14, 873–879 [Google Scholar]
- Nasrallah JB, Kao TH, Chen CH, Goldberg ML, Nasrallah ME. 1987. Amino acid sequence of glycoproteins encoded by three alleles of the S locus of Brassica oleracea . Nature 326, 617–619 [Google Scholar]
- Niikura S, Matsuura S. 2000. Genetic analysis of the reaction level of self-incompatibility to a 4% CO2 gas treatment in the radish (Raphanus sativus L.). Theoretical and Applied Genetics 101, 1189–1193 [Google Scholar]
- Nishio T, Kusaba M, Watanabe M, Hinata K. 1996. Registration of S alleles in Brassica campestris L. by the restriction fragment sizes of SLGs. Theoretical and Applied Genetics 92, 388–394 [DOI] [PubMed] [Google Scholar]
- Obayashi T, Kinoshita K, Nakai K, Shibaoka M, Hayashi S, Saeki M, Shibata D, Saito K, Ohta H. 2007. ATTED-II: a database of co-expressed genes and cis elements for identifying co-regulated gene groups in Arabidopsis . Nucleic Acids Research 35, D863–D869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ockendon DJ. 1978. Effects of hexan and humidity on self-incompatibility in Brassica oleracea . Theoretical and Applied Genetics 52, 113–117 [DOI] [PubMed] [Google Scholar]
- Okazaki K, Hinata K. 1987. Repressing the expression of self-incompatibility in crucifers by short-term high temperature treatment. Theoretical and Applied Genetics 73, 496–500 [DOI] [PubMed] [Google Scholar]
- Paran I, Zamir D. 2003. Quantitative traits in plants: beyond the QTL. Trends in Genetics 19, 303–306 [DOI] [PubMed] [Google Scholar]
- Ramchiary N, Nguyen VD, Li X, Hong CP, Dhandapani V, Choi SR, Yu G, Piao ZY, Lim YP. 2011. Genic microsatellite markers in Brassica rapa: development, characterization, mapping, and their utility in other cultivated and wild Brassica relatives. DNA Research 18, 305–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel MA, Chong YT, Haasen KE, Aldea-Brydges MG, Stone SL, Goring DR. 2009. Cellular pathways regulating responses to compatible and self-incompatible pollen in Brassica and Arabidopsis stigmas intersect at Exo70A1, a putative component of the exocyst complex. The Plant Cell 21, 2655–2671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarker RH, Elleman CJ, Dickinson HG. 1988. Control of pollen hydration in Brassica requires continued protein synthesis, and glycosylation is necessary for intraspecific incompatibility. Proceedings of the National Academy of Sciences, USA 85, 4340–4344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopfer CR, Nasrallah ME, Nasrallah JB. 1999. The male determinant of self-incompatibility in Brassica . Science 286, 1405–1412 [DOI] [PubMed] [Google Scholar]
- Schranz ME, Lysak MA, Mitchell-Olds T. 2006. The ABC’s of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends in Plant Science 11, 535–542 [DOI] [PubMed] [Google Scholar]
- Silva NF, Stone SL, Christie LN, Sulaman W, Nazarian KA, Burnett LA, Arnoldo MA, Rothstein SJ, Goring DR. 2001. Expression of the S receptor kinase in self-compatible Brassica napus cv. Wester leads to the allele-specific rejection of self-incompatible Brassica napus pollen. Molecular Genetics and Genomics 265, 552–559 [DOI] [PubMed] [Google Scholar]
- Stone SL, Anderson EM, Mullen RT, Goring DR. 2003. ARC1 is an E3 ubiquitin ligase and promotes the ubiquitination of proteins during the rejection of self-incompatible Brassica pollen. The Plant Cell 15, 885–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Arnoldo M, Goring DR. 1999. A break down of Brassica self-incompatibility in ARC1 antisense transgenic plants. Science 286, 1729–1731 [DOI] [PubMed] [Google Scholar]
- Suwabe K, Iketani H, Nunome T, Kage T, Hirai M. 2002. Isolation and characterization of microsatellites in Brassica rapa L. Theoretical and Applied Genetics 104, 1092–1098 [DOI] [PubMed] [Google Scholar]
- Suwabe K, Iketani H, Nunome T, Ohyama A, Hirai M, Fukuoka H. 2004. Characteristics of microsatellites in Brassica rapa genome and their potential. utilization for comparative genomics in Cruciferae. Breeding Science 54, 85–90 [Google Scholar]
- Suwabe K, Morgan C, Bancroft I. 2008. Integration of Brassica A genome genetic linkage map between Brassica napus and B. rapa. Genome 51, 169–176 [DOI] [PubMed] [Google Scholar]
- Suwabe K, Tsukazaki H, Iketani H, Hatakeyama K, Kondo M, Fujimura M, Nunome T, Fukuoka H, Hirai M, Matsumoto S. 2006. Simple sequence repeat-based comparative genomics between Brassica rapa and Arabidopsis thaliana: the genetic origin of clubroot resistance. Genetics 173, 309–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasaki T, Hatakeyama K, Suzuki G, Watanabe M, Isogai A, Hinata K. 2000. The S receptor kinase determines self-incompatibility in Brassica stigma. Nature 403, 913–916 [DOI] [PubMed] [Google Scholar]
- Takayama S, Isogai A, Tsunemoto C, Ueda Y, Hinata K, Okazaki K, Suzuki A. 1987. Sequences of S-glycoproteins, products of the Brassica campestris self-incompatibility locus. Nature 326, 102–104 [Google Scholar]
- Takayama S, Shiba H, Iwano M, Shimosato H, Che FS, Kai N, Watanabe M, Suzuki G, Hinata K, Isogai A. 2000. The pollen determinant of self-incompatibility in Brassica campestris . Proceedings of the National Academy of Sciences, USA 97, 1920–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama S, Shimosato H, Shiba H, Funato M, Che FS, Watanabe M, Iwano M, Isogai A. 2001. Direct ligand–receptor complex interaction controls Brassica self-incompatibility. Nature 413, 534–538 [DOI] [PubMed] [Google Scholar]
- Tatebe T. 1968. Studies on the physiological mechanism of self-incompatibility in Japanese radish. II. Breakdown of self-incompatibility by chemical treatments. Journal of the Japanese Society for Horitulcultural Science 37, 43–46 [Google Scholar]
- Tao G, Yang R. 1986. Use of CO2 and salt solution to overcome self-incompatibility of Chinese cabbage (B. campestris ssp. pekinensis ). Eucarpia Cruciferae Newsletter 11, 75–76 [Google Scholar]
- Upton G, Cook I. 1996. Understanding statistics. Oxford: Oxford University Press [Google Scholar]
- Van Ooijen JW. 2006. JoinMap® 4, software for the calculation of genetic linkage maps in experimental populations. Wageningen, The Netherlands: Kyazma BV.
- Van Ooijen JW. 2009. MapQTL® 4, software for the mapping of quantitative trait loci in experimental populations of diploid species. Wageningen, The Netherlands: Kyazma BV.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.