Abstract
Members of the TOPLESS gene family emerged recently as key players in gene repression in several mechanisms, especially in auxin perception. The TOPLESS genes constitute, in ‘higher-plant’ genomes, a small multigenic family comprising four to 11 members. In this study, this family was investigated in tomato, a model plant for Solanaceae species and fleshy fruits. Six open reading frames predicted to encode topless-like proteins (SlTPLs) containing the canonical domains (LisH, CTLH, and two WD40 repeats) were identified in the tomato genome. Nuclear localization was confirmed for all members of the SlTPL family with the exception SlTPL6, which localized at the cytoplasm and was excluded from the nucleus. SlTPL genes displayed distinctive expression patterns in different tomato organs, with SlTPL1 showing the highest levels of transcript accumulation in all tissues tested except in ripening fruit where SlTPL3 and SlTPL4 were the most prominently expressed. To gain insight into the specificity of the different TOPLESS paralogues, a protein–protein interaction map between TOPLESS and auxin/indole-3-acetic acid (Aux/IAA) proteins was built using a yeast two-hybrid approach. The PPI map enabled the distinction of two patterns: TOPLESS isoforms interacting with the majority of Aux/IAA, and isoforms with limited capacity for interaction with these protein partners. Interestingly, evolutionary analyses of the TOPLESS gene family revealed that the highly expressed isoforms (SlTPL1, SlTPL3, and SlTPL4) corresponded to the three TPL-related genes undergoing the strongest purifying selection, while the selection was much weaker for SlTPL6, which was expressed at a low level and encoded a protein lacking the capacity to interact with Aux/IAAs.
Key words: Aux/IAA, auxin signalling, co-repressor, multigenic family, protein–protein interactions, Solanum lycopersicum, tomato, TOPLESS.
Introduction
It is now well accepted that transcriptional co-repressors play crucial roles in a broad range of plant developmental processes (Liu and Karmarkar, 2008; Krogan and Long, 2009). In land plants, the Groucho (Gro)/Tup1 family of co-repressors includes TOPLESS/TOPLESS-RELATED (TPL/TPR) and LEUNIG/LEUNIG HOMOLOG (LUG/LUH) (Conner and Liu, 2000; Kieffer et al., 2006; Long et al., 2006). TPL proteins have been shown to be involved in multiple signalling pathways in higher plants, including hormone-signalling pathways (auxin, jasmonic acid, abscisic acid, and ethylene), meristem maintenance, floral induction, biotic stress, and circadian oscillator mechanism (Liu and Karmarkar, 2008; Szemenyei et al., 2008; Pauwels et al., 2010; Zhu et al., 2010, Causier et al., 2012a , b; Wang et al., 2013).
The first TPL gene was identified in Arabidopsis as responsible for the semi-dominant tpl-1 embryo development mutation resulting in altered polarity, ranging from fused cotyledons to complete replacement of the shoot with a second root (Long et al., 2002, 2006). Subsequently, the TPL family in Arabidopsis thaliana was found to comprise five members that seem to act redundantly (TPL, TPR1, TRP2, TRP3, and TRP4). Indeed, a quintuple loss of function, in which all five TPL/TPR genes were inactivated by mutation or RNA interference, is required to phenocopy the tpl-1 phenotype (Long et al., 2006).
It was established that, although TPL proteins are lacking a DNA-binding activity, they are incorporated into transcription complexes by interacting with transcription factors to repress gene expression in various processes. This inhibition of the expression of target genes is mediated by the recruitment of histone deacetylases into transcription complexes, and by changing the chromatin state from active to inactive (Long et al., 2006; Liu and Karmarkar, 2008; Krogan and Long, 2009; Krogan et al., 2012). Interaction between the TPL/TPR co-repressors and transcription factors depends on the Lissencephaly (LisH) and the C-terminal to LisH Homology (CTLH) domain of TPL (Szemenyei et al., 2008; Gallavotti et al., 2010), and on a small conserved protein motif found in transcription factors. This motif is known as the ethylene response factor-associated amphiphilic repression (EAR) domain (Ohta et al., 2001), with the consensus sequence (L/F)DLN(L/F)xP (Ohta et al., 2001; Hiratsu et al., 2004). Recently, the Arabidopsis TPL/TPR interactome framework revealed that the TPL co-repressors are able to interact with various transcription factors harbouring different repression domains (Causier et al., 2012a ). Among these TPL interactants, the transcriptional repressors involved in auxin signalling [i.e. auxin/indole-3-acetic acid (Aux/IAA) and auxin response factor (ARF) families] have been well documented. In Arabidopsis, the discovery that TPL is recruited by Aux/IAA proteins to suppress the expression of auxin-responsive genes in the absence of auxin revealed a crucial role for TPL in mediating the inhibitory effect of Aux/IAA on ARF-regulated transcription (Szemenyei et al., 2008). Large interactome studies in Arabidopsis identified 20 of the 29 AtIAA proteins as interacting partners of the TPL/TPRs (Arabidopsis Interactome Mapping Consortium, 2011; Causier et al., 2012a ). In addition, a large-scale analysis of the interaction between Aux/IAA and ARF in the Arabidopsis shoot apex revealed that the vast majority of the Aux/IAAs interacted with all the ARF activators and showed very limited interactions with ARF repressors (Vernoux et al., 2011). However, a recent study showed that repressive ARF proteins, such as ARF2 and ARF9, can interact directly with TPL/TPR proteins, suggesting a mechanism for repression implicating TPL/TPR co-repressors in both forms of ARF-mediated repression (Causier et al., 2012a ).
The release in recent years of several plant genome sequences has offered the possibility to investigate a large set of multigenic families at the genome scale. In this context, the tomato genome is of special interest, as (1) tomato has emerged as a model plant, for fleshy fruit development, and (2) tomato is a reference species for the Solanaceae family and also for the taxum of Asterids, particularly as the majority of sequenced dicot genomes belongs to Rosids (Sato et al. 2012). It is noteworthy that the structure of several multigenic families involved in auxin perception and responses have been examined in tomato (Kumar et al., 2011; Ren et al., 2011; Wu et al., 2011; Audran-Delalande et al., 2012; Kumar et al., 2012;Pattison & Catalá, 2012; Wu et al., 2012a , b), thus shaping an exhaustive picture of auxin signalization complementary to the Arabidopsis model plant. However, compared with the plant model Arabidopsis, the TPL gene family has so far been poorly described.
To characterize fully the molecular biology and evolution of the tomato TPL family and to understand its possible functions, we identified and characterized six SlTPL genes. Our analyses focused on the identification, evolutionary relationships, and expression patterns of each member of the tomato TPL family. Moreover, we used yeast two-hybrid (Y2H) approaches to establish the framework of TPL/Aux/IAA protein–protein interactions (PPIs). These results will provide a framework for further studies to better understand the potential functions of TPL proteins in tomato plants, especially during the flower and fruit development.
Materials and methods
Isolation and cloning of SlTPL genes
The full-length coding sequences of six SlTPLs were amplified from mature green fruit cDNA. The primers used were as follows: TPL1_attb1: 5′-ATGTCATCTCTCAGTAGAG AGCTT-3′ and TPL1_attb2: 5′-TCATCTTGGTGCTTGATCGGAGC-3′; TPL2_attb1: 5′-ATGTCTTCCTTGAGTAGGGAACTG -3′ and TPL2_attb2: 5′-TCACCTTGAAGGTGTTTCTGATG-3′; TPL3_attb1: 5′-ATGTCTTCTCTTAGCAGAGAATTG-3′ and TPL3_attb2: 5′-TCATCTTTGAACTTGGTCAGCAG-3′; TPL4_attb1: 5′-ATGACTTCTTTAAGCAGAGAGCTG-3′ and TPL4_attb2: 5′-C TACCTTGATGCTTGATCAAGACC-3′; TPL5_attb1: 5′-ATG AGGCATTTTGATGAAATGGT-3′ and TPL5_attb2: 5′-CT ACCTTTGAGGTTGATCT GAAT-3′; and TPL6_attb1: 5′-ATGT CTCTTAGTAAGGACCTTAT-3′ and TPL6_attb2: 5′-CTATATTG GTTGCTCAT TGGTAA-3′.
After amplification, the SlTPL genes were cloned into the pDONOR207 vector using the Gateway method (Invitrogen) and were fully sequenced.
Subcellular localization of SlTPL proteins
For localization of the SlTPL proteins, the SlTPL coding sequences were cloned using Gateway technology as a C-terminal fusion in frame with yellow fluorescent protein (YFP) into the pEarlyGate104 vector and expressed under the control of the 35S cauliflower mosaic virus promoter. The empty vector pEarleyGate104 was used as a control. Protoplasts were obtained from tobacco (Nicotiana tabacum) suspension-cultured BY-2-cells and transfected according to a method described previously (Leclercq et al., 2005). YFP localization by confocal microscopy was performed as described previously (Audran-Delalande et al., 2012).
Expression analysis of SlTPL genes
Total RNA extraction, removal of DNA contamination, cDNA generation of eight tomato tissues (root, stem, leaves, bud, flower, mature green fruit, breaker fruit, and red fruit), and quantitative reverse transcription-PCR (qRT-PCR) were performed according to methods previously described (Audran-Delalande et al., 2012; Pirrello et al., 2006). The primer sequences were as follows: TPL1F: 5′-TGTTCGT TCTAGGAGACTAACCAG-3′ and 5′-TPL1R: AAGACAAACCTTCCCTTC CGA-3′; TPL2F: 5′-CC TGTAAATACGCCT CTTGCT-3′ and TPL2R: 5′-ACTGGTTGG AATGGACTGTG-3′; TPL3F: 5′-CACTTTCTGCTCCAATAA CCT-3′ and TPL3R: 5′-TCCA TCTGTCAACCCAACTG-3′; TPL4 F: 5′-CCTTCTAACC CAAGCTCCAG-3′ and TPL4R: 5′-AT AAACTCCGCCATCAGTA AGTC-3′; TPL5F: 5′-CGTCTATT GTAACCCATCCA CTC-3′ and TPL5R: 5′-AGAAGTTACACCAT GAGGACCC-3′; and TPL6F: 5′-ACTG GACTAGCATTCTCT AACAC-3′ and TPL6R: 5′-TTGAATT CCACA CCACTATCTG AG-3′. Actin was used as an internal reference. The relative fold differences (with SlTPL6 as a reference gene) for each sample were calculated using the formula 2–ΔΔCt. Three independent RNA isolations were used for cDNA synthesis and, each cDNA sample was subjected to real-time PCR analysis in triplicate.
Bioinformatic analyses
SlTPL genes were searched using BLAST queries on the Genomic (Chromosome v.2.40) and transcript database (cDNA itag 2.4) available on the SGN website (http://solgenomics.net/tools/blast/index.pl). Exons and introns were deduced from the ITAG 2.3 annotation. For SlTPL5 (Solyc07g008040), the ‘predicted annotation’ missing the N-terminal extremity was completed with an additional exon (from position 2754093 to 2754173 on SL2.40ch07 chromosome 7 annotation). Protein domains were first predicted on the prosite database protein (http://prosite.expasy.org/). Prediction of the WD40 segments was refined using the PF00400.27 Pfam Hidden Markov Model with an i-value threshold at 0.1. For i-values > 0.1, the prediction of WD40 position was deduced from the sequence alignment of the different TPL isoforms.
Nuclear localization signal (NLS) analysis prediction was performed with ‘cNLS Mapper’ (http://nls-mapper.iab.keio.ac.jp/cgi-bin/NLS_Mapper_form.cgi) (Kosugi et al., 2009). NLS prediction scores >5.0 were considered positive.
Evolutionary analyses
Phylogenetic analyses and distance matrices were built using the MEGA5 package (Tamura et al., 2011). Full-length amino acid sequences were aligned using the ClustalW algorithm. For the overall phylogeny, an initial tree encompassing sequences from Physcomitrella patens, Selaginella moellendorffii, Oryza sativa, Zea mays, Sorghum bicolor, Arabidopsis thaliana, Solanum lycopersicon, Nicotiana benthamiana, Populus trichocarpa, Glycine max and Mimulus guttatus was performed using the neighbour-joining method. The percentage of replicate trees in which the associated taxa clustered together was calculated in the bootstrap test (500 replicates). The topology was further confirmed using the maximum-likelihood method. Ultimately, a simplified tree was performed by limiting the number of genomic sets as the topology remained unchanged. Trees were drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree.
The following genome annotations were used for phylogenetic analyses: Physcomitrella patens (Phypa1_1.FilteredModels; Rensing et al., 2008); Selaginella moellendorffii (Lycophyte Selmo1_GeneModels_FilteredModels3; Banks et al., 2011); A. thaliana (TAIR10; Swarbreck et al., 2008); Populus trichocarpa (Eudicot Populus.trichocarpa.v2.0; Tuskan et al., 2006); V. vinifera (12X March 2010 release, Glycine max Glyma1_pacId; Schmutz et al., 2010); O. sativa [MSU Rice Genome Annotation (Osa1) Release 6.1; Ouyang et al., 2007]; Z. mays (ZmB73_4a.53_working_translations; Schnable et al., 2009); Sorghum bicolor (Sorbi1_GeneModels_Sbi1_4_aa; Paterson et al., 2009); Solanum lycopersicon (ITAG2.3_release; Sato et al., 2012); Brassica rapa (Chiifu-401–42; Wang et al., 2011); Eucalyptus grandis (Egrandis_201; http://www.jgi.doe.gov/); M. guttatus (Mguttatus_140; http://www.jgi.doe.gov/); N. benthamiana (Niben.genome.v0.4.4; Pallas et al., 2012); Solanum tuberosum (PGSC_DM_v3.4; Xu et al., 2011).
Protein–protein interaction (PPI) assay of SlTPLs and SlIAAs by Y2H assay
Tomato TPL genes were amplified and cloned into the pDBD (BD-TPLs) vector (Clontech). Similarly, SlIAA target genes [IAA1 (JN379431), IAA3 (JN379433), IAA4 (JN379434), IAA7 (JN379435), IAA8 (JN379436), IAA9 (JN379437), IAA11 (JN379438), IAA12 (JN379439), IAA14 (JN379441), IAA15 (JN379442), IAA16 (JN379443), IAA17 (JN379444), IAA19 (JN379445), IAA22 (JN379447), IAA26 (JN379449), IAA27 (JN379450) and IAA29 (JN379451)] were inserted in pGAD (AD-IAAs) vectors (Clontech). Diploids were selected on medium lacking Trp and Leu, and interactions were validated by the use of HIS3 and ADE2 reporter genes on medium lacking Trp, Leu, His, and Ade. Manipulation and analysis of the Y2H assay followed the manufacturer’s instructions (Clontech Yeast Protocols Handbook), and all experiments were repeated three times independently. For SlTPL1 genes lacking LisH, the coding sequence was truncated at nucleotide position +112.
Results
Identification and cloning of TPL-related genes in the tomato genome
An in silico search was performed on the tomato genome and transcript databases (http://www.solgenomics.net/) using Arabidopsis TPL and TPR sequences as queries for BLAST searches. While the initial screen identified nine ORFs predicted to encode putative TPL-like proteins (SlTPLs), only six corresponded to full-length proteins containing all canonical motifs that define the TPL proteins (Table 1). The full-length cDNA of the six SlTPLs was further confirmed by RT-PCR amplification, indicating that the corresponding coding sequences range from 3396 to 3669bp with deduced protein sizes ranging from 1131 to 1222 aa (Table 1).
Table 1.
Main structural features of the tomato SlTPL family members
| Nomenclature | Gene | Predicted protein | Domains | |||||
|---|---|---|---|---|---|---|---|---|
| SlTPLs | iTAG Gene ID | Exons | Introns | Length (aa) | MW (kDa) | LisH | CTLH | WD-40 repeats |
| SlTPL1 | Solyc03g117360.2.1 | 25 | 24 | 1131 | 124.676 | 4–36 | 34–92 | 411–632/832/957 |
| SlTPL2 | Solyc08g076030.2.1 | 25 | 24 | 1136 | 124.60 | 4–36 | 34–92 | 341–668/834–959 |
| SlTPL3 | Solyc01g100050.2.1 | 25 | 24 | 1132 | 124.676 | 4–36 | 34–92 | 343–669/871–955 |
| SlTPL4 | Solyc03g116750.2.1 | 26 | 25 | 1133 | 124.318 | 4–36 | 34–92 | 413–634/839–964 |
| SlTPL5 | Solyc07g008040.2.1 | 24 | 23 | 1134 | 124.82 | 4–36 | 34–92 | 398–639/881/965 |
| SlTPL6 | Solyc08g029050.2.1 | 33 | 32 | 1222 | 134.181 | 3–35 | 33–91 | 531–664/934–1060 |
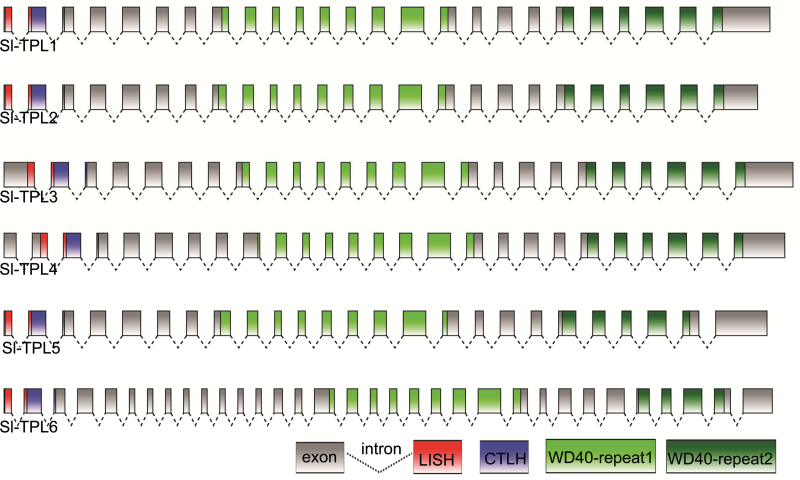
Structural analysis of the six SlTPL genes showed that they displayed similar numbers of introns (23–25) and exons (24–26), except for SlTPL6, which was longer than the other TPL members (Table 1). Pairwise comparison of the six SlTPL protein sequences showed that the percentage identity among family members ranged from 44 to 75%. Protein domain searches in the Pfam database (http://pfam.sanger.ac.uk/) indicated that all SlTPLs displayed the conserved LisH and CTLH domains and had two domains containing several WD40 repeats: WD40-repeat-1 and WD40-repeat-2 with seven and five WD40 segments, respectively (Fig. 1 and Supplementary Fig. S1 available at JXB online). The CTLH domain and the WD40-repeat-1 were separated by a proline-rich region.
Fig. 1.
Gene structure of the six tomato TPL genes. Grey boxes represent exons, dotted lines represent introns, the red box is the LisH domain, the blue box is the CTLH domain, the light green boxes are the WD40-repeat 1 and the dark green boxes are the WD40-repeat 2. The figure was produced using FancyGene software (http://bio.ieo.eu/fancygene/). (This figure is available in colour at JXB online.)
The tomato TPLs were distributed on four chromosomes: two SlTPLs (Solyc03g116750 and Solyc3g117360) on chromosome 3, two (Solyc08g076030.2.1 and Solyc08g029050.2.1) on chromosome 8, one (Solyc01g100050.2.1) on chromosome 1 and one (Solyc07g008040.2.1) on chromosome 7. There were three additional truncated TPL sequences lacking the LisH and CTLH domains, with two located on chromosome 3 (Solyc03g117370 and Solyc03g117410) and one on chromosome 1 (Solyc05g016070).
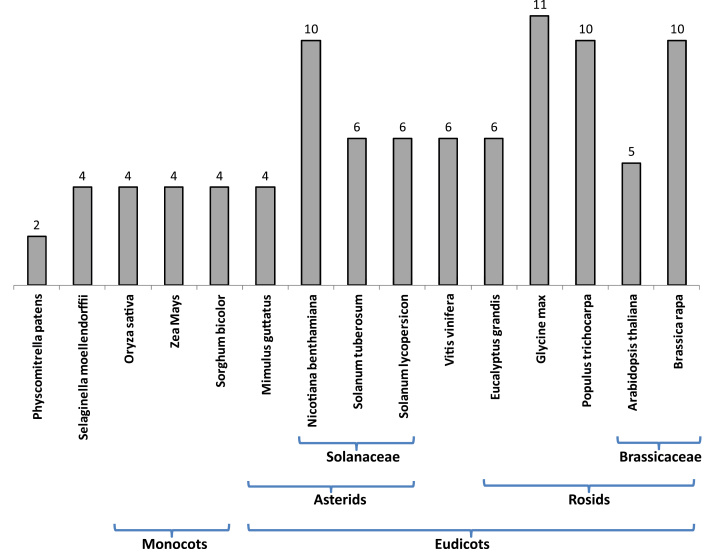
The number of ‘full-length’ TPL genes in tomato fell in the range found in other plant genomes, which varies in angiosperms from four members in monocots to 11 members in soybean (Fig. 2). It is noteworthy that a high number of isoforms is often observed in organisms having undergone recent whole-genome duplication or polyploidization events (e.g. G. max, N. benthamiana and B. rapa).
Fig. 2.
Inventory of TPL genes in different plant genomes. Only TPL genes containing the four canonical domains (LisH, CTLH and two WD40 repreats) were considered. The major taxons are shown below.
SlTPL nomenclature and phylogenetic analyses
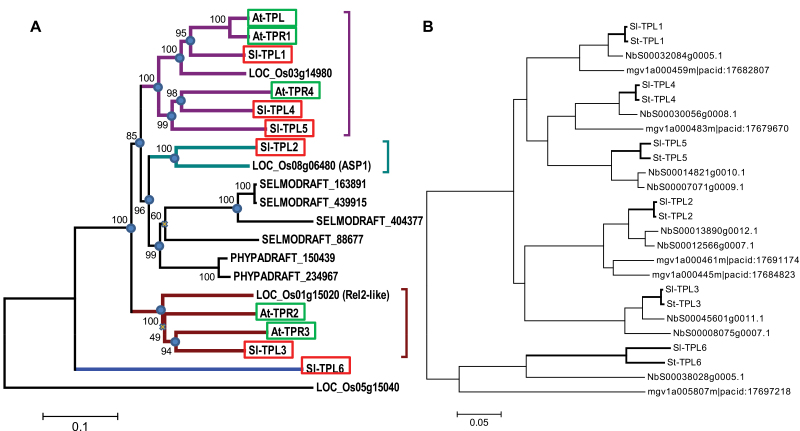
To adopt a nomenclature consensual with that of Arabidopsis TPL and TPR proteins, we carried out phylogenetic analyses on different TPL-like proteins or cDNAs from different plant sequenced genomes comprising moss, fern, and various angiosperm sequences (see Materials and methods). The phylogenetic trees (Fig. 3A) allowed the individualization of four branches. Three branches looked well defined in all dicot plants: the first branch contained AtTPL, AtTPL1, AtTPR4, Solyc3g117360.2.1 (named SlTPL1), Solyc03g117360.2.1 (named SlTPL4), and Solyc07g008040.2.1 (named SlTPL5); the second branch, absent in Arabidopsis yet present in Eucalyptus (Eucgr.K00093.1|PACid:23601479) and grapes (GSVIVT01024440001), contained Solyc08g076030 (named SlTPL2), rice ASP1 protein, and moss or lycophyte TPL-like proteins; and the third branch contained AtTPR2, AtTPR3, and Solyc01g100050.2.1 (named SlTPL3). Lastly, Solyc08g029050.2.1 (named SlTPL6) appeared as an outgroup branch in the phylogenetic tree (Fig. 3A). The robustness of the tree topology was assessed either with a bootstrap test (Fig. 3A) or by changing the number of genomes used in the phylogeny and the portion of the aligned sequence (N-terminal, C-terminal, or conserved domains) or the clustering method (neighbour-joining or maximum-likelihood method). The vast majority of the nodes presented in Fig. 3A remained unchanged.
Fig. 3.
Phylogenetic trees of some plant and tomato TPL proteins. (A) Representative phylogenetic tree of TPL proteins from land plants: moss (P. patens, PHYPADRAFT_xxx), lycophyte (Selaginella moellendorffii, SELMODRAFT_xxx), rice (LOC_Os-xxx), tomato (red boxes) and Arabidopsis (green boxes). The coloured brackets emphasize the main branches conserved among angiosperms. The present tree was obtained after alignment of full-length TPL sequences using ClustalW and clustering with the neighbour-joining method. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) are shown next to the branches. Phylogenetic analyses including additional genome sets (Z. mays, Sorghum bicolor, Populus trichocarpa, G. max, V. vinifera and M. guttatus) or using the maximum-likelihood clustering method displayed similar topologies, the majority of the nodes being conserved (blue circles) while only few nodes (yellow crosses) were unstable. (B) Phylogenetic tree of TPL proteins among Asterid and Solanaceaous species. The tree was built using sequences from four genomes: Solanum lycopersicon, Solanum tuberosum, N. benthamiana and M. guttatus. (This figure is available in colour at JXB online.)
To understand further the TPL phylogeny, and notably to characterize the SlTPL6 outgroup, the presence of TPL ‘orthologues’ was investigated in Asterid genomes belonging either to the Solanaceae family (Solanum tuberosum and N. benthamiana) or to the Lamiales order (M. guttatus). An SlTPL6 homologue was found in all Asterids, supporting the view that SlTPL6 homologues form a distinct clade (Fig. 3B). Within this SlTPL6 clade, the length of the branches suggested that these isoforms had evolved faster than other TPLs. This observation was supported by sequence divergences: the amino acid substitution rates calculated within the Solanaceae orthology groups varied from 2.6 to 6.3% for SlTPL1–5 and reaching 22.7% for the SlTPL6 (Table 2). Moreover, a neutrality test (dS/dN values) calculated on Solanaceae orthologues suggested that the purifying selection exerted by evolution on the SlTPL6 family is much weaker than the selection pressure exerted on other TPL genes.
Table 2.
Evolutionary features of TOPLESS-related genes in Solanaceous speciesMean distance was expressed as the proportion of amino acid or nucleic acids positions different after pairwise alignment. dS/dN values were calculated using the codon-based test of purifying selection performed on each pair of orthologous sequences from Solanum lycopersicon and Solanum tuberosum. The variance of the difference was computed using the bootstrap method (500 replicates). Analyses were conducted using the Nei and Gojobori (1986) method.
| SlTPL1 | SlTPL2 | SlTPL3 | SlTPL4 | SlTPL5 | SlTPL6 | ||
|---|---|---|---|---|---|---|---|
| Mean distance (Solanum/Nicotiana) | Amino acids | 0.026 | 0.041 | 0.032 | 0.029 | 0.063 | 0.227 |
| Nucleic acids | 0.055 | 0.050 | 0.054 | 0.054 | 0.067 | 0.154 | |
| Neutrality test (Solanum) | dS/dN | 7.08 | 6.66 | 6.98 | 7.62 | 6.19 | 3.645 |
Subcellular localization of SlTPLs
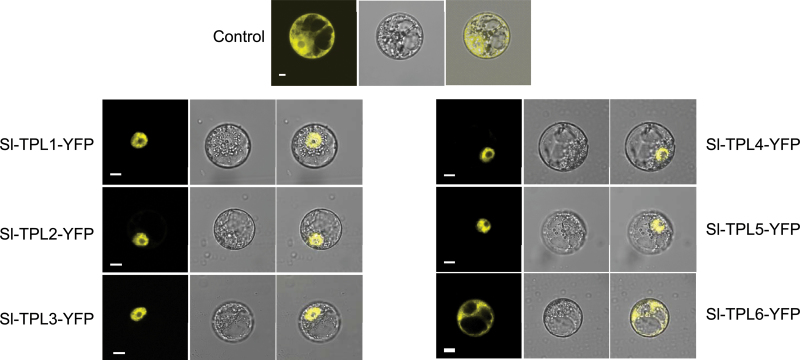
The subcellular localization of the SlTPL proteins was assessed by a transient expression assay in tobacco protoplasts using a translational fusion between each of the SlTPL proteins and YFP. Microscopy analysis showed that SlTPL1–5–YFP fusion proteins localized exclusively to the nucleus (Fig. 4) whereas SlTPL6 was localized at the cytoplasm and excluded from the nucleus. This result is in agreement with the in silico prediction of a conserved NLS for the five nuclear SlTPL1–5 proteins, while SlTPL6 NLS scores were below the 5.0 threshold value (Supplementary Table S1 available at JXB online). Altogether, the nuclear localization of the majority of SlTPLs was consistent with their putative role in transcriptional regulation activity.
Fig. 4.
Subcellular localization of tomato TPL proteins. SlTPL–YFP fusion proteins were transiently expressed in BY-2 tobacco protoplasts and subcellular localization was analysed by confocal laser-scanning microscopy. The merged pictures of the yellow fluorescence channel (left panels) and the corresponding bright field (middle panels) are shown (right panels). The empty vector pEarleyGate104 was used as a control. Bar, 10 μm. (This figure is available in colour at JXB online.)
Expression analyses
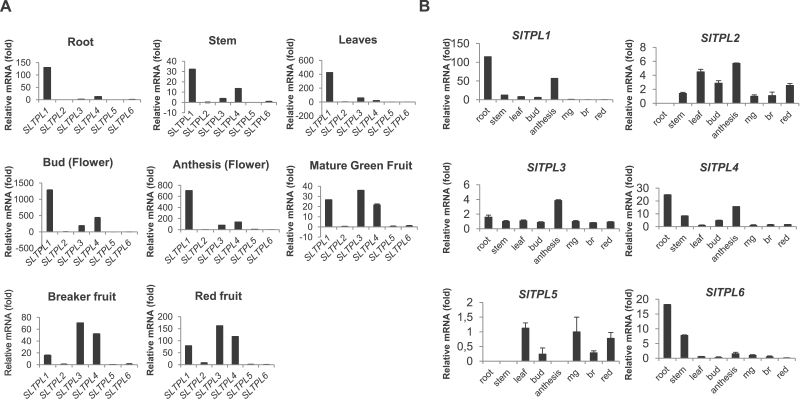
In order to study the spatio-temporal expression pattern of the six SlTPL genes, qRT–PCR was performed on eight different plant tissues and organs. Three SlTPL members (SlTPL1, SlTPL3, and SlTPL4) displayed significantly higher levels of expression than the three remaining paralogues. SlTPL1 and SlTPL4 were found to be highly expressed in flowers and vegetative tissues (roots, stems, and leaves) and in developing flowers (buds and during anthesis) but with reduced expression in ripening fruit, while SlTPL3 expression remained constant and high during fruit ripening (Fig. 5). This preferential expression of SlTPL1, SlTPL3, and SlTPL4 is coherent with their estimated expression in two public databases (RNAseq database: http://ted.bti.cornell.edu and ESTs database: http://solgenomics.net/). Although less expressed, SlTPL2 was found preferentially in leaves and developing flowers; the levels of SlTPL5 transcripts were low in all tissues; SlTPL6 expression was restricted to roots and stems (Fig. 5b).
Fig. 5.
Real-time PCR expression profiles of six tomato TPL genes. (A) Expression patterns of SlTPL genes in various tomato tissues. Relative mRNA levels of each SlTPL gene in different tissues were normalized against actin. The results were expressed using SlTPL6 as a reference (relative mRNA level 1). Values represent the best experiment among three independent biological repetitions. Bars indicate the standard deviation of three experimental repetitions. (B) Expression patterns in different tomato tissues of each SlTPL gene. The relative mRNA level of each SlTPL gene was normalized against actin. mg, Mature green fruit; br, breaker fruit; red, red fruit. The results were expressed using the mature green fruit as a reference (relative mRNA level 1). Values represent the best experiment among three independent biological repetitions. Bars indicate the standard deviation of three experimental repetitions.
Examination of PPIs in the framework of auxin mediation
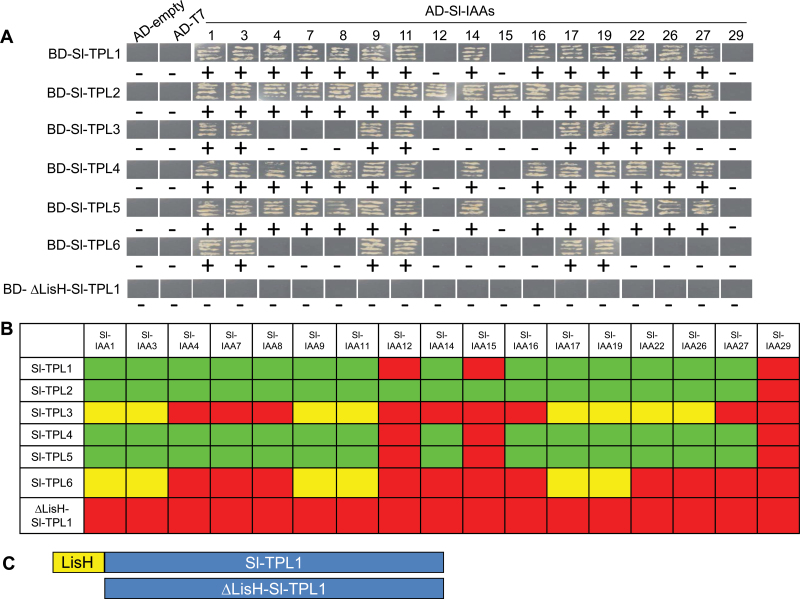
The differential expression of SlTPL genes evokes the critical question of functional redundancy within the TPL family. In a recent paper, Causier et al. (2012a ) compared the PPI patterns of different Arabidopsis TPL proteins using a high-throughput Y2H screen both on a whole-plant and on a transcription factor library. In the present work, we focused on the interactions with the Aux/IAA family by performing an exhaustive targeted analysis of Aux/IAA–TPL interactions. The six SlTPL proteins were fused to a binding domain (BD) and used as bait in a Y2H test with 17 different SlIAA proteins fused to an activating domain (AD). After monitoring the yeast growth on two auxotroph selective media, two patterns of TPL could clearly be defined (Fig. 6A, B): SlTPL1, SlTPL2, SlTPL4, and SlTPL5 interacted with the majority of SlIAAs and grew in all the selective media, and SlTPL3 and SlTPL6 exhibited only limited growth when co-expressed with Aux/IAA–AD fusion proteins. Contrary to other SlIAAs, SlIAA29 failed to show interaction with any of the SlTPLs. With the exception of SlIAA12 and SlIAA15, the Aux/IAAs did not harbour any obvious specificity towards the ‘TPL’ clade (SlTPL1, SlTPL4, and SlTPL5), sharing high similarity with AtTPL. In addition, SlTPL2, which belongs to a distinct clade of SlTPLs (1, 4, and 5), also exhibited a broad capacity to interact with the majority of SlIAAs. As a control, we performed a Y2H test with truncated SlTPL1 or SlTPL5 (ΔLisH-TPL) (Fig. 6C) lacking the LisH domain shown previously to be essential for TPL–WUS or TPL–Aux/IAA interactions (Kieffer et al., 2006; Szemenyei et al., 2008). Contrary to all SlTPLs BD fusions assayed, a complete lack of growth was observed when co-expressing BD–ΔLisH-TPL proteins with BD–Aux/IAAs (Fig. 6B).
Fig. 6.
PPI maps between SlTPLs and SlIAAs established by a Y2H screen. (A) Yeast growth of co-transformed BD–TPLs and AD–IAAs. The yeast clones grown on selected medium lacking Trp, Leu, His, and Ade (TLHA) were scratched again on a TLHA plate. After 3–4 d, the growth of the yeast strains confirmed a positive interaction, as shown. AD–empty vector and AD–T7 vector were used as negative controls. (B) Schematic representation of the interaction map between SlTPLs and SlIAAs. Green indicates that the yeast grew quickly, less than 4 or 5 d after co-transformation, indicating a strong interaction between the SlTPL and SlIAA partners. Yellow indicates that the yeast grew slowly 7–8 d after co-transformation, indicating a weak interaction between the tested SlTPL and SlIAA. Red indicates that there was no interaction detected between the tested SlTPLs and SlIAAs. (C) Truncated form of SlTPL1 protein lacking the N-terminal LisH domain N-terminal used as a negative control. (This figure is available in colour at JXB online.)
Discussion
The present study addressed the structural, evolutionary, and functional features of the tomato TPL family. TPL proteins have been primarily defined as a major component of the auxin transduction and response pathway, but the present data sustain the hypothesis of a functional diversification of these regulatory proteins. While mainly focusing on the TPL family in tomato, a plant model for Solanaceae and fleshy fruit research, the data also addressed the comparative features of this gene family within Plant kingdom at the evolutionary level, shedding new light on their functional diversification.
The structure of the SlTPL family is representative of that found in angiosperms where these proteins belong to a small multigenic family comprising five to 11 members. In the tomato, six full-length SlTPL genes were identified, as well as additional three pseudogenes with incomplete coding sequences. Among the six SlTPL genes, five were highly conserved (SlTPL1–5), while the last gene (SlTPL6) was more distant. With the exception of poplar genomes and genomes having undergone recent polyploidization (i.e. soybean, B. rapa, and N. benthamiana), the number of TPL isoforms ranges from four to six members, suggesting that the number of genes remains stable in this family and that usually, after a whole-genome duplication event, duplicated copies of TPL genes are not retained. The phylogenetic analysis of TPL genes enabled the distinction of three major clades gathering homologues in the majority of angiosperm genomes. The last clade, containing the distant SlTPL6, displays only clear homologues in closely related taxa (Asterids). Interestingly, highly diverging sequences of TPL-related proteins have also been found in other genomes such as the AtTPR-like gene (At2g25420; Causier et al., 2012b) and in poplar, but no clear relationship could be established with SlTPL6. Contrary to angiosperm TPL proteins, TPL from Physcomitrella patens and Selaginella moellendorffii clustered in a same branch, indicating the existence of ancestral divergences occurring before angiosperm radiation.
The functionality of SlTPL genes was addressed through three approaches: expression analysis, subcellular localization, and establishment of an interaction map between SlTPL and SlIAA proteins. The expression patterns of different SlTPLs revealed the tissue specificity of various isoforms and suggested a functional specialization of SlTPL isoforms. For example, SlTPL1 is highly expressed in vegetative organs (stems, roots) and flowers, while the expression of SlTPL3 and SlTPL4 prevails in fruit. Moreover, the overall intensity of gene expression evaluated by qPCR demonstrated a distinction between a group of three isoforms (SlTPL1, SlTPL3, and SlTPL4) that are highly expressed, SlTPL2, which is moderately expressed in the leaves and flowers, and third group made of two isoforms (SlTPL5 and SlTPL6) that displayed very low levels of expression. In agreement with our data, the prevalence of SlTPL1, SlTPL3, and SlTPL4 transcripts was also observed in expressed sequence tag (EST) and RNAseq expression databases (http://ted.bti.cornell.edu), whereas the expression of SlTPL6 was again found to be very low (no EST and few RNAseq reads). Interestingly, the overall expression level negatively correlated with the amino acid substitution rate. Indeed, after defining orthology groups among Solanaceous TPLs, we found that the highly expressed isoforms (SlTPL1, SlTPL3, and SlTPL4) showed the highest amino acid sequence conservation (<3.2% difference within Solanaceous sequences), while sequences were less conserved within the SlTPL6 orthology group (22.7% difference within Solanaceous sequences). The moderately expressed SlTPL2 and SlTPL5 displayed intermediate substitution rates (4 and 6% differences, respectively). This correlation was also supported by a neutrality test (dS/dN values) performed between potato and tomato pairs of othologues. The high substitution rate within the SlTPL6 orthology group was interpreted as an indication that the SlTPL6 subfamily undergoes a reduced purifying selection. By contrast, broadly expressed SlTPL isoforms are under a stronger purifying selection. Such a correlation between gene expression level and amino acid substitution rate has already been observed in genome-wide comparisons of expression patterns and protein evolution in Arabidopsis-related plants and in the Poaceae family (Wright et al., 2004; Slotte et al., 2011; Davidson et al., 2012). Indeed, this correlation is consistent with A. thaliana expression data (AtGenExpress), At-TPL being expressed more than other AtTPRs and AtTPL orthologues remaining highly conserved either in Arabidopsis lyrata or in B. rapa.
The subcellular localization established a second discrimination criterion among SlTPLs. YFP fusion proteins of SlTPL1–5 isoforms all migrated exclusively to the nucleus, as observed with other TPL proteins from Arabidopsis (Long et al., 2006), maize (Gallavotti et al., 2010), and rice (Yoshida et al., 2012). By contrast, the SlTPL6–YFP fusion protein displayed a divergent subcellular targeting, this isoform being targeted to the cytosol. This divergent localization is in line with the lower scores calculated by the NLS prediction tool for SlTPL6. This observation, in addition to the low expression level and the high substitution rate, supports the view of either a partial loss of functionality or divergent functionality regarding SlTPL6.
The first established function of TPL proteins is related to their role in auxin signalling via interaction with Aux/IAA partners (Szemenyei et al., 2008). To check whether this role is conserved among all SlTPLs isoforms and gain insight on either functional redundancy or potential functional diversification among family members in tomato, a comprehensive PPI study was carried out between all SlTPLs and SlIAA members using a Y2H screen. This targeted interactome study revealed two distinct patterns of interaction for tomato TPLs: four isoforms (SlTPL1, SlTPL2, SlTPL4, and SlTPL5) displayed a broad capacity for interaction with the majority of SlIAAs, and the remaining two isoforms (SlTPL3 and SlTPL6) showing a more restricted interaction capacity. It is noteworthy that a large number of SlIAAs showed positive interaction with SlTPLs, consistent with the outcome of Y2H screens performed in Arabidopsis where 20 out of the 29 AtAux/IAAs were able to interact with AtTPLs (Szemenyei et al., 2008; Arabidopsis Interactome Mapping Consortium, 2011; Causier et al., 2012a ). Interestingly, neither SlIAA29 nor its Arabidopsis homologue At IAA29 (AT4G32280.1) interacted with TPL proteins, although SlIAA29 exhibits a repressor activity (Audran-Delalande et al., 2012). On the other hand, the limited interaction capacity displayed by SlTPL6 adds another distinctive feature to this isoform, which has already diverged from other family members by its low expression level, high amino acid substitution rate and different subcellular localization. Altogether, the cumulative distinctive features support the idea that SlTPL6 has partially lost its ancestral function and may have gained new functionality.
In previous Y2H screens performed in Arabidopsis by Causier et al. (2012a ), AtTPR3 and AtTPR2, closely related to SlTPL3, both displayed the capacity to interact with various Aux/IAA proteins. However, a closer look at the interaction map published by Causier et al. (2012a ) could also suggest differences in specificity between AtTPL and AtTPR2 or AtTPR3, with the two latter notably interacting with partners displaying partial repression domains. Such hypothesis opens the possibility that At-TPR2, At-TPR3 and the closely related SlTPL3 display a specialization alternative to auxin signalling. The development of quantitative PPI methods such as Förster resonance energy transfer or surface plasmon resonance may provide deeper insight on discriminating interaction features among various TPL isoforms.
Functional redundancy among Arabidopsis TPL family members is supported by the absence of obvious phenotypes in single loss-of-function mutants of AtTPL/TPR genes and by the requirement for downregulation of all five AtTPL-TPRs in order to phenocopy the dominant mutation tpl-1 (Long et al., 2006). However, this assumption is in contrast to the situation prevailing in rice and maize, where genetic evidence seems to support a more specialized functionality for TPL genes. Thus, in rice (Yoshida et al., 2012), a single recessive mutation in Asp1, a TPL-like gene close to SlTPL2, exhibited several pleiotropic phenotypes, such as altered phyllotaxy and spikelet morphology. While these phenotypes suggest a close association of Asp1 with auxin action, they clearly reveal that the specialization of TPL-related proteins in some organisms can differ from that in Arabidopsis. Further evidence sustaining a diversified function for TPL proteins is provided by maize rel2 mutants affected in a TPL-like gene closely related to SlTPL3 and AtTPR3 (Gallavotti et al., 2010). A better clarification of the putative specialized functionality among tomato TPLs might be addressed by a reverse genetics approach. Simultaneous downregulation of SlTPL1 and SlTPL4 would uncover the importance of the TPL family in vegetative development and auxin action. Likewise, specific downregulation of SlTPL3 would be of particular interest to unravel the role of TPL co-repressors in flower and fruit biology.
Altogether, these data shed new light on structural, evolutionary, and some functional features of the tomato TPL gene family that suggest functional diversification of these regulatory proteins. Of particular interest, the setup of a comprehensive TPL–Aux/IAA interaction map and the differential subcellular targeting of some SlTPLs proteins would provide important clues towards designing appropriate strategies for the elucidation of both redundant and specific roles of TPL genes.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Fig. S1. Multiple sequence alignment of full-length SlTPL proteins.
Supplementary Table S1. NLS prediction scores computed with cNLS Mapper (Kosugi et al., 2009).
Acknowledgements
This work was carried out in the GBF laboratory, part of the TULIP ‘Laboratoire d’Excellence’ (LABEX) (ANR -10-LABX-41) and supported by the European Integrated Project EU-SOL (FOOD-CT-2006-016214). The work benefited from the networking activities within the European COST Action FA1106. YH and XW were supported by the China Scholarship Council.
Glossary
Abbreviations:
- AD
activating domain
- ARF
auxin response factor
- Aux/IAA
auxin/indole-3-acetic acid
- BD
binding domain
- EST
expressed sequence tag
- NLS
nuclear localization signal
- PPI
protein-protein interaction
- qRT-PCR
quantitative reverse transcription-PCR
- Y2H
yeast two-hybrid
- YFP
yellow fluorescent protein.
References
- Arabidopsis Interactome Mapping Consortium 2011. Evidence for network evolution in an Arabidopsis interactome map. Science 333, 601–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audran-Delalande C, Bassa C, Mila I, Regad F, Zouine M, Bouzayen M. 2012. Genome-wide identification, functional analysis and expression profiling of the Aux/IAA gene family in tomato. Plant and Cell Physiology 53, 659–672 [DOI] [PubMed] [Google Scholar]
- Banks JA, Nishiyama T, Hasebe M, et al. 2011. The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science 332, 960–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causier B, Ashworth M, Guo W, Davies B. 2012a. The TOPLESS interactome: a framework for gene repression in Arabidopsis. Plant Physiology 158, 423–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causier B, Lloyd J, Stevens L, Davies B. 2012b. TOPLESS co-repressor interactions and their evolutionary conservation in plants. Plant Signaling and Behavior 7, 325–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conner J, Liu Z. 2000. LEUNIG, a putative transcriptional corepressor that regulates AGAMOUS expression during flower development. Proceedings of the National Academy of Sciences, USA 97, 12902–12907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EA, Meyer AJ, Ellefson JW, Levy M, Ellington AD. 2012. An in vitro autogene. ACS Synthetic Biology 1, 190–196 [DOI] [PubMed] [Google Scholar]
- Gallavotti A, Long JA, Stanfield S, Yang X, Jackson D, Vollbrecht E, Schmidt RJ. 2010. The control of axillary meristem fate in the maize ramosa pathway. Development 137, 2849–2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsu K, Mitsuda N, Matsui K, Ohme-Takagi M. 2004. Identification of the minimal repression domain of SUPERMAN shows that the DLELRL hexapeptide is both necessary and sufficient for repression of transcription in Arabidopsis. Biochemical and Biophysical Research Communications 321, 172–178 [DOI] [PubMed] [Google Scholar]
- Kieffer M, Stern Y, Cook H, Clerici E, Maulbetsch C, Laux T, Davies B. 2006. Analysis of the transcription factor WUSCHEL and its functional homologue in Antirrhinum reveals a potential mechanism for their roles in meristem maintenance. Plant Cell 18, 560–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S, Hasebe M, Matsumura N, Takashima H, Miyamoto-Sato E, Tomita M, Yanagawa H. 2009. Six classes of nuclear localization signals specific to different binding grooves of importin alpha. Journal of Biological Chemistry 284, 478–485 [DOI] [PubMed] [Google Scholar]
- Krogan NT, Hogan K, Long JA. 2012. APETALA2 negatively regulates multiple floral organ identity genes in Arabidopsis by recruiting the co-repressor TOPLESS and the histone deacetylase HDA19. Development 139, 4180–4190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NT, Long JA. 2009. Why so repressed? Turning off transcription during plant growth and development. Current Opinion in Plant Biology 12, 628–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Agarwal P, Tyagi AK, Sharma AK. 2012. Genome-wide investigation and expression analysis suggest diverse roles of auxin-responsive GH3 genes during development and response to different stimuli in tomato (Solanum lycopersicum). Molecular Genetics and Genomics 287, 221–235 [DOI] [PubMed] [Google Scholar]
- Kumar R, Tyagi AK, Sharma AK. 2011. Genome-wide analysis of auxin response factor (ARF) gene family from tomato and analysis of their role in flower and fruit development. Molecular Genetics and Genomics 285, 245–260 [DOI] [PubMed] [Google Scholar]
- Leclercq J, Ranty B, Sanchez-Ballesta MT, Li Z, Jones B, Jauneau A, Pech JC, Latche A, Ranjeva R, Bouzayen M. 2005. Molecular and biochemical characterization of LeCRK1, a ripening-associated tomato CDPK-related kinase. Journal of Experimental Botany 56, 25–35 [DOI] [PubMed] [Google Scholar]
- Liu Z, Karmarkar V. 2008. Groucho/Tup1 family co-repressors in plant development. Trends in Plant Science 13, 137–144 [DOI] [PubMed] [Google Scholar]
- Long JA, Ohno C, Smith ZR, Meyerowitz EM. 2006. TOPLESS regulates apical embryonic fate in Arabidopsis. Science 312, 1520–1523 [DOI] [PubMed] [Google Scholar]
- Long JA, Woody S, Poethig S, Meyerowitz EM, Barton MK. 2002. Transformation of shoots into roots in Arabidopsis embryos mutant at the TOPLESS locus. Development 129, 2797–2806 [DOI] [PubMed] [Google Scholar]
- Nei M, Gojobori T. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Molecular Biology and Evolution 3, 418–426 [DOI] [PubMed] [Google Scholar]
- Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M. 2001. Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13, 1959–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang S, Zhu W, Hamilton J, et al. 2007. The TIGR Rice Genome Annotation Resource: improvements and new features. Nucleic Acids Research 35, D883–D887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallas V, Martinez G, Gomez G. 2012. The interaction between plant viroid-induced symptoms and RNA silencing. Methods in Molecular Biology 894, 323–343 [DOI] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Bruggmann R, et al. 2009. The Sorghum bicolor genome and the diversification of grasses. Nature 457, 551–556 [DOI] [PubMed] [Google Scholar]
- Pattison RJ, Catalá C. 2012. Evaluating auxin distribution in tomato (Solanum lycopersicum) through an analysis of the PIN and AUX/LAX gene families. The Plant Journal 70, 585–598 [DOI] [PubMed] [Google Scholar]
- Pauwels L, Barbero GF, Geerinck J, et al. 2010. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464, 788–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirrello J, Jaimes-Miranda F, Sanchez-Ballesta MT, Tournier B, Khalil-Ahmad Q, Regad F, Latche A, Pech JC, Bouzayen M. 2006. Sl-ERF2, a tomato ethylene response factor involved in ethylene response and seed germination. Plant and Cell Physiology 47, 1195–1205 [DOI] [PubMed] [Google Scholar]
- Ren Z, Li Z, Miao Q, Yang Y, Deng W, Hao Y. 2011. The auxin receptor homologue in Solanum lycopersicum stimulates tomato fruit set and leaf morphogenesis. Journal of Experimental Botany 62, 2815–2826 [DOI] [PubMed] [Google Scholar]
- Rensing SA, Lang D, Zimmer AD, et al. 2008. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319, 64–69 [DOI] [PubMed] [Google Scholar]
- Sato S, Tabata S, Hirakawa H, et al. 2012. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 635–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, et al. 2010. Genome sequence of the palaeopolyploid soybean. Nature 463, 178–183 [DOI] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, et al. 2009. The B73 maize genome: complexity, diversity, and dynamics. Science 326, 1112–1115 [DOI] [PubMed] [Google Scholar]
- Slotte T, Bataillon T, Hansen TT, St Onge K, Wright SI, Schierup MH. 2011. Genomic determinants of protein evolution and polymorphism in Arabidopsis. Genome Biology and Evolution 3, 1210–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarbreck D, Wilks C, Lamesch P, et al. 2008. The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Research 36, D1009–D1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szemenyei H, Hannon M, Long JA. 2008. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319, 1384–1386 [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution 28, 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan GA, Difazio S, Jansson S, et al. 2006. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313, 1596–1604 [DOI] [PubMed] [Google Scholar]
- Vernoux T, Brunoud G, Farcot E, et al. 2011. The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Molecular Systems Biology 7, 508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Kim J, Somers DE. 2013. Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription. Proceedings of the National Academy of Sciences, USA 110, 761–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang H, Wang J, et al. 2011. The genome of the mesopolyploid crop species Brassica rapa . Nature genetics 43, 1035–1039 [DOI] [PubMed] [Google Scholar]
- Wright TF, Johns PM, Walters JR, Lerner AP, Swallow JG, Wilkinson GS. 2004. Microsatellite variation among divergent populations of stalk-eyed flies, genus Cyrtodiopsis . Genetical research 84, 27–40 [DOI] [PubMed] [Google Scholar]
- Wu J, Liu S, He Y, Guan X, Zhu X, Cheng L, Wang J, Lu G. 2012a. Genome-wide analysis of SAUR gene family in Solanaceae species. Gene 509, 38–50 [DOI] [PubMed] [Google Scholar]
- Wu J, Peng Z, Liu S, He Y, Cheng L, Kong F, Wang J, Lu G. 2012b. Genome-wide analysis of Aux/IAA gene family in Solanaceae species using tomato as a model. Molecular Genetics and Genomics 287, 295–211 [DOI] [PubMed] [Google Scholar]
- Wu J, Wang F, Cheng L, Kong F, Peng Z, Liu S, Yu X, Lu G. 2011. Identification, isolation and expression analysis of auxin response factor (ARF) genes in Solanum lycopersicum . Plant Cell Reports 30, 2059–2073 [DOI] [PubMed] [Google Scholar]
- Xu X, Pan S, Cheng S, et al. 2011. Genome sequence and analysis of the tuber crop potato. Nature 475, 189–195 [DOI] [PubMed] [Google Scholar]
- Yoshida M, Ishii S, Fujii D, Otsuka S, Senoo K. 2012. Identification of active denitrifiers in rice paddy soil by DNA- and RNA-based analyses. Microbes and Environments 27, 456–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Xu F, Zhang Y, Cheng YT, Wiermer M, Li X. 2010. Arabidopsis resistance protein SNC1 activates immune responses through association with a transcriptional corepressor. Proceedings of the National Academy of Sciences, USA 107, 13960–13965 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.